Abstract
In many areas of China, tidal wetlands have been converted into agricultural land for rice cultivation. However, the consequences of land use changes for soil microbial communities are poorly understood. Therefore, we investigated bacterial and archaeal communities involved in inorganic nitrogen turnover (nitrogen fixation, nitrification, and denitrification) based on abundances and relative species richness of the corresponding functional genes along a soil chronosequence ranging between 50 and 2,000 years of paddy soil management compared to findings for a tidal wetland. Changes in abundance and diversity of the functional groups could be observed, reflecting the different chemical and physical properties of the soils, which changed in terms of soil development. The tidal wetland was characterized by a low microbial biomass and relatively high abundances of ammonia-oxidizing microbes. Conversion of the tidal wetlands into paddy soils was followed by a significant increase in microbial biomass. Fifty years of paddy management resulted in a higher abundance of nitrogen-fixing microbes than was found in the tidal wetland, whereas dominant genes of nitrification and denitrification in the paddy soils showed no differences. With ongoing rice cultivation, copy numbers of archaeal ammonia oxidizers did not change, while that of their bacterial counterparts declined. The nirK gene, coding for nitrite reductase, increased with rice cultivation time and dominated its functionally redundant counterpart, nirS, at all sites under investigation. Relative species richness showed significant differences between all soils with the exception of the archaeal ammonia oxidizers in the paddy soils cultivated for 100 and 300 years. In general, changes in diversity patterns were more pronounced than those in functional gene abundances.
INTRODUCTION
The influence of different types of changes in land use on microbial communities has been studied with growing interest in recent years due to the importance of soil microbes in geochemical nutrient turnover and soil health. This includes studies of the conversion of grassland to forest (6), changes in tillage management (4, 11, 38), and shifts in the intensity of land use in both agricultural (17) and forest (5, 19) ecosystems. Some articles have also addressed paddy ecosystems in particular and effects of temporary upland conversion (14). All these studies have shown significant shifts in the microbial community structure and function as a consequence of land use changes. However, since in most cases only effects shortly after land use changes were studied, it is not clear if the related changes were caused by the type of management or rather reflected a response of microbial communities toward the disturbance of a balanced ecosystem. To address this question, we compared four paddy soils that have been cultivated with rice for different time periods (50 to 2,000 years) with a tidal wetland, which typically represents the parent material for agricultural land reclamation in China (39).
Most studies published so far concerning the effects of land use changes on soil microbial communities have focused on alterations related to phylogenetic groups based on 16S rRNA sequences or on one functional marker. In modern rice cultivation, however, the yield-limiting factor is the availability of nitrogen (N) due to large losses in flooded soils (9, 18). Therefore, the objective of this study was to reconstruct functional microbial communities that are involved in key processes of the inorganic nitrogen cycle and to link these results to abiotic and biotic properties of the different soils of the chronosequence. We investigated functional gene abundances of bacteria and archaea performing nitrogen fixation, nitrification, and denitrification by quantifying genes encoding subunits of the nitrogenase (nifH), ammonia monooxygenase (amoA), nitrite reductase (nirK and nirS), and nitrous oxide reductase (nosZ). Since nifH, archaeal amoA, and nosZ genes turned out to be highly abundant and may therefore greatly contribute to the respective turnover processes, these genes were chosen for diversity analysis using terminal restriction fragment length polymorphism analysis (T-RFLP). For sampling, the beginning of the vegetation period was chosen to make the paddy soils more comparable with the tidal wetland, since at this time point agricultural activities like fertilizer or pesticide application, as well as the rhizosphere effect of rice plants, have been considered low.
During paddy soil evolution, different chemical and physical soil properties may influence soil microbial communities involved in nitrogen turnover. Directly after conversion of tidal wetlands into paddy soils, microbial communities might respond mainly to the leaching of marine salts by rainfall and the beginning of agricultural management. With ongoing paddy soil cultivation and continuous input of carbon and nitrogen to the system, microbes may adapt to the new conditions and become key players in certain functional traits. Long-term cultivation of rice with repeated tillage management fertilization and pesticide application, as well as effects by the plants (rhizosphere and litter), may strongly influence soil microbial communities and lead to a general increase in abundance and activity due to accumulation effects. Thus, we postulate that differences in microbial community structure and function along the chronosequence will be found.
MATERIALS AND METHODS
Site description and soil sampling.
The study sites are located in the area of Cixi, Zhejiang Province, People's Republic of China, a subtropical monsoon area with a mean annual temperature of 16.3°C and precipitation of 1,325 mm (40). We sampled a chronosequence of four paddy soils that have been under rice cultivation for approximately 50, 100, 300, and 2,000 years (P50, P100, P300, and P2000), as well as a tidal wetland (P0), which represents the parent material of land reclamation for agricultural use by sea dike building. The coordinates of the sampled sites are as follows: P0, 30°19′N, 121°09′E; P50, 30°11′N, 121°22′E; P100, 30°09′N, 121°21′E; P300, 30°06′N, 121°31′E; P2000, 30°05′N, 121°27′E. All five sites under investigation are located within a 40-km area.
The duration of rice cultivation at the respective sites was estimated according to well-documented points in time of sea dike construction (Cixi County Annals, abstracted information in Chinese, available at www.cixi.gov.cn), summarized by Cheng et al. (12). Thus, all paddy soils developed from comparable parent materials (tidal wetlands) and under the same ecological conditions. This is supported by the very similar textures among all age groups, which are strongly dominated by silt-sized particles (Table 1) and by a very similar mineral assemblage with comparable total contents, e.g., Al, Si, Ti, and Zr (data not shown). The small decrease in clay content (from 16 to 10%) is due to a displacement to deeper soil horizons caused by repeated flooding.
Table 1.
Characterization of the 5 examined soilsa
| Soil parameter | Value(s) for soil |
||||
|---|---|---|---|---|---|
| P0 | P50 | P100 | P300 | P2000 | |
| Soil texture (% sand, silt, clay) | 7.4, 80.4, 12.2 | 0.4, 83.6, 16.0 | 2.0, 81.2, 16.8 | 3.4, 81.2, 15.4 | 4.0, 85.1, 10.9 |
| pH | 8.1 (0.13) a | 7.6 (0.08) b | 7.6 (0.13) b | 7.5 (0.08) b | 7.3 (0.10) c |
| TOC (%) | 0.58 (0.17) a | 1.7 (0.14) b | 1.7 (0.16) b | 2.5 (0.16) c | 3.1 (0.11) d |
| TN (%) | 0.060 (0,012) a | 0.17 (0.014) b | 0.19 (0.015) c | 0.27 (0.020) d | 0.36 (0.019) e |
| WEOC (μg g−1 dw) | 9.5 (2.8) a | 17 (5.4) b | 18 (11) ab | 21 (8.3) ab | 14 (5.3) ab |
| WEON (μg g−1 dw) | 0.43 (0.19) a | 0.46 (0.77) a | 5.5 (6.6) a | BDLb | 0.36 (0.60) a |
| Nitrate (μg N g−1 dw) | 2.1 (0.70) a | 12 (1.2) ab | 8.3 (2.9) ab | 16 (6.5) b | 2.2 (1.7) a |
| Ammonium (μg N g−1 dw) | 0.42 (0.12) a | 6.0 (3.5) a | 25 (19) a | 27 (30) a | 22 (11) a |
| Cmic (μg g−1 dw) | 150 (58) a | 720 (120) b | 1,000 (330) b | 1,800 (780) b | 5,100 (1,300) c |
| Nmic (μg g−1 dry wt) | 39 (6.8) a | 28 (18) a | 110 (55) ab | 92 (36) ab | 150 (30) b |
| DNA content (μg g−1 dry wt) | 140 (35) a | 760 (24) bc | 630 (120) b | 810 (220) bc | 1,100 (140) c |
Soils (tidal wetland and paddy soils cultivated for 50, 100, 300, and 2,000 years) at 0- to 20-cm depth were analyzed using different parameters: soil texture, pH value (CaCl2), total organic carbon (TOC), and total nitrogen (TN), water extractable organic carbon (WEOC) and nitrogen (WEON), nitrate and ammonium concentrations, microbial biomass C and N, and DNA content. Standard deviations are given in parentheses (n = 5). Significant differences are indicated by different letters.
Below detection limit of 0.2 μg g−1 soil dry weight.
Since all fields are located in the same region and the agricultural management has been centrally controlled in China since 1949 by instructions of the Technical Service Bureau, comparable management has been performed for all sites. At all fields, rice was cultivated one time per year in summer using the same rice cultivars and an upland crop was cultivated one time per year in winter. Types, rates, and methods of application of fertilizers and pesticides were similar between the fields. The freshwater for flooding was not completely salt free but originated from the same river for all investigated sites, with comparable flooding regimes among the paddy soils driven by the interest for optimal crop yields (Zhihong Cao, Chinese Academy of Sciences, Nanjing, personal communication). All paddy fields are at least 10 km away from the sea; thus, direct flooding with marine water cannot occur. The topsoils are not influenced by groundwater due to the morphology of the soils. Only in the first years of reclamation might there have been an influence of salt being washed into freshwaterways, but this is part of the soil development and the shift from saltwater to freshwater conditions.
Soils were sampled at the beginning of July 2009, shortly after flooding. Rice plants on all paddy fields were at similar and early development stages (visual observation). Five independent field replicates in an area of 120 m2 were taken at each site with a soil auger from a 0- to 20-cm depth, each replicate being composed of seven individual soil cores (taken in a distance of 1 m2), which were pooled and homogenized to reduce heterogeneity. Aliquots (5 g) for DNA extraction were shock frozen in liquid nitrogen directly after sampling and stored at −80°C, whereas the remaining soil was stored at 4°C and analyzed in the following 2 weeks.
Soil physical and chemical properties.
Soils were extracted with 0.01 M CaCl2 at a soil-to-liquid ratio of 1:3. For determination of water extractable organic carbon (WEOC) and nitrogen (WEON), a total organic carbon analyzer, the Dimatoc 100 (Dimatec Analysentechnik GmbH, Germany), was used. Determination of ammonium (NH4+-N) and nitrate (NO3−-N) concentrations was done by continuous flow analysis with a photometric autoanalyzer (CFA-SAN Plus; Skalar Analytik, Germany). For the determination of microbial biomass carbon (Cmic) and nitrogen (Nmic), aliquots of the soils were fumigated with chloroform for 24 h prior to CaCl2 extraction (23). To measure total organic C (TOC) and N (TN), soil samples were frozen at −25°C and then lyophilized (FinnAqua Lyovac GT2; AMSCO Finn-Aqua GmbH, Germany). Homogenized samples were subjected to elemental analysis using an Elementar Vario EL III instrument in combustion mode. Carbonate carbon was determined using a total inorganic bound carbon (TIC) module attached to the Elementar analyzer, whereby carbonate was released from soils in a reaction chamber after addition of HCl and purged into the machine by a helium stream.
DNA extraction.
DNA of each of the 25 samples (5 sites × 5 replicates) was extracted in three subsamples from 0.5 g of soil with the FastDNA Spin kit for soil (MP Biomedicals) according to the protocol of the manufacturer. Quality and quantity of the DNA extracts were checked with a spectrophotometer (Nanodrop; PeqLab, Germany). Quantities of extracted DNA were very similar for all subsamples from one replicate, with an average variation of 15.6%. They were pooled and stored at −20°C until use.
Real-time PCR assay.
Quantitative real-time PCR was carried out on a 7300 real-time PCR system (Applied Biosystems, Germany) using SYBR green as a fluorescent dye. The respective reaction mixtures were composed as shown in Table 2.
Table 2.
Thermal profiles and primers used for real-time PCR quantification of different functional genesa
| Target gene | Source of standard (reference) | Thermal profile | No. of cycles | Primers (reference) | Primer concn (10 μM) | DMSOc |
|---|---|---|---|---|---|---|
| nifH | Sinorhizhobium meliloti | 95°C, 45 s; 55°C, 45 s; 72°C, 45 s | 40 | nifHF (30), nifHR (30) | 0.3 | |
| amoA AOA | Fosmid clone 54d9 (37) | 94°C, 45 s; 55°C, 45 s; 72°C, 45 s | 40 | amo19F (25), CrenamoA616r48x (32) | 0.5 | |
| amoA AOB | Nitrosomonas sp. | 94°C, 45 s; 58°C, 45 s; 72°C, 45 s | 40 | amoA1F (31), amoA2R (31) | 0.75 | |
| nirK | Azospirillum irakense | 95°C, 15 s; 63-58°C, 30 s; 72°C, 30 s | 5b | nirK876 (20), nirK5R (7) | 0.5 | 0.625 |
| 95°C, 15 s; 58°C, 30 s; 72°C, 30 s | 40 | |||||
| nirS | Pseudomonas stutzeri | 95°C, 45 s; 57°C, 45 s; 72°C, 45 s | 40 | cd3aF (26), R3cd (36) | 0.5 | 0.625 |
| nosZ | Pseudomonas fluorescens | 95°C, 30 s; 65-60°C, 30 s; 72°C, 30 s | 5b | nosZ2F (21), nosZ2R (21) | 0.5 | |
| 95°C, 15 s; 60°C, 15 s; 72°C, 30 s | 40 |
PCR mixtures consisted of Power Sybr green master mix (12.5 μl), BSA (3%, 0.5 μl), and template (2 μl, 0.5 to 2.6 ng μl−1), as well as primer and DMSO, as referenced in the table (in μl).
Touchdown: −1°C cycle−1.
DMSO, dimethyl sulfoxide.
Dilution series of the different DNA extracts were tested in a preexperiment with all soils to avoid inhibition of PCR, e.g., by coextracted humic substances. DNA extract dilutions of 1:128 turned out to be best suited (data not shown) and were used in three subsamples for the experiment. As standards, serial plasmid dilutions of the respective functional genes ranging from 101 to 107 gene copies μl−1 were used (sources of standards are shown in Table 2).
All PCR runs started with an initial enzyme activation step performed at 95°C for 10 min. The subsequent thermal profile was different for each gene, as shown in Table 2, followed by a melting curve, consisting of 95°C for 15 s, 60°C for 30 s, and a subsequent temperature increase until 95°C with a ramp rate of 0.03°C s−1.
Specificity of the amplified products was checked by the observation of a single melting peak and the presence of a unique band of the expected size in a 2% agarose gel stained with ethidium bromide. PCR efficiencies (Eff) were calculated from the standard curve by the formula Eff = [10(−1/slope) − 1] × 100% and accounted for 91.8 to 93.6% for nifH genes, 94.1 to 98.1% for archaeal amoA genes, 83.1 to 83.5% for bacterial amoA genes, 90.1 to 90.8% for nirK genes, 90.1 to 98.2% for nirS genes, and 93.7 to 97.2% for nosZ genes.
T-RFLP.
Diversity analysis by terminal restriction fragment length polymorphism analysis (T-RFLP) was performed with one functional gene of each examined process, nitrogen fixation (nifH), nitrification (amoA from ammonia-oxidizing archaea [AOA]), and denitrification (nosZ), using the primer pairs described for quantitative real-time PCR (Table 2). The forward primer was labeled with 5′-carboxyfluorescein. PCR amplifications were carried out with each of the 25 DNA samples in triplicate. PCR profiles were as follows: for nifH, according to real-time PCR (Table 1); for amoA AOA, 30 cycles, 45 s at 94°C, 45 s at 55°C, and 45 s at 72°C; for nosZ, touchdown PCR with 5 cycles consisting of 95°C for 15 s, 65°C for 30 s, and 72°C for 30 s, followed by 30 cycles at a 60°C annealing temperature.
The restriction enzymes AatII (nifH), MwoI (amoA AOA), and HpyCH4V (nosZ) were selected based on in silico T-RFLPs using the program REPK (restriction endonuclease picker) (15). Digestion was performed according to the manufacturer's protocol.
For T-RFLP migration, 15 ng of digested amplicons were mixed in triplicate with 0.25 μl Genome Lab DNA size standard 600 and 26.75 μl of Genome Lab sample loading solution (Beckman Coulter, Germany) and separated on a capillary electrophoresis sequencer (CEQ 2000 genetic analyzer; Beckman Coulter, Germany) run with the following program: denaturation for 2 min at 90°C, injection for 30 s at 2,000 V, and separation for 60 min at 4,800 V.
Chromatograms were analyzed with the CEQ 8000 genetic analysis system software (Beckman Coulter) using the quartic model as a size-calling algorithm (according to the Genome Lab fragment analysis protocol), with a slope threshold of 1, a relative peak height threshold of 10% (relative to the second-highest peak), and a confidence level of 95% to identify peaks. Amplified fragment length polymorphism analysis was done with a binning range of 1 bp. T-RFLP profiles of the three technical replicates for each sample were highly similar.
The data set was normalized to the percentage of the total peak height of a sample (2), excluding peaks smaller than 0.5%.
Statistical analysis.
Real-time PCR data and soil chemical parameters were subjected to analysis of variance (ANOVA) using the statistical software program SPSS 13.0. Normal distribution of the variables and homogeneity of variances were checked by the Kolmogorov-Smirnov test, box plot analysis, and the Levene test.
To analyze the terminal restriction fragment data, we used between-group analysis (BGA) (16) based on correspondence analysis.
For a global test of any difference between the groups, the between-groups inertia percentage was used as a test statistic. Based on 999 permutations, a P value was calculated. In case of a significant result (P < 0.05), we performed tests comparing all pairs of groups; the P values were adjusted for multiple comparisons by the method of Hommel.
All analyses were undertaken with the ADE4 package (13) within the R software environment (www.R-project.org).
RESULTS
Soil chemical properties.
TOC increased significantly with cultivation time of soils, from 0.58% in P0 up to 3.1% in P2000. The same trend, with significant differences between all soils, was found for TN (Table 1).
WEOC concentrations were higher in P50 than in P0. For WEON, no significant differences between the soil samples of all sites under investigation were found, with the exception of P300, for which it was below the detection limit. Also, ammonium and nitrate concentrations of the soil samples showed no significant differences between all five sites measured, with the exception of the nitrate concentration in the P300 soil, which was significantly higher than that in P0 and P2000 (Table 1).
Linear regression analysis with logarithmized age showed that microbial biomass C contents increased highly significantly with cultivation time of soils and paralleled increasing amounts of extracted DNA. The same trend was observed for microbial biomass N, but the difference was significant only between P0 (39 μg g−1 soil) and P2000 (150 μg g−1 soil) (Table 1).
Quantification of functional genes.
Due to the significant increase in microbial biomass from P0 to P2000 (Table 1), we normalized gene copy numbers to the amount of extracted DNA. Gene copy numbers of nifH were comparable between all five soils and ranged between 2.7 × 103 and 1.6 × 104 copies ng−1 extracted DNA. For soil samples from P50 only, a significantly higher copy number was measured than with the other four soils.
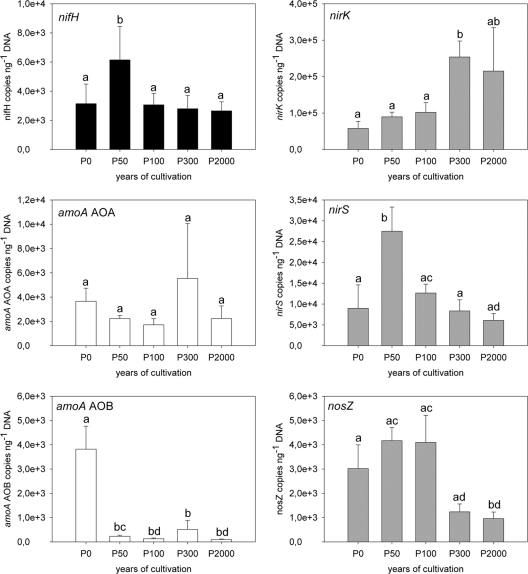
Gene copy numbers of ammonia-oxidizing archaea (amoA AOA) were higher in all samples than those of the bacterial counterpart (ammonia-oxidizing bacteria, amoA AOB), with the exception of P0, where values were comparable (for AOA, 3.7 × 103 copies ng−1 DNA; for AOB, 3.8 × 103 copies ng−1 DNA). AOA/AOB ratios increased significantly from P0 (1) to P2000 (23). For archaeal amoA gene copy numbers, no significant difference for all soils investigated was found (1.7 × 103 to 5.5 × 103 copies ng−1 DNA). In contrast, bacterial amoA gene copy numbers in the paddy soils were 9.9 × 101 (P2000) to 5.1 × 102 (P300) copies ng−1 DNA and, therefore, significantly lower than those for P0 (Fig. 1).
Fig. 1.
Copy numbers of the nifH, amoA AOA and AOB, nirK, nirS, and nosZ genes in the tidal wetland (P0) and the 50-, 100-, 300-, and 2,000-years-cultivated paddy soils (n = 5; error bars represent standard deviations). Significant differences are indicated by different letters.
In general, nirK gene copy numbers dominated over nirS genes by a factor of 3 to 35 in all soils investigated. nirK copy numbers in P300 were significantly higher than those in P0 (2.5 × 105, compared to 5.8 × 104 copies ng−1 DNA in P0). The nirS gene copy numbers were significantly higher in P50 than in P0, but there was no significant difference between P0 and the paddy soils different from P50. nosZ gene copy numbers were significantly lower in soil samples of P2000 than in those of P0, P50, and P100. Copy numbers for P300 were in between and showed neither a significant difference from those for P2000 nor from those for the other soils. In general, nosZ gene copy numbers were in the range of 103 copies ng−1 extracted DNA. The gene abundance results are summarized in Fig. 1.
Diversity analysis of functional genes.
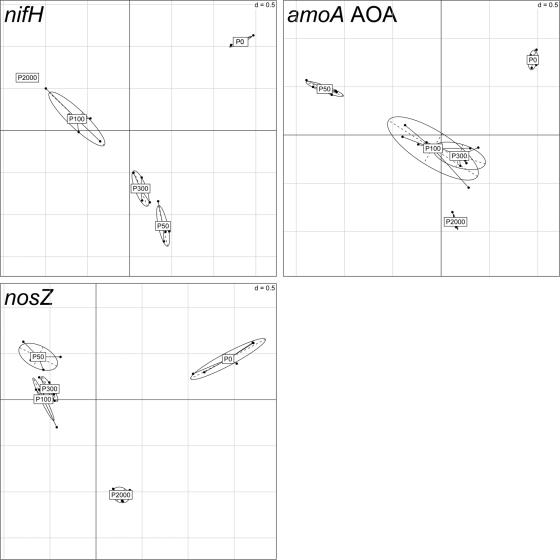
Statistical evaluation of genotype numbers for nifH, nosZ, and archaeal amoA genes by between-group analysis (BGA) showed a clustering according to the different soils for all genes under investigation, with the exception of P50, P100, and P300 for nosZ, all of which clustered together (Fig. 2). Also, no significant differences were found between P100 and P300 for amoA AOA.
Fig. 2.
Between-group analysis based on correspondence analysis of the T-RFLP data set for nifH, amoA AOA, and nosZ gene fragments. The first two axes explain 66.8 to 80.0% of variance. Symbols illustrate the five field replicates for each soil (P0, P50, P100, P300, and P2000). Ellipses surround the 5 replicates for each soil, showing that they cluster together.
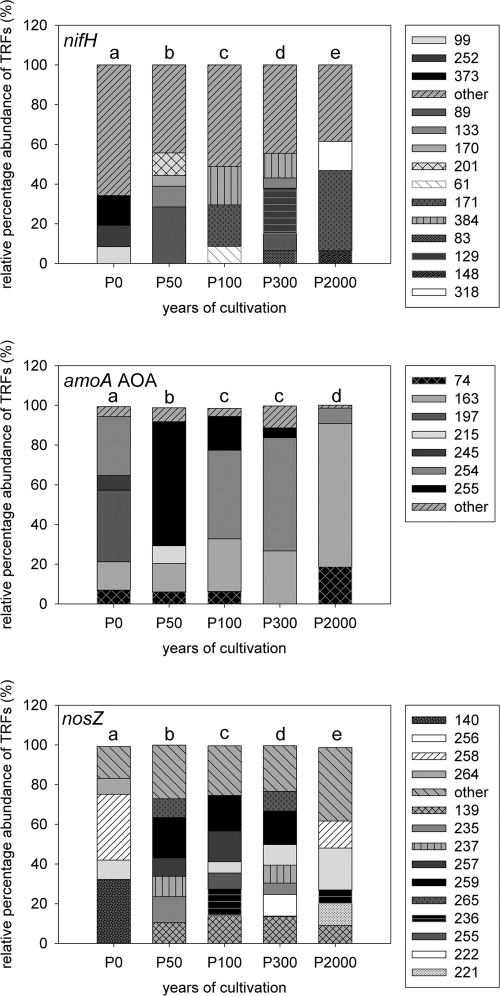
For nifH, the numbers of different terminal restriction fragments (T-RFs) in each soil were comparable and ranged between 21 and 23, with the exception of P2000, where only 15 T-RFs were detected and 1 very dominant T-RF was found, accounting for 40% of the total relative community richness (T-RF-171). This T-RF was also found in P100, with 21% of the total relative community richness. Dominant T-RFs in the other soils were different from each other. Statistical analysis comparing pairs of groups revealed significant differences between all five soils (Fig. 3 and Table 3).
Fig. 3.
Contributions of major T-RFs to total nifH, amoA AOA, and nosZ gene fragment diversity in the tidal wetland (P0) or 50-, 100-, 300-, or 2,000-year-cultivated paddy soils. T-RFs which contributed to less than 5% were summarized as “other.” Significant differences are indicated by different letters.
Table 3.
P values of pairwise comparisons for T-RFLP profiles of the functional genes nifH, amoA AOA, and nosZ, adjusted for multiple comparisons by the method of Hommela
| Gene and soil |
Pvalue for T-RFLP profile comparison |
||||
|---|---|---|---|---|---|
| P0 | P50 | P100 | P300 | P2000 | |
| nifH | |||||
| P0 | 1.000 | 0.008* | 0.008* | 0.008* | 0.008* |
| P50 | 1.000 | 0.008* | 0.008* | 0.008* | |
| P100 | 1.000 | 0.008* | 0.008* | ||
| P300 | 1.000 | 0.008* | |||
| P2000 | 1.000 | ||||
| amoA AOA | |||||
| P0 | 1.000 | 0.015* | 0.014* | 0.015* | 0.015* |
| P50 | 1.000 | 0.015* | 0.015* | 0.012* | |
| P100 | 1.000 | 0.192 | 0.015* | ||
| P300 | 1.000 | 0.012* | |||
| P2000 | 1.000 | ||||
| nosZ | |||||
| P0 | 1.000 | 0.008* | 0.008* | 0.008* | 0.008* |
| P50 | 1.000 | 0.008* | 0.008* | 0.008* | |
| P100 | 1.000 | 0.008* | 0.008* | ||
| P300 | 1.000 | 0.008* | |||
| P2000 | 1.000 | ||||
Significant impacts are marked by asterisks (P < 0.05).
T-RFLP analysis of the archaeal amoA genes showed a significant decrease in the genotype number from P0 (10) to P2000 (6). Dominant T-RFs in P0 were T-RF-197, -254, and -163, whereas T-RF-197 was unique for this soil. T-RF-163 became more dominant with cultivation time of soils and contributed to 72% of the total amoA AOA community in P2000. Other T-RFs, like T-RF-255, were present only in the paddy soils.
For nosZ, the total number of T-RFs in all paddy soils ranged between 19 and 21 but was lower in P0 (13). One dominant T-RF, T-RF-259, was found only in P50, P100, and P300. T-RF-139 was detected in all paddy soils but not in P0. The dominant T-RFs in P0 (T-RF-140 and -258; 32 and 33%, respectively) were not detected or were detected only in very small amounts in the paddy soils (1 to 14%) (Fig. 3).
DISCUSSION
Different chemical and physical properties could prove that the soils investigated developed in a chronological way. A significant loss of carbonates during paddy soil formation was found in paddy topsoils by considering the inorganic carbon concentrations (data not shown). A strong decalcification of the upper 20 cm was already observed after 50 years of rice cultivation (from 0.5% to 0.03%), whereas the complete decalcification of the total soil profile required almost 700 years of rice cultivation. In contrast, examination of neighboring upland fields of similar ages after 700 years of (nonpaddy) agricultural management showed only a decalcification of the upper 20 cm (L. Wissing et al., unpublished data). P2000 was completely free of carbonates, which may be a driver for the pH value being significantly lower than those of the other paddy fields (Table 1). These observations emphasize alteration of paddy soils due to the continuous rice paddy cultivation, with carbonates being leached by the periodical flooding and drainage.
The evolution of the paddy soils according to our age estimations was also confirmed by the continuously increasing TOC accumulation in the topsoils with cultivation time, from 1.7% in P50 to 3.1% in P2000, which is a possible driver for the increasing microbial biomass during paddy soil development (3, 29). This increasing TOC content may be caused by paddy soil management and especially by flooding of the fields during rice growth, since under waterlogged conditions, soil organic matter decomposition proceeds at lower rates than in well-drained, aerobic soils (27). Also, total nitrogen content of the soils increased chronologically with time in cultivation (Table 1).
Gene copy numbers and relative genotype richness of functional microbial communities involved in nitrogen transformation showed differences between the five soils investigated (P0, P50, P100, P300, and P2000). Low competition for free ammonia between plants and microbes in P0 might be a possible explanation for the high abundance of ammonia-oxidizing microbes in relation to the overall microbial biomass at this stage (significantly higher relative copy numbers of amoA AOB than were found in all paddy soils under investigation and comparable amounts for amoA AOA). Competition between plants and microbes for free ammonia has been shown in different studies (24, 33). Tanaka et al. (35) recently reported evidence that root exudates of rice can reduce nitrification rates in soil.
Gene copy numbers of archaeal ammonia oxidizers were constant at all phases of paddy soil development, indicating that they are able to adapt to changing environmental conditions (25). Another possible explanation, namely, that AOA just do not respond to these changes, can be excluded from the significantly changing T-RFLP pattern. A dominance of archaeal ammonia oxidizers over their bacterial counterparts in paddy soils has also been found in previous studies (8, 10).
Concerning alterations in the diversity pattern during paddy soil development, clear shifts in relative genotype richness for all genes under investigation (nifH, amoA AOA, and nosZ) were observed between P0 and the paddy soils. It could be assumed that microorganisms respond to the development from saltwater- to freshwater-influenced habitats at this first phase of paddy soil evolution. An influence of salinity on archaeal amoA community composition, for example, was suggested by Abell et al. (1).
After the conversion of the tidal wetland into paddy soils and the beginning of agricultural management, the high proportion of nitrogen-fixing microbes in P50 was very pronounced. In contrast, relative abundances of the dominant genes of nitrification (amoA AOA) and denitrification (nirK) in the paddy soils showed no significant differences between P0 and P50, indicating the important role of the plants as drivers for the nitrogen cycle, since they are competitors for ammonia and nitrate, as well as being donators of bioavailable carbon through exudation (34).
With ongoing rice cultivation, microbial biomass increased constantly (although the differences between P50, P100, and P300 were not significant) and reached its maximum in P2000. The relative abundance of nitrogen-fixing microbes at P100, P300, and P2000 was significantly lower than that for P50, probably due to ongoing fertilization. This hypothesis is confirmed by the relatively high abundance of nitrite reducers with the highest nirK copy numbers in P2000. These long-term fertilization effects have been also described by other authors (e.g., see reference 28). In general, nirK copy numbers were higher than nirS copy numbers in all soils investigated. A dominance of nirK over nirS in different soils was also found in previous studies (e.g., see reference 22).
Overall, our study indicated evidence of alterations in microbial communities involved in inorganic nitrogen cycling among the different soils investigated. We could clearly prove that (i) microbial communities between tidal wetland and paddy soils differ significantly and (ii) that during ongoing paddy soil cultivation, microbial communities adapt to the changes in soil structure and organic matter quality and specific phylotypes of selected functional groups become dominant. This addresses our introduced question, showing that changes are caused by the type of management and are not a response to the disturbance of a balanced ecosystem. In general, changes in the diversity pattern were more pronounced than those in functional gene copy numbers. The named soil properties changing along the chronosequence as a result of paddy soil evolution may be drivers for the observed shifts in microbial community function and diversity. However, it cannot be excluded that other between-site differences provide an explanation for the observed results, although agricultural management and water regimes were comparable between the different sites. This is a typical problem of field studies in general. To clarify, an additional experiment in the greenhouse under controlled conditions will be necessary. A closer link to activity and turnover rates would be possible by analysis of transcripts.
ACKNOWLEDGMENTS
We thank Gudrun Hufnagel for excellent technical support in measuring ammonia and nitrate concentrations. Many thanks also go to Adrian Ho and Peter Frenzel for their help during soil sampling. We also thank the reviewers of the manuscript for their valuable input.
Financial support was provided by the German Research Foundation DFG. This paper represents a contribution to the DFG FOR 995 biogeochemistry of paddy soil evolution project.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Abell G. C. J., et al. 2010. Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. ISME J. 4:286–300 [DOI] [PubMed] [Google Scholar]
- 2. Anderson M. J. 2003. Review of analysis of ecological communities (B McCune and J. B. Grace, 2002, MjM Software Design). J. Exp. Mar. Biol. Ecol. 289:303–305 [Google Scholar]
- 3. Anderson T.-H., Domsch K. H. 1989. Ratios of microbial biomass carbon to total organic carbon in arable soils. Soil Biol. Biochem. 21:471–479 [Google Scholar]
- 4. Attard E., et al. 2010. Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ. Microbiol. 12:315–326 [DOI] [PubMed] [Google Scholar]
- 5. Azul A. M., Sousa J. P., Agerer R., Martin M. P., Freitas H. 2010. Land use practices and ectomycorrhizal fungal communities from oak woodlands dominated by Quercus suber L. considering drought scenarios. Mycorrhiza 20:73–88 [DOI] [PubMed] [Google Scholar]
- 6. Berthrong S. T., Schadt C. W., Pineiro G., Jackson R. B. 2009. Afforestation alters the composition of functional genes in soil and biogeochemical processes in South American grasslands. Appl. Environ. Microbiol. 75:6240–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braker G., Fesefeldt A., Witzel K.-P. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Briones A. M., et al. 2002. Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice. Appl. Environ. Microbiol. 68:3067–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cassman K. G., et al. 1998. Opportunities for increased nitrogen-use efficiency from improved resource management in irrigated rice systems. Field Crops Res. 56:7–39 [Google Scholar]
- 10. Chen X. P., Zhu Y. G., Xia Y., Shen J. P., He J. Z. 2008. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10:1978–1987 [DOI] [PubMed] [Google Scholar]
- 11. Cheneby D., Brauman A., Rabary B., Philippot L. 2009. Differential responses of nitrate reducer community size, structure, and activity to tillage systems. Appl. Environ. Microbiol. 75:3180–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng Y.-Q., Yang L.-Z., Cao Z.-H., Ci E., Yin S. 2009. Chronosequential changes of selected pedogenic properties in paddy soils as compared with non-paddy soils. Geoderma 151:31–41 [Google Scholar]
- 13. Chessel D., Dufour A. B., Thioulouse J. 2004. The ade4 package-I-One-table methods. R News 4:5–10 [Google Scholar]
- 14. Chu H., Morimoto S., Fujii T., Yagi K., Nishimura S. 2009. Soil ammonia-oxidizing bacterial communities in paddy rice fields as affected by upland conversion history. Soil Sci. Soc. Am. J. 73:2026–2031 [Google Scholar]
- 15. Collins R. E., Rocap G. 2007. REPK: an analytical web server to select restriction endonucleases for terminal restriction fragment length polymorphism analysis. Nucleic Acids Res. 35:W58–W62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Culhane A. C., Perriere G., Considine E. C., Cotter T. G., Higgins D. G. 2002. Between-group analysis of microarray data. Bioinformatics 18:1600–1608 [DOI] [PubMed] [Google Scholar]
- 17. Dell E., Bowman D., Rufty T., Shi W. 2008. Intensive management affects composition of betaproteobacterial ammonia oxidizers in turfgrass systems. Microb. Ecol. 56:178–190 [DOI] [PubMed] [Google Scholar]
- 18. De Datta S., Buresh R. 1989. Integrated nitrogen management in irrigated rice. Adv. Soil Sci. 10:143–169 [Google Scholar]
- 19. Fraterrigo J. M., Balser T. C., Turner M. G. 2006. Microbial community variation and its relationship with nitrogen mineralization in historically altered forests. Ecology 87:570–579 [DOI] [PubMed] [Google Scholar]
- 20. Henry S., et al. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59:327–335 [DOI] [PubMed] [Google Scholar]
- 21. Henry S., Bru D., Stres B., Hallet S., Philippot L. 2006. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72:5181–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones C. M., Hallin S. 2010. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 4:633–641 [DOI] [PubMed] [Google Scholar]
- 23. Jörgensen R. G., Brookes P. C. 1991. Die Bestimmung der mikrobiellen Biomasse des Bodens mit der Fumigations-Extraktions-Methode, vol. 33, p. 511–514 Mitteilungen der Deutschen Bodenkundliche Gesellschaft, Oldenburg, Germany [Google Scholar]
- 24. Kaye J. P., Hart S. C. 1997. Competition for nitrogen between plants and soil microorganisms. Trends Ecol. Evol. 12:139–143 [DOI] [PubMed] [Google Scholar]
- 25. Leininger S., et al. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809 [DOI] [PubMed] [Google Scholar]
- 26. Michotey V., Mejean V., Bonin P. 2000. Comparison of methods for quantification of cytochrome cd1-denitrifying bacteria in environmental marine samples. Appl. Environ. Microbiol. 66:1564–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Neue H. U., Gaunt J. L., Wang Z. P., BeckerHeidmann P., Quijano C. 1997. Carbon in tropical wetlands. Geoderma 79:163–185 [Google Scholar]
- 28. Philippot L., Hallin S., Schloter M. 2007. Ecology of denitrifying prokaryotes in agricultural soil. Adv. Agron. 96:249–305 [Google Scholar]
- 29. Powlson D. S., Prookes P. C., Christensen B. T. 1987. Measurement of soil microbial biomass provides an early indication of changes in total soil organic matter due to straw incorporation. Soil Biol. Biochem. 19:159–164 [Google Scholar]
- 30. Rösch C., Mergel A., Bothe H. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rotthauwe J., Witzel K., Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schauss K., et al. 2009. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ. Microbiol. 11:446–456 [DOI] [PubMed] [Google Scholar]
- 33. Schimel J. P., Bennett J. 2004. Nitrogen mineralization: challenges of a changing paradigm. Ecology 85:591–602 [Google Scholar]
- 34. Schnurer J., Rosswall T. 1982. Fluorescein diacetate hydrolysis as a measure of total microbial activity in soil and litter. Appl. Environ. Microbiol. 43:1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tanaka J. P., Nardi P., Wissuwa M. 2010. Nitrification inhibition activity, a novel trait in root exudates of rice. AoB Plants 2010:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Throbäck I. N., Enwall K., Jarvis Å., Hallin S. 2004. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401–417 [DOI] [PubMed] [Google Scholar]
- 37. Treusch A. H., et al. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985–1995 [DOI] [PubMed] [Google Scholar]
- 38. Vargas Gil S., et al. 2009. Field trial assessment of biological, chemical, and physical responses of soil to tillage intensity, fertilization and grazing. Environ. Manag. 44:378–386 [DOI] [PubMed] [Google Scholar]
- 39. Yoon C. G. 2009. Wise use of paddy rice fields to partially compensate for the loss of natural wetlands. Paddy Water Environ. 7:357–366 [Google Scholar]
- 40. Zhang M., Lu H., Zhao X., Li R. 2004. A comparative study of fertility change of upland soil in Cixi County. Chin. J. Soil Sci. 35:91–93 [Google Scholar]