Abstract
With the advent of the -omics era, classical technology platforms, such as hyphenated mass spectrometry, are currently undergoing a transformation toward high-throughput application. These novel platforms yield highly detailed metabolite profiles in large numbers of samples. Such profiles can be used as fingerprints for the accurate identification and classification of samples as well as for the study of effects of experimental conditions on the concentrations of specific metabolites. Challenges for the application of these methods lie in the acquisition of high-quality data, data normalization, and data mining. Here, a high-throughput fingerprinting approach based on analysis of headspace volatiles using ultrafast gas chromatography coupled to time of flight mass spectrometry (ultrafast GC/TOF-MS) was developed and evaluated for classification and screening purposes in food fermentation. GC-MS mass spectra of headspace samples of milk fermented by different mixed cultures of lactic acid bacteria (LAB) were collected and preprocessed in MetAlign, a dedicated software package for the preprocessing and comparison of liquid chromatography (LC)-MS and GC-MS data. The Random Forest algorithm was used to detect mass peaks that discriminated combinations of species or strains used in fermentations. Many of these mass peaks originated from key flavor compounds, indicating that the presence or absence of individual strains or combinations of strains significantly influenced the concentrations of these components. We demonstrate that the approach can be used for purposes like the selection of strains from collections based on flavor characteristics and the screening of (mixed) cultures for the presence or absence of strains. In addition, we show that strain-specific flavor characteristics can be traced back to genetic markers when comparative genome hybridization (CGH) data are available.
INTRODUCTION
Metabolomic analysis, i.e., the measurement of (relative) concentrations of a large number of metabolites in a biological sample, is essential to come to a comprehensive understanding of living organisms, since phenotypic properties result from the history, genotype, environment, and their interactions (7). But even in the absence of a complete understanding of the causal chain leading to a particular metabolic profile, these profiles have the potential for use as fingerprints or biomarkers in a large variety of applications, like diagnosis in medicine (10) or in food sciences as biomarkers of taste and health properties (3). These applications depend on the ability to collect high-quality metabolomic data and on the proper normalization and analysis of the resulting high-dimensional data. The technology for collecting metabolome data has advanced rapidly in the past years (3). In particular, gas chromatography-mass spectrometry (GC-MS) and liquid chromatography (LC)-MS are increasingly utilized for profiling of biological samples because of their inherent robustness, sensitivity, and large dynamic range (23). However, the development of data analysis techniques is lagging behind, with the consequence that the novel techniques are probably not used to their full potential. Bottlenecks in data analysis concern the normalization and alignment of GC-MS data to allow the comparison of samples, the chemical identification of compounds, and the identification of biomarkers. To tackle the normalization and alignment problems, tools have been developed that facilitate user-assisted preprocessing of multiple mass spectra to correct retention time drifts that are inherent in chromatography (8, 14). Basically these packages perform baseline correction, smoothing, and alignment of mass spectra to enable peak comparison and subsequent multivariate analysis. Together with technological advances, such as ultrafast GC coupled to time-of-flight (TOF) detection of ions (19, 30), these software packages significantly reduce the time required to analyze and compare detectable compounds in large numbers of samples. One of the tools that is widely applied is MetAlign (http://www.metalign.wur.nl/UK/) (16), a package that initially was developed to support LC-MS-based metabolomics and was, for instance, used as such to study the metabolite profiles of Arabidopsis thaliana and fruits of the tomato plant (Lycopersicon esculentum) (2, 15).
Methods for the subsequent identification of biomarkers, by investigation of correlations of metabolome data with other sample properties, are sometimes included in the packages for data preprocessing. However, these are not suitable for all challenges in data mining and the researcher then depends on other methods. A “Swiss knife” among the methods for the detection of such correlations is the Random Forest algorithm. The Random Forest algorithm was introduced by Breiman (4) as a method for supervised or unsupervised data classification and regression. It has properties that make it very suitable as an exploratory tool for high-dimensional data gathered on relatively small sets of samples, like transcriptome or metabolome data. The method is relatively insensitive to noise and outliers, thereby avoiding overfitting. Also, data transformations like normalizations and rescaling are usually not necessary. Furthermore, the method indicates which of the variables are highly correlated with sample properties by so-called variable importance measures. Since only a small number of algorithm parameter values, to which the algorithm is not very sensitive, have to be set by the user, it is also a very user-friendly method. These properties have made the Random Forest algorithm a popular tool in life sciences, and in this paper we show how it can be applied to target differences in profiles of volatile compounds present in food products.
Mixed-culture fermentation represents an interesting case in which to apply metabolite profiling techniques, since the organoleptic properties of fermented food products are strongly influenced by microbial activity during and after fermentation and directly relate to the strains involved and their population dynamics. As a consequence, small changes in microbial composition or population dynamics may have a large effect on product characteristics and on flavors in particular, since small differences in the levels of some of these can have a significant impact on the aroma of the product (5, 11, 20). Hence, there is a clear need for fast and predictive methods to screen for flavor development in mixed-culture food fermentations (21, 24).
We investigated an ultrafast GC/TOF-MS approach to perform untargeted profiling of volatile compounds detected in food using MetAlign and subsequent mining of the data using the Random Forest algorithm. To reduce analysis times, samples were not concentrated and the usefulness of the approach for screening purposes was examined by analyzing the volatile compounds produced by various mono- and mixed cultures of lactic acid bacteria (LAB) during fermentation of milk.
MATERIALS AND METHODS
Ultrafast GC/TOF-MS.
A Thermo Finnigan Tempus ultrafast GC/TOF-MS system was used for separation and detection of volatile compounds in the headspace of fermented milk samples. Chromatography was performed using a capillary column (10 m by 0.18 mm; UFM PH-RTX200; Interscience, Breda, Netherlands) with a 0.4-μm film thickness. The column oven, which was cooled by injection of liquid carbon dioxide, was held at 10°C for 0.5 min, programmed to 200°C at 100°C/min, and then held at 200°C for 0.5 min. The injector, transfer line, and detector temperatures were kept constant at 250°C. Before injection, sample vials were transferred from a cooled sample tray (4°C) into a dynamic mini-oven and preheated for 5 min at 60°C. Static headspace samples of 200 μl were injected into the GC inlet using a 12:1 split ratio. The samples were run at a constant column flow rate of 2.0 ml/min. The mass range was scanned from 35 to 350 atomic mass units (amu) at a scan rate of 25 scans/s.
Culture conditions.
Streptococcus thermophilus strains CNRZ1066, LMG18311, and LMD9 and 40 Lactococcus lactis strains (Table 1) were precultured in GM-17 broth containing 1% glucose at 42°C and 30°C, respectively. Lactobacillus delbrueckii subsp. bulgaricus strain ATCC BAA-365 and Lactobacillus plantarum strain WCFS1 were precultured in MRS broth at 42 and 37°C, respectively. Cultures were adapted to growth in reconstituted ultra-heat-treated (UHT) skim milk by transferring 1% from the broth cultures. The lactobacilli and streptococci were incubated at 37°C for 24 h, and the lactococci were incubated at 30°C for 48 h. Acidification of each milk batch was recorded using the CINAC system (9), and CFU counts were determined by plating on selective agar media. Samples for ultrafast GC/TOF-MS analysis were prepared by transferring 5-ml fermented milk samples to 10-ml glass crimp-cap vials or by inoculating the milk within these vials for the lactococci. The vials were sealed immediately after inoculation or filling with magnetic crimp caps with Teflon inserts.
Table 1.
Origin and subspecies of Lactococcus lactis strains used in this study
| NIZO ID | Alternative ID | Subspecies | Isolation source |
|---|---|---|---|
| B1156 | Li-1 | lactis | Grass |
| B1157 | V4 | lactis | Raw sheep milk |
| B1173 | E34 | lactis | Silage |
| B1175 | N41 | lactis | Soil and grass |
| B1230 | N42 | lactis | Soil and grass |
| B1492 | MG1363 | lactis | Cheese starter |
| B1592 | DRA4 | lactis diacetylactis | Dairy starter A |
| B20 | ML8 | lactis | Unknown (dairy starter) |
| B2123 | LMG9446 | lactis | Frozen peas |
| B2124 | LMG9449 | lactis | Frozen peas |
| B2199 | K231 | lactis | White kimchi |
| B2202 | K337 | lactis | White kimchi |
| B2206 | P7266 | lactis | Litter of pasture grass |
| B2207 | P7304 | lactis | Litter of pasture grass |
| B2211 | NCIMB700895 | lactis | Unknown (dairy starter) |
| B2219 | KF7 | lactis | Alfafa sprouts |
| B2220 | KF24 | lactis | Alfafa sprouts |
| B2223 | KF67 | lactis | Grapefruit juice |
| B2226 | KF134 | lactis | Alfalfa and radish sprouts |
| B2229 | KF146 | lactis | Alfalfa and radish sprouts |
| B2230 | KF147 | lactis | Mung bean sprouts |
| B2236 | KF196 | lactis | Japanese kaiware shoots |
| B2238 | KF201 | lactis | Sliced mixed vegetables |
| B2244b | KF282 | lactis | Mustard and cress |
| B2244w | lactis | Mustard and cress | |
| B2249 | KW 10 | lactis | Kaanga wai |
| B2252 | FG2 | cremoris | Unknown (dairy starter) |
| B2418 | LMG6897T | cremoris | Cheese starter |
| B2424 | LMG14418 | lactis | Bovine milk |
| B2441 | IL1403 | lactis | Unknown |
| B24 | LMG8520 | hordniae | Leaf hopper |
| B26 | LMG8526 | lactis | Chinese radish seeds |
| B29 | ATCC19435T | lactis | Unknown (dairy starter) |
| B32 | SK11 | cremoris | Unknown (dairy starter) |
| B33 | AM2 | cremoris | Unknown (dairy starter) |
| B42 | HP | cremoris | Unknown (dairy starter) |
| B643 | ML3 | lactis | Unknown (dairy starter) |
| B644 | UC317 | lactis | Unknown (dairy starter) |
| B844 | M20 | lactis diacetylactis | Soil |
Data analysis.
Mass spectra were processed in MetAlign using parameters optimized for the spectra recorded on the Thermo Finnigan ultrafast GC/TOF-MS. Four replicate measurements were performed per sample condition (combination of bacteria). The resulting data sets, which consisted of ∼10,000 aligned time-mass peaks and their intensities in each sample, were reduced by analysis of variance (ANOVA) filtering. A one-way ANOVA test was applied to select only those time-mass peaks that displayed a statistically significant different signal in at least one of the sample conditions. The stringency of this filter was kept very low by using a P value cutoff of 0.5. Filtered data sets consisted of approximately 3,600 time-mass peaks. The Random Forest algorithm was applied on these data sets using an implementation of the algorithm in R (http://www.r-project.org/) written by A. Liaw and M. Wiener. A unique culture label for each of the mixtures was used as the response variable, whereas the 3,600 selected time-mass peak intensities were used as predictor variables. The mtry parameter (number of variables randomly sampled as candidates at each split) was set at the default value (square root of the number of variables), and the number of trees grown was set at 5,000. Sets of approximately 100 time-mass peaks with high importance in the resulting classifier were selected for further manual inspection. Using the Xcalibur software (Thermo Finnigan) and AMDIS (27), most of these time-mass peaks could be associated with high confidence to volatile compounds present in the NIST mass spectral library (http://www.nist.gov/srd/nist1a.htm). Subsequently, individual time-mass peak intensities were summed by compound.
RESULTS
Differential profiling of mono- and mixed-culture fermentations.
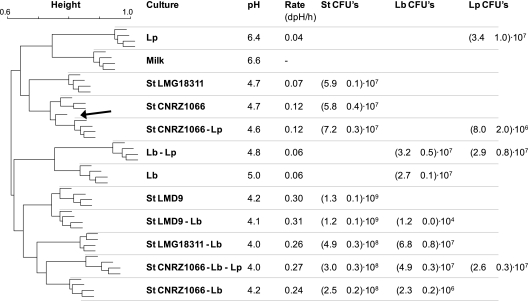
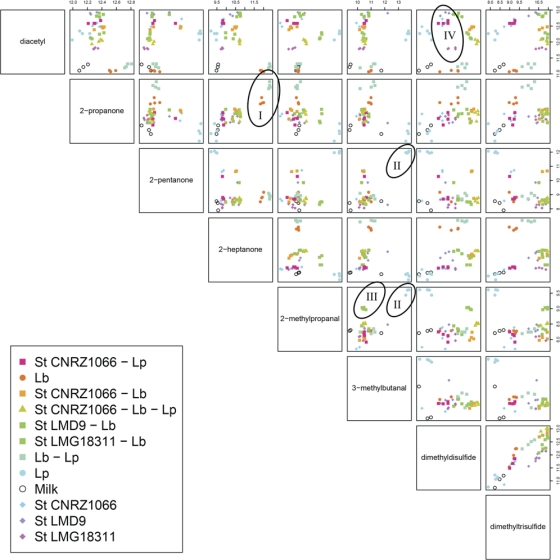
To assess whether the approach can be used for discrimination of cultures at the species and strain level based on volatile compounds produced, 11 single and mixed-culture fermentations were prepared. Batches containing 250 ml of preheated milk were inoculated with Lb. delbrueckii subsp. bulgaricus, Lb. plantarum, three strains of S. thermophilus, and mixtures of these (Fig. 1). The batches, including a control batch containing uninoculated milk, were incubated at 37°C in a water bath, and acidification of the cultures was recorded using the CINAC system. After 26 h, fermentations were stopped by cooling on ice and samples were taken to determine species-specific viable counts and to prepare for headspace analysis. The final pHs of the cultures varied between 4.0 for two of the cultures containing both yoghurt bacteria and 6.4 for the milk inoculated with Lb. plantarum only (Fig. 1). The culture containing S. thermophilus strain LMD9 and all mixed cultures including both S. thermophilus and Lb. delbrueckii subsp. bulgaricus showed the highest acidification rates. The volatile compounds present in each culture after fermentation were analyzed in quadruplicate using ultrafast GC/TOF-MS followed by alignment of the mass spectra and further processing of the resulting peak lists using Random Forest. To determine which mass peaks most significantly contributed to the distinction of the 12 different groups, the peak lists generated by MetAlign were first filtered by ANOVA, and the remaining peaks were used to build a Random Forest predictor. This allowed accurate prediction of nearly all samples. Only discrimination of the S. thermophilus CNRZ1066 culture from the mixed culture with Lb. plantarum seemed to be limited (Fig. 1). To identify the compounds corresponding to discriminatory mass peaks, they were traced back in the original spectra using the mass spectrometer software. Whenever possible, compounds were identified using the NIST library. Finally, a pair plot of the summed total ion count signals of relevant compounds was generated to visualize which compounds allowed discrimination of the different groups as well as to reveal possible relations between the levels of different compounds produced (Fig. 2). From this plot, compounds responsible for distinction of the flavor profiles of the different combinations of bacteria can be identified. Distinction of the cultures containing Lb. delbrueckii subsp. bulgaricus only or together with Lb. plantarum was mainly due to the higher levels of 2-heptanone and 2-propanone produced and the low level of diacetyl produced compared to the other cultures (Fig. 2, I). The presence of Lb. plantarum led to high levels of 2-methylpropanal, 3-methylbutanal, and 2-pentanone in milk but only in the absence of other lactic acid bacteria (Fig. 2, II). The level of 2-methylpropanal was also relatively high in the milk fermented with Lb. delbrueckii subsp. bulgaricus and S. thermophilus strain LMG18311 (Fig. 2, III). Separation of the three different strains of S. thermophilus was mainly due to differences in the levels of diacetyl produced (Fig. 2, IV, blue, lilac, and pink diamonds).
Fig. 1.
Hierarchical clustering using Random Forest sample proximities of mass peak profiles generated by ultrafast GC/TOF-MS analysis of volatile compounds in headspace samples. The tree represents 48 mono- and mixed cultures of lactic acid bacteria in milk, with uninoculated milk as a control, after 26 h incubation at 37°C. Each condition (12 conditions in total) was tested in quadruplicate to illustrate that differences in volatile compounds present after incubation are big enough to allow reproducible discrimination. ANOVA-filtered GC/TOF-MS peak data were used to construct a Random Forest classifier (see Materials and Methods). From the classifier, sample proximities were calculated, which are a measure of the similarity of samples. To display the results, a hierarchical tree based on these sample proximities was built, so that samples appearing close in the tree are those that had a close proximity in the Random Forest classifier. The tree shows that Random Forest is able to discriminate most sample types based on GC-MS peak data. With one exception (indicated with an arrow), all replicates cluster together. Acidification properties and species-specific CFU counts (CFU) of the cultures are presented in the right panel. St, S. thermophilus; Lp, Lb. plantarum WCFS1; Lb, Lb. delbrueckii subsp. bulgaricus ATCC BAA365.
Fig. 2.
Pair plot showing the differences in levels of volatile compounds detected in fermented milk cultures. The same data as in Fig. 1 were used. The compounds displayed are those corresponding to mass peaks of which the Random Forest analysis indicated that they were most important in discriminating sample types. Names of compounds in the squares on the diagonal indicate the compound displayed on the axes in the rows or columns of graphs intersecting with that square. Every pair of compounds is displayed once. The scales display the compound peak areas on a logarithmic scale, and the level increases from left to right (upper scale bar) and from bottom to top (right scale bar). The pair plot visualizes which compounds, or combinations of compounds, allow discrimination of individual groups. The pair plot shows, for instance, that a high concentration of 2-propanone is highly discriminative for the combination of Lb. plantarum and Lb. delbrueckii subsp. bulgaricus and pure Lb. delbrueckii subsp. bulgaricus cultures (I), whereas high concentrations of 2-pentanone, 2-methylbutanal, and 2-methylpropanal are characteristic of pure L. plantarum cultures (II). The pair plot can also be used to visualize correlations between different compounds. For instance, the levels of dimethyldisulfide and dimethyltrisulfide seem to be positively correlated. St, S. thermophilus; Lp, Lb. plantarum WCFS1; Lb, Lb. delbrueckii subsp. bulgaricus ATCC BAA365. Marked symbols I to IV are explained in Results.
Differential profiling of a Lactococcus lactis strain collection.
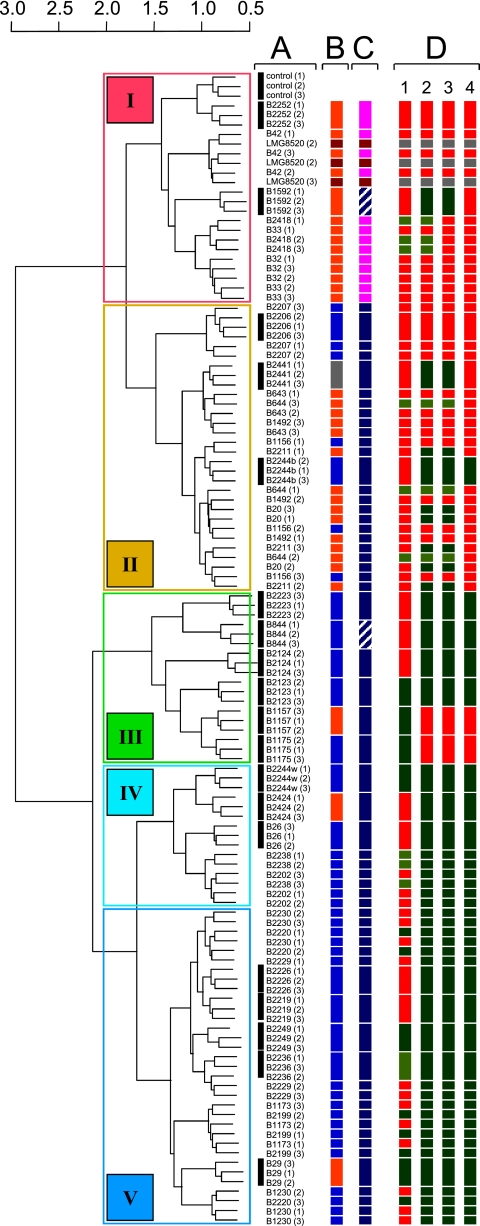
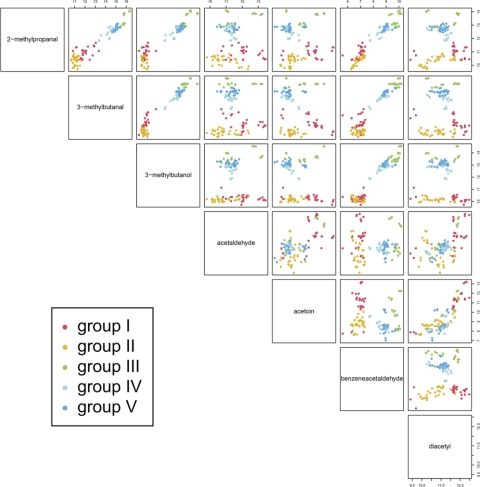
To examine whether the approach outlined here can also be used to distinguish strains in a collection of a single species, 40 Lc. lactis strains (Table 1) were cultured in triplicate in 10-ml crimp-cap vials containing 5 ml milk with additionally 2% glucose and 0.5% Casitone to ensure good growth of all strains, including lactose-deficient and nonproteolytic ones. An approach similar to that described above was chosen, except that we first performed an outlier identification. Before analyzing the aligned mass peaks with Random Forest, the mean absolute deviation for all peaks and all sample triplicates were calculated. Subsequently those outliers that were very far off the median value of the triplicates and for which the median signal value was >2,000 were repaired. By analyzing the resulting data set, Random Forest accurately predicted approximately 55% of the strain combinations. Hierarchical clustering based on Random Forest sample proximities revealed a significant correlation between the volatile compound profiles and strain origin (dairy/nondairy) as well as subspecies level (Fig. 3). A pair plot generated as described above, in which the samples were classified into 5 main groups (I to V) based on the results of the cluster analyses, showed that nearly all compounds contributed to the separation of the volatile compound profiles into two main groups (Fig. 3 and 4). In groups III to V, with predominantly nondairy strains, the strains produced relatively high levels of these compounds, while the levels were relatively low in groups I and II. Within the latter group, separation of Lc. lactis subsp. cremoris (into group I) was mainly due to the production of acetoin, but aldehydes and diacetyl also contributed significantly.
Fig. 3.
Hierarchical clustering using Random Forest sample proximities of mass peak profiles generated by ultrafast GC/TOF-MS analysis of volatiles compounds in headspace samples (in triplicate) of 40 Lc. lactis strains in milk. Colored Roman numerals I to V in the left panel correspond to the grouping used to illustrate the differences in levels and correlations of specific volatile compounds detected (Fig. 4). Column A, strains for which the triplicates cluster together are indicated with a black bar. Column B, isolation source: blue, nondairy; orange, dairy; dark red, insect (leaf hopper); gray, unknown. Column C, subspecies: dark blue, lactis; pink, cremoris; dark red, hordniae; striped white, diacetylactis (biovar of lactis). Column D, gene presence/absence based on pangenomic array signals of probes that target alpha-keto acid decarboxylase (kdcA) (1), indole-pyruvate decarboxylase (ipd) (2), keto-isovalerate decarboxylase (kivd) (3), and the 330-bp 3′ end of kivd corresponding to a deletion in ipd required for activity (4). Red, gene is absent (log signal intensity, <5.5); dark green, gene is present (log signal intensity, >6); green, gene is most likely present (log signal intensity, between 5.5 and 6). Strain LMG8520 (gray) was not included in this study.
Fig. 4.
Pair plot showing the differences in levels and correlations of volatile compounds detected in fermented milk cultures of 40 Lactococcus lactis strains. Groups correspond to those obtained from the hierarchical clustering in Fig. 3. For an explanation of pair plots, see the legend to Fig. 2.
To illustrate that flavor production levels can also be linked to gene content when whole-genome hybridization (CGH) data for the strains are available, we focused on 3-methylbutanal, which is a key flavor compound in many food products (25) and whose production by Lc. lactis is known to be strain dependent (26). Production of this compound and related aldehydes requires alpha-keto acid decarboxylase, encoded by either kdcA or kivd (6, 26). The presence or absence of these genes in each of the individual strains was evaluated using pangenomic microarray hybridization data (1). For all strains that produced high levels of 3-methylbutanal, the averaged hybridization signals of oligonucleotide probes on the microarray that targeted kdcA, kivd, or both were high enough to confirm the presence of the alpha-keto acid decarboxylase coding capacity (Fig. 3). Also, some of the strains that did not produce high levels of 3-methylbutanal were positive for the presence of kivd but for these, with only one exception, signals of probes that targeted the 3′ terminus of the gene thought to be essential to produce a functional alpha-keto acid decarboxylase (26) were low (Fig. 3).
DISCUSSION
Metabolomics of biological samples using mass spectrometry is a challenging field (7, 8, 14). In particular, GC-MS and LC-MS are increasingly utilized for differential profiling of biological samples because of their inherent robustness, sensitivity, and large dynamic range (23). As a result of recent advances in these technology platforms and development of tools for high-throughput processing of the data, many new applications are conceivable. One of these is volatile compound profiling of food products, for instance to assess the effects of altered process conditions or formulation on the organoleptic properties. We evaluated a nontargeted GC-MS-based approach to screen for differences in volatile compounds present in milk samples fermented by mono- and mixed cultures of lactic acid bacteria. The approach can for instance be used to assess the effects of the addition of adjunct cultures to well-characterized mixed-culture fermentations (18, 22), but it may serve many other applications where differential profiling of volatile compounds in food products may yield valuable information. We used ultrafast GC/TOF-MS to separate and detect volatile compounds, MetAlign to enable comparison of the data, and Random Forest to detect mass peaks that allowed discrimination of the samples.
To increase the sensitivity, volatile compounds in food products are usually first concentrated by solid-phase microextraction (SPME) or related techniques prior to GC-MS analysis (12, 17). We omitted concentration steps, since they are relatively time-consuming, and we were able to detect many compounds known to contribute to the flavor characteristics of fermented food without it, by using ultrafast GC/TOF-MS. Ultimately, after optimizing the GC-MS configuration, the time required to record one GC-MS chromatogram was approximately 7 min. When the CO2 cooling step, which was required for separation of some highly volatile compounds such as acetaldehyde and methanethiol, was omitted, this time could be reduced to 4 min. MetAlign has already been successfully applied in several studies, including a GC-MS-based screening of volatile compounds (29), and a new version of this metabolomics tool was launched recently (16). Since mass peaks that belong to one compound are separated in individual peaks with unique identifiers by this application, a tool for multivariate mass spectral reconstruction (MMSR) was also developed (29). However, the number of peaks that proved to be relevant for discrimination of our samples was relatively low, and therefore we performed this operation manually.
We showed that the approach outlined here is strong enough to discriminate different species and mixtures of lactic acid bacteria based on volatile compounds produced in milk and even different strains in a collection. The sensitivity of the approach largely depends on the volatile compounds produced or removed by individual species or strains and the levels of these compounds. For instance, the influence of Lb. delbrueckii subsp. bulgaricus on the volatile compound profile of milk fermented by a proteolytic S. thermophilus strain was still detectable while it was outnumbered by more than 4 orders of magnitude (Fig. 1). Remarkably, the levels of the methylated sulfides that allowed discrimination of these samples, and dimethyl trisulfide (DMTS) in particular, were relatively low for the monocultures of Lb. delbrueckii subsp. bulgaricus and S. thermophilus LMD9 (Fig. 2), which suggests that the increased levels in the mixed culture are the result of interaction between the two species. In line with this observation, high levels of aldehydes and 2-pentanone were observed only in the milk fermented with Lb. plantarum, not in mixed cultures containing this species. Clearly, the other species directly or indirectly affected the levels of these compounds by, for instance, limiting their production by Lb. plantarum or by reduction to the corresponding alcohols, which are less volatile. Interactions between strains were also observed for the formation of 2-heptanone by Lb. delbrueckii subsp. bulgaricus. Formation of this compound by this species has been reported (28), but unfortunately there is no information on how it is produced in lactic acid bacteria. All together, these data reinforce the increased awareness that bacteria could employ certain volatiles to influence other microorganisms (13).
We also showed that differences in flavor production levels may be linked to strain-specific genetic markers when genomic data of one or more of the strains is available. Production of high levels of 3-methylbutanal requires alpha-keto acid decarboxylase, which is encoded by the kdcA gene and is unique to some strains of Lc. lactis (26). When we focused on this enzyme alone, we could not find a correlation between the 3-methylbutanal production levels of the strains and signal intensities of probes that target the kdcA gene on a pangenomic Lc. lactis microarray (data not shown). However, when we included kivd in these analyses, a gene that also has been associated with production of aldehydes in Lc. lactis (6), we found that all 3-methylbutanal-producing strains harbor an alpha-keto acid decarboxylase gene, of which the gene sequence similarity for most strains was highest to kivd. Since some of the strains that produced low levels of 3-methylbutanal also seemed to be positive for kivd, the hybridization signals for probes that target the 3′ terminus of the gene were investigated separately. The gene sequence of kivd appeared to be highly similar to a putative indole pyruvate decarboxylase in Lc. lactis IL1403, and Smit et al. hypothesized that a 270-bp deletion at the 3′ terminus of this gene (with respect to kdcA) is essential for a functional alpha-keto acid decarboxylase (26). When the signals of the probes that target this part of the gene were considered, only one strain in the upper branch of the tree (low levels of 3-methylbutanal [Fig. 3]) appeared to be positive for the full kivd gene. A possible explanation could be that the enzyme in this strain is not functional or not expressed under the conditions tested.
The development of tools to facilitate differential profiling of biological samples using hyphenated mass spectrometry has widened the application window of these platforms toward high-throughput screening. To extract relevant information from large mass spectral data sets, alignment and data analysis are crucial steps that are still prone to improvement. We showed that with little effort a combination of a dedicated alignment package and the Random Forest algorithm can be applied to screen for relevant differences in volatile compounds present in fermented foods. The approach is universal and can be used for many applications where differences in volatile compounds need to be assessed.
ACKNOWLEDGMENTS
We acknowledge Jan van Riel for technical assistance, Herwig Bachmann for useful suggestions, and Kasper Hettinga for critical reading of the manuscript.
Footnotes
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Bayjanov J. R., et al. 2009. PanCGH: a genotype-calling algorithm for pangenome CGH data. Bioinformatics 25:309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bino R. J., et al. 2005. The light-hyperresponsive high pigment-2(dg) mutation of tomato: alterations in the fruit metabolome. New Phytologist 166:427–438 [DOI] [PubMed] [Google Scholar]
- 3. Blow N. 2008. Metabolomics: biochemistry's new look. Nature 455:697–700 [DOI] [PubMed] [Google Scholar]
- 4. Breiman L. 2001. Random forests. Mach. Learn. 45:5–32 [Google Scholar]
- 5. Cheng H. 2010. Volatile flavor compounds in yogurt: a review. Crit. Rev. Food Sci. Nutr. 50:938–950 [DOI] [PubMed] [Google Scholar]
- 6. de la Plaza M., Pelaez C., Requena T. 2009. Regulation of alpha-ketoisovalerate decarboxylase expression in Lactococcus lactis IFPL730. J. Mol. Microbiol. Biotechnol. 17:96–100 [DOI] [PubMed] [Google Scholar]
- 7. Fiehn O. 2001. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genomics 2:155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fiehn O. 2008. Extending the breadth of metabolite profiling by gas chromatography coupled to mass spectrometry. Trends Analyt. Chem. 27:261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fonseca F., Beal C., Corrieu G. 2000. Method of quantifying the loss of acidification activity of lactic acid starters during freezing and frozen storage. J. Dairy Res. 67:83–90 [DOI] [PubMed] [Google Scholar]
- 10. Holmes E., et al. 2008. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453:396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Imhof R., Glattli H., Bosset J. O. 1995. Volatile organic compounds produced by thermophilic and mesophilic single strain dairy starter cultures. Food Sci. Technol-Leb. 28:78–86 [Google Scholar]
- 12. Januszkiewicz J., Sabik H., Azarnia S., Lee B. 2008. Optimization of headspace solid-phase microextraction for the analysis of specific flavors in enzyme modified and natural Cheddar cheese using factorial design and response surface methodology. J. Chromatogr. A 1195:16–24 [DOI] [PubMed] [Google Scholar]
- 13. Kai M., et al. 2009. Bacterial volatiles and their action potential. Appl. Microbiol. Biotechnol. 81:1001–1012 [DOI] [PubMed] [Google Scholar]
- 14. Katajamaa M., Oresic M. 2007. Data processing for mass spectrometry-based metabolomics. J. Chromatogr. A 1158:318–328 [DOI] [PubMed] [Google Scholar]
- 15. Keurentjes J. J., et al. 2006. The genetics of plant metabolism. Nat. Genet. 38:842–849 [DOI] [PubMed] [Google Scholar]
- 16. Lommen A. 2009. MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 81:3079–3086 [DOI] [PubMed] [Google Scholar]
- 17. Mallia S., Fernandez-Garcis R., Bosset J. O. 2005. Comparison of purge and trap and solid phase microextraction techniques for studying the volatile aroma compounds of three European PDO hard cheeses. Int. Dairy J. 15:741–758 [Google Scholar]
- 18. Maragkoudakis P. A., et al. 2006. Production of traditional Greek yoghurt using Lactobacillus strains with probiotic potential as starter adjuncts. Int. Dairy J. 16:52–60 [Google Scholar]
- 19. Mastovska K., Lehotay S. J. 2003. Practical approaches to fast gas chromatography-mass spectrometry. J. Chromatogr. A 1000:153–180 [DOI] [PubMed] [Google Scholar]
- 20. Ott A., Fay L. B., Chaintreau A. 1997. Determination and origin of the aroma impact compounds of yogurt flavor. J. Agr. Food Chem. 45:850–858 [Google Scholar]
- 21. Pastink M. I., et al. 2008. Genomics and high-throughput screening approaches for optimal flavour production in dairy fermentation. Int. Dairy J. 18:781–789 [Google Scholar]
- 22. Randazzo C. L., Pitino I., De Luca S., Scifo G. O., Caggia C. 2008. Effect of wild strains used as starter cultures and adjunct cultures on the volatile compounds of the Pecorino Siciliano cheese. Int. J. Food Microbiol. 122:269–278 [DOI] [PubMed] [Google Scholar]
- 23. Robinson M. D., et al. 2007. A dynamic programming approach for the alignment of signal peaks in multiple gas chromatography-mass spectrometry experiments. BMC Bioinformatics 8:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smit B. A., et al. 2004. Development of a high throughput screening method to test flavour-forming capabilities of anaerobic micro-organisms. J. Appl. Microbiol. 97:306–313 [DOI] [PubMed] [Google Scholar]
- 25. Smit B. A., Engels W. J. M., Smit G. 2009. Branched chain aldehydes: production and breakdown pathways and relevance for flavour in foods. Appl. Microbiol. Biotechnol. 81:987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smit B. A., et al. 2005. Identification, cloning, and characterization of a Lactococcus lactis branched-chain alpha-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 71:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stein S. E. 1999. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectr. 10:770–781 [Google Scholar]
- 28. Tamime A. Y., Robinson R. K. 1999. Yoghurt science and technology, 2nd ed. Woodhead Publishing, Cambridge, United Kingdom [Google Scholar]
- 29. Tikunov Y., et al. 2005. A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 139:1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Williamson L. N., Bartlett M. G. 2007. Quantitative gas chromatography/time-of-flight mass spectrometry: a review. Biomed. Chromatogr. 21:664–669 [DOI] [PubMed] [Google Scholar]