Abstract
The microbial population and physicochemical process parameters of a sequencing batch reactor for nitrogen removal from urine were monitored over a 1.5-year period. Microbial community fingerprinting (automated ribosomal intergenic spacer analysis), 16S rRNA gene sequencing, and quantitative PCR on nitrogen cycle functional groups were used to characterize the microbial population. The reactor combined nitrification (ammonium oxidation)/anammox with organoheterotrophic denitrification. The nitrogen elimination rate initially increased by 400%, followed by an extended period of performance degradation. This phase was characterized by accumulation of nitrite and nitrous oxide, reduced anammox activity, and a different but stable microbial community. Outwashing of anammox bacteria or their inhibition by oxygen or nitrite was insufficient to explain reactor behavior. Multiple lines of evidence, e.g., regime-shift analysis of chemical and physical parameters and cluster and ordination analysis of the microbial community, indicated that the system had experienced a rapid transition to a new stable state that led to the observed inferior process rates. The events in the reactor can thus be interpreted to be an ecological regime shift. Constrained ordination indicated that the pH set point controlling cycle duration, temperature, airflow rate, and the release of nitric and nitrous oxides controlled the primarily heterotrophic microbial community. We show that by combining chemical and physical measurements, microbial community analysis and ecological theory allowed extraction of useful information about the causes and dynamics of the observed process instability.
INTRODUCTION
The ever-increasing arsenal of molecular and biochemical methods for microbial ecology has shaped our appreciation of biotechnological systems, e.g., in wastewater treatment, as complex, diverse, and dynamic microbial ecosystems. With the understanding that these systems can rival classic aquatic or terrestrial ecosystems in complexity came the realization that their effective design and operation may benefit from or even require an understanding of ecological theory (28).
The microbial ecosystem in many biotechnological systems consists of diverse microbial populations linked through a multitude of enzymatic processes. While understanding the structure, functions, and dynamics of the microbial communities is of fundamental concern for process optimization, such systems are so complex that determining all interactions is very challenging, if not impossible (28). Microbial community fingerprints such as denaturing gradient gel electrophoresis (DGGE) and automated ribosomal intergenic spacer analysis (ARISA) can be useful tools to assess the influences of process operation on the overall microbial community structure without knowing the abundance or identity of individual species (17, 46). While microbial community fingerprinting allows fast and simple qualitative characterization of microbial communities, statistical tools are required to elucidate the interdependencies of microbial populations and the environmental conditions. In wastewater treatment, microbial processes have usually been described with mechanistic models focusing on known mechanisms of limitation and inhibition of the most important microbial groups. This approach has its limitations for complex systems, because too many kinetic parameters have to be known to accurately model the microbial dynamics (18). Alternatively, statistical approaches such as canonical ordination that are frequently used in ecology can be applied to identify driving forces of the microbial community structure in the studied system. In addition to such gradient analysis, the rapid and often step-like changes in system parameters led us to analyze our data for evidence for ecological regime shifts. The theory of ecological regime shifts suggests that (some) ecological systems may have a tendency for rapid (catastrophic) change in ecosystem species composition and function, interpreted as a switch to an alternate stable state as certain variables cross a threshold or in response to stress events (32). This property thus arises from the interaction of internal processes and external forcing (8). The concept of regime shifts has been explored in general ecology to a considerable extent (8, 13, 32). In microbial ecology, biotechnology, and wastewater engineering, this is not the case, and indeed, evidence for regime shifts in purely microbial communities is currently missing (4). The prevailing view is still that microbial communities are flexible systems that adjust mostly deterministically to the external environmental conditions (11). The idea that “everything is everywhere, the environment selects” (3) was long considered one of the foundations of microbial ecology (11), although this view is increasingly being questioned (26, 44). The traditional perspective thus predicts that microbial communities along temporal or spatial gradients of environmental parameters will be determined exclusively by these gradients and not by history or a specific local microbial community. Also, disturbances are seen as temporal phenomena according to this perspective, since high dispersal and a large local seed bank are thought to result in resilient communities that will eventually return to an optimal state. There is increasing evidence that this may not always be the case and that disturbance can directly affect microbially mediated ecosystem processes (1). The importance of a comprehensive understanding of the role of microbial community composition and the need to apply ecological theory not only to microbial ecology in general (30) but also to engineered microbial reactors have consequently become increasingly evident (5, 28).
In this study, a combined nitrification/anammox reactor for urine treatment is used as an example. The process consists of two basic steps: first, aerobic ammonium-oxidizing bacteria (AOB) oxidize about 50% of the ammonium (NH4+) to nitrite (NO2−; equation 1), and then anammox bacteria (AMX) oxidize the NH4+ with NO2− to molecular nitrogen (N2; equation 2).
| (1) |
| (2) |
Combined nitrification/anammox in a single reactor has proven to be simple and stable, if the oxygen (O2) concentration is controlled well (21, 41).
Nitrification/anammox processes are particularly interesting for NH4+-rich wastewaters that do not contain sufficient biodegradable organic substrate for organoheterotrophic denitrification. The source-separated urine used in this study had an average biodegradable organic substrate content of 1.5 g chemical oxygen demand (COD)/g NH4+ N, which is too low for complete organoheterotrophic denitrification (1.71 g COD/g NH4+ N) but still sufficiently high to support considerable growth of denitrifying bacteria (DNB) and aerobic organoheterotrophic bacteria (AHB).
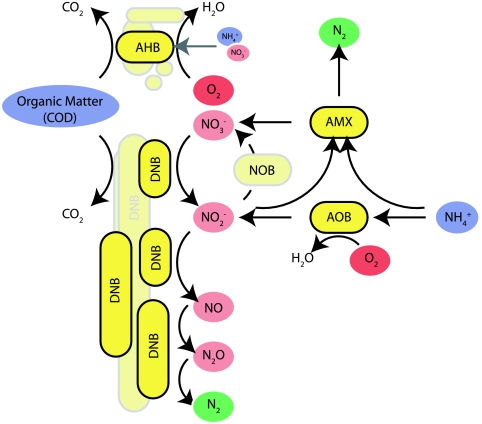
Figure 1 shows a simplified scheme of the main bacterial groups and their nitrogen and organic matter conversion patterns. The bacterial groups potentially involved in the nitrogen conversion are AOB, AMX, DNB, and, if process control is insufficient, nitrite-oxidizing bacteria (NOB). AMX convert about 11% of their total nitrogen substrate (NH4+ and NO2−) into nitrate (NO3−) (35) as a result of reverse electron flow. This NO3− as well as NO2− can be reduced by DNB. Environmental conditions and the kinetic properties of the bacterial groups determine their relative abundance. O2, nitrite/nitrous acid (NO2−/HNO2), and ammonium/ammonia (NH4+/NH3) are of special importance for inhibition and limitation effects. Table 1 summarizes some typical literature values for relevant kinetic parameters. The data show that NOB growth can be suppressed if O2 concentrations are very low, because NOB have a lower affinity to O2 than AOB and AHB. The data also illustrate that AMX have strong kinetic disadvantages. Their growth rate is very low, and they are (reversibly) inhibited by traces of O2 (10, 36). AHB and DNB grow about 100 times faster than AMX, and their yield (with respect to electrons transferred) is 10 times higher. Previous studies reported that in solutions with high COD/NH4+ ratios, anammox activity can be suppressed by heterotrophic bacteria (40). Imbalances in the microbial community or substrate limitations can result in the accumulation of the denitrification intermediates NO2−, nitric oxide (NO), and nitrous oxide (N2O). While N2O is mainly of environmental concern, NO is toxic for microorganisms (27).
Fig. 1.
Microbial functional groups and processes that dominate the nitrogen cycle and O2 consumption in a combined nitrification/anammox reactor with a high COD/NH4+ ratio. AHB primarily live by aerobic respiration and utilize organic matter as an electron donor and carbon source. Inorganic nitrogen (NO3−, NH4+) may be taken up as a nutrient but is not involved in catabolic processes. DNB live mostly or exclusively by anaerobic respiration using nitrate or one, several, or all of the intermediates of the denitrification pathway as electron acceptors. NOB are suppressed by low O2 in the study system.
Table 1.
Kinetic parameters of bacteria potentially involved in nitrogen removal from urinea
| Bacterial group | Growth rate (day−1) | Yield (g COD mol−1 e−) | O2 half-saturation constant (Monod kinetics) (mg liter−1) | Concn for 50% HNO2 inhibition (mg liter−1) | NH3 concn (mg liter−1) |
|---|---|---|---|---|---|
| AMXb | 0.065 | 0.40 | 0.04d | 100e | 770g |
| AOBc | 0.77 | 0.34 | 0.3 | 0.21f | 540 |
| NOBc | 1.08 | 0.29 | 1.1 | 0.26 | —h |
| AHB (aerobic)c | 7.1 | 3.4 | 0.08 | NDi | ND |
| DNB (nitrate reduction)c | 2.6 | 3.2 | ND | ND | |
| DNB (nitrite reduction)c | 1.5 | 3.2 | ND | ND |
These are typical parameters from a wide range of literature values.
Strous et al., 32°C (35).
Wiesmann, 20°C, assuming 1 g COD/g organic dry matter (45).
Complete inhibition at 0.04 mg liter−1. Strous et al. (36).
Inhibition at NO2− concentration of >100 mg liter−1.
Lochtman (1995) in Hellinga et al. (20).
Dapena-Mora et al. (9).
Stüven et al. (38). In mixed systems, NH2OH (ammonium oxidation intermediate) rather than NH3 inhibits NOB.
ND, not determined.
The complex interactions between the different bacterial groups, the competition for nitrite between AMX and DNB, and the potential for system instabilities due to accumulation of toxic intermediates make the nitrification/anammox treatment of urine an ideal subject with which to explore the use of microbial community analysis and ecological theory. In the present study, we analyzed a comprehensive data set for indications of regime shift behavior in the microbial ecosystem of a bioreactor and discuss possible implications of regime shifts in microbial systems.
MATERIALS AND METHODS
A large number of parameters have been measured or calculated in this work. An overview and additional explanations are given in Table S1 in the supplemental material.
Reactor operation.
The experimental system was a sequencing batch reactor (SBR) designed for treating diluted urine with a combined nitrification/anammox process by alternating between aerobic and anoxic phases. Throughout the duration of the monitoring period, the reactor was operated with the goal of achieving or maintaining a high nitrogen elimination rate (NER). The reactor setup is described in detail by Udert et al. (39). Briefly, the experiments were conducted in an SBR with a total liquid volume of 6.7 liters. A Simatic S5-100 apparatus was used for process control (Siemens, Munich, Germany). The durations of the aerobic and anoxic phases were operator controlled (times between 15 and 60 min were chosen), with the proportion of the total duration of anoxic phase (P_anox) versus aerobic phase per cycle varying between 34% and 83%. The airflow (Q_air) was kept sufficiently low to always keep the O2 concentration below 0.2 mg O2 liter−1. The number of aerobic and anoxic phases and therefore the total duration of each cycle depended on an operator-controlled pH threshold (pH_set). In each cycle, following sedimentation, between 1.3 and 1.8 liters of liquid (V_Exch) was replaced with 5-times-diluted anthropogenic urine collected from the NoMix system in the Eawag main building (39). An overview of the most important changes in operating parameters over the observation period is given in Fig. S1 in the supplemental material.
Chemical parameters. (i) Continuous measurements.
Electrodes were used to measure pH (HA405-DXK-S8/225; Mettler Toledo, Greifensee, Switzerland), O2 concentration (O2), and electric conductivity (Cond) (TriOxmatic 700 and Tetracon 325, both sensors; WTW, Weilheim, Germany). pH, O2, electric conductivity, and temperature (Temp) were continuously monitored with a data logger (Memograph S; Endress+Hauser, Weil am Rhein, Germany). For this paper, we did not quantify the N2O concentrations. It was shown that the increase of the O2 sensor baseline concentration is linearly correlated with the N2O concentration in the reactor (demineralized water was aerated with different mixtures of 100,000 ppm N2O in N2 and pure N2 gas; unpublished data). We used this to calculate a noncalibrated indicator variable for N2O accumulation, N2O_ind, with the units mg O2 liter−1 (value of interference on the O2 sensor).
(ii) Sampling-based analyses.
Beginning on 15 February 2008, the reactor was regularly monitored for further chemical parameters: samples were regularly taken from the inflow (on average, every 2.5 days) and analyzed for NH4+ (NH4_in) and dissolved COD (COD_in). The reactor outflow was sampled at the same time and analyzed for NH4+, NO2−, and NO3− (NH4_out, NO2_out, and NO3_out, respectively). The COD in the outflow was measured on selected dates. COD and NH4+ were measured photometrically with vial tests (Hach Lange, Düsseldorf, Germany). NO2− and NO3− were analyzed either with vial tests (Hach Lange) or on an ion chromatograph (Metrohm, Herisau, Switzerland). A small systematic deviation of the NO2− and NO3− vial test results was corrected to match the ion chromatograph measurements. NO3− vial tests were corrected for cross-sensitivity to NO2−. All samples were filtered (pore size, 0.7 mm; GF/F; Whatman, Middlesex, United Kingdom), measured immediately (photometric measurements), or stored for less than 7 days at 4°C (for analysis on an ion chromatograph). Starting on 22 September 2008, total suspended solids (TSS) were determined in the mixed reactor and in the outflow according to standard methods (2). These values were used to calculate the sludge age (SA). The biodegradable fraction of the dissolved COD in the influent was determined by subtracting the measured dissolved COD in the effluent from the measured dissolved COD in the influent.
Microbial community analysis. (i) Sampling.
Eleven samples for microbial community analysis were taken throughout the observation period from 15 April 2008 to 15 April 2009 (samples C to N). Two additional samples taken prior to the beginning of the regular chemical monitoring, on 25 September 2007 and 21 November 2007 (samples A and B), are considered only for microbial community analysis. The sampling dates are given in Table S2 in the supplemental material.
(ii) Nucleic acid extraction.
At each time point, 1-ml samples of suspended biomass were taken from the reactor with a micropipette, mixed with 10% of a nucleic acid preserving solution (95% ethanol, 5% buffered phenol [7]), centrifuged, and decanted, and the pellets were frozen at −80°C until further processing. DNA was extracted with a bead-beating method described previously (24) with some modifications. Briefly, 0.5 ml sample and 1.5 ml lysis buffer (50 mM Tris [pH 8], 50 mM EDTA, 50 mM sodium chloride) were lysed by bead beating (3 times for 80 s each time at 3,000 rpm using 0.2 g 106-μm- and 0.2 g 150- to 212-μm-diameter beads), followed by chemical and enzymatic lysis with 10 mg ml−1 lysozyme (final concentration) for 10 min and with 1% SDS and 3,600 U ml−1 proteinase K for 30 min. DNA was purified by phenol-chloroform extraction and precipitation in isopropanol.
(iii) DNA quantification and quality control.
DNA was quantified by standard measurements of light adsorption at 260- and 280-nm wavelengths using a Nanodrop spectrophotometer (Nanodrop Products, Wilmington, DE) or by detection of fluorescence with a picogreen DNA quantification kit (Invitrogen, Basel, Switzerland) according to the manufacturer's instructions. Fluorescence was determined on a Synergy HT plate reader (Bio-Tek Instruments, Inc., Winooski, VT). DNA integrity was checked by gel electrophoresis in 1% agarose gels. Extracts with poor 260-nm wavelength/280-nm wavelength ratios were cleaned using a QIAquick gel extraction kit (Qiagen, Basel, Switzerland) according to the manufacturer's instructions. Prior to use in PCR, extracted nucleic acids were diluted to 10 or 2 ng μl−1 in nuclease-free water (Qiagen).
(iv) Phylogenetic community characterization.
DGGE band sequencing (short sequence reads) and a nearly full-length amplicon clone library sequencing approach (long reads) were combined to obtain an overview of the microbial community compositions. DGGE was performed using general bacterial primers 341F-GC and 534R as described in reference 6 using a DCode DGGE system (Bio-Rad Laboratories AG, Reinach, Switzerland) for gel electrophoresis. For sequencing, all distinct band types were cut from three different gels containing samples from the period from 25 November 2007 to 15 April 2009. Cut bands were eluted in nuclease-free water and reamplified using the same primers described above but without the GC clamp and following the procedures described previously (6). Successful PCR products were purified using a QIAquick PCR purification kit (Qiagen), and commercial sequencing was performed by Microsynth (Balgach, Switzerland). Out of 17 band types cut for sequencing, 10 sequences were obtained; other bands failed to amplify or sequences were unreadable, possibly indicating comigrating bands. DGGE patterns were analyzed using the Quantity One software package (Bio-Rad).
Nearly full-length 16S rRNA gene amplicons from the sample obtained on 25 September 2007 were obtained by PCR using general bacterial primers 27F (3′-AGA GTT TGA TCM TGG CTC AG-5′) and 1492R (TAC GGY TAC CTT GTT ACG ACT T) (25). Products were cloned using a pGEM-T Easy vector systems cloning kit (Promega, Madison, WI) according to the manufacturer's instructions, followed by plasmid preparation with a GenElute HP plasmid miniprep kit (Sigma-Aldrich, St. Louis, MO), also according to the manufacturer's instructions. Thirty-eight clones were screened with DGGE as described above. Eight clones representing unique DGGE phylotypes were selected for sequencing.
All PCRs were prepared with Taq polymerase and buffer obtained from MP Biomedical (Solon, OH), and primers were synthesized by Microsynth (Balgach, Switzerland).
(v) Phylogenetic classification.
Sequences were assigned to the taxonomical hierarchy using the Classifier tool on the Ribosomal Database Project 2 website (http://rdp.cme.msu.edu/classifier/) (42). Sequences were checked for chimera formation using the Bellerophone (version 3) program (http://greengenes.lbl.gov/cgi-bin/nph-bel3_interface.cgi). One nearly full-length sequence was a suspected chimera and was eliminated from the data set. Where multiple DGGE band sequences were obtained for the same band type or where sequences were highly similar (>97% similarity in overlapping segments), only the sequence of the best quality was deposited in GenBank.
(vi) Microbial community fingerprinting.
Although we also analyzed the DGGE fingerprints, we relied on ARISA as the main microbial community fingerprinting method. ARISA was performed as described previously (12, 47), with minor modifications. The ribosomal intergenic spacer was amplified using primers 1406f (labeled with fluorescent dye 6-carboxyfluorescein) and 23S. PCR mixtures (1 or 0.5 μl) were denatured for 3 min at 95°C with 9 μl HiDi formamide and 0.5 μl LIZ1200 size standard (both from Applied Biosystems, Carlsbad, CA) and placed on ice immediately afterwards. Fragment size analysis was performed on an ABI 3130xl capillary sequencer, using a 50-cm 16-capillary array and POP7 polymer (Applied Biosystems). Fragment analysis was performed with GeneMapper software. Only peaks with sizes between 350 and 1,250 bp were considered for analysis, and the minimum peak area in fluorescence units (FU) was 150 FU.
(vii) qPCR.
The temporal dynamics of the bacterial functional groups of the nitrogen cycle were analyzed using a quantitative real-time PCR (qPCR) approach. The nitrogen cycle functional genes amoA, nirS, nirK, and nosZ were analyzed using the primers and protocols of Geets et al (16), with some changes. Reaction mixtures contained 12.5 μl 2× SYBR green master mix (Applied Biosystems), 0.3 μM each primer, and bovine serum albumin at a final concentration of 0.1 mg ml−1 in a final volume of 25 μl. For nosZ, KCl was added to increase the concentration by 25 mM. The thermal cycling program for amoA, nirS, nirK, and nosZ was as described in Geets et al (16): 50°C for 2 min, 94°C for 10 min, and 40 cycles of 95°C for 60 s and annealing at 50°C for 60 s and 60°C for 60 s.
AMX-specific 16S rRNA gene qPCR was performed using primers Pla46rc and Amx0368aA18 (33). qPCRs were performed as described above, but 2 mM MgCl2 was added to the reaction mixture. The thermal cycling program was as before, but with an annealing temperature of 56°C.
General bacterium-specific 16S rRNA gene qPCR was performed using primers 519f and 907r (25, 37). PCRs were as described above for functional genes, but the PCR mixtures contained each primer at a final concentration of 0.8 μM. The thermal cycling program was as before, but with an annealing temperature of 55°C.
All qPCR analyses were performed on an Applied Biosystems 7500 Fast real-time PCR system. Standard curves were prepared from dilutions of reference PCR amplicons cloned into pGEM-T Easy vectors and quantified as described above. Copy numbers of functional group targets are expressed as abundance according to the percentage of the general bacterial 16S rRNA gene copy numbers. Abundance as calculated here is operationally defined and may be distorted by different copy numbers of target genes in bacterial genomes. All samples were tested in duplicate or triplicate on each 96-well plate, and at least two independently prepared plates were analyzed per target. The amplification efficiency of all reactions was 95% or higher, except for nosZ, where one run had a lower efficiency of 88%.
Statistical analysis.
All errors given are standard deviations. Linear regression analyses (least-squares method) were calculated and plotted in Excel software. Regime-shift analysis was performed using the method described by Rodionov (31) using the Sequential Regime Shift Detection program (version 2.1; http://www.beringclimate.noaa.gov/regimes/).
Heat map cluster analysis (Euclidean distance, average linkage method) of ARISA data was performed using the R program (version 2.9.1; R Development Core Team, 2006) function heatmap.2 from the gplots package (43).
Constrained and unconstrained ordination analysis and Bray similarity matrix calculation were conducted in R with the packages BiodiversityR (23) and Vegan (29).
The environmental data set used in constrained ordination (redundancy analysis [RDA]) was created with samples from the chemical sampling date closest to the DNA sampling date (0 to 6 days offset; 0.7 days, on average). Alternative methods for data set assembly (period averages) were tested but rejected because of their lower predictive power.
Three groups of variables were considered for RDA. (i) Group 1 consists of process control parameters, which are operator-defined parameters used for process control, i.e., the pH value that triggers a new cycle (pH_set), proportion of anoxic conditions (including sedimentation) relative to total duration of the cycle (P_anox), airflow rate (Q_air), and exchange volume (V_Exch). (ii) Group 2 consisted of environmental variables, which comprised variables for the properties of the inflow or the physical environment not directly under operator control: chemical oxygen demand (COD_in), concentration of NH4+ in the inflow (NH4_in), electrical conductivity (Cond; using the maximum value per cycle), and temperature (T). (iii) Group 3 included response variables, which are parameters that depend on the process control, the environmental variables, and the microbial activity. They include NO2−, NO3−, and NH4+ in the outflow (NO2_out, NO3_out, and NH4_out, respectively), cycle duration (determined by pH set point and proton production in the reactor [Cycle]), maximum O2 concentration during aerobic phase (O2), and the N2O indicator (N2O_ind). Derived parameters, i.e., parameters that are calculated from one or more of the above-mentioned measured variables (e.g., NER) were not considered for RDA, as they would violate the assumption of independent variables. RDA was performed once with the variables in groups 1 and 2 and separately with the variables in group 3. For each environmental data set, we performed forward selection of constraining variables using the ordistep function (Vegan). The statistical model was built from the selected variables. The significance of the model and the contribution of individual constraints were tested by permutation analysis.
Inspired by the moving window analysis described by Wittebolle et al. (46), an analysis of community similarity over time was conducted using the Bray distance metric (B) (equation 3) calculated on the basis of ARISA peak area (FU) data in R.
| (3) |
where xik and xjk the area of peak k in samples i and j, respectively, and S is the total number of peaks.
Nucleotide sequence accession numbers.
The nearly full-length sequences are available in GenBank under accession numbers HQ917036 to HQ917042. DGGE band sequences were deposited in GenBank under accession numbers HQ917043 to HQ917052.
RESULTS
Reactor performance.
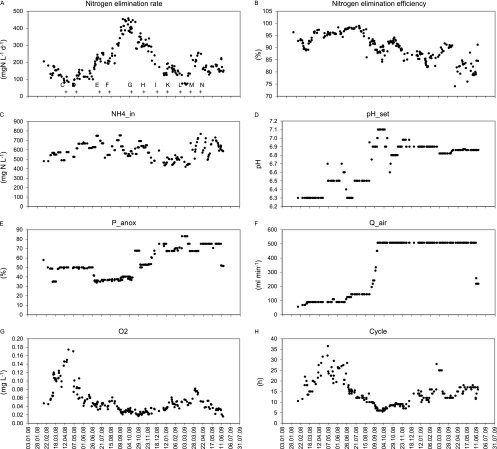
At the beginning of the observation period in February 2008, the reactor had been in continuous operation for 10 months (39). From the beginning of June to the end of September 2008, the NER (Fig. 2A) increased continuously from <100 to >430 mg N liter−1 day−1. Beginning in late October 2008, a marked and sustained decrease in NER to values of 165 ± 35 mg N liter−1 day−1 was observed.
Fig. 2.
Time series for several process indicators. Nitrogen elimination rate (A) and nitrogen elimination efficiency (B) are measures for the reactor performance. NH4+ in the influent is an independent environmental variable (C). pH_set (D), P_anox (E), and Q_air (F) are independent process parameters controlled by the operator. O2 (G) and Cycle (H) are response variables which depend on environmental variables and process parameters. The plus signs in panel A indicate the sampling dates for microbial community analysis, and letters indicate sample designations. Samples A and B (not shown) were taken in 2007.
Nitrogen elimination efficiency was >80% throughout the study period and remained >94% between 11 April 2008 and 25 August 2008 (Fig. 2B). The highest efficiencies were measured in early August 2008 and decreased continuously after that.
Environmental variables.
The concentrations in the influent did not show strong variations: NH4+ concentrations in the inflow (NH4_in; Fig. 2C) averaged 590 ± 90 mg NH4 N liter−1, the concentration of dissolved COD was 960 ± 110 mg O2 liter−1 (proportion of biodegradable COD, 89% ± 16%), the pH was 8.7 ± 0.2, and the average electric conductivity was 6.1 ± 0.4 mS cm−1 (25°C). However, NH4_in showed a slight seasonal behavior, with lower concentrations in winter and higher concentrations in summer (Fig. 2C). The average temperature in the laboratory varied seasonally, with higher values in summer and lower values in winter, but the temperature variation in the reactor (22.8 ± 1.5°C) was small.
Process control parameters.
Process control parameters were adjusted at various times to improve the performance of the SBR (also see Fig. S1 in the supplemental material). The increase in NER was achieved mainly by gradually raising the pH set point (pH_set; Fig. 2D) from 6.3 to 7.1, which resulted in shorter cycles and thus higher urine throughput. Maximum NERs were achieved in September and October 2008, with pH_set adjusted to 7.0 and 7.1. During the same time period the airflow rate (Q_air) was gradually increased from 140 ml min−1 to 510 ml min−1 (Fig. 2F), primarily for the purpose of improving conditions for AOB. The increase in Q_air had little effect on the measured maximum dissolved O2 concentrations (O2; Fig. 2G), which was always below 0.2 mg liter−1 and decreased from April to the end of October 2008 to values as low as 0.02 mg liter−1. P_anox (Fig. 2E) was kept at approximately 50% until the end of June 2008 and was then lowered to approximately 35 to 40% until 13 October 2008. Subsequently, higher values up to 83% were used. The exchange volume (V_Exch; data not shown) increased from 1.3 liters to 1.5 liters and finally 1.8 liters between 22 September 2008 and 20 October 2008 but was reset to 1.3 liters on 22 October 2008 and not changed any more. The short period with increased V_Exch had only a limited effect on the sludge age (SA), which remained high (>30 days) for most (10 out of 18) of the sampling events during that period as well as for the entire remaining observation period (156 out of 173).
Response variables.
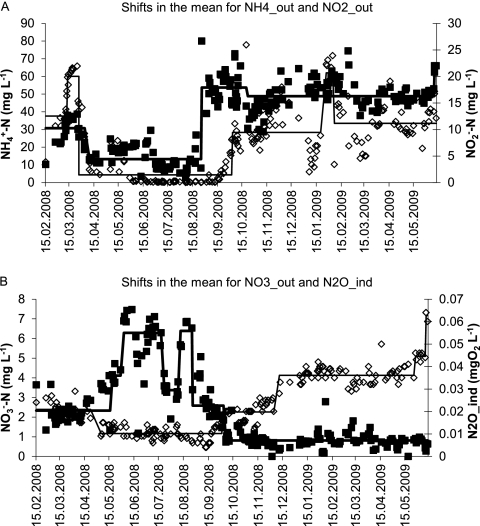
NH4+ was the dominant nitrogen species in the outflow (75%, on average). NO2− reached maximum concentrations of 25 mg N liter−1. NO3− in the outflow remained below 8 mg N liter−1 at any time. All outflow concentrations (NH4_out, NO3_out, NO2_out) and the indicator variable for N2O (N2O_ind) were subjected to statistical regime-shift analysis, in order to determine phases of relative stability (Fig. 3). The regime-shift analysis of NH4_out (Fig. 3A) indicated that after an initial drop, this parameter remained in a stable regime (despite decreasing cycle duration; Fig. 2H), extending from mid-April to mid-August 2008. During this period, NH4+ concentrations mostly varied between 10 and 20 mg NH4+ N liter−1. This phase ended with a strong regimen shift on 26 August 2008, with a sudden increase to >50 mg NH4+ N liter−1. This is assumed to be due to an increase of the pH set point from 6.5 to 6.95 on 26 August 2008 (Fig. 2D). Since AOB lower the pH during ammonia oxidation, a higher pH_set decreases the amount of ammonia oxidized. The higher ammonia level was maintained with some variations until the end of the study period.
Fig. 3.
Regime-shift analysis of the temporal trends in the concentrations of NH4+ (NH4_out; black squares) and NO2− (NO2_out; empty diamonds) in the outflow (A) and nitrate (NO3_out; black squares) in the outflow and the maximum N2O in the rector extracted from O2 sensor data (N2O_ind; empty diamonds) (B). Solid lines indicate statistically significant regimes determined using sequential regime-shift detection with the parameters probability equal to 0.001, cutoff length equal to 10, and Huber parameter equal to 1.
NO2_out exhibited a similar pattern as NH4_out (Fig. 3A); an early regime of elevated concentrations was followed by a stable phase with low values. From the beginning of June 2008 to mid-September 2008, NO2_out concentrations were mostly lower than 1 mg N liter−1. Values began to increase in mid-September, and the regime-shift analysis placed the significant shift to a new regime with higher NO2_out concentrations on 25 September 2008, a month after the shift in NH4_out. NO2_out was slightly more variable than NH4_out, but a similar regime with elevated concentrations became established until the end of the study period. The indicator for N2O accumulation (N2O_ind) also began to increase in mid-September, and the statistically significant shift to increased N2O accumulation was detected for 2 October 2008. N2O_ind shifted again to a regime of even higher accumulation on 11 December 2008 (Fig. 3B). NO3_out fluctuated strongly during the first 8 months of the observation period. During the period from June to August 2008, several episodes with relatively high NO3− concentrations (6 to 7.4 mg NO3− N liter−1) occurred. By 26 August 2008, NO3_out had decreased and a stable regime was established with an average concentration of 0.8 mg NO3− N liter−1 that lasted until the end of the study period (Fig. 3B).
In summary, all nitrogen-related system response variables showed a statistically significant regime shift between mid-August and the beginning of October 2008.
Analysis and dynamics of bacterial community. (i) Phylogenetic characterization.
The sequences retrieved from the clone library and DGGE band sequencing were classified as belonging to the Flavobacteria; Bacteroidia; Clostridia; Alpha-, Beta-, and Gammaproteobacteria; Sphingobacteria; Bacilli; and Acidobacteria. The detected organisms had a wide range of substrate preferences, which ranged from aerobic (e.g., Flavobacteria) to facultatively anaerobic (Burkholderiales, Simplicispira) and strictly anaerobic, mostly fermentative species (Anaerovorax, Bacteroidales). A number of sequences were similar to those of known nitrate reducers/denitrifiers (Simplicispira, Pleomorphomonas, Petrimonas, Thauera). Haliscomenobacter is a known filamentous activated sludge bacterium which is associated with O2-deficient situations (15). While our sequencing effort was not exhaustive, the analysis revealed that organoheterotrophs were abundant members of the microbial community in the reactor.
(ii) Dynamics of functional microbial groups.
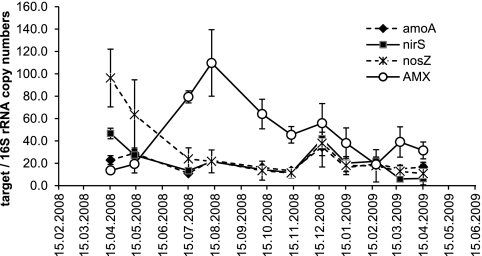
qPCR revealed a continued presence of all major functional microbial groups involved in the nitrogen cycle that were analyzed (Fig. 4). AMX were initially present in low numbers (this was also the case for the samples from 2007; data not shown), with a pronounced abundance maximum on 11 August 2008 (Fig. 4). The amoA gene, representing AOB, did not show a pronounced dynamic. For nirS and nosZ, representing different process steps of denitrification by DNB, we observed decreases in their abundance of 71 and 79%, respectively, during the period between 15 April 2008 and 15 July 2008. During this period we observed a strong increase in AMX. The abundance of nirK was below 1% throughout (data not shown).
Fig. 4.
Quantitative PCR analysis of bacterial functional groups of the nitrogen cycle. AOB are represented by the ammonium monooxygenase gene (amoA), DNB are represented by the nitrite reductase gene (nirS [nirK was present at <1% throughout]) and the nitrous oxide reductase gene (nosZ), and AMX were detected using a group-specific primer set for the AMX 16S rRNA gene. All data were normalized against qPCR using general bacterial 16S rRNA gene primers.
(iii) Community dissimilarity.
Community turnover was visualized by plotting the Bray dissimilarities of ARISA fingerprints for consecutive sampling dates (Fig. 5). The analyses based on DGGE (data not shown) agreed remarkably well with those based on ARISA (R2 = 0.62, P < 0.01), considering the different principles, genetic targets, and specific biases of each method. Two spikes in dissimilarity indicated a community shift between the August and October 2008 samples and between the February and March 2009 samples, separated by a period of high stability.
Fig. 5.
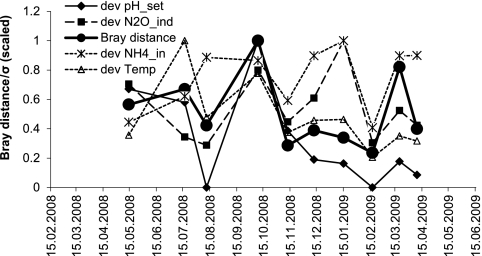
Time series analysis of Bray distance based on ARISA community fingerprints compared to time series of standard deviations (dev, σ) plotted for important driving variables according to redundancy analysis. The Bray distance graph plots the Bray dissimilarity metric of each sample to the previous sample. The time series of standard deviation plots the standard deviation of all measurements from the time interval between the date closest to the microbial sampling date and the previous microbial sampling date. All series are scaled to the highest value. Solid lines, significant correlations (P < 0.05) with Bray dissimilarity; dashed lines, no significant correlation.
(iv) Cluster analysis.
The heat map cluster analysis of ARISA profiles (Fig. 6) demonstrated the good reproducibility of the analyzed technical replicates with 1 and 0.5 μl DNA, as these are largely indistinguishable. Using an arbitrarily set cutoff (Fig. 6), we designated 3 main clusters: The samples from 2007 (A and B) form the first cluster, and the 2008 samples up to August 2008 (C to F) form cluster 2. Cluster 3, which consists of all later samples (G to N), is clearly separated from both cluster 1 and cluster 2. The strongest shift in the community thus occurred between the times of collection of samples F (11 August 2008) and G (9 October 2008). The two latest samples (M and N) formed a separate subcluster (cluster 3b).
Fig. 6.
Heat map representation and hierarchical clustering of ARISA community fingerprints. Row labels indicate the sampling dates (see Fig. 2A) and the DNA volume used in ARISA (1 = 1 liter, 2 = 0.5 liter), and columns correspond to ARISA peaks (phylotypes). Darker shading indicates a higher relative ARISA peak area, an indication of high relative abundance. Numbers on the right are cluster designations.
The clustering of the ARISA peaks indicated operational taxonomic units (OTUs) with similar behavior over the time course (Fig. 6). It is evident that the shift between clusters 1 and 2 not only was due to a strong shift in the dominance structure of abundant OTUs (left side of the heat map in Fig. 6), including the disappearance of some key OTUs (e.g., peak P54), but also is reflected in the appearance of previously undetected OTUs (right side of the heat map).
(v) Drivers of community dynamics.
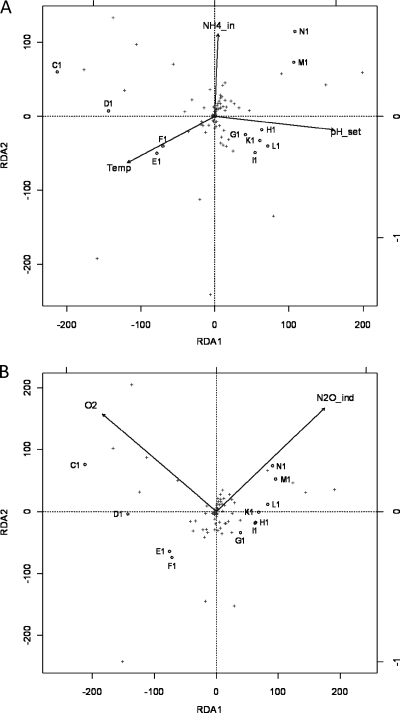
The effect of key variables on the bacterial community was studied using a constrained ordination, RDA. Ordinations were carried out once with process control parameters and environmental variables (process variables; Fig. 7A) and once with response variables (Fig. 7B) as the environmental data set. Forward selection of constraints selected pH_set, NH4_in, and Temp from the process variables data set. O2 and N2O_ind were selected from the response variables. However, it should be noted that N2O_ind, NO2_out, and NH4_out had similar vectors in RDA, and the use of any one of these parameters results in a statistically significant model. Forward selection from a combination data set of process and response variables resulted in selection of the same variables selected from the process variables alone. The models using process variables and response variables as constraints explained 69 and 66% of the total variance on the two primary axes, respectively. Permutation analysis (n = 1,000) revealed that the ordinations were highly significant (P < 0.001). All individual constraints contributed significantly (P < 0.05 to P < 0.01). In the process variable ordination (Fig. 7A), pH_set was mainly associated with the first ordination axis, and the period of the regime shift (from F to G) was mostly associated with a shift of the community along this axis, while the community dynamics prior to the regime shift may also have been affected by Temp. It is noteworthy that the samples taken after the proposed regime shift from October 2008 to February 2009 (G to L) cluster very closely together, indicating high community stability in this phase. NH4_in contributed most strongly to axis 2 and appears to be related to the community development in the last two samplings (N and M).
Fig. 7.
Triplots of redundancy analysis of ARISA community structure data using operational parameters and independent environmental variables (A) or system response (output) variables as the constraining data set (B). Empty circles, samples labeled according to sampling date; plus signs, ARISA peaks, representing OTUs. Arrows, constraining variables of the environmental data set.
In the RDA with response variables, N2O_ind and O2 were equally related to both axes. The community development in the pre-regime-shift phase is best explained by O2, while the regime shift itself (shift from samplings F to G) and subsequent changes are best explained by N2O_ind.
The influence of the parameters pH_set, NH4_in, Temp, and N2O_ind was analyzed further by overlaying the analysis of community dissimilarity with plots of the standard deviation calculated for the period between the current and the previous microbial sampling (Fig. 5). The result supports the ordination in indicating a key role for pH_set. The variability in pH_set was significantly correlated with the extent of change in the bacterial community (R2 = 0.49, P < 0.05), while this was not the case, e.g., for NH4_in or Temp (P > 0.05). Variability in community composition was also linked to variability in the nitrogen outflow Ntot_out (sum of NH4_out, NO2_out, and NO3_out; R2 = 0.59 and P < 0.01; data not shown).
Similar results were obtained with unconstrained ordination (data not shown).
DISCUSSION
Evidence for a regime shift.
Ecological regime shifts are defined as large and rapid shifts from one stable ecosystem state to another (32). In theory, such shifts can be attributed to the existence of alternative stable system states for a given set of control factors (8, 32). For purely microbial ecosystems, evidence for alternate stable states is still largely missing (4). Scheffer and Carpenter discuss typical indications for ecological regime shifts and possible sources of evidence in field data in their review (32). Typical observations indicating an ecological regime shift include (i) jumps in time series, (ii) clear differences in species composition or abundance, (iii) a change in the relationship between system control parameters and response variables or between response variables, (iv) the resilience of the system against a return to the previous state, and (v) the absence of strong stress events.
Our data provide multiple lines of evidence that an ecological regime shift was observed. (i) Strong shifts in the response variables NH4_out, NO2_out, NO3_out, and N2O_ind were visible in our data and were confirmed by a statistical analysis (sequential regime-shift detection [31]). The regimen shifts were statistically highly significant (P < 0.001) for each of these parameters during the period from mid-August to mid-October 2008 (Fig. 3).
(ii) The bacterial community structure showed dynamic changes throughout most of the observation period, but ordination and cluster analysis (Fig. 6 and 7) and time series analysis of community dissimilarity (Fig. 5) indicated that the most dramatic shift in the community structure occurred between August and October 2008. This coincides with the period when the reactor reached its peak performance and began to accumulate NO2−. It also coincides with other observed regime shifts in the nitrogen outflow data. Finally, we observed a strong decrease of the AMX abundance from its peak in August 2008. DNB gene abundance (nirS, nosZ) had already reached a low level before the shift and continued to decline during this period. According to the phylogenetic analysis, the general bacterial community was dominated by heterotrophs. This indicates that the entire microbial community and not only the functional groups directly involved in nitrogen transformations was affected by the regime shift. The detected heterotrophic genera are well-known to have different physiological properties which correspond well to the SBR environment, with niches created by the fluctuating availability of O2 and oxidized nitrogen compounds as external electron acceptors and the availability of various organic carbon substrates in urine. Our data thus point to the potential value of general microbial community analysis as a tool for monitoring engineered microbial systems.
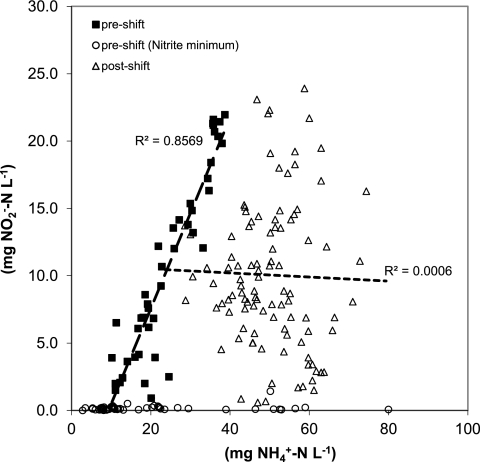
(iii) Correlations between variables were affected by the proposed regime shift: e.g., an analysis of the correlation between NO2_out and NH4_out showed a strong correlation prior to and a complete lack of correlation after the period of regime shifts (Fig. 8).
Fig. 8.
Correlation between outflow concentrations of NH4+ and NO2− before (black squares) and after (white triangles) the observed main regime shifts in nitrite and NH4+. The initially highly significant correlation (P < 0.001) breaks down completely after the regime shift. For a period of time before and during the regime shift, nitrite levels were at or below the detection limit; these values are not used in the regression (white circles).
(iv) Both the time series of community dissimilarity (Fig. 5) and ordination analysis (Fig. 7) clearly showed that following the shift, the community experienced an extended period of high stability from October 2008 to February 2009, despite continued fluctuations in system control parameters, environmental variables, and response variables. Compared to the dynamics during the first half of the observation period, this relative stability was also observed in the qPCR data (Fig. 4). This is an indication that the system had settled into a new stable state. However, we did not attempt to move operating parameters back to values from before the performance maximum within the observation period.
(v) Changes in control parameters, such as the nearly simultaneous changes of pH_set and Q_air, are strongly indicated to be related to the observed shifts in the microbial community. However, it is not clear to what extent these changes constitute strong stress events: raising pH_set represented a reduction of pH-related stress (e.g., for AOB). However, the resulting decrease in cycle duration and sludge age probably represented a stress event for AMX, although these changes were gradual (Fig. 2H) and temporary. While the control parameter Q_air was increased considerably during the regime-shift period, with the idea of improving conditions for AOB and thus NH4+ conversion rates, the O2 concentrations always remained in the subinhibitory range, showing that the microbial community was capable of absorbing the additional O2. In addition, raising Q_air did not have a strong effect on the cumulative air volume per cycle (V_air; data not shown), which describes the total amount of the electron acceptor oxygen available per cycle. This parameter showed a roughly linear increase from an initial value of about 20 liters to an average of 96 ± 22 liters, reached approximately in January 2009, and afterwards fluctuated in that range until the end of the monitoring period. Due to its dependence on Cycle, P_anox, and Q_air, this parameter is dependent on both control and system response parameters and was therefore not included in the ordination analysis, despite its obvious relevance.
Causes of regime shift.
The regime shift between July and October 2008 resulted in an unfavorable system state. NER was greatly reduced from its peak value, NO2− accumulated in the system and was present in the outflow, and an increase of N2O_ind shows that the system started producing the greenhouse gas N2O and, possibly, also toxic NO. pH_set, Temp, and N2O_ind were indicated by the ordination analysis to be possible drivers of the community dynamics between July and October 2008. In the following, we will discuss why changes of pH_set, Temp, and N2O_ind might have triggered the strong changes in microbial community structure and function.
(i) pH_set.
The higher pH_set was expected to increase the activity of AOB, which are inhibited at lower pH values. qPCR indicated that AOB did not increase in number in response, but this does not necessarily rule out a higher AOB activity. The higher AOB activity could have caused the increase of the NO2− concentration, which in turn supported the growth of DNB that use NO2− as an electron acceptor. While the new conditions favored certain DNB, AMX activity decreased due to the higher NO2− and AHB had to compete with DNB for organic substances. This change could largely explain the observations made with ARISA and DGGE, which are mainly due to changes in the heterotrophic community.
By increasing pH_set, the cycle duration was significantly reduced (Fig. 2D and H). A possible consequence might have been the outwashing of slow-growing organisms such as AMX. However, qPCR analysis revealed that while the abundance of AMX indeed decreased, they were never lost from the system and their abundance remained above levels present at the beginning of the observation period (Fig. 4). Thus, their inability to increase in abundance again was not due to losing them from the system.
(ii) Temp.
All microbial activity increases with temperature, as long as the temperature does not exceed the physiological range. Between July and October 2008, the temperature steadily decreased from a maximum of 26.6°C to a minimum of 21.9°C. The concomitant decrease of the maximum growth rate can be assumed to have been most critical for slow growing bacteria, especially AMX. As mentioned above, however, AMX were never completely lost.
(iii) N2O_ind.
Besides N2O, another intermediate of denitrification, NO, is also known to interfere with oxygen measurements (14) and could have contributed to N2O_ind. In contrast to N2O, NO production not only is critical for the environment but also affects microorganisms. NO is generally known to be inhibiting or toxic for microorganisms involved in the nitrogen cycle, but it is also an electron acceptor for some denitrifying bacteria (48). The decrease of nosZ abundance (Fig. 4) during the phase of low NO2− concentrations (June to August 2008) may have contributed to the strong increase in N2O_ind after the system switched back to NO2− accumulation. There are only a few studies specifically on the effect of NO on AMX. However, previous studies (22, 34) showed that “Candidatus Brocadia fulgida” and “Candidatus Brocadia anammoxidans” tolerated NO concentrations of 600 ppm and 3,500 ppm, respectively, and even higher AMX activities in the presence of NO were reported. We therefore assume that AMX might not be negatively affected by NO, but it is very likely that NO influenced other microbial communities involved in nitrogen cycling and thus contributed to the community change.
In summary, we hypothesize that the concomitant change of an important process control parameter (pH_set), a strongly influential environmental parameter (Temp), and a reinforcing response parameter (N2O_ind) caused a fundamental change of the microbial community and nitrogen cycle processes. This combination of external forcings and internal processes is typical for regime shifts (8). A final conclusion about the actual cause of the regime shift cannot be drawn in this paper, since targeted experiments are required to test the different hypotheses.
Implications of regime shifts for microbial reactors.
The occurrence of regime shifts in wastewater treatment systems could have strong implications on the use of models based on microbial kinetics, such as activated sludge model (ASM) 3 (19). In these models, microbial kinetics are modeled on the basis of constant Monod parameters, such as the half-saturation constant K and the maximum growth rate μmax. Adaptations of the bacteria to new environmental conditions are modeled by using saturation and inhibition terms only. Regime shifts, however, can change the microbial population so significantly that the constant kinetic parameters would not be applicable anymore. To account for regime shifts in Monod parameter-based mechanistic models, the number of bacterial groups would have to be increased, and with them, the number of kinetic parameters would have to be increased. The need for a high number of reliable kinetic parameters, however, limits the applicability of large mechanistic models. Statistical and ecological model approaches might be viable alternatives for complex biological processes such as nitrification/anammox treatment of urine, but further work is required to be able to assess their full potential.
Conclusions.
A 400% increase of NER was achieved in the studied SBR for partial nitrification/anammox over the course of 5 months by raising pH_set and by increasing aeration. However, continued operation resulted in a low NER and NO2− and N2O accumulation. Analysis of the chemical and biological data indicated that the system behavior can be described as an ecological regime shift. This regime shift could not be successfully reversed during the observation period, and the microbial community remained stable over an extended period of time, indicating that the system had shifted to a new stable state marked by reduced AMX and increased DNB activity.
The concept of ecological regime shifts and the existence of alternate stable states in microbial communities have so far received little attention in microbial ecology or environmental engineering. In this paper, we have, for the first time, presented evidence that complex, purely microbial communities may experience regime shifts with marked effects on ecosystem processes. The concept of regime shifts provides a new perspective on process instabilities in engineered microbial systems. Alternative stable states could underlie other observations in wastewater treatment, such as bulking and foaming. An understanding of the stability landscapes of microbial reactors and the mechanistic consequences associated with them could provide important insights for the development of stable and efficient processes.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge funding by Eawag and the Swiss National Science Foundation, project 200021_125133.
We thank M. S. M. Jetten and M. Schmid from Radboud University Nijmegen for providing positive controls for AMX PCR. We express our thanks to Nathanaël Legeard, Ashish Sahu, Anna Kraj, Karin Rottermann, and Claudia Bänninger for support in the laboratory.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Allison S. D., Martiny J. B. H. 2008. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105:11512–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. American Public Health Association 2005. Standard methods for the examination of water & wastewater, 21st ed. American Public Health Association, American Water Works Association, and Water Environment Federation, Washington, DC [Google Scholar]
- 3. Baas-Becking L. G. M. 1934. Geobiologie of inleiding tot de milieukunde. W. P. van Stockum and N. V. Zoon, The Hague, Netherlands [Google Scholar]
- 4. Botton S., van Heusden M., Parsons J. R., Smidt H., van Straalen N. 2006. Resilience of microbial systems towards disturbances. Crit. Rev. Microbiol. 32:101–112 [DOI] [PubMed] [Google Scholar]
- 5. Briones A., Raskin L. 2003. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr. Opin. Biotechnol. 14:270–276 [DOI] [PubMed] [Google Scholar]
- 6. Buesing N., Filippini M., Bürgmann H., Gessner M. O. 2009. Microbial communities in contrasting freshwater marsh microhabitats. FEMS Microbiol. Ecol. 69:84–97 [DOI] [PubMed] [Google Scholar]
- 7. Bürgmann H., Meier S., Bunge M., Widmer F., Zeyer J. 2005. Effects of model root exudates on structure and activity of a soil diazotroph community. Environ. Microbiol. 7:1711–1724 [DOI] [PubMed] [Google Scholar]
- 8. Collie J. S., Richardson K., Steele J. H. 2004. Regime shifts: can ecological theory illuminate the mechanisms? Prog. Oceanogr. 60:281–302 [Google Scholar]
- 9. Dapena-Mora A., et al. 2007. Evaluation of activity and inhibition effects on anammox process by batch tests based on the nitrogen gas production. Enzyme Microb. Technol. 40:859–865 [Google Scholar]
- 10. Egli K., et al. 2001. Enrichment and characterization of an anammox bacterium from a rotating biological contactor treating ammonium-rich leachate. Arch. Microbiol. 175:198–207 [DOI] [PubMed] [Google Scholar]
- 11. Finlay B. J., Maberly S. C., Cooper J. I. 1997. Microbial diversity and ecosystem function. Oikos 80:209–213 [Google Scholar]
- 12. Fisher M. M., Triplett E. W. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Folke C., et al. 2004. Regime shifts, resilience, and biodiversity in ecosystem management. Annu. Rev. Ecol. Evol. Syst. 35:557–581 [Google Scholar]
- 14. Fux C., Velten S., Carozzi V., Solley D., Keller J. 2006. Efficient and stable nitritation and denitritation of ammonium-rich sludge dewatering liquor using an SBR with continuous loading. Water Res. 40:2765–2775 [DOI] [PubMed] [Google Scholar]
- 15. Gaval G., Pernelle J.-J. 2003. Impact of the repetition of oxygen deficiencies on the filamentous bacteria proliferation in activated sludge. Water Res. 37:1991–2000 [DOI] [PubMed] [Google Scholar]
- 16. Geets J., et al. 2007. Real-time PCR assay for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Appl. Microbiol. Biotechnol. 75:211–221 [DOI] [PubMed] [Google Scholar]
- 17. Gilbride K. A., Lee D. Y., Beaudette L. A. 2006. Molecular techniques in wastewater: understanding microbial communities, detecting pathogens, and real-time process control. J. Microbiol. Methods 66:1–20 [DOI] [PubMed] [Google Scholar]
- 18. Gujer W. 2006. Activated sludge modelling: past, present and future. Water Sci. Technol. 53:111–119 [DOI] [PubMed] [Google Scholar]
- 19. Gujer W., Henze M., Mino T., van Loosdrecht M. 1999. Activated sludge model no. 3. Water Sci. Technol. 39:183–193 [Google Scholar]
- 20. Hellinga C., Van Loosdrecht M. C. M., Heijnen J. J. 1999. Model based design of a novel process for nitrogen removal from concentrated flows. Math. Comp. Model. Dyn. 5:351–371 [Google Scholar]
- 21. Joss A., et al. 2009. Full-scale nitrogen removal from digester liquid with partial nitritation and anammox in one SBR. Environ. Sci. Technol. 43:5301–5306 [DOI] [PubMed] [Google Scholar]
- 22. Kartal B., et al. 2010. Effect of nitric oxide on anammox bacteria. Appl. Environ. Microbiol. 15:6304–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kindt R., Coe R. 2005. Tree diversity analysis: a manual and software for common statistical methods for ecological and biodiversity studies. World Agroforestry Centre, Nairobi, Kenya: http://www.worldagroforestry.org/resources/databases/tree-diversity-analysis [Google Scholar]
- 24. Kleikemper J., et al. 2002. Activity and diversity of sulfate-reducing bacteria in a petroleum hydrocarbon-contaminated aquifer. Appl. Environ. Microbiol. 68:1516–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–147 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 26. Logue J. B., Bürgmann H., Robinson C. T. 2008. Progress in the ecological genetics and biodiversity of freshwater bacteria. Bioscience 58:103–113 [Google Scholar]
- 27. Mancinelli R. L., McKay C. P. 1983. Effects of nitric oxide and nitrogen dioxide on bacterial growth. Appl. Environ. Microbiol. 46:198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McMahon K. D., Martin H. G., Hugenholtz P. 2007. Integrating ecology into biotechnology. Curr. Opin. Biotechnol. 18:287–292 [DOI] [PubMed] [Google Scholar]
- 29. Oksanen J., et al. 2010. Vegan: community ecology package. R package, version 1.17-3. The R Project for Statistical Computing, Vienna, Austria [Google Scholar]
- 30. Prosser J. I., et al. 2007. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5:384–392 [DOI] [PubMed] [Google Scholar]
- 31. Rodionov S. N. 2004. A sequential algorithm for testing climate regime shifts. Geophys. Res. Lett. 31:L09204 [Google Scholar]
- 32. Scheffer M., Carpenter S. R. 2003. Catastrophic regime shifts in ecosystems: linking theory to observation. Trends Ecol. Evol. 18:648–656 [Google Scholar]
- 33. Schmid M. C., et al. 2005. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 71:1677–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmidt I., et al. 2002. Anaerobic ammonia oxidation in the presence of nitrogen oxides (NOx) by two different lithotrophs. Appl. Environ. Microbiol. 68:5351–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strous M., Heijnen J. J., Kuenen J. G., Jetten M. S. M. 1998. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl. Microbiol. Biotechnol. 50:589–596 [Google Scholar]
- 36. Strous M., Van Gerven E., Zheng P., Kuenen J. G., Jetten M. S. M. 1997. Ammonium removal from concentrated waste streams with the anaerobic ammonium oxidation (anammox) process in different reactor configurations. Water Res. 31:1955–1962 [Google Scholar]
- 37. Stubner S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen(TM) detection. J. Microbiol. Methods 50:155–164 [DOI] [PubMed] [Google Scholar]
- 38. Stüven R., Vollmer M., Bock E. 1992. The impact of organic matter on nitric oxide formation by Nitrosomonas europaea. Arch. Microbiol. 158:439–443 [Google Scholar]
- 39. Udert K. M., Kind E., Teunissen M., Jenni S., Larsen T. A. 2008. Effect of heterotrophic growth on nitritation/anammox in a single sequencing batch reactor. Water Sci. Technol. 58:277–284 [DOI] [PubMed] [Google Scholar]
- 40. Van Hulle S. W. H., et al. 2010. Engineering aspects and practical application of autotrophic nitrogen removal from nitrogen rich streams. Chem. Eng. J. 162:1–20 [Google Scholar]
- 41. Vlaeminck S. E., et al. 2009. Nitrogen removal from digested black water by one-stage partial nitritation and anammox. Environ. Sci. Technol. 43:5035–5041 [DOI] [PubMed] [Google Scholar]
- 42. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Warnes G. R., et al. 2010. gplots: various R programming tools for plotting data, 2.8.0 ed. The R Project for Statistical Computing, Vienna, Austria [Google Scholar]
- 44. Whitaker R. J., Grogan D. W., Taylor J. W. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976–978 [DOI] [PubMed] [Google Scholar]
- 45. Wiesmann U. 1994. Biological nitrogen removal from wastewater. Adv. Biochem. Eng. Biotechnol. 51:113–154 [DOI] [PubMed] [Google Scholar]
- 46. Wittebolle L., Vervaeren H., Verstraete W., Boon N. 2008. Quantifying community dynamics of nitrifiers in functionally stable reactors. Appl. Environ. Microbiol. 74:286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yannarell A. C., Kent A. D., Lauster G. H., Kratz T. K., Triplett E. W. 2003. Temporal patterns in bacterial communities in three temperate lakes of different trophic status. Microb. Ecol. 46:391–405 [DOI] [PubMed] [Google Scholar]
- 48. Zumft W. G. 1993. The biological role of nitric oxide in bacteria. Arch. Microbiol. 160:253–264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.