Abstract
Subsurface amendments of slow-release substrates (e.g., emulsified vegetable oil [EVO]) are thought to be a pragmatic alternative to using short-lived, labile substrates for sustained uranium bioimmobilization within contaminated groundwater systems. Spatial and temporal dynamics of subsurface microbial communities during EVO amendment are unknown and likely differ significantly from those of populations stimulated by soluble substrates, such as ethanol and acetate. In this study, a one-time EVO injection resulted in decreased groundwater U concentrations that remained below initial levels for approximately 4 months. Pyrosequencing and quantitative PCR of 16S rRNA from monitoring well samples revealed a rapid decline in groundwater bacterial community richness and diversity after EVO injection, concurrent with increased 16S rRNA copy levels, indicating the selection of a narrow group of taxa rather than a broad community stimulation. Members of the Firmicutes family Veillonellaceae dominated after injection and most likely catalyzed the initial oil decomposition. Sulfate-reducing bacteria from the genus Desulforegula, known for long-chain fatty acid oxidation to acetate, also dominated after EVO amendment. Acetate and H2 production during EVO degradation appeared to stimulate NO3−, Fe(III), U(VI), and SO42− reduction by members of the Comamonadaceae, Geobacteriaceae, and Desulfobacterales. Methanogenic archaea flourished late to comprise over 25% of the total microbial community. Bacterial diversity rebounded after 9 months, although community compositions remained distinct from the preamendment conditions. These results demonstrated that a one-time EVO amendment served as an effective electron donor source for in situ U(VI) bioreduction and that subsurface EVO degradation and metal reduction were likely mediated by successive identifiable guilds of organisms.
INTRODUCTION
Soil and groundwater contamination by uranium-bearing wastes is a pervasive problem at U mining and processing sites around the world (1). Uranium in the oxidized state, U(VI), is highly soluble in water, is toxic, and has the potential to migrate and contaminate groundwater and surface water sources (19). When reduced to U(IV), sparingly soluble U-bearing precipitates are formed, and the mobility of uranium in the subsurface is diminished. Therefore, reductive immobilization has been proposed as a practical approach to minimizing uranium transport from contaminated sites (12).
Reduction of U(VI) to U(IV) is catalyzed by a variety of microorganisms, including iron- and sulfate-reducing bacteria (SRB) and some fermentative bacteria (23, 46). In situ biostimulation using subsurface electron donor amendments has been explored previously in several different locations and subsurface environments (4, 22, 39, 48), and diminution of U(VI) in groundwater to levels below the U.S. EPA drinking water standard (30 μg liter−1) (44) has been achieved (49). All of these field experiments used either ethanol or acetate amendments to specifically target the metabolic capabilities of rather narrow groups of U(VI)-reducing microorganisms (e.g., Geobacter and Desulfovibrio) that may or may not be abundant under natural conditions but quickly respond to such amendments (4, 21, 49). Due to the labile nature of these electron donors, treatments using acetate and ethanol yield U bioimmobilization within only a limited aquifer volume near the injection wells (48). Furthermore, without frequent amendments (i.e., daily to weekly), reducing conditions cannot be maintained, and U(IV) is reoxidized and remobilized (49, 50). Reoxidation remains one of the largest impediments to in situ subsurface strategies for the bioremediation of uranium.
Biostimulation with electron donors, such as glycerol polylactate (17) and emulsified vegetable oil (EVO) (6, 51), has been recognized as being cost-effective for anaerobic bioremediation of various organic and inorganic contaminants (7, 27). The effectiveness of these substrates derives from their retarded flow in groundwater systems, high energy density, and relatively slow degradation, which provides a sustained source of electrons in the treatment zone (7). To date, slow-release electron donor substrates have not been tested for field-scale uranium bioremediation, and therefore dynamics of in situ microbial communities during EVO amendments are unknown.
The objectives of this study were to spatially and temporally assess subsurface microbial populations as a biostimulated zone transitioned through reduction and reoxidation phases after EVO injection, correlate microbial community changes with geochemical processes, and ultimately improve the ability to predict the long-term effectiveness of EVO-based remediation strategies. The field experiment was conducted at the U.S. Department of Energy Oak Ridge Integrated Field Research Challenge (ORIFRC) study site, Oak Ridge, TN. EVO was injected into a shallow aquifer within a uranium-bearing contaminant plume that was subsequently monitored for over 9 months. Results indicated that the microbial community proceeded through a logical sequence of changes that correspond well with the known physiologies of the dominant organisms and the resulting geochemical changes observed in the groundwater system. These data have allowed us to develop a testable theoretical model of microbial community interactions and function during EVO biostimulation that will guide future experiments.
MATERIALS AND METHODS
Subsurface stimulation experiment.
A field-scale biostimulation experiment was initiated by injecting EVO as an electron donor amendment to the subsurface at area 2 of the ORIFRC (www.esd.ornl.gov/orifrc/), as shown in Fig. S1 in the supplemental material. Contaminants detected at area 2 were previously characterized to be mainly U (3.8 to 9.1 μM), sulfate (1.0 to 1.2 mM), and nitrate (0.2 to 1.5 mM) in groundwater and U at 300 mg/kg or higher in soil-saprolite (31). Prior to the experiment, the hydrology of the site was characterized by injecting a conservative tracer (bromide) solution (3,400 liters of 450 mg Br− liter−1) into three adjacent wells on 10 December 2008. The monitoring wells in this study were selected based on their connection to the three injection wells. The EVO mixture used was the product SRS (Terra Systems, Inc., Wilmington, DE), containing 60% (wt/wt) soybean oil, 6% food-grade surfactant, 0.3% yeast extract, and 0.05% (NH4)3PO4 in water. An EVO solution was prepared by rigorously mixing SRS with groundwater pumped from the site to produce a 20% (vol/vol) SRS emulsion. A total of 3,400 liters of the groundwater-SRS emulsion was injected evenly into the same three adjacent wells over 2 h on 9 February 2009. Groundwater samples for microbial community analyses were taken from 8 monitoring wells, 7 of which were down-gradient from the injection zone and 1 of which was up-gradient, at intervals designed to bracket the major geochemical and redox changes associated with the experiment (−28 days and +4, 17, 31, 80, 140, and 269 days relative to injection). During the test period, the temperature of groundwater varied from 16 to 21°C (see the supplemental material). Samples of groundwater were filtered onto 142-mm-diameter membranes (8-μm prefilter, 0.2-μm sample filter) that were preserved at −80°C for nucleic acid analyses. Although groundwater and sediment microbial populations may differ, we chose to sample groundwater communities in this study, as sediment sampling was not possible over the temporal and spatial scales of the experiment. Groundwater chemistry sample collection and analysis methods were the same as those described in previous publications (48–50) and are further described in the supplemental material and in more detail in a forthcoming manuscript (D. B. Watson, W.-M. Wu, T. Mehlhorn, J. Earles, K. Lowe, T. M. Gihring, G. Zhang, F. Zhang, J. Phillips, S. D. Kelly, M. Boyanov, B. P. Spalding, C. W. Schadt, K. M. Kemner, C. S. Criddle, P. M. Jardine, and S. C. Brooks, unpublished data).
PCR amplification and pyrosequencing of 16S rRNA gene fragments.
Frozen 0.2-μm filters were cut aseptically into small pieces (10-cm2 total filter area per sample) that were processed using PowerSoil DNA isolation kits (MO BIO Laboratories Inc., Carlsbad, CA) as per the manufacturer's instructions. Bacterial and archaeal 16S rRNA gene amplicons for pyrosequencing were generated from DNA samples according to Vishnivetskaya et al. (45) and Porat et al. (34), respectively. PCR amplicons were purified using an Agencourt AMPure system (Beckman Coulter Inc., Brea, CA) according to the manufacturer's instructions, and a portion of each product was evaluated for size and purity using an Agilent 2100 bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA). The 16S rRNA gene amplicons were sequenced using a 454 Life Sciences genome sequencer FLX system following 454 protocols (454 Life Sciences-Roche, Branford, CT). Raw 16S rRNA sequences were trimmed and sorted using the RDP Pyrosequencing Pipeline online tools (13). Average read lengths were 249 for archaea and 207 for bacteria. The quality control procedure implemented through the RDP tools excluded all sequences that (i) had homopolymers of >8 bp in length, (ii) had ambiguous base calls, and (iii) contained any mismatches to the forward or reverse primers. A total of 268,708 bacterial (mean = 4,886 reads per sample; n = 55 samples) and 32,421 archaeal (mean = 1,544 reads; n = 21 samples) pyrosequencing reads were used in subsequent analyses. We have made the sequences available for download through the IFRC project website at http://public.ornl.gov/orifc.
Sequence clustering and taxonomic classification.
Sequences from individual libraries were aligned using the RDP Pyrosequencing Pipeline aligner (13), and all bacterial and archaeal sequences were merged into two respective alignments. Sequences were assigned to operational taxonomic units (OTUs) using the RDP Pipeline complete linkage clustering tool set at 3% distance. Bacterial and archaeal OTUs were assigned taxonomic information by first selecting representative reads using the RDP Pipeline dereplicate tool and then applying the RDP classifier (47). Taxonomic assignments with RDP classifier confidence values of less than 50% at the phylum, class, family, or genus level were considered unclassified at that level. Those bacterial OTUs that were unclassified to at least the genus level, but represented greater than 0.01% of all reads, were also taxonomically positioned using greengenes classifier assignments following the simrank identity thresholds of DeSantis et al. (15). The combined RDP and greengenes classification approach resulted in reliable taxonomic assignments for 80.9% and 59% of bacterial sequences at the phylum and family levels, respectively.
Archaeal OTUs were classified using RDP according to the same thresholds as described above. Due to the relatively small number of archaeal OTUs (n = 811), it was feasible to also manually verify taxonomic assignments for all OTUs by parsimony insertion of sequences within the greengenes curated phylogenetic tree (14) using the software package ARB (26, 30). Taxonomic affiliations were recorded for those sequences that were inserted into tree clades identified by Hugenholtz annotations. After both the RDP classifier and ARB parsimony insertion methods were applied, 69.4% of archaeal sequences could be assigned to recognized families, and 30.0% of archaeal sequences were identified as members of previously described environmental clone groups within the greengenes taxonomic tree.
To further identify the most abundant and dynamic bacterial OTUs, RDP complete linkage clustering results were used to calculate the relative abundance of each OTU in each sample, normalized for variable library sizes. Relative abundances of each OTU were also averaged across all samples, and OTUs were ranked according to their mean relative abundance to determine the dominant OTUs. Standard deviations of OTU relative abundances across all samples were used as a measure of the variability of OTU distribution in space and time. The most abundant families or environmental clone groups were identified by summing the relative abundances of all OTUs assigned to the same classification and then ranking the total relative abundance of those groups across all samples in space and time.
Similarity-based comparisons and diversity indices.
Comparisons of community structure overlap between samples were performed based on the fractions of individuals belonging to shared OTUs. To avoid artifacts due to sample size dependency, each individual library was equalized to 2,500 (bacteria) or 650 (archaea) reads by randomly resampling sequences from the full-size libraries. Alignments and complete linkage clustering at 3% distance were performed on the equalized libraries using the RDP Pipeline. OTU clustering results were then imported into MOTHuR (38), where unweighted-pair group method with arithmetic mean grouping was used with abundance-based Sorenson distance measures. Community diversity and richness within each sample library were estimated using Simpson's and Margalef index calculations, respectively, on the libraries normalized for sequence number.
Quantification of bacterial and archaeal 16S rRNA genes.
Real-time PCR was performed on groundwater DNA extracts (described above) using iQ SYBR green supermix (Bio-Rad Laboratories, Hercules, CA). Bacterial 16S rRNA gene copies were quantified using 5 μM (each) primers 338F and 518R (33) (see Table S1 in the supplemental material) under the following thermal conditions: 95°C for 180 s, followed by 40 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 60 s. Plates were read after each 72°C extension step. After thermal cycling, a melt curve was analyzed from 65 to 95°C with readings every 0.5°C. Escherichia coli genomic DNA, analyzed at 7 different dilutions between 1.1 × 104 and 1.1 × 107 16S rRNA gene copies per reaction, was used for a standard curve. Archaeal 16S rRNA gene copy concentrations were determined using primers 915F and 1059R (see Table S1) at 10 μM each. Thermal cycle conditions were 95°C for 180 s, followed by 45 cycles of 95°C for 30 s, 61°C for 30 s, and 72°C for 30 s. Melt curve parameters were the same as for bacteria. Methanococcus maripaludis genomic DNA was analyzed at 7 different concentrations between 7.2 × 103 and 7.2 × 106 16S rRNA gene copies per reaction to generate a standard curve. Triplicate reactions were run for each standard and experimental sample, positive- and negative-control reactions were performed in duplicate for each primer set, and the resulting copy numbers were averaged. PCR-quality water was used in negative-control reactions.
Quantification of bacterial 16S rRNA and dsrB mRNA copies.
Total RNA was extracted from groundwater filters according to Chin et al. (10). RNA extracts used to generate dsrB cDNA were enriched for mRNA using MICROBExpress bacterial mRNA purification and DNase kits (Ambion, San Francisco, CA). RNA extracts for 16S rRNA cDNA were untreated. Reverse transcription reactions for dsrB and 16S rRNA were then performed using MultiScribe reverse transcriptase kits (Applied Biosystems Inc., Foster City, CA) with the dsr4R and 1492R reverse primers (see Table S1 in the supplemental material), respectively, at 0.2 μM. Primer, deoxynucleoside triphosphates (dNTPs), and RNA were combined and heated at 70°C for 2 min. Buffer, MgCl2, and enzymes were then added and heated to 25°C for 10 min, 48°C for 90 min, and then 95°C for 5 min.
Real-time PCR for quantifying dsrB in cDNA was conducted using a Power SYBR green PCR master mix (Applied Biosystems Inc.). Reactions were run for 50 cycles of 94°C for 30 s, 60°C for 20 s, 72°C for 30 s, and a fluorescent reading at 79°C for 15 s. Each reaction mixture contained the primers dsr2060F and dsr4R (see Table S1 in the supplemental material) at 400 nM each. Amplicons from cloned Desulfotomaculum orientis dsrAB genes were used for standard curves at five concentrations between 107 and 104 copies μl−1. TaqMan assay kits (Ambion) were used for real-time PCR quantification of 16S cDNA copy numbers according to Nadkarni et al. (32), with bacterial primers 331F and 797R and probe 506. All real-time PCRs on dsrB and 16S rRNA cDNA unknowns, standards, and controls were performed in triplicate, and the resulting copy numbers were averaged. PCR-quality water was used in negative-control reactions.
Multivariate analyses of community and geochemical data.
The BIO-ENV procedure (11) within the program Primer 6 (Primer-E Ltd., Ivybridge, United Kingdom) was used to identify the set of environmental variables that best explained the correlation between measured environmental parameters and community measurements. Groundwater physicochemical data (see Table S5 in the supplemental material) were transformed to best approximate a normal distribution according to Ryan-Joiner tests (36) and were standardized to a mean of zero with unit variance. Abundances of 16S rRNA gene pyrosequencing reads classified at the OTU or family level were normalized for each sample, log transformed, and then used to generate a distance matrix based on Bray-Curtis measures. The best Spearman rank correlation between matrices of Euclidean distances of environmental factors and Bray-Curtis distance measures of family abundances was then determined. Dissolved acetate, aluminum, calcium, iron, manganese, nitrate, sulfate, and uranium data were selected for canonical correspondence analysis (CCA) (43) with 16S rRNA gene abundance data. CCA was performed using the program CANOCO 4.5 (Microcomputer Power, Ithaca, NY).
RESULTS
Environmental measures and evolution of groundwater geochemistry.
After EVO injection, reductions of nitrate, Mn(IV), Fe(III), sulfate, and U(VI) occurred sequentially in down-gradient monitoring wells. Within 16 days, the nitrate, sulfate, and U(VI) concentrations in the down-gradient wells were diminished substantially relative to preinjection concentrations, whereas concentrations were largely unchanged in the up-gradient well FW215 (Table 1; also see Fig. S3 in the supplemental material). Sulfate and nitrate concentrations began to rebound by 80 to 135 days after injection, whereas uranium levels remained low until 135 to 269 days after EVO injection. Aqueous iron concentrations increased by over 1 order of magnitude in all down-gradient wells within 16 days and then again decreased to preinjection levels by 269 days. Acetate was detected in most down-gradient wells 4 days after EVO injection and increased to peak levels by 16 to 31 days before declining to below detection limits again by 269 days, indicating that EVO was consumed (see Fig. S3).
Table 1.
Groundwater geochemical data in control (FW215) and monitoring wells during select time points associated with microbiological sample collectiona
| Well ID | Time point (days) | pH | Acetate (μM) | Nitrate (μM) | Sulfate (μM) | Iron (μM) | Uranium (μM) |
|---|---|---|---|---|---|---|---|
| FW215 | −18 | 6.84 | BD | 107 | 1,230 | 1.85 | 5.46 |
| 4 | 6.78 | BD | 85.7 | 1,160 | 3.16 | 5.11 | |
| 16 | 6.52 | BD | 304 | 1,020 | 2.46 | 5.04 | |
| 31 | 6.08 | BD | 348 | 1,040 | 1.21 | 4.79 | |
| 80 | 6.74 | BD | 457 | 1,040 | 3.10 | 4.94 | |
| 135 | 6.52 | BD | 510 | 995 | 0.72 | 5.19 | |
| 269 | 6.15 | BD | 43.7 | 45.9 | 2.87 | 4.81 | |
| MLSG4 | −18 | 6.74 | BD | 59.2 | 1,250 | 12.0 | 8.51 |
| 4 | 6.71 | 212 | 60.3 | 631 | 73.0 | 9.56 | |
| 16 | 6.31 | 1,450 | BD | 90.0 | 46.2 | 0.25 | |
| 31 | 6.13 | 646 | 16.3 | 237 | 25.7 | 0.61 | |
| 80 | 6.71 | 446 | BD | 623 | 6.41 | 3.37 | |
| 135 | 6.44 | BD | 59.6 | 730 | 9.65 | 3.92 | |
| 269 | 6.14 | BD | 760 | 1,140 | 20.9 | 2.57 | |
| MLSA3 | −18 | 6.88 | BD | 336 | 986 | 0.23 | 5.10 |
| 4 | 6.82 | BD | 8.55 | 1,240 | 0.46 | 9.05 | |
| 16 | 6.45 | 270 | BD | 582 | 58.4 | 3.75 | |
| 31 | 6.48 | 1,120 | 15.0 | 8.54 | 247 | 1.05 | |
| 80 | 6.81 | 559 | BD | 3.69 | 198 | 0.43 | |
| 135 | 6.55 | 1,740 | 1.82 | 47.1 | 201 | 0.40 | |
| 269 | 6.83 | BD | 298 | 1,040 | 77.6 | 3.80 | |
| FW216 | −18 | NA | NA | NA | NA | NA | NA |
| 4 | 6.65 | 50.1 | 8.48 | 470 | 79.9 | 9.46 | |
| 16 | 6.69 | 656 | BD | 346 | 33.6 | 2.09 | |
| 31 | 5.96 | 1,400 | 16.7 | 161 | 3.33 | 0.37 | |
| 80 | 6.92 | BD | 111 | 1,060 | 1.29 | 6.37 | |
| 135 | 6.66 | 42.0 | 155 | 871 | 2.19 | 5.16 | |
| 269 | 6.81 | BD | 228 | 1,010 | 0.89 | 4.89 | |
| MLSB3 | −18 | 6.84 | BD | 41.9 | 1,240 | 0.23 | 8.68 |
| 4 | 6.64 | 387 | BD | 689 | 44.7 | 9.53 | |
| 16 | 6.80 | 1,570 | BD | 7.00 | 53.8 | 0.59 | |
| 31 | 6.39 | 1,720 | 16.6 | 14.1 | 17.3 | 0.23 | |
| 80 | 6.64 | 529 | 10.3 | 289 | 5.41 | 0.96 | |
| 135 | 6.34 | 1,140 | BD | 184 | 29.4 | 1.15 | |
| 269 | 6.17 | BD | BD | 834 | 13.4 | 3.25 | |
| GP01 | −18 | 6.83 | BD | 472 | 1,030 | 0.23 | 5.36 |
| 4 | 6.66 | 9.80 | 80.7 | 286 | 114 | 9.09 | |
| 16 | 6.54 | 244 | 298 | 360 | 138 | 2.65 | |
| 31 | 6.72 | 1,600 | 13.4 | 11.3 | 45.6 | 0.22 | |
| 80 | 7.01 | BD | BD | 622 | 20.6 | 2.60 | |
| 135 | 6.77 | BD | 221 | 1,470 | 1.16 | 6.74 | |
| 269 | 6.60 | BD | 33.1 | 417 | 0.89 | 5.03 | |
| FW202 | −18 | 6.92 | BD | 207 | 1,170 | 0.23 | 10.5 |
| 4 | 6.79 | BD | 460 | 1,460 | 2.32 | 8.99 | |
| 16 | 6.83 | 1,290 | BD | 13.0 | 267 | 1.37 | |
| 31 | 6.64 | 1,290 | 25 | 8.22 | 124 | 0.85 | |
| 80 | 6.81 | 251 | BD | 661 | 25.6 | 1.00 | |
| 135 | 6.73 | BD | 162 | 1,080 | 21.8 | 6.08 | |
| 269 | 6.78 | BD | 572 | 138 | 0.89 | 8.15 | |
| GP03 | −18 | 6.87 | BD | 168 | 1,140 | 0.23 | 10.2 |
| 4 | 6.75 | 13.6 | BD | 896 | 85.3 | 11.4 | |
| 16 | 6.45 | 1,450 | BD | 37.2 | 142 | 0.41 | |
| 31 | 6.48 | 2,820 | 27.2 | 22.4 | 144 | 0.61 | |
| 80 | 6.71 | 738 | 227 | 101 | 54.5 | 0.12 | |
| 135 | 7.02 | 642 | 7.39 | 129 | 15.3 | 0.17 | |
| 269 | 6.94 | BD | 560 | 945 | 0.18 | 7.02 |
The subsurface temperature change varied from 16°C (May) to 21°C (November). ID, identifier; NA, not analyzed; BD, below detection limit (acetate, <0.01 mM; nitrate, <0.005 mM).
Effect of EVO amendment on groundwater bacterial community structures.
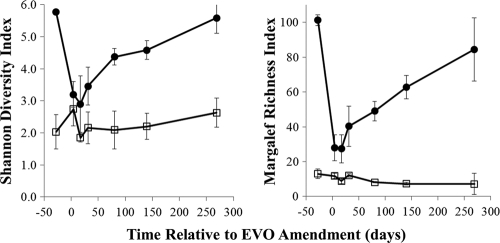
Amendment of EVO to the subsurface initiated a cascade of shifts in the microbial community structure and resulted in long-term modification of the subsurface microbial community (see Fig. 1 to 5). One prominent effect of the amendment was a rapid decrease in bacterial diversity, as assayed by rRNA gene sequence libraries (Fig. 1). Bacterial richness and diversity were highest prior to EVO injection, were lowest immediately after injection, and slowly rebounded over the 9-month duration of the experiment. Concurrently, the number of bacterial 16S rRNA and rRNA gene copies increased by up to 2 orders of magnitude after EVO addition (see Fig. 5), an indication of stimulated biomass production.
Fig. 1.
Time series of bacterial (filled circles) and archaeal (open squares) Margalef richness indices and Shannon diversity indices averaged for all analyzed libraries at each time point. Error bars represent 1 standard deviation.
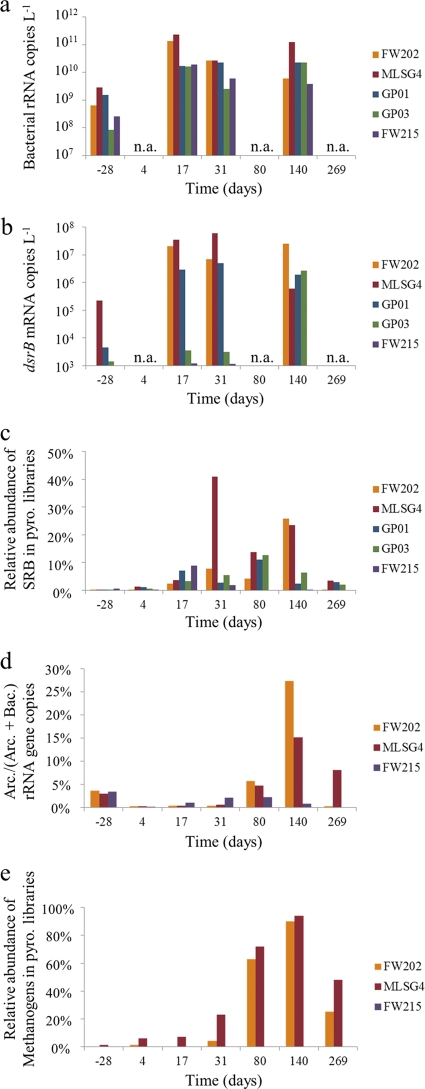
Fig. 5.
Characterization of sulfate-reducing and methanogenic populations using dsrB and 16S rRNA gene analyses. (a) Number of bacterial 16S rRNA copies detected in groundwater using quantitative PCR on RNA extracts. (b) Number of dsrB mRNA transcript copies detected in groundwater using quantitative PCR on RNA extracts. (c) Sum of the relative abundances of 16S rRNA genes detected by pyrosequencing classified as members of the Desulfobacteraceae, Desulfovibrionaceae, and Desulfobulbaceae. (d) Fraction of archaeal 16S rRNA gene copies, relative to the sum of archaeal and bacterial gene copies, detected in groundwater using quantitative PCR on DNA extracts. (e) Sum of the relative abundances of 16S rRNA genes detected by pyrosequencing classified as members of known methanogen groups (Methanobacteraceae, Methanoregulaceae, Methanospirillaceae, and Methanosarcinaceae). n.a., not analyzed; pyro., pyrosequencing.
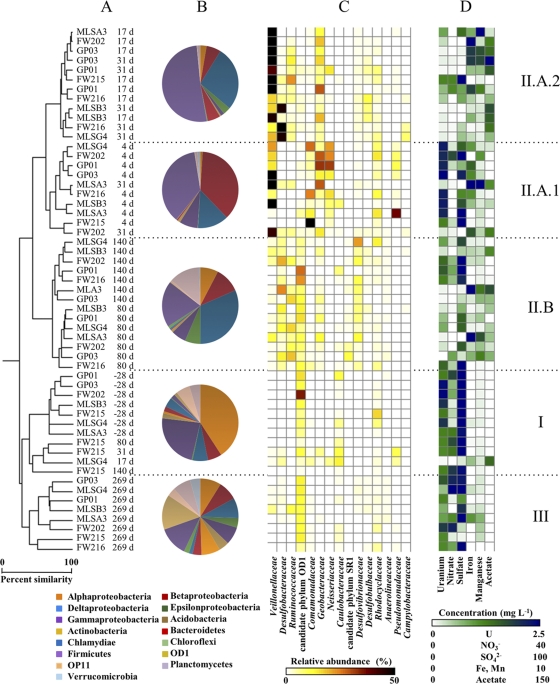
The subsurface bacterial community was observed to undergo shifts in community structure that were broadly grouped into three categories: native, contaminated groundwater (cluster I; prior to injection and up-gradient wells); active EVO degradation (cluster II; 4 to 180 days postinjection, down-gradient wells); and the establishment of new, postinjection bacterial communities (cluster III; 269 days postinjection) (Fig. 2). The microbial communities in cluster I samples were dominated by Alpha- and Gammaproteobacteria (Fig. 2). Despite a strong proteobacterial presence, there were few dominant taxa at or below the family level, consistent with the relatively high richness and diversity observed for these samples (Fig. 1).
Fig. 2.
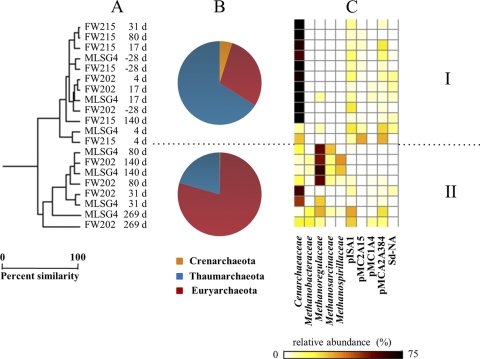
Comparisons of groundwater bacterial community structures and composition during the EVO experiment. Sample names indicate the monitoring well identifier (ID) and the sampling time point relative to EVO amendment. (A) Samples clustered according to abundance-based Sorenson's indices of 16S rRNA gene libraries normalized to 2,500 reads each. d, days. (B) Pie charts indicate the mean relative abundance of 16S rRNA gene pyrosequencing reads, classified at the division or class level, for samples corresponding to the indicated clades. (C) The heat map indicates the sum of the relative abundance of those 16S rRNA gene reads classified as members of the indicated taxonomic groups. Relative abundances for the 15 most abundant families or candidate phyla are shown for each sample. (D) The heat map indicates the concentrations of various dissolved constituents in groundwater samples collected at the same time as the microbiological samples.
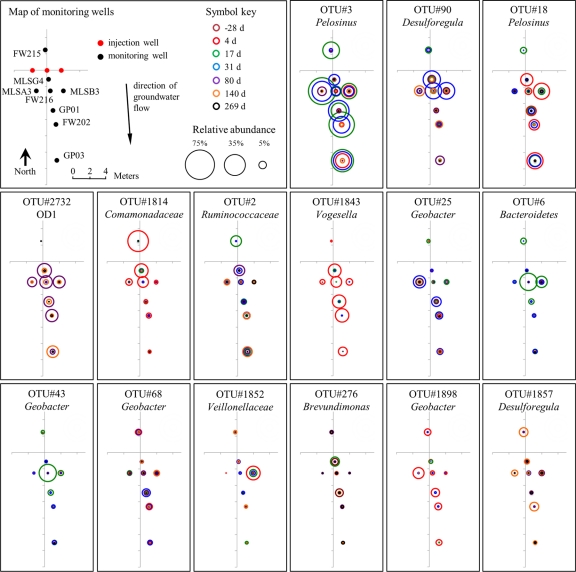
The bacterial community in the groundwater was significantly altered subsequent to EVO amendment, and time since injection was a key predictor of community structure (Fig. 2). Three subclusters within cluster II samples were identified: clusters II.A.1 (primarily 4 days postinjection), II.A.2 (primarily 17 to 31 days postinjection), and II.B (80 to 140 days postinjection). Sequence libraries of groundwater samples grouping together in cluster II.A.1 were dominated by sequences of Firmicutes and Proteobacteria, with sequences from the families Veillonellaceae (Firmicutes), Comamonadaceae (Betaproteobacteria), Geobacteraceae (Deltaproteobacteria), Neisseriaceae (Betaproteobacteria), Rhodocyclaceae, and Pseudomonadaceae (Gammaproteobacteria) predominating. OTUs belonging to Pelosinus, Comamonadaceae, and Vogesella were particularly abundant at 4 days postinjection (Fig. 3). It should be noted that due to the volume of EVO injected, some of the amendment was forced to travel against the prevailing hydrological gradient during injection, and changes in the up-gradient FW215 bacterial community due to EVO were also apparent at 4 and 17 days (Fig. 2).
Fig. 3.
Temporal and spatial distributions of the 15 most abundant bacterial OTUs detected in groundwater. Relative abundances of sequences belonging to each OTU, detected in each of the 8 monitoring wells at 7 different time points, are represented according to circle diameter. Circle colors indicate the duration (days) relative to EVO injection.
After 17 to 31 days, sequence libraries of groundwater samples grouping together in cluster II.A.2 were predominantly comprised of Veillonellaceae, Desulfobacteraceae (Deltaproteobacteria), Ruminococcaceae (Firmicutes), Geobacteraceae, and Rhodocyclaceae. The majority of sequences at these time points were derived from only a small variety of OTUs, including those identified as members of Pelosinus, Desulforegula, Bacteroidetes, and Geobacter (Fig. 3). Sequences from the families Veillonellaceae and Desulfobacteraceae continued to dominate the 80-day and 140-day samples (cluster II.B), although members of the Desulfovibrionaceae (Deltaproteobacteria) and Desulfobulbaceae (Deltaproteobacteria) and candidate phylum OD1 were also abundant (Fig. 2 and 3). Groundwater samples from 269 days postinjection formed cluster group III and were most similar to preinjection groundwater samples, though significantly divergent (Fig. 2). Members of the candidate phylum OD1 were in relatively high abundance; however, cluster group III samples were otherwise relatively diverse.
Characterization of SRB abundance and activity.
Quantitative PCR analyses of dsrB gene transcripts showed relatively low dsrB mRNA copy numbers in the preinjection samples (see Fig. 5). After 17 days, dsrB transcript abundances had increased by over 2 orders of magnitude in wells FW202, MLSG4, and GP01, reaching between 2.9 × 106 and 3.5 × 107 mRNA copies liter−1. Samples from GP03, the monitoring well farthest from the injection site, showed a more delayed response until 140 days, when the dsrB copy level increased to 2.7 × 106 copies liter−1. Copies of dsrB mRNA in the up-gradient well FW215 remained below or slightly above the limit of detection during the entire experiment (see Table S4 in the supplemental material). Two peaks in the ratio of dsrB mRNA to 16S rRNA copies were observed: a maximum in well MLSG4 at 31 days and a peak in well FW202 at 140 days (see Table S4). These maxima in dsrB transcript concentrations, and ratios of dsrB mRNA to 16S rRNA, corresponded with time points having the highest relative abundance of 16S rRNA gene pyrosequencing reads classified as Desulfobacteraceae, Desulfovibrionaceae, and Desulfobulbaceae, the dominant families of known SRB detected in this experiment (see Fig. 5).
Response of the archaeal community.
A distinct separation of archaeal communities based on time and location relative to EVO injection was evident from OTU cluster analyses and quantitative PCR (Fig. 4 and 5). Prior to EVO amendment, and over the initial 31 days, Archaea comprised only 0.2 to 4.0% of all 16S rRNA gene copies (Fig. 5). Members of the Cenarchaeaceae family within the Thaumarchaeota phylum dominated (cluster group I; Fig. 4), and archaeal 16S rRNA gene copy numbers remained nearly constant (see Table S4 in the supplemental material). During the later phase of the experiment (80 to 269 days), archaeal 16S rRNA gene copy numbers increased and reached maxima of 15% to 27% of all 16S rRNA gene copies detected at 140 days (Fig. 5; also see Table S4). The down-gradient samples (group II; Fig. 4) were dominated by members of the Methanobacteriaceae, Methanoregulaceae, Methanosarcinaceae, and Methanospirillaceae (Euryarchaeota phylum), which comprised over 90% of all archaeal pyrosequencing 16S rRNA gene reads at 140 days (Fig. 5). Despite these drastic shifts in community compositions, archaeal diversity indices were largely unchanged over the course of the experiment (Fig. 1), which likely reflects the overall simplicity of the archaeal community compared to that of the bacteria.
Fig. 4.
Comparisons of groundwater archaeal community structures and composition during the EVO experiment. (A) Samples clustered according to abundance-based Sorenson's indices of 16S rRNA gene libraries normalized to 650 reads each. Sample names are the monitoring well ID and the sampling time point relative to SRS amendment. (B) Pie charts indicate the mean relative abundances of 16S rRNA gene pyrosequencing reads, classified at the division level, for samples corresponding to the indicated clades. (C) The heat map indicates the sum of the relative abundance of those 16S rRNA gene reads classified as members of the indicated taxonomic groups. Relative abundances for the 10 most abundant families or environmental clone groups are shown for each sample.
Correlations between microbial communities and groundwater geochemistry.
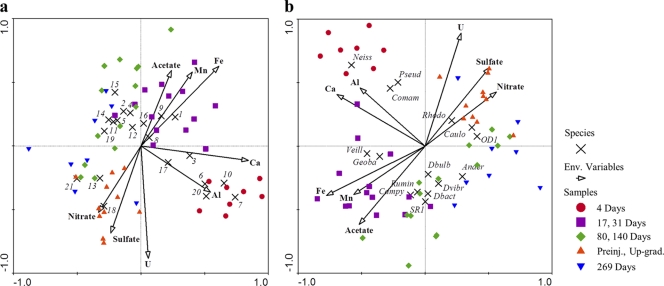
CCA ordination showed that temporal variations in bacterial community compositions were associated with specific geochemical conditions (Fig. 6). For example, members of the Geobacteraceae family and Geobacter OTUs were correlated positively with higher aqueous Fe and Mn concentrations (Fig. 6), indicators of active metal reduction. Members of Veillonellaceae, Geobacteraceae, Campylobacteraceae, Ruminococcaceae, candidate phylum SR1, Desulfobulbaceae, Desulfovibrionaceae, and Desulfobacteraceae were correlated positively with higher concentrations of dissolved iron, manganese, and acetate and negatively with uranium, sulfate, and nitrate (Fig. 6). Sequences from groups of putative nitrate reducers (Neisseriaceae, Pseudomonadaceae, and Comamonadaceae) were not statistically correlated with lower nitrate concentrations. However, although samples were collected during an active denitrification phase (4 days) and nitrate levels were significantly lower than those under preinjection conditions, nitrate was not yet entirely depleted (Table 1). Several groups of OTUs were negatively correlated with higher levels of dissolved uranium, including Geobacter, Desulforegula, Desulfobacteraceae, OD1, and Ruminococcaceae (Fig. 6).
Fig. 6.
Ordination diagrams from canonical correspondence analyses of bacterial abundances (response variables indicated by black crosses) and geochemistry data (illustrated by vectors). (a) Relative abundance of dominant OTUs. (b) Relative abundances of dominant families. Samples are represented by colored symbols coded according to clustering results shown in Fig. 2. OTU abbreviations: 1, OTU 3 (Pelosinus); 2, OTU 90 (Desulforegula); 3, OTU 18 (Pelosinus); 4, OTU 2 (Ruminococcaceae); 5, OTU 2732 (OD1); 6, OTU 1814 (Comamonadaceae); 7, OTU 1843 (Vogesella); 8, OTU 6 (Bacteroidetes); 9, OTU 25 (Geobacter); 10, OTU 43 (Geobacter); 11, OTU 273 (Proteobacteria); 12, OTU 68 (Geobacter); 13, OTU 276 (Brevundimonas); 14, OTU 294 (Proteobacteria); 15, OTU 100 (Ruminococcaceae); 16, OTU 2990 (Neisseriaceae); 17, OTU 1852 (Veillonellaceae); 18, OTU 2762 (unclassified); 19, OTU 1857 (Desulfobacteraceae); 20, OTU 1898 (Geobacter); 21, OTU 17077 (Rhodospirillaceae). Family abbreviations: Anaer, Anaerolineaceae; Campy, Campylobacteraceae; Caulo, Caulobacteraceae; Comam, Comamonadaceae; Dbact, Desulfobacteraceae; Dbulb, Desulfobulbaceae; Dvibr, Desulfovibrionaceae; Geoba, Geobacteraceae; Neiss, Neisseriaceae; OD1, candidate phylum OD1; Pseud, Pseudomonadaceae; Rhodo, Rhodocyclaceae; Rumin, Ruminococcaceae; SR1, candidate phylum SR1; Veill, Veillonellaceae.
DISCUSSION
Effects of EVO stimulation on microbial community complexity.
The dramatic decreases in diversity of 16S rRNA gene OTUs after EVO injection, concurrent with increased microbial biomass (rRNA genes) and activity (rRNA gene and dsrB transcripts), strongly suggest that the EVO amendment resulted in the rapid selection for a limited number of taxa rather than a broad stimulation of the larger community as observed after ethanol amendments in the ORIFRC subsurface (9, 21). This change likely resulted from a selective environment that favors the relatively few taxa having the ability to degrade the complex vegetable oil substrate. This rapid stimulation of specific taxa within the down-gradient communities, in concert with an expansion in the functional diversity of specific gene sets involved in dissimilatory reduction processes, suggests that the native subsurface groundwater community in area 2, where the pH is circumneutral, retains a substantial complexity that can respond rapidly to the ecological disturbance and new selective environment represented by the biostimulation event. Ecologically and evolutionarily, this response might be considered similar to the effects of a disturbance, in that the disturbance regime sets up a specific set of selective conditions and pressures on the community and the organisms and traits that inevitably thrive in the postdisturbance regime may differ dramatically from the preexisting steady-state community and may or may not return to similar composition and function depending on the functional redundancy of the community (3, 5).
Proposed model of subsurface EVO degradation and metal bioreduction.
Anaerobic microbial degradation of the vegetable oil amendment was expected to follow a sequence analogous to reactions observed in wastewater treatment facilities. In these environments, triglycerides are initially broken down by lipid hydrolysis to release long-chain fatty acids (LCFAs) and glycerol. LCFAs are then incompletely oxidized to produce H2 and either acetate (from even-carbon FAs) or acetate plus propionate (odd-carbon FAs). In the absence of sulfate, anaerobic LCFA oxidation is not thermodynamically favorable and requires a syntrophic relationship between LCFA oxidizers (e.g., Syntrophomonas) and acetate- and hydrogen-consuming methanogens (41). A similar syntrophic mechanism has been proposed for subsurface microbial oxidation of LCFAs in oil reservoirs (18). However, where sulfate is present, such as in the ORIFRC subsurface, SRB can outcompete syntrophic acetogens and methanogens for LCFAs, acetate, and H2 (42). LCFA oxidation under iron-reducing conditions has been reported (28), and where Fe(III) and Mn(IV) are bioavailable, metal-reducing microbes are also expected to compete for acetate and H2 produced by LCFA-oxidizing bacteria.
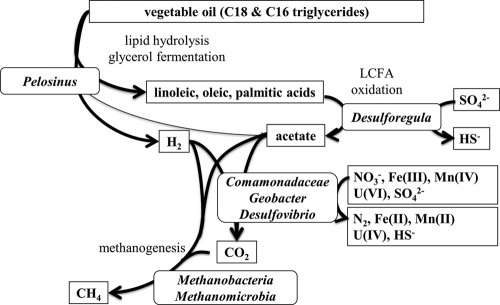
Based on the observed timing and changes in community structures, OTU abundances and closest relatives with known physiologies, the geochemical evolution of the groundwater, and anticipated substrate degradation pathways, we constructed a conceptual model of EVO degradation and metal reduction at the ORIFRC site (Fig. 7). The stimulation and predominance of members of the Pelosinus-like organisms (Veillonellaceae family) after EVO injection (Fig. 2 and 3) suggest that these organisms may have been involved in the degradation of the vegetable oil and possibly the minor yeast extract component and were key drivers of subsurface geochemical conditions. Members of the Veillonellaceae family (formerly the Acidaminococcaceae) are generally recognized as anaerobes capable of growth on complex media, and the sole described Pelosinus isolate (P. fermentans) ferments various defined substrates (40). Triglyceride hydrolysis by P. fermentans has not been studied previously; however, many other members of the Veillonellaceae have lipase activity (e.g., Anaerovibrio lipolyticus) (20) and can ferment glycerol residues of diglycerides (e.g., Anaerosinus glycerini) (37), which also releases LCFAs for further later oxidation. The correlation of Veillonellaceae sequences with acetate production (Fig. 2 and 6) also suggests that these organisms either generated acetate directly as a fermentation product or provided LCFAs to organisms performing LCFA degradation via β-oxidation to produce acetate and hydrogen. Future efforts to isolate and characterize the predominant Pelosinus-like organisms from the ORIFRC are necessary to confirm this physiology.
Fig. 7.
Conceptual model of EVO degradation and metal reduction in subsurface environments based on the relative abundance of representative abundant OTUs and known physiologies of closely allied species or genera.
We propose that under anaerobic conditions with bioavailable sulfate, iron, and manganese in the subsurface, LCFA oxidation is mediated by sulfate- or metal-reducing bacteria and not the commonly recognized consortia of Syntrophomonas and methanogens. Members of the Desulforegula genus were likely the organisms primarily responsible for LCFA degradation to acetate and H2. The described species within the Desulforegula genus (D. conservatrix) grows only by sulfate reduction coupled with LCFA oxidation to acetate (35) and does not utilize many short-chain fatty acids (such as acetate) or other typical fermentation products (e.g., ethanol or lactate). Therefore, the preponderance of Desulforegula spp. during EVO degradation and acetate production (Fig. 2 and 6) and their phylogenetic association with LCFA-oxidizing SRB suggest that Desulforegula spp. are important in sustaining reducing conditions by oxidizing LCFA and providing hydrogen and acetate (and likely some propionate derived from odd-chain-length LCFAs) that is utilized by other organisms. In subsurface ecosystems without abundant sulfate, the EVO degradation pathway may be mediated by iron-reducing LCFA oxidizers (28) instead of Desulforegula-like organisms. Cultures of Desulforegula-like organisms which are capable of LCFA oxidation of palmitic and oleic acids with concurrent sulfate reduction have now been obtained from the EVO stimulated zone (DSM 24590 [T. M. Gihring and C. W. Schadt, unpublished results]). HS− liberated from Desulforegula and other sulfate reducers may also play a significant role in the indirect reduction of Fe(III) and U(VI) in this experiment.
A clear succession of groundwater microbial community structures, concurrent with nitrate, manganese, iron, sulfate, and uranium reduction following EVO amendment, was evident from combined 16S rRNA gene sequencing, quantitative PCR, and multivariate statistical results (Fig. 2 and 6; Table 1). Many (but not all) members of the Neisseriaceae, Pseudomonadaceae, and Comamonadaceae families are known for dissimilatory nitrate reduction and, given their early abundance in our data set, were possibly important in mediating rapid denitrification immediately after EVO injection (Table 1; Fig. 2 and 3). Previous experiments have also shown that these groups are associated with nitrate reduction at the ORIFRC site (2, 8). The rapid response of denitrifying organisms after EVO injection and the peak in populations prior to significant acetate production suggest that these populations may have used the small amount of yeast extract additive in the SRS product as a carbon and energy source, rather than the vegetable oil.
As H2 and acetate became available, it appears that the phylogenetic and functional diversity of microbial communities expanded, as many more organisms are known to grow using H2 or acetate than using triglycerides or LCFAs. In our proposed model, a variety of sulfate- and metal-reducing bacteria are likely to consume acetate and H2 derived from LCFAs, as long as bioavailable sulfate, Fe(III), and Mn(IV) were not limiting. Based on their prevalence during the metal reduction phase (Fig. 2, 3, and 5), members of the Geobacteraceae responded positively to EVO amendment and are likely also critical in mediating metal reduction. Sulfate reduction in the ORIFRC subsurface after EVO amendment is also coincident with the abundance of members of the Desulfovibrionaceae and Desulfobulbaceae (Fig. 2 and 4), groups that often utilize hydrogen but not acetate (24, 25) as an electron donor. A sustained H2 source that outlasted the supply of bioavailable nitrate, Fe(III), and Mn(IV) may have resulted in an overall increased relative importance of sulfate reducers during the course of U bioremediation compared to that from bioreduction experiments using acetate or ethanol amendments. Prolonged predominance of members of the Desulfovibrionaceae and Geobacteraceae, known for U(VI) reduction (29, 46), was likely highly beneficial to sustaining U(VI)-reducing conditions during biostimulation with EVO. Indeed, isolates of both a previously undescribed Geobacter sp. and another Desulfovibrio sp. corresponding to the OTUs observed here at >99% 16S rRNA gene sequence identity have been purified from enrichments obtained from the manipulation zone and show physiologies consistent with our model assumptions in laboratory tests (DSM 24454 and DSM 24453 [Gihring and Schadt, unpublished]).
The stimulation of methanogens (Fig. 3 to 5) suggests that despite the general reoxidation of the test area and continuous inflow of nitrate- and sulfate-rich groundwater, flooding the system with electron donor produced persistent reducing zones in the subsurface. The driving of electrons to methane in this system also had the consequence of losing some of the potential electrons associated with the EVO injection to a functional group incapable of U(VI) reduction. Sequences closely related to 16S rRNA genes of H2-oxidizing methanogens dominated (Methanobacteraceae, Methanoregulaceae, and Methanospirillaceae) compared to those of known acetoclastic methanogens (Methanosarcinaceae) (Fig. 4), suggesting that H2 availability was more important in controlling methanogenesis than acetate. Concentrations of methane measured in the injection wells also increased during the experiment—in most cases increasing to >500 parts per trillion by volume (pptv), and this was a strong indicator of active methanogenesis (Watson et al., unpublished). Archaeal communities have not previously been studied extensively at uranium-contaminated sites, and our results underline the importance of understanding archaeal community dynamics during future U bioremediation.
Community changes during aquifer reoxidation.
Oxidation of bioreduced U(IV) was expected following the consumption of EVO because numerous studies have demonstrated the potential for reoxidation of bioreduced U(IV) after substrate consumption and exposure to oxygen- and nitrate-bearing groundwater (22, 49, 50). Despite a gradual increase in diversity during the reoxidation phase (80 to 269 days) (Fig. 1) and a shift away from community compositions and structures established during U(VI) reduction (Fig. 2 and 6), bacterial communities after 269 days remained dissimilar to the preinjection samples (Fig. 2). Clearly the archaeal communities also did not reestablish their preinjection structure (Fig. 4). The relatively distant relationship between bacterial communities at 269 days and the preinjection time point may be indicative of a community specifically adapted to reoxidation conditions. For example, one might expect Fe(II)- and Mn(II)-oxidizing microbes to thrive in the presence of oxygen and bioavailable Fe(II) and Mn(II) that may not otherwise have been abundant without EVO amendment. However, sequences related to known iron-oxidizing bacteria were not abundant in any of the 16S rRNA gene libraries. At this time, the role of OD1 microbes in subsurface contaminant cycling remains unknown (16), although it is possible that members of the OD1 phylum and the Ruminococcaceae family, both relatively abundant during reoxidation (Fig. 2 and 3), were stimulated by the death and decay of copious microbial biomass generated after EVO amendment. To our knowledge, this lasting effect on in situ subsurface microbial community compositions due to a one-time substrate amendment has not been demonstrated previously.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Stephens and S. Mueller for field and logistical support, as well as T. J. Phelps and 3 anonymous reviewers for their critical reviews of the manuscript.
Research was sponsored by the U.S. Department of Energy, Office of Science, Biological and Environmental Research as part of the Oak Ridge Integrated Field Research Challenge (IFRC) project. Oak Ridge National Laboratory is managed by UT Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Present address: Institute of Tibetan Plateau Research, Chinese Academy of Sciences, P.O. Box 2871, Beijing 100085, China.
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Abdelouas A., Lutze W., Nuttall H. E. 1999. Uranium contamination in the subsurface; characterization and remediation. Rev. Mineral. Geochem. 38:433–473 [Google Scholar]
- 2. Akob D. M., Mills H. J., Kostka J. E. 2007. Metabolically active communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59:95–107 [DOI] [PubMed] [Google Scholar]
- 3. Allison S. D., Martiny J. B. H. 2008. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105:11512–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson R. T., et al. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atlas R. M., Horowitz A., Krichevsky M., Bej A. K. 1991. Response of microbial populations to environmental disturbance. Microb. Ecol. 22:249–256 [DOI] [PubMed] [Google Scholar]
- 6. Borden R. C. 2007. Concurrent bioremediation of perchlorate and 1,1,1-trichloroethene in an emulsified oil barrier. J. Contamin. Hydrol. 94:13–33 [DOI] [PubMed] [Google Scholar]
- 7. Borden R. C., Rodriguez B. X. 2006. Evaluation of slow release substrates for anaerobic bioremediation. Bioremediat. J. 10:59–69 [Google Scholar]
- 8. Brodie E. L., et al. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cardenas E., et al. 2008. Microbial communities in contaminated sediments associated with bioremediation of uranium to submicromolar levels. Appl. Environ. Microbiol. 74:3718–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chin K. J., Sharma M. L., Russell L. A., O'Neill K. R., Lovley D. R. 2008. Quantifying expression of a dissimilatory bisulfite reductase gene in petroleum-contaminated marine harbor sediments. Microb. Ecol. 55:489–499 [DOI] [PubMed] [Google Scholar]
- 11. Clarke K. R., Ainsworth M. 1993. A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser. 92:205–219 [Google Scholar]
- 12. Coates J. D., Anderson R. T. 2000. Emerging techniques for anaerobic bioremediation of contaminated environments. Trends Biotechnol. 18:408–412 [DOI] [PubMed] [Google Scholar]
- 13. Cole J. R., et al. 2009. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeSantis T. Z., et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeSantis T. Z., et al. 2007. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb. Ecol. 53:371–383 [DOI] [PubMed] [Google Scholar]
- 16. Elshahed M. S., et al. 2005. Metagenomic analysis of the microbial community at Zodletone Spring (Oklahoma): insights into the genome of a member of the novel candidate division OD1. Appl. Environ. Microbiol. 71:7598–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faybishenko B., et al. 2008. In situ long-term reductive bioimmobilization of Cr(VI) in groundwater using hydrogen release compound. Environ. Sci. Technol. 42:8478–8485 [DOI] [PubMed] [Google Scholar]
- 18. Grabowski A., Blanchet D., Jeanthon C. 2005. Characterization of long-chain fatty-acid-degrading syntrophic associations from a biodegraded oil reservoir. Res. Microbiol. 156:814–821 [DOI] [PubMed] [Google Scholar]
- 19. Hazen T., Tabak H. 2005. Developments in bioremediation of soils and sediments polluted with metals and radionuclides: 2. field research on bioremediation of metals and radionuclides. Rev. Environ. Sci. Biotechnol. 4:157–183 [Google Scholar]
- 20. Henderson C. 1971. A study of the lipase produced by Anaerovibrio lipolytica, a rumen bacterium. J. Gen. Microbiol. 65:81–89 [DOI] [PubMed] [Google Scholar]
- 21. Hwang C., et al. 2009. Bacterial community succession during in situ uranium bioremediation: spatial similarities along controlled flow paths. ISME J. 3:47–64 [DOI] [PubMed] [Google Scholar]
- 22. Istok J. D., et al. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Technol. 38:468–475 [DOI] [PubMed] [Google Scholar]
- 23. Kostka J. E., Green S. J. 2011. Microorganisms and processes linked to uranium reduction and immobilization, p. 117–138 In Stolz J. F., Oremland R. S. (ed.), Microbial metal and metalloid metabolism: advances and applications. ASM Press, Washington, DC [Google Scholar]
- 24. Kuever J., Rainey F. A., Widdel F. 2005. Family I. Desulfovibrionaceae fam. nov., p. 926–938 In Brenner D. J., et al. (ed.), Bergey's manual of systematic bacteriology. Springer-Verlag, New York, NY [Google Scholar]
- 25. Kuever J., Rainey F. A., Widdel F. 2005. Family II. Desulfobulbaceae fam. nov., p. 988–992 In Brenner D. J., et al. (ed.), Bergey's manual of systematic bacteriology. Springer-Verlag, New York, NY [Google Scholar]
- 26. Liu Z., Lozupone C., Hamady M., Bushman F. D., Knight R. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Löffler F., Edwards E. 2006. Harnessing microbial activities for environmental cleanup. Curr. Opin. Biotechnol. 17:274–284 [DOI] [PubMed] [Google Scholar]
- 28. Lovley D. R. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55:259–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lovley D. R., Roden E. E., Phillips E. J. P., Woodward J. C. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar. Geol. 113:41–53 [Google Scholar]
- 30. Ludwig W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moon J.-W., et al. 2006. Physiochemical and mineralogical characterization of soil-saprolete cores from a field research site, Tennessee. J. Environ. Qual. 35:1731–1741 [DOI] [PubMed] [Google Scholar]
- 32. Nadkarni M. A., Martin F. E., Jacques N. A., Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range universal probe and primers set. Microbiology 148:257–266 [DOI] [PubMed] [Google Scholar]
- 33. Øvreås L., Torsvik V. 1998. Microbial diversity and community structure in two different agricultural soil communities. Microb. Ecol. 36:303–315 [DOI] [PubMed] [Google Scholar]
- 34. Porat I., et al. 2010. Characterization of archaeal community in contaminated and uncontaminated surface stream sediments. Microb. Ecol. 60:784–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rees G. N., Patel B. K. C. 2001. Desulforegula conservatrix gen. nov., sp. nov., a long-chain fatty acid-oxidizing, sulfate-reducing bacterium isolated from sediments of a freshwater lake. Int. J. Syst. Evol. Microbiol. 51:1911–1916 [DOI] [PubMed] [Google Scholar]
- 36. Ryan T. A., Joiner B. L. Normal probability plots and tests for normality. 1976 technical report, Department of Statistics. The Pennsylvania State University, University Park, PA [Google Scholar]
- 37. Schauder R., Schink B. 1989. Anaerovibrio glycerini sp. nov., an anaerobic bacterium fermenting glycerol to propionate, cell matter, and hydrogen. Arch. Microbiol. 152:473–478 [Google Scholar]
- 38. Schloss P. D., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Senko J. M., Istok J. D., Suflita J. M., Krumholz L. R. 2002. In-situ evidence for uranium immobilization and remobilization. Environ. Sci. Technol. 36:1491–1496 [DOI] [PubMed] [Google Scholar]
- 40. Shelobolina E. S., et al. 2007. Geobacter pickeringii sp. nov., Geobacter argillaceus sp. nov. and Pelosinus fermentans gen. nov., sp. nov., isolated from subsurface kaolin lenses. Int. J. Syst. Evol. Microbiol. 57:126–135 [DOI] [PubMed] [Google Scholar]
- 41. Sousa D. Z., Smidt H., Alves M. M., Stams A. J. M. 2007. Syntrophomonas zehnderi sp. nov., an anaerobe that degrades long-chain fatty acids in co-culture with Methanobacterium formicicum. Int. J. Syst. Evol. Microbiol. 57:609–615 [DOI] [PubMed] [Google Scholar]
- 42. Sousa D. Z., Alves J. I., Alves M. M., Smidt H., Stams A. J. M. 2009. Effect of sulfate on methanogenic communities that degrade unsaturated and saturated long-chain fatty acids (LCFA). Environ. Microbiol. 11:68–80 [DOI] [PubMed] [Google Scholar]
- 43. Ter Braak C. J. F. 1986. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:1167–1179 [Google Scholar]
- 44. U.S. Environmental Protection Agency 2009. National primary drinking water regulations. EPA 816-F-09-0004. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 45. Vishnivetskaya T. A., et al. 2011. Mercury and other heavy metals influence bacterial community structure in low-order Tennessee streams. Appl. Environ. Microbiol. 77:302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wall J. D., Krumholz L. R. 2006. Uranium reduction. Annu. Rev. Microbiol. 60:149–166 [DOI] [PubMed] [Google Scholar]
- 47. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu W.-M., et al. 2006. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer. 2: U(VI) reduction and geochemical control of U(VI) bioavailability. Environ. Sci. Technol. 40:3986–3995 [DOI] [PubMed] [Google Scholar]
- 49. Wu W.-M., et al. 2007. In situ bioreduction of uranium (VI) to submicromolar levels and reoxidation by dissolved oxygen. Environ. Sci. Technol. 41:5716–5723 [DOI] [PubMed] [Google Scholar]
- 50. Wu W.-M., et al. 2010. Effects of nitrate on the stability of uranium in the bioreduced region of the subsurface. Environ. Sci. Technol. 44:5104–5111 [DOI] [PubMed] [Google Scholar]
- 51. Zhang F., et al. 2010. Kinetic analysis and modeling of oleate and ethanol stimulated uranium (VI) bio-reduction in contaminated sediments under sulfate reduction conditions. J. Hazard. Mater. 183:482–499 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.