Abstract
Soil microbial community characterization is increasingly being used to determine the responses of soils to stress and disturbances and to assess ecosystem sustainability. However, there is little experimental evidence to indicate that predictable patterns in microbial community structure or composition occur during secondary succession or ecosystem restoration. This study utilized a chronosequence of developing jarrah (Eucalyptus marginata) forest ecosystems, rehabilitated after bauxite mining (up to 18 years old), to examine changes in soil bacterial and fungal community structures (by automated ribosomal intergenic spacer analysis [ARISA]) and changes in specific soil bacterial phyla by 16S rRNA gene microarray analysis. This study demonstrated that mining in these ecosystems significantly altered soil bacterial and fungal community structures. The hypothesis that the soil microbial community structures would become more similar to those of the surrounding nonmined forest with rehabilitation age was broadly supported by shifts in the bacterial but not the fungal community. Microarray analysis enabled the identification of clear successional trends in the bacterial community at the phylum level and supported the finding of an increase in similarity to nonmined forest soil with rehabilitation age. Changes in soil microbial community structure were significantly related to the size of the microbial biomass as well as numerous edaphic variables (including pH and C, N, and P nutrient concentrations). These findings suggest that soil bacterial community dynamics follow a pattern in developing ecosystems that may be predictable and can be conceptualized as providing an integrated assessment of numerous edaphic variables.
INTRODUCTION
Soil microbial community structure and composition measures are increasingly being used to assess ecosystem responses to anthropogenic disturbances and to provide an indicator of ecosystem recovery (30, 41, 60). However, in comparison to plant communities, there is limited experimental evidence that predictable patterns in the microbial community structure or composition occur during secondary succession (18, 37) or ecosystem restoration (27, 33). Microbial communities are able to respond more rapidly than plant communities to changes in environmental conditions and may provide an early indication of the recovery trajectory (29). However, the high level of sensitivity to numerous environmental factors can also result in long-term shifts (in the order of decades or more) in the microbial community structure in rehabilitated ecosystems (33). Following extreme disturbances, such as mining, even best-practice rehabilitation programs may be expected to leave a soil legacy in terms of some alteration to the soil organophysicochemical environment.
Edaphic factors that are purported to be significant drivers of soil microbial community structure include soil pH (20, 52, 61); the quantity, quality, and availability of soil carbon (C) (5, 11, 47) and nitrogen (N) (48, 53), soil water (17, 28), texture (12), and mineralogy (23). These factors may exert an influence on the microbial community structure simultaneously and produce interactive and feedback effects (1). Thus, microbial community structure measures could be conceptualized as an integrated assessment of numerous soil and ecosystem characteristics. However, a comprehensive characterization of soil microbial community dynamics during ecosystem restoration has been limited by the enormous microbial diversity within soils (59, 64).
In southwestern Australia, bauxite mining within the jarrah (Eucalyptus marginata) forest has created a mosaic landscape of rehabilitation forest in various states of succession alongside nonmined forest. This industrial-scale mining and rehabilitation program (covering an area of 13,000 ha to date) with a documented management history of over 40 years (36) enables the utilization of a chronosequence (space-for-time substitution) design. The existence of gradients in soil chemical (pH, total C [Ctot], and total N [Ntot]) and biological (microbial biomass and activity) characteristics in these rehabilitation sites was established previously (4). The two central hypotheses tested in this study were (i) that the mining disturbance would change the soil microbial community structure and (ii) that the community structure would recover over time and become more similar to nonmined forest soils with rehabilitation age. Considering the sensitivity of soil microbial community structures to edaphic variables, we also sought to elucidate which soil characteristics might be most important in driving microbial successional change.
MATERIALS AND METHODS
Study area.
The study sites were located in the northern jarrah (Eucalyptus marginata) forest region of Western Australia, approximately 110 km south-southeast (SSE) of Perth (32°38′S, 116°06′E). The jarrah forest is a dry sclerophyll type growing in a Mediterranean-type climate in highly weathered, lateritic, sandy soils with low concentrations of major nutrients such as N and P (42). Bauxite mining has been conducted in the area since 1963, and currently, Alcoa of Australia clears, mines, and rehabilitates around 550 ha of forest per year (36). Detailed descriptions of Alcoa's mining and rehabilitation practices were reported elsewhere previously (25, 36). Briefly, mining involves the complete removal of vegetation and surface soils (average removal of soil up to a 40-cm depth) in order to access the bauxite ore. Rehabilitation involves the re-landscaping of the mined site; return of the topsoil; surface contour ripping; seeding with native overstorey species (with the exception of pre-1988 sites, in which some nonnative tree species were used) and understorey species, including N2-fixing legumes; and fertilization with diammonium phosphate (DAP). The concentration of legumes in the understorey seed mix and fertilizer application rates have changed over time, and both have been decreased in more recent rehabilitations. In this study, the vegetated rehabilitation sites had been seeded with a legume density of between 0.6 and 1.0 kg ha−1 and fertilized with 500 kg ha−1 DAP. Ripping produces a distinct surface microtopography (termed mounds and furrows) known to impact litter accumulation (58) and topsoil characteristics (39).
Soil sampling and soil characteristics.
Thirty sites were selected, encompassing five replicates of four rehabilitation ages postmining (0, 6, 14, and 18 years old) and five replicates of two site vegetation types of jarrah forest prevalent in the region and commonly targeted for mining. The site vegetation forest types (S and TS), classified according to the Havel system (31), are all dominated by a jarrah and marri (Corymbia calophylla) overstorey but with differences in understorey species composition. These floristic differences are related to topography and soil characteristics, with T-type sites being commonly found on loamier soils in areas with higher levels of rainfall than S-type sites. The management history and vegetation characteristics of the sampled sites were described previously (4), and a map of the sampling locations is provided in Fig. S1 in the supplemental material. Composite soil samples were collected from three plots per site, positioned to encompass topographical variation. Within rehabilitation areas, soil from the highest and lowest points of the mounds and furrows, respectively, was collected separately, to a depth of 5 cm, as previous studies have shown that soil nutrients in jarrah forest are concentrated in the top 5 cm (26). Soil cores were collected at random points within each plot to a weight of 3 kg and combined (i.e., composite sample of 9 kg). Soil was sieved (<4 mm) and stored at 4°C before biochemical characterizations or frozen at −40°C for DNA extraction.
The soil texture, water-holding capacity, electrical conductivity (EC), pH (CaCl2), total C and N, bicarbonate-extractable (available) P, inorganic N, soluble organic C (Csol org), soluble organic N (Nsol org), and microbial biomass C (Cmic) concentrations were described previously (4). This study utilized a subset of the previously described experimental design, and the relevant soil characteristics are given in Table S1 in the supplemental material.
Automated ribosomal intergenic spacer analysis (ARISA).
Total soil DNA was extracted from 0.8 g of each composite 0- to 5-cm soil sample by using the UltraClean soil DNA isolation kit (Mo Bio Laboratories, Inc.). The manufacturer's instructions were modified to perform cell lysis by using a Mini Bead Beater (BioSpec Products, Inc.) at 2,500 rpm for 2 min. The bacterial intergenic spacer region between the 16S and 23S rRNA genes was amplified by using 6-carboxyfluorescein (FAM)-labeled forward primer S-D-Bact-1522-b-S-20 and reverse primer L-D-Bact-132-a-A-18 (45). The two fungal intergenic spacer regions spanning the 5.8S rRNA gene were amplified by using fungal-specific FAM-labeled forward primer ITS1-F (21) and universal reverse primer ITS4 (62). PCR cycling conditions were described previously by Gleeson et al. (22, 23). Triplicate PCR amplification mixtures were pooled and cleaned with a Wizard PCR Preps DNA purification system (Promega Corporation, Australia).
Intergenic fragment lengths were determined by using an ABI 3730 automated sequencer with 20-bp to 1,200-bp size standards, using GeneMapper v4.0 software (Applied Biosystems). Fragments smaller than 200 bp and larger than 1,200 bp were excluded from the profiles. Profiles of ribotype abundances (based on peak heights) were created by using the program RiboSort (55) within the statistical package R, version 2.6.0 (13). Fragment sizes that differed by less than 0.5 bp were considered to be identical ribotypes. Only fragments with fluorescence greater than 1% of the total fluorescence summed across all samples were included. The majority of bacterial ARISA fragments were 300 to 820 bp in length, which is typical of soil bacterial automated ribosomal intergenic spacer analysis (ARISA) profiles (51). The majority of fungal ARISA fragments were 500 to 800 bp in length.
PhyloChip microarray analyses.
A subset of the DNA extracts, including furrow soils of each rehabilitation age and S-type nonmined forest soil, were used for microarray analyses. These extracts were selected as representative of the larger sample set, based on the ARISA profiles. Three out of five field replicates were randomly selected for the PCR amplification of bacterial 16S rRNA genes using primers 27F and 1492R (63). Replicate PCR amplifications were performed for each sample (MasterCycler; Eppendorf) using eight different annealing temperatures between 48°C and 58°C to encompass a range of primer-template specificities, using cycling conditions described previously (16). Replicate PCR products were pooled, cleaned, and concentrated by ethanol precipitation.
For application onto the PhyloChip, 1,000 ng of bacterial 16S rRNA gene amplicons was fragmented, biotin labeled, and hybridized as described previously (6). The PhyloChip can resolve 8,434 bacterial taxa using an average of 24 perfect-match-mismatch probe pairs per taxon (7). For a taxon to be reported as being present in a sample, 90% of the probe pairs in its set must have been positive. The criteria used to score a probe pair as positive were described previously (16). Hybridization scores for each taxon, which are an averages of the differences between the perfect-match and mismatch fluorescent intensities of all probe pairs (excluding the highest and lowest), were normalized by using the fluorescence intensity of internal standards (6) and log transformed to represent the relative abundance of each taxon. The relative abundance data for each taxon within a phylum or class were summed to allow comparisons between samples at higher taxonomic ranks.
Statistical analyses.
Overall differences in soil characteristics were tested by one-way analysis of variance using GenStat v12 (VSN International Ltd., Hemel Hempstead, United Kingdom). All multivariate statistical routines were conducted by using PRIMER 6 and PERMANOVA+ (Primer-E Ltd., Plymouth, United Kingdom) (14). Multivariate analyses of ARISA and PhyloChip profiles were based on Bray-Curtis dissimilarities on log-transformed data, standardized by the sample sum. The Bray-Curtis measure was chosen because it is unchanged by the inclusion or exclusion of variables that are jointly absent between samples. Analyses of ARISA profile data transformed to presence or absence were also performed. To visualize differences between treatments, ordinations were performed by principal coordinate (PCO) analysis. Tests of the null hypothesis that there are no differences among a priori-defined groups were performed by permutational multivariate analysis of variance (PERMANOVA) (2). The significances of the treatments “age” and “position” and their interaction were tested within rehabilitation sites. Differences between nonmined sites and each rehabilitation age (with mound and furrow separate) were tested by pairwise comparisons.
Relationships between changes in the microbial community structure and individual soil characteristics were analyzed by using distance-based multivariate multiple regression (DistLM). Two-sided significance tests were used to determine whether a correlation was significantly different from zero. Soil characteristics were then subjected to a forward-selection procedure to develop a model to explain the variance in profile data, taking into account the covariance between soil characteristics. Pearson correlations of individual soil variables with PCO axes were also performed.
RESULTS
Bacterial and fungal community structure by ARISA.
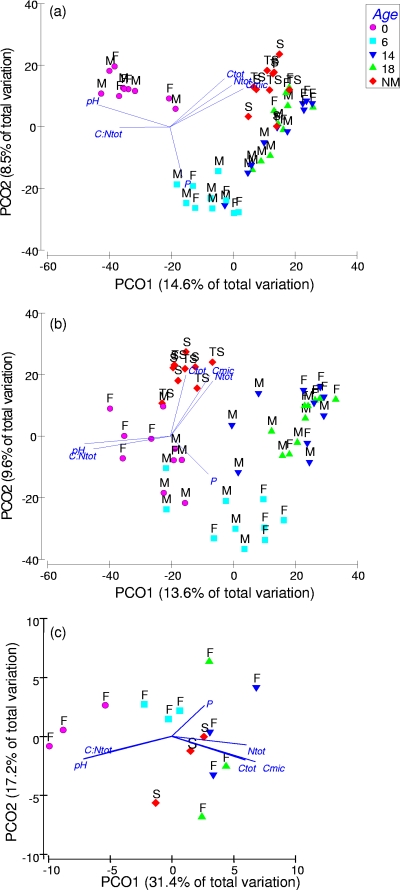
Rehabilitation age and microtopographical position were found to be significant factors affecting soil bacterial and fungal community structures (P < 0.05). All rehabilitation soils had significantly different bacterial and fungal community structures compared to those of both nonmined forest reference soils (P < 0.05), but the vegetation types of the two nonmined site were not different from each other (P > 0.1). The average Bray-Curtis similarity between rehabilitation and nonmined forest soils was 21% for 0-year-old rehabilitation soils, increasing to 30% for 14-year-old rehabilitation soils (and decreasing again to 27% for 18-year-old rehabilitation soils). All nonmined forest soils had an average Bray-Curtis similarity of 40% to each other. This suggests a trend of increasing similarity of bacterial community structures in 0- to 14-year-old rehabilitation soils compared to those of nonmined forest soils, which is also evident in the ordination (Fig. 1a). This trend was also evident with data transformed to a binary matrix, i.e., the use of composition information only. Rehabilitation age was also a significant factor affecting soil fungal community structure (Fig. 1b). However, based on the comparison of Bray-Curtis similarities, there was no clear trend of increasing similarity between rehabilitation and nonmined forest soils with age. The average Bray-Curtis similarity in fungal community structure between all rehabilitation ages and nonmined forest soils was 16%, with nonmined forest soils having an average similarity to each other of 30%.
Fig. 1.
Principal coordinate (PCO) analysis of bacterial community structure by ARISA (a) and fungal community structure by ARISA (b) of a jarrah forest rehabilitation chronosequence and nonmined reference soils, and bacterial community structure by microarray analysis of a sample subset (rehabilitation soils from furrows and nonmined S-type forest only) (c). Labels for a and b are as follows: M, mound soils; F, furrow soils; NM, nonmined soils of S and TS forest site vegetation types. Vectors show Pearson correlations with six selected soil characteristics. Abbreviations are as follows: Ctot, total C; Ntot, total N; Cmic, microbial biomass C; P, available (Colwell) P.
Bacterial community analysis by microarray.
A total of 2,673 bacterial taxa were detected, and the total richness within each phylum is given in Table S2 in the supplemental material. All nine phyla that typically dominate 16S rRNA gene libraries from soils (34) were represented. Other phylum-level lineages that have been found in soil clone libraries elsewhere were also detected by the microarray analysis, such as members of the Chlorobi, Cyanobacteria, BRC1, Nitrospirae, OP10, termite group I, TM6, and TM7 (Table S2). The trend in bacterial community structure detected by the microarray analysis mirrored that detected by ARISA profiling, suggesting an increase in similarity to nonmined forest soils with rehabilitation age (Fig. 1c). PERMANOVA tests, using all taxa as individual variables, suggested that 0- and 6-year-old rehabilitation soils were significantly different from the nonmined forest soil (P < 0.05) but that 14- and 18-year-old rehabilitation soils were not.
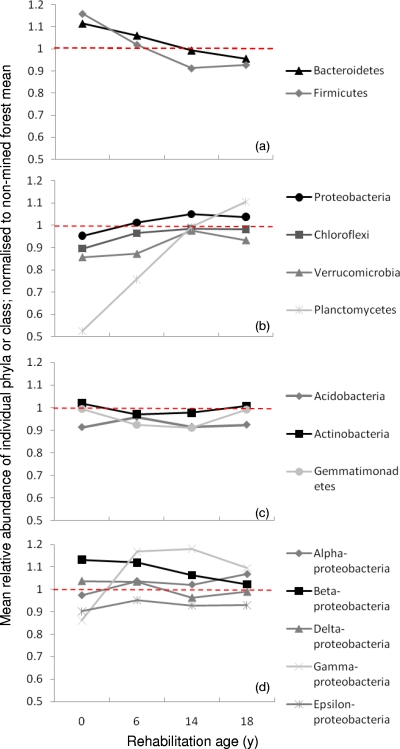
The changes in the relative abundance of the major soil bacterial phyla followed three distinct trends with respect to rehabilitation age: (i) decreasing (Bacteroidetes and Firmicutes), (ii) increasing (Chloroflexi, Planctomycetes, Proteobacteria, and Verrucomicrobia), or (iii) no change or no consistent trend (Acidobacteria, Actinobacteria, and Gemmatimonadetes); (Fig. 2 a to c). The first two trends both contributed to an increase in similarity to the nonmined forest soil with rehabilitation age. The phylum Proteobacteria is the most well-represented phylum in culture collections, in rRNA gene databases, and on the PhyloChip (with 1,172 probe sets). Analyzed at the class level, the largest change with rehabilitation age occurred for the Gammaproteobacteria, which increased during early rehabilitation, and the Betaproteobacteria, which decreased with rehabilitation age (Fig. 2d).
Fig. 2.
Changes in mean relative abundance of individual soil bacterial phyla exhibiting a decreasing trend (a), increasing trend (b), or no change or trend (c) with rehabilitation age and of classes of Proteobacteria (d) in a jarrah forest rehabilitation chronosequence, determined by microarray analysis of 16S rRNA genes using the PhyloChip. Data have been normalized to the mean relative abundance of each phylum or class found within nonmined reference soil, represented by the dotted line at y = 1.
Relationships between microbial community profiles and soil characteristics.
The soil chemical and biological characteristics exhibited a gradient with rehabilitation age, with distinct heterogeneity between microtopographical positions (see Table S1 in the supplemental material). These soil characteristics were described previously (4) but can be summarized as exhibiting three broad trends: (i) increasing with rehabilitation age and becoming more similar to those of nonmined forest soils (Ctot, Ntot, Csol org, Nsol org, Cmic, Cmic/Corg [microbial quotient], and water holding capacity [WHC]), (ii) decreasing with age and becoming less similar to those of nonmined forest soils (pH and C/Ntot), and (iii) no differences between nonmined forest and rehabilitation soils at any age (inorganic N and C/Nsol org). The exception was available P, the level of which was low in nonmined forest and 0-year-old rehabilitation soils (collected prefertilization) and highest in 6-year-old rehabilitation soils and then decreased with age.
Significant relationships were found between almost all soil characteristics tested individually and the bacterial and fungal community structure profiles. However, there were many significant correlations between the measured characteristics (see Table S3 in the supplemental material). Pearson correlations with the PCO axes (Fig. 1a to c) demonstrated that bacterial and fungal community structure changes were negatively correlated with the decline in soil pH and the C/Ntot ratio and positively correlated with the increase in soil organic matter (Ctot and Ntot) and microbial biomass (Cmic). Only available P displayed a negative correlation with PCO axis 2 and represents the separation in microbial community structures between fertilized rehabilitation soils and unfertilized soils (0-year-old rehabilitation and nonmined forest soils). Forward-selection models identified between six and nine soil variables, which explained up to 36% of the variance in the ARISA profiles and 65% of the variance in the microarray data (Table 1). Three soil variables, pH, microbial biomass, and total C, were significant explanatory variables in the forward-selection models of all three microbial community profiles.
Table 1.
Distance-based multivariate multiple regression (DistlmF) showing relationships between soil characteristics and bacterial and fungal community structures by ARISA and bacterial community structure by microarraya
| Test and variable | P | Prop. (%) |
|---|---|---|
| Bacterial ARISA | ||
| Cmic/Corg | <0.005 | 10.9 |
| Ctot | <0.005 | 15.3 |
| Ntot | <0.005 | 20.0 |
| P | <0.005 | 23.7 |
| Cmic | <0.005 | 27.1 |
| pH | <0.005 | 29.8 |
| Csol org | <0.1 | 31.9 |
| Ninorg | <0.1 | 33.9 |
| % clay + silt | <0.1 | 36.0 |
| Fungal ARISA | ||
| pH | <0.005 | 11.0 |
| Cmic | <0.005 | 15.6 |
| C/Ntot | <0.005 | 19.0 |
| Cmic/Corg | <0.005 | 21.9 |
| Ctot | <0.005 | 25.1 |
| Ntot | <0.005 | 27.9 |
| Bacterial microarray | ||
| pH | <0.005 | 25.3 |
| EC | <0.005 | 39.2 |
| P | <0.1 | 45.9 |
| Cmic | <0.1 | 52.5 |
| Csol org | <0.1 | 58.5 |
| Ctot | <0.1 | 65.2 |
Results from a forward-selection model with only variables that contributed significantly to the model are shown. The significance of the relationships (P) and the cumulative percentage of variance explained (Prop.) is shown. Abbreviations are as follows: EC, electrical conductivity; Ctot, total C; Ntot, total N; Csol org, soluble organic C, Cmic, microbial biomass C; Cmic/Corg, microbial quotient; Ninorg, inorganic N (ammonium plus nitrate); P, available (Colwell) P.
DISCUSSION
Bauxite mining in the jarrah forest of southwestern Australia is known to result in microbial biomass declines in topsoil (losses of more than 80% were estimated by comparisons with nonmined forest soils) and alterations of several soil physicochemical characteristics (4). The hypothesis in this study that the mining-induced disturbance, which involves a number of soil perturbations, such as increased temperature, desiccation, physical disruption, and a loss of organic matter, would also alter soil microbial community structure was supported. The successional change in the microbial community structure is likely to be driven by the availability of limiting resources and the ability of populations to utilize these resources (under altered physicochemical conditions), as hypothesized for plant community succession (57). There are many factors influencing resource availability to the soil microbial populations in this rehabilitation chronosequence. Nonetheless, we hypothesized that as the rehabilitation area matures, the soil microbial community structure would become more similar to that of the surrounding nonmined forest soils. This hypothesis was supported by a trend of increasing similarity in bacterial community structure to that of nonmined forest at between 0 and 14 years of rehabilitation, with a Bray-Curtis similarity of up to 26%. Comparisons within nonmined forest soils only suggested that the maximum Bray-Curtis similarity achievable in the rehabilitation soil was around 37%.
The similarity in bacterial community structure between 14-year-old rehabilitation sites and nonmined forests is higher than the 14% Bray-Curtis similarity reported previously for similar site comparisons of vegetation structures (44). Similarities in vegetation structure within nonmined forest sites averaged 34%, but no age-related trend in the rehabilitation vegetation structure toward that of the nonmined forest was found. This finding supports the suggestion that soil microbial community structure comparisons may provide an earlier indicator of the recovery trajectory than vegetation structure comparisons (29). However, the time frame for the detection of recovery trends in the soil bacterial community following extreme disturbance is still beyond a decade. This time frame is similar to that required for microbial biomass recovery in rehabilitation forest soils and roughly comparable to indications elsewhere of 20 to 30 or more years for bacterial community recovery after a disturbance (27, 33, 43).
Significant relationships were found between most of the soil characteristics measured and the microbial community profiles. Soil characteristics varied in response to rehabilitation age (e.g., microbial and organic C and the water-holding capacity), changes in vegetation structure (e.g., declines in the soil C/N ratio and pH as a consequence of the high legume density in rehabilitation sites), and fertilization practices (e.g., high available P in 6- and 14-year-old rehabilitation soils, which also favors N2 fixation by legumes and contributes to the observed declines in the C/N ratio and pH). The covariation of many soil variables in developing ecosystems makes it difficult to assess the significance of individual soil variables despite the use of forward-selection models. Nonetheless, the number and extent of correlations between the microbial community structure and the soil variables support the conceptualization of microbial community structure as an integrated assessment of the edaphic environment.
Unlike the bacterial community, the fungal community structure did not exhibit a trend of increasing similarity to that of nonmined forest soils, with overall lower levels of similarity. The fungal primers used in this study were reported previously to amplify predominantly basidiomycetes and ascomycetes (21, 35) and therefore are likely to include ectomycorrhizal fungi, which are known to be associated with many jarrah forest plant species (8), and free-living saprophytes but not the arbuscular mycorrhizal fungi. Previously, it was reported that the richness of ectomycorrhizal fungal species recovers during jarrah forest rehabilitation, but the species composition remains different (24). Thus, analyses of fungal community structures in soil (this study) and in root tips and sporocarps (24) have both indicated that differences between rehabilitation and nonmined forests are likely to persist for more than 16 to 18 years. This is the first study to show a relationship between the soil fungal (and, to a lesser extent, bacterial) community structure and differences in available P as a consequence of the fertilization of these ecosystems. Elsewhere, increases in soil P availability have also been associated with declines of root-associated fungal diversity (9) and changes in the whole soil fungal community structure (40). Longer-term shifts in the soil fungal community structure, compared to the bacterial community structure, may also be more directly linked to the persistent differences in vegetation structures between rehabilitation and nonmined forests (44), due to fungal mycorrhizal associations. There is also some evidence that fungi, in particular mycorrhizal fungi, may have less efficient dispersal and colonizing abilities than bacteria, which may also contribute to a slower recovery of fungal communities following disturbances that involve vegetation removal (32).
To determine whether there are predictable successional patterns in soil microbial communities of relevance to a broader range of postdisturbance ecosystems, it is necessary to identify changes in groups of phylogenetically or functionally related populations. It has recently been considered that bacteria at high taxonomic ranks, such as the phylum or class level, may display ecological coherence (19, 49, 50). Ecological coherence implies that despite the physiological diversity between bacteria within a phylum, there may be some general life strategies that have evolved in one phylum that distinguish it from other phyla. In order to identify whether high-taxonomic-level successional patterns could be identified in this study, we utilized a high-density microarray approach, which allows the identification of almost 104 taxa and can detect variations in abundance over 5 orders of magnitude (7). The microarray approach is potentially subject to PCR bias, as is the case for all endpoint PCR approaches. However, potential biases were minimized in this study by combining replicate PCRs, using the minimum number of amplification cycles possible (56), using fast temperature ramping during cycling (38), and using log-transformed abundance data (10). While the microarray approach is limited to the identification of known taxa for which probes have been designed, previous comparisons with clone library compositions have confirmed the comprehensive coverage provided by the PhyloChip (6, 15).
Previously, we have shown that the microbial quotient (Cmic/Corg) is low in early rehabilitation (4). This may favor r-strategist populations, indicative of an uncrowded environment with higher resource availability (3). Conditions would shift in favor of K-strategists as the rehabilitation site matures and the microbial quotient recovers. The existence of such a trend was supported by the changes in the relative abundance of members of the Bacteroidetes and the Betaproteobacteria, many of which (although not all) exhibit r-strategist attributes (19). Conversely, the lack of changes in the relative abundance of members of the Acidobacteria or Actinobacteria, many of which exhibit K-strategist attributes, did not fit this trend. However, knowledge of the ecological niches of many bacterial phyla remains limited, and classification as r- or K-strategists may not always be relevant (19). Physiological attributes, other than growth metabolic strategies, may also significantly influence a microorganism's competitive ability. For example, the higher relative abundance of members of the Firmicutes in early rehabilitation is likely related to their ability to form spores (e.g., the Bacilli and Clostridia). Other phyla, such as the Verrucomicrobia and Planctomycetes, have few cultivated members, and little is known of their growth or other ecological attributes (46, 54). Further exploration of the response of phyla, or other high taxonomic groupings, to stress or disturbance is needed to understand their ecological roles. Nevertheless, the microarray analysis in this study revealed different successional patterns for individual bacterial phyla in rehabilitation forest soils following bauxite mining and supported the trend found by community structure profiling of an increased similarity to nonmined forest soils with rehabilitation age.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Australian Research Council under the Linkage Program scheme with the industry partner Alcoa of Australia and a UWA Faculty of Natural and Agricultural Sciences start-up grant to support the collaboration with the Lawrence Berkeley National Laboratory. Support for D.B.G. was provided by an Australian Research Council discovery grant (DP0985832). Part of this work was supported by the U.S. Department of Energy under contract no. DE-AC02-05CH11231 and by Laboratory Directed Research and Development awards to E.L.B.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Allison V. J., Yermakov Z., Miller R. M., Jastrow J. D., Matamala R. 2007. Using landscape and depth gradients to decouple the impact of correlated environmental variables on soil microbial community composition. Soil Biol. Biochem. 39:505–516 [Google Scholar]
- 2. Anderson M. J. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26:32–46 [Google Scholar]
- 3. Andrews J. H.(ed.). 1991. Comparative ecology of microorganisms and macroorganisms. Springer-Verlag, New York, NY [Google Scholar]
- 4. Banning N. C., Grant C. D., Jones D. L., Murphy D. V. 2008. Recovery of soil organic matter, organic matter turnover and nitrogen cycling in a post-mining forest rehabilitation chronosequence. Soil Biol. Biochem. 40:2021–2031 [Google Scholar]
- 5. Bardgett R. D., et al. 1999. Below-ground microbial community development in a high temperature world. Oikos 85:193–203 [Google Scholar]
- 6. Brodie E. L., et al. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brodie E. L., et al. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brundrett M. C., Abbott L. K. 1991. Roots of jarrah forest plants. 1. Mycorrhizal associations of shrubs and herbaceous plants. Aust. J. Bot. 39:445–457 [Google Scholar]
- 9. Burke D. J., Lopez-Gutierrez J. C., Smemo K. A., Chan C. R. 2009. Vegetation and soil environment influence the spatial distribution of root-associated fungi in a mature beech-maple forest. Appl. Environ. Microbiol. 75:7639–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao Y., Williams D. D., Williams N. E. 1999. Data transformation and standardization in the multivariate analysis of river water quality. Ecol. Appl. 9:669–677 [Google Scholar]
- 11. Carney K., Matson P. 2005. Plant communities, soil microorganisms, and soil carbon cycling: does altering the world belowground matter to ecosystem functioning? Ecosystems 8:928–940 [Google Scholar]
- 12. Carson J. K., Rooney D., Gleeson D. B., Clipson N. 2007. Altering the mineral composition of soil causes a shift in microbial community structure. FEMS Microbiol. Ecol. 61:414–423 [DOI] [PubMed] [Google Scholar]
- 13. Chambers J. M. 2008. Software for data analysis: programming with R. Springer, New York, NY [Google Scholar]
- 14. Clarke K. R., Gorley R. N. 2006. PRIMER v6: user manual/tutorial. Primer-E Ltd., Plymouth, United Kingdom [Google Scholar]
- 15. Cruz-Martinez K., et al. 2009. Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. ISME J. 3:738–744 [DOI] [PubMed] [Google Scholar]
- 16. DeAngelis K. M., et al. 2009. Selective progressive response of soil microbial community to wild oat roots. ISME J. 3:168–178 [DOI] [PubMed] [Google Scholar]
- 17. Drenovsky R. E., Vo D., Graham K. J., Scow K. M. 2004. Soil water content and organic carbon availability are major determinants of soil microbial community composition. Microb. Ecol. 48:424–430 [DOI] [PubMed] [Google Scholar]
- 18. Felske A., Wolterink A., Van Lis R., De Vos W. M., Akkermans A. D. L. 2000. Response of a soil bacterial community to grassland succession as monitored by 16S rRNA levels of the predominant ribotypes. Appl. Environ. Microbiol. 66:3998–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fierer N., Bradford M. A., Jackson R. B. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364 [DOI] [PubMed] [Google Scholar]
- 20. Fierer N., Jackson R. B. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gardes M., Bruns T. D. 1993. ITS primers with enhanced specificity for Basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113–118 [DOI] [PubMed] [Google Scholar]
- 22. Gleeson D., Clipson N., Melville K., Gadd G., McDermott F. 2005. Characterization of fungal community structure on a weathered pegmatitic granite. Microb. Ecol. 50:360–368 [DOI] [PubMed] [Google Scholar]
- 23. Gleeson D., McDermott F., Clipson N. 2006. Structural diversity of bacterial communities in a heavy metal mineralized granite outcrop. Environ. Microbiol. 8:383–393 [DOI] [PubMed] [Google Scholar]
- 24. Glen M., et al. 2008. Ectomycorrhizal fungal communities of rehabilitated bauxite mines and adjacent, natural jarrah forest in Western Australia. Forest Ecol. Manage. 255:214–225 [Google Scholar]
- 25. Grant C. D. 2006. State-and-transition successional model for bauxite mining rehabilitation in the jarrah forest of Western Australia. Restor. Ecol. 14:28–37 [Google Scholar]
- 26. Grierson P. F., Adams M. A. 2000. Plant species affect acid phosphatase, ergosterol and microbial P in a jarrah (Eucalyptus marginata Donn ex Sm.) forest in south-western Australia. Soil Biol. Biochem. 32:1817–1827 [Google Scholar]
- 27. Gros R., Monrozier L. J., Faivre P. 2006. Does disturbance and restoration of alpine grassland soils affect the genetic structure and diversity of bacterial and N2-fixing populations? Environ. Microbiol. 8:1889–1901 [DOI] [PubMed] [Google Scholar]
- 28. Hackl E., Zechmeister-Boltenstern S., Bodrossy L., Sessitsch A. 2004. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 70:5057–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harris J. 2009. Soil microbial communities and restoration ecology: facilitators or followers? Science 325:573–574 [DOI] [PubMed] [Google Scholar]
- 30. Harris J. A. 2003. Measurements of the soil microbial community for estimating the success of restoration. Eur. J. Soil Sci. 54:801–808 [Google Scholar]
- 31. Havel J. 1975. Site-vegetation mapping in the northern jarrah forest (Darling Range). 1. Definition of site-vegetation types. Bulletin 86. Forest Department Western Australia, Perth, Australia [Google Scholar]
- 32. Hedlund K., et al. 2004. Trophic interactions in changing landscapes: responses of soil food webs. Basic Appl. Ecol. 5:495–503 [Google Scholar]
- 33. Jangid K., et al. 2010. Development of soil microbial communities during tallgrass prairie restoration. Soil Biol. Biochem. 42:302–312 [Google Scholar]
- 34. Janssen P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kasel S., Bennett L. T., Tibbits J. 2008. Land use influences soil fungal community composition across central Victoria, south-eastern Australia. Soil Biol. Biochem. 40:1724–1732 [Google Scholar]
- 36. Koch J. M. 2007. Alcoa's mining and restoration process in South Western Australia. Restor. Ecol. 15:S11–S16 [Google Scholar]
- 37. Kuramae E. E., et al. 2010. Microbial secondary succession in a chronosequence of chalk grasslands. ISME J. 4:711–715 [DOI] [PubMed] [Google Scholar]
- 38. Kurata S., et al. 2004. Reevaluation and reduction of a PCR bias caused by reannealing of templates. Appl. Environ. Microbiol. 70:7545–7549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lalor B. M., Cookson W. R., Murphy D. V. 2007. Comparison of two methods that assess soil community level physiological profiles in a forest ecosystem. Soil Biol. Biochem. 39:454–462 [Google Scholar]
- 40. Lauber C. L., Strickland M. S., Bradford M. A., Fierer N. 2008. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40:2407–2415 [Google Scholar]
- 41. Lewis D. E., et al. 2010. Soil functional diversity analysis of a bauxite-mined restoration chronosequence. Microb. Ecol. 59:710–723 [DOI] [PubMed] [Google Scholar]
- 42. McArthur W. M. 2004. Reference soils of south-western Australia. Department of Agriculture, Perth, Western Australia, Australia [Google Scholar]
- 43. McKinley V. L., Peacock A. D., White D. C. 2005. Microbial community PLFA and PHB responses to ecosystem restoration in tallgrass prairie soils. Soil Biol. Biochem. 37:1946–1958 [Google Scholar]
- 44. Norman M. A., Koch J. M., Grant C. D., Morald T. K., Ward S. C. 2006. Vegetation succession after bauxite mining in Western Australia. Restor. Ecol. 14:278–288 [Google Scholar]
- 45. Normand P., Ponsonnet C., Nesme X., Neyra M., Simonet P. 1996. ITS analysis of prokaryotes, p. 1–12In Akkermans A. D., van Elsas J. D., deBruijn E. I.(ed.), Molecular microbial ecology manual. Kluwer Academic Press, Amsterdam, Netherlands [Google Scholar]
- 46. Nunes da Rocha U., Van Overbeek L., Van Elsas J. D. 2009. Exploration of hitherto-uncultured bacteria from the rhizosphere. FEMS Microbiol. Ecol. 69:313–328 [DOI] [PubMed] [Google Scholar]
- 47. Pennanen T., et al. 2004. Community-level responses of metabolically-active soil microorganisms to the quantity and quality of substrate inputs. Soil Biol. Biochem. 36:841–848 [Google Scholar]
- 48. Pennanen T., et al. 1999. Structure of the microbial communities in coniferous forest soils in relation to site fertility and stand development stage. Microb. Ecol. 38:168–179 [DOI] [PubMed] [Google Scholar]
- 49. Philippot L., et al. 2010. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 8:523–529 [DOI] [PubMed] [Google Scholar]
- 50. Philippot L., et al. 2009. Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ. Microbiol. 11:3096–3104 [DOI] [PubMed] [Google Scholar]
- 51. Ranjard L., et al. 2001. Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl. Environ. Microbiol. 67:4479–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rousk J., et al. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4:1340–1351 [DOI] [PubMed] [Google Scholar]
- 53. Ruppel S., Torsvik V., Daae F. L., Ovreas L., Ruhlmann J. 2007. Nitrogen availability decreases prokaryotic diversity in sandy soils. Biol. Fert. Soils 43:449–459 [Google Scholar]
- 54. Sangwan P., Kovac S., Davis K. E. R., Sait M., Janssen P. H. 2005. Detection and cultivation of soil verrucomicrobia. Appl. Environ. Microbiol. 71:8402–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Scallan U., Liliensiek A., Clipson N., Connolly J. 2008. Ribosort: a program for automated data preparation and exploratory analysis of microbial community fingerprints. Mol. Ecol. Res. 8:95–98 [DOI] [PubMed] [Google Scholar]
- 56. Suzuki M., Giovannoni S. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tilman D. 1985. The resource-ratio hypothesis of plant succession. Am. Nat. 125:827–852 [Google Scholar]
- 58. Todd M. C. L., Grierson P. F., Adams M. A. 2000. Litter cover as an index of nitrogen availability in rehabilitated mine sites. Aust. J. Soil Res. 38:423–434 [Google Scholar]
- 59. Torsvik V., Ovreas L. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240–245 [DOI] [PubMed] [Google Scholar]
- 60. van Dijk J., et al. 2009. Can differences in soil community composition after peat meadow restoration lead to different decomposition and mineralization rates? Soil Biol. Biochem. 41:1717–1725 [Google Scholar]
- 61. Wakelin S. A., et al. 2008. Habitat selective factors influencing the structural composition and functional capacity of microbial communities in agricultural soils. Soil Biol. Biochem. 40:803–813 [Google Scholar]
- 62. White T. J., Bruns T., Lee S., Taylor J. 1990. Amplification and direct sequencing of fungal rRNA genes for phylogenetics, p. 315–322In Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, NY [Google Scholar]
- 63. Wilson K. H., Blitchington R. B., Greene R. C. 1990. Amplification of bacterial 16S ribosomal DNA with PCR. J. Clin. Microbiol. 28:1942–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Youssef N. H., Elshahed M. S. 2008. Diversity rankings among bacterial lineages in soil. ISME J. 3:305–313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.