Abstract
Protozoa constitute the earliest branch of the eukaryotic lineage, and several groups of protozoans are serious parasites of humans and other animals. Better understanding of biochemical pathways that are either in common with or divergent from those of higher eukaryotes is integral in the defense against these parasites. In yeast and humans, the posttranslational methylation of arginine residues in proteins affects myriad cellular processes, including transcription, RNA processing, DNA replication and repair, and signal transduction. The protein arginine methyltransferases (PRMTs) that catalyze these reactions, which are unique to the eukaryotic kingdom of organisms, first become evident in protozoa. In this review, we focus on the current understanding of arginine methylation in multiple species of parasitic protozoa, including Trichomonas, Entamoeba, Toxoplasma, Plasmodium, and Trypanosoma spp., and discuss how arginine methylation may play important and unique roles in each type of parasite. We mine available genomic and transcriptomic data to inventory the families of PRMTs in different parasites and the changes in their abundance during the life cycle. We further review the limited functional studies on the roles of arginine methylation in parasites, including epigenetic regulation in Apicomplexa and RNA processing in trypanosomes. Interestingly, each of the parasites considered herein has significantly differing sets of PRMTs, and we speculate on the importance of this diversity in aspects of parasite biology, such as differentiation and antigenic variation.
INTRODUCTION
The posttranslational methylation of proteins was first demonstrated by Paik and Kim in the late 1960s (103–106). The field of protein methylation was underappreciated, however, until the mid-1990s, when more extensive investigations revealed the enzymes responsible for protein methylation (48, 84). The transfer of methyl groups from the methyl donor S-adenosylmethionine (AdoMet) to proteins has been documented on carboxyl groups (cysteine, leucine, glutamate, aspartate) and nitrogen groups (glutamine, histidine, lysine, arginine, and N-terminal alpha-amino). A major research focus to date has been the roles of histone lysine and arginine methylation, which function in regulating transcription activation or repression (73, 76, 127). However, arginine methylation is a widespread posttranslational modification, occurring on multiple classes of proteins in both the cytoplasm and nucleus (13), and many recent studies have addressed the functions of this modification on nonhistone proteins (reviewed in references 7 and 80). Protein arginine methylation appears to be an evolutionarily ancient modification. Although absent in prokaryotes, methylarginine proteins and enzymes responsible for their synthesis have been reported in earlier diverged organisms (reviewed in reference 4). Here, we provide an introduction to protein arginine methylation and discuss the current state of knowledge regarding the synthesis and functions of this modification in parasitic protozoa.
PROTEIN ARGININE METHYLATION: ENZYMES AND FUNCTIONS
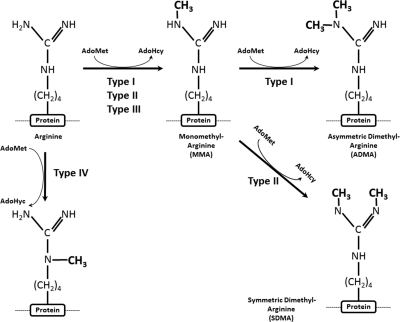
The transfer of methyl groups from AdoMet to arginine residues in proteins is carried out by a family of seven beta fold (Rossmann fold) enzymes known as protein arginine methyltransferases (PRMTs) (4, 7, 49, 70). PRMTs are further classified into four types, designated I to IV, according to the terminal arginine modification produced by each type of enzyme. The first catalytic step of arginine methylation involves the transfer of a single methyl group from AdoMet to the terminal nitrogen group of arginine, yielding omega-NG-monomethylarginine (MMA). The type I, type II, and type III PRMTs all catalyze this first methyl transfer reaction (Fig. 1), and type III PRMTs can catalyze only the production of MMA. Type I PRMTs subsequently transfer a second methyl group to the same terminal nitrogen, yielding omega NG-NG-asymmetric dimethylarginine (ADMA). Alternately, type II PRMTs transfer a second methyl group to the adjacent terminal nitrogen, yielding omega NG-NG-symmetric dimethylarginine (SDMA). Lastly, type IV PRMTs catalyze the transfer of only a single methyl group to the internal (guanidino) nitrogen of arginine (Fig. 1). To date, type IV PRMTs have been demonstrated only in the yeasts Saccharomyces cerevisiae and Candida albicans but may also be present in plants as well based on sequence similarities (90, 99) (Table 1). The vast majority of PRMT research has focused on three archetype organisms, the yeasts S. cerevisiae and Schizosaccharomyces pombe and humans (Table 1). Unicellular yeasts exhibit a trend apparent in most eukaryotes, which is the presence of a type I PRMT (Hmt1 in S. cerevisiae; Rmt1 in S. pombe) and a type II PRMT (Hsl7 in S. cerevisiae; Skb1 in S. pombe) (Table 1). Another type I PRMT, Rmt3, a homologue of the human PRMT3, has also been demonstrated in S. pombe (Table 1). In addition, S. pombe may also possess a type IV PRMT, but this has not been demonstrated. In humans, there are currently nine confirmed PRMTs, the majority of which are type I enzymes (PRMT1, -2, -3, -4 [also known as CARM1], -6, and -8). Humans also possess a single type II enzyme (PRMT5) and a third enzyme whose designation as a type II or III PRMT is still in question (PRMT7). Finally, the enzymatic activity of PRMT9 has not been explored, but it may be a type II or III PRMT based on sequence homology (7) (Table 1). The existence of multiple type I PRMTs in humans can be partially explained by enzyme redundancy (40, 68, 138). In addition, the substrate specificity of some human PRMTs is confined through subcellular localization or cell type (7, 49). For example, HsPRMT1 and HsPRMT8 are 72% identical; however, the N terminus of PRMT8 alters its enzymatic activity and confers localization to the cell membrane through posttranslational myristoylation (78, 122). Regardless, the human PRMT1 is the workhorse of the type I group, accounting for 85% of the methylation reactions in mammalian cells (137). Protein arginine methylation was long considered to be an irreversible modification, and this subject is still not resolved. An initial report of direct histone arginine demethylation (23) has not been reproduced (50, 145). It is clear, however, that the arginine methylation status of a protein can be modulated indirectly by protein arginine deiminases. This class of enzymes modulates protein function by catalyzing the deimination of arginine and monomethylarginine residues to citrulline, which are then unable to serve as substrates for PRMTs (29, 144, 148). Conversely, a dimethylated arginine cannot be citrullinated, so the two processes of arginine methylation and citrullination negatively regulate each other (115).
Fig. 1.
Protein arginine methylation reactions. Using the methyl donor S-adenosyl methionine (AdoMet), protein arginine methyltransferases (PRMTs) catalyze the transfer to the terminal (ω) nitrogen, yielding monomethylarginine (MMA) and the by-product S-adenosyl homocysteine (AdoHcy). This initial reaction is carried out by PRMT types I, II, and III. Subsequently, type I PRMTs synthesize ADMA by catalyzing a second methyl group addition on the same terminal nitrogen, while type II PRMTs transfer a second methyl group to the adjacent terminal nitrogen, yielding SDMA. Type IV PRMTs catalyze the transfer of a single methyl group to the internal (δ) nitrogen, in a reaction that is currently thought to be confined to yeast.
Table 1.
Established PRMTs in yeasts and humans
| Organism | PRMT name | Gene identifier | PRMT type | Size (no. of amino acids/mol mass [kDa]) | Note | Reference(s) |
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | Hmt1/Rmt1 | NP_009590 | I | 348/40 | 42 | |
| Hsl7 | NP_009691 | II | 827/95 | 79, 121 | ||
| Rmt2 | NP_010753 | IV | 412/47 | 99 | ||
| Schizosaccharomyces pombe | Rmt1 | NP_594825 | I | 340/39 | 146 | |
| Rmt2 | NP_594160 | IVa | 357/41 | 146 | ||
| Rmt3 | NP_595572 | I | 543/62 | 5 | ||
| Skb1 | NP_595936 | II | 645/73 | 113 | ||
| Homo sapiens | HsPRMT1 | NP_938074 | I | 353/41 | 84 | |
| HsPRMT2 | NP_001526 | I | 433/49 | 74 | ||
| HsPRMT3 | NP_005779 | I | 531/60 | Zn finger required for substrate recognition | 138 | |
| CARM1 (HsPRMT4) | NP_954592 | I | 608/66 | 24 | ||
| HsPRMT5 | NP_006100 | II | 637/73 | 18 | ||
| HsPRMT6 | NP_060607 | I | 375/42 | 40 | ||
| HsPRMT7 | NP_061896 | II/IIIa | 692/78 | Contains duplication of active domains | 77, 93 | |
| HsPRMT8 | NP_062828 | I | 394/45 | 78 | ||
| HsPRMT9 (4q31) | NP_612373 | a | 845/95 | Contains duplication of active domains | 6 |
Type of PRMT activity undetermined.
Protein arginine methylation modulates multiple cellular pathways in yeasts and humans. As stated above, much of the research in this area has focused on transcriptional regulation and the effects of arginine methylation on the exposed tails of histones H3 and H4. Methylation of arginine residues in histones has been linked to transcriptional activation or to indirect repression of transcription by subsequent activation of lysine methylation at adjacent sites (53, 54). For thorough reviews of histone methylation, refer to several existing reviews on this subject (76, 132, 148). In addition, several nonhistone substrates of arginine methylation have been reported to modulate transcription, either as transcriptional coactivators with histones (1) or as DNA binding transcription factors (72, 95, 96, 151). The largest family of nonhistone proteins that are arginine methylated is RNA binding proteins (RBPs) (17, 85, 102, 110, 129). One reason for the abundance of arginine-methylated RBPs is the common occurrence in this class of proteins of glycine-arginine-rich (GAR) domains, which commonly serve as PRMT substrates (64, 85, 97). Arginine methylation of RBPs affects multiple pathways, including pre-mRNA splicing (12, 16, 25, 100), RNA stability (81), translation (5, 22, 116, 125), and small RNA pathways (65, 141). Other cellular pathways that are affected at several points by arginine methylation include DNA damage and repair (14, 15, 33, 47, 60), cell signaling (10, 11, 41, 57, 58, 71), and organelle biogenesis (107, 147, 152).
Protein arginine methylation generally alters protein function by influencing protein-protein interactions, protein-nucleic acid interactions, and protein trafficking. The addition of a methyl group to arginine, while not changing its charge, does alter the side chain shape, thereby increasing hydrophobicity and steric hindrance while also removing a potential hydrogen donor group (7). Therefore, arginine methyl proteins are often altered in their abilities to bind other protein partners or nucleic acids. For example, arginine methylation directly disrupts the binding of Sam68 to SH3 (8). Conversely, arginine methylation of several proteins is required for their binding to Tudor domain proteins (16, 27, 142). Direct effects on nucleic acid binding are less common. However, the arginine methylation of several RBPs, including the HIV Rev and nucleocapsid proteins, results in diminished RNA binding (55, 56, 117). Arginine methylation also greatly impacts the subcellular localization of proteins and RNA (3, 46, 52, 83, 124, 130). The first reported instance of this phenomenon was in yeast lacking Hmt1p, where the nuclear export of two RNA binding proteins, Npl3 and Hrp1, is blocked (124). Arginine methylation can also facilitate nuclear import of proteins such as hnRNPA2 and RNA helicase A (98, 130). Lastly, arginine methylation may impact subsequent posttranslational modification of a protein. For example, methylation of histone H3R2 antagonizes H3K4 methylation, and H4R3 methylation facilitates H4K9/K14 acetylation (53, 82). In summary, the effects of arginine methylation on target proteins can be profound, and many of these are still being elucidated.
CLASSES OF PRMTs IN PARASITIC PROTOZOA
In this review, we focus on several groups of parasitic protozoa of human health importance, including the amitochondrial metamonads (Giardia, Trichomonas) and archamoebae (Entamoeba), the Apicomplexa (Plasmodium, Toxoplasma), and the kinetoplastids (Trypanosoma, Leishmania). Overall, the current understanding of arginine methylation in these organisms is limited. Using several published reports and genome mining of parasite databases, we compiled a list of putative PRMT enzymes in each group (Table 2). Those parasites that contain PRMTs resemble yeasts in that they harbor at least one type I PRMT with homology to human (Hs) PRMT1 and at least one type II PRMT with homology to HsPRMT5 (Table 2). Outside this generalization, however, there are vast differences in the number and potential types of PRMTs in each parasitic group.
Table 2.
PRMTs in parasitic protozoaa
| Organism | Gene identifier | Published name | Inferred PRMT type | Activity type | Closest human homologue (% identity/similarity) | Size (no. of amino acids/mol mass [kDa]) | Note | Reference |
|---|---|---|---|---|---|---|---|---|
| Giardia lamblia | NA | NA | NA | NA | NA | NA | No identifiable PRMT-encoding genes | |
| Trichomonas vaginalis | TVAG_048280 | NA | I | ND | PRMT1 (22/28) | 331/37 | ||
| TVAG_225950 | NA | I | ND | PRMT1 (22/28) | 331/37 | Duplication of | ||
| TVAG_028100 | NA | I | ND | PRMT1 (37/40) | 320/37 | TVAG_048280 | ||
| TVAG_199700 | NA | I | ND | PRMT1 (44/47) | 327/37 | |||
| TVAG_433490 | NA | I | ND | PRMT1 (45/48) | 327/37 | |||
| TVAG_045760 | NA | I | ND | PRMT1 (36/42) | 320/37 | |||
| TVAG_254540 | NA | II | ND | PRMT5 (23/32) | 436/50 | |||
| TVAG_096150 | NA | II | ND | PRMT5 (27/39) | 528/60 | |||
| Entamoeba histolytica | EHI_202470 | NA | I | ND | PRMT1 (41/48) | 332/38 | ||
| EHI_152460 | NA | I | ND | PRMT1 (37/46) | 328/38 | |||
| EHI_105780 | EhPRMT1 | I | ND | PRMT1 (37/42) | 319/37 | |||
| EHI_159180 | NA | I | ND | PRMT1 (25/32) | 367/43 | |||
| EHI_158560 | NA | II | ND | PRMT5 (32/44) | 586/69 | |||
| Trypanosoma brucei | Tb927.1.4690 | TbPRMT1 | I | I | PRMT1 (48/54) | 345/39 | 109 | |
| Tb927.10.3560 | TbPRMT3 | I | ND | PRMT3 (20/33) | 389/42 | Lacks Zn finger | 108 | |
| Tb927.10.640 | TbPRMT5 | II | II | PRMT5 (24/31) | 784/87 | 108 | ||
| Tb927.5.3960 | TbPRMT6 | I | I | PRMT6 (28/31) | 368/41 | 38 | ||
| Tb927.7.5490 | TbPRMT7 | III | III | PRMT7 (13/28) | 390/44 | Lacks C-terminal duplication | 37 | |
| Plasmodium falciparum | PF14_0242 | PfPRMT1 | I | I | PRMT1 (37/51) | 402/47 | 35 | |
| PF08_0092 | PfCARM1 | I | ND | PRMT3 (17/27) | 912/108 | Lacks Zn finger | ||
| PF13_0323 | PfPRMT5 | II | ND | PRMT5 (30/44) | 724/86 | |||
| Toxoplasma gondii | GT1_030400 | TgPRMT1 | I | ND | PRMT1 (46/57) | 392/44 | 120 | |
| GT1_074560 | TgCARM1 | I | I | CARM1 (28/48) | 660/72 | 120 | ||
| GT1_001320 | NA | I | ND | PRMT3 (24/29) | 802/88 | Weak Zn finger | ||
| GT1_126490 | NA | II | ND | PRMT5 (24/39) | 979/107 | |||
| GT1_073730 | NA | Ib | ND | NA | 1305/139 | Unknown homologue, unusually large size |
NA, not applicable; ND, not determined.
Weak homology.
Giardia.
According to several genomic studies, Giardia and the related metamonads represent the most simple of eukaryotes, lacking the typical mitochondria and Golgi complexes found in other eukaryotes (30, 34, 126). Additionally, many genes and systems are believed to have been lost as the organisms transitioned to a parasitic lifestyle (94). In terms of protein methylation, at least six SET-like lysine methyltransferases can be identified in the Giardia genome database (131). In striking contrast, we were unable to identify any apparent PRMT homologues in the current annotation of the Giardia database (GiardiaDB) (Table 1), marking this as the only known eukaryotic group lacking PRMTs. Accordingly, to our knowledge, there are no reports of methylarginine proteins in this parasite. Barring the inclusion of a noncanonical PRMT in Giardia, it appears that the downstream effects of arginine methylation required for other eukaryotes are dispensable in Giardia.
Trichomonas.
The sexually transmitted parasite Trichomonas vaginalis, like Giardia, represents a group that underwent vast gene loss upon transition to a parasitic life cycle (20, 59). Also similar to Giardia, Trichomonas lacks a mitochondria or a true Golgi apparatus. However, Trichomonas differs substantially from Giardia in that it harbors at least eight putative PRMT enzymes in its genome (TrichDB) (Table 1). At least six of these enzymes appear to be homologues of the type I HsPRMT1, with various degrees of identity. TVAG_048280 and TVAG_225950 are 99% identical, indicating a true duplication event, which is expected considering the high number of transposable elements and repetitive genes evident in this organism (20). However, the remaining four type I PRMTs are dissimilar enough to raise the question of whether each serves a different function during the parasite life cycle. The T. vaginalis genome also contains two potential type II PRMTs, which are dissimilar enough to suggest that the enzymes play separate functional roles (Table 2). The putative T. vaginalis PRMTs have not been examined experimentally to date.
Entamoeba.
The intestinal parasite Entamoeba histolytica resembles Giardia and Trichomonas in its lack of a mitochondrion but differs in the presence of a rudimentary Golgi-like apparatus (19). In a trend similar to Trichomonas, E. histolytica also contains multiple homologues of HsPRMT1, two of which (EHI_202470 and EHI_152460) share more sequence similarity with the HsPRMT1 enzyme than does the annotated EhPRMT1 enzyme (EHI_105780) (Table 2). Although both E. histolytica and T. vaginalis have multiple type I PRMTs, none of these enzymes contain characteristics that would differentiate them from being purely a PRMT1 homologue. That is, all of the type I enzymes in these species display the highest homology to HsPRMT1, as opposed to other human type I enzymes, and they lack distinguishing features of some human type I PRMTs, such as the Zn finger present in HsPRMT3 or the N-terminal myristoylation signal present in HsPRMT8. In addition to its four putative type I PRMTs, the E. histolytica genome also encodes an apparent type II PRMT (EHI_158560), consistent with the common trend that all eukaryotes with PRMTs in their genomes have at least one type I and one type II enzyme. The activities of these putative PRMTs have not yet been demonstrated.
Plasmodium.
The genome of Plasmodium falciparum (PlasmoDB) encodes only three putative PRMTs, including homologues of HsPRMT1 and HsPRMT5. This represents considerably fewer PRMTs than observed in the other parasitic protozoa presented here and is similar to the number of PRMTs found in the archetypical yeasts (Tables 1 and 2). Of the three putative P. falciparum PRMTs (PfPRMT), one has been experimentally confirmed to have PRMT activity, PfPRMT1 (35). Consistent with its sequence, PfPRMT1 is a type I enzyme. It contains an extended N terminus that is essential for enzyme activity and is present in both the cytoplasm and nucleus of the parasite. In vitro, PfPRMT1 is capable of methylating the transcriptionally activating histone H4R3, as well as histone H2A, although whether this enzyme can affect transcription in vivo is currently unknown. Several proteins involved in RNA metabolism are also in vitro substrates of PfPRMT1. The other two putative PfPRMTs were annotated PfPRMT5 and PfCARM1 due to sequence homologies, although the enzyme annotated PfCARM1 is actually quite diverged from other known CARM1 homologues, including that found in Toxoplasma gondii. In fact, this enzyme exhibits higher homology to HsPRMT3, although it lacks a zinc finger (Table 2) (120). Both PfPRMT5 and PfCARM1 contain stretches of asparagine residues, a common trend in the low-complexity, highly A/T-rich P. falciparum genome (112).
Toxoplasma.
T. gondii contains five putative PRMTs in its genome (ToxoDB), two of which, TgPRMT1 and TgCARM1, have been experimentally shown to possess PRMT activity (120). These two enzymes exhibit substrate specificities similar to those of their human homologues in that TgPRMT1 methylates histone H4R3 and TgCARM1 methylates H3R17 using in vitro methylation assays. The addition of ATP and proteins with SWI/SNF activity significantly stimulated the ability of TgCARM1 to methylate nucleosomal histones in vitro, suggesting that this enzyme may act in combination with chromatin remodeling enzymes in vivo. TgCARM1 is likely homo-hexameric and localizes to the parasite nucleus. Functional studies implicate TgCARM1-mediated methylation in gene regulation and parasite development (see below). Of the three remaining putative TgPRMTs, one has significant homology to the type II HsPRMT5 (Table 2). Another gene (GT1_001320) has homology to HsPRMT3 and possesses a weak homology zinc finger, consistent with its being a PRMT3 homologue. The remaining TgPRMT (GT1_073730) is the most unique of this group due to its large size (139 kDa) and lack of significant homology to any other characterized PRMT in the literature, although it has very weak homology (<10%) in its conserved methyltransferase domain to PRMT6 and as such may be a type I PRMT (Table 2). The activity of this noncanonical TgPRMT and its functions in the life cycle of T. gondii present a fascinating subject for future research.
Kinetoplastids.
The kinetoplastid parasites (Trypanosoma brucei, Trypanosoma cruzi, and the Leishmania spp.) are the group of parasitic protozoa in which the most research on arginine methylation has been conducted to date. In this review, we will use the causative agent of African sleeping sickness, T. brucei, as the model for kinetoplastids, as the biochemical data are derived from this organism and the set of PRMT-encoding genes found in T. brucei is highly conserved in the other kinetoplastid parasite genomes (TriTrypDB). The T. brucei genome encodes five putative PRMTs, including obvious homologues of the type I HsPRMT1 and type II HsPRMT5 (Table 1). Like HsPRMT1, TbPRMT1 exhibits type I PRMT activity, and upon its knockdown through RNA interference (RNAi), a global reduction in ADMA was observed (109). Thus, TbPRMT1 is responsible for the vast majority of ADMA formation in vivo, similar to what is observed in mammalian cells (137). In vitro, TbPRMT1 appears to prefer GAR domain-containing substrates (109). T. brucei also contains a type II PRMT homologue, TbPRMT5, that displays a broad substrate range, including both GAR domain and non-GAR domain targets (108). TbPRMT5 associates in vivo with a DEAD-box RNA helicase (108) and is capable of methylating this enzyme in vitro (S. Menon and L. Read, unpublished data), suggesting that TbPRMT5 may affect RNA metabolism.
In addition to TbPRMT1, the T. brucei genome encodes two other type I PRMTs. However, in contrast to Entamoeba and Trichomonas, the three trypanosome type I enzymes do not constitute a family of PRMT1 homologues. Rather, each of the type I TbPRMTs displays clear homology to a different human type I enzyme (Table 2). One of these exhibits the highest homology to HsPRMT3. TbPRMT3 has not been characterized biochemically, although its sequence clearly positions it as a putative type I enzyme. In humans, PRMT3 interacts with its major known substrate, the ribosomal protein RPS2, through its N-terminal zinc finger, subsequently facilitating ribosome assembly (39, 135). Interestingly, TbPRMT3 lacks a zinc finger, as does the PRMT3 homologue from the fungus Aspergillus (140), suggesting that TbPRMT3 may exhibit a different substrate specificity than its human homologue. Yeast PRMT3 also displays methyltransferase-independent functions in ribosome assembly (111). The absence of the substrate binding zinc finger, in combination with nonconserved residues in the THW and double E loops in TbPRMT3, suggests that this enzyme might also have functions independent of methyltransferase activity.
The remaining two PRMTs in the kinetoplastids are intriguing, as they appear to be absent in other parasitic protozoa. We recently characterized the T. brucei homologues of HsPRMT6 and HsPRMT7 (37, 38). Like HsPRMT6, TbPRMT6 is a type I enzyme with an apparently narrow substrate specificity, and it undergoes automethylation, although the consequence of this is currently unknown (38). In vitro, TbPRMT6 apparently lacks the ability to methylate GAR domain-containing targets. Rather, it is capable of methylating bovine histones, raising the possibility that it might modify trypanosome histones in vivo. RNAi studies of TbPRMT6 indicate a vital role in the T. brucei life cycle (see below).
The trypanosome PRMT7 enzyme is the most unique of all kinetoplastid PRMTs (37). First, unlike HsPRMT7, which has a C-terminal duplication of the enzymatic domain that is essential for its activity, TbPRMT7 possesses only a single enzymatic cluster, making it roughly half the size of the human enzyme (Tables 1 and 2). Second, both GST-TbPRMT7 expressed in E. coli and TAP-TbPRMT7 expressed in T. brucei catalyze the formation of only MMA, marking this enzyme as the first exclusively type III PRMT demonstrated. Recently, a similar finding of a type III enzyme was also reported in the nematode Caenorhabditis elegans (136). In contrast, the type of activity catalyzed by HsPRMT7 is unresolved, having been reported as either type II or type III (77, 93). TbPRMT7 is also a very robust enzyme with a wide substrate range. It is estimated that TbPRMT7 is at least 30 times more active than its human homologue in terms of methyl groups transferred/hour/microgram of enzyme, making it one of the most active PRMTs described to date. In vitro, TbPRMT7 catalyzes methyltransfer to bovine histones, myelin basic protein, and multiple T. brucei RNA binding proteins, including the non-GAR domain protein MRP2 and the GAR domain proteins RBP16, TbRGG1, and TbRGG2, suggesting that it may be involved in numerous cellular processes in vivo. The presence in T. brucei of a very active PRMT that synthesizes only MMA is unique and suggests two potential functions of such an enzyme. First, MMA may be a more commonly used terminal methyl mark in kinetoplastids than other organisms. Although typically considered an intermediate in ADMA and SDMA synthesis, MMA has been reported in yeast to be an activating signal on histone H3(R2), compared to ADMA, which is commonly inhibitory at this position (66, 67). Likewise, MMA on the mammalian epidermal growth factor receptor was recently shown to affect downstream signaling (51). The second, but not mutually exclusive, possibility is the action of synergism between the type III TbPRMT7 with the other type I and type II trypanosome PRMTs. In this scenario, the highly active TbPRMT7 would deposit the initial MMA mark, which another type I or II PRMT could recognize as a substrate for catalysis of the subsequent ADMA or SDMA. Several human PRMTs have been demonstrated to have either processive or distributive enzymatic mechanisms (74, 75, 101), suggesting that different PRMTs have various abilities to recognize MMA in order to create ADMA or SDMA. Therefore, in the simplified trypanosomes, a distinct process may exist, involving a two-enzyme mechanism to facilitate the final dimethylarginine mark. Further kinetic studies as well as genetic knockout of the TbPRMT7 gene are necessary to finalize a definitive trypanosome model. Overall, trypanosomes may present a simplified model for studying the role(s) of MMA.
FUNCTIONS OF PROTEIN ARGININE METHYLATION IN PARASITE LIFE CYCLES
The presence of different sets of PRMT-encoding genes in the parasites discussed here suggests that protein arginine methylation makes distinct contributions during the life cycles of these organisms. Regulation of protein function through arginine methylation may be especially important during the dramatic life cycle transitions that these parasites undergo as they move through the environment, their vectors, and their mammalian hosts. The complex lifestyles of parasitic organisms require massive changes in gene regulation and morphology, processes in which protein arginine methylation could feasibly play a role. Below, we summarize the currently limited information regarding developmental regulation of PRMTs and their functions in parasitic protozoa. We reference transcriptomic studies to address changing levels of PRMT-encoding RNAs during parasite life cycles. However, keep in mind that this may not necessarily reflect enzyme activity, since protein levels can differ from RNA levels and because PRMT activity can be modulated by association with regulatory proteins in the absence of changes in PRMT levels (62, 84, 118, 128).
Entamoeba.
To our knowledge, no PRMT substrates have been identified, nor have any functional studies regarding protein arginine methylation been performed in Entamoeba. Transcriptomic data indicate that RNA levels of most Entamoeba histolytica PRMTs are unchanged during development. However, transcripts encoding the type I PRMT, EHI_202470, decrease substantially at early time points following intestinal E. histolytica infection in mice, suggesting a potential role in adaptation to different environmental niches (AmoebaDB). Histone lysine methylation is reportedly involved in regulation of the virulence factor amoebaphore A (Ehap-a), whose transcription is silenced following demethylation of H3K4, suggesting a role of methylation in pathogenesis (2). Because a typically repressive arginine at H3R2 is also conserved in E. histolytica, methylation at this arginine residue could feasibly alter the methylation at H3K4 and thus pathogenesis.
Plasmodium.
The levels of RNAs encoding PfPRMTs are not substantially changed during development, with the exception of a slight (∼30%) decrease for PfPRMT5 in the schizont stage (PlasmoDB). However, Western blot analysis of PfPRMT1 revealed developmentally regulated expression of this protein, which is present at very low levels in ring stages and highest levels in late trophozoites (35). Several methylarginine modifications on P. falciparum histones were detected in two mass spectrometry studies (92, 139). These include both mono- and dimethylated versions of H3R17, H4R3, and H3.3R17. As these same marks are often transcriptionally activating in other organisms, further research is needed to determine if these same sites are activating in the malaria parasite. H3R17 levels, which can potentially be attributed to PfCARM1 activity, were unchanged during development. In contrast, dynamic dimethylation of H4R3, which is mediated by PfPRMT1 in vitro, was observed during the parasite life cycle, with high levels of H4R3 observed in late trophozoites and schizonts (35). These data suggest a conserved role for histone arginine methylation in epigenetic gene regulation in Plasmodium along with other posttranslational modifications (28, 35, 91).
Toxoplasma.
In the human host, Toxoplasma gondii exists in the rapidly growing tachyzoite form and the latent cyst form termed bradyzoites. Transcriptomic data are available for four of the five putative Toxoplasma PRMTs, all of which appeared unchanged at the RNA level during the life cycle. However, two functional studies have focused on the role of histone arginine methylation in T. gondii gene regulation and development (43, 120). Saksouk et al. (120) reported that several attempts to create null mutations of TgCARM1 were unsuccessful, and even transient expression of enzyme-dead mutant TgCARM1 in tachyzoites resulted in parasites with decreased viability. These data strongly suggest that TgCARM1 is essential in tachyzoites, consistent with the perinatal lethality of CARM1 knockout in mice (120, 149). Treatment of extracellular tachyzoites with AMI-1, a PRMT inhibitor that affects TgCARM1 but not TgPRMT1, induced stage conversion to bradyzoite forms. Induced differentiation was correlated with a 50% decrease in global H3R17 methylation. Methylation of H3R17 T. gondii was also examined by comprehensive chromatin immunoprecipitation with microarray technology (ChIP-chip) studies of chromosome 1 (43). Gissot et al. detected dimethylated H3R17 at a specific subset of promoters, specifically at 4 of 52 genes exhibiting modified histone peaks (43). Interestingly, ESTs for all 4 of these H3R17-containing genes are present in both tachyzoites and bradyzoites, which is unusual since only 26 of 91 predicted genes on the chromosome 1 microarray have ESTs in both stages. This observation might suggest that H3R17 methylation is involved in tachyzoite to bradyzoite differentiation. While the specific role of TgCARM1-mediated H3R17 methylation still remains to be clarified, together these data point to this modification as an important regulator of signals for life cycle transitions in T. gondii (120). In addition, regulation of differentiation by TgCARM1-mediated methylation of nonhistone substrates remains to be explored.
Trypanosoma brucei.
Three of the five T. brucei PRMTs (TbPRMTs 1, 5, and 6) are unchanged at the RNA level during development or transition from log to stationary phase (63, 114). In contrast, TbPRMT3 and TbPRMT7 RNA levels are decreased ∼60% during the transition from log growth to stationary growth in the insect vector midgut procyclic form (PF), and TbPRMT7 RNA increases ∼30% during differentiation from stumpy mammalian bloodstream forms (BF) to PF cells. To begin to define PRMT function in T. brucei, we knocked down each of the five individual PRMTs in the T. brucei genome using RNAi in the two major life cycle stages that can propagated in culture, the insect PF and the mammalian slender BF (37, 38, 108, 109) (J. Fisk, D. Tomasello, and L. Read, unpublished data). No growth defects were observed in TbPRMT3-, TbPRMT5-, or TbPRMT7-depleted cells or in TbPRMT1-depleted PF. The absence of a growth effect upon knockdown of these enzymes may reflect the partial depletion of protein levels by RNAi. Indeed, several attempts to knock out both alleles of TbPRMT7 in BF have been unsuccessful (J. Fisk and L. Read, unpublished data), suggesting that this enzyme may be essential for growth and that the reduced levels of this very robust enzyme that remain following RNAi are sufficient to maintain cell function in vitro. Redundant functions among some of the five TbPRMTs are also possible; several PRMTs are not essential in mammals and yeasts, indicating redundancy (89, 134, 150). In contrast to the studies described above, RNAi-mediated knockdown of TbPRMT1 in BF does result in a slow growth phenotype, consistent with the report of a growth defect and abnormal cell cycle progression noted for TbPRMT1 knockdowns in a chromosome 1-wide RNAi screen in BF (133). The essential function(s) of this enzyme in BF T. brucei awaits discovery.
In T. brucei, the most profound effect on growth and morphology was observed upon depletion of TbPRMT6 (38). Both PF and BF cells ablated for TbPRMT6 grow slowly under normal culture conditions, and low serum stress in PF greatly exacerbates this effect. In addition to a growth defect, TbPRMT6 knockdown cells exhibit dramatic defects in cytokinesis with differential phenotypes in PF and BF (38). Thus, despite its apparently weak activity and narrow substrate range in vitro, TbPRMT6 appears to methylate an essential target in vivo. Western blot analysis of TbPRMT6 in subcellular fractions of PF T. brucei reveals a predominant cytoplasmic localization, with a small nuclear component. Interestingly, mass spectrometry of TbPRMT6-associated proteins in PF T. brucei revealed numerous histones and components of the nuclear pore complex, suggesting that these classes of proteins might serve as TbPRMT6 substrates in vivo. Whether TbPRMT6 modulates gene expression through histone methylation will be a particular area of future interest, and this putative function is consistent with our finding that bovine histones were the only identified TbPRMT6 substrate in vitro. To date, no arginine methylation has been detected on T. brucei histones, although a noncanonical methylated site was identified at H4R53 in the related T. cruzi, and this site is conserved in T. brucei (31, 61, 88). In addition, the major histone arginine residue that is methylated by PRMT6 in other organisms, namely, the H3R2, is conserved in T. brucei and other kinetoplastids (54, 61, 88). Analyses of histone modifications in T. brucei to date were performed using Edman degradation (61, 88), and recently developed mass spectrophotometric techniques for detection of methylarginines (143) may be useful for identification of previously overlooked methylarginine residues in histones of T. brucei. If so, it will be of interest to determine whether these sites are altered in TbPRMT6 knockdown cells.
In order to gain a broader understanding of the roles of arginine methylation in trypanosomes, we have used mass spectrometry to identify arginine methyl proteins in PF T. brucei. Early matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) experiments identified RBP16, a mitochondrial RNA binding protein that functions in RNA stabilization and editing, as an arginine methyl protein (44, 119). Methylation of RBP16 has also been detected in Leishmania donovani (44, 119). In T. brucei, RBP16 contains at least three arginine residues that can undergo methylation, and differentially methylated forms of the protein are present. Knockdown of TbPRMT1 abolished methylation at two of three sites examined, demonstrating that RBP16 is an in vivo TbPRMT1 substrate (44). Both TbPRMT1 knockdown and overexpression of nonmethylatable RBP16 lead to destabilization of the mitochondrial ND4 RNA, which is also destabilized by knockdown of RBP16 itself. Thus, methylation of RBP16 is critical for some aspects of its function in RNA stabilization. Conversely, the role of RBP16 in RNA editing was unaffected by its methylation status, suggesting that arginine methylation can differentially affect RBP16 functions. Subsequent biochemical experiments demonstrated that methylation of RBP16 facilitates its association with macromolecular complexes within the trypanosome mitochondrion (45). RBP16 harboring arginine to lysine mutations at three methylatable positions fails to associate with these higher-ordered structures, an effect apparently due to perturbation of protein-protein interactions. The loss of these RBP16-containing protein complexes is correlated with the impaired ability of the protein to stabilize specific RNAs, suggesting that methylation-specific protein-protein interactions may be important for this RBP16 function. The identities of methylarginine-specific protein binding partners of RBP16 are currently not known. However, it will be of great interest to identify proteins that bind in a methylarginine-sensitive manner since trypanosomes appear to contain very few of the Tudor domain proteins known to bind methylarginines in other species (27, 143). Arginine methylation also differentially affects the ability of RBP16 to bind to different classes of RNAs, increasing its association with mRNA but, conversely, decreasing its binding to guide RNAs. Therefore, limited studies of one trypanosome methyl protein reveal profound impacts of this modification on protein function.
More recently, we are utilizing a sensitive electron-transfer dissociation/collision-induced dissociation fragmentation method (32, 143) to examine the arginine methyl proteome of T. brucei. To date, we have identified over 850 arginine methyl proteins, originating from multiple cellular compartments and organelles. These include RNA binding proteins and modifying enzymes, DNA replication and repair proteins, signal transduction components, and several classes of metabolic enzymes. Approximately 200 of these proteins are mitochondrially localized and are predicted to affect many aspects of mitochondrial function (J. Fisk, J. Qu, and L. Read, unpublished data). Future efforts will be directed toward determining the cellular and biochemical consequences of arginine methylation in a subset of these proteins in T. brucei.
PERSPECTIVES
Multiple genes encoding putative PRMTs were readily identified in all of the species that we examined, with the exception of Giardia spp. Thus, arginine methylation is widespread throughout evolution. A common theme in parasitic protozoa, as in all PRMT-containing eukaryotes, is the presence of homologues of the type I HsPRMT1 and the type II HsPRMT5 in each species. The invariable presence of both of these enzyme classes indicates that ADMA and SDMA serve separate and essential purposes in these organisms. As PRMT1 accounts for the majority of PRMT activity in mammalian cells, it is not surprising that its archetype is found in each of these early branching eukaryotes. What is unusual is the propensity of some groups of parasitic protozoa to have multiple copies of a PRMT1-like enzyme (e.g., 6 in T. vaginalis and 4 in E. histolytica; Table 2). In human cells, there are six type I PRMTs, each with distinct sequence characteristics, substrates, and subcellular/tissue localizations that discriminate them from each other. In contrast, the PRMT1-like enzymes in T. vaginalis and E. histolytica lack distinct features enabling them to be differentiated from PRMT1 homologues. While some may simply have arisen through genetic duplication, the majority of PRMT1 homologues in Trichomonas and Entamoeba spp. have enough dissimilarity to suggest that they have distinct functions. The roles of these enzymes in parasite biology await discovery.
One potentially important role for arginine methylation in parasitic protozoa is in epigenetic control of gene expression through modification of histones. As noted above, histone arginine methylation has been reported in Toxoplasma and Plasmodium and likely affects life cycle transitions in the former. Importantly, in the deadly human parasite P. falciparum, epigenetic regulation is also critical in modulating the expression of variant surface antigens (reviewed in references 28 and 91). var genes encode the variant PfEMP proteins that function in both immune evasion and in pathogenesis through their propensity to bind host cell surface receptors. Although epigenetic regulation of var gene expression has been attributed solely to changes in histone acetylation to date, histone arginine methylation could play an important role, potentially by modulating acetylation of nearby lysine residues. In the kinetoplastid parasite, T. brucei, epigenetic regulation also factors prominently in antigenic variation (36). Histone lysine acetylation and methylation are important in variant surface glycoprotein (VSG) transcription, and the latter affects VSG switching in BF T. brucei. While histone arginine methylation has not yet been reported in T. brucei, it is tempting to speculate that this modification could play a role in regulating VSG expression and/or switching by modulating the creation of adjacent lysine methyl and acetyl marks. Another potential role for arginine methylation in kinetoplastids is in cell cycle control. Histone acetylates and deacetylases, as well as lysine methyltransferases, have been shown to play a role in cell cycle progression in T. brucei. The cell cycle defects that we observed in TbPRMT6 knockdown cells (38) could result from secondary alterations in previously reported histone modifications and/or through direct effects of histone arginine methyl marks.
Posttranslational modifications of RBPs by arginine methylation may be especially important in kinetoplastids, as these org anisms regulate gene expression primarily through posttranscriptional processes in which RBPs are prominent (26, 69, 86, 123). RBPs and RNA helicases have been reported to function in RNA decay, RNA editing, differentiation, and cell cycle control in T. brucei. As noted above, we have identified numerous RBPs and RNA helicases in the T. brucei arginine methylome, and several proteins that function in mitochondrial RNA editing and stability are PRMT substrates in vitro (37, 44, 108, 109). The ease with which T. brucei is genetically manipulated will allow us to define the roles of RBP arginine methylation in many critical aspects of the trypanosome life cycle.
Finally, the presence of multiple PRMTs in a wide range of parasitic protozoa raises the possibility of targeting these enzymes for chemotherapeutic attack. Anti-PRMT drugs are currently being developed for use as cancer therapeutics (9, 21, 87). Future studies on the functions of divergent PRMTs with unique features in parasitic protozoa could aid in drug development in addition to providing important insights into parasite biology.
ACKNOWLEDGMENTS
This work was supported by NIH awards R01 AI060260 to L.K.R. and F32 AI07718501 to J.C.F.
Footnotes
Published ahead of print on 17 June 2011.
REFERENCES
- 1. An W., Kim J., Roeder R. G. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735–748 [DOI] [PubMed] [Google Scholar]
- 2. Anbar M., et al. 2005. Involvement of a short interspersed element in epigenetic transcriptional silencing of the amoebapore gene in Entamoeba histolytica. Eukaryot. Cell 4:1775–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aoki K., Ishii Y., Matsumoto K., Tsujimoto M. 2002. Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 30:5182–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachand F. 2007. Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryot. Cell 6:889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bachand F., Silver P. A. 2004. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 23:2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bedford M. T. 2007. Arginine methylation at a glance. J. Cell Sci. 120:4243–4246 [DOI] [PubMed] [Google Scholar]
- 7. Bedford M. T., Clarke S. G. 2009. Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bedford M. T., et al. 2000. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275:16030–16036 [DOI] [PubMed] [Google Scholar]
- 9. Bissinger E. M., et al. 2011. Acyl derivatives of p-aminosulfonamides and dapsone as new inhibitors of the arginine methyltransferase hPRMT1. Bioorg. Med. Chem. 19:3717–3731 [DOI] [PubMed] [Google Scholar]
- 10. Blanchet F., Schurter B. T., Acuto O. 2006. Protein arginine methylation in lymphocyte signaling. Curr. Opin. Immunol. 18:321–328 [DOI] [PubMed] [Google Scholar]
- 11. Boisvert F.-M., Chenard C. A., Richard S. 2005. Protein interfaces in signaling regulated by arginine methylation. Sci. STKE 2005:re2. [DOI] [PubMed] [Google Scholar]
- 12. Boisvert F.-M., et al. 2002. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J. Cell Biol. 159:957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boisvert F.-M., Cote J., Boulanger M.-C., Richard S. 2003. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics 2:1319–1330 [DOI] [PubMed] [Google Scholar]
- 14. Boisvert F.-M., Dery U., Masson J.-Y., Richard S. 2005. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19:671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boisvert F.-M., Rhie A., Richard S., Doherty A. J. 2005. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 4:1834–1841 [DOI] [PubMed] [Google Scholar]
- 16. Brahms H., Meheus L., de Brabandere V., Fischer U., Luhrmann R. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7:1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brahms H., et al. 2000. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 275:17122–17129 [DOI] [PubMed] [Google Scholar]
- 18. Branscombe T. L., et al. 2001. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276:32971–32976 [DOI] [PubMed] [Google Scholar]
- 19. Bredeston L. M., Caffaro C. E., Samuelson J., Hirschberg C. B. 2005. Golgi and endoplasmic reticulum functions take place in different subcellular compartments of Entamoeba histolytica. J. Biol. Chem. 280:32168–32176 [DOI] [PubMed] [Google Scholar]
- 20. Carlton J. M., et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castellano S., et al. 2010. Design, synthesis and biological evaluation of carboxy analogues of arginine methyltransferase inhibitor 1 (AMI-1). ChemMedChem 5:398–414 [DOI] [PubMed] [Google Scholar]
- 22. Ceman S., Blackwell E., Zhang X. 2010. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 19:1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang B., Chen Y., Zhao Y., Bruick R. K. 2007. JMJD6 is a histone arginine demethylase. Science 318:444–447 [DOI] [PubMed] [Google Scholar]
- 24. Chen D., et al. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174–2177 [DOI] [PubMed] [Google Scholar]
- 25. Cheng D., Cote J., Shaaban S., Bedford M. T. 2007. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25:71–83 [DOI] [PubMed] [Google Scholar]
- 26. Clayton C., Shapira M. 2007. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 156:93–101 [DOI] [PubMed] [Google Scholar]
- 27. Cote J., Richard S. 2005. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 280:28476–28483 [DOI] [PubMed] [Google Scholar]
- 28. Cui L., Miao J. 2010. Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum. Eukaryot. Cell 9:1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cuthbert G. L., et al. 2004. Histone deimination antagonizes arginine methylation. Cell 118:545–553 [DOI] [PubMed] [Google Scholar]
- 30. Dacks J. B., Doolittle W. F. 2001. Reconstructing/deconstructing the earliest eukaryotes: how comparative genomics can help. Cell 107:419–425 [DOI] [PubMed] [Google Scholar]
- 31. da Cunha J. P. C., Nakayasu E. S., de Almeida I. C., Schenkman S. 2006. Post-translational modifications of Trypanosoma cruzi histone H4. Mol. Biochem. Parasitol. 150:268–277 [DOI] [PubMed] [Google Scholar]
- 32. Duan X., et al. 2009. A straightforward and highly efficient precipitation/on-pellet digestion procedure coupled with a long gradient nano-LC separation and Orbitrap mass spectrometry for label-free expression profiling of the swine heart mitochondrial proteome. J. Proteome Res. 8:2838–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. El-Andaloussi N., et al. 2007. Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. FASEB J. 21:26–34 [DOI] [PubMed] [Google Scholar]
- 34. Embley T. M., Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440:623–630 [DOI] [PubMed] [Google Scholar]
- 35. Fan Q., Miao J., Cui L. 2009. Characterization of PRMT1 from Plasmodium falciparum. Biochem. J. 421:107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Figueiredo L. M., Cross G. A. M., Janzen C. J. 2009. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat. Rev. Microbiol. 7:504–513 [DOI] [PubMed] [Google Scholar]
- 37. Fisk J. C., et al. 2009. A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei. J. Biol. Chem. 284:11590–11600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fisk J. C., et al. 2010. TbPRMT6 is a type I protein arginine methyltransferase that contributes to cytokinesis in Trypanosoma brucei. Eukaryot. Cell 9:866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frankel A., Clarke S. 2000. PRMT3 is a distinct member of the protein arginine N-methyltransferase family. Conferral of substrate specificity by a zinc-finger domain. J. Biol. Chem. 275:32974–32982 [DOI] [PubMed] [Google Scholar]
- 40. Frankel A., et al. 2002. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 277:3537–3543 [DOI] [PubMed] [Google Scholar]
- 41. Ganesh L., et al. 2006. Protein methyltransferase 2 inhibits NF-kappaB function and promotes apoptosis. Mol. Cell. Biol. 26:3864–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gary J. D., Lin W. J., Yang M. C., Herschman H. R., Clarke S. 1996. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J. Biol. Chem. 271:12585–12594 [DOI] [PubMed] [Google Scholar]
- 43. Gissot M., Kelly K. A., Ajioka J. W., Greally J. M., Kim K. 2007. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 3:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goulah C. C., Pelletier M., Read L. K. 2006. Arginine methylation regulates mitochondrial gene expression in Trypanosoma brucei through multiple effector proteins. RNA 12:1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goulah C. C., Read L. K. 2007. Differential effects of arginine methylation on RBP16 mRNA binding, guide RNA (gRNA) binding, and gRNA-containing ribonucleoprotein complex (gRNP) formation. J. Biol. Chem. 282:7181–7190 [DOI] [PubMed] [Google Scholar]
- 46. Green D. M., et al. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 277:7752–7760 [DOI] [PubMed] [Google Scholar]
- 47. He W., et al. 2011. A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res. 39:4719–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Henry M. F., Silver P. A. 1996. A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol. Cell. Biol. 16:3668–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Herrmann F., Pably P., Eckerich C., Bedford M. T., Fackelmayer F. O. 2009. Human protein arginine methyltransferases in vivo—distinct properties of eight canonical members of the PRMT family. J. Cell Sci. 122:667–677 [DOI] [PubMed] [Google Scholar]
- 50. Hong X., et al. 2010. Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 107:14568–14572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hsu J. M., et al. 2011. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat. Cell Biol. 13:174–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hung M. L., Hautbergue G. M., Snijders A. P., Dickman M. J., Wilson S. A. 2010. Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1. Nucleic Acids Res. 38:3351–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hyllus D., et al. 2007. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 21:3369–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Iberg A. N., et al. 2008. Arginine methylation of the histone H3 tail impedes effector binding. J. Biol. Chem. 283:3006–3010 [DOI] [PubMed] [Google Scholar]
- 55. Invernizzi C. F., et al. 2007. Arginine methylation of the HIV-1 nucleocapsid protein results in its diminished function. AIDS 21:795–805 [DOI] [PubMed] [Google Scholar]
- 56. Invernizzi C. F., Xie B., Richard S., Wainberg M. A. 2006. PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA. Retrovirology 3:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Iwasaki H., et al. 2010. Disruption of protein arginine N-methyltransferase 2 regulates leptin signaling and produces leanness in vivo through loss of STAT3 methylation. Circ. Res. 107:992–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Iwasaki H., Yada T. 2007. Protein arginine methylation regulates insulin signaling in L6 skeletal muscle cells. Biochem. Biophys. Res. Comm. 364:1015–1021 [DOI] [PubMed] [Google Scholar]
- 59. Iyer L. M., Anantharaman V., Wolf M. Y., Aravind L. 2008. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 38:1–31 [DOI] [PubMed] [Google Scholar]
- 60. Jansson M., et al. 2008. Arginine methylation regulates the p53 response. Nat. Cell Biol. 10:1431–1439 [DOI] [PubMed] [Google Scholar]
- 61. Janzen C. J., et al. 2006. Unusual histone modifications in Trypanosoma brucei. FEBS Lett. 580:2306–2310 [DOI] [PubMed] [Google Scholar]
- 62. Jelinic P., Stehle J.-C., Shaw P. 2006. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 4:e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jensen B. C., Sivam D., Kifer C. T., Myler P., Parsons M. 2009. Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics 10:482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim S., et al. 1997. Identification of N(G)-methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry 36:5185–5192 [DOI] [PubMed] [Google Scholar]
- 65. Kirino Y., et al. 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 11:652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kirmizis A., et al. 2009. Distinct transcriptional outputs associated with mono- and dimethylated histone H3 arginine 2. Nat. Struct. Mol. Biol. 16:449–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kirmizis A., et al. 2007. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449:928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kleinschmidt M. A., Streubel G., Samans B., Krause M., Bauer U.-M. 2008. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 36:3202–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kramer S., et al. 2010. The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J. Cell Sci. 123:699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Krause C. D., et al. 2007. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Ther. 113:50–87 [DOI] [PubMed] [Google Scholar]
- 71. Krones-Herzig A., et al. 2006. Signal-dependent control of gluconeogenic key enzyme genes through coactivator-associated arginine methyltransferase 1. J. Biol. Chem. 281:3025–3029 [DOI] [PubMed] [Google Scholar]
- 72. Kwak Y. T., et al. 2003. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell 11:1055–1066 [DOI] [PubMed] [Google Scholar]
- 73. Lachner M., Jenuwein T. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286–298 [DOI] [PubMed] [Google Scholar]
- 74. Lakowski T. M., Frankel A. 2009. Kinetic analysis of human protein arginine N-methyltransferase 2: formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem. J. 421:253–261 [DOI] [PubMed] [Google Scholar]
- 75. Lakowski T. M., Frankel A. 2008. A kinetic study of human protein arginine N-methyltransferase 6 reveals a distributive mechanism. J. Biol. Chem. 283:10015–10025 [DOI] [PubMed] [Google Scholar]
- 76. Lee D. Y., Teyssier C., Strahl B. D., Stallcup M. R. 2005. Role of protein methylation in regulation of transcription. Endocrine Rev. 26:147–170 [DOI] [PubMed] [Google Scholar]
- 77. Lee J.-H., et al. 2005. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J. Biol. Chem. 280:3656–3664 [DOI] [PubMed] [Google Scholar]
- 78. Lee J., Sayegh J., Daniel J., Clarke S., Bedford M. T. 2005. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 280:32890–32896 [DOI] [PubMed] [Google Scholar]
- 79. Lee J. H., et al. 2000. Hsl7p, the yeast homologue of human JBP1, is a protein methyltransferase. Biochem. Biophys. Res. Comm. 274:105–111 [DOI] [PubMed] [Google Scholar]
- 80. Lee Y. H., Stallcup M. R. 2009. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol. Endocrinol. 23:425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li H., et al. 2002. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 277:44623–44630 [DOI] [PubMed] [Google Scholar]
- 82. Li X., et al. 2010. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood 115:2028–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li Y.-J., Stallcup M. R., Lai M. M. C. 2004. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 78:13325–13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lin W. J., Gary J. D., Yang M. C., Clarke S., Herschman H. R. 1996. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 271:15034–15044 [DOI] [PubMed] [Google Scholar]
- 85. Liu Q., Dreyfuss G. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 15:2800–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lukes J., Hashimi H., Verner Z., Cicova Z. 2010. The remarkable mitochondrion of trypanosomes and related flagellates, p. 227–252In de Souza W. (ed.), Structures and organelles in pathogenic protists , vol. 17 Springer-Verlag, Berlin, Germany [Google Scholar]
- 87. Mai A., et al. 2007. Synthesis and biological validation of novel synthetic histone/protein methyltransferase inhibitors. ChemMedChem 2:987–991 [DOI] [PubMed] [Google Scholar]
- 88. Mandava V., et al. 2007. Histone modifications in Trypanosoma brucei. Mol. Biochem. Parasitol. 156:41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. McBride A. E., Weiss V. H., Kim H. K., Hogle J. M., Silver P. A. 2000. Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function. Cofactor binding and substrate interactions. J. Biol. Chem. 275:3128–3136 [DOI] [PubMed] [Google Scholar]
- 90. McBride A. E., et al. 2007. Protein arginine methylation in Candida albicans: role in nuclear transport. Eukaryot. Cell 6:1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Merrick C. J., Duraisingh M. T. 2010. Epigenetics in Plasmodium: what do we really know? Eukaryot. Cell 9:1150–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Miao J., Fan Q., Cui L., Li J. 2006. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene 369:53–65 [DOI] [PubMed] [Google Scholar]
- 93. Miranda T. B., Miranda M., Frankel A., Clarke S. 2004. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem. 279:22902–22907 [DOI] [PubMed] [Google Scholar]
- 94. Morrison H. G., et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926 [DOI] [PubMed] [Google Scholar]
- 95. Mowen K. A., Schurter B. T., Fathman J. W., David M., Glimcher L. H. 2004. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell 15:559–571 [DOI] [PubMed] [Google Scholar]
- 96. Mowen K. A., et al. 2001. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell 104:731–741 [DOI] [PubMed] [Google Scholar]
- 97. Najbauer J., Johnson B. A., Young A. L., Aswad D. W. 1993. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J. Biol. Chem. 268:10501–10509 [PubMed] [Google Scholar]
- 98. Nichols R. C., et al. 2000. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 256:522–532 [DOI] [PubMed] [Google Scholar]
- 99. Niewmierzycka A., Clarke S. 1999. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 274:814–824 [DOI] [PubMed] [Google Scholar]
- 100. Ohkura N., Takahashi M., Yaguchi H., Nagamura Y., Tsukada T. 2005. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J. Biol. Chem. 280:28927–28935 [DOI] [PubMed] [Google Scholar]
- 101. Osborne T. C., Obianyo O., Zhang X., Cheng X., Thompson P. R. 2007. Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry 46:13370–13381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Pahlich S., Zakaryan R. P., Gehring H. 2006. Protein arginine methylation: cellular functions and methods of analysis. Biochim. Biophys. Acta 1764:1890–1903 [DOI] [PubMed] [Google Scholar]
- 103. Paik W. K., Kim S. 1969. Enzymatic methylation of histones. Arch. Biochem. Biophys. 134:632–637 [DOI] [PubMed] [Google Scholar]
- 104. Paik W. K., Kim S. 1968. Protein methylase I. Purification and properties of the enzyme. J. Biol. Chem. 243:2108–2114 [PubMed] [Google Scholar]
- 105. Paik W. K., Kim S. 1969. Protein methylation in rat brain in vitro. J. Neurochem. 16:1257–1261 [DOI] [PubMed] [Google Scholar]
- 106. Paik W. K., Paik D. C., Kim S. 2007. Historical review: the field of protein methylation. Trends Biochem. Sci. 32:146–152 [DOI] [PubMed] [Google Scholar]
- 107. Pang C. N., Gasteiger E., Wilkins M. R. 2010. Identification of arginine- and lysine-methylation in the proteome of Saccharomyces cerevisiae and its functional implications. BMC Genomics 11:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Pasternack D. A., Sayegh J., Clarke S., Read L. K. 2007. Evolutionarily divergent type II protein arginine methyltransferase in Trypanosoma brucei. Eukaryot. Cell 6:1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Pelletier M., Pasternack D. A., Read L. K. 2005. In vitro and in vivo analysis of the major type I protein arginine methyltransferase from Trypanosoma brucei. Mol. Biochem. Parasitol. 144:206–217 [DOI] [PubMed] [Google Scholar]
- 110. Pelletier M., et al. 2001. Arginine methylation of a mitochondrial guide RNA binding protein from Trypanosoma brucei. Mol. Biochem. Parasitol. 118:49–59 [DOI] [PubMed] [Google Scholar]
- 111. Perreault A., Gascon S., D'Amours A., Aletta J. M., Bachand F. 2009. A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis. J. Biol. Chem. 284:15026–15037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pizzi E., Frontali C. 2001. Low-complexity regions in Plasmodium falciparum proteins. Genome Res. 11:218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Pollack B. P., et al. 1999. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 274:31531–31542 [DOI] [PubMed] [Google Scholar]
- 114. Queiroz R., Benz C., Fellenberg K., Hoheisel J. D., Clayton C. 2009. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics 10:495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Raijmakers R., et al. 2007. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J. Mol. Biol. 367:1118–1129 [DOI] [PubMed] [Google Scholar]
- 116. Ren J., et al. 2010. Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J. Biol. Chem. 285:12695–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Rho J., Choi S., Jung C.-R., Im D.-S. 2007. Arginine methylation of Sam68 and SLM proteins negatively regulates their poly(U) RNA binding activity. Arch. Biochem. Biophys. 466:49–57 [DOI] [PubMed] [Google Scholar]
- 118. Robin-Lespinasse Y., et al. 2007. hCAF1, a new regulator of PRMT1-dependent arginine methylation. J. Cell Sci. 120:638–647 [DOI] [PubMed] [Google Scholar]
- 119. Rosenzweig D., Smith D., Myler P. J., Olafson R. W., Zilberstein D. 2008. Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics 8:1843–1850 [DOI] [PubMed] [Google Scholar]
- 120. Saksouk N., et al. 2005. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 25:10301–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sayegh J., Clarke S. G. 2008. Hsl7 is a substrate-specific type II protein arginine methyltransferase in yeast. Biochem. Biophys. Res. Comm. 372:811–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sayegh J., Webb K., Cheng D., Bedford M. T., Clarke S. G. 2007. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J. Biol. Chem. 282:36444–36453 [DOI] [PubMed] [Google Scholar]
- 123. Schwede A., et al. 2009. The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res. 37:5511–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Shen E. C., et al. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12:679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Shin H. S., et al. 2009. Arginine methylation of ribosomal protein S3 affects ribosome assembly. Biochem. Biophys. Res. Comm. 385:273–278 [DOI] [PubMed] [Google Scholar]
- 126. Simpson A. G., Roger A. J. 2002. Eukaryotic evolution: getting to the root of the problem. Curr. Biol. 12:R691–R693 [DOI] [PubMed] [Google Scholar]
- 127. Sims R. J., III, Nishioka K., Reinberg D. 2003. Histone lysine methylation: a signature for chromatin function. Trends Genet. 19:629–639 [DOI] [PubMed] [Google Scholar]
- 128. Singh V., et al. 2004. DAL-1/4.1B tumor suppressor interacts with protein arginine N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate substrates in vitro and in vivo. Oncogene 23:7761–7771 [DOI] [PubMed] [Google Scholar]
- 129. Smith J. J., et al. 1999. Unusual sites of arginine methylation in poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J. Biol. Chem. 274:13229–13234 [DOI] [PubMed] [Google Scholar]
- 130. Smith W. A., Schurter B. T., Wong-Staal F., David M. 2004. Arginine methylation of RNA helicase A determines its subcellular localization. J. Biol. Chem. 279:22795–22798 [DOI] [PubMed] [Google Scholar]
- 131. Sonda S., et al. 2010. Epigenetic mechanisms regulate stage differentiation in the minimized protozoan Giardia lamblia. Mol. Microbiol. 76:48–67 [DOI] [PubMed] [Google Scholar]
- 132. Stallcup M. R. 2001. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 20:3014–3020 [DOI] [PubMed] [Google Scholar]
- 133. Subramaniam C., et al. 2006. Chromosome-wide analysis of gene function by RNA interference in the African trypanosome. Eukaryot. Cell 5:1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Swiercz R., Cheng D., Kim D., Bedford M. T. 2007. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J. Biol. Chem. 282:16917–16923 [DOI] [PubMed] [Google Scholar]
- 135. Swiercz R., Person M. D., Bedford M. T. 2005. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 386:85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Takahashi Y., et al. 2011. The C. elegans PRMT-3 possesses a type III protein arginine methyltransferase activity. J. Recept. Signal Transduct. Res. 31:168–172 [DOI] [PubMed] [Google Scholar]
- 137. Tang J., et al. 2000. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275:7723–7730 [DOI] [PubMed] [Google Scholar]
- 138. Tang J., Gary J. D., Clarke S., Herschman H. R. 1998. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J. Biol. Chem. 273:16935–16945 [DOI] [PubMed] [Google Scholar]
- 139. Trelle M. B., Salcedo-Amaya A. M., Cohen A. M., Stunnenberg H. G., Jensen O. N. 2009. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J. Proteome Res. 8:3439–3450 [DOI] [PubMed] [Google Scholar]
- 140. Trojer P., et al. 2004. Histone methyltransferases in Aspergillus nidulans: evidence for a novel enzyme with a unique substrate specificity. Biochemistry 43:10834–10843 [DOI] [PubMed] [Google Scholar]
- 141. Vagin V. V., Hannon G. J., Aravin A. A. 2009. Arginine methylation as a molecular signature of the Piwi small RNA pathway. Cell Cycle 8:4003–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Vagin V. V., et al. 2009. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 23:1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wang H., et al. 2009. Accurate localization and relative quantification of arginine methylation using Nanoflow liquid chromatography coupled to electron transfer dissociation and Orbitrap mass spectrometry. J. Am. Soc. Mass Spectrom. 20:507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Wang Y., et al. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306:279–283 [DOI] [PubMed] [Google Scholar]
- 145. Webby C. J., et al. 2009. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325:90–93 [DOI] [PubMed] [Google Scholar]
- 146. Wood V., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880 [DOI] [PubMed] [Google Scholar]
- 147. Wu C. C., et al. 2004. Organellar proteomics reveals Golgi arginine dimethylation. Mol. Biol. Cell 15:2907–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wysocka J., Allis C. D., Coonrod S. 2006. Histone arginine methylation and its dynamic regulation. Front. Biosci. 11:344–355 [DOI] [PubMed] [Google Scholar]
- 149. Yadav N., et al. 2003. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 100:6464–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Yoshimoto T., et al. 2006. The arginine methyltransferase PRMT2 binds RB and regulates E2F function. Exp. Cell Res. 312:2040–2053 [DOI] [PubMed] [Google Scholar]
- 151. Zhao X., et al. 2008. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 22:640–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zhou Z., et al. 2010. PRMT5 regulates Golgi apparatus structure through methylation of the golgin GM130. Cell Res. 20:1023–1033 [DOI] [PubMed] [Google Scholar]