Abstract
We have performed a genomic characterization of a kinetoplastid protist living within the amoebozoan Neoparamoeba pemaquidensis. The genome of this “Ichthyobodo-related organism” was found to be unexpectedly large, with at least 11 chromosomes between 1.0 and 3.5 Mbp and a total genome size of at least 25 Mbp.
TEXT
Kinetoplastids are an important group of eukaryotic microbes named by virtue of their shared possession of a conspicuous mass of DNA—the “kinetoplast”—inside the mitochondrion (21, 31). They are remarkable for their bizarre suite of biochemical features (e.g., spliced leader [SL] trans splicing and mitochondrial RNA editing) (1, 5, 21) and are best known as pathogens: the so-called “tritryps” are responsible for mass mortality and morbidity in humans and other animals (1, 14, 31). The kinetoplastids also contain a variety of other parasites, such as the fish pathogen Cryptobia, as well as many free-living groups (e.g., Bodo) (31). Phylogenetic studies suggest that within kinetoplastids, parasitic lineages have evolved from free-living ones on at least four occasions (31). Despite many genomics-enabled advances for the tritryp pathogens (e.g., see references 2, 14, and 18), a comprehensive evolutionary framework for understanding the biology of these important pathogens is currently lacking.
The kinetoplastid endosymbiont of Neoparamoeba pemaquidensis, an amoebozoan that causes disease in fish (e.g., Atlantic salmon) and invertebrates such as lobster (20, 27), represents an apparent example of “recent” adaptation to intracellularity. We refer to this enigmatic endosymbiont as the “Ichthyobodo-related organism” (IRO) based on 18S ribosomal DNA (rDNA) analyses showing its close affinity to Ichthyobodo (6, 10–12). In this study we characterized genes from N. pemaquidensis and its kinetoplastid endosymbiont and carried out the first investigation of the chromosomes of both organisms using pulsed-field gel electrophoresis (PFGE) and Southern blotting.
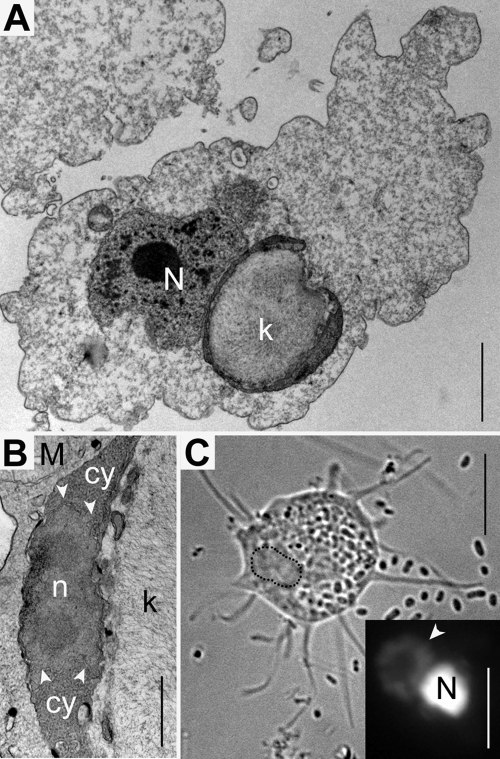
N. pemaquidensis strains CCAP 1560/4 and 1560/5 and Neoparamoeba branchiphila AFSM3/II were cultured as described previously (13). Using standard transmission electron microscopy (TEM) and 4′,6-diamidino-2-phenylindole (DAPI) staining protocols, N. pemaquidensis strain 1560/4 was mostly found to possess 1 (and only occasionally 2) oval or round IRO per cell, each 4 to 6 μm in diameter. The IRO was typically very closely associated with the host cell nucleus, with their surfaces often appearing to be in direct contact (Fig. 1A and C). This is consistent with previous reports based on studies of multiple strains of Neoparamoeba (e.g., see references 10, 12, 13, and 17). Within the IRO, a single mitochondrion was found to occupy more than half of the endosymbiont cell volume, with its distinctive kinetoplast DNA (kDNA) appearing as a complex fibrillar structure in TEM micrographs (Fig. 1A and B). The close proximity of the IRO to the amoeba host nucleus was also apparent under DAPI staining (Fig. 1C). The IRO nucleus stained weakly with DAPI relative to the amoeba nucleus, suggesting that the host and endosymbiont nuclei differ greatly in terms of their DNA content (Fig. 1C).
Fig. 1.
Morphological observation using TEM and DAPI staining of Neoparamoeba pemaquidensis CCAP1560/4 and its kinetoplastid endosymbiont (i.e., the IRO). (A) Cross section of the host amoeba cell, showing its nucleus (N) and the IRO with its distinctive kinetoplast DNA (k). Scale bar = 2 μm. (B) Close-up view of the IRO nucleus (n) with its double membrane envelope (arrowheads). Two nucleoli are visible. The cytoplasm of the IRO (cy) is rich with ribosomes. Scale bar = 500 nm. (C) Differential interference contrast (DIC) image showing an N. pemaquidensis cell with extended pseudopods. Bacteria are apparent outside and inside the cell. Scale bar = 10 μm. The inset box shows an enlarged DAPI-stained view of the region outlined by the dotted line. The position of the IRO relative to the amoeba nucleus (N) is indicated by an arrowhead. Scale bar for inset box = 5 μm.
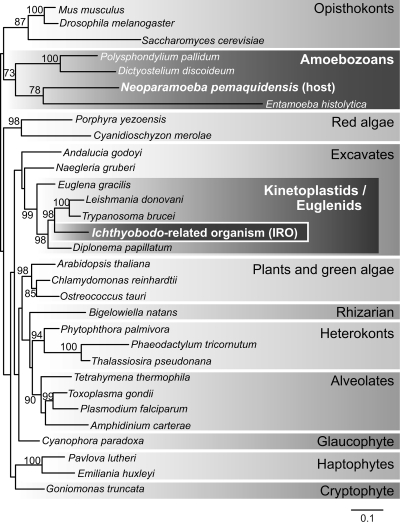
In order to gain insight into the evolutionary origins of Neoparamoeba and the IRO, we amplified six protein genes from both the host and its endosymbiont using PCR and/or reverse transcriptase PCR (RT-PCR) (alpha-tubulin [aTub], beta-tubulin [bTub], elongation factor 2 [EF2], heat shock protein 90 [hsp90], an RNA polymerase I subunit [Rpa1], and an RNA polymerase II subunit [Rpb1]; primer sequences are available upon request). PCR products were cloned into suitable vectors, and several clones per amplicon were sequenced on a CEQ8000 capillary DNA sequencer (Beckman Coulter, Inc., Fullerton, CA). Protein alignments were analyzed individually (data not shown) and as a single concatenate. Trees inferred from the concatenated data set showed the IRO branching robustly with the tritryps Leishmania and Trypanosoma, to the exclusion of two other euglenozoans, Diplonema and Euglena (Fig. 2). We also amplified and sequenced the SL RNA gene from the IRO genome and confirmed the presence of an SL mini-exon addition to IRO alpha- and beta-tubulin mRNAs by RT-PCR and 5′ rapid amplification of cDNA ends (RACE)-PCR (data not shown). Additional RACE-PCR experiments showed that the host-derived alpha-tubulin mRNA does not undergo mini-exon addition. These results suggest that SL trans splicing takes place in the kinetoplastid endosymbiont but not in its host. No spliceosomal introns were found in any of the IRO protein genes sequenced herein, which are among the first to be determined, so it remains to be seen whether both cis and trans splicing occurs in these organisms, as has been shown for euglenoids and other kinetoplastids (3).
Fig. 2.
Phylogeny of Neoparamoeba pemaquidensis CCAP1560/4 and its Ichthyobodo-related organism (IRO). The phylogenetic tree shown was constructed using maximum likelihood (ML) from a concatenated data set including the alpha-tubulin, beta-tubulin, elongation factor 2, and heat shock protein 90 proteins (Rpb1 and Rpa1 sequences were excluded due to the partial nature of the N. pemaquidensis and IRO sequences). ML analysis was performed using the RAxML software program, ver. 7.0.4 (33), with the WAG+Gamma+I model, and bootstrap analysis was based on 1,000 replicates with the CAT model. The amino acid alignment consisted of 31 operational taxonomic units (OTUs) and 942 unambiguously aligned positions. The tree was rooted arbitrarily. Bootstrap support values are shown if > 70%. The scale bar indicates an inferred number of amino acid substitutions per site.
Consideration of the Neoparamoeba host- and IRO-derived sequences revealed interesting differences in base composition. The G+C content of the IRO genes was 51.0 to 65.5% for the protein coding genes sequenced in our study, and the previously sequenced ribosomal intergenic spacer regions (ITS1 and -2) showed 68.1% G+C when considered together. In stark contrast, sequences derived from the Neoparamoeba host were 49.8 to 55.2% G+C in protein-coding genes and were much lower overall in G+C content, especially in noncoding regions, such as the internal transcribed spacer (ITS) (21.5%). Although low G+C content is often seen in endosymbiont and organellar genomes, the N. pemaquidensis host and IRO genomes exhibit the opposite pattern.
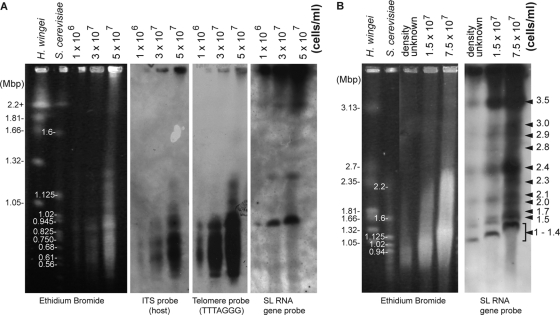
In addition to changes in base composition, adaptation to a life of intracellularity often leads to a reduction in genome size. This phenomenon is commonly seen in plastids and mitochondria (15, 32), bacterial endosymbionts, such as Buchnera (25, 26, 30), and eukaryotic intracellular pathogens, such as microsporidia (7, 8, 35). A strong signal for coevolution of Neoparamoeba-IRO pairs is clear from analyses of rDNA sequences (6, 11), and given that Neoparamoeba is invariably found with at least one IRO residing within it, the relationship between the two is very likely obligate (6). How big is the IRO nuclear genome, and where does this enigmatic organism reside along the continuum between transient endosymbiont and organelle? To address this question, we used pulsed-field gel electrophoresis (PFGE) to investigate the structure of the host and endosymbiont genomes of N. pemaquidensis. PFGE plugs were prepared using the general procedure described by Tanifuji et al. (34), with N. pemaquidensis prey bacteria being removed by filtration with a 3-μm pore polycarbonate membrane (Sterlitech, Kent, WA) prior to plug formation. A strong “smear” of similarly sized chromosomes in the range of ∼0.45 to 1 Mbp was apparent under a variety of different PFGE conditions and ethidium bromide stainings and with different PFGE plug concentrations (Fig. 3A and B). In order to differentially target host and IRO chromosomes, nonradioactive Southern hybridization analyses were performed using digoxigenin (DIG)-labeled probes against the predicted telomeric repeat of the N. pemaquidensis host [(TTTAGGG)n], the host rDNA ITS, and the SL RNA gene of the IRO. Southern hybridizations using host ITS and telomere probes detected strong signals between 0.45 and 1.2 Mbp, a pattern that was also seen with a host 28S rDNA probe (data not shown). These results suggest that the host genome contains many chromosomes in the 0.45- to 1.2-Mbp range.
Fig. 3.
Karyotype analysis of Neoparamoeba pemaquidensis CCAP1560/4 and its IRO. Ethidium bromide-stained pulsed-field gels are shown alongside Southern blots using probes designed against the N. pemaquidensis internal transcribed spacer (ITS) sequence and telomeric repeat (TTTAGGG), as well as the IRO spliced-leader (SL) RNA gene. Southern hybridizations were performed using the Roche DIG System (Roche Diagnostics, Mannheim, Germany). Hansenula wingei and Saccharomyces cerevisiae were used as DNA size markers. Cell densities of DNA plugs of Neoparamoeba pemaquidensis CCAP1560/4 are shown, as are the chromosome sizes of the markers. (A) Pulsed-field gel run with 1% agarose, 0.5× Tris-borate-EDTA (TBE), 14°C, a run time of 24 h at 6 V/cm, 120° angle, and a 60- to 120-s pulse time. (B) A 1% agarose pulsed-field gel, 0.5× TBE, 14°C, 48-h running time, 3 V/cm, 106° angle, and 500-s pulse time.
Chromosomes of the N. pemaquidensis IRO were detected in Southern hybridization analyses using an SL RNA gene probe. Euglenoids and dinoflagellates are known to possess SL RNA genes in high copy number (2, 18, 28, 36), and our results suggest that the same is true for the N. pemaquidensis IRO. To our surprise, hybridizations using the SL RNA gene probe against membranes derived from multiple independent PFGE runs and using various plug cell concentrations revealed a total of 11 distinct hybridizing bands between ∼1.0 and 3.5 Mbp; particularly strong signals were detected at ∼1.0, 2.4, and 3.5 Mbp (Fig. 3). Taking into account the fact that the chromosomal band migration distance was found to vary depending on PFGE plug cell density (see Fig. 3B; this is often the case in PFGE), these 11 bands total ∼25 Mbp. We consider this to be a tentative absolute lower bound for the size of the N. pemaquidensis IRO nuclear genome. This estimate must be considered in the context of several important caveats, including (i) the likelihood of comigrating chromosomes in our PFGE analyses, (ii) interchromosomal variation in the SL RNA gene copy number (and thus variation in hybridization intensity), and (iii) the possibility that not all IRO chromosomes contain an SL RNA gene.
Across the breadth of eukaryotic diversity, nuclear genome size varies tremendously (16), but even free-living organisms, such as the red-alga Cyanidioschyzon merolae and the green alga Ostreococcus tauri, can have very “small” genomes (16.5 Mbp and 12.6 Mbp, respectively) (9, 22). Trypanosomatid haploid genomes are in the range of 35 to 55 Mbp (14), and based on the preliminary data presented herein, it is possible that the IRO genome is similar in size. Our base composition analyses are also significant. A reduction in G+C content is also a hallmark feature of many (but not all) reduced genomes (15, 23, 24, 26, 29, 32), but our newly determined IRO protein gene sequences exhibit a G+C content between 55% and 65%, similar to that seen in other kinetoplastid taxa, including the free-living bodonid Bodo saltans (19). In fact, the rDNA ITS of the N. pemaquidensis IRO is 68.1% G+C over 188 bp, more G+C rich than the ITS of the prokinetoplastid Ichthyobodo necator (40.8% G+C) and the metakinetoplastids Bodo caudatus (49.2% G+C), Trypanosoma brucei, and Leishmania major (33.6 to 49.0%) (2, 4, 18). In sum, while the N. pemaquidensis IRO nuclear genome is very likely not “large,” there is currently no evidence to suggest that it has undergone significant reductive evolution in response to what is clearly a life of obligate intracellularity. In concert with investigation of the Neoparamoeba nuclear genome, determining the sequence of the IRO genome will provide a framework for better understanding Neoparamoeba pathogenesis and the nature of this interesting host-endosymbiont relationship.
Nucleotide sequence accession numbers.
Genes of Neoparamoeba and the IRO, encoding the following proteins, were sequenced: alpha-tubulin, beta-tubulin, elongation factor 2, heat shock protein 90, an RNA polymerase I subunit, and an RNA polymerase II subunit; GenBank accession numbers are AB505628 to JF262536 to -42, JF262544 to -53, JF419568 to -72, FJ706693 to -98, and JF706718. Sequence of the SL RNA gene from the IRO genome has been deposited in GenBank under accession numbers JF262543 and JF441171.
Acknowledgments
This work was supported by an operating grant (ROP85016) from the Canadian Institutes of Health Research (CIHR) regional Partnership Program, together with the Nova Scotia Health Research Foundation (NSHRF). G.T. and E.K. are supported by the Tula Foundation. J.M.A. holds a CIHR New Investigator Award, and J.M.A. and J.L. are Fellows of the Canadian Institute for Advanced Research, Program in Integrated Microbial Biodiversity. J.L. is supported by the Czech Ministry of Education and the Praemium Academiae award.
We thank Mary Ann Trevors for assistance with TEM preparation.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Barrett M. P., et al. 2003. The trypanosomiases. Lancet 362:1469–1480 [DOI] [PubMed] [Google Scholar]
- 2. Berriman M., et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422 [DOI] [PubMed] [Google Scholar]
- 3. Breckenridge D. G., Watanabe Y., Greenwood S. J., Gray M. W., Schnare M. N. 1999. U1 small nuclear RNA and spliceosomal introns in Euglena gracilis. Proc. Natl. Acad. Sci. U. S. A. 96:852–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Callahan H. A., Litaker R. W., Noga E. J. 2002. Molecular taxonomy of the suborder Bodonina (order Kinetoplastida), including the important fish parasite, Ichthyobodo necator. J. Eukaryot. Microbiol. 49:119–128 [DOI] [PubMed] [Google Scholar]
- 5. Campbell D. A., Sturm N. R., Yu M. C. 2000. Transcription of the kinetoplastid spliced leader RNA gene. Parasitol. Today. 16:78–82 [DOI] [PubMed] [Google Scholar]
- 6. Caraguel C. G. B., et al. 2007. Microheterogeneity and coevolution: an examination of rDNA sequence characteristics in Neoparamoeba pemaquidensis and its prokinetoplastid endosymbiont. J. Eukaryot. Microbiol. 54:418–426 [DOI] [PubMed] [Google Scholar]
- 7. Cornman R. S., et al. 2009. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. Plos. Pathog. 5:e1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corradi N., Pombert J. F., Farinelli L., Didier E. S., Keeling P. J. 2010. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat. Commun. 1:77 doi: 10.1038/ncomms1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Derelle E., et al. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U. S. A. 103:11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dyková I., Fiala I., Lom J., Lukeš J. 2003. Perkinsiella amoebae-like endosymbionts of Neoparamoeba spp., relatives of the kinetoplastid Ichthyobodo. Eur. J. Protistol. 39:37–52 [Google Scholar]
- 11. Dyková I., Fiala I., Pecková H. 2008. Neoparamoeba spp. and their eukaryotic endosymbionts similar to Perkinsela amoebae (Hollande, 1980): coevolution demonstrated by SSU rRNA gene phylogenies. Eur. J. Protistol. 44:269–277 [DOI] [PubMed] [Google Scholar]
- 12. Dyková I., Figueras A., Peric Z. 2000. Neoparamoeba Page, 1987: light and electron microscopic observations on six strains of different origin. Dis. Aquat. Organ. 43:217–223 [DOI] [PubMed] [Google Scholar]
- 13. Dyková I., et al. 2005. Neoparamoeba branchiphila n. sp., and related species of the genus Neoparamoeba Page, 1987: morphological and molecular characterization of selected strains. J. Fish Dis. 28:49–64 [DOI] [PubMed] [Google Scholar]
- 14. El-Sayed N. M., et al. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404–409 [DOI] [PubMed] [Google Scholar]
- 15. Gray M. W., Lang B. F., Burger G. 2004. Mitochondria of protists. Annu. Rev. Genet. 38:477–524 [DOI] [PubMed] [Google Scholar]
- 16. Gregory T. R. 2005. The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Ann. Bot. (Lond.) 95:133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hollande A. 1980. Identification du parasome (Nebenkern) de Janickina pigmentifera a un symbionte (Perkinsella amoebae nov gen-nov sp.) apparente aux flagelles kinetoplastidies. Protistologica 16:613–625 [Google Scholar]
- 18. Ivens A. C., et al. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackson A. P., Quail M. A., Berriman M. 2008. Insights into the genome sequence of a free-living kinetoplastid: Bodo saltans (Kinetoplastida: Euglenozoa). BMC Genomics 9:594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee L. E. J., et al. 2006. High yield and rapid growth of Neoparamoeba pemaquidensis in co-culture with a rainbow trout gill-derived cell line RTgill-W1. J. Fish Dis. 29:467–480 [DOI] [PubMed] [Google Scholar]
- 21. Lukeš J., et al. 2002. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell 1:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsuzaki M., Misumi O., Shin T., et al. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653–657 [DOI] [PubMed] [Google Scholar]
- 23. McCutcheon J. P., McDonald B. R., Moran N. A. 2009. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. Plos. Genet. 5:e1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore C. E., Archibald J. M. 2009. Nucleomorph genomes. Annu. Rev. Genet. 43:251–264 [DOI] [PubMed] [Google Scholar]
- 25. Moran N. A., McCutcheon J. P., Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190 [DOI] [PubMed] [Google Scholar]
- 26. Moran N. A., McLaughlin H. J., Sorek R. 2009. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323:379–382 [DOI] [PubMed] [Google Scholar]
- 27. Mullen T. E., et al. 2004. Paramoebiasis associated with mass mortality of American lobster Homarus americanus in Long Island Sound, USA. J. Aquat. Anim. Health 16:29–38 [Google Scholar]
- 28. Murthy V. K., Dibbern K. M., Campbell D. A. 1992. PCR amplification of mini-exon genes differentiates Trypanosoma cruzi from Trypanosoma rangeli. Mol. Cell Probe 6:237–243 [DOI] [PubMed] [Google Scholar]
- 29. Nakabachi A., et al. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267. [DOI] [PubMed] [Google Scholar]
- 30. Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86 [DOI] [PubMed] [Google Scholar]
- 31. Simpson A. G. B., Stevens J. R., Lukeš J. 2006. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 22:168–174 [DOI] [PubMed] [Google Scholar]
- 32. Smith D. R. 2009. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol. Biol. 71:627–639 [DOI] [PubMed] [Google Scholar]
- 33. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 34. Tanifuji G., Erata M., Ishida K., Onodera N., Hara Y. 2006. Diversity of secondary endosymbiont-derived actin-coding genes in cryptomonads and their evolutionary implications. J. Plant Res. 119:205–215 [DOI] [PubMed] [Google Scholar]
- 35. Texier C., Vidau C., Vigues B., El Alaoui H., Delbac F. 2010. Microsporidia: a model for minimal parasite-host interactions. Curr. Opin. Microbiol. 13:443–449 [DOI] [PubMed] [Google Scholar]
- 36. Zhang H., et al. 2007. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. U. S. A. 104:4618–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]