Abstract
The Ras-like GTPase Rab11 is implicated in multiple aspects of intracellular transport, including maintenance of plasma membrane composition and cytokinesis. In metazoans, these functions are mediated in part via coiled-coil Rab11-interacting proteins (FIPs) acting as Rab11 effectors. Additional interaction between Rab11 and the exocyst subunit Sec15 connects Rab11 with exocytosis. We find that FIPs are metazoan specific, suggesting that other factors mediate Rab11 functions in nonmetazoans. We examined Rab11 interactions in Trypanosoma brucei, where endocytosis is well studied and the role of Rab11 in recycling well documented. TbSec15 and TbRab11 interact, demonstrating evolutionary conservation. By yeast two-hybrid screening, we identified additional Rab11 interaction partners. Tb927.5.1640 (designated RBP74) interacted with both Rab11 and Rab5. RBP74 shares a coiled-coil architecture with metazoan FIPs but is unrelated by sequence and appears to play a role in coordinating endocytosis and recycling. A second coiled-coil protein, Tb09.211.4830 (TbAZI1), orthologous to AZI1 in Homo sapiens, interacts exclusively with Rab11. AZI1 is restricted to taxa with motile cilia/flagella. These data suggest that Rab11 functions are mediated by evolutionarily conserved (i.e., AZI1 and Sec15) and potentially lineage-specific (RBP74) interactions essential for the integration of the endomembrane system.
INTRODUCTION
Trafficking of molecules to and from the cell surface is a critical cellular function (52). Multiple endocytic recycling pathways are present in metazoans and fungi (16). Endocytic cargo is targeted to multiple destinations, and two distinct routes for return of proteins to the plasma membrane via recycling pathways are known: a Rab4-dependent constitutive route for rapid recycling (8, 53, 75) and a separate Rab11-dependent pericentriolar pathway with slower kinetics (81). Trafficking through the Rab11 pathway is important for sorting biosynthetic and endocytosed material (43, 79, 84), but Rab11 is also a key mediator of additional transport pathways. In particular, the importance of Rab11 to maintaining cell polarity has been demonstrated in several studies. For example, in mammalian cells Rab11 isoforms regulate transcytosis of IgA in epithelia (17), delivery of material to the cleavage furrow during cytokinesis (30, 72), cell migration by promoting integrin transport to specialized domains within the plasma membrane (45), and ciliogenesis as part of the ciliary targeting complex (59). This high degree of functional complexity is achieved by mutually exclusive recruitment of a range of Rab11 effector proteins (62, 69). It remains unclear how these additional roles are related to the central role the GTPase plays in endocytic recycling (78).
With the exceptions of myosin Vb and Sec15, a component of the exocyst complex, known Rab11 effectors in mammalian cells belong to a family of FIPs, defined by the presence of a conserved C-terminal Rab11-binding box and extensive coiled-coil segments mediating formation of helical dimers. FIPs provide a platform for recruitment of additional endocytic factors (35, 80). This macromolecular assemblage facilitates interactions of Rab11 with multiple aspects of trafficking (reviewed in references 46 and 70). In particular, several studies suggest that the two class I FIPs, Rip11/FIP5 and FIP2, are involved in selective sorting and targeting of a subpopulation of plasma membrane receptors through pericentriolar recycling endosomes (9, 17, 18, 56, 68). Both may act via regulating vesicle binding to the correct molecular motor, coordinating traffic of cargo along microtubule and actin cytoskeletal elements (36, 51, 74). The class II effectors, FIP3 and FIP4, or Arfophilins, act cooperatively with Arf and Rab11 GTPases to coordinate actin remodeling and the delivery of new material to the cleavage furrow during cytokinesis (40, 41, 85).
Although Rab4 and Rab11 are present across representative organisms of all eukaryotic supergroups and hence were present in the LECA, Rab4 is prone to secondary loss and many effectors of small GTPases are poorly conserved (22). It is possible that Rab effector diversity forms the basis for functional specialization among Rab11 pathways in different organisms and could provide considerable differentiation in recycling mechanisms between taxa, despite the near-universal retention of Rab11. Alternatively, poor sequence conservation between orthologues may confound identification of relationships by in silico approaches, especially for divergent taxa. Here, we used the protozoan Trypanosoma brucei, a member of the eukaryotic Excavata supergroup, to gain further insight into Rab11 function in divergent organisms and to assess the levels of divergence and similarity across eukaryotes.
T. brucei is an extracellular parasite infecting many mammalian hosts, and maintenance of the plasma membrane composition is essential for survival (2, 35, 39). Changes to the cell surface accompany adaptation to environmental challenges, including alternating hosts. The molecular mechanisms underlying this adaptation involve life-cycle-dependent differentiation of the endocytic system (33, 49, 64). Unusually, endocytosis is AP-2 independent and exclusively clathrin and Rab5 dependent (2, 22, 35, 38). All recycling in the bloodstream-form trypanosome is Rab11 dependent and, unlike what is seen for metazoans, does not involve Rab4 to a major degree (33, 35, 39). While trypanosome Rab11 is essential (39), it is unclear how Rab11 integrates with the endocytic system or if it participates in the full range of processes described in higher eukaryotes.
One possible approach to understanding the molecular mechanisms behind Rab11 function is to characterize the factors with which it interacts. We used a combination of in silico and yeast two-hybrid screening strategies to identify trypanosome Rab11 effectors and demonstrated both evolutionarily conserved and novel interactions.
MATERIALS AND METHODS
Abbreviations.
AP-2, adaptor complex-2; AZI1, 5-azacytidine-induced 1; BSA, bovine serum albumin; BSF, bloodstream form; CCD, charge-coupled device; ConA, concanavalin A; DAPI, 4′,6-diamidino-2-phenylindole; ER, endoplasmic reticulum; FACS, fluorescence-activated cell sorting; FIP, Rab11-family interacting protein; FITC, fluorescein isothiocyanate; GFP, green fluorescent protein; HA, hemagglutinin; LECA, last eukaryotic common ancestor; PBS, phosphate-buffered saline; PCF, procyclic form; PFA, paraformaldehyde; PFR, paraflagellar rod; RBD, Rab11-binding domain; RBP74, Rab11-binding protein of 74 kDa; RNAi, RNA interference; RT-PCR, reverse transcriptase-PCR; SD, synthetic defined; SMB, single-marker bloodstream form; TGN, trans-Golgi network; vPBS, Voorheis's modified PBS; VSG, variant surface glycoprotein; YFP, yellow fluorescent protein.
Sequence homology searches.
For the present study, we selected representative taxa of five of the six major eukaryotic supergroups. We restricted our BLAST analyses to completed genomes, and when possible we included organisms with and without flagella/cilia. Homo sapiens data were obtained from NCBI (www.ncbi.nlm.nih.gov). Drosophila melanogaster data were from FlyBase (www.flybase.org), Caenorhabditis elegans data were obtained from WormBase (www.wormbase.org). Nematostella vectensis, Monosiga brevicollis, Chlamydomonas reinhardtii, Ostreococcus tauri, Thalassiosira pseudonana, Phytophthora ramorum, and Naegleria gruberi data were obtained from the Joint Genome Initiative (genome.jgi-psf.org). Arabidopsis thaliana, Cryptococcus neoformans, Theileria parva, Tetrahymena thermophila, and Trichomonas vaginalis data were from TIGR (www.tigr.org). Dictyostelium discoideum, Entamoeba histolytica, Plasmodium falciparum, Eimeria tenella, Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major data were obtained from geneDB (www.genedb.org). Toxoplasma gondii data were from ToxoDB (www.toxodb.org), Cryptosporidium parvum data were from CryptoDB (www.cryptodb.org), and Cyanidioschyzon merolae data were retrieved from the C. merolae genome BLAST server (merolae.biol.s.u-tokyo.ac.jp). Paramecium tetraurelia data were from the Paramecium database (paramecium.cgm.cnrs-gif.fr/). Saccharomyces cerevisiae data were from the Saccharomyces Genome Database (www.yeastgenome.org/), and Batrachochytrium dendrobatidis data were from the Broad Institute (www.broadinstitute.org/annotation/genome/batrachochytrium_dendrobatidis).
Cells and routine culture.
T. brucei BSF and PCF cells were routinely cultured in HMI9 and SDM79 media, respectively, supplemented with 10% fetal bovine serum and antibiotics as described previously (19).
Yeast two-hybrid screening of a T. brucei genomic library.
As a bait for the screen, the dominant-active GTP-locked mutant form of Rab11, Rab11Q66L, was amplified from a pXS5 construct containing Rab11QL using the primers R11F1 (AGTCGAATTCATGGAAGACATGAACCTTACG) and R11R1 (CGTAGGATCCTTAACAGCACCCGCCACTCGCCTTTCC) (67) and subcloned into the pGBKT7 plasmid of the Matchmaker system (Clontech). The pGBKT7-Rab11QL construct was used to transform AH109 Saccharomyces cerevisiae. The transcriptional activity of the bait was tested by growth in SD −Trp/−His medium, and no transcriptional activity was detected in these conditions. A T. brucei genomic library (kind gift of Ralph Schwarz, Marburg, Germany) was cloned into pGADT7 and screened by transformation of AH109 yeast expressing pGBKT7-Rab11QL. Transformants were plated on SD −Trp/−Leu/−His medium. After incubation for a period of 72 to 96 h at 30°C, colonies were recovered and DNA from each positive clone was extracted and sequenced. In order to eliminate false positives, isolated library prey plasmids were transformed into Y187 yeast and crossed with AH109 yeast, carrying either the empty plasmid or the bait plasmid. Activation of the reporter gene was assessed according to growth in SD −Trp/−Leu/−His or SD −Trp/−Leu/−His/−Ade medium.
Yeast two-hybrid mating assays.
QL mutant isoforms of members of the trypanosome Rab family were cloned into the bait plasmid pGBKT7. The following full-length and truncated versions chosen for convenient restriction sites were prepared for the positive library clones: RBP74 was amplified using the primers RBP74F1 (ATATGAATTCATGCGCCCCAAC) and RBP74R1 (ATCGGGATCCTCAGTAGGTTGTG) and cloned into pGADT7 and pGBKT7; RBP74 (residues 234 to 532), N-terminal RBP74 (residues 1 to 453), and the C-terminal fragment (residues 532 to 663) were subcloned from pGBKT7-RBP74 into pGADT7; TbAZI1 was amplified using the primers TbAZI1F1 (GCTAGAATTCTTTGGCATGGATG) and TbAZI1R1 (GTAAGGATCCGTGTCGCAACATCC) and cloned into pGADT7 and pGBKT7; and a C-terminal TbAZI1 fragment (residues 328 to 660) was subcloned from pGBKT7-TbAZI1 into pGADT7. A C-terminal fragment of TbSec15 (residues 205 to 1003) was amplified using the following primers: Sec15F1 (ATTTGAATTCGACGGGGAACAGAGTAGC) and Sec15R1 (TTCCCTCGAGCTATAAAGTTCGCTTTAGC) and cloned into pGADT7. AH109 yeast transformed with bait constructs were crossed with Y187 yeast expressing the prey constructs. Diploids were grown on double-dropout medium and transferred to SD −Trp/−Leu/−His medium. Plates displaying colony growth were scanned, and images were prepared with Adobe Photoshop.
In vitro coimmunoprecipitation.
For immunoprecipitation assays, we used the full-length RBP74 and TbAZI1 cloned into the pGADT7 plasmid (as described above) in frame and C terminal to an HA tag, downstream of the T7 promoter. Full-length QL mutants TbRab1AQL, TbRab5AQL, and TbRab11QL cloned into the pGBKT7 plasmid in frame and C terminally to a myc tag, downstream of the T7 promoter, were prepared as described above. 35S-methionine-labeled and HA-tagged RBP74 and TbAZI1, and 35S-methionine-labeled myc-tagged TbRab proteins were in vitro synthesized using the TNT-coupled transcription/translation kit (Promega). The in vitro-translated proteins were used for coimmunoprecipitation assays to demonstrate direct interactions. Equivalent volumes of HA-tagged TbRab11 and TbAZI1 effectors were mixed with myc-tagged TbRab1AQL, TbRab5AQL, or TbRab11QL and incubated with monoclonal anti-HA antibody for 1 h at ambient temperature. Protein A agarose beads were added to the mixture and incubation continued for another 1 h. The beads were washed three times with 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100 supplemented with complete protease inhibitor cocktail (Roche). Bound material was eluted by boiling in Laemmli loading buffer for SDS-PAGE analysis, and detection was by autoradiography.
Plasmids and transfections.
Full-length RBP74, TbAZI1, and TbSec15 were amplified by PCR using the following primers: RBP74F2 (CTTTAAGCTTCGCGCCCCAACTCG), RBP74R2 (CATAGGATCCCCTCGCATTGAAGG), TbAZI1F2 (ATCGAAGCTTAAGTTTGGCATGG), TbAZI1R2 (CTTAGGATCCCAATGTATCTATCGG), Sec15F2 (CATTAAGCTTTGGGGCGGATAGAGG), and Sec15R2 (TTCCCTCGAGCTATAAAGTTCGCTTTAGC). TbAZI1 N-terminal (residues 1 to 328) and C-terminal (residues 328 to 660) fragments were generated by subcloning from pGBKT7-TbAZI1. Fragments were inserted downstream of a YFP tag or two HA tags in the plasmids pHD1034 and pXS519. An Amaxa nucleofector was used to transfect BSF cells with NotI-digested pHD1034-RBP74 and pHD1034-TbAZI1 constructs and XhoI-digested pXS519-TbSec15p following the manufacturer's procedure (49). Cells expressing the YFP-tagged or HA-tagged recombinant proteins were selected in the presence of either 0.2 μg ml−1 puromycin or 2.5 μg ml−1 Geneticin for pHD1034 constructs or pXS519 constructs, respectively.
RNA interference.
A 414-bp fragment of Tb927.5.1640 (RBP74) and a 502-bp fragment of Tb09.211.4830 (TbAZI1) were selected and verified using RNAit software to knock down Tb09.211.4830 and Tb927.4.1640 gene products by using RNAi (71). These fragments were PCR amplified from T. brucei genomic DNA using the following primers: RBP74RNAiF (CGTACTCGAGCTCAAAGAAGGC), RBP74R (CATTGGATCCCACGTCATTGAGC), TbAZI1RNAiF (CTTACTCGAGGCTTCATGTCTTACG), and TbAZI1R (CGTAGGATCCGCTCTAACGACTGG). The PCR products were inserted into the p2T7TABlue plasmid and p2T7177 plasmid. Tetracycline-responsive SMB cells were transfected with NotI-digested p2T7TA-RBP74 and p2T7TA-TbAZI1 constructs by using an Amaxa nucleofector (24). PTT procyclic cells were transfected with NotI-digested p2T7177-TbAZI1 (24). Single clones were selected in the presence of 5 μg ml−1 hygromycin and 2.5 μg ml−1 Geneticin in bloodstream forms or in the presence of 25 μg ml−1 hygromycin, 25 μg ml−1 Geneticin, and 5 μg ml−1 phleomycin for procyclic forms. For growth curves, cultures were carried out in triplicate in the presence or absence of tetracycline at 1 μg ml−1. Cell density was measured using a Z2 Coulter Counter (Beckman). All PCR clones and fusions used in this study were confirmed by DNA sequencing.
Quantitative real-time RT-PCR.
We employed real-time RT-PCR to quantify a change in expression levels of RBP74 and TbAZI1 mRNA and to validate the RNAi cell lines. RNA of bloodstream- and procyclic-form Lister 427 cells or the RBP74 and TbAZI1 RNAi cell lines were prepared using the RNeasy kit (Qiagen). To measure transcript levels of RBP74 and TbAZI1, quantitative RT-PCR was carried out as described elsewhere (49) using the following primers; RBP74QRTF (GACGCTGCAAAACAACTTGA), RBP74QRTR (CCTTTCGTAACGATGCAAGC), TbAZI1QRTF (GGATGCATGTGCTAGATTCG), and TbAZI1QRTR (AGCCTGGAGCTCTATGGTGA).
Western blotting.
For protein electrophoresis, lysates of 106 to 107 cells grown to log phase were used. SDS-PAGE and Western analysis were performed as described previously. Primary antibody working dilutions were 1:10,000 for mouse anti-HA (Santa Cruz Biotechnology), 1:2,000 for rabbit anti-TbRab11 antibodies, and 1:10,000 for rabbit anti-BiP antibody (kindly provided by James D. Bangs). Detection was carried out following standard procedures.
Subcellular fractionation.
Cells expressing HA-TbAZI1 were grown to log phase, washed twice in PBS, and resuspended in cold hypotonic lysis buffer (10 mM Tris-HCl [pH 7.5] plus protease inhibitors). Cell lysates were obtained by incubating cells on ice for 5 min and later centrifuged at 14,000 rpm at 4°C for 10 min to separate membrane-associated from soluble material. The pellet was further washed with cold hypotonic lysis buffer and resuspended in sample buffer. Supernatant and pellet fractions were equally loaded on an SDS-PAGE gel and analyzed by Western blotting as described above.
Immunofluorescence.
Log-phase cells were fixed in 3% PFA in vPBS, adhered to poly-l-lysine slides (Sigma), and permeabilized with 0.1% Triton X-100. Cells were blocked in PBS with 20% fetal bovine serum, and slides were incubated with relevant antibodies and mounted with Vectashield solution as described elsewhere. The dilutions used for primary antibodies were as follows: 1:1,000 for mouse anti-HA, 1:250 for TbRab5A (20) and TbRab11A (44), and 1:50 for mouse anti-BBA4, mouse anti-L8C4, and mouse L3B2 (kindly provided by Keith Gull, University of Oxford). Cells were analyzed with either a standard wide-field Nikon Eclipse with a Hamamatsu CCD camera or a Leica TCS-NT confocal microscope with a 100×/1.4 numerical aperture objective, both with optically matched blocks. Immunofluorescence staining images were acquired with Metamorph software (Molecular Devices, V.6) or Leica confocal software (Leica Microsystems) and processed in Photoshop (Adobe).
Concanavalin A staining of the flagellar pocket.
Cells expressing HA-TbAZI1 were grown to log phase and washed once with serum-free HMI-9 medium. Cells were then resuspended to a density of 107 cells ml−1 and incubated at 4°C for 20 min in serum-depleted medium supplemented with 1% BSA. To label the flagellar pocket, FITC-conjugated ConA was added to a final concentration of 0.1 mg ml−1 (Vectalabs), and cells were incubated for a further 20 min at 4°C. Samples were further processed for immunofluorescence analysis as described above.
Concanavalin A and transferrin endocytosis.
Cells transfected with the p2T7-TbAZI1 RNAi plasmid were grown to logarithmic phase and induced for 48 h; as a control, we used uninduced cultures. Cells were then analyzed for concanavalin A and transferrin uptake by FACS as described elsewhere (27).
RESULTS
Sec15-Rab11 interaction and subcellular localization are conserved.
We attempted to identify evolutionarily conserved Rab11-interacting proteins by comparative genomics. However, BLAST analyses of complete genomes (see Materials and Methods) representing species from all major eukaryotic supergroups retrieved orthologues only of members of the FIP family of Rab11-interacting proteins in Metazoa (data not shown). Proteins comprising this family are structurally diverse and share only a Rab11 RBD (pfam: PF09311) at the C terminus. We next asked if the FIP RBD occurred independently in additional endocytic factors by running hmmsearch (hmmer .janelia.org) with pfam RBD domain against predicted proteomes encompassing all major eukaryotic supergroups with fully sequenced genomes. This approach detected only the RBD in previously identified Rab11 effectors, and again these were restricted to the Metazoa (see Table S1 in the supplemental material). Thus, the FIP family of Rab11-interacting proteins is probably metazoan specific. Likewise, class V myosins are restricted to Metazoa and fungi (25, 66). These data indicate that some Rab11 effectors are not conserved, suggesting evolutionary plasticity within the Rab11 system (12).
In contrast to the restricted occurrence of FIP factors, comparative genomics has identified an ancestral endocytic factor core system (10, 11, 54) including Sec15, a component of the exocyst complex. Sec15 interacts directly with Rab11 in mammalian cells and Drosophila melanogaster (50, 90) and was likely present in the LECA (48). To determine if an interaction between Sec15 and Rab11 was conserved, we analyzed Sec15-Rab11 binding in T. brucei by using the yeast two-hybrid system. A fragment (residues 205 to 1003) of Tb11.02.4970, the trypanosome Sec15 orthologue, was cloned downstream of the GAL4 activation domain in the pGADT7 vector and introduced into Y187 yeast cells (Fig. 1A). Diploid cells expressing combinations of bait and prey proteins were produced by crossing Y187 cells expressing Sec15 with AH109 yeast cells containing either the Rab11QL or Rab11SN isoform fused to the DNA-binding domain. To control for transcriptional activity, we used the DNA-binding and activation domains alone. Similar to its mammalian counterpart, Sec15 in T. brucei was found to interact with Rab11, and this interaction was specific to the QL form of the GTPase, i.e., the GTP-bound conformation, as no growth in selective media was detected in cells expressing the GDP-bound form Rab11SN (Fig. 1A).
Fig. 1.
Sec15 and Rab11 homologues interact in T. brucei. (A) A 799-amino-acid fragment encompassing an N-terminal deletion of TbSec15 was expressed downstream of the activation domain (AD) in a yeast two-hybrid system. A mating yeast assay of TbSec15 against Rab11QL or Rab11SN (GTP and GDP locked configurations), fused to the binding domain (BD) in the yeast two-hybrid system, indicates a specific interaction of TbSec15 with the QL dominant-active GTP form of Rab11. (B) Procyclic-form parasites expressing GFP-tagged TbSec15 were fixed and prepared for immunofluorescence analysis using anti-GFP antibody. Left, merge of TbSec15-GFP (green) and DAPI fluorescence (blue); right, fluorescence channels merged with the transmitted light image. TbSec15-GFP distributes next to the flagellar pocket, the only site for exocytosis in T. brucei. Note the presence of a “C”-shaped profile for TbSec15, which is observed in the majority of stained cells. Bar, 2 μm. (C) Location of YFP-TbSec15 in bloodstream-form parasites compared to the locations of endosomes by immunostaining for Rab11 and Rab5. YFP-TbSec15 is visualized in green (white arrows), and rabbit polyclonal antibodies against Rab11 (left) and Rab5 (right) are in red. DNA was visualized with DAPI (blue, right panels, merge). Bar, 2 μm.
We next addressed the subcellular location of Sec15. We expressed an N-terminal YFP-Sec15 chimera in bloodstream-form cells where endocytosis is most active (49, 64). YFP-Sec15 intimately associated with the flagellar pocket, the only site for endocytosis and exocytosis in this organism (Fig. 1C). This distribution is consistent with that of the exocyst in mammalian cells and yeast, to sites of active exocytosis at the plasma membrane, and also encompasses the region of the trypanosome cell containing most Rab11 (reviewed in reference 42). Juxtaposition of Rab11 and Sec15 location was confirmed by immunofluorescence (Fig. 1C). The degree of overlap was somewhat variable (Fig. 1C), but 78% (n > 100) of cells exhibited contact between the immunofluorescence patterns of the two proteins. Significantly, there was no contact or overlap with Rab5, suggesting a presence of Sec15 at sites where recycling endosomes, but not early endosomes, are located (Fig. 1C). Further, a YFP C-terminal genomically tagged version of Sec15 in procyclic cells, which were expressed at endogenous levels, revealed a clear crescent-shaped localization, suggesting that it may be localized to substantial portions of the cytoplasmic face of the trypanosome flagellar pocket (Fig. 1B). We would not anticipate a high degree of colocalization for Rab11 and Sec15, as both proteins are known to play major roles in additional functions, i.e., Sec15 in exocytosis and Rab11 in both recycling and sorting pathways, while Rab interactions in many systems are known to be transient. Likely, these two proteins collaborate only briefly, potentially at the point where Rab11 endosomes dock and fuse with the flagellar pocket membrane, intersecting with the exocytic pathway.
Analysis of YFP-Sec15 location at different points in the cell cycle shows that during the course of cell division a subpopulation of Sec15 is also found at the new flagellar pocket, suggesting either that exocytosis is active in the newly formed flagellar pocket or that TbSec15 functions in flagellum biogenesis, as recently suggested for mammalian cells (59) (Fig. 1B). Overall, these data indicate that the Rab11-Sec15 interaction and localization is conserved between trypanosomes and Metazoa.
Identification of additional Rab11-interacting proteins, RBP74 and TbAZI1.
To identify additional trypanosome Rab11 effectors, we performed a yeast two-hybrid screen using a T. brucei genomic library. We previously demonstrated that the Q66L mutation within the G3 (82) motif of the GTP binding site of Rab11 acts as a dominant-active mutant in recycling of surface immune complexes and transferrin in vivo (67). We therefore used the Rab11Q66L mutant as bait to isolate proteins that specifically interacted with the GTP-bound form of Rab11. We verified expression of trypanosome Rab11 in yeast cells by Western blotting (see Fig. S1 in the supplemental material). The empty vector displayed no Rab11 expression, as expected (data not shown). In screening ∼2 × 106 transformants, we obtained five positive clones. One of these clones comprised a 1,683-nucleotide fragment corresponding to the Tb927.5.1640 open reading frame and spanning amino acids 103 to 664. Two further clones carried identical 964-nucleotide fragments from the Tb09.211.4830 open reading frame, spanning amino acids 219 to 540. The final two clones incorporated the same 1,023-nucleotide fragment encoding a highly repetitive sequence from open reading frame Tb11.01.2880, which we have previously identified in a proteomics analysis of the nuclear envelope (13). We chose to not pursue this last protein any further. We named Tb927.5.1640 RBP74 for Rab11-binding protein, molecular mass of 74 kDa, and Tb09.211.4830 as TbAZI1 for its homology to AZI1 in Homo sapiens (see Fig. S2B in the supplemental material).
Analysis of the RBP74 and TbAZI1 sequences by using hmmpfam (hmmer.janelia.org) did not reveal any domains with a highly significant score. A relevant Pfam motif with a low score, E-value 0.72, was a Rab5-binding domain in RBP74 spanning residues 248 to 413. This motif is present in members of the Rabaptin family of Rab5-interacting proteins (91), and the significance of this is increased by the observation that the motif is contained within the region mediating interaction with Rab5A (see below).
BLAST searches against representative genomes for all major eukaryotic supergroups only identified a RBP74 homologue in T. cruzi (see Fig. S2A in the supplemental material). The failure to detect homologues in other taxa implies a trypanosomatid-specific function, and also limited representation within kinetoplastida, as it was not detected in Leishmania major. By contrast, TbAZI1 orthologues were confirmed in L. major and T. cruzi and broader searches of genome databases identified AZI1 orthologues in most taxa that retain a flagellar apparatus. We were unable to identify AZI1 orthologues in any plant genomes except the flagellate Chlamydomonas reinhardtii, or in the majority of the fungal group, the exception being Batrachochytrium dendrobatidis, in agreement with the loss of the flagellar apparatus in the early stages of fungal evolution (58). Indeed, AZI1 was identified in several comparative genomic analyses of the C. reinhardtii genome as an uncharacterized gene likely associated with ciliary function and probably related to motility due to its absence from C. elegans, an organism with only sensory cilia (55, 61, 77). Not surprisingly, we also found AZI1 to be missing in species that present extreme variations in the molecular structures found in most flagella (Fig. 2), for example the stramenopile Thalassiosira pseudonana and the malarial parasite Plasmodium falciparum (4). The distribution of AZI1 is similar to the distribution observed for cytoplasmic dynein 2, a motor protein associated with intraflagellar transport (83) and a family of centriolar proteins, Pix, required for assembly only of cilia or flagella that undulate (86).
Fig. 2.
Distribution of TbAZI1 across selected representatives of the Eukaryota. Left, names of organisms used in the analysis, including representatives of each supergroup and, where possible, examples of organisms with a ciliary/flagellar apparatus. See Materials and Methods for sources of these data.
We then attempted to verify the direct interaction of both proteins with Rab11 by in vitro immunoprecipitation, using the GTPase-deficient GTP-bound form, Rab11QL. 35S- methionine-radiolabeled HA-tagged RBP74 and TbAZI1 and myc-tagged Rab11QL were prepared by in vitro transcription/translation. As control binding partners, we also made in vitro-synthesized myc-Rab1AQL and myc-Rab5AQL. Coimmunoprecipitation with anti-HA antibodies confirmed a specific interaction between RBP74 and Rab11AQL (Fig. 3A). Interestingly RBP74 also binds Rab5AQL (Fig. 3A), an early endosomal Rab GTPase whose location is intimately associated with that of Rab11 (44). Further, Rab5A functions upstream of Rab11 within the endosomal/recycling pathways in T. brucei (67). Despite repeated attempts to coimmunoprecipitate TbAZI1 and Rab11QL, we failed to detect an interaction using this approach. Interaction between TbAZI1 and Rab11 was validated functionally later (see below).
Fig. 3.
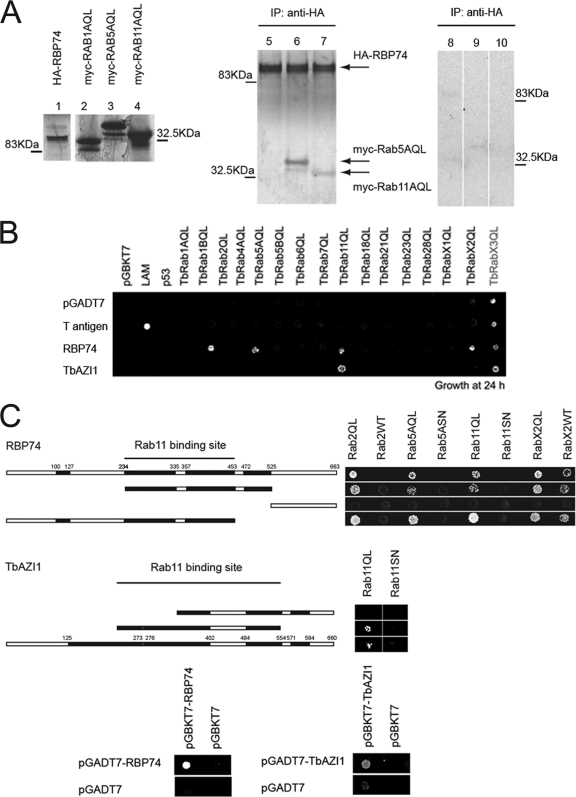
Identification of two novel Rab11-interacting proteins and mapping of the Rab11 interaction domain. (A) Autoradiography of HA-tagged RBP74 alone (lane 1) and myc-tagged Rab1A, Rab5, and Rab11 (lanes 2 to 4), in vitro expressed and labeled with 35S-methionine. Rab5 and Rab11 coimmunoprecipitate (IP) with HA-RBP74 with a mouse anti-HA antibody (lanes 6 and 7, respectively). Rab1A showed no evidence of interaction with HA-TbRBP74 (negative control, lane 5 in autoradiograph). Lanes 8 to 10 are negative controls for nonspecific bead binding of the Rab proteins; myc-Rab1, myc-Rab5, and myc-Rab11, respectively, all immunoprecipitated with anti-HA antibody. Note that these lanes are from the same autoradiogram but are rearranged for display purposes. (B) Schematic representation of the yeast two-hybrid mating assay of RBP74 and TbAZI1 against members of the Rab subfamily from T. brucei. Activation of the reporter gene, HIS, was evaluated by growth of cross-mated yeast under medium-stringency conditions (SD −histidine, −leucine, −tryptophan plates). Gray indicates weak interaction and black strong interactions. (C) Interaction mating assay of yeast expressing fragments of RBP74 (upper panels) and TbAZI1 (lower panels) against selected trypanosome Rabs. Cross-mated diploid yeast cells were grown under the same conditions as in panel A. Growth in triple-dropout medium (−histidine, −leucine, −tryptophan) indicates a positive interaction. Stripes in the schematic represent coiled-coil segments. Bottom panels, Analysis of RBP74 and TbAZI1 homodimerization by yeast two-hybrid mating assay. Yeast expressing RBP74 and TbAZI1 downstream of the DNA-binding domain or the activation domain in the bait and prey vectors were crossed to form diploid cells expressing both bait and prey proteins. Growth under medium-stringency conditions for RBP74 (SD −histidine, −leucine, −tryptophan) (left panels) or high-stringency conditions for TbAZI1 (SD −adenine, −histidine, −leucine, −tryptophan) (right panels) indicates a positive interaction.
The finding that Rab5AQL coimmunoprecipitated with RBP74 raised the possibility that RBP74 and TbAZI1 may bind additional trypanosome Rab proteins, a possibility that is clearly of interest for understanding the coordination between Rab functions in trypanosomes. To directly address this possibility, we used a yeast two-hybrid mating assay for both RBP74 and TbAZI1 against the complete complement of 16 T. brucei Rab and Rab-like GTPases (1). We found that whereas TbAZI1 was specific for Rab11QL, RBP74 also bound Rab5AQL (Fig. 3B), confirming the results obtained in the in vitro coimmunoprecipitation. In addition, RBP74 bound the GTP forms of two ER-Golgi-associated Rab proteins: Rab2QL, which localizes to the cis face of the Golgi complex (15), and RabX2QL, which localizes to the trans-Golgi network (21, 65) (Fig. 3B). Interestingly, Rab2 and Rab11 are part of the same Rab subfamily, which may explain the cross-recognition with RBP74 (1). However, sequence relatedness cannot account for all specificity, as Rab5 and RabX2 are phylogenetically quite distinct from Rab11 and Rab2. The ability of Rab effectors to interact with more than one Rab GTPase within connecting pathways is not unprecedented (26, 57), and the lack of detectable interactions between RBP74 or TbAZI1 and additional Rabs indicates that both proteins are able to discriminate among different members of the Rab family.
Mapping the Rab-binding domain of RBP74 and TbAZI1.
We next sought to map the Rab-binding domain within the amino acid sequences of both effectors. Truncated versions of both proteins were analyzed for their abilities to bind to the panel of Rab GTPases that interacted with the full-length proteins. Yeast two-hybrid interactions showed that the N-terminal deletion mutant of RBP74 (containing residues 532 to 663) failed to interact with any of the tested GTPases. Conversely, RBP74 constructs encompassing residues 1 to 453 or residues 234 to 532 displayed specificity for the dominant-active forms of the Rab proteins with which RBP74 interacts. Some binding to wild-type RabX2 was also observed (Fig. 3C, upper panels). Likewise, while full-length TbAZI1 and the library fragment spanning residues 219 to 540 showed specificity for the active form of Rab11, deletion of the N-terminal region abolished this interaction (Fig. 3C, lower panels). Significantly, the association is GTP dependent for both RBP74 and TbAZI1, as neither was able to bind the GDP forms of its respective GTPase partners (Fig. 3C). There is no evidence for homology within the Rab-binding regions of TbAZI1 and RBP74, which suggests that they interact via distinct mechanisms.
RBP74 and TbAZI1 contain long coiled-coil segments and are able to self-associate.
A feature common among Rab effectors is the presence of extensive coiled-coil domains that frequently mediate the formation of homo- and heterodimers. In FIPs, these segments also encompass the Rab-binding domain at the carboxyl terminus (80). We used the COILS program to predict regions likely to adopt a coiled-coil conformation in both RBP74 and TbAZI1. Several coiled-coil segments were found distributed throughout the predicted RBP74 and TbAZI1 polypeptides (Fig. 3C). Given the extensive coiled-coil structure of RBP74 and TbAZI1, and the propensity of GTPase-interacting factors to dimerize, we assessed the self-interaction potential of RBP74 and TbAZI1. Yeast two-hybrid analysis indicate that RBP74 and TbAZI1 are indeed able to homodimerize (Fig. 3C, lower panels). However, no direct interaction between RBP74 and TbAZI1 could be detected (data not shown).
RBP74 is upregulated in bloodstream forms and distributes to endosomes.
Increased Rab11 expression in T. brucei correlates with upregulation of endocytic activity seen in bloodstream forms, where Rab11 compartments are found in close juxtaposition to Rab5A early endosomes (44). To assess whether RBP74 is also developmentally regulated, we quantified the expression of RBP74 at the transcript level by quantitative RT-PCR. We found that RBP74 mRNA abundance in bloodstream form was at least 3-fold higher than levels found in the insect form (Fig. 4A), suggesting a stage-specific requirement for RBP74.
Fig. 4.
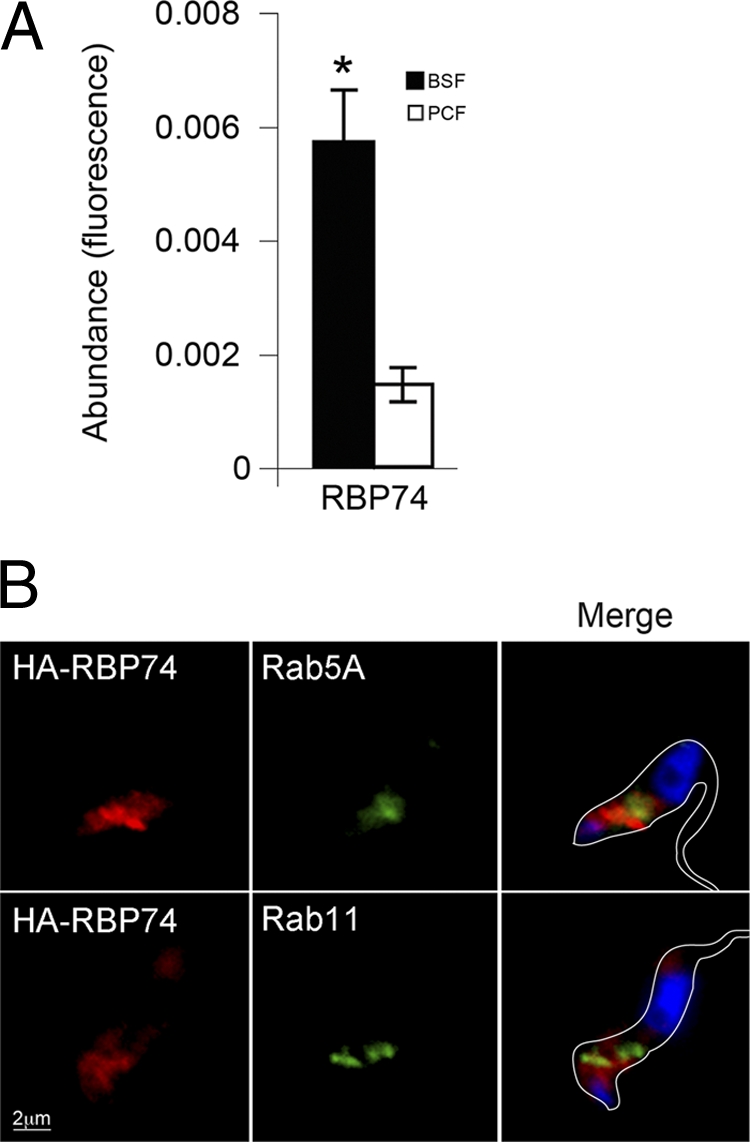
RBP74 transcript levels and endocytic location. (A) Abundance of RBP74 mRNA in bloodstream (filled bars) and insect (open bars) forms was assayed by quantitative RT-PCR using tubulin as an internal standard. RBP74 is significantly upregulated in the bloodstream form (*, P < 0.05). Error bars are standard errors from experiments performed in triplicate. (B) Location of HA epitope-tagged RBP74 in bloodstream cells compared to the location of early and recycling endosomes by immunostaining of Rab5A or Rab11. Left panels, RBP74 stained with mouse anti-HA antibody (red); center panels, rabbit polyclonal anti-Rab5 (upper panel) or anti-Rab11 (lower panel) (green). DNA was visualized with DAPI (blue, right panels, merge). Bar, 2 μm.
In order to investigate the intracellular distribution of RBP74 in bloodstream forms, we expressed an N-terminally HA-tagged form of the protein. RBP74 was distributed in the posterior end of the parasite between the nucleus and the kinetoplast, where the majority of endomembrane organelles reside in this organism (Fig. 4B). Colocalization studies with anti-Rab11 and anti-Rab5 antibodies also showed some juxtaposition between the HA-tagged variant of RBP74 and Rab5 and Rab11 but clearly not colocalization (Fig. 4B). This location is consistent with Rab11 and Rab5 distribution in the bloodstream-form trypanosome where Rab11 is found on endosomal structures that are intimately associated with Rab5 and, similarly to Sec15, is consistent with a transient interaction between Rab11/Rab5 and RBP74 (44).
RBP74 knockdown does not affect endocytosis of ConA or transferrin turnover.
The interaction of RBP74 with both Rab5A and Rab11 suggested a role at the intersection of the endosomal/recycling pathways. We examined the effect in bloodstream-form cells of RBP74 knockdown on endocytic trafficking. Although the ablation of RBP74 expression caused a rapid cell proliferation defect detectable within 24 h (see Fig. S3A in the supplemental material), we did not observe any effect on ConA or transferrin uptake (see Fig. S3C). Neither could we detect an effect on transferrin turnover (see Fig. S3D). These observations could be explained by incomplete knockdown of the protein (see Fig. S3B), by some redundancy in the role of RBP74, or by assay insensitivity. Although BLAST analysis against the T. brucei-predicted proteome failed to identify any proteins with significant similarity to RBP74, we cannot rule out some redundancy in recycling pathways. However, it does not appear that RBP74 is an essential player in maintaining endocytosis or recycling pathways in trypanosomes, at least for ConA and transferrin.
TbAZI1 is equally expressed in bloodstream and procyclic forms and locates at the base of the flagellum.
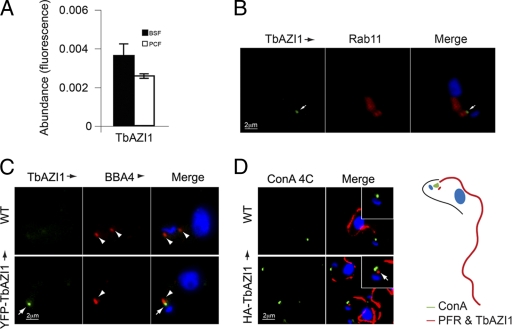
In contrast to what is seen for RBP74, TbAZI1 mRNA is expressed at similar levels in bloodstream and insect stage cells (Fig. 5A), suggesting a function required by both parasite forms. As the flagellum/flagellar pocket is also expressed in both stages, this is in agreement with TbAZI1 location, via HA- and YFP-tagged TbAZI1, close to the Rab11-positive structures present in both bloodstream and procyclic forms (Fig. 5 and data not shown). Indeed, when TbAZI1 location in bloodstream forms was studied, TbAZI1 distributed to a discrete region juxtaposed to Rab11 characteristic of both bloodstream and insect stages and distinct from RBP74, and it closely associated to the basal body and juxtaposed to the flagellar pocket (Fig. 5B). Contact between Rab11- and TbAZI1-positive structures is very common but likely is due to both having multiple interactions and functions, and collaboration takes place only in a minority of these contacts. When staining cells expressing YFP-TbAZI1 with BBA4 antibody specific for the basal body (87), we found TbAZI1 directly adjacent to the basal body (Fig. 5C). We then sought to determine the orientation of HA-TbAZI1 with respect to the flagellar pocket and kinetoplast. We used concanavalin A at 4°C to specifically stain the lumen of the flagellar pocket in cells expressing HA-TbAZI1 and counterstained with L8C4 antibody against the PFR2 component of the PFR (47). TbAZI1 was observed as a small spot close to the base of the PFR and adjacent to the flagellar pocket in all cells analyzed (n = 25) (Fig. 5D).
Fig. 5.
TbAZI1 transcript levels and endocytic location. (A) Abundance of TbAZI1 mRNA in bloodstream (filled bars) and insect (open bars) forms was assayed by quantitative RT-PCR using tubulin as an internal standard, as previously. TbAZI1 is equally expressed in both insect and mammalian stages. Error bars are standard error from experiments performed in triplicate. (B) Location of HA epitope-tagged TbAZI1 by immunostaining with mouse anti-HA antibody (green, white arrows). Cells were counterstained with rabbit polyclonal anti-Rab11 (center panels, red), DNA was visualized with DAPI (blue, right panels, merge). (C) Wild-type (WT) bloodstream-form cells or cells expressing YFP-TbAZI1 (green, white arrows) were fixed and prepared for immunofluorescence. Cells were then stained with mouse BBA4 antibody that labels the basal body (red, arrowheads). DNA was visualized with DAPI (blue, right panels, merge). (D) Wild-type cells or cells expressing HA-TbAZI1 were incubated with concanavalin A (green) at 4°C for 20 min to stain the lumen of the flagellar pocket. Cells were then processed for immunofluorescence analysis and counterstained with mouse antibody L8C4 specific for the paraflagellar rod (red) and anti-HA antibody for HA-TbAZI1 (red dot in lower panels, arrow). Note the essentially identical locations of HA-TbAZI1 (B and D) and YFP-TbAZI1 (C), eliminating the possibility of mistargeting based on the presence of the epitope tag. Bars, 2 μm.
TbAZI1 levels are Rab11 dependent.
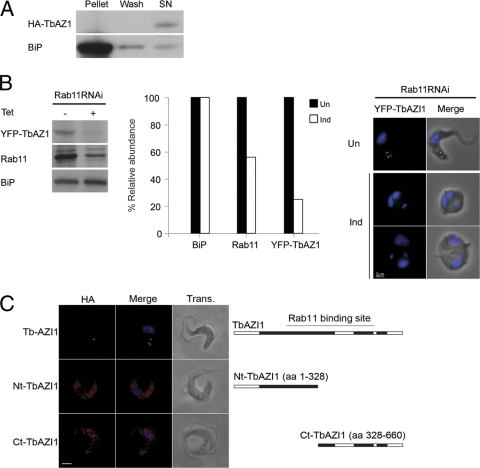
HA-TbAZI1 was found in the cytosolic fraction under hypotonic lysis (Fig. 6A), showing that the protein is not robustly docked with membranes, despite localization near the flagellar pocket. To further demonstrate a functional connection between Rab11 and TbAZI1, we next determined whether TbAZI1 targeting required Rab11 and also if location is conditional on the presence of the Rab-binding domain.
Fig. 6.
TbAZI1 is a downstream effector of Rab11. (A) Hypotonic lysis of bloodstream cells expressing HA-tagged TbAZI1. Log-phase BSF cells expressing HA-TbAZI1 were lysed in a hypotonic buffer as described in Materials and Methods. Equivalent amounts of the supernatant and pellet fractions were fractionated by SDS-PAGE and analyzed by Western blotting. TbAZI1 was exclusively found in the cytosolic fraction (SN). To control for excessive lysis or fragmentation of membranes, we analyzed distribution of BiP, an ER lumenal protein. BiP was recovered in the pellet fraction, with only minor quantities in the wash or cytosolic fractions, indicating minimal release of microsomal lumenal contents. (B) Interaction between Rab11 and TbAZI1 at the protein level. Cells expressing YFP-TbAZI1 were treated with 1 μg ml−1 tetracycline for 24 h for induction of RNAi against Rab11. Cell lysates were then analyzed by Western blotting following SDS-PAGE. There is a clear decrease in Rab11 levels upon RNAi induction as expected (middle of left panel), but also in levels of TbAZI1 (top of left panel). No effect was seen on BiP, used as the loading control (bottom of left panel). The histogram represents quantitation of Western blot data by using ImageJ (National Institutes of Health); filled bars represent uninduced samples and open bars represent levels in induced cells. Rab11-dependent localization and abundance of TbAZI1 were confirmed by immunofluorescence analysis of uninduced and induced Rab11 RNAi cells, showing that the loss of Rab11 expression upon the induction of Rab11 RNAi causes a decrease in TbAZI1 protein levels (right panels). All images were accumulated under identical exposure conditions. (C) Bloodstream-form cells expressing HA-tagged full-length TbAZI1 (upper panels), an amino terminal (Nt) TbAZI1 fragment (residues 1 to 328) (middle panels), or a carboxyl terminal (Ct) TbAZI1 fragment (residues 328 to 660) (lower panels) were incubated with anti-HA antibody (red) and further processed for immunofluorescence analysis. Truncation of the region known to contain the Rab11-binding site results in loss of the location observed for the full-length TbAZI1. Images were accumulated under identical exposure conditions. aa, amino acids. Bars, 2 μm.
Suppression of Rab11 expression produced a concomitant decrease in levels of epitope-tagged TbAZI1 by both Western blotting and immunofluorescence (Fig. 6B). Conversely, TbAZI1 knockdown did not produce any discernible effects on Rab5A and Rab11 levels from Western blot analysis (see Fig. S4, bottom panels, in the supplemental material). Immunofluorescence microscopy and Western blots of cells expressing HA-tagged N-terminal or C-terminal fragments of TbAZI1, which disrupt contiguous expression of the central Rab11-binding region, showed that these truncated proteins were expressed at higher levels and had lost the localization observed for the full-length protein (Fig. 6C; also see Fig. S5 in the supplemental material). Therefore, these data suggest that TbAZI1 and Rab11 functionally interact, confirming the yeast two-hybrid analysis, and specifically, that TbAZI1 targeting and possibly copy number regulation are Rab11 dependent and require the presence of the Rab-interacting domain.
TbAZI1 is required for the later steps of cytokinesis.
Knockdown of Rab11 in T. brucei results in severe proliferative defects, and by 18 h a high proportion of cells present a phenotype similar to the BigEye phenotype observed upon knockdown of the clathrin heavy chain (2). This morphology follows the loss of cell polarity and likely results from a block in endocytic pathways, but with ongoing exocytosis (Fig. 6B) (39). The rapid development of this imbalance in trafficking of material to and from the plasma membrane hampers the evaluation of any additional roles for Rab11. Thus, study of TbAZI1 as a Rab11 downstream effector is of interest.
Comparative genomics suggests that TbAZI1 is a protein involved in flagellar motility (see above and references 55 and 61). The loss of Rab11 expression produces a lethal phenotype in both procyclic and bloodstream stages, likely the result of the broad effect that Rab11 exerts on several pathways (39). RNAi of components of the flagellum reveals more restricted morphological defects in the segregation of basal bodies and cell division, ultimately affecting the viability of bloodstream parasites but not of the insect-stage procyclic forms (5). Hence, as a first step to dissect the role of TbAZI1, whether in trafficking through recycling pathways or in connecting Rab11 to a flagellum-related function, we produced a knockdown of TbAZI1 in the bloodstream and procyclic forms. Ablation of TbAZI1 expression in procyclic forms did not produce any significant effects on cell viability or cytokinesis (Fig. 7A). In contrast, a decrease in mRNA levels of TbAZI1 in BSF cells correlated with a proliferation defect that manifested at early times after RNAi induction and was prominent by 48 h (Fig. 7B). Analyses of ConA and transferrin uptake by FACS, as measures of receptor-mediated endocytosis, indicated the induction of TbAZI1 RNAi produced a 25% decrease in endocytosis of these ligands in bloodstream forms (see Fig. S6, upper panels, in the supplemental material). The recycling of fluorescent transferrin was unaffected, but as this probe is extensively degraded, the recycling may not be wholly dependent on vesicle-mediated mechanisms (see Fig. S6, lower panels). Examination of TbAZI1 RNAi-induced cells by light microscopy revealed an elevated number of cells that had initiated, but not completed, cytokinesis. We did not observe a loss in cell polarity or enlargement of the flagellar pocket, both characteristics of the loss of Rab11 expression (Fig. 6B and 7D). Staining of nuclei and kinetoplasts with DAPI indicated that while cytokinesis is blocked, mitosis proceeds normally (Fig. 7D). Moreover, we observed similar numbers of cells at each cytokinesis stage (initiation, furrow ingression, and abscission), denoting a general effect in progression through cytokinesis (Fig. 7D). The migration of the kinetoplasts was also affected as measured in 2K2N cells (Fig. 7E). These data support the hypothesis that TbAZI1 plays a role in a flagellum-related function despite having no effect on the overall structure of the flagellum, as observed by staining of the flagellum attachment zone and the paraflagellar rod in induced cells with a cytokinesis block (see Fig. S7 in the supplemental material). This is not without precedent; the knockdown of proteins that selectively affect motility while the structural integrity of the flagellum is maintained also creates a compromise in cytokinesis and viability in BSF cells (5).
Fig. 7.
TbAZI1 knockdown produces a block in cytokinesis. (A) Growth curves of p2T7-177-TbAZI1 PTT cells. Cell cultures were grown in triplicate in the presence (open squares) or absence (filled squares) of tetracycline (1 μg ml−1) and maintained under mid-logarithmic growth conditions by periodic dilution (indicated by breaks in the growth curves). Cell density was assessed by using a Coulter Counter. Data points are mean values ± standard error. TbAZI1 knockdown by RNAi was confirmed by real-time RT-PCR. The histogram shows the loss of TbAZI1 transcript 24 h after induction. (B) Growth curves of p2T7TA-TbAZI1 SMB cells. Cell cultures were grown in triplicate in the presence (open squares) or absence (filled squares) of tetracycline (1 μg ml−1). Cell density was assessed by using a Coulter Counter. Data points are mean values ± standard error. TbAZI1 knockdown by RNAi was confirmed by real-time RT-PCR. The histogram shows the loss of TbAZI1 transcript 24 h after induction. (C) Histogram representing cell cycle analysis of TbAZI1 RNAi cells and the accumulation of abnormal cellular forms. Uninduced and induced cells were fixed and mounted with Vectashield containing DAPI to stain for DNA. Numbers of nuclei and kinetoplasts per cell were counted at 24, 48, and 72 h; at least 200 cells were counted for each time point. (D) TbAZI1 knockdown cells manifest a delay in cytokinesis. RNAi cells were induced for 24 h, and then uninduced and induced cells were fixed and DAPI stained. Mitotic cells at different stages of cytokinesis were assessed by light microscopy. Numbers represent cells that have replicated their DNA but not initiated cytokinesis (upper panels), cells that have started ingression of the cleavage furrow (middle panels), and cells that have not yet undergone abscission (bottom panels). Two hundred cells were counted. (E) Distance between kinetoplasts was measured in at least 30 2K2N cells by using Metamorph (P < 0.05). Filled bars represent uninduced cultures, and open bars represent induced cell cultures. Bars, 2 μm.
DISCUSSION
Rab proteins represent a core component of the vesicle transport specificity machine and integrate multiple activities encompassing cytoskeletal interactions, SNARE-mediated vesicle fusion, and signal transduction (6, 32, 76). However, despite huge diversity across the eukaryotes and the manner in which many organisms deploy their endomembrane systems, a comparatively small, conserved core of Rab proteins appears to be sufficient for life. Remarkably, core Rabs perform similar functions in diverse taxa (22). In general, the functional specialization of small GTPases is mediated by alterations to protein interactomes (23); for example, metazoan Rho proteins interact with ∼70 distinct guanine exchange factors (29, 60, 73), while Rab5 has ∼40 probable partners (14, 34, 89). This high level of functional elaboration may be restricted to animals and fungi, suggesting lineage-specific mechanisms (23). We selected trypanosome Rab11 to investigate evolutionary novelty associated with small GTPase function and also to increase understanding of how Rab11 functions in trypanosomes (24, 39, 44). In mammalian cells, Rab11 has a rich interaction network, including the exocyst (Sec15), cytoskeletal motors (myosin V), which are FIPs, which are homodimeric coiled-coil proteins (35, 46, 70, 88, 90). We asked which elements of the Rab11 interactome are conserved, and which may be specific to trypanosomes. We found evidence for conservation of Rab11 interactions but also potential examples of metazoan and trypanosomatid-specific aspects.
Sec15 was identified as a conserved Rab11 interaction partner, confirmed by both yeast two-hybrid screening and juxtaposition between Sec15 and Rab11 locations. While colocalization in trypanosomes is less impressive than that in mammalian cells (28), Rab11 in trypanosomes associates with the flagellar pocket membrane only during final delivery of recycling endosomes to the cell surface, while other portions of the pocket membrane are engaged in exocytic transport (33), making this unsurprising. Also, we examined endogenous antigen expression rather than virus-mediated overexpression as done for mammalian cells, which may have increased the overlap between these antigens (28). Significantly, the presence of Sec15 across much of the flagellar pocket membrane, as suggested by the crescent-shaped staining pattern, indicates that a substantial portion of the flagellar pocket is capable of exocytic activity and argues against polarization within this organelle, consistent with earlier analysis suggesting nonpolarity of endocytosis (28). We were unable to localize Sec15 at the ultrastructural level despite repeated attempts. Therefore, the Rab11-Sec15 interaction is likely ancient and probably present in the LECA, while the FIP family are restricted to the Metazoa. Significantly, class V myosins (36) demonstrate a taxonomic distribution similar to that of FIPs, while ARF GTPases, which also interact with FIPs, must also interact with nonconserved factors, in agreement with the lineage-specific evolution of ARF families (3).
We identified two candidate effectors, RBP74 and TbAZI1, by using a yeast two-hybrid assay. Both share Rab effector architecture, i.e., long coiled-coil regions, and an ability to homodimerize. Evidence that TbAZI1 interacts with Rab11 comes from two-hybrid screening, including the specific interaction with the GTP form of Rab11 and interaction with a central Rab-binding domain, a “kissing” juxtaposition between both antigens, comparative genomics evidence for flagellar association, cosuppression under RNAi, and dependence of targeting of TbAZI1 on Rab11 expression and retention of the Rab-binding domain. The phylogenetic distribution of AZI1, including species where the ciliary/flagellar apparatus has been lost or modified during evolution, indicate that TbAZI1 is limited to organisms with canonical motile cilia/flagella, consistent with LECA possessing a cilium/flagellum (7, 55, 61) and with expansive loss of cilium/flagellum-related genes from nonflagellates (83, 86). Significantly, TbAZI1 binds Rab11 exclusively among the trypanosome Rab complement and locates to a domain close to the base of the PFR, adjacent to the basal body and the anterior face of the flagellar pocket, suggesting some role in flagellum biogenesis and maintenance, implicating Rab11 in this process in trypanosomes for the first time (33). The dependence of TbAZI1 targeting on Rab11 expression and retention of the intact Rab-binding domain provides functional confirmation of interaction and evidence that TbAZI1 functions downstream of Rab11. In trypanosomes, Rab11 knockdown is lethal in both insect and bloodstream stages, leading to a loss of cell polarity from, at least in part, imbalance in polarized membrane trafficking (39). Of relevance is that while the flagellum is required for cytokinesis in bloodstream cells, insect forms can complete new rounds of cell division even when components required for flagellar integrity are suppressed (5). In agreement with a role associated with flagellar maintenance, TbAZI1 expression is required for bloodstream cell proliferation but not procyclic forms, while the impact of TbAZI1 knockdown on endocytosis and recycling was comparatively minor and less prominent than for Rab11. Moreover, while TbAZI1 does not appear necessary for the generation or maintenance of the flagellum per se, knockdown in bloodstream parasites does retard proliferation, cytokinesis, and basal body/kinetoplast segregation. Conversely, this phenotype was not observed in the insect stage. In higher eukaryotes, compartmentalization of cilia/flagella relies on traffic of material from a subpopulation of Golgi-derived vesicles into this compartment via the ciliary pore complex, in a process likely to involve Rab8 and Rab11; critically, Rab8 is absent from trypanosomatids (59, 63). Despite growing evidence for several pathways regulating the selective targeting of material to the ciliary pore complex, little is known about the final steps, i.e., the direct recruitment of molecules to the ciliary/flagellar membrane. It does not appear that TbAZI1 is essential for this process.
Significantly, RBP74 is potentially restricted to trypanosomatids. Yeast two-hybrid screening uncovers interactions of RBP74 with GTP-bound Rab5 and Rab11, but not Rab4, consistent with the configuration of endocytic and recycling pathways in T. brucei and implicating RBP74 in coordination of endocytosis and recycling (37, 39, 67). RBP74 interaction with Rab11 is further supported by specific interaction with the GTP form of Rab11 and the requirement for a central rab-binding domain, similar expression profiles for TbAZI1 and Rab11, i.e., increased in BSF versus PCF cells, and in vitro pulldowns. An extensive localization for RBP74, probably due to interaction with at least Rab5 and potentially additional factors, makes this less informative than for TbAZI1, albeit the protein is restricted to the endosomal region of the cell. Induction of RBP74 RNAi in bloodstream cells, where transcript levels are upregulated, showed the RBP74 is required for proliferation, but we found no evidence for a specific cell cycle block. Knockdown of RBP74 did not lead to the BigEye phenotype associated with both Rab5 and Rab11 knockdowns, nor was endocytosis of transferrin and ConA significantly affected. Hence, the precise role of RBP74 was not uncovered despite the evidence for participation in Rab11 activity. Given that RNAi did result in the loss of 80% of the RBP74 mRNA, we consider inefficient RNAi an unlikely explanation. Rather, we suspect redundancy, that RBP74 is involved in uptake of specific cargo, or that RBP74 acts in some other Rab11 pathway that was undetected in the assays used here. As there are no obvious paralogues of RBP74 in T. brucei, distinct proteins would have to support Rab5 and Rab11 if RBP74 is redundant. While RBP74 demonstrates no clear homology to FIPs, the conservation of coiled-coil and discrete RBD architecture between these proteins is provocative and does raise the issue of divergent or convergent evolution. Divergent evolution is potentially supported by the very weak domain similarity to Rabaptin5 and our demonstration that proteins can share clear functional relationships despite absence of significant sequence similarity among over 20 trypanosome nucleoporins (13); in the present case, this is more challenging to demonstrate, as there are multiple FIPs, with diverse functions, so a discriminating localization or functional assay is not possible. Clearly, this is a very important issue, but the development of more sophisticated informatics tools is needed before this can be resolved satisfactorily.
The emerging view of how eukaryotic trafficking pathways arose predicts a complex LECA, where essentially all major pathways were already established (10). Subsequent evolution led to innovation of unique factors in various taxa. For Rab proteins, function is critically associated with interactions between the GTPase and its effectors. Interestingly, conservation of Sec15 and AZI1 suggests a considerable selective pressure to retain elements of Rab11 function associated with the flagellum together with the canonical role in recycling and exocytic pathways.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by program and project grants from the Wellcome Trust (to M.C.F.) which are gratefully acknowledged.
We thank Keith Gull (University of Oxford) and James Bangs (University of Wisconsin—Madison) for generous gifts of antibodies.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Ackers J. P., Dhir V., Field M. C. 2005. A bioinformatic analysis of the RAB genes of Trypanosoma brucei. Mol. Biochem. Parasitol. 141:89–97 [DOI] [PubMed] [Google Scholar]
- 2. Allen C. L., Goulding D., Field M. C. 2003. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 22:4991–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berriman M., et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422 [DOI] [PubMed] [Google Scholar]
- 4. Briggs L. J., Davidge J. A., Wickstead B., Ginger M. L., Gull K. 2004. More than one way to build a flagellum: comparative genomics of parasitic protozoa. Curr. Biol. 14:R611–612 [DOI] [PubMed] [Google Scholar]
- 5. Broadhead R., et al. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440:224–227 [DOI] [PubMed] [Google Scholar]
- 6. Cai H., Reinisch K., Ferro-Novick S. 2007. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 12:671–682 [DOI] [PubMed] [Google Scholar]
- 7. Cavalier-Smith T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52:297–354 [DOI] [PubMed] [Google Scholar]
- 8. Conti F., Sertic S., Reversi A., Chini B. 2009. Intracellular trafficking of the human oxytocin receptor: evidence of receptor recycling via a Rab4/Rab5 “short cycle.” Am. J. Physiol. Endocrinol Metab. 296:E532–E542 [DOI] [PubMed] [Google Scholar]
- 9. Cullis D. N., Philip B., Baleja J. D., Feig L. A. 2002. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J. Biol. Chem. 277:49158–49166 [DOI] [PubMed] [Google Scholar]
- 10. Dacks J. B., Field M. C. 2007. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci. 120:2977–2985 [DOI] [PubMed] [Google Scholar]
- 11. Dacks J. B., Poon P. P., Field M. C. 2008. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc. Natl. Acad. Sci. U. S. A. 105:588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dacks J. B., Peden A. A., Field M. C. 2009. Evolution of specificity in the eukaryotic endomembrane system. Int. J. Biochem. Cell Biol. 41:330–340 [DOI] [PubMed] [Google Scholar]
- 13. DeGrasse J. A., et al. 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell Proteomics 8:2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deininger K., et al. 2008. The Rab5 guanylate exchange factor Rin1 regulates endocytosis of the EphA4 receptor in mature excitatory neurons. Proc. Natl. Acad. Sci. U. S. A. 105:12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dhir V., Goulding D., Field M. C. 2004. TbRAB1 and TbRAB2 mediate trafficking through the early secretory pathway of Trypanosoma brucei. Mol. Biochem. Parasitol. 137:253–265 [DOI] [PubMed] [Google Scholar]
- 16. Doherty G. J., McMahon H. T. 2009. Mechanisms of endocytosis. Annu. Rev. Biochem. 78:857–902 [DOI] [PubMed] [Google Scholar]
- 17. Ducharme N. A., et al. 2007. Rab11-FIP2 regulates differentiable steps in transcytosis. Am. J. Physiol. Cell Physiol. 293:C1059–C1072 [DOI] [PubMed] [Google Scholar]
- 18. Fan G. H., Lapierre L. A., Goldenring J. R., Sai J., Richmond A. 2004. Rab11-family interacting protein 2 and myosin Vb are required for CXCR2 recycling and receptor-mediated chemotaxis. Mol. Biol. Cell 15:2456–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Field H., Field M. C. 1997. Tandem duplication of rab genes followed by sequence divergence and acquisition of distinct functions in Trypanosoma brucei. J. Biol. Chem. 272:10498–10505 [DOI] [PubMed] [Google Scholar]
- 20. Field H., Farjah M., Pal A., Gull K., Field M. C. 1998. Complexity of trypanosomatid endocytosis pathways revealed by Rab4 and Rab5 isoforms in Trypanosoma brucei. J. Biol. Chem. 273:32102–32110 [DOI] [PubMed] [Google Scholar]
- 21. Field H., Sherwin T., Smith A. C., Gull K., Field M. C. 2000. Cell-cycle and developmental regulation of TbRAB31 localisation, a GTP-locked Rab protein from Trypanosoma brucei. Mol. Biochem. Parasitol. 106:21–35 [DOI] [PubMed] [Google Scholar]
- 22. Field M. C., Gabernet-Castello C., Dacks J. B. 2007. Reconstructing the evolution of the endocytic system: insights from genomics and molecular cell biology. Adv. Exp. Med. Biol. 607:84–96 [DOI] [PubMed] [Google Scholar]
- 23. Field M. C., O'Reilly A. J. 2008. How complex is GTPase signaling in trypanosomes? Trends Parasitol. 24:253–257 [DOI] [PubMed] [Google Scholar]
- 24. Field M. C., Horn D., Carrington M. 2008. Analysis of small GTPase function in trypanosomes. Methods Enzymol. 438:57–76 [DOI] [PubMed] [Google Scholar]
- 25. Foth B. J., Goedecke M. C., Soldati D. 2006. New insights into myosin evolution and classification. Proc. Natl. Acad. Sci. U. S. A. 103:3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fukuda M., Kanno E., Ishibashi K., Itoh T. 2008. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol. Cell Proteomics 7:1031–1042 [DOI] [PubMed] [Google Scholar]
- 27. Gabernet-Castello C., Dacks J. B., Field M. C. 2009. The single ENTH-domain protein of trypanosomes; endocytic functions and evolutionary relationship with epsin. Traffic 10:894–911 [DOI] [PubMed] [Google Scholar]
- 28. Gadelha C., et al. 2009. Membrane domains and flagellar pocket boundaries are influenced by the cytoskeleton in African trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 106:17425–17430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garcia-Mata R., Burridge K. 2007. Catching a GEF by its tail. Trends Cell Biol. 17:36–43 [DOI] [PubMed] [Google Scholar]
- 30. Giansanti M. G., Belloni G., Gatti M. 2007. Rab11 is required for membrane trafficking and actomyosin ring constriction in meiotic cytokinesis of Drosophila males. Mol. Biol. Cell 18:5034–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reference deleted.
- 32. Grosshans B. L., Ortiz D., Novick P. 2006. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U. S. A. 103:11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grunfelder C. G., et al. 2003. Endocytosis of a glycosylphosphatidylinositol-anchored protein via clathrin-coated vesicles, sorting by default in endosomes, and exocytosis via RAB11-positive carriers. Mol. Biol. Cell 14:2029–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haas A. K., Fuchs E., Kopajtich R., Barr F. A. 2005. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 7:887–893 [DOI] [PubMed] [Google Scholar]
- 35. Hales C. M., et al. 2001. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276:39067–39075 [DOI] [PubMed] [Google Scholar]
- 36. Hales C. M., Vaerman J. P., Goldenring J. R. 2002. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J. Biol. Chem. 277:50415–50421 [DOI] [PubMed] [Google Scholar]
- 37. Hall B. S., Pal A., Goulding D., Field M. C. 2004. Rab4 is an essential regulator of lysosomal trafficking in trypanosomes. J. Biol. Chem. 279:45047–45056 [DOI] [PubMed] [Google Scholar]
- 38. Hall B., Allen C. L., Goulding D., Field M. C. 2004. Both of the Rab5 subfamily small GTPases of Trypanosoma brucei are essential and required for endocytosis. Mol. Biochem. Parasitol. 138:67–77 [DOI] [PubMed] [Google Scholar]
- 39. Hall B. S., et al. 2005. Developmental variation in Rab11-dependent trafficking in Trypanosoma brucei. Eukaryot. Cell 4:971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hickson G. R., et al. 2003. Arfophilins are dual Arf/Rab 11 binding proteins that regulate recycling endosome distribution and are related to Drosophila nuclear fallout. Mol. Biol. Cell 14:2908–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horgan C. P., Walsh M., Zurawski T. H., McCaffrey M. W. 2004. Rab11-FIP3 localises to a Rab11-positive pericentrosomal compartment during interphase and to the cleavage furrow during cytokinesis. Biochem. Biophys. Res. Commun. 319:83–94 [DOI] [PubMed] [Google Scholar]
- 42. Hsu S. C., TerBush D., Abraham M., Guo W. 2004. The exocyst complex in polarized exocytosis. Int. Rev. Cytol. 233:243–265 [DOI] [PubMed] [Google Scholar]
- 43. Innamorati G., Le Gouill C., Balamotis M., Birnbaumer M. 2001. The long and the short cycle. Alternative intracellular routes for trafficking of G-protein-coupled receptors. J. Biol. Chem. 276:13096–13103 [DOI] [PubMed] [Google Scholar]
- 44. Jeffries T. R., Morgan G. W., Field M. C. 2001. A developmentally regulated rab11 homologue in Trypanosoma brucei is involved in recycling processes. J. Cell Sci. 114:2617–2626 [DOI] [PubMed] [Google Scholar]
- 45. Jones M. C., Caswell P. T., Norman J. C. 2006. Endocytic recycling pathways: emerging regulators of cell migration. Curr. Opin. Cell Biol. 18:549–557 [DOI] [PubMed] [Google Scholar]
- 46. Junutula J. R., et al. 2004. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 279:33430–33437 [DOI] [PubMed] [Google Scholar]
- 47. Kohl L., Sherwin T., Gull K. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46:105–109 [DOI] [PubMed] [Google Scholar]
- 48. Koumandou V. L., Dacks J. B., Coulson R. M., Field M. C. 2007. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol. Biol. 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koumandou V. L., Natesan S. K., Sergeenko T., Field M. C. 2008. The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics 9:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Langevin J., et al. 2005. Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev. Cell 9:365–376 [DOI] [PubMed] [Google Scholar]
- 51. Lapierre L. A., et al. 2001. Myosin vb is associated with plasma membrane recycling systems. Mol. Biol. Cell 12:1843–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lemmon S. K., Jones E. W. 1987. Clathrin requirement for normal growth of yeast. Science 238:504–509 [DOI] [PubMed] [Google Scholar]
- 53. Leterrier C., Bonnard D., Carrel D., Rossier J., Lenkei Z. 2004. Constitutive endocytic cycle of the CB1 cannabinoid receptor. J. Biol. Chem. 279:36013–36021 [DOI] [PubMed] [Google Scholar]
- 54. Leung K. F., Dacks J. B., Field M. C. 2008. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9:1698–1716 [DOI] [PubMed] [Google Scholar]
- 55. Li J. B., et al. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117:541–552 [DOI] [PubMed] [Google Scholar]
- 56. Lindsay A. J., McCaffrey M. W. 2002. Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J. Biol. Chem. 277:27193–27199 [DOI] [PubMed] [Google Scholar]
- 57. Lindsay A. J., et al. 2002. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J. Biol. Chem. 277:12190–12199 [DOI] [PubMed] [Google Scholar]
- 58. Liu Y. J., Hodson M. C., Hall B. D. 2006. Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom Fungi inferred from RNA polymerase II subunit genes. BMC Evol. Biol. 6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mazelova J., et al. 2009. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 28:183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Meller N., Merlot S., Guda C. 2005. CZH proteins: a new family of Rho-GEFs. J. Cell Sci. 118:4937–4946 [DOI] [PubMed] [Google Scholar]
- 61. Merchant S. S., et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Meyers J. M., Prekeris R. 2002. Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J. Biol. Chem. 277:49003–49010 [DOI] [PubMed] [Google Scholar]
- 63. Nachury M. V., et al. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129:1201–1213 [DOI] [PubMed] [Google Scholar]
- 64. Natesan S. K., Peacock L., Matthews K., Gibson W., Field M. C. 2007. Activation of endocytosis as an adaptation to the mammalian host by trypanosomes. Eukaryot. Cell 6:2029–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Natesan S. K., et al. 2009. The trypanosome Rab-related proteins RabX1 and RabX2 play no role in intracellular trafficking but may be involved in fly infectivity. PLoS One 4:e7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Odronitz F., Kollmar M. 2007. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8:R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pal A., Hall B. S., Jeffries T. R., Field M. C. 2003. Rab5 and Rab11 mediate transferrin and anti-variant surface glycoprotein antibody recycling in Trypanosoma brucei. Biochem. J. 374:443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Prekeris R., Klumperman J., Scheller R. H. 2000. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell 6:1437–1448 [DOI] [PubMed] [Google Scholar]
- 69. Prekeris R., Davies J. M., Scheller R. H. 2001. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 276:38966–38970 [DOI] [PubMed] [Google Scholar]
- 70. Prekeris R. 2003. Rabs, Rips, FIPs, and endocytic membrane traffic. ScientificWorldJournal 3:870–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Redmond S., Vadivelu J., Field M. C. 2003. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 128:115–118 [DOI] [PubMed] [Google Scholar]
- 72. Riggs B., et al. 2003. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J. Cell Biol. 163:143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rossman K. L., Der C. J., Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180 [DOI] [PubMed] [Google Scholar]
- 74. Schonteich E., et al. 2008. The Rip11/Rab11-FIP5 and kinesin II complex regulates endocytic protein recycling. J. Cell Sci. 121:3824–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sonnichsen B., De Renzis S., Nielsen E., Rietdorf J., Zerial M. 2000. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 149:901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 [DOI] [PubMed] [Google Scholar]
- 77. Swoboda P., Adler H. T., Thomas J. H. 2000. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell 5:411–421 [DOI] [PubMed] [Google Scholar]
- 78. Ullrich O., Reinsch S., Urbe S., Zerial M., Parton R. G. 1996. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135:913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Varthakavi V., et al. 2006. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic 7:298–307 [DOI] [PubMed] [Google Scholar]
- 80. Wallace D. M., Lindsay A. J., Hendrick A. G., McCaffrey M. W. 2002. Rab11-FIP4 interacts with Rab11 in a GTP-dependent manner and its overexpression condenses the Rab11 positive compartment in HeLa cells. Biochem. Biophys. Res. Commun. 292:909–915 [DOI] [PubMed] [Google Scholar]
- 81. Wang F., Chen X., Zhang X., Ma L. 2008. Phosphorylation state of μ-opioid receptor determines the alternative recycling of receptor via Rab4 or Rab11 pathway Mol. Endocrinol. 22:1881–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wennerberg K., Rossman K. L., Der C. J. 2005. The Ras superfamily at a glance. J. Cell Sci. 118:843–846 [DOI] [PubMed] [Google Scholar]
- 83. Wickstead B., Gull K. 2007. Dyneins across eukaryotes: a comparative genomic analysis. Traffic 8:1708–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wilcke M., et al. 2000. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-golgi network. J. Cell Biol. 151:1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wilson G. M., et al. 2005. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol. Biol. Cell 16:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Woodland H. R., Fry A. M. 2008. Pix proteins and the evolution of centrioles. PLoS One 3:e3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Woods A., et al. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500 [DOI] [PubMed] [Google Scholar]
- 88. Wu S., Mehta S. Q., Pichaud F., Bellen H. J., Quiocho F. A. 2005. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat. Struct. Mol. Biol. 12:879–885 [DOI] [PubMed] [Google Scholar]
- 89. Zerial M., McBride H. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117 [DOI] [PubMed] [Google Scholar]
- 90. Zhang X. M., Ellis S., Sriratana A., Mitchell C. A., Rowe T. 2004. Sec15 is an effector for the Rab11 GTPase in mammalian cells. J. Biol. Chem. 279:43027–43034 [DOI] [PubMed] [Google Scholar]
- 91. Zhu G., et al. 2004. Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat. Struct. Mol. Biol. 11:975–983 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.