Abstract
The overexpression of the MDR1 gene, which encodes a multidrug efflux pump of the major facilitator superfamily, is a frequent cause of resistance to the widely used antimycotic agent fluconazole and other toxic compounds in the pathogenic yeast Candida albicans. The zinc cluster transcription factor Mrr1 controls MDR1 expression in response to inducing chemicals, and gain-of-function mutations in MRR1 are responsible for the constitutive MDR1 upregulation in fluconazole-resistant C. albicans strains. To understand how Mrr1 activity is regulated, we identified functional domains of this transcription factor. A hybrid protein consisting of the N-terminal 106 amino acids of Mrr1 and the transcriptional activation domain of Gal4 from Saccharomyces cerevisiae constitutively induced MDR1 expression, demonstrating that the DNA binding domain is sufficient to target Mrr1 to the MDR1 promoter. Using a series of C-terminal truncations and systematic internal deletions, we could show that Mrr1 contains multiple activation and inhibitory domains. One activation domain (AD1) is located in the C terminus of Mrr1. When fused to the tetracycline repressor TetR, this distal activation domain induced gene expression from a TetR-dependent promoter. The deletion of an inhibitory region (ID1) located near the distal activation domain resulted in constitutive activity of Mrr1. The additional removal of AD1 abolished the constitutive activity, but the truncated Mrr1 still could activate the MDR1 promoter in response to the inducer benomyl. These results demonstrate that the activity of Mrr1 is regulated in multiple ways and provide insights into the function of an important mediator of drug resistance in C. albicans.
INTRODUCTION
Infections by the pathogenic yeast Candida albicans are commonly treated with the antifungal agent fluconazole, which inhibits ergosterol biosynthesis. C. albicans can develop resistance to fluconazole by different mechanisms, which often are combined to result in clinically relevant, high-level resistance (20). Besides mutations in the target enzyme that decrease its affinity for the drug, alterations in gene expression are a frequent cause of fluconazole resistance. The constitutive upregulation of ergosterol biosynthesis genes and the overexpression of multidrug efflux pumps of the ABC transporter and major facilitator superfamilies all result in increased azole resistance. Zinc cluster proteins, a family of transcription factors that is unique to the fungal kingdom (17), play a central role in the regulation of genes involved in drug resistance. In C. albicans, Upc2 controls the expression of ergosterol biosynthesis genes (16, 34), Tac1 regulates the expression of the ABC transporters CDR1 and CDR2 (4), and Mrr1 controls the expression of the major facilitator MDR1 (22). These transcription factors mediate the upregulation of their respective target genes in response to inducing stimuli. In addition, gain-of-function mutations in Upc2, Tac1, and Mrr1 result in the constitutive activation of the transcription factors and overexpression of their target genes in fluconazole-resistant strains (1–4, 6, 7, 11, 12, 22, 33, 37). However, it is currently not understood how these transcriptional regulators achieve an activated state under inducing conditions or after the acquisition of gain-of-function mutations.
The zinc cluster proteins are defined by a conserved DNA binding motif, which consists of six cysteine residues that coordinate two zinc atoms (Zn2Cys6). Most zinc cluster transcription factors have their DNA binding domain at the N terminus, a large negative regulatory domain in the middle of the protein, and an activation domain at the C terminus (17). The transcriptional activity of zinc cluster proteins can be regulated in different ways, but many of them are activated by the binding of inducing molecules (17, 23). These can be metabolites, as for transcription factors regulating genes involved in nutrient utilization and biosyntheses (e.g., Put3 and Leu3), or xenobiotics, which activate the pleiotropic drug resistance regulators Pdr1 and Pdr3 in Saccharomyces cerevisiae and Candida glabrata. Pdr1 and Pdr3 are functionally related to Tac1 and Mrr1 of C. albicans, because they also control the expression of multidrug efflux pumps. It has been shown that the binding of drugs and other toxic chemicals to the negative regulatory domain of Pdr1/Pdr3 results in a conformational change that enables the C-terminal activation domain to interact with the Med15 subunit of the mediator complex and recruit RNA polymerase II to the promoters of Pdr1/Pdr3 target genes (35). Mutations or deletions within the negative regulatory region result in constitutive activity of Pdr1/Pdr3 and some other zinc cluster transcription factors, indicating that this inhibitory domain maintains the transcription factors in an inactive state in the absence of inducing signals (17).
The C. albicans drug resistance regulators Tac1 and Mrr1 also activate their target genes in response to inducing chemicals. For example, the Tac1-dependent upregulation of the ABC transporters CDR1 and CDR2 is efficiently induced by fluphenazine and estradiol (5), and MDR1 expression is induced by benomyl and hydrogen peroxide in an Mrr1-dependent fashion (13, 22, 28). Whether these inducers directly bind to the zinc cluster transcription factors or mediate their activation in an indirect manner remains to be established. The hydrogen peroxide-induced MDR1 upregulation seems to be mediated by the bZip transcription factor Cap1. When activated by oxidative stress, Cap1 accumulates in the nucleus, where it binds to the MDR1 promoter and, together with Mrr1, induces the expression of the efflux pump (28, 32, 36). The antimitotic drug benomyl also causes oxidative stress and activates Yap1, the Cap1 ortholog in S. cerevisiae (15). Cap1 contributes to, but is not essential for, benomyl-induced MDR1 expression, indicating that benomyl promotes MDR1 expression via Cap1 as well as in a Cap1-independent fashion, possibly by directly activating Mrr1 (32).
No functional domains of Mrr1 have been experimentally determined yet, although the DNA binding domain can be predicted by the presence of the zinc cluster motif in the N terminus. Mrr1 is a relatively large protein of more than 1,100 amino acids, and it can be assumed that it is subject to multiple regulatory inputs that may affect its interaction with other proteins, binding to the promoters of its target genes, and/or transcriptional activation. Mrr1 cooperates with at least two other transcription factors that bind to the MDR1 promoter, Cap1 and the MADS box transcription factor Mcm1, to induce the expression of the efflux pump. Both Mrr1 and Cap1 are necessary for the induction of MDR1 expression in response to hydrogen peroxide, while Mcm1 is dispensable under these conditions. On the other hand, a hyperactive Mrr1 containing a gain-of-function mutation can bind to and activate the MDR1 promoter in the absence of Cap1, but it requires Mcm1 to induce MDR1 expression (19, 32).
Gain-of-function mutations that result in constitutive activity of Mrr1 are located in different regions of the protein and are thought to affect amino acid residues that are important to keep the transcription factor in an inactive state in the absence of an inducer (21). Almost all gain-of-function mutations identified so far are located outside domains that can be predicted by sequence analysis, the DNA binding domain containing the Zn2Cys6 motif and a conserved domain known as the middle homology region. To provide a basis for understanding how Mrr1 is activated in response to inducing conditions and after the acquisition of gain-of-function mutations, we undertook a systematic analysis of this transcription factor with the aim of identifying its functional domains.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans wild-type strain SC5314 (10), the mrr1Δ mutants SCMRR1M4A and SCMRR1M4B (22), and strain CAG48MRR1M4B, an mrr1Δ mutant that contains a PMDR1-caGFP reporter fusion (22), were used as hosts for the integration of the DNA constructs described below. All strains were stored as frozen stocks with 15% glycerol at −80°C and subcultured on YPD agar plates (10 g yeast extract, 20 g peptone, 20 g glucose, 15 g agar per liter) at 30°C. Strains were routinely grown in YPD liquid medium at 30°C in a shaking incubator. For the induction of the MDR1 promoter by benomyl, overnight cultures of the reporter strains were diluted 10−2 in fresh YPD medium and grown for 3 h at 30°C. After adding 50 μg/ml benomyl, the cultures were incubated for an additional hour and the fluorescence of the cells was quantified by fluorescence-activated cell sorter (FACS) analysis.
Plasmid constructions.
The plasmids used in this study are listed in Table 1. pZCF36K2, pZCF36K3, pZCF36E2, and pZCF36EH2 have been described previously (22, 32, 33). Derivatives of pZCF36K2 encoding C-terminally truncated Mrr1 proteins were generated by PCR with primers introducing a stop codon at the desired positions (primer sequences are available upon request). Derivatives of pZCF36K2 with internal deletions also were generated by PCR. In these cases, the N-terminal and C-terminal parts of MRR1 were connected by an introduced NarI site encoding a Gly-Ala linker or by a BamHI site encoding a Gly-Ser linker.
Table 1.
Plasmids used in this study
| Name | Description of encoded protein |
|---|---|
| Control plasmids | |
| pZCF36K2 | Wild-type Mrr1 (amino acids 1-1108) |
| pZCF36K3 | Hyperactive Mrr1 with P683S gain-of-function mutation |
| pZCF36E2 | Wild-type Mrr1 expressed from the ADH1 promoter |
| pZCF36EH2 | HA-tagged Mrr1 expressed from the ADH1 promoter |
| pTET52 | TetR expressed from the ADH1 promoter |
| pTET53 | TetR-Gal4AD fusion expressed from the ADH1 promoter |
| Plasmids encoding Mrr1DB-Gal4AD fusion proteins (expressed from the ADH1 promoter) | |
| pZCF36DBH2 | HA-tagged Gal4AD fused to Mrr1 amino acids 1-128 |
| pZCF36DBH4 | HA-tagged Gal4AD fused to Mrr1 amino acids 1-121 |
| pZCF36DBH5 | HA-tagged Gal4AD fused to Mrr1 amino acids 1-106 |
| pZCF36DBH6 | HA-tagged Gal4AD fused to Mrr1 amino acids 1-90 |
| Plasmids encoding TetR-Mrr1 fusion proteins (expressed from the ADH1 promoter) and containing a Ptet-caGFP reporter fusion | |
| pZCF3667-1108-TETR | Mrr1 amino acids 67-1108 fused to TetR |
| pZCF3691-1108-TETR | Mrr1 amino acids 91-1108 fused to TetR |
| pZCF36104-1108-TETR | Mrr1 amino acids 104-1108 fused to TetR |
| pZCF36122-1108-TETR | Mrr1 amino acids 122-1108 fused to TetR |
| pZCF36144-1108-TETR | Mrr1 amino acids 144-1108 fused to TetR |
| pZCF36168-1108-TETR | Mrr1 amino acids 168-1108 fused to TetR |
| pZCF36269-1108-TETR | Mrr1 amino acids 269-1108 fused to TetR |
| pZCF36524-1108-TETR | Mrr1 amino acids 524-1108 fused to TetR |
| pZCF361001-1108-TETR | Mrr1 amino acids 1001-1108 fused to TetR |
| pZCF361051-1108-TETR | Mrr1 amino acids 1051-1108 fused to TetR |
| pZCF3667-526-TETR | Mrr1 amino acids 67-526 fused to TetR |
| pZCF36269-812-TETR | Mrr1 amino acids 269-812 fused to TetR |
| Plasmids encoding C-terminally truncated Mrr1 | |
| pZCF36K2ΔC1066 | Mrr1 amino acids 1-1066 |
| pZCF36K2ΔC1040 | Mrr1 amino acids 1-1040 |
| pZCF36K2ΔC1031 | Mrr1 amino acids 1-1031 |
| pZCF36K2ΔC1015 | Mrr1 amino acids 1-1015 |
| pZCF36K2ΔC1001 | Mrr1 amino acids 1-1001 |
| pZCF36K2ΔC955 | Mrr1 amino acids 1-955 |
| pZCF36K2ΔC952 | Mrr1 amino acids 1-952 |
| pZCF36K2ΔC948 | Mrr1 amino acids 1-948 |
| pZCF36K2ΔC947 | Mrr1 amino acids 1-947 |
| pZCF36K2ΔC946 | Mrr1 amino acids 1-946 |
| pZCF36K2ΔC945 | Mrr1 amino acids 1-945 |
| pZCF36K2ΔC944 | Mrr1 amino acids 1-944 |
| pZCF36K2ΔC933 | Mrr1 amino acids 1-933 |
| pZCF36K2ΔC911 | Mrr1 amino acids 1-911 |
| pZCF36K2ΔC812 | Mrr1 amino acids 1-812 |
| pZCF36K2ΔC706 | Mrr1 amino acids 1-706 |
| pZCF36K2ΔC366 | Mrr1 amino acids 1-366 |
| Plasmids encoding Mrr1 with internal deletions | |
| pZCF36K2Δ342-381 | Mrr1 lacking amino acids 342-381 |
| pZCF36K2Δ873-896 | Mrr1 lacking amino acids 873-896 |
| pZCF36K2Δ101-150 | Mrr1 lacking amino acids 101-150 |
| pZCF36K2Δ151-200 | Mrr1 lacking amino acids 151-200 |
| pZCF36K2Δ201-250 | Mrr1 lacking amino acids 201-250 |
| pZCF36K2Δ251-300 | Mrr1 lacking amino acids 251-300 |
| pZCF36K2Δ301-350 | Mrr1 lacking amino acids 301-350 |
| pZCF36K2Δ351-400 | Mrr1 lacking amino acids 351-400 |
| pZCF36K2Δ401-450 | Mrr1 lacking amino acids 401-450 |
| pZCF36K2Δ451-500 | Mrr1 lacking amino acids 451-500 |
| pZCF36K2Δ501-550 | Mrr1 lacking amino acids 501-550 |
| pZCF36K2Δ551-600 | Mrr1 lacking amino acids 551-600 |
| pZCF36K2Δ601-650 | Mrr1 lacking amino acids 601-650 |
| pZCF36K2Δ651-700 | Mrr1 lacking amino acids 651-700 |
| pZCF36K2Δ701-750 | Mrr1 lacking amino acids 701-750 |
| pZCF36K2Δ751-800 | Mrr1 lacking amino acids 751-800 |
| pZCF36K2Δ801-850 | Mrr1 lacking amino acids 801-850 |
| pZCF36K2Δ851-900 | Mrr1 lacking amino acids 851-900 |
| pZCF36K2Δ901-950 | Mrr1 lacking amino acids 901-950 |
| pZCF36K2Δ951-1000 | Mrr1 lacking amino acids 951-1000 |
| pZCF36K2Δ1001-1050 | Mrr1 lacking amino acids 1001-1050 |
| pZCF36K2Δ951-1050 | Mrr1 lacking amino acids 951-1050 |
| pZCF36K2Δ129-1000 | Mrr1 lacking amino acids 129-1000 |
| pZCF36K2Δ129-1050 | Mrr1 lacking amino acids 129-1050 |
| pZCF36DB-751-952 | Mrr1 amino acids 751-952 fused to amino acids 1-128 |
| pZCF36DB-851-952 | Mrr1 amino acids 851-952 fused to amino acids 1-128 |
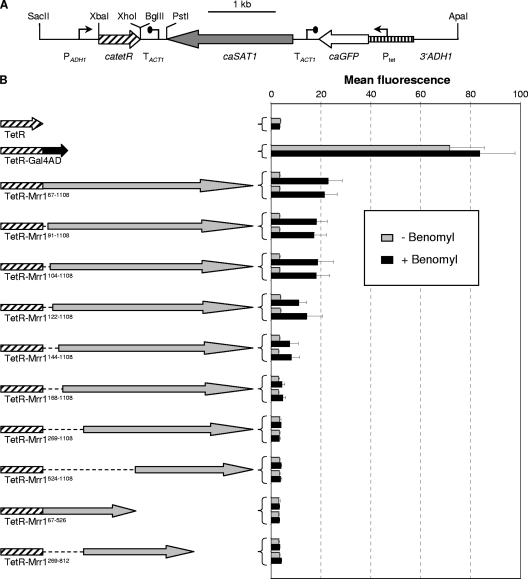
To generate plasmids encoding fusions of the tetracycline repressor protein TetR to various parts of Mrr1, the tetR gene first was amplified from pTET17 (24) with the primers TETR1 (5′-ATATGTCTAGATTAGATAAAAGTAAAGTGATTAACAGCGCATTAGAGTTGCTTAATGAGGTC-3′) and TETR2 (5′-GGCTGCTCGAGGACCCACTTTCACATTTAAG-3′), thereby changing the single CTG codon in tetR, which would be mistranslated as serine instead of leucine in C. albicans, to TTG (boldface). The PCR product containing the Candida-adapted tetR gene (catetR) was digested with XbaI and XhoI and cloned in the vector pBluescript to obtain pTET49. The transcription termination sequence of the C. albicans ACT1 gene (TACT1) was amplified with the primers ACT37 (5′-GGTCCTCGAGTTAGAGATCTAAATTCTGGAAATCTGG-3′; XhoI and BglII sites are underlined) and ACT38 (5′-ATATGGGCCCTGCAGACATTTTATGATGGAATGAATGGG-3′; overlapping ApaI and PstI sites are underlined) and cloned in the XhoI/ApaI-digested pTET49 to yield pTET50. An XbaI-PstI fragment from pTET50 containing the catetR and TACT1 sequences then was used to replace the corresponding fragment in pTET51, a derivative of pNIM1 (24) in which the BglII site behind the caGFP (GFP) reporter gene had been destroyed by filling in the overhanging ends and religation. The resulting plasmid, pTET52, contains the catetR gene extended by two serine codons under the control of the ADH1 promoter and caGFP under the control of the Tet promoter (see Fig. 2A). The insertion of a SalI-BamHI fragment from pGAL4AD2 (24), containing a Candida-adapted gene encoding the C-terminal activation domain of S. cerevisiae Gal4 (caGAL4AD), between the XhoI and BglII sites of pTET52 resulted in pTET53, which encodes a TetR-Gal4AD fusion protein. Different parts of the MRR1 coding region were amplified by PCR with suitable primers (sequences are available upon request) and cloned as XhoI-BglII fragments in frame with the catetR coding sequence in pTET52 to express the desired TetR-Mrr1 fusion proteins.
Fig. 2.
(A) Map of the insert from plasmid pTET52 containing the Candida-adapted tetR gene (catetR) under the control of the ADH1 promoter (PADH1) and the caGFP reporter gene under the control of a TetR-dependent promoter (Ptet). Relevant restriction sites used for the construction of pTET52 and derivatives (see Materials and Methods) are indicated. Fusions of TetR with the Gal4 activation domain or different portions of Mrr1 were generated by inserting the corresponding DNA fragments between the unique XhoI and BglII sites behind catetR. The flanking ADH1 sequences served for the integration of the cassettes into the ADH1 locus by homologous recombination. (B) MDR1 promoter activity in transformants of strain SC5314 expressing the indicated TetR-Mrr1 fusion proteins and containing the Ptet-caGFP reporter fusion. Strains were grown in the absence or presence of benomyl, and the mean fluorescence of the cells was determined by flow cytometry. The results obtained with two independent transformants are shown in each case (means and standard deviations from three experiments). Strains expressing a TetR-Gal4AD fusion protein or unfused TetR served as positive and negative controls, respectively.
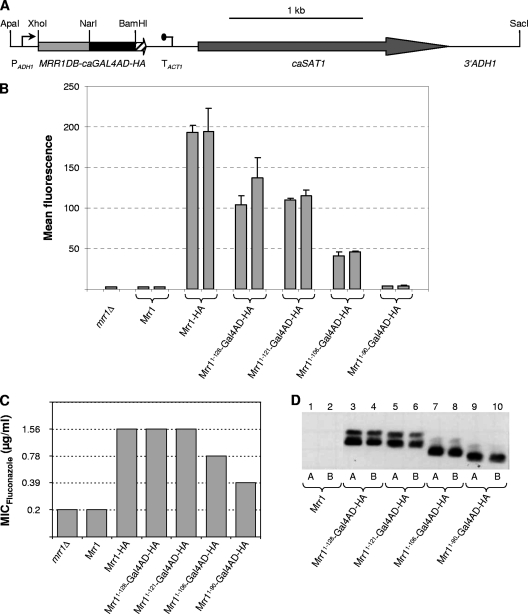
To generate fusions of the MRR1 DNA binding domain (MRR1DB) to the GAL4 activation domain, the N-terminal region of MRR1 encoding amino acids 1 to 128 was amplified with the primers ZCF36-21 (5′-TCTAACTCGAGAAAAATGTCAATTGCCACCACCC-3′) and ZCF36-22 (5′-GTTGGGCGCCGGCAACAGTATTGTCTGGTG-3′), and the PCR product was digested at the introduced XhoI and NarI sites (underlined). The GAL4 activation domain was amplified from plasmid pTET53 with a primer binding in the tetR region and primer GAL5 (5′-ATATAGATCTTTACTCTTTTTTTGGGTTTGGTGG-3′), digested at an internal NarI site and at the BglII site (underlined) behind the stop codon and cloned together with the XhoI-NarI MRR1DB fragment in XhoI/BglII-digested pADH1E1 (26). In the resulting plasmid pZCF36DB1, the MRR1DB-GAL4AD fusion is placed under the control of the ADH1 promoter. To add a 3× hemagglutinin (HA) tag, the MRR1DB-GAL4AD fragment was amplified from pZCF36DB1 with primers ZCF36-21 and GAL6 (5′-ATATGGATCCCTCTTTTTTTGGGTTTGGTGG-3′), which introduced a BamHI site (underlined) in place of the stop codon, and cloned in pBluescript to yield pZCF36DB3. A BamHI-SacI fragment from pZCF36H2 (32), containing the 3× HA tag, the ACT1 transcription termination sequence, the caSAT1 selection marker, and the C-terminal part of the ADH1 gene, then was ligated with the BamHI/SacI-digested pZCF36DB3 to obtain pZCF36DBH1. The insert from this plasmid was cloned in the XhoI/SacI-digested pZCF36DB1 to generate pZCF36DBH2, which encodes a fusion protein encompassing amino acids 1 to 128 of Mrr1, the Gal4 activation domain, and a C-terminal 3× HA tag (Fig. 1A). Shorter fragments encoding the first 121, 106, or 90 amino acids of Mrr1 were obtained by PCR with suitable primers and used to replace the MRR1DB fragment in pZCF36DBH2, thereby generating plasmids pZCF36DBH4, pZCF36DBH5, and pZCF36DBH6, respectively.
Fig. 1.
(A) Map of the insert from plasmid pZCF36DBH2, in which the 3× HA-tagged GAL4 activation domain (caGAL4AD-HA) is fused to the MRR1 DNA binding domain (MRR1DB, codons 1 to 128 of MRR1). Relevant restriction sites used for the construction of the plasmid and excision of the cassette (see Materials and Methods) are indicated. PADH1, ADH1 promoter; TACT1, transcription termination sequence of the ACT gene; caSAT1, Candida-adapted nourseothricin resistance marker; 3′ADH1, sequence from the C-terminal part of the ADH1 gene. The flanking ADH1 sequences (PADH1 and 3′ADH1) served for integration into the ADH1 locus by homologous recombination. (B) MDR1 promoter activity in an mrr1Δ mutant carrying a PMDR1-caGFP reporter fusion (strain CAG48MRR1M4B) and in transformants expressing wild-type Mrr1, an HA-tagged, hyperactive Mrr1, or the indicated Mrr1DB-Gal4AD fusion proteins. Cells were grown to log phase, and their mean fluorescence was determined by flow cytometry. The results obtained with two independent transformants are shown in each case (means and standard deviations from three experiments). (C) MICs of fluconazole for the mrr1Δ mutants SCMRR1M4A and SCMRR1M4B and transformants expressing wild-type Mrr1, HA-tagged Mrr1, or the indicated Mrr1DB-Gal4AD fusion proteins. Identical results were obtained with two independent series of strains. (D) Detection of the indicated HA-tagged Mrr1DB-Gal4AD fusion proteins in cell extracts of the corresponding strains by Western immunoblotting with an anti-HA antibody. Cell extracts of strains expressing untagged, wild-type Mrr1 were used as negative controls. A and B denote transformants of strains SCMRR1M4A and SCMRR1M4B, respectively. All samples were analyzed on the same gel, but some irrelevant lanes between lanes 2 and 3 have been removed from the figure.
C. albicans transformation.
C. albicans strains were transformed by electroporation (14) with the gel-purified inserts from the plasmids described above. Nourseothricin-resistant transformants were selected on YPD agar plates containing 200 μg/ml nourseothricin (Werner Bioagents, Jena, Germany) as described previously (27). The correct integration of each construct was confirmed by Southern hybridization using the flanking sequences as probes.
Isolation of genomic DNA and Southern hybridization.
Genomic DNA from C. albicans was isolated as described previously (27), digested with appropriate restriction enzymes, separated on a 1% agarose gel, and, after ethidium bromide staining, transferred by vacuum blotting onto a nylon membrane and fixed by UV cross-linking. Southern hybridization with enhanced chemiluminescence-labeled probes was performed with the Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare, Braunschweig, Germany) according to the instructions of the manufacturer.
FACS analysis.
Fluorescence-activated cell sorter (FACS) analysis was performed with a FACSCalibur cytometry system equipped with an argon laser emitting at 488 nm (Becton Dickinson, Heidelberg, Germany). Fluorescence was measured on the FL1 fluorescence channel equipped with a 530-nm band-pass filter. Twenty thousand cells were analyzed per sample and were counted at a flow rate of 500 cells per s. Fluorescence data were collected by using logarithmic amplifiers. The mean fluorescence (arbitrary values) was determined with CellQuest Pro (Becton Dickinson) software.
Western immunoblot analysis.
YPD overnight cultures of the strains expressing Mrr1DB-Gal4AD-HA fusion proteins were diluted to an optical density at 600 nm (OD600) of 0.2 and grown at 30°C to an OD600 of 1.0. Cells were collected by centrifugation, washed twice in water, and broken by vortexing for 10 min at 4°C with 300 μl 0.5-mm glass beads in 300 μl breaking buffer (100 mM Tris-Cl [pH 7.5], 200 mM NaCl, 20% glycerol, 5 mM EDTA, 4% Complete EDTA-free protease inhibitor cocktail [Roche Diagnostics GmbH, Mannheim, Germany], 0.1% β-mercaptoethanol). Samples were centrifuged at 13,000 rpm for 5 min at 4°C, the supernatant was collected, and the protein concentration was quantified with a NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, MA). Extracts were boiled for 5 min, and 110 μg protein of each sample was separated on an SDS–12% polyacrylamide gel. Proteins were transferred onto a nitrocellulose membrane with a Trans-Blot SD semidry transfer apparatus (Bio-Rad, Munich, Germany). The HA-tagged fusion proteins were detected with a rat anti-HA monoclonal antibody (anti-HA-peroxidase, high affinity [3F10]; Roche Diagnostics GmbH, Mannheim, Germany) at a dilution of 1:500 using a chemiluminescence detection system (GE Healthcare UK Limited, Chalfont, United Kingdom) under conditions recommended by the manufacturer.
Fluconazole susceptibility tests.
To determine the fluconazole susceptibilities of the strains, a 2-fold dilution series of fluconazole was prepared in the assay medium, starting from an initial concentration of 100 μg/ml. Susceptibility tests were carried out in high-resolution medium (14.67 g HR medium [Oxoid GmbH, Wesel, Germany], 1 g NaHCO3, 0.2 M phosphate buffer [pH 7.2]) using a previously described microdilution method (29).
RESULTS
The DNA binding domain is sufficient to target Mrr1 to the MDR1 promoter.
Like most zinc cluster transcription factors, Mrr1 contains the predicted DNA binding domain in its N terminus, with the CX2CX6CX5-12CX2CX6-8C motif that is characteristic for this protein family extending from amino acids 31 to 59. As mentioned in the introduction, Mrr1 cooperates with other transcription factors to induce MDR1 expression in response to toxic chemicals or after the acquisition of gain-of-function mutations (19, 32). It therefore seemed possible that other regulators are required to recruit Mrr1 to the MDR1 promoter. To determine whether the DNA binding domain is sufficient to target Mrr1 to the MDR1 promoter or if additional regions of the protein are necessary, we fused the N-terminal 128 amino acids of Mrr1 to the transcriptional activation domain of the Gal4 protein from S. cerevisiae and tested the ability of the hybrid protein to activate a PMDR1-caGFP reporter fusion (Fig. 1A and B). Reporter strains expressing full-length, wild-type Mrr1 or a 3× HA-tagged, constitutively active Mrr1 were used for comparison. As reported previously (32), the wild-type Mrr1 did not activate the MDR1 promoter in the absence of an inducer, whereas MDR1 was constitutively expressed in strains containing the HA-tagged, hyperactive Mrr1. The hybrid Mrr1DB-Gal4AD protein also could activate the MDR1 promoter, indicating that the N-terminal part of Mrr1 containing the DNA binding domain is sufficient to target the transcription factor to the MDR1 promoter.
To delimit the minimal region that is required for the recognition of the MDR1 promoter by a hybrid Mrr1 protein containing a heterologous activation domain, we created Mrr1DB-Gal4AD fusions in which the Mrr1 DNA binding domain was sequentially shortened from its C-terminal end. The N-terminal 106 amino acids of Mrr1 still were sufficient to allow the activation of the MDR1 promoter by the hybrid protein, albeit at a reduced level compared to that of proteins containing 128 or 121 N-terminal amino acids of Mrr1. In contrast, a fusion protein containing only the first 90 amino acids of Mrr1 did not significantly activate MDR1 expression (Fig. 1B). The ability of the various Mrr1DB-Gal4AD hybrid proteins to activate the MDR1 promoter correlated with their capacity to increase fluconazole resistance when expressed in mrr1Δ mutants of strain SC5314 (Fig. 1C). Of note, a slight increase in fluconazole resistance also was observed in cells expressing the shortest hybrid protein, indicating that it retained some ability to activate other Mrr1 target genes that also contribute to fluconazole resistance (32). Western immunoblot analysis with an anti-HA antibody showed that all hybrid proteins were expressed at similar levels (Fig. 1D). The HA-tagged Mrr1DB-Gal4AD fusion proteins were detected as a double band, which may be due to a protein modification, but this has not been studied further. These results demonstrate that the first 106 amino acids in the N terminus of Mrr1 are sufficient to target the transcription factor to the MDR1 promoter.
Transcriptional activity of TetR-Mrr1 fusion proteins.
To investigate if Mrr1 could confer transcriptional activity upon a heterologous DNA binding protein, we deleted the DNA binding domain from Mrr1 and fused the remainder of the protein (amino acids 67 to 1108) to the Tet repressor (TetR) from Escherichia coli. For this purpose, a cassette was designed in which a Candida-adapted tetR gene (catetR) or a desired catetR fusion gene is expressed from the ADH1 promoter. The cassette also contains the caGFP reporter gene under the control of a TetR-regulated promoter to allow the quantification of transcriptional activation by the TetR fusion proteins (Fig. 2 A). When expressed in strain SC5314, the hybrid TetR-Mrr1 fusion protein could activate the Tet promoter in the presence of benomyl (Fig. 2B). This result demonstrated that Mrr1 retained its transcription activation function and dependence on the inducer benomyl when its own DNA binding domain was replaced by TetR. To identify the transcriptional activation domain(s) of Mrr1, overlapping Mrr1 fragments (amino acids 67 to 526, 269 to 812, and 524 to 1108) were fused to TetR. However, none of these hybrid proteins could activate the Tet promoter (Fig. 2B, last three constructs). We therefore generated additional TetR fusion proteins in which the Mrr1 part was sequentially shortened from its N terminus. The ability of a TetR-Mrr1 hybrid protein to activate the Tet promoter was already strongly reduced when 143 amino acids from the N terminus of Mrr1 were missing, and hardly any activity could be observed with a fusion protein lacking the N-terminal 167 amino acids of Mrr1 (Fig. 2B). Therefore, the Mrr1 fragments used in the tested hybrid proteins were not useful for the delimitation of a transcriptional activation domain in Mrr1.
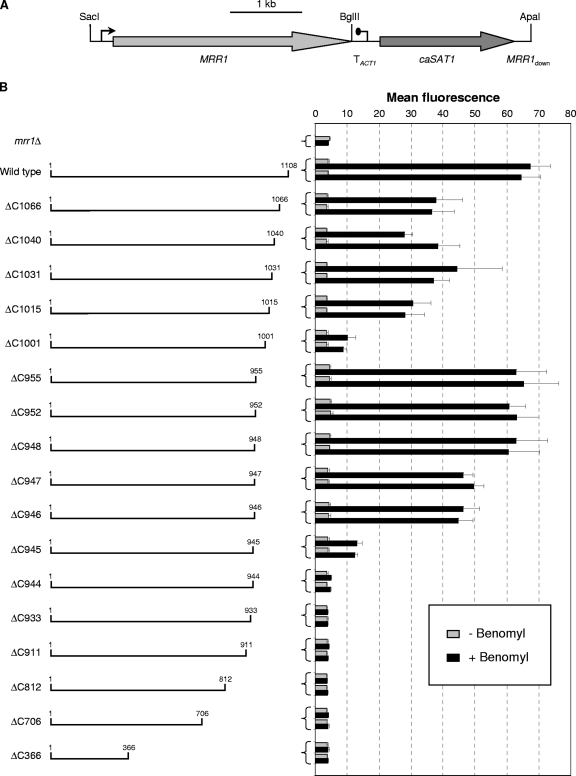
Analysis of C-terminally truncated Mrr1 proteins.
To further delimit the regions of Mrr1 that are required for its activity, we generated Mrr1 derivatives that were sequentially truncated from the C terminus and tested their ability to activate a PMDR1-caGFP reporter fusion (Fig. 3). The deletion of the last 42 (ΔC1066) to 93 (ΔC1015) amino acids reduced the activity of Mrr1, but the truncated derivatives were still well inducible by benomyl. A strong decrease in Mrr1 activity was observed when the C-terminal 107 amino acids were lacking (ΔC1001). Interestingly, the deletion of 46 additional amino acids (ΔC955) restored Mrr1 activity to wild-type levels. A further truncation to amino acid 944 completely abolished the ability of Mrr1 to activate the MDR1 promoter. Similarly, all tested shorter versions of Mrr1 were nonfunctional. None of the C-terminal truncations resulted in a constitutively active protein, because MDR1 induction occurred only in the presence of benomyl. These results suggested that the C-terminal 107 amino acids of Mrr1 contain an activation domain that is required for its efficient induction by benomyl. However, this domain is dispensable when the region located between amino acids 955 and 1001 is absent. Additional activation and regulatory domains must be located within the first 948 amino acids, because Mrr1 lacking the last 160 amino acids was fully able to activate the MDR1 promoter in a benomyl-dependent fashion.
Fig. 3.
(A) Map of the insert from plasmid pZCF36K2, which contains the wild-type MRR1 gene and served as the basis for the generation of MRR1 genes with C-terminal truncations and internal deletions. (B) MDR1 promoter activity in transformants of the reporter strain CAG48MRR1M4B expressing a wild-type copy of MRR1 or C-terminally truncated derivatives (the first and last amino acids in the encoded proteins are indicated). Strains were grown in the absence or presence of benomyl, and the mean fluorescence of the cells was determined by flow cytometry. The results obtained with two independent transformants are shown in each case (means and standard deviations from three experiments). The parental strain CAG48MRR1M4B (mrr1Δ) was included as a control.
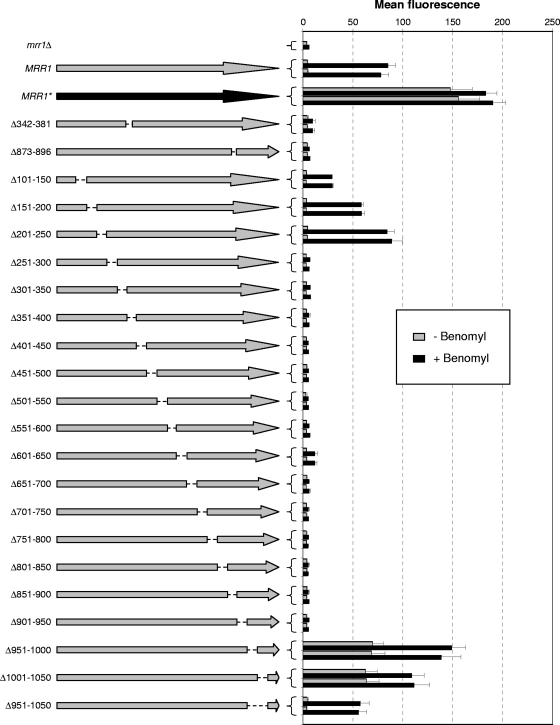
Analysis of Mrr1 proteins with internal deletions.
With the aim of identifying regulatory domains in Mrr1, we generated derivatives lacking internal regions. Mrr1 contains several hot spots where gain-of function mutations, which render the transcription factor constitutively active, occur in MDR1-overexpressing, fluconazole-resistant strains. Assuming that the absence of these regions would similarly result in a constitutively active protein, we first generated Mrr1 derivatives in which two of these hot spots were deleted. However, mutated proteins lacking amino acids 342 to 381 or amino acids 873 to 896 were nonfunctional and failed to induce the MDR1 promoter (Fig. 4). We therefore constructed a series of Mrr1 proteins with systematic short deletions of 50 amino acids, starting from amino acid 101. The deletion of amino acids 101 to 150 and 151 to 200 only reduced, but did not abolish, the benomyl-induced activation of the MDR1 promoter, and an Mrr1 protein lacking amino acids 201 to 250 behaved like wild-type Mrr1 in these assays. In contrast, all subsequent deletions from amino acids 251 to 950 eliminated Mrr1 activity, except for Δ601-650, which retained a slight inducibility. Strikingly, the deletion of either amino acids 951 to 1000 or amino acids 1001 to 1050 resulted in a constitutively active protein that activated the MDR1 promoter in the absence of the inducer benomyl. This constitutive activity was lost when the complete region between amino acids 951 and 1050 was deleted, but the mutated protein still was inducible by benomyl. These results indicated that the region between amino acids 951 and 1050 of Mrr1 contains an inhibitory domain that keeps the transcription factor in an inactive state in the absence of an inducer.
Fig. 4.
MDR1 promoter activity in transformants of the reporter strain CAG48MRR1M4B expressing a wild-type copy of MRR1 (gray arrow), the hyperactive MRR1P683S allele (MRR1*; black arrow), or MRR1 with the indicated internal deletions (amino acids missing from the encoded proteins are indicated to the left). Strains were grown in the absence or presence of benomyl, and the mean fluorescence of the cells was determined by flow cytometry. The results obtained with two independent transformants are shown in each case (means and standard deviations from three experiments). The parental strain CAG48MRR1M4B (mrr1Δ) was included as a control.
Identification of a C-terminal activation domain in Mrr1.
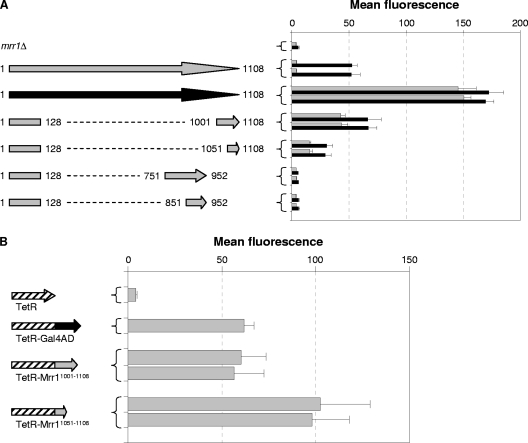
A comparison of the results obtained with the C-terminally truncated Mrr1 proteins and the proteins containing internal deletions provided valuable clues about the functional domains of the transcription factor. Mrr1 proteins that were truncated near amino acids 950 (ΔC955 to ΔC948) exhibited wild-type inducibility by benomyl but no constitutive activity (Fig. 3). Interestingly, the addition of the C-terminal 108 amino acids to the first 950 amino acids of Mrr1 conferred constitutive activity upon the transcription factor (Fig. 4, Δ951-1000). This result indicated that the last 108 amino acids of Mrr1 contain an activation domain that is constitutively active in the absence of an inhibitory domain located between amino acids 951 and 1000. Similarly, a truncated Mrr1 ending at amino acid 1001 had lost most of its activity (Fig. 3, ΔC1001), but the addition of the last 58 amino acids to the first 1,000 amino acids of Mrr1 also rendered the transcription factor constitutively active (Fig. 4, Δ1001-1050), suggesting that the activation domain is located between amino acids 1051 and 1108. The fact that the fusion of the C-terminal 58 amino acids to the first 950 amino acids of Mrr1 did not result in constitutive activity (Fig. 4, Δ951-1050) may be due to specific constraints of the hybrid protein, e.g., because the activation domain requires a flexible linker to achieve an active conformation in the absence of an inducer.
To test whether the C terminus of Mrr1 indeed contains an activation domain, we fused the last 58 or 108 amino acids of Mrr1 to its DNA binding domain (amino acids 1 to 128) and evaluated the ability of the resulting proteins to activate the MDR1 promoter. As can be seen in Fig. 5 A, both proteins constitutively activated the MDR1 promoter, although the shorter protein displayed somewhat lower activity than the longer version. In a complementary approach, we also fused the C-terminal 58 and 108 amino acids to TetR and tested the capacity of the hybrid proteins to induce the Tet promoter (Fig. 5B). Indeed, both TetR-Mrr1 fusion proteins activated the TetR-dependent promoter at least as efficiently as a TetR-Gal4AD fusion protein, confirming that the C-terminal 58 amino acids of Mrr1 contain a transcriptional activation domain. The fact that the shorter C-terminal Mrr1 fragment, which contained only 58 amino acids, exhibited lower activity than the 108-amino-acid fragment when fused to the Mrr1 DNA binding domain but higher activity than the 108-amino-acid fragment when fused to TetR most likely is explained by structural properties or slightly different stabilities of the fusion proteins.
Fig. 5.
(A) MDR1 promoter activity in transformants of the reporter strain CAG48MRR1M4B expressing a wild-type copy of MRR1 (gray arrow), the hyperactive MRR1P683S allele (black arrow), or fusions of the MRR1 DNA binding domain (codons 1 to 128) to different C-terminal or internal regions of MRR1 (amino acids contained in the encoded proteins are indicated). Strains were grown in the absence or presence of benomyl, and the mean fluorescence of the cells was determined by flow cytometry. The results obtained with two independent transformants are shown in each case (means and standard deviations from three experiments). The parental strain CAG48MRR1M4B (mrr1Δ) was included as a control. (B) MDR1 promoter activity in transformants of strain SC5314 expressing the indicated TetR-Mrr1 fusion proteins and containing the Ptet-caGFP reporter fusion. Strains were grown to log phase, and the mean fluorescence of the cells was determined by flow cytometry. The results obtained with two independent transformants are shown in each case (means and standard deviations from three experiments). Strains expressing a TetR-Gal4AD fusion protein or unfused TetR served as positive and negative controls, respectively.
As mentioned above, the C-terminal activation domain of Mrr1 was dispensable when the inhibitory domain located between amino acids 951 and 1050 was deleted, and a C-terminally truncated Mrr1 ending at amino acid 948 retained wild-type inducibility but lost its activity when it was further truncated to amino acid 944 (Fig. 3). This observation indicated that Mrr1 contains another transcriptional activation domain that is located proximal to amino acid 948. To delimit this second activation domain, we fused an Mrr1 fragment extending from amino acids 851 to 952 and a longer fragment extending from amino acids 751 to 952 to the Mrr1 DNA binding domain. However, the resulting fusion proteins were unable to induce the MDR1 promoter (Fig. 5A). It therefore seems that the second activation domain includes extended regions of Mrr1, which may overlap with inhibitory sequences and cannot be easily identified by fusing shorter protein fragments to a DNA binding domain.
DISCUSSION
In this study, we dissected the C. albicans zinc cluster transcription factor Mrr1 with the aim of identifying functional domains that are required to target it to the promoter of the MDR1 efflux pump, to activate transcription in response to the inducer benomyl, and to retain it in an inactive state in the absence of inducing conditions. We found that the N-terminal region of Mrr1 containing the DNA binding domain was sufficient to promote MDR1 expression when it was fused to a heterologous transcription activation domain, demonstrating that the C-terminal 900 amino acids are dispensable for targeting Mrr1 to the MDR1 promoter. This result does not exclude the possibility that the DNA binding domain of Mrr1 interacts with other proteins that may recruit it to the MDR1 promoter. In this respect, it is worth noting that an Mrr1-Gal4AD fusion protein that contained only the first 90 amino acids of Mrr1, including the Zn2Cys6 motif, did not significantly activate the MDR1 promoter but still conferred slightly increased fluconazole resistance. Mrr1 controls additional genes besides MDR1 that also contribute to fluconazole resistance (32). Therefore, the requirements for Mrr1-induced gene expression seem to differ between Mrr1 target genes. Depending on the conditions, Mrr1 cooperates with Cap1 and/or Mcm1 to induce MDR1 expression (19, 32), although such a cooperation may not necessarily involve a physical interaction between the proteins. Attempts to copurify Mrr1-interacting proteins with a TAP-tagged Mrr1 were unsuccessful (our unpublished results). The DNA binding domain of Mrr1 extends beyond the zinc cluster motif (amino acids 31 to 59), in line with the fact that this motif often is followed by additional important regions, the linker domain and a dimerization domain, in other members of the family (17).
Transcriptional activation domains of zinc cluster proteins often are located at their C terminus (17). Unlike the DNA binding domain, they are not defined by specific primary sequences and therefore must be experimentally determined. This can be done by fusing them to a DNA binding domain and assessing the capacity of the hybrid protein to activate a suitable reporter gene. We have used the Tet repressor from E. coli as a DNA binding domain to identify transcriptional activation domains of Mrr1 and determined the ability of TetR-Mrr1 fusion proteins to activate a TetR-dependent promoter that was previously established in our laboratory (24). By this approach we could identify an activation domain (AD1) that is located within the C-terminal 58 amino acids of Mrr1 (Fig. 5B). In the wild-type Mrr1, AD1 did not stimulate MDR1 expression in the absence of an inducer due to the presence of an inhibitory domain (ID1) located nearby between amino acids 951 and 1050. When portions of this inhibitory domain were deleted (Δ951-1000 or Δ1001-1050), Mrr1 became constitutively active in the absence of an inducer (Fig. 4). This constitutive activity depended on AD1, because it was lost when AD1 was additionally deleted (ΔC955 to ΔC946) (Fig. 3B). Interestingly, several Mrr1 fragments containing both AD1 and ID1 were unable to activate transcription when fused to TetR even in the presence of the inducer benomyl (TetR-Mrr1168-1108, TetR-Mrr1269-1108, TetR-Mrr1524-1108) (Fig. 2B). These results indicate that in the presence of ID1, AD1 requires additional regions of Mrr1 that extend into the N-terminal part of the protein. In line with this, all small deletions between amino acids 250 and 950 abolished the ability of Mrr1 to induce the MDR1 promoter (Fig. 4). It is likely that the absence of any of these regions prevents Mrr1 from achieving an inducible conformation or renders the protein unstable.
Our results also demonstrate that Mrr1 contains further activation and regulatory domains, because C-terminally truncated proteins lacking AD1 and ID1 (ΔC955 to ΔC946) could efficiently activate the MDR1 promoter in a benomyl-dependent fashion (Fig. 3B). A second activation domain seems to involve amino acid residues proximal to position 948, because Mrr1 ending at amino acid 948 exhibited wild-type inducibility, whereas the removal of the next four amino acids (ΔC944) rendered the transcription factor inactive. However, we were unable to determine the extent of this activation domain, because fusions of amino acids 851 to 952 or amino acids 751 to 952 to the Mrr1 DNA binding domain did not result in transcriptional activity in either the absence or in the presence of the inducer benomyl. It is possible that the second activation domain is located near an inhibitory region and/or requires further N-terminally located regions of Mrr1 to induce transcription. ID1, or at least parts of it, apparently also affects the second activation domain, because the activity of a truncated protein lacking AD1 (ΔC1001) was strongly reduced, and its inducibility by benomyl only was restored to wild-type levels when ID1 was completely removed (Fig. 3B).
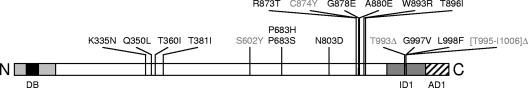
Figure 6 gives an overview on the functional domains of Mrr1 that were identified in the present study and the location of all known gain-of-function mutations that result in constitutive activity of the transcription factor. It is striking that four different gain-of-function mutations found in MDR1-overexpressing strains, two amino acid substitutions and two small in-frame deletions, are located within ID1, between amino acids 993 and 1005. The affected amino acids therefore are important for the functionality of ID1, and their mutation has an effect similar to that of the deletion of ID1. On the contrary, all other gain-of-function mutations found in MDR1-overexpressing, fluconazole-resistant strains are located in regions whose deletion resulted in a nonfunctional rather than a hyperactive transcription factor. Therefore, these regions are important for transcriptional activation but also contain amino acid residues that prevent Mrr1 from becoming active under noninducing conditions.
Fig. 6.
Location of identified functional domains of Mrr1. The Mrr1 protein is represented as a linear bar from the N to the C terminus. The light gray area (DB; amino acids 1 to 106) contains the DNA binding domain and represents the region that was sufficient for the activation of the MDR1 promoter when fused to the Gal4 activation domain. The zinc cluster motif (amino acids 31 to 59) is indicated by black shading. The hatched area (AD1; amino acids 1051 to 1108) depicts the distal activation domain 1, which could activate the TetR-dependent promoter when fused to TetR. The dark gray area (ID1; amino acids 951 to 1050) was defined as inhibitory domain 1, because the deletion of amino acids 951 to 1000 or 1001 to 1050 resulted in constitutive activity of Mrr1. The locations of all known activating mutations found in MDR1-overexpressing C. albicans strains are indicated. Gain-of-function mutations found at the corresponding positions in Mrr1 of C. dubliniensis are indicated in gray letters.
Many zinc cluster proteins contain a conserved domain, known as the middle homology region (MHR), which regulates their transcriptional activity. The deletion of this region in Hap1 or Leu3 results in constitutive activity of the transcription factors (9, 25). In addition, several gain-of-function mutations in Pdr1 of C. glabrata are located in the MHR (8). The middle homology region also is present in Mrr1, from amino acids 560 to 664 (31). Interestingly, none of the deletions in this region resulted in constitutive activity of Mrr1. Rather, mutated Mrr1 proteins lacking amino acids 551 to 600 or amino acids 601 to 650 could not or could not efficiently induce the MDR1 promoter in response to benomyl (Fig. 3). Only one gain-of-function mutation in the MHR has been found in Mrr1 of Candida dubliniensis, at amino acid 595 (corresponding to S602 in Mrr1 of C. albicans) (33). The precise function of the MHR in Mrr1 therefore is unclear, but apparently it is important both for the functionality of the transcription factor and for regulating its activity.
Of note, no gain-of-function mutation has been described so far in the distal activation domain of Mrr1. In contrast, several such mutations have been found in the confirmed or putative C-terminal activation domains of Tac1 of C. albicans, Pdr1 of C. glabrata, and Pdr1 and Pdr3 of S. cerevisiae (31). These mutations probably relieve the transcription factors from repression by their inhibitory domains, and it is possible that a systematic search would identify similar mutations in Mrr1. In fact, the C-terminal tagging of Mrr1 with a 3× HA epitope resulted in constitutive activity of the transcription factor (32) (Fig. 1B), presumably by relieving AD1 from inhibition by ID1.
In addition to providing important insights into the functional domains of Mrr1, our present work also introduces some novel tools for the functional analysis of transcription factors in C. albicans. The cassette contained in plasmid pZCF36DBH2 (Fig. 1A) can be used to fuse potential DNA binding domains of other transcription factors to the Gal4 activation domain by inserting them in place of the Mrr1 DNA binding domain between the unique XhoI and NarI sites. After the integration of the resulting constructs into a suitable C. albicans host strain, the fusion proteins are constitutively expressed from the ADH1 promoter and can be tested for their ability to induce the expression of the target genes of the transcription factor and resulting phenotypes. An even more valuable tool is provided by the TetR fusion cassette contained in plasmid pTET52 (Fig. 2A), which allows the experimental identification of transcriptional activation domains. Gene fragments from transcription factors can be inserted between the unique XhoI and BglII sites of this plasmid for in-frame fusion with the catetR gene. After insertion into the C. albicans genome, the encoded TetR fusion proteins are expressed from the ADH1 promoter, and their transcriptional activity can be conveniently tested by their ability to induce the expression of the caGFP reporter gene from the TetR-dependent promoter. As the reporter gene is present in the same cassette, the activity of the hybrid proteins can be quantified in any host strain without the necessity of additional genetic manipulations. A one-hybrid system based on LexA from Staphylococcus aureus has already been established for C. albicans and used for the identification of transcription factor activation domains (18, 30). The TetR-based system described in our present study offers an alternative approach, and the presence of the reporter gene and the dominant caSAT1 selection marker in the same cassette allows the highly flexible use of the system.
ACKNOWLEDGMENTS
This study was supported by the Deutsche Forschungsgemeinschaft (DFG grant MO 846/3 and SFB 630) and the National Institutes of Health (NIH grant AI058145).
Footnotes
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Coste A., et al. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell 6:1889–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coste A., et al. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coste A. T., Crittin J., Bauser C., Rohde B., Sanglard D. 2009. Functional analysis of cis- and trans-acting elements of the Candida albicans CDR2 promoter with a novel promoter reporter system. Eukaryot. Cell 8:1250–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coste A. T., Karababa M., Ischer F., Bille J., Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Micheli M., Bille J., Schueller C., Sanglard D. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197–1214 [DOI] [PubMed] [Google Scholar]
- 6. Dunkel N., Blaß J., Rogers P. D., Morschhäuser J. 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69:827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dunkel N., et al. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 7:1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferrari S., et al. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 5:e1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friden P., Reynolds C., Schimmel P. 1989. A large internal deletion converts yeast LEU3 to a constitutive transcriptional activator. Mol. Cell. Biol. 9:4056–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 11. Heilmann C. J., Schneider S., Barker K. S., Rogers P. D., Morschhäuser J. 2010. An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob. Agents Chemother. 54:353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoot S. J., Smith A. R., Brown R. P., White T. C. 2011. An A643V amino acid substitution in Upc2p contributes to azole resistance in well-characterized clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 55:940–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karababa M., Coste A. T., Rognon B., Bille J., Sanglard D. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 48:3064–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Köhler G. A., White T. C., Agabian N. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lucau-Danila A., et al. 2005. Early expression of yeast genes affected by chemical stress. Mol. Cell. Biol. 25:1860–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. MacPherson S., et al. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49:1745–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. MacPherson S., Larochelle M., Turcotte B. 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70:583–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martchenko M., Levitin A., Whiteway M. 2007. Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryot. Cell 6:291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mogavero S., Tavanti A., Senesi S., Rogers P. D., Morschhäuser J. 2011. Differential requirement of the transcription factor Mcm1 for activation of the Candida albicans multidrug efflux pump MDR1 by its regulators Mrr1 and Cap1. Antimicrob. Agents Chemother. 55:2061–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morschhäuser J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587:240–248 [DOI] [PubMed] [Google Scholar]
- 21. Morschhäuser J. 2010. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 47:94–106 [DOI] [PubMed] [Google Scholar]
- 22. Morschhäuser J., et al. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Näär A. M., Thakur J. K. 2009. Nuclear receptor-like transcription factors in fungi. Genes Dev. 23:419–432 [DOI] [PubMed] [Google Scholar]
- 24. Park Y.-N., Morschhäuser J. 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 4:1328–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfeifer K., Kim K. S., Kogan S., Guarente L. 1989. Functional dissection and sequence of yeast HAP1 activator. Cell 56:291–301 [DOI] [PubMed] [Google Scholar]
- 26. Reuß O., Morschhäuser J. 2006. A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol. Microbiol. 60:795–812 [DOI] [PubMed] [Google Scholar]
- 27. Reuß O., Vik Å., Kolter R., Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 28. Rognon B., Kozovska Z., Coste A. T., Pardini G., Sanglard D. 2006. Identification of promoter elements responsible for the regulation of MDR1 from Candida albicans, a major facilitator transporter involved in azole resistance. Microbiology 152:3701–3722 [DOI] [PubMed] [Google Scholar]
- 29. Ruhnke M., et al. 1994. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J. Clin. Microbiol. 32:2092–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Russell C. L., Brown A. J. 2005. Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gcn4 is a transcriptional activator. Fungal Genet. Biol. 42:676–683 [DOI] [PubMed] [Google Scholar]
- 31. Sanglard D., Coste A., Ferrari S. 2009. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 9:1029–1050 [DOI] [PubMed] [Google Scholar]
- 32. Schubert S., et al. 2011. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob. Agents Chemother. 55:2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schubert S., Rogers P. D., Morschhäuser J. 2008. Gain-of-function mutations in the transcription factor MRR1 are responsible for overexpression of the MDR1 efflux pump in fluconazole-resistant Candida dubliniensis strains. Antimicrob. Agents Chemother. 52:4274–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Silver P. M., Oliver B. G., White T. C. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3:1391–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thakur J. K., et al. 2008. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452:604–609 [DOI] [PubMed] [Google Scholar]
- 36. Znaidi S., et al. 2009. Identification of the Candida albicans Cap1p regulon. Eukaryot. Cell 8:806–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Znaidi S., et al. 2007. The zinc cluster transcription factor Tac1p regulates PDR16 expression in Candida albicans. Mol. Microbiol. 66:440–452 [DOI] [PubMed] [Google Scholar]