Abstract
Fluconazole is a commonly used antifungal drug that inhibits Erg11, a protein responsible for 14α-demethylation during ergosterol synthesis. Consequently, ergosterol is depleted from cellular membranes and replaced by toxic 14α-methylated sterols, which causes increased membrane fluidity and drug permeability. Surface-grown and planktonic cultures of Candida albicans responded similarly to fluconazole at 0.5 mg/liter, showing reduced biomass formation, severely reduced ergosterol levels, and almost complete inhibition of hyphal growth. There was no evidence of cell leakage. Mass spectrometric analysis of the secretome showed that its composition was strongly affected and included 17 fluconazole-specific secretory proteins. Relative quantification of 14N-labeled query walls relative to a reference standard mixture of 15N-labeled yeast and hyphal walls in combination with immunological analysis revealed considerable fluconazole-induced changes in the wall proteome as well. They were, however, similar for both surface-grown and planktonic cultures. Two major trends emerged: (i) decreased incorporation of hypha-associated wall proteins (Als3, Hwp1, and Plb5), consistent with inhibition of hyphal growth, and (ii) increased incorporation of putative wall repair-related proteins (Crh11, Pga4, Phr1, Phr2, Pir1, and Sap9). As exposure to the wall-perturbing drug Congo red led to a similar response, these observations suggested that fluconazole affects the wall. In keeping with this, the resistance of fluconazole-treated cells to wall-perturbing compounds decreased. We propose that fluconazole affects the integrity of both the cellular membranes and the fungal wall and discuss its potential consequences for antifungal therapy. We also present candidate proteins from the secretome for clinical marker development.
INTRODUCTION
Although Candida albicans is normally dwelling in the human flora as a harmless commensal, it is also capable of causing disease, especially when the immune system is weakened. The resulting infections range from superficial infections of the mucosal layers to life-threatening systemic infections. A common way to treat this fungal overload is by azole administration, predominantly in the form of fluconazole (FCZ). Azoles execute their antifungal activity mainly by inhibiting the cytochrome P450 enzyme Erg11, which is required for 14α-demethylation of lanosterol during ergosterol biosynthesis (46). This results in the replacement of ergosterol by methylated sterols, such as 14α-methyl-3,6-diol. The presence of an additional 6-OH group disturbs membrane packing and decreases membrane rigidity, resulting in water penetration and increased drug uptake (1, 15). The azole-induced membrane changes evoke numerous other responses in the cell. For example, ergosterol depletion has also been found to inactivate vacuolar ATPase (V-ATPase) function, impairing vacuolar acidification (70). Furthermore, hyphal development is affected (49). In addition, it has been suggested that azole treatment evokes oxidative and nitrosative stress (6). Finally, transcriptional data suggest that azoles could have an impact on the wall proteome and secretome of C. albicans (13, 38).
Secreted proteins, like hydrolytic enzymes, are mainly involved in the destruction of tissue and nutrient uptake (54). Together with the wall proteins, they are acting at the interface between the host and the fungus. Wall proteins are also required for the adherence to plastic surfaces and tissues, formation of biofilms, and tissue invasion. Furthermore, they promote nutrient uptake and defense against the host immune system and play a pivotal role in maintaining the integrity of the cell wall (28). Cell wall integrity is crucial for cell survival since it acts as an opposing force to the intracellular turgor pressure preventing the cell from bursting under hypo-osmotic conditions. It is also fundamental for maintaining cell shape, bud formation, and proper cell division. Since a strong wall is essential, adequate maintenance and regulation are required. External factors that generate cell wall stress as well as cell cycle progression lead to (local) wall weakening, which in turn activates the cell wall integrity (CWI) pathway (reviewed in reference 36). For instance, stretching of the plasma membrane, hypo-osmotic shock, exposure to the wall-degrading enzyme preparation Zymolyase or to wall-perturbing compounds such as calcofluor white, which binds chitin, and Congo red, which preferentially perturbs wall construction in the neck region (29), lead to activation of the CWI pathway (17, 26, 42, 47). Another trigger is oxidative stress (4, 47). Upon alterations in the fungal wall, plasma membrane-bound sensor proteins get activated and trigger mitogen-activated protein kinase (MAPK) signaling cascades that end in the activation of transcription factors involved in wall maintenance. Consequently, the transcription of wall biosynthetic and wall repair genes increases, and the wall gets reinforced by higher chitin levels (44). In C. albicans the MAPKs Mkc1 and Cek1 are involved in the response to various forms of wall stress (31, 47, 52).
In this study we used mass spectrometry (MS) to systematically identify and quantify the effects of fluconazole on the wall proteome and secretome of C. albicans. Surface-grown and planktonic cultures showed similar major fluconazole-induced changes in their wall proteomes. The secretome of planktonic cultures also showed considerable changes, including the appearance of many fluconazole-specific proteins. Interestingly, the incorporation levels of four wall proteins in surface-grown cultures were considerably higher than in planktonic cultures, both in the absence and presence of fluconazole, suggesting a role for these proteins in surface growth. Considerable increases in the relative abundance of proteins associated with the cell wall integrity response were observed. This was accompanied by decreased resistance against wall-perturbing agents. Additionally, the abundances of a similar set of wall proteins increased upon exposure to the wall-perturbing compound Congo red. In summary, our data show that fluconazole compromises fungal wall integrity and causes major changes in the proteomes of the wall and the culture medium.
MATERIALS AND METHODS
Strains and growth conditions.
Chemicals were obtained from Sigma-Aldrich, unless stated otherwise. C. albicans SC5314 (21) was precultured in liquid YPD medium (10 g/liter yeast extract, 20 g/liter peptone, and 20 g/liter glucose) and incubated overnight at 30°C and 200 rpm in a rotary shaker. The preculture was spun down and washed in phosphate-buffered saline (PBS) buffer to remove the medium and either used to inoculate liquid cultures or to inoculate petri dishes containing semisolid low-agarose medium. For experiments with liquid cultures, 50 ml of YNB-S medium (6.7 g/liter yeast nitrogen base [YNB], 20 g/liter sucrose, and 75 mM MOPSO [3-(N-morpholino)-2-hydroxypropanesulfonic acid] set to pH 7.4) was inoculated with the preculture to an optical density at 600 nm (OD600) of 0.05. For drug-treated cultures, 0.5 mg/liter (1.6 μM) fluconazole from a 1 g/liter stock solution or 2 mg/liter (2.9 μM) Congo red was added. The liquid cultures were incubated for 18 h at 37°C and 200 rpm. For experiments with agarose-grown cultures, about 6 × 106 cells of the preculture were plated on low-agarose medium containing 1.7 g/liter YNB medium without ammonium sulfate, 5 g/liter porcine stomach mucin as a sole nitrogen source, 3 g/liter agarose, 5 mM glucose, and 75 mM MOPSO set to pH 7.4 and with or without 0.5 mg/liter FCZ. The petri dishes were incubated for 18 h at 37°C.
Analysis of morphology, biomass, and ergosterol content.
For analysis of the morphology after 18 h of fluconazole treatment, the cells were visualized by light microscopy and photographed. For determination of the biomass, liquid cultures were spun down after 18 h of growth, and the pellet was dried at 60°C overnight and weighed. Since cells grew invasively on a semisolid surface, the whole content of the plates was first solubilized in 6 M guanidine thiocyanate at 60°C. Subsequently, the cells were spun down, and the pellet was dried and weighed. For analyzing the ergosterol content, the cells were cultivated as described above. Cells grown on low-agarose plates were washed off the surface and collected. The extraction procedure was performed as described previously (9). Two milliliters of the wash-off was quenched with 6 ml of ice-cold methanol, the remaining part was dried, and the biomass was determined. For liquid cultures, 2 ml was quenched directly with 6 ml of ice-cold methanol. The remaining part of the culture was used for biomass determination. Six milliliters of petroleum ether (boiling point, 40 to 60°C) was added to the 2 ml of cells suspended in 6 ml of methanol. After the sample was vortexed for 1 min and centrifuged (2 min at 3,000 rpm), the upper petroleum ether phase was transferred into an N2-flushed tube. The whole procedure was repeated once again, and the upper phases were combined. When the petroleum ether was completely evaporated, the sample was resuspended with a glass rod in 60 μl of ethanol, and 10 μl was injected onto a LiChrosorb RP-18 high-performance liquid chromatography (HPLC) column (Chrompack; Bergen op Zoom, The Netherlands). Methanol was used as the mobile phase, and the flow rate was 2 ml/min at 50°C. Ergosterol was detected at 290 nm, and an ergosterol standard was used to calculate the measured amounts. The ergosterol content was normalized to biomass dry weight.
Cell wall isolation.
For cell wall isolation, cells were harvested in the same way as for the biomass determination. Walls were isolated as previously described (16). In short, cells were first washed five times with cold demineralized water. Subsequently, they were resuspended in a 10 mM Tris-HCl buffer, pH 7.5, together with a protease inhibitor cocktail, and disintegrated with the help of 0.25- to 0.50-mm-diameter glass beads using a Bio-Savant Fast Prep 120 machine (Qbiogene, Montreal, Canada). After the crude walls were washed five times with 1 M NaCl and five times with demineralized water, they were boiled four times for 10 min in SDS extraction buffer (150 mM NaCl, 2% [wt/vol] SDS, 100 mM Na-EDTA, 100 mM β-mercaptoethanol, 50 mM Tris-HCl, pH 7.8). To remove the buffer, walls were washed with demineralized water repeatedly, freeze-dried, and stored at −80°C.
Analysis of fluconazole-induced changes of the C. albicans secretome.
The supernatant of the liquid cultures was centrifuged once again at 5,000 rpm for 10 min to remove remaining cells and was concentrated by 10-kDa cutoff filters (Amicon Ultra-15 Centrifugal filter units; Millipore, Billerica, MA). Using a bicinchoninic acid (BCA) assay and bovine serum albumin as a standard (56), the amount of secreted proteins was quantified and normalized to biomass dry weight. The concentrated proteins were reduced and alkylated and digested with trypsin as described for wall proteins. The peptide concentration was determined at 205 nm by a NanoDrop spectrophotometer (Isogen Life Science, IJsselstein, The Netherlands), and peptides were diluted to a final concentration of 75 ng/μl in 0.1% trifluoroacetic acid and 1% formic acid. The tryptic peptide mixtures were analyzed with liquid chromatography (LC)-nano-electrospray ionization tandem MS (ESI-MS/MS) using a quadrupole time of flight (QTOF) mass spectrometer (Micromass, Whyttenshawe, United Kingdom) coupled to an Ultimate 2000 nano-HPLC system (LC Packings, Amsterdam, The Netherlands). Data acquisition and processing details have been described previously (57). Five independent biological samples with three technical replicates each were analyzed for the two conditions: the cell cultures with (FCZ) and without (control) fluconazole. Proteins were identified by matching the processed MS/MS data with the complete C. albicans protein database (8) using the MASCOT (Matrix Science, United Kingdom) search engine. For each identified protein, the peptide count was summed over all five analyses of the FCZ and control samples (37). The FCZ/control ratio of the total number of tryptic peptide identifications for each individual protein (peptide count) was taken as an indication for the change in the corresponding protein level.
15N-labeled reference culture.
C. albicans from a preculture was used to inoculate a second preculture in YNB-S medium with 15N-labeled ammonium sulfate (15N content of >99%; Spectra Stable Isotopes), buffered with 75 mM tartaric acid at pH 4, and incubated overnight at 30°C. Tartaric acid was used because it has two pKa values close to pH 4 (4.37 and 3.02 at 37°C) and is not metabolized by C. albicans (our observations). The 15N-labeled preculture was used to inoculate two cultures of 600 ml of 15N-labeled YNB-S medium, either buffered with 75 mM tartaric acid at pH 4 to favor yeast growth or with 75 mM MOPSO at pH 7.4 for hyphal induction, to an OD600 of 0.1. The cultures were incubated at 37°C and 200 rpm for 18 h. After 18 h the 15N-labeled cells were spun down, the pellets were combined, and the walls were subsequently isolated, divided in aliquots, freeze-dried, and stored at −80°C.
Sample preparation for relative quantification.
14N-labeled walls from planktonic cultures or low-agarose plates were mixed with 15N-labeled reference walls in a ratio of 1:1 based on their dry weights. The combined walls with their covalently attached proteins were reduced with 10 mM dithiothreitol in 100 mM NH4HCO3 (1 h at 55°C), followed by alkylation with 65 mM iodoacetamide in 100 mM NH4HCO3 for 45 min at room temperature in the dark. After samples were quenched with 55 mM dithiothreitol in 100 mM NH4HCO3 for 5 min in the dark, they were washed six times with 50 mM NH4HCO3. The walls were digested with trypsin gold (Promega, Madison, WI) at 37°C for 18 h. After 18 h the walls were spun down, and the supernatant containing the tryptic peptides was transferred to a new tube. Subsequently, the peptides were desalted by a C18 tip column (Varian, Palo Alto, CA), and their concentrations were measured at 205 nm with a NanoDrop ND-1000 spectrophotometer (Isogen Life Science, IJsselstein, The Netherlands).
Wall proteome analysis of Congo red-treated cells.
Cells from liquid cultures supplemented with and without 2 mg/liter Congo red were harvested by centrifugation, and walls were isolated, reduced, and alkylated as described above. Further sample processing, data acquisition, and processing were carried out as described for secretome analysis except that the samples contained 25 ng/μl of tryptic peptides.
FTMS data acquisition, processing, and relative quantification.
A total of 800 ng of the mixtures of 14N- and 15N-labeled peptides in a solution of 0.1% trifluoroacetic acid was loaded on an Ultimate 3000 (Dionex, Sunnyvale, CA) HPLC system with a PepMap100 C18 (5-μm, 100 Å, 300-μm internal diameter by 5 mm) precolumn and a PepMap100 C18 (5 μm, 100 Å, 300-μm internal diameter by 250 mm) analytical column (Dionex, Sunnyvale, CA) coupled to an ApexQ FTMS (Bruker Daltonik, Bremen, Germany) equipped with a 7T magnet and a CombiSource instrument. Experimental details on data acquisition and processing were previously described (59; also C. J. Heilmann et al., unpublished data). Proteins were identified based on their 14N/15N-labeled peptide pairs, with the corresponding 14N/15N isotope abundance ratio as the protein level relative to the reference standard. Three or four independent biological replicates were analyzed per condition for semisolid surface- and liquid-grown cultures, respectively. First, the peptide 14N/15N isotope abundance ratios were averaged over the corresponding peptides for each protein. Then, resulting protein 14N/15N isotope abundance ratios were averaged over the replicates. Finally, the FCZ/control ratio was calculated for each protein as the FCZ 14N/15N protein isotope abundance ratio divided by the control 14N/15N protein isotope abundance ratio.
Immunoblot analysis of Hwp1 and Rbt5.
Glycosylphosphatidylinositol (GPI)-anchored proteins were released from freeze-dried walls as described previously (58). Cell walls (4 mg) were incubated overnight with 2.5 μl of recombinant Trichoderma harzianum endo-β-1,6-glucanase (10) and 2 μl of protease inhibitor cocktail (Sigma, St. Louis, MO) in 50 mM sodium phosphate buffer, pH 5.5, at 37°C. The released wall proteins were first separated on a 3 to 8% Tris-acetate polyacrylamide gradient gel (Invitrogen, San Diego, CA) and then transferred to an Immobilon polyvinylidene difluoride membrane (Millipore, Billerica, MA). Immunoblot analysis with polyclonal Hwp1 (60) or Pga10 (67) antiserum was performed in 5% (wt/vol) milk powder in PBS buffer for 2 h. After thorough washing with PBS, the membrane was incubated with goat anti-rabbit antiserum that was conjugated to peroxidase (Thermo Fisher Scientific, Waltham, MA) in 5% (wt/vol) milk powder in PBS. Proteins were visualized using enhanced chemiluminescence (ECL) detection reagent (GE Healthcare, Waukesha, WI).
qRT-PCR.
For quantitative reverse transcription-PCR (qRT-PCR) of Orf19.7104 and ACT1, as a reference gene, liquid cultures were inoculated from a preculture to an OD600 of 0.05 and incubated at 37°C without and with 0.5 mg/liter FCZ. After 4, 6, and 18 h the cultures were harvested. For each time point and condition, two independent biological replicates were analyzed. Total RNA was isolated using an RNeasy kit (Qiagen) and then treated with DNase (Ambion). cDNA was generated from 280 ng of RNA with Oligo(d)T12-18 primers using a SuperScript First-Strand Synthesis System (Invitrogen). PCRs were carried out with 1× Power SYBR green PCR Mastermix (Applied Biosystems), 8 μl of cDNA (derived from 10 ng of RNA), and 6 pmol of each primer (Orf19.7104 5′ to 3′ forward primer, TTG TCC TTT CCT TAG CCG GTA TAG, and 5′ to 3′ reverse primer, TTG CTT GTG GAG GTG TAT CAA GA; ACT1 5′ to 3′ forward primer, AGC TTT GTT CAG ACC AGC TGA TT, and 5′ to 3′ reverse primer, AGT TGA AAG TGG TTT GGT CAA TAC C). PCRs were run in technical duplicates on a 7300 Real-time PCR System (Applied Biosystems). Primer specificity was verified by determining a dissociation profile. The mRNA abundance of Orf19.7104 was normalized with respect to ACT1 levels.
Analyzing the effects of cell wall-perturbing agents.
C. albicans was grown in liquid YPD medium at 30°C overnight. Two microliters of 1/10 serial dilutions was spotted onto low-agarose plates containing YNB-S medium at pH 7.4. The plates were supplemented with either 0.5 mg/liter (1.6 μM) FCZ, calcofluor white (100 mg/liter), Congo red (200 mg/liter), and sodium dodecyl sulfate (100 mg/liter) or a combination of 0.5 mg/liter FCZ with each stress agent. After incubation for 2 days at 37°C, the plates were photographed.
RESULTS
FCZ influences surface-grown and planktonic cultures similarly.
The cell wall and its covalently attached proteins as well as the secreted proteins are of fundamental importance for fungal virulence and fitness. Since transcriptional studies have shown that azoles affect the transcript levels of several wall protein-encoding genes and also of some genes that code for proteins isolated from the culture medium (13, 38), we wanted to quantify the effect of fluconazole on the secretome and wall proteome of C. albicans. We chose two distinct ways of culturing C. albicans in the presence or absence of 0.5 mg/liter FCZ. The first setup is intended to mimic mucosal surfaces (59), where C. albicans frequently colonizes. It uses low-agarose plates, with mucin as the sole source of nitrogen and a low glucose concentration (59). After inoculation and incubation, the plates were solubilized in order to harvest cells that grew invasively into the surface. The other setup consisted of liquid cultures, using ammonium sulfate as a nitrogen source and 2% sucrose as a carbon source to avoid glucose repression (57). We selected a concentration of 0.5 mg/liter (1.6 μM) fluconazole, which severely inhibited the ergosterol synthesis (97 and 92% for planktonic and surface-grown cells, respectively) (Table 1). Unexpectedly, growth of planktonic and especially surface-grown cultures was considerably less reduced (74 and 31%, respectively) (Table 1). This suggests that nearly complete growth inhibition by fluconazole, as observed at higher fluconazole concentrations, is caused not only by inhibition of the cytochrome P450 enzyme Erg11, and hence abrogated demethylation, but also by an effect of fluconazole on other processes. Conceivably, at a higher concentration, fluconazole could inhibit another cytochrome P450 enzyme, Erg5, which is responsible for a later step in ergosterol synthesis that involves Δ22-desaturation in the side chain (34). It is further known that at higher azole concentrations, desaturation and elongation of fatty acids are inhibited (19, 66), further perturbing cellular membranes and consequently growth. In both experimental setups, azole-treated cultures grew predominantly in the yeast form with some clusters of nondissociated yeast cells. Few pseudohyphae and hyphae were present compared to the control, which consisted of a mixture of yeast, pseudohyphal, and hyphal cells (data not shown). The almost complete inhibition of hyphal growth at the fluconazole concentration used suggests that it is not so much associated with the only moderately decreased growth rate as with ergosterol content (23, 40). Table 1 further shows that control cells and FCZ-treated cells release about 1 μg of protein per mg of biomass (Table 1). This corresponds to about 0.25% of total cellular protein, based on the assumption that the protein content of yeast cells accounts for ∼40% of the total biomass (28). These results indicate that fluconazole at the concentration used does not cause significant cell leakage and that transwall transfer of cytosolic proteins by vesicular transport (3), at least under our experimental conditions, does not strongly contribute to the protein content of the medium. Importantly, the wall proteomes of surface-grown and planktonic cultures were largely similar (Fig. 1; also see below for further discussion). In view of the similarities between the results obtained with both approaches and the technical difficulties involved in processing agarose-grown cells, most experiments were carried out using planktonic cultures.
Table 1.
Morphology, biomass, ergosterol content, and amount of medium proteins of C. albicans grown in planktonic and surface-grown cultures with or without 0.5 mg/liter FCZ for 18 ha
| Growth condition and treatment | Morphology | Biomass (mg/ml) | Ergosterol content (μg/mg of biomass) | Amt of protein in medium (μg/mg of biomass) |
|---|---|---|---|---|
| Planktonic | ||||
| Control | Yeast + hyphae | 2.23 ± 0.09 | 2.63 ± 0.93 | 0.83 ± 0.33 |
| FCZ | Mainly yeast, few (pseudo)hyphae | 0.81 ± 0.28 | 0.08 ± 0.04 | 1.00 ± 0.05 |
| Surface-grown | ||||
| Control | Yeast + hyphae | 0.59 ± 0.03 | 1.69 ± 0.56 | |
| FCZ | Mainly yeast, few (pseudo)hyphae | 0.41 ± 0.04 | 0.14 ± 0.06 |
The values represent the means ± standard deviations. All data are from at least three independent biological replicates, measured independently. The amount of secreted medium proteins was determined only for planktonic cultures.
Fig. 1.
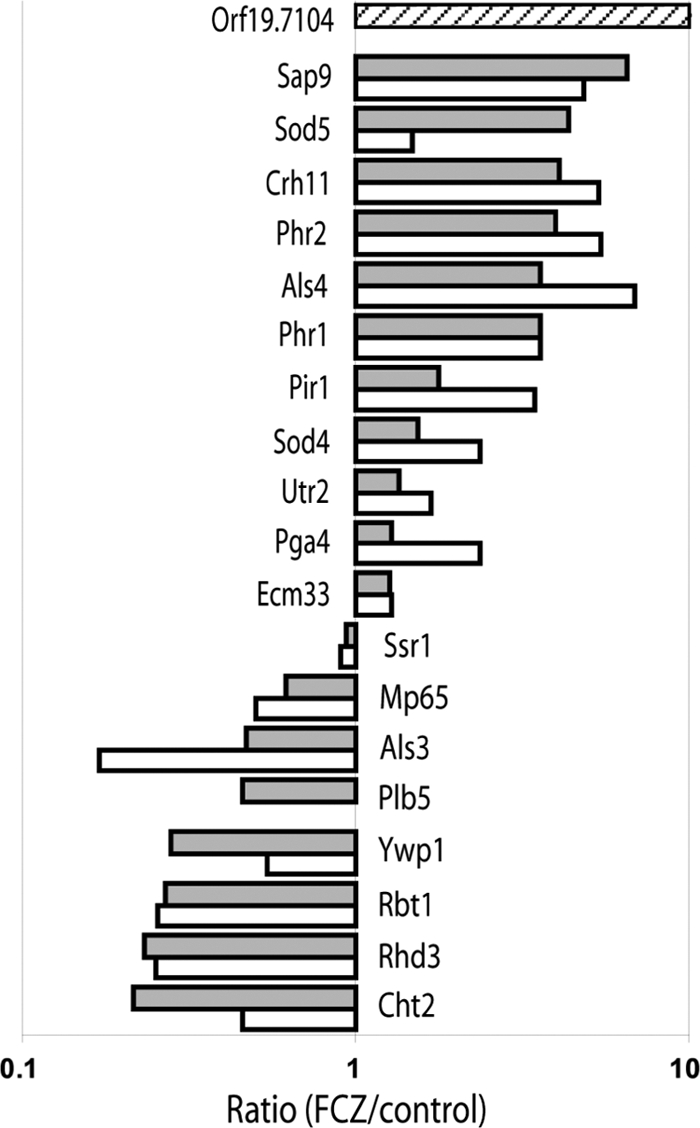
Relative changes in the wall proteome of C. albicans induced by fluconazole, expressed as the protein ratio FCZ/control. Gray bars, planktonic cultures; white bars, surface-grown cultures; striped bar, Orf19.7104, which was exclusively identified in planktonic cultures of FCZ-treated cells and was given an arbitrary ratio of 10. Ratios are plotted on a logarithmic scale. Values of >1 represent increased protein levels, and values of <1 represent decreased protein levels after 18 h of incubation with 0.5 mg/liter FCZ compared to the control.
FCZ strongly influences the composition of the secretome.
The tryptic digests of the medium proteins of planktonic C. albicans cultures grown with (FCZ) or without (control) fluconazole were analyzed by LC-MS/MS. Five independent biological FCZ and control replicates were each analyzed three times. For each identified protein, the peptide count was summed over all analyses of the FCZ and control samples. For all identified proteins the peptide counts are listed in Table 2, where the FCZ/control peptide count ratio is an indication of the relative change in the corresponding protein level.
Table 2.
Changes in the secretome of planktonic cultures of C. albicans induced by FCZ as indicated by the total number of identifications of tryptic peptides from secretory proteins
| Growth condition(s) and protein | Peptide counta |
FCZ/control ratio | |
|---|---|---|---|
| FCZ culture | Control culture | ||
| FCZ-treated cultures only | |||
| Cfl2c | 1 | ND | |
| Cht1 | 2 | ND | |
| Ece1 | 1 | ND | |
| Op4 | 3 | ND | |
| Orf19.1239 | 6 | ND | |
| Orf19.1765c | 1 | ND | |
| Orf19.1766 | 2 | ND | |
| Orf19.3499 | 6 | ND | |
| Orf19.6553c | 1 | ND | |
| Pbr1c | 2 | ND | |
| Pga7b | 2 | ND | |
| Pga46b | 4 | ND | |
| Phr2b | 7 | ND | |
| Plb3b | 2 | ND | |
| Sap9b | 5 | ND | |
| Sod5b | 1 | ND | |
| Sur7c | 3 | ND | |
| Control cultures only | |||
| Als3b | ND | 5 | |
| Fgr41b | ND | 1 | |
| Hex1 | ND | 1 | |
| Pga45b | ND | 2 | |
| Pra1 | ND | 1 | |
| Rbt1b | ND | 1 | |
| Sap5 | ND | 1 | |
| FCZ-treated and control cultures | |||
| Phr1b | 67 | 1 | 67 |
| Crh11b | 12 | 2 | 6.0 |
| Sod4b | 6 | 1 | 6.0 |
| Dag7 | 27 | 9 | 3.0 |
| Rbt4 | 11 | 4 | 2.8 |
| Ecm33b | 25 | 10 | 2.5 |
| Gca1 | 5 | 2 | 2.5 |
| Tos1 | 56 | 30 | 1.9 |
| Bgl2 | 27 | 15 | 1.8 |
| Rbe1 | 13 | 8 | 1.6 |
| Pga4b | 8 | 5 | 1.6 |
| Ssr1b | 9 | 6 | 1.5 |
| Utr2b | 23 | 17 | 1.4 |
| Ywp1b | 16 | 13 | 1.2 |
| Pir1 | 7 | 6 | 1.2 |
| Xog1 | 57 | 50 | 1.1 |
| Sun41 | 48 | 46 | 1.0 |
| Rbt5b | 3 | 3 | 1.0 |
| Mp65 | 55 | 66 | 0.8 |
| Coi1 | 29 | 35 | 0.8 |
| Sim1 | 27 | 35 | 0.8 |
| Cht3 | 28 | 41 | 0.7 |
| Scw11 | 36 | 53 | 0.7 |
| Msb2c | 4 | 10 | 0.4 |
| Cht2b | 1 | 4 | 0.2 |
| Eng1 | 2 | 19 | 0.1 |
The values represent the number of tryptic peptides. Five independent biological samples with three technical replicates each were analyzed for both conditions. For more detailed information and information about proteins without secretion signal found in the control and in FCZ-treated cultures, see Tables S2 and S3, respectively, in the supplemental material. ND, not detected.
GPI wall protein.
Transmembrane sensor protein in the plasma membrane participating in the Cek1 pathway.
In total, 50 secretory proteins (Table 2) and 34 proteins with predicted intracellular localization (see Tables S1 and S2 in the supplemental material) were identified in the growth medium under both conditions. The intracellular proteins probably originate from damaged cells, dying mother cells, apoptotic cells, nonconventional secretion (48), or vesicular transwall transport (3). Among the secretory proteins, 25 contain just a signal peptide for secretion, 5 proteins possess a secretion signal followed by one or more transmembrane domains, and 20 proteins have a secretion signal and a GPI attachment signal. GPI proteins are normally expected to be associated with the plasma membrane or the wall, but in agreement with our own and other previous studies (24, 57, 61), they were also detected in the growth medium. Because the cultures are shaken to ensure sufficient aeration, some wall proteins possibly get accidentally released and washed out from the wall before being incorporated or get released after their incorporation during isotropic growth and the accompanying wall remodeling or during local cell wall loosening at the incipient bud site. Loss of cell wall material at the site of cytokinesis, which requires severance of the wall between daughter and mother cell, seems likely. Another possibility is that shearing forces resulting from the use of shaking cultures might tear off wall proteins. In addition, similar to the yapsins in Candida glabrata, the yapsin-like C. albicans Sap9 could be involved in releasing wall proteins into the medium (27). The abundances of most GPI proteins in the culture medium were influenced by the FCZ treatment, and an elevated concentration in the growth medium correlated in most cases with an enrichment in the wall, and a lower concentration correlated with the opposite (ranked correlation analysis, RP of 0.85; P = 0.00003) (Fig. 1); hence, they will be discussed during the relative quantification of the wall proteome. Among the non-GPI secretory proteins, the levels of several proteins were strongly affected by FCZ while the abundance of others seemed rather stable, including a previously identified core set of proteins consisting of five (trans)glycosylases (Cht3, Mp65, Scw11, Sim1, and Sun41) as well as Coi1 and Tos1 (57). These proteins are consistently and abundantly present in the medium, and neither FCZ treatment nor various other growth conditions seem to have a notable impact on their secretion.
Several proteins isolated from the culture medium might be involved in fluconazole-related stress responses. Op4, although described as an opaque cell-specific protein, was found among the FCZ-specific non-GPI proteins. As it has been identified before in the secretome of yeast cells grown at pH 4 (57), this might indicate that this protein is not opaque cell-specific but can be expressed in white-type cells, depending on environmental conditions. In addition, we verified five predicted open reading frames (ORFs) on the protein level in FCZ-treated cultures (Table 2), like Orf19.1239, which is a predicted protein in assemblies 19, 20, and 21 of the Candida genome and was found exclusively in the medium of FCZ-stressed cells. Orf19.3499 is regulated by Tsa1 (64), a protein required to cope with oxidative stress. Taking into account the number of its peptide identifications (27 versus 9) (Table 2), Dag7 levels were increased by the FCZ treatment. Relevantly, a mutation in this gene confers hypersensitivity to toxic ergosterol analogues (68), suggesting a protective function for this protein during azole stress. Sur7 was found exclusively in FCZ-stressed cells. It contains a signal peptide as well as four putative transmembrane domains and is located in the plasma membrane. Two of the peptides identified are derived from putative extracellular domains while one originates from a predicted intracellular domain. It is probably involved in cell wall synthesis (5), suggesting that it could play a role in a cell wall stress response. In addition to Sur7, we found four other transmembrane proteins in the medium: Cfl2, Msb2, Orf19.1765, and Orf19.6553. Msb2 has an N-terminal signal peptide and is a glycosylation sensor protein that transfers its signal through the Cek1 MAP kinase pathway (52). Upon activation, the extracellular domain of Msb2 is cleaved off and released into the medium (57, 65). We detected peptides only from the extracellular domain (see Tables S1 and S2 in the supplemental material). Interestingly, the peptide count of Msb2 decreased from 10 to 4 in fluconazole-exposed cultures (Table 2), suggesting that the Msb2 MAP kinase pathway does not play a major role in the response to fluconazole. Except for Msb2, all transmembrane proteins were identified only in the FCZ-treated cultures. The finding of several transmembrane proteins in the medium of FCZ-stressed cultures could be due to increased membrane fluidity caused by FCZ (1). Interestingly, the levels of the endo-1,3-β-glucanase Eng1 and the chitinase Cht2 were negatively influenced by FCZ. The transcript levels of the corresponding genes are also repressed by caspofungin (38). The lower abundances of Eng1 and Cht2 are probably related to diminished cell separation during FCZ treatment (see also the next section).
FCZ-induced changes in wall protein incorporation.
Quantitative changes of the wall proteome of C. albicans grown in surface-grown and planktonic cultures with (FCZ) or without (control) fluconazole were analyzed by LC-Fourier transform mass spectrometry (FTMS). For this, the FCZ and control cell walls were mixed 1:1 with the reference standard mixture of metabolically 15N-labeled yeast and hyphal walls and then digested. Proteins were identified based on their 14N/15N-labeled peptide pairs, with the corresponding peptide 14N/15N isotope abundance ratios. The protein 14N/15N isotope abundance ratios were calculated as the averaged peptide 14N/15N isotope abundance ratios and represent the protein levels relative to the reference standard. (59; also Heilmann et al., unpublished). Relative standard errors for the individual 14N/15N isotope abundance ratios per replica ranged from 0.5% to 54%, with a mean of 7%. Next, the protein 14N/15N isotope abundance ratios were averaged over all corresponding replicas. Relative standard errors for the averaging of the individual protein 14N/15N isotope abundance ratios ranged from 2% to 74%, with a mean of 12%. These errors combine statistical experimental errors with the biological variation. Since both sets of standard errors are similar, it is concluded that the biological variation over the replicas is comparatively small. This justifies discussion of the biological basis of the protein quantification results. Finally, for the planktonic and semisolid agarose-grown cultures, the FCZ/control ratio was calculated for each protein as the FCZ 14N/15N protein isotope abundance ratio divided by the corresponding control 14N/15N protein isotope abundance ratio. The FCZ/control ratios represent the changes in the protein levels induced by fluconazole. Representative examples for the calculation of the protein ratios (strongly decreased, strongly increased, and showing relatively minor changes) are presented in Table 3 while a comprehensive overview of all data is presented in Table S3 in the supplemental material. The FCZ/control ratios for all identified wall proteins are depicted in Fig. 1.
Table 3.
Three representative examples of relative quantification of wall proteins of planktonic C. albicans cultures with and without fluconazole
| Protein name and position (aa)a | Protein function (length [aa]) | Tryptic peptide | FCZ 14N/15N ratio by replicateb |
Control 14N/15N ratio by replicateb |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | R1 | R2 | R3 | R4 | |||
| Cht2 | Chitinase, GH18 domain (583) | |||||||||
| 99–121 | TVLLSLGGGVGDYGFSDVASATK | 0.22 | 0.19 | 0.22 | 0.22 | 1.08 | 1.19 | 1.03 | 0.82 | |
| 122–129 | FADTLWNK | 0.18 | 0.21 | 0.24 | 0.22 | 1.26 | 1.25 | 1.09 | 0.84 | |
| 178–200 | NYFLSAAPQBPYPDASLGDLLSK | 0.26 | 0.22 | 0.27 | 0.25 | 1.26 | 1.15 | ND | 0.91 | |
| 240–258 | LFVGVPATSNIAGYVDTSK | 0.24 | 0.20 | 0.24 | 0.21 | 1.07 | 1.16 | 1.02 | 0.78 | |
| 259–267 | LSSAIEEIK | 0.26 | 0.20 | 0.23 | 0.24 | 1.06 | 1.13 | 0.81 | 0.88 | |
| 268–290 | BDSHFAGVSLWDASGAWLNTDEK | ND | ND | ND | ND | 1.20 | low | ND | 0.78 | |
| 291–299 | GENFVVQVK | 0.16 | 0.20 | 0.24 | 0.23 | 1.04 | 1.17 | 0.98 | 0.83 | |
| Avg per replicate | 0.21 | 0.20 | 0.24 | 0.23 | 1.14 | 1.17 | 0.98 | 0.83 | ||
| Avg per condition | 0.22 ± 0.08 | 1.03 ± 0.01 | ||||||||
| FCZ/control ratio | 0.22 ± 0.08 | |||||||||
| Crh11 | Transglycosylase (453) | |||||||||
| 22–28 | DTBNPLK | 1.38 | 8.18 | 6.97 | ND | ND | ND | ND | ND | |
| 29–45 | SSDBSPVPALGSSFLEK | 3.87 | ND | 4.79 | 6.20 | 0.79 | 1.45 | 1.40 | 1.27 | |
| 46–58 | FDNGLGPHFESLK | 2.93 | 2.74 | 3.52 | 5.01 | 0.59 | 1.11 | 1.26 | 1.08 | |
| 46–59 | FDNGLGPHFESLKK | 3.23 | 3.11 | 5.42 | 9.08 | low | 1.05 | ND | ND | |
| 60–75 | QGTIDSGSNGLSLTMK | 4.74 | ND | ND | low | ND | ND | ND | ND | |
| 77–84 | RFDNPSFK | ND | 3.60 | 4.14 | 5.57 | 0.78 | 1.48 | 1.15 | low | |
| 78–84 | FDNPSFK | 3.48 | 3.86 | 4.92 | 5.95 | 0.74 | 1.45 | ND | 1.16 | |
| 85–93 | SNFYIMFGR | 3.52 | 3.52 | 4.31 | 5.20 | 0.83 | 1.43 | 1.26 | 1.13 | |
| 148–158 | GGYHDIANPLK | 2.22 | 2.38 | 3.62 | ND | 0.25 | 0.49 | ND | ND | |
| 159–169 | DYHTYVIDWTK | 3.51 | 3.52 | 4.50 | 5.62 | 0.85 | 1.39 | 1.30 | 1.15 | |
| 170–182 | DAVTWSVDGSVIR | 4.82 | 3.43 | 4.09 | 6.02 | 1.36 | 1.37 | 1.36 | 1.22 | |
| 236–246 | SVLVADYSSGK | 3.59 | 3.64 | 4.53 | 5.31 | 0.84 | 1.43 | 1.34 | 1.17 | |
| 247–261 | QYSYSDQSGSWESIK | ND | low | 7.76 | 8.81 | ND | ND | ND | ND | |
| 271–279 | YDQAQDDIK | 6.82 | ND | ND | ND | ND | ND | ND | ND | |
| Avg per replicate | 3.44 | 3.59 | 4.75 | 6.15 | 0.72 | 1.21 | 1.29 | 1.17 | ||
| Avg per condition | 4.48 ± 0.13 | 1.10 ± 0.63 | ||||||||
| FCZ/control ratio | 4.05 ± 0.64 | |||||||||
| Ecm33 | Role in wall integrity (423) | |||||||||
| 139–168 | TGLTAGITSAESVVISDTGLSSLTGINVFK | ND | 0.55 | 0.50 | ND | ND | ND | ND | 0.60 | |
| 246–262 | VELAELTSIGNSLTINK | 0.58 | 0.56 | 0.56 | 0.67 | 0.43 | 0.45 | 0.53 | 0.51 | |
| 263–273 | NDDLTELDFPK | 0.54 | 0.62 | 0.57 | 0.64 | 0.43 | 0.49 | 0.45 | 0.49 | |
| 276–290 | TIGGALQISDNSELR | 0.86 | ND | 0.49 | 0.65 | 0.33 | 0.32 | 0.50 | 0.48 | |
| 291–297 | SFSGFPK | 0.67 | 0.67 | 0.67 | 0.61 | 0.49 | 0.50 | 0.48 | 0.51 | |
| 322–329 | VSGGFILK | 0.33 | 0.59 | 0.58 | 0.61 | 0.43 | 0.45 | 0.48 | 0.46 | |
| 334–341 | LSBSAFNK | 0.60 | 0.61 | 0.67 | 0.64 | 0.52 | 0.48 | 0.53 | 0.51 | |
| Avg per replicate | 0.57 | 0.60 | 0.57 | 0.64 | 0.43 | 0.44 | 0.49 | 0.51 | ||
| Avg per condition | 0.60 ± 0.02 | 0.47 ± 0.01 | ||||||||
| FCZ/control ratio | 1.26 ± 0.03 | |||||||||
aa, amino acids. For full information about all proteins from planktonic and surface-grown cultures see Table S3 in the supplemental material.
R1 to R4, replicates 1 to 4; ND, not detected in the 14N-labeled cultures; low, peptides of an intensity too low to be suitable for quantification. Average values and the FCZ/control ratio are means ± standard errors.
Figure 1 clearly shows that, except for Orf19.7104, wall proteins induced by FCZ in surface-grown cultures are induced in planktonic cultures and vice versa. The abundance of Plb5 in surface-grown cultures was also reduced by FCZ, but since there were too few peptides identified for reliable quantification, the data were not included in Fig. 1. Indeed, when we performed a ranked correlation analysis, we found a correlation RP of 0.85 (P = 6.7 × 10−6) between the two data sets. Nevertheless, surface-grown and planktonic cultures show an important difference with respect to the incorporation levels of the wall proteins Als4, Pir1, Rhd3, and Sod5. Although their FCZ/control ratios were similar in both experimental setups (Fig. 1), showing that their incorporation levels are affected by fluconazole independent of how the cells were grown, direct comparison of their wall incorporation levels to the corresponding levels in the 15N-labeled reference culture showed that Als4, Pir1, Rhd3, and Sod5 were present at much higher levels in surface-grown cultures than in planktonic cultures (median value, 8-fold increase; range, 2.4- to 17-fold) (see Table S4 in the supplemental material). As the MAP kinase Mkc1 becomes phosphorylated in cells growing on a semisolid surface (31), these wall proteins are possibly controlled by Mkc1 and could play a role in contact-dependent growth.
By relative quantification, we could identify wall proteins that were increased by FCZ stress (Als4, Crh11, Pga4, Phr1, Phr2, Pir1, Sap9, Sod4, and Sod5) or decreased (Als3, Cht2, Mp65, Rbt1, Rhd3, Plb5, and Ywp1). Consistent with the observed changes in abundance, peptide coverage also tended to increase or decrease according to protein abundance (Table 3). Only a few proteins showed less than a 2-fold change upon FCZ treatment (Ecm33, Ssr1, and Utr2) in both setups. Orf19.7104 was exclusively identified in walls from planktonic cultures of FCZ-treated cells and in walls from the reference culture, although only by one peptide pair. It is a predicted open reading frame in assemblies 19, 20, and 21 with sequence similarities to mucins (45). It contains an N-terminal secretion signal but no putative GPI attachment site. Intriguingly, it contains an amino acid motif (DGQSQ, amino acids 160 to 164) that is similar to the one found in the Pir repeats in C. albicans and Saccharomyces cerevisiae and that is believed to be involved in cross-linking wall proteins to β-glucans (18, 28). Unexpectedly, when we measured the relative transcript levels of Orf19.7104 by qRT-PCR with respect to ACT1, we found that in the presence of fluconazole they were systematically about 5- to 6-fold lower than in the control culture (0.20, 0.125, and 0.059 in the control cultures compared to 0.038, 0.021, and 0.012 in the fluconazole-treated cultures after 4, 6, and 18 h of culturing, respectively). Possibly, transcript stability, translation efficiency, or the efficiency of wall incorporation of Orf19.7104 changes during azole stress.
We found two proteins without secretion signal, Tdh3 and Ssa2, in the wall preparations of FCZ-treated and control cells. Although it remains elusive how these proteins reach the cell surface and are attached there, they were previously found associated with the fungal wall (22, 39). However, for Tdh3 and Ssa2 the observed peptide ratios varied more strongly than those for the other wall proteins (see Table S3 in the supplemental material). Since Thd3 and Ssa2 are very abundant intracellular proteins (both have a high codon bias index of 0.83 and 0.74, respectively) (see the Candida Genome Database), it seems possible that they were not fully removed during wall isolation, possibly because of incomplete cell breakage.
Hwp1 does not contain tryptic peptides of a suitable size detectable by mass spectrometry, and Rbt5 shares a peptide with other proteins of the CFEM-family (Csa1, Csa2, Pga7, and Pga10), making it unsuitable for quantification purposes. In addition, although one Rbt5-specific peptide was detected (see Table S1 in the supplemental material), it was very often of low intensity in the query or the reference culture. We therefore decided to use immunoblot analysis (Fig. 2). Hwp1 was previously identified as a hyphal-enriched wall protein (60), and consistent with these findings, its wall level was increased in our control cultures, where hyphae are more abundant. On the other hand, Rbt5 was more abundant after FCZ treatment, in agreement with a transcriptional study that found RBT5 induced upon azole stress (38). Rbt5 is an iron acquisition protein and involved in hemoglobin or heme utilization (67). Several enzymes of the ergosterol biosynthesis pathway are induced during azole stress (38), like ERG11, which needs iron for its function. Hence, it seems likely that iron requirements during FCZ treatment increase, explaining the induction of Rbt5. In spite of previous findings that indicated a membrane and not a wall localization of Rbt5 (67), we do consistently detect it in the wall of yeasts as well as hyphae under a variety of growth conditions. Moreover, Rbt5 was already shown to be wall localized before (58). Taken together, these data indicate that Rbt5 is located both in the plasma membrane and in the cell wall.
Fig. 2.
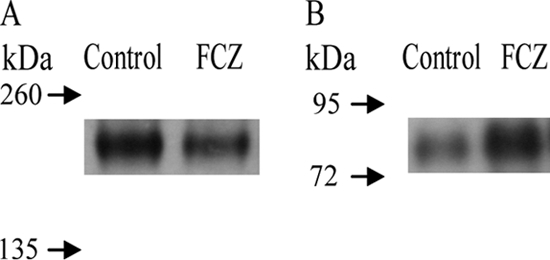
Immunoblot analysis of Hwp1 (A) and Rbt5 (B). Cells were grown for 18 h without (control) or with (FCZ) 0.5 mg/liter fluconazole. The GPI wall proteins were released from isolated walls by β-1,6-glucanase. For detection of Hwp1 and Rbt5, proteins isolated from 40 or 160 μg of walls, respectively, were subjected to immunoblot analysis.
Fluconazole causes cell wall stress.
Since FCZ treatment results in increased fluidity of the plasma membrane (1), it seems possible that this could indirectly affect cell wall integrity, for example, by affecting the activity of plasma membrane-located cell wall construction enzymes or by affecting endocytosis. Fluconazole-treated C. albicans cells contain considerably less glucan in their walls, suggesting severe alterations (51). Chitin levels are probably increased to compensate for FCZ-induced wall perturbations (44). For S. cerevisiae the membrane-perturbing agents chlorpromazine and chitosan, indeed, trigger the CWI pathway (26, 69). In addition, protein kinase C (Pkc1), a key enzyme in the response to cell wall stress, plays a pivotal role in the tolerance of drugs that affect the cell membrane in C. albicans (32), and phosphorylation of the MAP kinase Mkc1 increases upon exposure to the membrane-perturbing agent chlorpromazine (31). We identified several wall proteins that were induced by fluconazole stress and that are required for cell wall cross-linking and integrity (Crh11, Pga4, Phr1, Phr2, Pir1, and Sap9). Additionally, we found that the changes in the transcript levels of several wall protein-encoding genes when C. albicans is challenged with sublethal concentrations of caspofungin (increased transcript levels of ALS1, CRH11, PGA13, PHR1, PHR2, and SAP9, and a decreased transcript level of CHT2 [12]) correctly predicted the directions of the changes in the corresponding wall protein levels upon treatment with FCZ (Fig. 1). We therefore tested the wall-perturbing agents calcofluor white, Congo red, and SDS without and in combination with fluconazole. Figure 3 shows that fluconazole treatment renders C. albicans hypersensitive to all three agents, supporting the notion that fluconazole compromises cell wall integrity. We next analyzed the wall protein profile of Congo red-treated cells by QTOF mass spectrometry using a Congo red concentration of 2 mg/liter, resulting in a biomass decrease of 32% at the end of the culturing period. Like fluconazole, Congo red inhibited hyphal growth (data not shown). Table 4 shows that exposure to Congo red increased the abundances of a set of wall proteins that are mainly involved in cell wall maintenance (Crh11, Phr1, Phr2, Pir1, Sap9, and Utr2) and furthermore of Rbt5, Rhd3, Pga31, and Sod5, whereas the incorporation level of the hyphal growth-associated wall protein Als3 strongly declined. Apart from Rhd3, the levels of which were reduced during fluconazole stress, and Pga31, which was not identified by relative quantification using FTMS, the incorporation levels of all these proteins were influenced in a similar way by fluconazole (Fig. 1), offering further support to the notion that fluconazole causes cell wall stress.
Fig. 3.
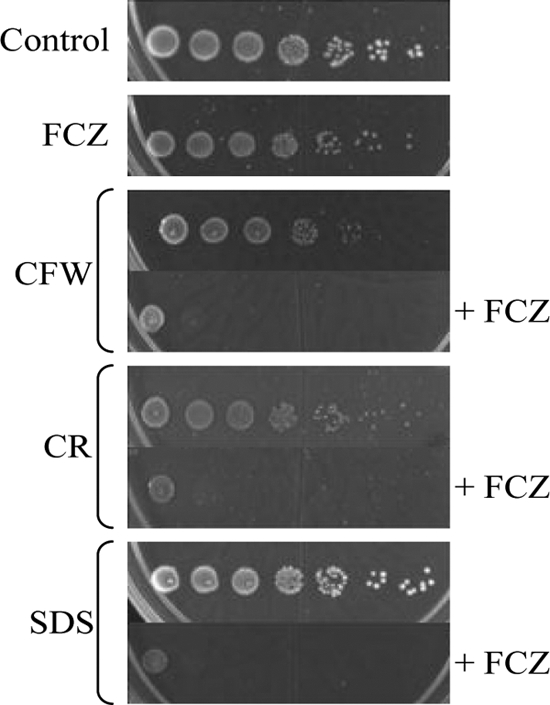
C. albicans spot assays on low-agarose plates incubated for 2 days at 37°C. Where indicated, plates were supplemented with 0.5 mg/liter FCZ, 100 mg/liter calcofluor white (CFW), 200 mg/liter Congo red (CR), 100 mg/liter SDS, or a combination of FCZ and one of the other drugs.
Table 4.
Changes in the wall proteome of planktonic cultures of C. albicans induced by Congo red as indicated by the total number of identifications of tryptic peptides from secretory proteins
| Protein | Peptide counta |
CR/control ratiob | |
|---|---|---|---|
| CR (7 runs) | Control (8 runs) | ||
| Pga31 | 14 | ND | |
| Sap9 | 6 | ND | |
| Pir1 | 6 | 1 | 6.9 |
| Phr2 | 12 | 3 | 4.6 |
| Sod5 | 18 | 9 | 2.3 |
| Utr2 | 22 | 11 | 2.3 |
| Phr1 | 33 | 17 | 2.2 |
| Rhd3 | 21 | 11 | 2.2 |
| Rbt5 | 7 | 4 | 2.0 |
| Crh11 | 51 | 33 | 1.8 |
| Ecm33 | 35 | 23 | 1.7 |
| Mp65 | 19 | 14 | 1.6 |
| Ywp1 | 7 | 8 | 1.0 |
| Cht2 | 20 | 25 | 0.9 |
| Pga4 | 9 | 12 | 0.9 |
| Sod4 | 12 | 22 | 0.6 |
| Ssr1 | 7 | 15 | 0.5 |
| Als3 | ND | 8 | |
Number of tryptic peptides. Four biological replicates were analyzed for both conditions. Each biological replicate was analyzed twice (giving a maximum of 8 runs), except for Congo red, where biological replicate 1 was analyzed only once (7 runs). Congo red (CR) was used at 2 mg/liter. For sequence information of the peptides identified in control and in FCZ-treated cultures, see Tables S5 and S6, respectively, in the supplemental material. ND, not detected.
The ratios are normalized for the number of runs.
DISCUSSION
As the proteins of the secretome and the wall proteome act at the interface of the fungus and its host, we wanted to investigate the fluconazole-induced changes in protein abundances in both subproteomes. In steady-state-like (exponential phase) cells of S. cerevisiae, a statistically significant but only limited correlation between protein and transcript levels has been found (Spearman rank correlation coefficient, Rs, of 0.57; R2 = 0.32) (20). This implies that in many cases and certainly under non-steady-state conditions there is no linear relationship between transcript levels and protein abundances and that transcript levels can only predict in which directions protein abundances are expected to move (14). Precise estimates of protein abundances are, however, important in themselves because they determine, for example, the output of metabolic pathways or the adhesive capacity of a cell. Precise estimates are also required for systems biology-based approaches (14).
Our proteomics data represent the first systematic overview of the changes induced by azole stress in the protein composition of the secretome and in the incorporation levels of wall proteins and greatly extend our current knowledge. The experimental strategy for relative quantification of the wall proteome of C. albicans was based on using [15N]ammonium sulfate as the sole nitrogen source to thoroughly label two large reference cultures grown under yeast- and hypha-promoting conditions, respectively. The walls from both cultures were then isolated, mixed, and stored. This combinatorial approach ensures a wide representation of differentially expressed wall proteins in the reference walls. The mixture of 15N-labeled reference walls can then be used to study the relative abundance of wall proteins in 14N-labeled query walls obtained from cultures grown under various environmental conditions, such as in the absence or presence of fluconazole (this paper) or of different hyphal growth-inducing compounds (C. J. Heilmann et al., unpublished observations), obtained from infected tissues or organs, or obtained from a time course experiment. Importantly, using the same reference walls in a series of experiments makes it possible to construct a library of relative abundances of wall proteins and to calculate and analyze retrospectively relative abundances between all experimental conditions tested.
Many individual medium protein levels and wall protein levels were strongly affected by the FCZ treatment. The wall proteome changes were similar in both surface-grown and planktonic cultures (ranked correlation, RP, of 0.85), indicating that the results obtained with planktonic cultures are to a large extent representative for surface-grown cultures. However, the incorporation levels of Als4, Pir1, Rhd3, and Sod5 were specifically increased in surface-grown cultures both in the absence and presence of fluconazole, suggesting that surface growth could stimulate their wall incorporation levels. To the limited extent that they are available, the directional changes in protein levels predicted by transcriptional studies of azole-stressed cultures (13, 38) tend to be consistent with our results. In agreement with the transcriptional results of these studies, we found increased levels of Als4, Crh11, Phr1, Phr2, Pir1, Rbt5 (using immunoblot analysis), and Sap9 in the presence of fluconazole and a decrease in abundance of Hwp1 (immunoblot analysis). However, we observed upon fluconazole exposure a decrease in the wall level for Ywp1 and an increase for Orf19.7104, both of which conflict with the transcriptional data (13; also this paper), possibly because of differences in growth conditions or because of posttranscriptional control mechanisms, such as observed for the wall adhesin Epa3 of C. glabrata, for example, which is released into the medium of postexponential phase cells in a yapsin-dependent manner (27).
Fluconazole affects the wall proteome by altering morphology and wall integrity.
Two major trends emerge from our study of the fluconazole-driven changes in the wall proteome, both of which were also observed during Congo red-induced stress (Fig. 4). First, fluconazole inhibits hyphal growth and strongly promotes growth in the yeast form (23; also this study), raising the question of whether this morphological change evokes some of the observed changes. We found that azole treatment strongly decreased the levels of Als3, Hwp1, and Plb5, all of them strongly associated with the hyphal morphotype (25, 60, 62; also Heilmann et al., unpublished). The wall levels of Mp65 and Rbt1 were also lower, consistent with lower transcript levels of the corresponding genes in yeast cultures than in hyphal cultures (11, 35). Together, these data indicate that some of the FCZ-induced changes are morphotype associated and result from the inhibition of hyphal growth and the promotion of yeast growth. Similar to fluconazole cell treatment, Congo red treatment also caused inhibition of hyphal growth and promoted growth in the yeast mode. Second, some of the observed changes in relative abundances seem to be correlated with cell wall stress (12). Consistent with this, cell wall-perturbing agents sensitized C. albicans to the effect of fluconazole (Fig. 3). As shown by our relative quantification data in Fig. 1 and Table 3, the levels of several transglycosidases (Crh11, Pga4, Phr1, and Phr2) and the putatively β-1,3-glucan cross-linking protein Pir1 were increased by FCZ stress. Congo red treatment also increased the levels of these proteins (Table 4). Although peptide levels of Crh11 are only 1.8-fold increased in the presence of Congo red, comparing the actual number of peptide identifications (58.3 from walls of Congo red-treated cultures versus 33 from the control, normalized to eight runs) indicates that this increase is significant and corroborates this observation. Crh11, together with Crh12 and Utr2, belongs to a family of proteins that were found to serve a crucial role in cell wall biogenesis (50). Also in agreement with transcriptional data (38), it seems that within this family Crh11 is mainly responsible for counteracting stress elicited by azole treatment. Phr1 and Phr2 are two closely related transglycosidases and are expressed oppositely with respect to pH (43, 53). Mutants in either phr1Δ or phr2Δ display an abnormal cell wall at elevated or low pH, respectively. Intriguingly, and consistent with the trends predicted by previous transcriptional studies (12, 13, 38), levels of both proteins were increased when C. albicans was challenged with FCZ although all our experiments were performed at pH 7.4 and also buffered to avoid acidification (63). It seems that under regular growth conditions, either Phr1 or Phr2— depending on the pH—is sufficient for a proper cell wall, but upon cell wall stress both proteins might be induced as part of a compensatory response to wall defects. Also PIR1 is required for cell wall maintenance (41), and we find it upregulated upon FCZ treatment. Both FCZ- and Congo red-induced stress resulted in increased levels of Sap9, a GPI-modified yapsin-like aspartyl protease that participates in maintaining cell wall integrity (2, 55). Interestingly, in S. cerevisiae wall stress caused by Congo red and other wall-perturbing compounds leads to increased transcript levels of YPS1, which is the closest homolog in S. cerevisiae of C. albicans SAP9, and this is mediated by the cell wall integrity pathway, whereas disruption of YPS1 results in a much lower resistance to Congo red (30). These observations suggest that C. albicans SAP9 and S. cerevisiae YPS1 share similar functions and might be similarly regulated. In addition to these two major trends, some FCZ-induced changes in the wall proteome are probably due to other effects of FCZ. For example, FCZ has been found to cause nitrosative and oxidative stress (6). This could explain the increased levels of the superoxide dismutases Sod4 and Sod5 in the walls of FCZ-treated cultures. Exposure to Congo red led to an increase in abundance of Sod5 as well (Table 4). Finally, phosphorylation of the MAP kinase Mkc1, which plays a role in the cell wall integrity pathway, has been shown to increase upon both membrane and cell wall stress (31, 47), suggesting a role for Mkc1 in the observed changes in the wall proteome and the secretome.
Fig. 4.
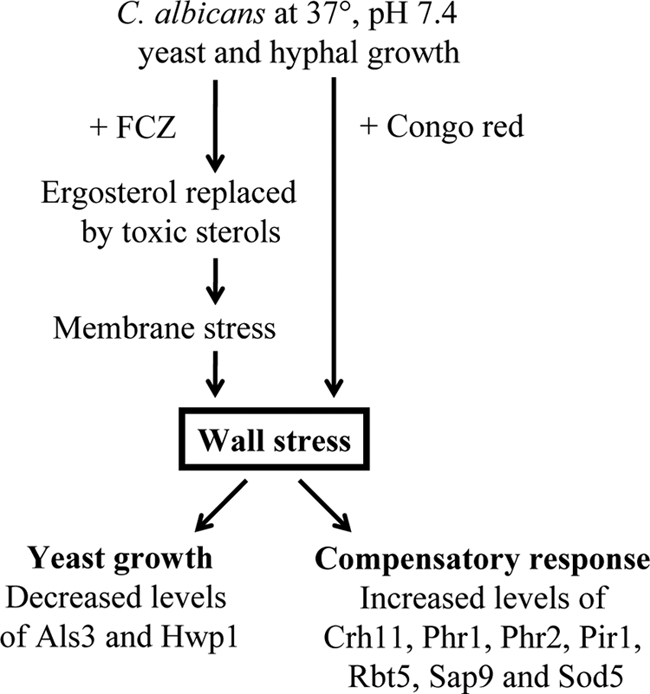
Proposed scheme of the response of C. albicans to fluconazole and Congo red.
Potential clinical markers from the secretome.
Providing immunocompromised patients with an early diagnosis of invasive Candida infections is an ongoing problem since usually time-consuming blood cultures are first required, thus delaying life-saving therapies. Diagnosis based on the detection of antibodies against fungal antigens poses the problem of how to distinguish between antibodies generated due to harmless colonization of the mucosa or due to a serious invasive infection. Additionally, immunocompromised patients might produce low levels of antibodies leading to false-negative results (reviewed in reference 33). The detection of the actual antigens partially avoids these problems; nevertheless, due to relatively rapid clearance of some of them, their levels need to be high enough. To serve as a potential diagnostic marker candidate, several requirements should be fulfilled, including extracellular localization and high abundance under different environmental conditions depending on the site of infection. We have already previously reported about a core set of proteins that seems to be abundantly secreted under a variety of growth conditions (57). Also, treatment with the antifungal drug fluconazole did not seem to strongly influence the secretion of Cht3, Coi1, Mp65, Scw11, Sim1, Sun41, and Tos1. Indeed, Mp65 seems to be a promising marker (7). In addition, Xog1 also seems an abundant secretory protein (Table 2) and could thus function as a potential marker of early infections.
Collectively, our findings provide valuable information for understanding the mode of action of azole drugs as well as for alternative therapeutic strategies and for potential infection markers.
Supplementary Material
ACKNOWLEDGMENTS
F.M.K. was supported by the EU Programme FP7-214004-2 FINSysB. A.G.S. and C.J.H. are grateful for the support by all members of the FINSysB consortium.
We are grateful to and Sepehr Mohamady for his help with the wall protein analysis of Congo red-treated cultures. We thank D. Kornitzer and P. Sundstrom for the C. albicans Pga10 and Hwp1 antiserum, respectively.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 27 May 2011.
REFERENCES
- 1. Abe F., Usui K., Hiraki T. 2009. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry 48:8494–8504 [DOI] [PubMed] [Google Scholar]
- 2. Albrecht A., et al. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281:688–694 [DOI] [PubMed] [Google Scholar]
- 3. Albuquerque P. C., et al. 2008. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol. 10:1695–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alic N., Higgins V. J., Pichova A., Breitenbach M., Dawes I. W. 2003. Lipid hydroperoxides activate the mitogen-activated protein kinase Mpk1p in Saccharomyces cerevisiae. J. Biol. Chem. 278:41849–41855 [DOI] [PubMed] [Google Scholar]
- 5. Alvarez F. J., Douglas L. M., Rosebrock A., Konopka J. B. 2008. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol. Biol. Cell 19:5214–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arana D. M., Nombela C., Pla J. 2010. Fluconazole at subinhibitory concentrations induces the oxidative- and nitrosative-responsive genes TRR1, GRE2 and YHB1, and enhances the resistance of Candida albicans to phagocytes. J. Antimicrob. Chemother. 65:54–62 [DOI] [PubMed] [Google Scholar]
- 7. Arancia S., Sandini S., Cassone A., De Bernardis F., La Valle R. 2004. Construction and use of PCR primers from a 65 kDa mannoprotein gene for identification of C. albicans. Mol. Cell Probes 18:171–175 [DOI] [PubMed] [Google Scholar]
- 8. Arnaud M. B., et al. 2007. Sequence resources at the Candida genome database. Nucleic Acids Res. 35:D452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bekker M., et al. 2010. The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J. Bacteriol. 192:746–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bom I. J., et al. 1998. A new tool for studying the molecular architecture of the fungal cell wall: one-step purification of recombinant trichoderma beta-(1-6)-glucanase expressed in Pichia pastoris. Biochim. Biophys. Acta 1425:419–424 [DOI] [PubMed] [Google Scholar]
- 11. Braun B. R., Head W. S., Wang M. X., Johnson A. D. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bruno V. M., et al. 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Copping V. M., et al. 2005. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 55:645–654 [DOI] [PubMed] [Google Scholar]
- 14. Cox J., Mann M. 2011. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80:273–299 [DOI] [PubMed] [Google Scholar]
- 15. Cruz M. C., et al. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21:546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Groot P. W., et al. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3:955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Nobel H., et al. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146:2121–2132 [DOI] [PubMed] [Google Scholar]
- 18. Ecker M., Deutzmann R., Lehle L., Mrsa V., Tanner W. 2006. Pir proteins of Saccharomyces cerevisiae are attached to beta-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 281:11523–11529 [DOI] [PubMed] [Google Scholar]
- 19. Georgopapadakou N. H., Dix B. A., Smith S. A., Freudenberger J., Funke P. T. 1987. Effect of antifungal agents on lipid biosynthesis and membrane integrity in Candida albicans. Antimicrob. Agents Chemother. 31:46–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghaemmaghami S., et al. 2003. Global analysis of protein expression in yeast. Nature 425:737–741 [DOI] [PubMed] [Google Scholar]
- 21. Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 22. Gil-Navarro I., et al. 1997. The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J. Bacteriol. 179:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ha K. C., White T. C. 1999. Effects of azole antifungal drugs on the transition from yeast cells to hyphae in susceptible and resistant isolates of the pathogenic yeast Candida albicans. Antimicrob. Agents Chemother. 43:763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hiller E., Heine S., Brunner H., Rupp S. 2007. Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation. Eukaryot. Cell 6:2056–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoyer L. L., Payne T. L., Bell M., Myers A. M., Scherer S. 1998. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 33:451–459 [DOI] [PubMed] [Google Scholar]
- 26. Kamada Y., Jung U. S., Piotrowski J., Levin D. E. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559–1571 [DOI] [PubMed] [Google Scholar]
- 27. Kaur R., Ma B., Cormack B. P. 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. U. S. A. 104:7628–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klis F. M., Sosinska G. J., de Groot P. W., Brul S. 2009. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 9:1013–1028 [DOI] [PubMed] [Google Scholar]
- 29. Kopecka M., Gabriel M. 1992. The influence of Congo red on the cell wall and (1-3)-beta-d-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch. Microbiol. 158:115–126 [DOI] [PubMed] [Google Scholar]
- 30. Krysan D. J., Ting E. L., Abeijon C., Kroos L., Fuller R. S. 2005. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot. Cell 4:1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumamoto C. A. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. U. S. A. 102:5576–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. LaFayette S. L., et al. 2010. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 6:piie1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lain A., et al. 2008. Use of recombinant antigens for the diagnosis of invasive candidiasis. Clin. Dev. Immunol. 2008:721950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lamb D. C., et al. 1999. Purification, reconstitution, and inhibition of cytochrome P-450 sterol Δ22-desaturase from the pathogenic fungus Candida glabrata. Antimicrob. Agents Chemother. 43:1725–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. La Valle R., et al. 2000. Generation of a recombinant 65-kilodalton mannoprotein, a major antigen target of cell-mediated immune response to Candida albicans. Infect. Immun. 68:6777–6784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levin D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu H., Sadygov R. G., Yates J. R., III 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76:4193–4201 [DOI] [PubMed] [Google Scholar]
- 38. Liu T. T., et al. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lopez-Ribot J. L., Alloush H. M., Masten B. J., Chaffin W. L. 1996. Evidence for presence in the cell wall of Candida albicans of a protein related to the hsp70 family. Infect. Immun. 64:3333–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin S. W., Konopka J. B. 2004. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot. Cell 3:675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez A. I., et al. 2004. Role of Pir1 in the construction of the Candida albicans cell wall. Microbiology 150:3151–3161 [DOI] [PubMed] [Google Scholar]
- 42. Mensonides F. I., Brul S., Klis F. M., Hellingwerf K. J., Teixeira de Mattos M. J. 2005. Activation of the protein kinase C1 pathway upon continuous heat stress in Saccharomyces cerevisiae is triggered by an intracellular increase in osmolarity due to trehalose accumulation. Appl. Environ. Microbiol. 71:4531–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muhlschlegel F. A., Fonzi W. A. 1997. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol. Cell. Biol. 17:5960–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Munro C. A., et al. 2007. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 63:1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murillo L. A., et al. 2005. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot. Cell 4:1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mysiakina I. S., Funtikova N. S. 2007. The role of sterols in morphogenetic processes and dimorphism in fungi. Mikrobiologiia 76:5–18 [PubMed] [Google Scholar]
- 47. Navarro-Garcia F., Eisman B., Fiuza S. M., Nombela C., Pla J. 2005. The MAP kinase Mkc1p is activated under different stress conditions in Candida albicans. Microbiology 151:2737–2749 [DOI] [PubMed] [Google Scholar]
- 48. Nombela C., Gil C., Chaffin W. L. 2006. Non-conventional protein secretion in yeast. Trends Microbiol. 14:15–21 [DOI] [PubMed] [Google Scholar]
- 49. Odds F. C., Cockayne A., Hayward J., Abbott A. B. 1985. Effects of imidazole- and triazole-derivative antifungal compounds on the growth and morphological development of Candida albicans hyphae. J. Gen. Microbiol. 131:2581–2589 [DOI] [PubMed] [Google Scholar]
- 50. Pardini G., et al. 2006. The CRH family coding for cell wall glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans. J. Biol. Chem. 281:40399–40411 [DOI] [PubMed] [Google Scholar]
- 51. Pfaller M., Riley J. 1992. Effects of fluconazole on the sterol and carbohydrate composition of four species of Candida. Eur. J. Clin. Microbiol. Infect. Dis. 11:152–156 [DOI] [PubMed] [Google Scholar]
- 52. Roman E., Cottier F., Ernst J. F., Pla J. 2009. Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot. Cell 8:1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saporito-Irwin S. M., Birse C. E., Sypherd P. S., Fonzi W. A. 1995. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15:601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schaller M., Borelli C., Korting H. C., Hube B. 2005. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48:365–377 [DOI] [PubMed] [Google Scholar]
- 55. Schild L., et al. 2011. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot. Cell 10:98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith P. K., et al. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76–85 [DOI] [PubMed] [Google Scholar]
- 57. Sorgo A. G., et al. 2010. Mass spectrometric analysis of the secretome of Candida albicans. Yeast 27:661–672 [DOI] [PubMed] [Google Scholar]
- 58. Sosinska G. J., et al. 2008. Hypoxic conditions and iron restriction affect the cell-wall proteome of Candida albicans grown under vagina-simulative conditions. Microbiology 154:510–520 [DOI] [PubMed] [Google Scholar]
- 59. Sosinska G. J., et al. 2011. Mass spectrometric quantitation of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology 157:136–146 [DOI] [PubMed] [Google Scholar]
- 60. Staab J. F., Ferrer C. A., Sundstrom P. 1996. Developmental expression of a tandemly repeated, proline-and glutamine-rich amino acid motif on hyphal surfaces on Candida albicans. J. Biol. Chem. 271:6298–6305 [DOI] [PubMed] [Google Scholar]
- 61. Stead D. A., et al. 2010. Impact of the transcriptional regulator, Ace2, on the Candida glabrata secretome. Proteomics 10:212–223 [DOI] [PubMed] [Google Scholar]
- 62. Theiss S., et al. 2006. Inactivation of the phospholipase B gene PLB5 in wild-type Candida albicans reduces cell-associated phospholipase A2 activity and attenuates virulence. Int. J. Med. Microbiol. 296:405–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsuboi R., Matsuda K., Ko I. J., Ogawa H. 1989. Correlation between culture medium pH, extracellular proteinase activity, and cell growth of Candida albicans in insoluble stratum corneum-supplemented media. Arch. Dermatol. Res. 281:342–345 [DOI] [PubMed] [Google Scholar]
- 64. Urban C., et al. 2005. The moonlighting protein Tsa1p is implicated in oxidative stress response and in cell wall biogenesis in Candida albicans. Mol. Microbiol. 57:1318–1341 [DOI] [PubMed] [Google Scholar]
- 65. Vadaie N., et al. 2008. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J. Cell Biol. 181:1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vanden Bossche H. 1985. Biochemical targets for antifungal azole derivatives: hypothesis on the mode of action. Curr. Top. Med. Mycol. 1:313–351 [DOI] [PubMed] [Google Scholar]
- 67. Weissman Z., Kornitzer D. 2004. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 53:1209–1220 [DOI] [PubMed] [Google Scholar]
- 68. Xu D., et al. 2007. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 3:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zakrzewska A., Boorsma A., Brul S., Hellingwerf K. J., Klis F. M. 2005. Transcriptional response of Saccharomyces cerevisiae to the plasma membrane-perturbing compound chitosan. Eukaryot. Cell 4:703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang Y. Q., et al. 2010. Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 6:e1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


