Abstract
In the rice blast fungus Magnaporthe oryzae, the PMK1 mitogen-activated protein (MAP) kinase gene regulates appressorium formation and infectious growth. Its homologs in many other fungi also play critical roles in fungal development and pathogenicity. However, the targets of this important MAP kinase and its interacting genes are not well characterized. In this study, we constructed two yeast two-hybrid libraries of M. oryzae and screened for Pmk1-interacting proteins. Among the nine Pmk1-interacting clones (PICs) identified, two of them, PIC1 and PIC5, were selected for further characterization. Pic1 has one putative nuclear localization signal and one putative MAP kinase phosphorylation site. Pic5 contains one transmembrane domain and two functionally unknown CTNS (cystinosin/ERS1p repeat) motifs. The interaction of Pmk1 with Pic1 or Pic5 was confirmed by coimmunoprecipitation assays. Targeted gene deletion of PIC1 had no apparent effects on vegetative growth and pathogenicity but resulted in a significant reduction in conidiation and abnormal germ tube differentiation on onion epidermal cells. Deletion of PIC5 led to a reduction in conidiation and hyphal growth. Autolysis of aerial hyphae became visible in cultures older than 4 days. The pic5 mutant was defective in germ tube growth and appressorium differentiation. It was reduced in appressorial penetration and virulence on the plant. Both PIC1 and PIC5 are conserved in filamentous ascomycetes, but none of their orthologs have been functionally characterized. Our data indicate that PIC5 is a novel virulence factor involved in appressorium differentiation and pathogenesis in M. oryzae.
INTRODUCTION
Rice blast caused by the heterothallic ascomycete Magnaporthe oryzae (anamorph Pyricularia grisea) is one of the most severe fungal diseases of rice throughout the world (36, 41). The fungus uses the enormous turgor pressure generated inside a highly specialized infection structure known as appressorium (AP) for plant penetration (6, 11). After penetration, the bulbous invasive hyphae grow biotrophically inside plant cells (31). At late stages, blast lesions are developed on rice plants, and the pathogen produces numerous conidia to reinitiate the infection cycle.
In the past 2 decades, several signal transduction pathways involved in surface recognition, appressorium formation, and infectious growth have been characterized in M. oryzae (5, 36, 42). The cyclic AMP (cAMP) signaling pathway is known to be important for surface recognition and appressorium turgor generation (25, 37, 45). The PMK1 mitogen-activated protein (MAP) kinase gene plays an important role in appressorium development and infectious growth. PMK1 is orthologous to the FUS3 and KSS1 genes in Saccharomyces cerevisiae. The pmk1 deletion mutant is defective in appressorium formation and infectious growth, but it still recognizes hydrophobic surfaces and responds to exogenous cAMP (43). Another MAP kinase (MAPK) gene essential for plant infection in M. oryzae is MPS1 (44), an ortholog of S. cerevisiae SLT2. The mps1 mutant is nonpathogenic and defective in appressorial penetration (44). Deletion of MCK1, an MEK kinase gene functioning upstream from Mps1, resulted in similar defects with the mps1 mutant (13). Studies in other plant pathogenic fungi, including Fusarium graminearum, F. oxysporum, Ustilago maydis, Cochliobolus heterostrophus, Claviceps purpurea, and Colletotrichum lagenarium, also have shown the importance of the cAMP signaling and MAP kinase pathways in regulating different plant infection processes (1a, 7, 12, 17, 22, 24, 26, 32, 35, 40, 52).
In M. oryzae, a number of upstream components involved in the activation of Pmk1 MAP kinase have been identified, including the MST50, MST11, MST7, MGB1, and RAS2 genes (27, 29, 48). Mst50 functions as an adaptor protein that binds with both the Mst7 MEK and Mst11 MEK kinases. Mst7 also directly interacts with Mst11 in yeast two-hybrid and coimmunoprecipitation (co-IP) assays (48). However, the interaction of Pmk1 with Mst7, Mst11, or Mst50 was not detectable in yeast two-hybrid assays (48). In co-IP assays with proteins isolated from appressoria, Mst7 and Pmk1 interact with each other via the MAP kinase docking site and docking region. Therefore, it has been hypothesized that the Mst7-Pmk1 interaction is relatively transient and that the weak interaction between Mst7 and Pmk1 may be stabilized or facilitated by additional components of the Pmk1 MAP kinase pathway during appressorium formation (50).
Of the downstream targets of Pmk1, to date Mst12 is the only transcription factor that is known to weakly interact with Pmk1 in yeast two-hybrid assays (30). Unlike PMK1, MST12 is dispensable for appressorium formation. However, appressoria formed by the mst12 mutant fail to penetrate and infect rice seedlings. The mst12 mutant is defective in appressorial penetration and cytoskeleton reorganization in mature appressoria (28). Studies in C. lagenarium also indicate that CST12 is dispensable for appressorium formation but essential for plant infection (38). In U. maydis, the Prf1 transcription factor gene functions downstream from both the cAMP signaling and MAP kinase pathways (14). However, M. oryzae and other filamentous ascomycetes lack a distinct homolog of Prf1.
To further characterize the PMK1 MAP kinase pathway, in this study we constructed two yeast two-hybrid libraries and screened for genes that interacted with Pmk1. Two of the Pmk1-interacting genes, PIC1 and PIC5, were selected for detailed characterization. Results from this study indicate that some of these Pmk1-interacting clones may be involved in surface attachment and appressorium morphogenesis in M. oryzae.
MATERIALS AND METHODS
Strains and culture conditions.
All the wild-type (WT) and mutant strains of M. oryzae were routinely cultured on complete medium (CM), 5× YEG (0.5% yeast extract and 2% glucose), or oatmeal agar (OA) plates as described previously (19, 46). For DNA, RNA, and protein isolation, vegetative hyphae were harvested from 2-day-old liquid 5× YEG or CM cultures. Growth rate and conidiation were assayed with CM and OA cultures (18). For fungal transformation, transformants were selected on TB3 medium (0.3% yeast extract, 0.3% Casamino Acids, and 20% glucose) with 200 μg/ml zeocin (Invitrogen, Carlsbad, CA) or 250 μg/ml hygromycin B (Calbiochem, La Jolla, CA). For the cell wall integrity test, vegetative hyphae harvested from 2-day-old CM cultures were digested with 5 mg/ml lytic enzyme (Sigma-Aldrich, St. Louis, MO) for 40 min at 30°C as described previously (44).
Construction and screening of the yeast two-hybrid libraries.
RNA samples used for library construction were isolated from vegetative hyphae under nitrogen starvation (−N) and appressoria formed on plastic surfaces for 36 h as described previously (8, 20). Total RNAs were isolated with the TRIzol reagent (Invitrogen), and poly(A)+ RNAs were purified with the Oligotex mRNA isolation kit (Qiagen Inc., Valencia, CA) following the instructions provided by the manufacturers. The cDNA libraries were constructed with the HybridZAP-2.1 XR library construction kit (Stratagene, La Jolla, CA). The PMK1 gene was amplified with primers MKBF (5′-GATGAATTCATGTCTCGCGCCAATCC-3′) and MKBR (5′-GATCTCGAGTTACCGCATAATTTCCTC-3′) and cloned between the EcoRI and XhoI sites of pBD-GAL4 (Stratagene) as the bait construct. The yeast strain YRG-2 (ura3 trp1 leu2 his3) provided by Stratagene was used for library screening. Trp+ Leu+ colonies were collected and assayed for growth on SD-Trp-Leu-His plates and LacZ activities as described previously (6a, 50). Yeast transformants expressing the MST11 bait-MST50 prey and PMK1 bait-MST50 prey constructs (29, 48) were used as the positive and negative controls, respectively.
co-IP and Western blot analysis.
The PIC1-3xFLAG and PIC5-3xFLAG constructs were generated with the yeast gap repair approach (1, 2) and confirmed by sequencing analysis. The resulting fusion constructs were transformed into protoplasts of strain 70-15. Transformants expressing the PIC1-3xFLAG and PIC5-3xFLAG constructs were identified by PCR and confirmed by Western blot analysis with an anti-FLAG antibody (Sigma-Aldrich). For co-IP assays, total proteins were isolated from vegetative hyphae as described previously (2) and incubated with anti-FLAG M2 beads (Sigma-Aldrich). Western blots of proteins eluted from the M2 beads were detected with the anti-Pmk1 (2), anti-FLAG, and anti-actin (Sigma-Aldrich) antibodies with the ECL Supersignal System (Pierce, Rockford, IL).
Appressorium formation, penetration, and plant infection assays.
Conidia were harvested from 10-day-old OA cultures with sterile distilled water and filtered through one layer of Miracloth. For appressorium formation and penetration assays, freshly harvested conidia were resuspended to 5 × 104 conidia/ml in sterile water. Drops (30 μl) of conidium suspension were placed on glass coverslips (Fisher Scientific Co., Pittsburgh, PA), onion epidermal strips, or rice leaf sheaths. Appressorium formation and development of invasive hyphae were examined after incubation in a moisture chamber for 24 or 48 h (15, 16, 39). Effects of treatments with 5 mM cAMP, 10 μM 1,16-hexadecanediol (Sigma-Aldrich), and bee waxes were assayed as described previously (20). For plant infection assays, conidia were resuspended to 5 × 104 conidia/ml in 0.25% gelatin. Two-week-old seedlings of rice cultivar Nipponbare and 8-day-old seedlings of barley cultivar Golden Promise were used for spray or injection inoculation (28, 39).
The PIC1 and PIC5 gene replacement constructs and mutants.
The ligation PCR approach (51) was used to generate the PIC1 and PIC5 gene replacement constructs. Approximately 1-kb upstream and downstream flanking sequences of PIC1 were amplified by PCR with primer pairs PIC1UF (5′-GCTGGTTGTGTACCGATACTG-3′)/PIC1UR (5′-TCAGGCGCGCCAAAGCGACTGAGCTGATCAC-3′) and PIC1DF (5′-TCAGGCCGGCCGATGAGCCAAAGCTGGAGAAG-3′)/PIC1DR (5′-GTAGCCCTGCTGCCAGATCCAAG-3′), respectively. The flanking sequences of PIC5 were amplified with primer pairs PIC5UF (5′-CCGACACCAAGCAGTTGATGT-3′)/PIC5UR (5′-TCAGGCGCGCCCTTGTGTGTAATCGGCTGTAC-3′) and PIC5DF (5′-TCAGGCCGGCCTCCTGACTGTAATAGAGTGCG-3′)/PIC5DR (5′-CGAGATAGTCTTGGTAGTTTG-3′). The resulting PCR products of PIC1 or PIC5 were digested with FseI or AscI and ligated with the hph cassette released from pCX63 (51). The PIC1 and PIC5 gene replacement constructs were amplified from the ligation products and transformed into protoplasts of strain 70-15 (3). Putative pic1 and pic5 mutants were identified by PCR and further confirmed by Southern blot analyses. For complementation assays, the full-length PIC1 and PIC5 genes were amplified and cloned into the bleomycin-resistant vector pYP1 (47) by the yeast in vivo recombination approach (2) and transformed into the pic1 and pic5 mutants, respectively. The PIC1 and PIC5 genes were fused in frame with the GFP reporter gene carried on vector pYP1 (47).
Generation of the PIC1-GFP and PIC5-GFP fusion constructs.
For generating the PIC1-GFP and PIC5-GFP fusion constructs, 2.3-kb and 4.4-kb fragments of the PIC1 and PIC5 genes containing the promoter regions, respectively, were amplified and cotransformed with XhoI-digested pKB04 into S. cerevisiae strain XK1-25 (2). Plasmids pHZ10 and pHZ11 were rescued from Trp+ yeast transformants and confirmed by sequencing analysis to contain the in-frame PIC1-GFP and PIC5-GFP fusion constructs. Zeocin-resistant transformants were isolated after transformation of the pic1 and pic5 mutants with pHZ10 and pHZ11, respectively. The resulting transformants were examined for GFP signals under an epifluorescence microscope and the expression of full-length GFP fusion proteins with a monoclonal anti-GFP antibody (Roche, Indianapolis, IN).
RESULTS
Construction of yeast two-hybrid libraries.
RNA samples isolated from vegetative hyphae grown under nitrogen starvation conditions and appressoria formed on hydrophobic surfaces (36 h) were used to construct the yeast two-hybrid cDNA libraries. The nitrogen starvation (−N) and appressorium (AP) libraries consisted of 5.7 × 109 and 1.25 × 1010 PFU/ml primary clones, respectively. The average insertion sizes were estimated to be 0.70 kb and 0.83 kb, respectively. Both libraries were amplified and preserved at −80°C.
PIC1 and PIC5 interact with PMK1.
For library screening, the PMK1 bait construct pCX38 was cotransformed into yeast strain YRG-2 with plasmid DNA isolated from amplified −N and AP cDNA libraries. A total of 9 Pmk1-interacting clones (named PICs) (Table 1) were identified and confirmed by retransformation into yeast. Five of them encode hypothetical proteins with no known functional domains. One of them, PIC8, encodes a protein homologous to nmrA of Aspergillus nidulans that is a negative regulator of nitrogen metabolism (33). PIC9 is similar to the ribosome organization protein gene BRX1 in S. cerevisiae (9).
Table 1.
Putative Pmk1-interacting genes identified by yeast two-hybrid assays
| Clone | Librarya | Gene | Description |
|---|---|---|---|
| PIC1 | −N, AP | MGG_11168.6 | Hypothetical protein with a nuclear localization signal and a MAP kinase phosphorylation site |
| PIC2 | AP | MGG_00299.6 | Hypothetical protein with no known domain |
| PIC3 | AP | MGG_04598.6 | Hypothetical protein with no known domain |
| PIC4 | AP | MGG_04655.6 | Hypothetical protein with no known domain |
| PIC5 | AP | MGG_08600.6 | Hypothetical protein with one transmembrane domain and two CTNS motifs |
| PIC6 | AP | MGG_06000.6 | Hypothetical protein with a DUF1761 domain |
| PIC7 | −N | MGG_02503.6 | Homologous to NcGrp78, a 78-kDa glucose-regulated protein with a signal peptide and an Hsp70 domain |
| PIC8 | −N | MGG_00156.6 | Homologous to the nitrogen metabolite repression regulator protein NmrA |
| PIC9 | AP | MGG_01078.6 | Ribosome biogenesis protein BRX1 |
AP, appressorium library; −N, nitrogen starvation library.
Two of them, PIC1 (MGG_11168.6) and PIC5 (MGG_08600.6), were selected for further characterization in this study. In yeast two-hybrid assays, both PIC1 and PIC5 strongly interacted with PMK1 (Fig. 1A). PIC1 encodes an 832-amino-acid protein that contains one putative nuclear localization signal (NLS) and one putative MAP kinase phosphorylation site. PIC5 encodes a 271-amino-acid protein that has one transmembrane (TM) domain and two putative CTNS (cystinosin/ERS1p repeat) motifs (IPR006603). The function of the CTNS motif is not clear. It was defined by the repetitive motif present between the transmembrane helices of the cystinosin protein encoded by the human CTNS gene. Both PIC1 and PIC5 are conserved in filamentous ascomycetes but lack distinct homologs in the budding or fission yeast. The Pic1 protein shares 19% and 21% identity with its homologs from Neurospora crassa and F. graminearum, respectively. Compared with PIC1, PIC5 homologs are more conserved in filamentous ascomycetes. Its homologs from N. crassa and F. graminearum share 70% and 65% identity, respectively, with Pic5.
Fig. 1.
Yeast two-hybrid and co-IP assays for the interaction of PMK1 with PIC1 and PIC5. (A) Yeast transformants expressing the PMK1 bait and PIC1 or PIC5 prey constructs were assayed for growth on SD-Leu-Trp-His (SD-His) plates and β-galactosidase (LacZ) activities. Positive and negative controls are marked with + and −. (B) Western blot analysis with total proteins isolated from M. oryzae transformants expressing the PIC1-3xFLAG and PIC5-3xFLAG constructs and proteins eluted from the anti-FLAG M2 beads. The presence of Pmk1 was detected with an anti-Pmk1 antibody (2). Total proteins isolated from the wild-type M. oryzae strain (70-15) and detection with an anti-actin antibody were included as controls.
To confirm the interaction of Pmk1 with Pic1 and Pic5, the PIC1-3xFLAG and PIC5-3xFLAG constructs were generated and transformed into the wild-type strain 70-15. Transformants HZA6 and HZB8, expressing the PIC1-3xFLAG and PIC5-3xFLAG constructs, respectively, were identified by PCR and confirmed by Western blot analysis with an anti-FLAG antibody (data not shown). When detected with an anti-Pmk1 antiserum (2), total proteins isolated from transformants HZA6 and HZB8 had a 42-kDa band of the expected Pmk1 size (Fig. 1B). In proteins eluted from anti-FLAG M2 beads, the same band was detected with the anti-Pmk1 antibody but its intensity was much higher in transformant HZA6 than in transformant HZB8 (Fig. 1B). Because the Pmk1 band had similar intensities in total proteins isolated from HZB8 and HZA6, results from the co-IP assays indicate that Pmk1 interacted with both Pic1 and Pic5 in vivo but that the interaction between Pic5 and Pmk1 may be relatively weaker than the Pic1-Pmk1 interaction.
Targeted gene deletion of PIC1 and PIC5 in M. oryzae.
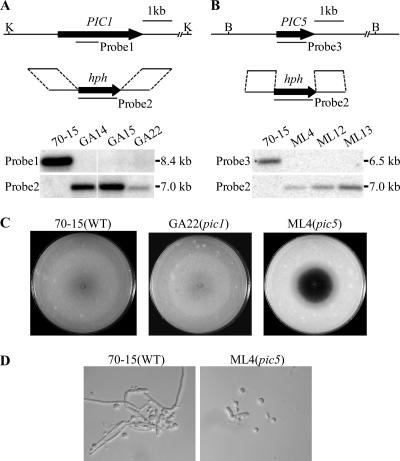
To determine their functions in M. oryzae, the PIC1 and PIC5 gene replacement constructs (Fig. 2A and B) were generated and transformed into strain 70-15. Three putative deletion mutants of each gene (Table 2) were identified and further confirmed by Southern blot analysis. When hybridized with a PIC1 fragment (probe 1), 70-15 had a single 8.4-kb KpnI band that was absent in pic1 mutants GA14, GA15, and GA22 (Fig. 2A). When hybridized with the hygromycin phosphotransferase (hph) gene (probe 2), a 7.0-kb band was detected in transformants GA14, GA15, and GA22 but not in 70-15 (Fig. 2A).
Fig. 2.
Targeted deletion of the PIC1 and PIC5 genes. (A) Schematic diagram of the PIC1 gene and gene replacement construct and Southern blots of KpnI-digested genomic DNA of the wild type (70-15) and pic1 mutants (GA14, GA15, and GA22) hybridized with probe 1 and probe 2. (B) Schematic diagram of the PIC5 gene replacement constuct and Southern blots of BamHI-digested DNA of 70-15 and pic5 mutants (ML4, ML12, and ML13) hybridized with probe 2 and probe 3. (C) Oatmeal agar cultures of 70-15, GA22, and ML4. Photographs were taken after incubation for 10 days. The central part of the ML4 colony underwent autolysis and became darkly pigmented. (D) Hyphae harvested from 2-day-old CM cultures of 70-15 and mutant ML4 were digested with 5 mg/ml lytic enzyme for 40 min. The pic5 mutant produced abundant spheroplasts and had almost no hyphal fragments left.
Table 2.
Wild-type and mutant strains of Magnaporthe oryzae used in this study
| Strain | Genotype description | Source or reference |
|---|---|---|
| 70-15 | Wild type (MAT1-1 AVR-Pita) | Chao and Ellingboe (3) |
| GA14 | pic1 mutant | This study |
| GA15 | pic1 mutant | This study |
| GA22 | pic1 mutant | This study |
| ML4 | pic5 mutant | This study |
| ML12 | pic5 mutant | This study |
| ML13 | pic5 mutant | This study |
| HZA6 | Transformant of 70-15 expressing PIC1-3xFLAG | This study |
| HZB8 | Transformant of 70-15 expressing PIC5-3xFLAG | This study |
| MC93 | Complemented transformant of GA22 (pic1/PIC1-GFP) | This study |
| MF2 | Complemented transformant of ML4 (pic5/PIC5-GFP) | This study |
Similar approaches were used to identify and confirm the pic5 deletion mutants ML4, ML12, and ML13 (Table 2). When Southern blots of genomic DNA samples digested with BamHI were hybridized with a PIC5 fragment (probe 3), 70-15 had a 6.5-kb band that was absent in the pic5 mutants ML4, ML12, and ML13 (Fig. 2B). When hybridized with the hph gene as the probe, the pic5 mutants had a 7.0-kb BamHI band that was absent in 70-15 (Fig. 2B).
PIC1 is dispensable for growth and virulence but plays a role in conidiation.
All three pic1 mutants had similar phenotypes, although only data for GA15 and GA22 are presented below. The pic1 mutant had a normal growth rate and produced typical grayish colonies on oatmeal agar plates (Fig. 2C). Conidium morphology was normal, but conidiation was reduced approximately 74% in the pic1 mutant compared with the wild type (Table 3). When assayed for appressorium formation on artificial hydrophobic surfaces, mutant GA22 had no defects in germination and formed normal appressoria. However, on onion epidermal cells, many of the pic1 conidia produced more than one appressorium on either branched germ tubes or multiple germ tubes that emerged from the same conidium compartment (Fig. 3A). Under the same conditions, this phenomenon was rarely observed in the wild type. Nevertheless, the pic1 mutant had no obvious changes in virulence. On rice leaves sprayed with pic1 conidia, similar amounts of blast lesions were observed 7 days postinoculation (dpi) on leaves sprayed with the wild-type and pic1 mutant strains (Fig. 3B).
Table 3.
Growth rate, conidiation, and penetration efficiency of the pic1 and pic5 mutants
| Strain | Growth rate (mm/day) | Conidiation (×105 spores/plate) | Penetration (%)a |
|---|---|---|---|
| 70-15 (WT) | 3.4 ± 0.1 | 5.8 ± 6.4 | 53.1 ± 6.9 |
| GA22 (pic1) | 3.2 ± 0.1 | 1.5 ± 5.8 | 51.2 ± 5.2 |
| ML4 (pic5) | 2.7 ± 0.1 | 1.8 ± 5.2 | 6.5 ± 1.9 |
| MC93 (pic1/PIC1) | 3.4 ± 0.1 | 5.6 ± 5.0 | 60.2 ± 7.8 |
| MF2 (pic5/PIC5) | 3.4 ± 0.1 | 5.4 ± 4.8 | 52.2 ± 6.4 |
Percentage of appressoria penetrated onion epidermal cells at 24 h postinoculation. Mean and standard deviations were calculated with results from three replicates.
Fig. 3.
Appressorium formation and infection assays with the pic1 mutant. (A) Conidia of the wild-type 70-15, pic1 mutant strains GA15 and GA22, and complemented transformant MC93 were incubated on glass coverslips (upper panels) and onion epidermal cells (lower panel) for 24 h. Bar = 10 μm. (B) Rice leaves sprayed with conidia from 70-15, pic1 mutants GA15 and GA22, and complemented strain MC93.
The pic5 mutant is defective in colony morphology and appressorium differentiation.
Unlike the pic1 mutant, the pic5 mutant had defects in colony morphology. Although only data for ML4 are presented below, the three pic5 mutants (Table 1) generated in this study had similar phenotypes. When cultured on oatmeal agar plates, the pic5 mutant had a reduced growth rate (Table 3). It also was reduced in the production of aerial hyphae (Fig. 2C) and conidiation (Table 3). After 4 days, autolysis of aerial hyphae was observed in the pic5 mutant (Fig. 2C), indicating that the pic5 mutant had defects in cell wall integrity. To confirm this observation, we tested the sensitivity of vegetative hyphae of the pic5 mutant to cell-wall-degrading enzymes. After digestion with 5 mg/ml lytic enzyme for 40 min, the pic5 mutant produced abundant spheroplasts. Hyphal fragments were rarely observed (Fig. 2D). Under the same conditions, the wild type produced fewer spheroplasts and still had many hyphae or hyphal fragments (Fig. 2D), indicating increased sensitivity to lytic enzyme in the pic5 mutant.
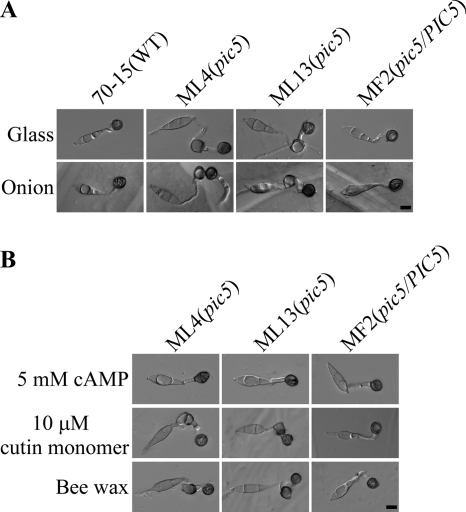
When assayed for appressorium formation on glass coverslips, 79.3% ± 7.0% of pic5 conidia were defective in appressorium formation (Fig. 4A). Chains of appressoria or appressorium-like structures were formed on germ tubes, which may be derived from appressoria formed on germinating appressoria. In the wild type, melanized appressoria were formed apically on the unbranched germ tubes (Fig. 4A). Under the same conditions, no further growth or germination from appressoria formed on hydrophobic surfaces were observed in the wild type. Germ tube growth and appressorium formation also were abnormal in the pic5 mutant on the surface of onion epidermal cells (Fig. 4A). We then assayed appressorium formation on artificial surfaces treated with exogenous cAMP, a cutin monomer, and bee waxes. Germ tube growth and appressorium formation became normal in the pic5 mutant in the presence of 5 mM cAMP (Fig. 4B). Treatments with 10 μM 1,16 hexadecanediol and bee waxes had no obvious effects on the defects of the pic5 mutant in appressorium formation (Fig. 4B). These results indicate that the pic5 mutant may be defective in surface attachment and sensing for chemical and physical signals of plant surface. This defect could be suppressed by cAMP treatment that may increase the intracellular cAMP level and bypass the recognition for surface signals.
Fig. 4.
Appressorium formation assays with the pic5 mutant. (A) Conidia from the wild-type 70-15, pic5 mutants ML4 and ML13, and complemented transformant MF2 (pic5/PIC5) were incubated on glass coverslips and onion epidermal cells for 24 h. Over 70% of the pic5 germ tubes formed more than one appressorium on individual germ tubes. (B) Appressoria formed by the pic5 mutant strains ML4 and ML13 on glass coverslips in the presence of 5 mM cAMP, 10 μM cutin monomer 1,16-hexadecanediol, and bee waxes after incubation for 24 h. The defect of the pic5 mutant in appressorium formation was suppressed by 5 mM cAMP. Bar = 10 μm.
The pic5 mutant is reduced in virulence on the plant.
To determine whether PIC5 is involved in plant infection, rice seedlings were sprayed with conidia from the pic5 mutant. On leaves inoculated with conidia of the wild-type 70-15 and complemented transformant MF2 (pic5/PIC5), similar numbers of typical rice blast lesions were formed 7 dpi (Fig. 5A). Under the same conditions, only a few lesions were observed on rice leaves sprayed with conidia of the pic5 mutant (Fig. 5A). To further confirm these observations, we repeated infection assays with barley seedlings. While barley leaves sprayed with 70-15 and complemented strain MF2 developed abundant lesions 6 dpi, only a few lesions were observed on barley leaves inoculated with the pic5 mutant (Fig. 5B). Similar results were obtained in injection infection assays. On leaves inoculated with the same amounts of conidia, the wild type caused more lesions than did the pic5 mutant surrounding the wounding sites (Fig. 5C). These data indicate that the deletion of PIC5 results a significant reduction in virulence.
Fig. 5.
Infection assays with the pic5 mutant. Rice (A) and barley (B) leaves sprayed with conidia from the wild-type strain 70-15, pic5 mutant strains ML4 and ML13, and complement transformant MF2 (pic5/PIC5). Inoculation with 0.25% gelatin solution was used as the control. The pic5 mutant caused fewer lesions compared to the complemented transformant. (C) Injection infection assays. Rice leaves inoculated with 70-15 developed more lesions than those inoculated with ML4.
The PIC5 gene plays an important role in appressorium penetration.
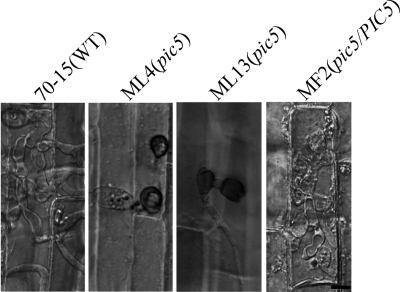
Because the pic5 mutant formed appressoria but had a reduced virulence, we assayed appressorial penetration in the pic5 mutant. On the surface of rice leaf sheaths, melanized appressoria were formed by the pic5 mutant. However, similar to what was observed on artificial surfaces, the pic5 mutant often formed chains of appressoria or appressorium-like structures (Fig. 6). While the majority of the appressoria formed by the wild-type 70-15 and complemented transformant MF2 penetrated epidermal cells of rice leaf sheaths and formed branching invasive hyphae at 48 h (Fig. 6), successful penetration and development of invasive hyphae were rarely observed in the pic5 mutant (Table 3; Fig. 6). Similar results were obtained in penetration assays with onion epidermal cells (data not shown). Appressoria formed by the pic5 mutant appeared to be defective in appressorial penetration and differentiation of invasive hyphae, which may be related to the reduction in the virulence.
Fig. 6.
Penetration assays with rice leaf sheaths. Extensive invasive hyphae were developed by the wild type (70-15) and the pic5 complemented transformant (MF2) by 48 h after inoculation. Most of the appressoria formed by the pic5 mutant strains ML4 and ML13 failed to penetrate or had only limited growth of primary invasive hyphae. Bar = 10 μm.
Expression and subcellular localization of PIC1-GFP and PIC5-GFP fusion proteins.
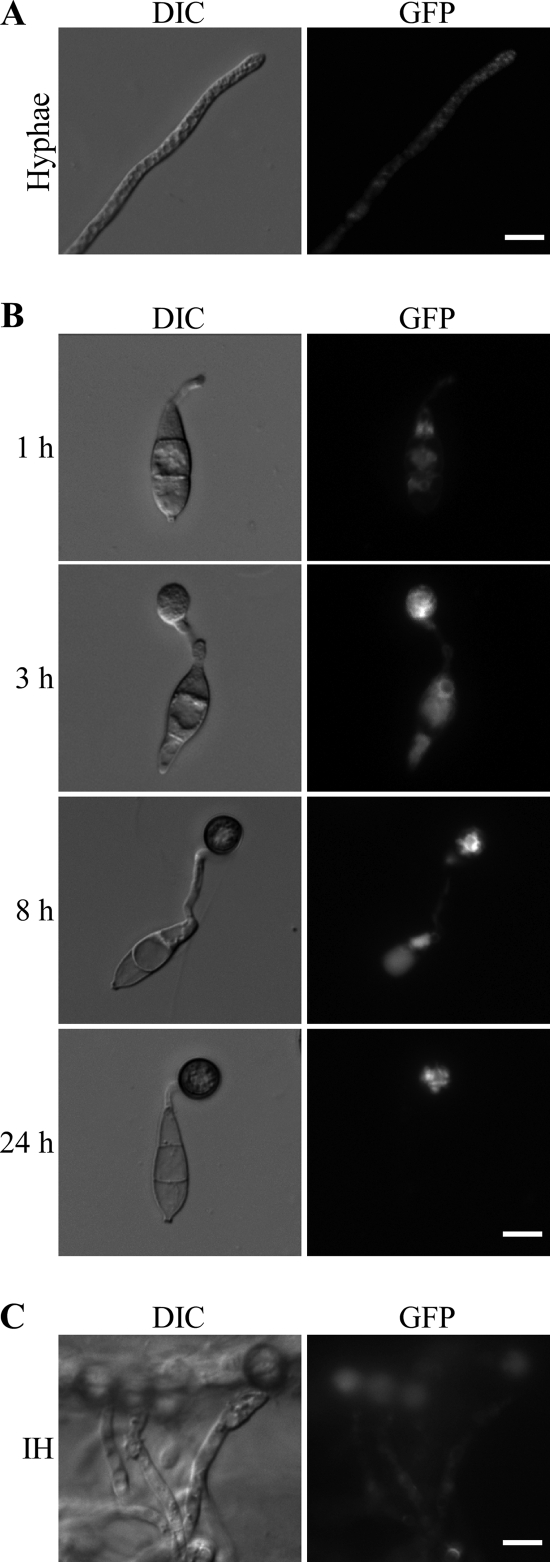
To determine their expression profiles, we generated PIC1-GFP and PIC5-GFP fusion constructs and transformed them into the pic1 and pic5 knockout mutants GA22 and ML4, respectively. On Western blots of proteins isolated from transformants expressing the PIC1-GFP and PIC5-GFP fusion constructs, 118-kDa and 56-kDa bands of the predicted Pic1-GFP and Pic5-GFP fusion proteins, respectively, were detected with an anti-GFP antibody (data not shown). These results demonstrated that full-length Pic1-GFP and Pic5-GFP fusion proteins were expressed in these transformants. In the PIC1-GFP transformant MC93 (Table 2), weak GFP signals were observed in the vegetative hyphae, conidia, appressoria, and invasive hyphae (data not shown). Transformant MC93 was normal in conidiation (Table 3) and appressorium formation on onion epidermal cells (data not shown), indicating that the expression of PIC1-GFP complemented the defects of the pic1 mutant. However, no specific subcellular localization pattern was observed for the Pic1-GFP fusion proteins.
In transformant MF2 (Table 2), expressing the PIC5-GFP construct, GFP signals were detected in the cytoplasm of vegetative hyphae (Fig. 7A), germinating conidia, and young germ tubes (Fig. 7B). On hydrophobic surfaces, single melanized appressoria were formed apically on germ tubes of transformant MF2, indicating that the expression of PIC5-GFP complemented the defects of the pic5 mutant in appressorium formation. By 8 h, fluorescent signals were still visible in the germ tubes and conidia but significantly enhanced in young appressoria. By 24 h, faint GFP signals were still observed in the cytoplasm of mature appressoria but not in the germ tubes or conidia (Fig. 7B). Unlike Pmk1 (2), nuclear localization was not observed in the PIC1-GFP and PIC5-GFP transformants MC93 and MF2. We also examined GFP signals in invasive hyphae formed by transformant MF2 in onion epidermal cells. No specific distribution patterns could be recognized, although weak fluorescent signals appeared to be in the cytoplasm of invasive hyphae (Fig. 7C).
Fig. 7.
Expression and localization of PIC5-GFP. (A) Vegetative hyphae of transformant MF2 expressing the PIC5-GFP fusion construct. (B) Conidia from transformant MF2 were incubated on glass coverslips at room temperature for 1, 3, 8, and 24 h. (C) Invasive hyphae (IH) formed by transformant MF2 in onion epidermal cells. The same field was examined under differential interference contrast (DIC) (left panels) and epifluorescence microscopy (right panels). Bar = 10 μm.
DISCUSSION
In M. oryzae, the PMK1 MAP kinase pathway is important for appressorium formation and plant infection (43, 49). PMK1 is constitutively expressed but its expression is increased during appressorium formation and young conidium development (2). The Pmk1-GFP fusion protein localizes to the nucleus in appressoria. To further characterize the Pmk1 pathway, in this study we constructed yeast two-hybrid libraries and identified nine putative Pmk1-interacting genes. Whereas six of the PIC genes were isolated from the appressorium library, two were identified in the nitrogen starvation library. PIC1 was the only one that that was identified in both libraries. Four of these genes, PIC2, PIC4, PIC7, and PIC9, have one putative MAP kinase docking site. Some of these proteins may be involved in stimulating or stabilizing the Mst7-Pmk1 interaction during appressorium formation. In these screens, the upstream MST7 MEK and downstream MST12 transcription factor were not identified as Pmk1-interacting clones. It has been reported that the Pmk1-Mst7 interaction was not detectable in yeast two-hybrid assays (48). For Mst12, its weak interaction with Pmk1 (30) may be too transient to be detected in the yeast two-hybrid library screening. However, it is also possible that Pmk1 interacts only with full-length Mst12. The average insert size of these two yeast two-hybrid libraries is less than 0.9 kb, which is smaller than the 2,148-bp MST12 open reading frame (ORF).
The difference in the phenotypes of the pmk1 and mst12 mutants has led to the hypothesis that additional transcription factors other than MST12 must exist in M. oryzae for regulating appressorium formation. Unfortunately, no putative transcription factor genes other than the nmrA ortholog were identified among the PIC genes. Pic1 is the only one with a putative NLS but it lacks a DNA binding motif. For the nmrA ortholog, its function in nitrogen metabolism and interaction with Pmk1 remain to be determined. Several transcription factor genes have been reported to play critical roles in appressorium formation in M. oryzae (15a, 27a). However, none of them has been shown to be functionally related to the Pmk1 pathway. Although the pth12 and con7 mutants were reported to fail to form appressoria (15a, 27a), deletion of PTH12 or CON7 in Guy11 or 70-15 in our laboratory resulted in a reduction in appressorium formation and virulence but did not block appressorium formation (J.-R. Xu, unpublished data). Transcription factors regulated by Pmk1 for appressorium formation may be expressed at a relatively low level, or their association with PMK1 is too transient to be identified in yeast two-hybrid assays.
The PIC1 and PIC5 genes were selected for functional characterization because they had the strongest interaction with PMK1 in yeast two-hybrid assays. In co-IP assays, the interaction of Pmk1 with PIC1 and PIC5 was confirmed. Although their interactions with Pmk1 were similar in yeast two-hybrid assays, PIC1 appeared to have a stronger interaction with Pmk1 than did PIC5 in co-IP assays (Fig. 1B). Structurally, PIC1 and PIC5 lack any common domains or motifs. While Pic1 has one putative NLS and one putative MAPK phosphorylation site, Pic5 contains one transmembrane domain and two CTNS motifs. Both PIC1 and PIC5 lack distinct homologous genes in the fission and budding yeasts, but their homologs are well conserved in filamentous ascomycetes that have been sequenced, including N. crassa and F. graminearum (4, 10). However, none of the PIC1 and PIC5 homologs have been functionally characterized in fungi. PIC5 shares limited homology (mainly in the PQ-loop repeats) with the mannose-P-dolichol utilization defect 1 protein of mammalian cells (34).
Unlike the pmk1 mutant (43), the pic1 and pic5 mutants still produced melanized appressoria. For the pic1 mutant, no obvious defects were observed in plant infection and appressorium formation on artificial surfaces. Interestingly, germ tubes of the pic1 mutant tended to branch and form multiappressoria on onion epidermal cells. In the mst11ΔRAD mutant of M. oryzae, germ tubes that formed appressoria often branch and form additional appressoria (48). One hypothesis is that deletion of the RAS association domain (RAD) may result in the defect of the feedback inhibition of germ tube branching and the formation of additional appressoria when one appressorium is formed on the germ tube tips. The PIC1 gene may function downstream from Pmk1 and play a role in this feedback inhibition. However, the formation of branching germ tubes and multiple appressoria in the mst11ΔRAD mutant (48) is not a surface-dependent event. The difference between artificial hydrophobic surfaces and onion epidermal cells may be related to surface hydrophobicity, hardness, and chemical signals that are known to play key roles in inducing appressorium formation. Therefore, it will be interesting to further characterize the defects of the pic1 mutant in the production of multiple appressoria.
The pic5 mutant was defective in appressorium formation on both plant and artificial surfaces (Fig. 4A). The vast majority (>70%) of appressorium-like structures appeared to germinate and form other appressoria by 24 h (Fig. 4). To our knowledge, no mutants with similar defects have been reported in M. oryzae. PIC5 may function in the terminal commitment of deformed germ tubes to form appressoria that are arrested in the G1 stage. Deletion of PIC5 may result in a defect in G1 arrest and failure to prevent the further growth or germination of developing appressoria. However, it is also possible that appressoria formed by the pic5 mutant failed to attach tightly to the surface. Appressorium formation was aborted when the internal turgor lifted up the developing appressorium, which resulted in further growth. Treatments with bee waxes and a cutin monomer had no effect on the defect of the pic5 mutant in appressorium formation, further indicating that that PIC5 may be involved in surface attachment but not in the recognition of physical or chemical signals of the surface. When treated with cAMP, that is, in contrast to cutin monomers or waxes, an intracellular messenger rather than an extracellular signal, appressorium formation became normal in the pic5 mutant. The difference between cAMP and cutin monomers or waxes is that the former is an intracellular messenger, not an extracellular signal molecule. Unlike the pic5 mutant, which formed chains of appressorium-like structures, the pmk1 mutant was blocked in appressorium formation on artificial hydrophobic surfaces (43). However, the pmk1 mutant still responds to cAMP treatment. On hydrophilic surfaces, exogenous cAMP stimulates the formation of subapical swollen bodies in the pmk1 mutant (43). In M. oryzae, cAMP signaling may be involved in both surface attachment and surface sensing.
Unlike the pic1 mutant, the pic5 mutant displayed a significant reduction in plant penetration and virulence. Appressorium penetration efficiency was reduced to 6.5% in the pic5 mutant. In comparison with the wild type and the complemented transformant, the pic5 mutant was reduced approximately 6-fold in terms of the number of lesions formed on rice or barley leaves. Because turgor generation is dependent on the carbon reserve in conidium compartments, the formation of chains of appressoria or appressorium-like structures will reduce appressorium turgor and penetration efficiency. Therefore, the reduction of the pic5 mutant in virulence may be directly related to its defects in appressorium formation and penetration. However, the pic5 mutant has a reduced growth rate. It remains possible that PIC5 also plays a role in the differentiation and growth of invasive hyphae after penetration. In M. oryzae, both PMK1 and MST12 are known to be essential for infectious growth.
Colonies of the pic5 mutant produced fewer aerial hyphae and underwent autolysis in cultures older than 10 days (Fig. 2C). Vegetative hyphae of the mutant also had increased sensitivity to lytic enzyme (Fig. 2D), suggesting that deletion of PIC5 resulted in a defect in cell wall integrity. In M. oryzae, the MPS1 MAP kinase cascade is known to be important for regulating cell wall integrity and plant infection (13, 42). However, deletion of RLM1, one of its downstream transcription factors, had no obvious cell wall defects (23). It is possible that PIC5 is also functionally related to MPS1. Further characterization of the PIC5 gene may lead to better understanding of possible cross talk between the PMK1 and MPS1 pathways.
In transformants expressing the PIC1-GFP and PIC5-GFP fusion constructs, weak GFP signals could be detected in vegetative hyphae, conidia, germ tubes, appressoria, and invasive hyphae. PIC1 and PIC5 appeared to have similar expression patterns. However, PIC1 was identified in both the −N and AP libraries but PIC5 was isolated only in the AP library. The expression level of PIC5 may be relatively low in vegetative hyphae. In M. oryzae, PMK1 is constitutively expressed, but its expression is increased during appressorium formation (2). Pmk1-GFP fusion proteins localize to the nucleus during appressorium formation. In PIC1-GFP and PIC5-GFP transformants, nuclear localization of GFP signals was not observed in conidia, appressoria, or vegetative or invasive hyphae. Pic5 has one putative transmembrane domain. However, GFP signals were mainly observed in the cytoplasm in the pic5/PIC5-GFP transformant. The interaction between Pmk1 and Pic5 likely occurs in the cytoplasm, which may interfere with the localization of Pmk1 to the nucleus. Unlike Pic5, Pic1 has a putative nucleus localization signal. Our results indicate that the predicted NLS of Pic1 is not functional or that Pic1 localizes to the nucleus only at a specific stage.
Overall, although PIC1 and PIC5 are dispensable for the initiation and formation of melanized appressoria, both of them appear to be involved in the late stages of appressorium formation. PIC1 may be involved in the feedback inhibition of germ tube branching or the production of secondary germination from the same conidium compartment after the formation of an appressorium apically under certain conditions. PIC5 is important for arresting the further growth of appressoria or preventing appressoria from germinating. In addition, PIC5 may be functionally related to the PMK1 MAP kinase in infectious growth after penetration. These results indicate that isolating and characterizing the PMK1-interacting genes are a useful approach to identify genes important for appressorium formation and function. It will be interesting to further characterize other PIC genes and to determine their functional relationship with Pmk1.
ACKNOWLEDGMENTS
We thank Larry Dunkle and Charles Woloshuk at Purdue University for critical reading of this work.
This work was supported by grants to J.-R.X. from the National Research Initiative of the USDA CSREES (#2007-35319-102681) and the USDA National Institute of Food and Agriculture (#2010-65110-20439).
Footnotes
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Bourett T. M., Sweigard J. A., Czymmek K. J., Carroll A., Howard R. J. 2002. Reef coral fluorescent proteins for visualizing fungal pathogens. Fungal Genet. Biol. 37:211–220 [DOI] [PubMed] [Google Scholar]
- 1a. Brachmann A., Schirawski J., Muller P., Kahmann R. 2003. An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22:2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bruno K. S., Tenjo F., Li L., Hamer J. E., Xu J. R. 2004. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3:1525–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chao C. C. T., Ellingboe A. H. 1991. Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can. J. Bot. 69:2130–2134 [Google Scholar]
- 4. Cuomo C. A., et al. 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317:1400–1402 [DOI] [PubMed] [Google Scholar]
- 5. Dean R. A. 1997. Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35:211–234 [DOI] [PubMed] [Google Scholar]
- 6. de Jong J. C., McCormack B. J., Smirnoff N., Talbot N. J. 1997. Glycerol generates turgor in rice blast. Nature 389:244–245 [Google Scholar]
- 6a. Ding S., et al. 2010. The Tigl HDAC complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae. Plant Cell 22:2495–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Di Pietro A., Garcia-MacEira F. I., Meglecz E., Roncero M. I. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39:1140–1152 [PubMed] [Google Scholar]
- 8. Ebbole D. J., et al. 2004. Gene discovery and gene expression in the rice blast fungus, Magnaporthe grisea: analysis of expressed sequence tags. Mol. Plant Microbe Interact. 17:1337–1347 [DOI] [PubMed] [Google Scholar]
- 9. Eisenhaber F., Wechselberger C., Kreil G. 2001. The Brix domain protein family - a key to the ribosomal biogenesis pathway? Trends Biochem. Sci. 26:345–347 [DOI] [PubMed] [Google Scholar]
- 10. Galagan J. E., et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868 [DOI] [PubMed] [Google Scholar]
- 11. Howard R. J., Ferrari M. A., Roach D. H., Money N. P. 1991. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. U. S. A. 88:11281–11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jenczmionka N. J., Maier F. J., Losch A. P., Schafer W. 2003. Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase GPMK1. Curr. Genet. 43:87–95 [DOI] [PubMed] [Google Scholar]
- 13. Jeon J., et al. 2008. A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol. Plant Microbe Interact. 21:525–534 [DOI] [PubMed] [Google Scholar]
- 14. Kaffarnik F., Muller P., Leibundgut M., Kahmann R., Feldbrugge M. 2003. PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 22:5817–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kankanala P., Czymmek K., Valent B. 2007. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19:706–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a. Kim S., et al. 2009. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 5:e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koga H. 1994. Hypersensitive death, autofluorescence, and ultrastructural-changes in cells of leaf sheaths of susceptible and resistant near-isogenic lines of rice (PI-Z(T)) in relation to penetration and growth of Pycularia oryza. Can. J. Bot. 72:1463–1477 [Google Scholar]
- 17. Lev S., Sharon A., Hadar R., Ma H., Horwitz B. A. 1999. A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. U. S. A. 96:13542–13547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li L., Ding S. L., Sharon A., Orbach M., Xu J. R. 2007. Mir1 is highly upregulated and localized to nuclei during infectious hyphal growth in the rice blast fungus. Mol. Plant Microbe Interact. 20:448–458 [DOI] [PubMed] [Google Scholar]
- 19. Li L., Xue C. Y., Bruno K., Nishimura M., Xu J. R. 2004. Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant Microbe Interact. 17:547–556 [DOI] [PubMed] [Google Scholar]
- 20. Liu W., et al. 2011. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 7:e1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reference deleted.
- 22. Mayorga M. E., Gold S. E. 1999. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34:485–497 [DOI] [PubMed] [Google Scholar]
- 23. Mehrabi R., Ding S., Xu J. R. 2008. MADS-box transcription factor Mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot. Cell 7:791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mey G., Oeser B., Lebrun M. H., Tudzynski P. 2002. The biotrophic, non-appressorium-forming grass pathogen Claviceps purpurea needs a FUS3/PMK1 homologous mitogen-activated protein kinase for colonization of rye ovarian tissue. Mol. Plant Microbe Interact. 15:303–312 [DOI] [PubMed] [Google Scholar]
- 25. Mitchell T. K., Dean R. A. 1995. The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell 7:1869–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muller P., Aichinger C., Feldbrugge M., Kahmann R. 1999. The MAP kinase Kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34:1007–1017 [DOI] [PubMed] [Google Scholar]
- 27. Nishimura M., Park G., Xu J. R. 2003. The G-beta subunit MGB1 is involved in regulating multiple steps of infection-related morphogenesis in Magnaporthe grisea. Mol. Microbiol. 50:231–243 [DOI] [PubMed] [Google Scholar]
- 27a. Odenbach D., et al. 2007. The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea. Mol. Microbiol. 64:293–307 [DOI] [PubMed] [Google Scholar]
- 28. Park G., Bruno K. S., Staiger C. J., Talbot N. J., Xu J. R. 2004. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol. Microbiol. 53:1695–1707 [DOI] [PubMed] [Google Scholar]
- 29. Park G., et al. 2006. Multiple upstream signals converge on an adaptor protein Mst50 to activate the PMK1 pathway in Magnaporthe grisea. Plant Cell 18:2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park G., Xue G. Y., Zheng L., Lam S., Xu J. R. 2002. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 15:183–192 [DOI] [PubMed] [Google Scholar]
- 31. Perfect S. E., Green J. R. 2001. Infection structures of biotrophic and hemibiotrophic fungal plant pathogens. Mol. Plant Pathol. 2:101–108 [DOI] [PubMed] [Google Scholar]
- 32. Ruiz-Roldan M. C., Maier F. J., Schafer W. 2001. PTK1, a mitogen-activated-protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity of Pyrenophora teres on barley. Mol. Plant Microbe Interact. 14:116–125 [DOI] [PubMed] [Google Scholar]
- 33. Stammers D. K., et al. 2001. The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J. 20:6619–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strausberg R. L., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U. S. A. 99:16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takano Y., et al. 2000. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant Microbe Interact. 13:374–383 [DOI] [PubMed] [Google Scholar]
- 36. Talbot N. J. 2003. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57:177–202 [DOI] [PubMed] [Google Scholar]
- 37. Thines E., Weber R. W. S., Talbot N. J. 2000. MAP kinase and protein kinase A-dependent mobilizationof triacylglycerol and glycogen during appressorium tugor generation by Magnaporthe grisea. Plant Cell 12:1703–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsuji G., Fujii S., Tsuge S., Shiraishi T., Kubo Y. 2003. The Colletotrichum lagenarium Ste12-like gene CST1 is essential for appressorium penetration. Mol. Plant Microbe Interact. 16:315–325 [DOI] [PubMed] [Google Scholar]
- 39. Tucker S. L., et al. 2004. A fungal metallothionein is required for pathogenicity of Magnaporthe grisea. Plant Cell 16:1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Urban M., Mott E., Farley T., Hammond-Kosack K. 2003. The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 4:347–359 [DOI] [PubMed] [Google Scholar]
- 41. Valent B., Chumley F. G. 1991. Molecular genetic analysis of the rice blast fungus Magnaporthe grisea. Annu. Rev. Phytopathol. 29:443–467 [DOI] [PubMed] [Google Scholar]
- 42. Xu J. R. 2000. MAP kinases in fungal pathogens. Fungal Genet. Biol. 31:137–152 [DOI] [PubMed] [Google Scholar]
- 43. Xu J. R., Hamer J. E. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10:2696–2706 [DOI] [PubMed] [Google Scholar]
- 44. Xu J. R., Staiger C. J., Hamer J. E. 1998. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. U. S. A. 95:12713–12718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu J. R., Urban M., Sweigard J. A., Hamer J. E. 1997. The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Mol. Plant Microbe Interact. 10:187–194 [Google Scholar]
- 46. Xue C. Y., et al. 2002. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 14:2107–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., Choi Y. E., Zou X., Xu J. R. 2011. The FvMK1 mitogen-activated protein kinase gene regulates conidiation, pathogenesis, and fumonisin production in Fusarium verticillioides. Fungal Genet. Biol. 48:71–79 [DOI] [PubMed] [Google Scholar]
- 48. Zhao X., Kim Y., Park G., Xu J. R. 2005. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 17:1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao X., Mehrabi R., Xu J.-R. 2007. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 6:1701–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao X. H., Xu J. R. 2007. A highly conserved MAPK-docking site in Mst7 is essential for Pmk1 activation in Magnaporthe grisea. Mol. Microbiol. 63:881–894 [DOI] [PubMed] [Google Scholar]
- 51. Zhao X. H., Xue C., Kim Y., Xu J. R. 2004. A ligation-PCR approach for generating gene replacement constructs in Magnaporthe grisea. Fungal Genet. Newsl. 51:17–18 [Google Scholar]
- 52. Zheng L., Campbell M., Murphy J., Lam S., Xu J. R. 2000. The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 13:724–732 [DOI] [PubMed] [Google Scholar]