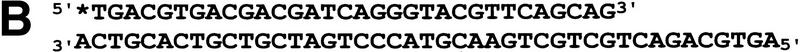Abstract
Werner syndrome (WS) is the hallmark premature aging disorder in which affected humans appear older than their chronological age. The protein WRNp, defective in WS, has helicase function, DNA-dependent ATPase, and exonuclease activity. Although WRNp functions in nucleic acid metabolism, there is little or no information about the pathways or protein interactions in which it participates. Here we identify Ku70 and Ku86 as proteins that interact with WRNp. Although Ku proteins had no effect on ATPase or helicase activity, they strongly stimulated specific exonuclease activity. These results suggest that WRNp and the Ku complex participate in a common DNA metabolic pathway.
Keywords: Werner syndrome, Ku proteins, WRNp, exonuclease activity
Werner syndrome (WS) is a human premature aging disorder characterized by the early display of many of the signs and symptoms that are associated with the normal aging process. Cells from WS patients also show a number of phenotypic changes typical of chronologically older normal cells including increased chromosomal abnormalities and rapid onset of cellular senescence (Martin 1997). WS cells are not generally sensitive to DNA-damaging agents but are hypersensitive to the carcinogen 4 nitroquinoline (4NQO) and topoisomerase I-specific inhibitor, campthothecin (Lebel and Leder 1998). The WS gene WRN (Yu et al. 1996) is a member of the RecQ family that includes the RecQ4 and BLM genes, defects that are responsible for the premature aging and cancer prone phenotypes of Rothmund-Thomson (Kitao et al. 1999) and Bloom syndrome (Ellis 1997), respectively. These syndromes are characterized by a high degree of genomic instability, including chromosomal breaks, multiple large deletions, and translocations (Moser et al. 1999). WRNp has helicase activity and an associated DNA-dependent ATPase activity (Gray et al. 1997; Brosh et al. 1999). WRNp also has an exonuclease function located in its amino-terminal region. Two laboratories have identified the directionality of the exonuclease to be 3′ → 5′ (Huang et al. 1998; Kamath-Loeb et al. 1998; Shen et al. 1998), whereas one laboratory reported a 5′ → 3′ directionality (Suzuki et al. 1999).
The enzymatic functions of WRNp indicate a role in nucleic acid metabolism, but information about the pathways in which it participates is lacking. In this study we used a chromatographic procedure to isolate proteins that bind to WRNp. This approach identified Ku70 and Ku86. This physical interaction between the Ku86/70 complex was verified by immunoprecipitation from cell extracts. Moreover, the Ku86/70 complex functionally interacts with WRNp, strongly stimulating the 3′ → 5′ exonuclease activity of WRNp.
Results
The carboxyl terminus of WRNp interacts physically with the Ku86/70 complex
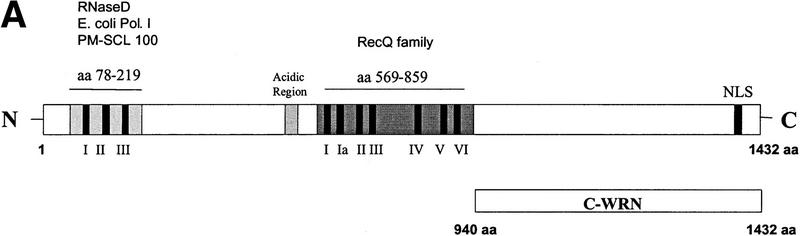
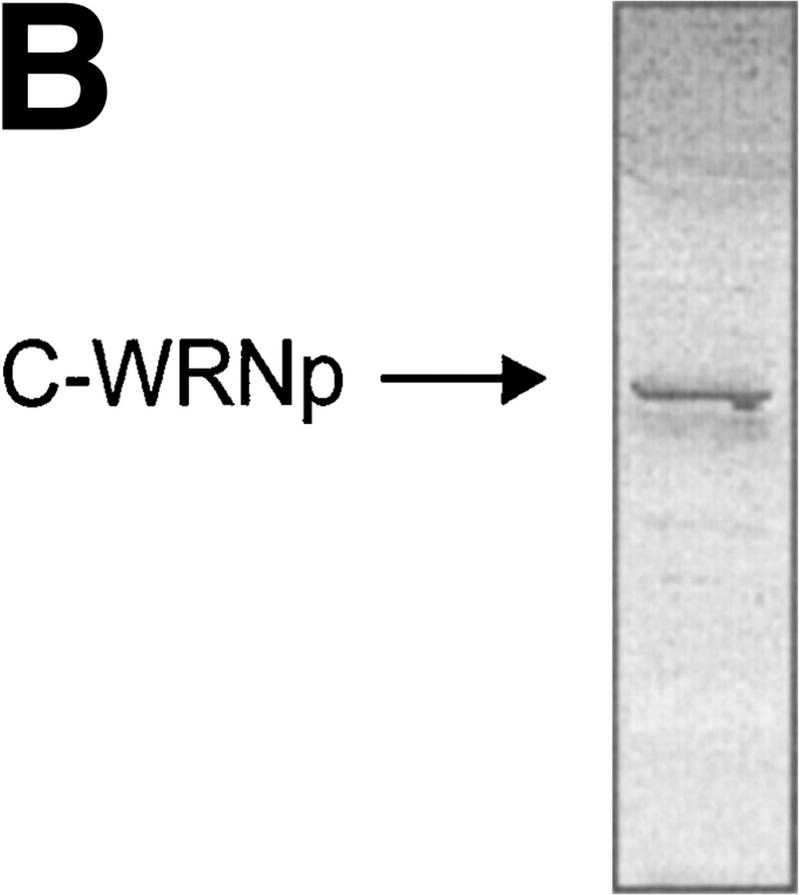
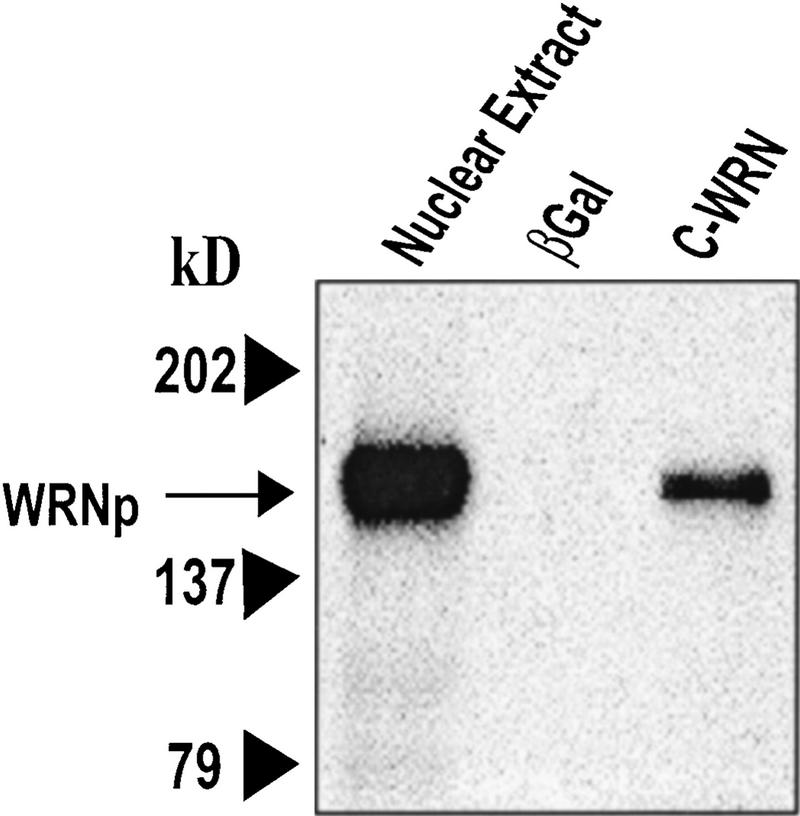
The exonuclease domain of WRNp is located in the amino-terminal end of the protein and the helicase/ATPase domain in its central part (Fig. 1A). Because the carboxyl terminus (C-WRNp) of WRNp lacks homology to known functional domains, we searched for interactions with this portion of the protein. The carboxyl terminus was cloned into an Escherichia coli expression vector and the recombinant protein was expressed and purified (Fig. 1B). This fragment was immobilized on an agarose bead matrix, and specific proteins were identified by chromatographing HeLa nuclear extract through the column. Columns either without protein attached or with excess β-galactosidase were used to identify proteins that bound nonspecifically. Proteins were eluted with increasing salt concentrations. Radioactively labeled extract was used to identify a 160-kD band on SDS-PAGE, which was shown to be full-length WRNp by Western blot (Fig. 2). This result suggests dimerization or a higher order assembly of WRNp, an observation that has not been reported previously.
Figure 1.
Purification of the carboxyl terminus of Werner protein. (A) Schematic of C-WRNp fragment. The amino terminus contains the 3 conserved exonuclease motifs. The central region contains the conserved 7 motif helicase domain. (B) SDS-PAGE with Coomassie staining of C-WRNp.
Figure 2.
Specific proteins bound to C-WRNp. C-WRNp binds to full-length WRNp (∼160 kD). Western blot with anti-WRNp antibody. (Lane 1) Nuclear extract, positive control; (lane 2) β-galactosidase column eluate; (lane 3) C-WRNp column eluate.
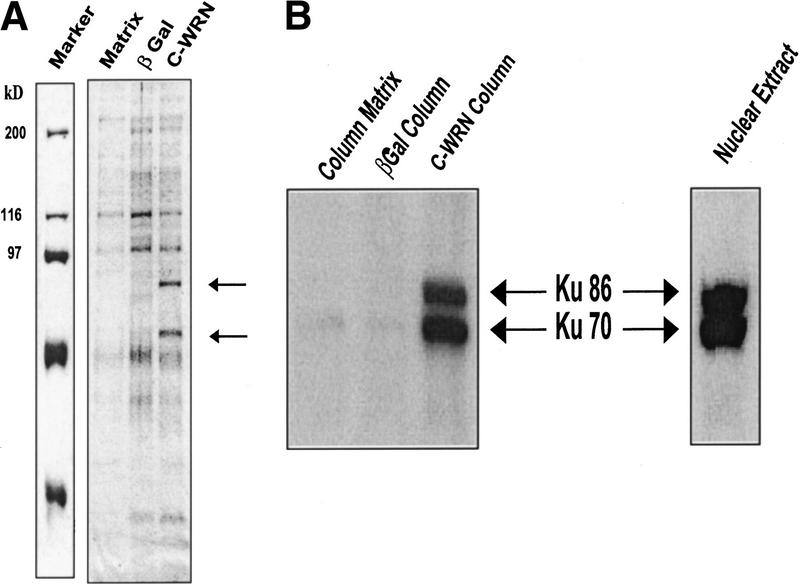
Two strong bands (Fig. 3A, arrows) were observed in the 0.2-M salt eluate at ∼70 and 90 kD, which were not present in the eluate from the β-galactosidase or matrix columns. These bands were excised and identified by matrix-assisted laser desorption ionization mass spectrometry (MALDI–MS) to be the Ku70 and Ku86 proteins. The identities of the Ku proteins were verified by Western blot (Fig. 3B) using antibodies against Ku70 and Ku86. As there was no binding of Ku70 or Ku86 to either the β-galactosidase or matrix alone column (Fig. 3B), the C-WRN interactions with Ku proteins appears to be specific. Fluorimager analysis demonstrated that the recombinant C-WRNp and Ku proteins contained no DNA, ruling out that the interactions were mediated via DNA (data not shown).
Figure 3.
Ku proteins interact with C-WRNp. (A) Two proteins tightly associated with C-WRNp were identified by Coomassie staining of samples analyzed by SDS-PAGE. (Lane 1) Marker; (lane 2) no protein column; (lane 3) β-galactosidase column; (lane 4) C-WRNp column. (Arrows) Proteins that associated with C-WRNp specifically. (B) Western analysis using antibodies against Ku86 and Ku70 identify the two specific proteins as the Ku proteins. Antibodies against Ku86 and Ku70 were obtained from Oncogene Research Products (Cambridge, MA) and used to sequentially probe the membrane.
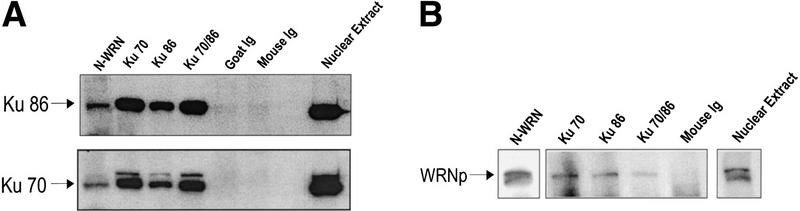
To confirm this interaction in vivo and to investigate whether the Ku proteins bind to full-length WRNp, an antibody against the amino terminus of WRNp was generated and used to immunoprecipitate full-length WRNp and associated proteins from cell extracts. The antibody immunoprecipitated WRNp (Fig. 4B, lane 1) and coimmunoprecipitated Ku86 and Ku70 (Fig. 4A, lane 1). Immunoprecipitation of the HeLa cell extracts with antibodies to Ku70, Ku86, and Ku86/70 showed interaction by Western blot with Ku86 and Ku70 as expected (Fig. 4A) and with full-length WRNp (Fig 4B). The nuclear extract served as a control to locate the Ku bands. This interaction was also verified by immunoprecipitation of purified proteins (M. Cooper, unpubl.). Furthermore, WRNp was not immunoprecipitated from a WS fibroblast cell extract with a Ku antibody (data not shown).
Figure 4.
WRNp and Ku complex coimmunoprecipitate. (A) Polyclonal antibody raised against the amino terminus of WRN (amino acids 1–600, designated N-WRN) was raised in mouse. The blot was sequentially probed with the Ku86 and Ku70 antibodies. (Lane 1) Anti-WRN mouse polyclonal antibody immunoprecipitate; (lane 2) anti-Ku70 mouse monoclonal antibody immunoprecipitate; (lane 3) anti-Ku86 mouse monoclonal antibody immunoprecipitate; (lane 4) anit-Ku70/86 goat polyclonal antibody immunoprecipitate; (lane 5) goat immunoglobin precipitate; (lane 6) mouse immunoglobin precipitate; (lane 7) blank; (lane 8) HeLa nuclear extract, unprecipitated, to mark the positions of Ku86 and Ku70. (B) Immunoprecipitation of WRNp by antibodies specific for N-WRN, Ku70, Ku86, and the Ku86/70 heterodimer. The insert labeled WRN (lane 1) shows the anti-N-WRN antibody immunoprecipitate probed with polycolonal antibodies against the carboxyl terminus of WRN (prepared similarly to the anti-N WRN antibody), demonstrating that the anti-N-WRN antibody is specific for full-length WRN. (Lane 2) Anti-Ku70 mouse mAb immunoprecipitate; (lane 3) anti-Ku86 mouse mAb immunoprecipitate; (lane 4) anti-Ku86/70 goat polyclonal antibody immunoprecipitate; (lane 5) mouse immunoglobulin immunoprecipitate; (lane 6) HeLa nuclear extract, unprecipitated, to mark the position of WRNp.
Ku complex has no effect on WRNp helicase or ATPase activities
Next, we investigated whether the physical interaction between WRNp and the Ku86/70 complex might mediate a change in WRNp activity. WRNp is a processive DNA-dependent ATPase that prefers ssDNA as the DNA effector (Orren et al. 1999; Brosh et al. 1999). The ATPase activity (kcat) of WRNp (7 nm) in the absence of Ku86/70 was determined to be 139/min, consistent with previously published data (Orren et al. 1999; Brosh et al. 1999). In control reactions, Ku86/70 (70 nm, heterotrimer) alone did not demonstrate any detectable ATP hydrolysis over background. When Ku86/70 and WRNp were preincubated together with M13 ssDNA and reactions were initiated with [3H]ATP, the kcat for ATP hydrolysis by WRNp was 152/min, indicating that Ku protein does not significantly modulate ATP hydrolysis catalyzed by WRNp.
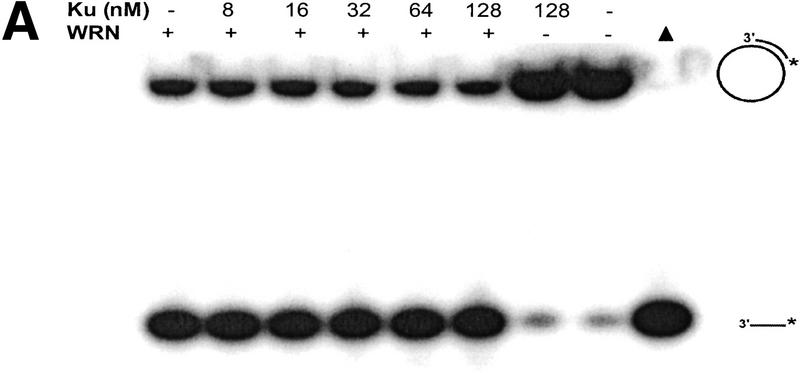
The helicase activity of WRNp was examined on a M13 28-bp partial duplex DNA substrate in the presence or absence of Ku86/70 (Fig. 5A). Reaction products were analyzed by nondenaturing gel electrophoresis. WRNp (15 nm monomer) alone unwound ∼70% of the helicase substrate in a 30-min reaction (Fig. 5A, lane 1), whereas Ku86/70 (128 nm) alone did not detectably unwind the double-stranded (ds) DNA substrate (Fig. 5A, lane 7). When WRNp and Ku86/70 were preincubated together with the dsDNA substrate and reactions were initiated with ATP, Ku86/70 (8–128 nm) had no effect on WRNp helicase activity (Fig. 5A, lanes 2–6). Moreover, Ku86/70 (8–128 nm) did not stimulate WRNp helicase activity on the 28-bp partial duplex substrate with an amount of WRNp (3 nm monomer) that unwinds only 5% of the duplex DNA substrate (A. Machwe and D. Orren, unpubl.). These results demonstrate that Ku86/70 neither stimulates nor inhibits WRN helicase activity on short DNA duplexes.
Figure 5.
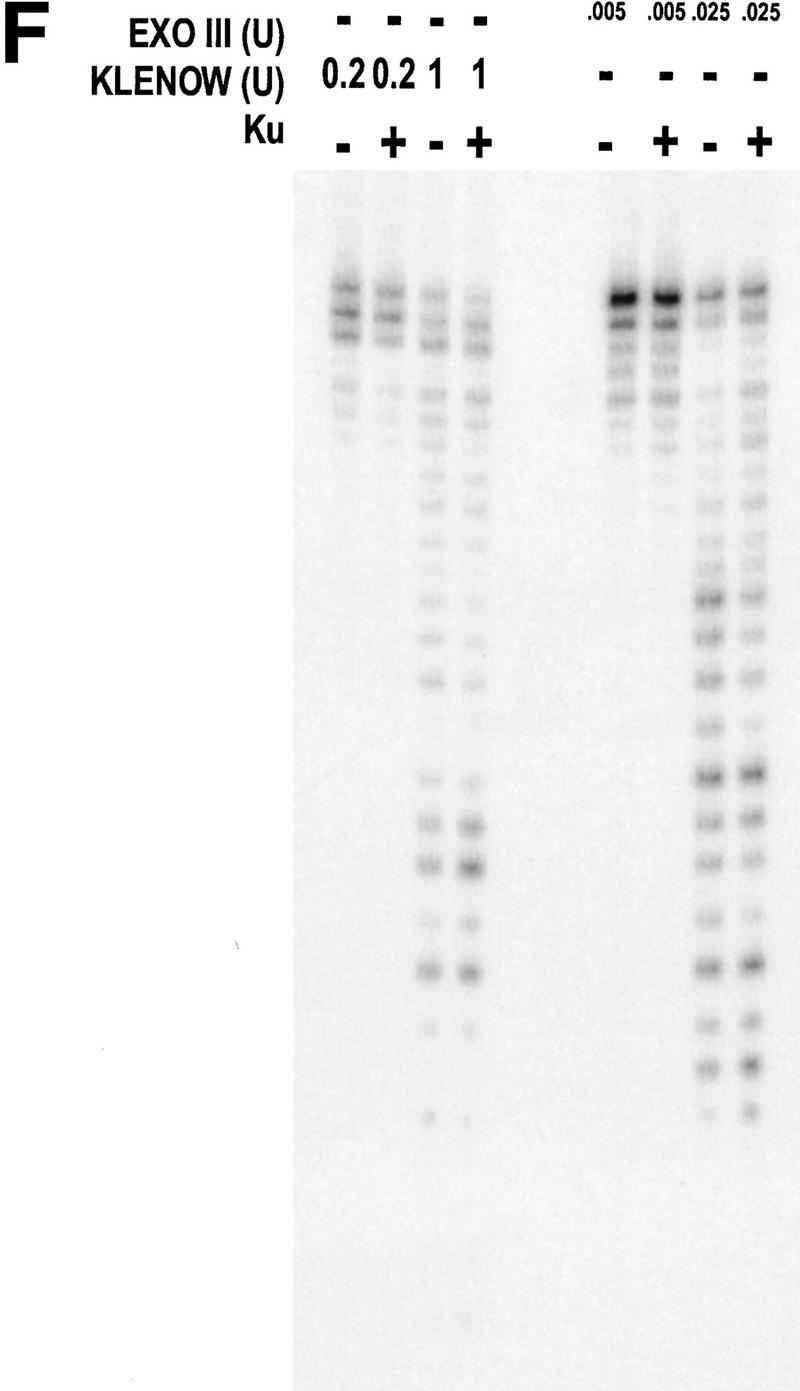
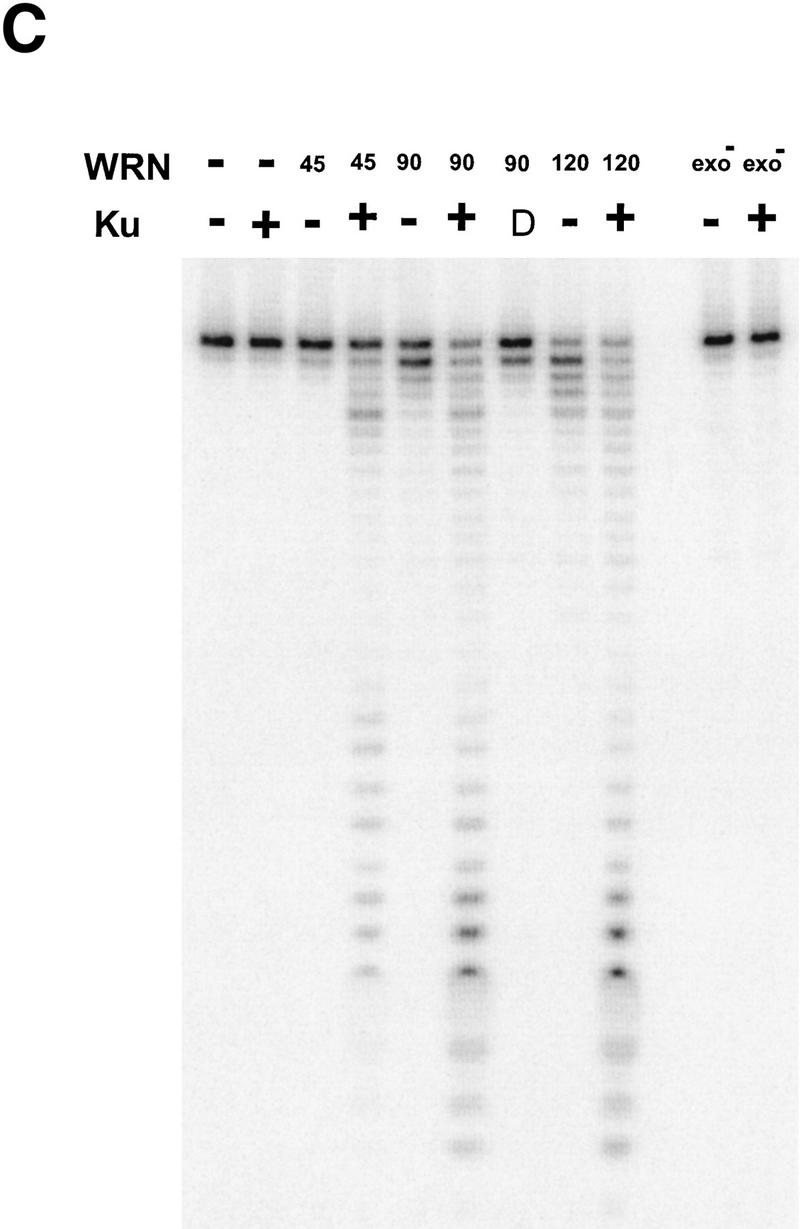
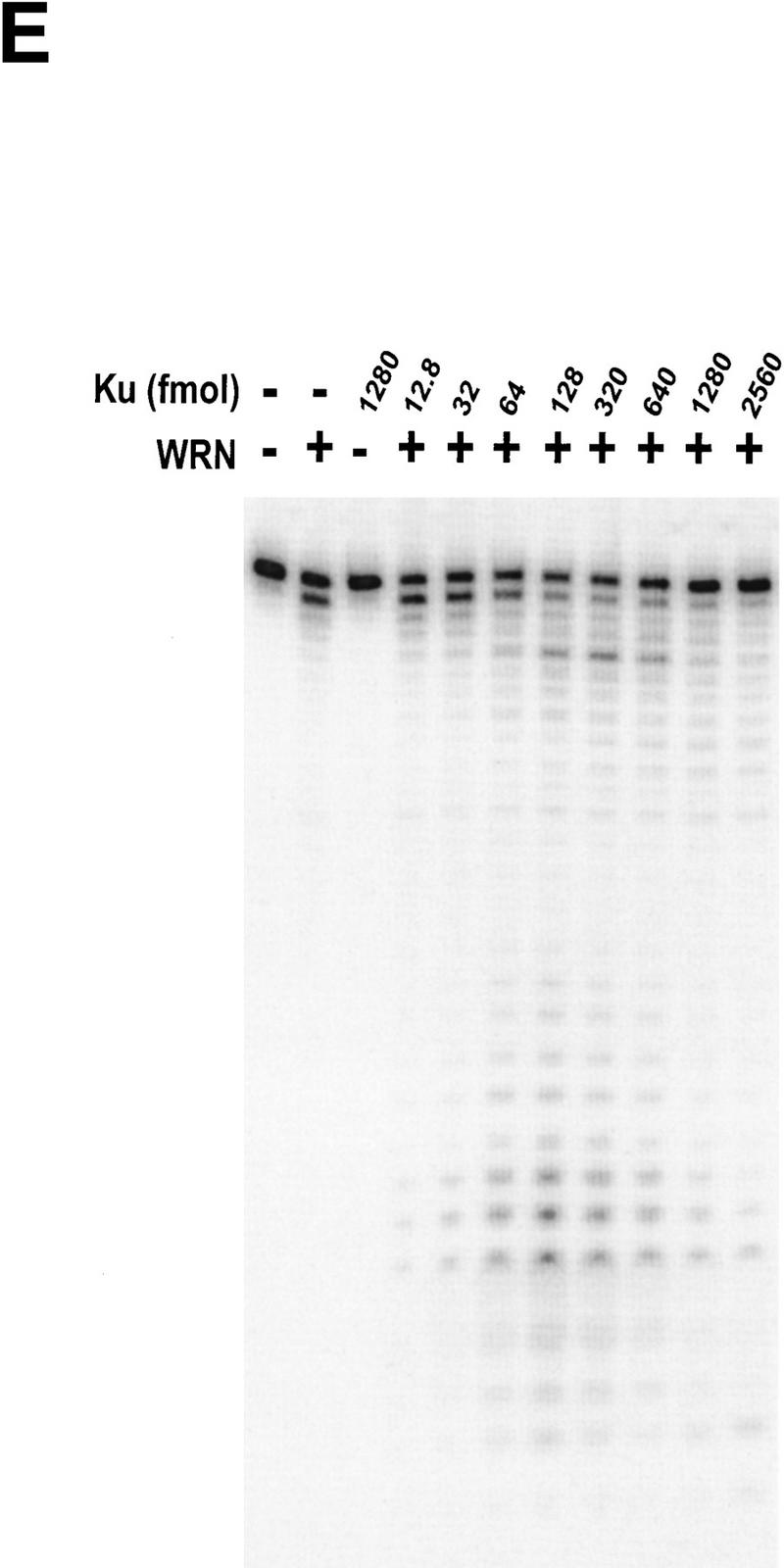
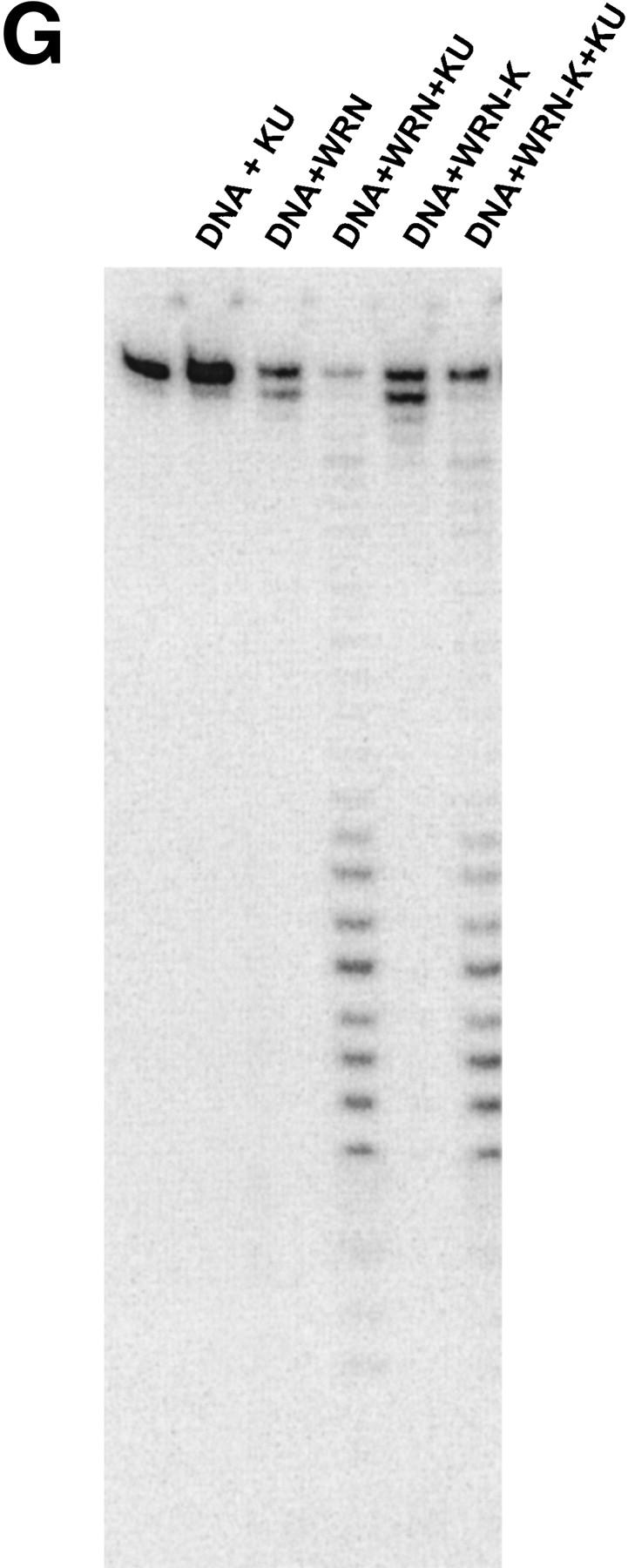
WRN exonuclease, but not helicase, is stimulated by Ku86/70. (A) WRNp (15 nm monomer) was incubated with the 28-bp partial duplex in the presence of the indicated concentrations of Ku86/70 under standard helicase reactions (Brosh et al. 1999). Lanes show increasing concentrations of Ku86/70. (Lane 7) No WRNp added, shows that Ku proteins do not unwind this substrate. (_) Heat-denatured control. (B) The exonuclease substrate containing a 5′ overhang. (C) Ku stimulation of WRN exonuclease at different concentrations of WRN (fmoles); Ku was 64 fmoles (10 ng). (D) Heat denaturation of Ku86/70. (Exo-) Mutant WRNp with no exonuclease activity. (D) Scans of lanes 5 and 6. (E) Ku86/70 stimulation of WRN exonuclease (at 45 fmoles), Ku titration. (F) Ku does not stimulate Exonuclease III or Klenow. (G) Ku stimulates WRN [wild-type and helicase mutant (Brosh et al. 1999)] exonuclease (both at 120 fmoles) in the absence of ATP.
Ku70/86 stimulates WRN exonuclease activity
WRNp has been reported previously to be an exonuclease with either 3′ → 5′ or 5′ → 3′ directionality. Our studies agree with the notion that WRNp is a 3′ → 5′ exonuclease with preference for 3′ recessed ends (Fig. 5; D. Orren, unpubl.). To examine the effect of Ku86/70 on the exonuclease activity of WRNp, a DNA substrate with one blunt end and one recessed 3′ end was designed (Fig. 5B) that is not detectably unwound by the helicase activity of WRNp (data not shown). Exonuclease digestion of this substrate by WRNp in the presence or absence of Ku86/70 is shown in Figure 5C. Ku86/70 alone does not have nuclease activity on this substrate. At low concentrations of WRNp alone, digestion from the 3′ recessed end is limited. At higher concentrations of WRNp, there is extensive 3′ → 5′ exonuclease activity (Fig. 5C, lane 8, 120 fmoles). When Ku86/70 is added with WRNp, the extent of digestion into the substrate is increased dramatically (Fig. 5C,D). Heat inactivation of Ku86/70 completely abolishes this stimulatory effect (Fig. 5C). Moreover, a WRNp with a mutation at a conserved amino acid (E84A) in its nuclease domain (Huang et al. 1998) has no detectable 3′ → 5′ exonuclease activity whether Ku86/70 is present or not (Fig. 5C). These results show that addition of Ku86/70 stimulates the 3′–5′ exonuclease activity of WRNp. The stimulation of wild-type WRNp exonuclease activity occurs over a broad range of Ku86/70 concentrations, and appears to be optimal at approximately equimolar ratios (Fig. 5E). In contrast, the addition of Ku86/70 does not detectably stimulate the 3′ → 5′ exonuclease activities of other exonucleases, prokaryotic exonuclease III or the Klenow fragment of DNA polymerase I (Fig. 5F). Thus, this stimulation of exonuclease activity is specific for WRNp.
The Ku complex has been reported to have some DNA unwinding activity (Tuteja et al. 1994; Yoo and Dynan 1998), although other studies find no unwinding (Tuteja et al. 1994). We did not observe any unwinding of a M13 partial duplex substrate by Ku86/70 (Fig. 5A) or of the dsDNA substrate (Fig. 5B) used for exonuclease studies (data not shown). A possible source of this controversy might arise from its tight binding to WRNp, which could result in copurification of WRNp in Ku preparations.
The possibility existed that Ku86/70, in the presence of ATP, caused a local unwinding of the substrate that would then stimulate the exonuclease activity of WRNp. To test this possibility, we performed an exonuclease assay in the absence of ATP (Fig. 5F). Ku86/70 still greatly stimulates the WRN exonuclease activity under those conditions. This stimulation is also observed in a WRNp K mutant, devoid of helicase activity (Brosh et al. 1999) (Fig. 5G, lanes 5,6). These results rule out the possibility that either WRNp or Ku unwinding activity is involved in the stimulation of WRN exonuclease.
Discussion
We have shown that the carboxyl terminus of the WRNp interacts strongly with the Ku86/70 complex. This physical interaction was also observed in cell extracts and between the purified proteins. We ascertained that no DNA was present under these conditions, thus excluding the hypothesis that this interaction could occur via a DNA bridge. The Ku86/70 complex strongly stimulated the exonuclease activity of WRNp, although it had no effect on its DNA-dependent ATPase or helicase activity. This stimulatory effect is mediated by the interaction between WRNp and Ku86/70 and serves to dramatically enhance the processivity of the exonuclease function of WRNp. The functional effect of this interaction occurs at the amino terminus, where the exonuclease activity is located. It is possible that there are other binding sites between Ku86/70 and WRNp. The interaction with Ku stimulates the exonuclease activity of WRNp via a direct molecular binding without any local unwinding of DNA by the Ku86/70 complex.
Several studies have shown that Ku86/70 binds to DNA ends and other types of discontinuity in dsDNA (Smith and Jackson 1999). It is a tightly associated heterodimer that, together with the ∼470-kD catalytic subunit, DNA–PKcs, form the DNA-dependent protein kinase (Featherstone and Jackson 1999). This complex is involved in repairing DNA double-strand breaks (DSBs) formed during V(D)J recombination or caused, for example, by endogenous oxidative byproducts, ionizing radiation, or certain chemotherapeutic drugs. A current model proposes that the complex joins the DNA ends, whereas a helicase unwinds the DNA duplex locally at the break, permitting annealing of the broken strands at sites of microhomology (Smith and Jackson 1999). The unannealed DNA ends then might be trimmed by an exonuclease before gap-filling and ligation by a polymerase and ligase (Yoo and Dynan 1998). No specific helicase or exonuclease that is involved in this process in mammalian cells has as yet been identified. Such exonuclease activity could be performed by WRNp, and the WRNp helicase activity may also participate in the process. WRNp and Ku could act together in recombination and/or DSB repair pathways. Our results suggest that Ku might help direct and stimulate WRN exonuclease activity on DNA ends.
There is good evidence for a role of the WRNp in DNA replication. A prolonged S phase has been observed in WS cells (Poot et al. 1992). The WRNp is 66% homologous to the Xenopus laevis FFA1 protein (foci-forming activity 1), which is essential for the formation of replication origins and RPA aggregation (Yan et al. 1998). We have reported previously that WRNp interacts physically and functionally with RPA (Brosh et al. 1999). Furthermore, the WRNp is a part of the replication complex (Lebel et al. 1999). The Ku86/70 and DNA–PK complex is also involved in DNA replication (Ruiz et al. 1999), and thus the main interaction between the Ku complex and WRNp may be via both of their roles in the replication process.
WS cells are not generally hypersensitive to DNA damaging agents. They are, however, sensitive to the carcinogen 4NQO (Gebhart et al. 1988) and to certain topoisomerase inhibitors such as the topisomerase I inhibitor, camptothecin (Lebel and Leder 1998). One study reports a repair deficiency for higher dose X-ray-induced damage (Weirich-Schwaiger et al. 1994), but others do not. The genomic instability in WS cells (Martin 1997) and the high frequency of deletions and translocations could be compatible with a DSB repair defect. We are currently investigating this in more detail, but as no cell lines with functional domain knockout in the WRN gene are yet available, many primary WS cell lines need to be analyzed.
If WRNp and Ku86/70 act in the same pathway, deficiencies in either component would be expected to have similar biochemical and cellular phenotypes. This notion is supported by a recent characterization of the Ku86 knockout mouse. The mouse has a senescent phenotype and, like WS patients, shows early onset of many age-related changes and a shortened life span (Vogel et al. 1999). Furthermore, cells deficient in WRNp, Ku70, or Ku86 undergo premature replicative senescence (Gu et al. 1997) and have elevated levels of chromosomal abnormalities. These similarities combined with our experimental results suggest that WRNp and Ku86/70 function in a common pathway, most likely in replication, recombination, or DSB repair. Our data might argue that a defective exonuclease function could lead to an aging phenotype.
Materials and methods
Maldi–MS
The matrix-assisted laser desorption ionization mass spectrometry (MALDI–MS) was performed by the HHMI/Keck facility at Yale University, where information is posted on the website: http://info.med.yale.edu/wmkeck.
Preparation of WRNp antibodies
Purified denatured N-WRN (100 μg) was injected intraperitoneal on day 0, and booster injections were administered at day 21 and day 28. Serum was collected from tail bleeds and used for immunoprecipitation.
Purification of WRNp and Ku complex
Recombinant human Ku was expressed in insect cells using baculoviral constructs and 70- and 86-kD subunits were amplified by PCR from cDNAs (a gift of D. Capra, University of Washington School of Medicine, Seattle). The 70-kD subunit was modified such that the amino acid sequence LEIEGRHHHHHH was placed in front of the stop codon at the carboxyl terminus. Both coding sequences were placed in pFASTBAC1, their sequence verified correct as reported, and high titer viral stocks for each subunit made by the BAC to BAC system (Life Technologies Incorporated, Bethesda, MD). SF-9 cells were coinfected with viruses for both subunits. An extract from an infection of ∼0.5 liters of Sf9 cells (∼1 × 109 cells) was prepared and heterodimeric Ku purified by successive chromatography of Ku-containing fractions over a Ni–NTA Superflow (Qiagen) column, a Mono Q (Pharmacia) column, and a native DNA cellulose (Sigma) column as described previously (Ramsden and Gellert 1998). The peak fraction was dialyzed against a buffer containing 25 mm Tris-HCl (pH 8.0), 150 mm KCl, 10% glycerol, 2 mm DTT, frozen in small aliquots on liquid nitrogen, and stored at −80°C. The final fraction from such preparations was determined to be a 1:1 heterodimer, >98% pure, as determined by chromatography on an S200 column (Pharmacia) and Coomassie staining of protein run on a SDS–polyacrylamide gel. The activity of the protein preparation was tested by gel-shift assay and stimulation of mammalian ligase (data not shown).
The WRNp was purified as described (Orren et al. 1999). A series of extra chromatographic steps were included to secure a high degree of purity of the protein (Orren et al. 1999).
Purification of the carboxyl terminus of WRNp
The gene encoding human WRNp (kindly provided by Dr. Junko Oshima, University of Washington Medical School, Seattle) was subjected to PCR of the carboxyl terminus (the region encoding amino acids 940–1432 and designated C-WRN). The product was subcloned into pET 30a (Novagen, Madison, WI) and confirmed by restriction analysis and sequence as the carboxyl terminus of WRN. The clone was then overexpressed in E. coli (BL21-DE3) and grown as described previously (Cooper et al. 1999). Culture flasks containing induced cells were stored on ice for 10 min, and the cells were pelleted. The pellet was frozen on liquid nitrogen and stored at −80°C overnight. The pellet was resuspended in 300 ml of ice cold lysis buffer (50 mm HEPES at pH 7.8, 0.2 m NaCl, 0.1% Triton X-100, 1 mg/ml lysozyme, 5 mm β-mercaptoethanol, 1 mm PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, and 10 mg/ml aprotinin). The sample was subjected to two 10-sec cycles of sonication on ice. The lysate containing C-WRNp was clarified at 20,000g for 1 hr at 4°C and filtered through a 0.22-μm filter. The lysate was loaded onto a 35-ml SP Sepharose column (Pharmacia). The column was washed with 10-column volumes of lysis buffer and eluted on a 0.2–0.5 m NaCl gradient. The fractions containing C-WRNp of ∼90% purity were pooled and loaded onto a 5-ml Ni2+–NTA resin column (Qiagen). The sample was sequentially washed with 10-column volumes of binding buffer (50 mm NaH2PO4, 0.3 m NaCl, 0.1% Triton X-100, 10 mm imidazole, and 5 mm β-mercaptoethanol) and 10 column volumes of wash buffer (binding buffer with a final concentration of 20 mm imidazole). The bound C-WRN protein was then eluted with a 20 mm–1 m imidazole gradient. Fractions containing purified C-WRN protein were pooled and dialyzed against 50 mm HEPES (pH 7.5), 0.4 m NaCl, 1 mm EDTA, and 2 mm DTT for 1 hr at 4°C. The purified C-WRN was concentrated by membrane filtration (Amicon), frozen on liquid nitrogen, and stored at −80°C. The typical yield of protein from 1 liter of culture was 14 mg.
Affinity chromatography to C-WRN
Purified C-WRNp (1.5 mg) was immobilized to an activated 4% beaded agarose matrix by reductive amination of Schiff bases (Pierce, Rockford, IL) as per the manufacturer's instructions. Control columns containing a fivefold excess of β-galactosidase (7.5 mg) or matrix only were prepared similarly. The procedures described previously (Hughes and Baldacci 1997; Fahrmann et al. 1998) were modified. Columns were equilibrated in 10 column volumes of binding buffer (50 mm HEPES at pH 7.4, 0.1 m NaCl, 0.05% Triton X-100, 1 mm EDTA, 1 mm Na vanadate, 1 mm NaF, 1 mm DTT, 1 mm PMSF, 10 μg/ml leupeptin, 10 μ/ml pepstatin A, and 10 μg/ml aprotinin. Nuclear extract (20 mg) prepared from HeLa cells was sequentially incubated for 1 hr at 4°C with the no protein column, the β-galactosidase column, and the C-WRNp column, respectively. Proteins adsorbed to the columns were washed with 10 column volumes of binding buffer to remove weakly bound proteins. The columns were then step eluted with binding buffer containing 0.2 m NaCl and 1 m NaCl to identify tight binding proteins. The eluted fractions were dialyzed and concentrated by membrane filtration (Amicon), lyophilized, and samples were analyzed by SDS-PAGE (10%) followed by either Coomassie staining or Western blot. All columns were prepared fresh for each experiment and all results were duplicated.
Immunoprecipitation assays
Immunoprecipation of WRNp was performed in IP buffer (50 mm HEPES at pH 7.4, 0.1 m NaCl, 0.05% Triton X-100, 1 mm EDTA, 1 mm Na vanadate, 1 mm NaF, 1 mm DTT, 1 mm PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, and 10 μg/ml aprotinin). Briefly, HeLa nuclear extract (500 μg) was precleared with either mouse or goat immunoglobin and preswollen protein A/G beads for 1 hr at 4°C. The precleared extract was incubated with the anti-N-WRN antibody for 1–2 hr at 4°C and preswollen protein A/G beads added. The complex was pelleted and washed four times with IP buffer. The precipitates were subjected to SDS-PAGE (10%) and Western blot analysis.
ATPase assay
ATPase assay reaction mixtures (20 μl) contained 40 mm Tris (pH 7.4), 4 mm MgCl2, 5 mm DTT, M13mp18 ssDNA (30 μm nucleotide phosphate), 0.8 mm [3H]ATP (42 cpm/pmole), WRN protein (7 nm, monomer) and/or Ku protein (70 nm heterodimer). WRN and Ku proteins were preincubated for 2 min at 24°C in ATPase reaction buffer containing M13 ssDNA effector. Reactions were initiated by the addition of [3H]ATP and incubated at 24°C for 10 min. Samples (5 μl) were removed and evaluated by TLC as described previously (Matson and Richardson 1983). Less than 20% of the substrate ATP was consumed in the reaction over the entire time course of the experiment.
Helicase assay
Helicase assay reaction mixtures (20 μl) contained 40 mm Tris (pH 7.4), 4 mm MgCl2, 5 mm DTT, 2 mm ATP, WRN helicase (15 nm, monomer), and the indicated amount of Ku86/70 protein. The concentration of the 28-bp partial duplex helicase substrate in the reaction mixture was ∼2 μm (nucleotide phosphate). WRN and Ku proteins were preincubated with partial duplex DNA substrate for 2 min at 24°C before reactions were initiated by the addition of ATP and incubated at 24°C for 30 min. Reactions were terminated by the addition of 10 μl of 50 mm EDTA, 40% glycerol, 0.9% SDS, 0.1% bromophenol blue, and 0.1% xylene cyanol. The products of helicase were resolved on a 12% nondenaturing polyacrylamide gel. Radiolabeled DNA species in polyacrylamide gels were visualized by a PhosphorImager or film autoradiography and quantitated using ImageQuant software (Molecular Dynamics). The percent helicase substrate unwound was calculated by the following formula: % Displacement = 100 × P/(S + P). P is the product volume and S is the substrate volume. The values for P and S have been corrected after subtracting background values in the no enzyme and heat-denatured controls, respectively.
Exonuclease assay
Single-stranded, partially complementary DNA oligonucleotides (32 and 43 nucleotides) were obtained from GIBCO BRL, and the 32 nucleotide (7 pmoles) was 5′ labeled with [γ-32P]ATP (60 μCi, 3000 Ci/mmole) and polynucleotide kinase (10 units) using standard conditions. For construction of a dsDNA substrate with one blunt end and one 3′-recessed (5′ overhang) end (Fig. 5B), labeled 32-mers were mixed with a twofold excess of unlabeled 43-mer, heated together at 90°C for 5 min, then cooled slowly to 25°C. The annealed dsDNA was then separated from unannealed and excess single-stranded oligonucleotides by nondenaturing polyacrylamide (12%) gel electrophoresis. Intact dsDNA substrates were recovered using a Qiaex II gel extraction kit (Qiagen) and stored at 4°C. WRNp exonuclease assays were carried out in buffer containing 40 mm Tris (pH 8.0), 4 mm MgCl2, 1 mm ATP, 0.1 mg/ml BSA, and 5 mm DTT. Klenow exonuclease assays were carried out in 50 mm Tris (pH 7.4), 10 mm MgCl2, and 1 mm DTT; Exo III reactions were done in 67 mm Tris (pH 7.4), 0.66 mm MgCl2, and 1 mm β-mercaptoethanol. DNA substrates (3 fmoles/reaction) were incubated with Klenow (0.2–1.0 units), Exo III (0.005–0.025 units), WRNp (45–120 fmoles), or WRNp–exo− (180 fmoles) and, where indicated, with Ku70/Ku86 complex (12.8–1280 fmoles) for 1 hr at 37°C. The reactions (10 μl volume) were stopped by addition of an equal volume of formamide loading buffer (80% formamide, 0.5× TBE, 0.1% xylene cyanol, and 0.1% bromophenol blue). The digestion products of these reactions were separated on 15% denaturing polyacrylamide gels and visualized with a PhosphorImager (Molecular Dynamics). Quantitative comparison of individual lanes and generation of line graphs was accomplished using ImageQuant software.
Acknowledgments
We thank J. Campisi for the WRN exonuclease mutant baculovirus. D.R. was a recipient of a grant, RPG GMC-98562 from the American Cancer Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL vbohr@nih.gov; FAX (410) 558-8157.
References
- Brosh RM, Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J Biol Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- Cooper MP, Balajee AS, Bohr VA. The C-terminal domain of p21 inhibits nucleotide excision repair In vitro and In vivo. Mol Biol Cell. 1999;10:2119–2129. doi: 10.1091/mbc.10.7.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA. DNA helicases in inherited human disorders. Curr Opin Genet Dev. 1997;7:354–363. doi: 10.1016/s0959-437x(97)80149-9. [DOI] [PubMed] [Google Scholar]
- Fahrmann M, Mohlig M, Schatz H, Pfeiffer A. Purification and characterization of a Ca2+/calmodulin-dependent protein kinase II from hog gastric mucosa using a protein-protein affinity chromatographic technique. Eur J Biochem. 1998;255:516–525. doi: 10.1046/j.1432-1327.1998.2550516.x. [DOI] [PubMed] [Google Scholar]
- Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- Gebhart E, Bauer R, Schinzel M, Ruprecht KW, Jonas JB. Spontaneous and induced chromosomal instability in Werner syndrome. Hum Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng HL, Sekiguchi JM, Frank K, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′ → 5′ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes P, Baldacci G. A DNA helicase purified by replication protein A (RPA) affinity chromatography from mouse FM3A cells. Nucleic Acids Res. 1997;25:3881–3888. doi: 10.1093/nar/25.19.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Shen JC, Loeb LA, Fry M. Werner Syndrome Protein. Ii. characterization of the integral 3′ → 5′ DNA exonuclease. J Biol Chem. 1998;273:34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc Natl Acad Sci. 1998;95:13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M, Spillare EA, Harris CC, Leder P. The werner syndrome gene product Co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- Martin GM. The pathobiology of the Werner syndrome. FASEB J. 1997;11:A1449–A1449. [Google Scholar]
- Matson SW, Richardson CC. DNA-dependent nucleoside 5′-triphosphatase activity of the gene 4 protein of bacteriophage T7. J Biol Chem. 1983;258:14009–14016. [PubMed] [Google Scholar]
- Moser MJ, Oshima J, Monnat RJ., Jr WRN mutations in Werner syndrome. Hum Mutat. 1999;13:271–279. doi: 10.1002/(SICI)1098-1004(1999)13:4<271::AID-HUMU2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: High affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cells. Exp Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Gellert M. Ku protein stimulates DNA end joining by mammalian DNA ligases: A direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MT, Matheos D, Price GB, Zannis-Hadjopoulos M. OBA/Ku86: DNA binding specificity and involvement in mammalian DNA replication. Mol Biol Cell. 1999;10:567–580. doi: 10.1091/mbc.10.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner Syndrome Protein. I. dna helicase and dna exonuclease reside on the same polypeptide. J Biol Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes & Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Shiratori M, Goto M, Furuichi Y. Werner syndrome helicase contains a 5′ → 3′ exonuclease activity that digests DNA and RNA strands in DNA/DNA and RNA/DNA duplexes dependent on unwinding. Nucleic Acids Res. 1999;27:2361–2368. doi: 10.1093/nar/27.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Tuteja R, Ochem A, Taneja P, Huang NW, Simoncsits A, Susic S, Rahman K, Marusic L, Chen J. Human DNA helicase II: A novel DNA unwinding enzyme identified as the Ku autoantigen. EMBO J. 1994;13:4991–5001. doi: 10.1002/j.1460-2075.1994.tb06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich-Schwaiger H, Weirich HG, Gruber B, Schweiger M, Hirsch-Kauffmann M. Correlation between senescence and DNA repair in cells from young and old individuals and in premature aging syndromes. Mutat Res. 1994;316:37–48. doi: 10.1016/0921-8734(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Yan H, Chen CY, Kobayashi R, Newport J. Replication focus-forming activity 1 and the Werner syndrome gene product. Nat Genet. 1998;19:375–378. doi: 10.1038/1263. [DOI] [PubMed] [Google Scholar]
- Yoo S, Dynan WS. Characterization of the RNA binding properties of Ku protein. Biochemistry. 1998;37:1336–1343. doi: 10.1021/bi972100w. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]