Abstract
Trypanosoma brucei rhodesiense is the causative agent of human African sleeping sickness. While the closely related subspecies T. brucei brucei is highly susceptible to lysis by a subclass of human high-density lipoproteins (HDL) called trypanosome lytic factor (TLF), T. brucei rhodesiense is resistant and therefore able to establish acute and fatal infections in humans. This resistance is due to expression of the serum resistance-associated (SRA) gene, a member of the variant surface glycoprotein (VSG) gene family. Although much has been done to establish the role of SRA in human serum resistance, the specific molecular mechanism of SRA-mediated resistance remains a mystery. Thus, we report the trafficking and steady-state localization of SRA in order to provide more insight into the mechanism of SRA-mediated resistance. We show that SRA traffics to the flagellar pocket of bloodstream-form T. brucei organisms, where it localizes transiently before being endocytosed to its steady-state localization in endosomes, and we demonstrate that the critical point of colocalization between SRA and TLF occurs intracellularly.
INTRODUCTION
Trypanosoma brucei brucei causes the veterinary disease Nagana, but it is unable to establish infections in humans. Human resistance to T. brucei brucei infection is due to the presence of a trypanolytic component of human serum, which provides innate immunity against infection. This component is a minor subfraction of high-density lipoproteins (HDLs) called the trypanosome lytic factor 1 (TLF-1) (16, 29). Like all HDLs, TLF-1 contains apolipoprotein A-I (apoA-I), as well as two unique, primate-specific proteins, apolipoprotein L-I (apoL-I) (39) and haptoglobin-related protein (Hpr) (34), that confer lytic activity to the particle. This toxic class of HDLs is internalized in T. brucei brucei via receptor-mediated endocytosis and is ultimately targeted to the lysosome, where it initiates low-pH-dependent killing (8, 15, 24, 33, 37, 39).
While TLF-1 is toxic to T. brucei brucei, T. brucei rhodesiense is resistant to TLF-mediated killing and causes the acute form of human African trypanosomiasis (HAT). The mechanism of resistance to TLF-1 remains to be fully elucidated; however, it is well established that the resistance phenotype of T. brucei rhodesiense is due to the expression of the serum resistance-associated (SRA) protein. Most human isolates of T. brucei rhodesiense have been found to express SRA (7), and loss of SRA expression leads to susceptibility to TLF-1 toxicity (23). Furthermore, transfection of the SRA gene into susceptible T. brucei brucei cell lines confers resistance to TLF-1 killing (25, 41).
SRA is a member of the VSG gene family and is predicted to share similar structures and posttranslational modifications with VSGs and the trypanosome transferrin receptor (TfR), another VSG family member (5, 23). Trypanosome VSGs and TfR are glycosylated cell surface proteins that are anchored to the plasma membrane via the glycosylphosphatidylinositol (GPI) lipid anchor (31, 36). Both VSGs and TfR are continually trafficked to and from the cell surface via the flagellar pocket by robust secretory and recycling pathways (9, 14, 19). The GPI anchor attachment is typically associated with cell surface proteins and has been shown to be involved in the trafficking of these proteins (1, 38). Previous studies have reported that SRA is intracellularly localized, despite being a VSG family protein with a predicted GPI anchor attachment site (25, 39). SRA has also been found to bind TLF-1 via direct interaction with apoL-I and to colocalize intracellularly (25, 39).
In this study, we show for the first time that SRA traffics to the flagellar pocket before rapid uptake into cytoplasmic vesicles, which we now identify as early endosomes. We also find that lysosomal localization of SRA is fleeting and is detectable only when protein degradation is inhibited. Deletion of the GPI anchor addition site disrupts flagellar pocket localization of the protein but is not required for trafficking to the endosomes or colocalization with TLF-1. Furthermore, loss of SRA trafficking to the flagellar pocket does not result in increased susceptibility to TLF-1, suggesting that the critical point of interaction of toxin and inhibitor is not at the cell surface. Finally, we show that a trypanosome cysteine protease is involved in rapid TLF-1 turnover in SRA-expressing T. brucei brucei transfectants, indicating that the mechanism of SRA-mediated resistance to TLF-1 killing may involve accelerated degradation and destabilization of the TLF-1 particle.
MATERIALS AND METHODS
Cell culture.
Bloodstream-form T. brucei brucei cultures were grown in HMI-9 medium (with fetal bovine serum [FBS]; Gemini Bio-products, West Sacramento, CA) with Serum Plus medium supplement (SAFC Biosciences, Lenexa, KS).
Construction of reporter genes.
All SRA gene constructs were cloned into the pURAN trypanosome expression vector as previously described (25). The SRA-Ty construct was transfected into both T. brucei brucei 427-221 and T. brucei brucei 060R cells to generate T. brucei brucei 427-221 SRA-Ty transfectants and T. brucei brucei 060R SRA-Ty transfectants, respectively. The T. brucei brucei 427-221 SRA-Ty cell line was previously generated and characterized (25). T. brucei brucei 060R cells were derived from a TLF-1-resistant cell line lacking the T. brucei haptoglobin/hemoglobin receptor (TbHpHbR). T. brucei brucei 221 SRAΔGPI cells were generated by transfection into the T. brucei brucei 221 cells. The construct is shown below schematically in Fig. 4A. The sequence was generated by PCR amplification of the full-length SRA-Ty coding sequence by using the following primers: 5′-SRAΔGPI, CCTGCAGG ATG CCC CGA AAT TCG GGC CGG; 3′ SRAΔGPI, GGCGCGCC TTA TTT GGA TTC TTT TCC TTC CC. PCR-amplified products were ligated into the TA cloning vector (Invitrogen, Eugene, OR). The SRAΔGPI insert was digested from the cloning vector, using the EcoRI restriction endonuclease. The gel-purified insert was then ligated into the pURAN expression vector, which was linearized by BsiWI restriction digest prior to transfection. To generate an epitope-tagged T. brucei Rab5a construct, trypanosome genomic DNA was amplified using the following primers: 5′ Rab5aHA, CCTGCAGG ATG TAC CCA TAC GAC GTC CCA GAC TAC GTC CCA GAC TAC GCT ATG TCG GTG TCA GCG ACA CCA; 3′ Rab5aHA, GGCGCGCC TTA TCA GCA GGC ACA CCC GCC TTC. The full-length coding sequence of TbRab5a (accession no. U24678) was engineered with the hemagglutinin (HA) epitope (YPYDVPDYA) at the N terminus. Primers were synthesized with 5′ SbfI and 3′ AscI overhangs for directional cloning into the pTub-phleo expression vector (30). In the Rab5aHA primers listed above, restriction sites are underlined, with start and termination codons shown in boldface, and the epitope tag sequence is shown in italics.
Fig. 4.
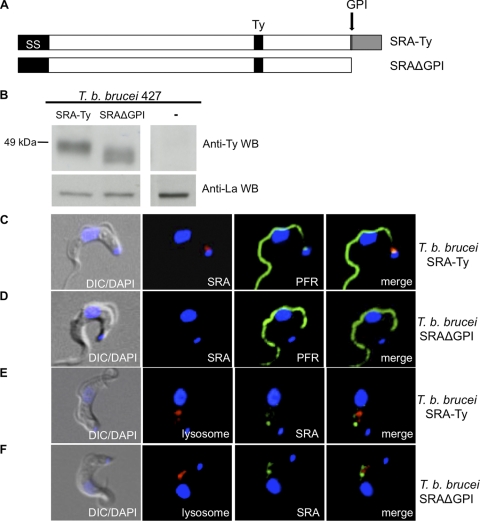
Loss of the GPI anchor disrupts flagellar pocket localization. (A) Schematic diagram of SRA-Ty and SRAΔGPI. (B) Western blot of SRA-expressing T. brucei brucei transfectants probed with anti-Ty (SRA) and anti-La (La protein loading control). (C and D) Binding of Alexa Fluor 594 anti-Ty (SRA) antibody at 3°C in SRA-Ty cells (C) and SRAΔGPI cells (D) is shown in red. PFR staining is shown in green. (E and F) SRA-Ty (E) and SRAΔGPI (F) cells were stained for SRA (green) and lysosomal localization (red).
Transfections.
Transfections were performed using the Lonza nucleofection system according to the manufacturer's instructions. A total of 1 × 107 cells were resuspended in 100 μl of the Lonza nucleofection solution containing 10 μg of linearized plasmid DNA. The cell suspension was subjected to nucleofection using the preprogrammed setting X-001. Transfected cells were then placed in HMI-9 medium for 24 h before selection of clonal cultures (2.5 μg/ml G418 for SRA constructs; 3 μg/ml phleomycin for the Rab5a construct). The solutions provided with the Lonza nucleofection kit are of unknown composition.
Immunoblotting. (i) Western blotting.
Total cell lysates for Western blotting were obtained and analyzed as previously described (25). The anti-Ty antibody was directly conjugated to horseradish peroxidase (HRP) by using Pierce EZ-link activated peroxidase (Thermo Fisher Scientific, Rockford, IL) and used at a dilution of 1:5,000. Rat monoclonal anti-HA–biotin (Roche Diagnostics, Indianapolis, IN) was used at a dilution of 1:1,000, with streptavidin-HRP conjugate (Invitrogen, Camarillo, CA) used for secondary detection at 1:5,000.
(ii) Immunofluorescence assays.
All cells were fixed in 1% paraformaldehyde (PFA) on ice for 15 min followed by a brief methanol treatment at −20°C, unless otherwise stated. Under nonpermeabilizing conditions, cells were fixed in 1% PFA in HMI-9 medium, pH 7.5, for 15 min on ice. Fixed cells were washed once in phosphate-buffered saline (PBS), then resuspended in PBS, 10% FBS for 1 h. The following antibody dilutions were used: mouse anti-Ty at 1:1,000; rabbit anti-TbCatL at 1:4,000; mouse anti-paraflagellar rod (PFR) at 1:1,000, rat anti-HA-biotin at 1:100. Primary antibody staining was carried out for 45 min. For secondary antibody staining, slides were incubated with the appropriate Alexa Fluor 488 or Alexa Fluor 594 mouse or rabbit immunoglobulin G (Invitrogen, Eugene, OR) for 30 min for all primary antibodies except anti-HA-biotin. Secondary detection for anti-HA-biotin was carried out using streptavidin-Alexa Fluor 594 conjugate (Invitrogen, Eugene, OR). 4′,6′-diamidino-2-phenylindole (DAPI) was added after final washes. Serial image z-stacks were acquired through oil immersion optics at ×63 magnification, with exposure times kept constant for each experiment. Imaging was carried out using a Zeiss Axio Observer inverted microscope equipped with an AxioCam HSm camera and analyzed with the AxioVision v4.6 software (Zeiss). A single stack is shown for each experiment, with individual channels contrasted to the same extent for each image set and merged using Adobe Photoshop CS2 v9.0. Polyclonal anti-TbCatL was kindly provided by Jay Bangs, University of Wisconsin, Madison. The anti-paraflagellar rod antibody was generously provided by Diane McMahon-Pratt, New Haven, CT.
Binding and uptake assays. (i) Binding assays.
Cells were collected and resuspended in chilled HMI-9 medium, 10 μg/ml hemoglobin at 3 × 107cells/ml and allowed to equilibrate to a 3°C water bath for binding assays. Alexa Fluor-conjugated ligands were added to cell suspensions and incubated for 15 min at 3°C and then washed 3 times in ice-cold PBS-1% glucose (PBS-G). Cells were fixed by resuspension in 1% PFA for 15 min on ice. Cells were then processed for immunofluorescence as previously described. Alexa Fluor-conjugated ligands were labeled according to the manufacturer's directions (Invitrogen, Eugene, OR) and used at the following concentrations: Alexa Fluor 594–anti-Ty, 0.2 μg/ml; Alexa Fluor 594–transferrin, 50 μg/ml; Alexa Fluor 488–TLF, 5 μg/ml.
(ii) Uptake assays.
Cells were collected and resuspended in HMI-9 medium with 10 μg/ml hemoglobin at 3 × 107cells/ml. Cells were incubated with Alexa Fluor 488-TLF at 37°C for 30 min and then washed 3 times in ice cold PBS-G, fixed, and analyzed by microscopy as previously described. Cells were also analyzed by flow cytometry using a Beckman Coulter CyAn ADP flow cytometer and FlowJo v7.5 software.
Binding-chase assays.
Binding was carried out as previously described. After washing, cells were resuspended in warm HMI-9 medium and then placed in a 37°C water bath for 1 min. PFA at 2% was added to cell suspensions for a final concentration of 1% PFA, in order to fix cells and halt uptake at 1 min. Cells were processed for immunostaining as previously described.
Protease inhibition assays.
Cells were treated with 20 μM FMK024 or left untreated for 1 h at 37°C. For immunofluorescence assays, cells were then fixed with 1% PFA and prepared for immunostaining as previously described. For Western blotting, protease inhibition was halted by cell lysis through addition of an equal volume of 10% tricholoroacetic acid (TCA) for TCA precipitation. Lysates were incubated overnight at 4°C, precipitated by addition of isopropanol, and then centrifuged at 13,000 rpm for 15 min. Pellets were washed 3 times with cold (−20°C) acetone. Protein pellets were then dissolved in PBS, and protein concentrations were determined in a Bradford assay. Samples were prepared for Western blotting as previously described. Fifteen micrograms of protein was analyzed for each condition.
TLF-1 purification and lysis assays.
Human blood was obtained from healthy, fasted donors, and plasma was separated by low-speed centrifugation (3,500 rpm for 10 min) and subsequent high-speed centrifugation (10,000 rpm for 5 min) of the supernatant to pellet residual red blood cells. HDLs were purified as previously described (16). After collection of the HDL fraction of plasma (1.26 g/ml), trypanolytic HDLs were isolated from this fraction by affinity purification using monoclonal antibodies against Hpr. Antibodies were coupled to Pierce Affigel resin based on the manufacturer's recommendations (Thermo Fisher Scientific, Rockford, IL). Eluates were dialyzed against phosphate-buffered saline–75 μM EDTA (PBSE) at 4°C and stored in single-use aliquots at −80°C. Susceptibility to TLF-1 was determined in 2-h lysis assays as previously described (16).
RESULTS
SRA traffics to the flagellar pocket.
Since VSG family proteins (VSG and TfR) localize to the trypanosome cell surface membrane, this led us to reexamine the cellular localization of SRA. Previous localization studies showed that SRA does not localize to the trypanosome cell surface coat like VSG but is distributed within intracellular vesicles localized between the kinetoplast and nucleus (25, 39). We used the previously characterized cell line in which Ty epitope-tagged SRA was expressed (T. brucei brucei SRA-Ty) (25) (Fig. 1A). Northern blot analysis of SRA-Ty mRNA transcript levels in T. brucei brucei SRA-Ty transfectants were found to be comparable to SRA transcript levels in T. brucei rhodesiense (see Fig. S1A in the supplemental material). Intracellular localization may be the result of direct sorting to a stable vesicular pool or secondary sorting following transient localization at the cell surface. We therefore examined the flagellar pocket, a specialized region of the cell surface membrane to which the trypanosome transferrin receptor localizes. In two-dimensional images, the flagellar pocket can best be delineated as the region between the kinetoplast DNA and the posterior end of the PFR. The basal bodies are directly adjacent to the kinetoplast and are also useful as organellar markers for the flagellar pocket (Fig. 1B). In order to examine this specific cell surface region, we carried out live cell binding studies at 3°C by using Alexa Fluor 594-conjugated transferrin and Alexa Fluor 594-conjugated anti-Ty (SRA). At this temperature, we found that endocytosis was strongly inhibited and receptor-bound transferrin was detected in the flagellar pocket (Fig. 1B). Under 3°C binding conditions, 93.8 ± 5.3% (mean ± standard error of the mean [SEM]) of transferrin-labeled cells showed distinct flagellar pocket localization of transferrin (Table 1). To determine whether SRA localized to the flagellar pocket, we incubated live T. brucei brucei SRA-Ty cells at 3°C with Alexa Fluor 594-conjugated anti-Ty antibody followed by fixation with paraformaldehyde and then we visualized the localization by fluorescence microscopy. As with transferrin, these live cell binding studies also showed flagellar pocket localization of bound Alexa Fluor 594 anti-Ty, indicating that SRA traffics to the cell surface of trypanosomes but its distribution is limited to the flagellar pocket (Fig. 1C).
Fig. 1.
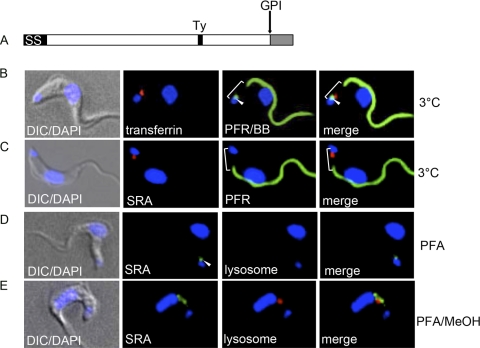
SRA traffics to the flagellar pocket. (A) Diagram of SRA, indicating the predicted positions of the N-terminal signal peptide (SS), GPI anchor addition site (D388), C-terminal peptide (gray), and Ty epitope tag. (B and C) SRA traffics to the flagellar pocket. Live cell binding studies at 3°C with Alexa Fluor 594-transferrin (red) (B) or Alexa Fluor 594 anti-Ty (SRA) (red) (C). After binding and fixation, basal bodies were stained with anti-YL1/2 (green) (B), and the paraflagellar rod was stained with anti-PFR (green) (B and C). The white bracket shows the flagellar pocket region, and the arrowheads denote the positions of the basal bodies. (D and E) Cells were fixed under nonpermeabilizing (D) and permeabilizing (E) conditions and stained for SRA with anti-Ty (green). Cells were also stained for the lysosomal compartment with anti-TbCatL (red). Image acquisition was carried out at the same exposure, and images were contrasted to the same extent.
Table 1.
Alexa Fluor 594-transferrin labeling of trypanosomes at 3°C
| Distribution of labeled cells | % of cellsa |
|---|---|
| Total cells | |
| % labeled | 91.1 ± 4.7 |
| % unlabeled | 8.9 ± 4.6 |
| Transferrin-labeled cells | |
| % flagellar pocket | 93.8 ± 5.3 |
| % internal | 6.2 ± 5.3 |
Quantitation of Alexa Fluor 594-transferrin labeling of cells at 3°C. Results are presented as the means (± SEM) of 3 binding assays. The total number of cells was 100.
In addition to live cell binding studies, we examined cells that were fixed under nonpermeabilizing conditions for flagellar pocket-localized SRA. Immunostaining of fixed, nonpermeabilized cells with anti-Ty revealed SRA localization to be restricted to the flagellar pocket (Fig. 1D). Cells were also immunostained with the anti-T. brucei cathepsin L (TbCatL) antibody, an intracellular marker for the trypanosome lysosomal protease cathepsin L (trypanopain) (3, 4), which colocalizes with the known lysosomal marker p67 (see Fig. S1B in the supplemental material). Lysosomal staining was not detectable, thereby confirming that the fixed cells were impermeable to antibodies under those conditions and the observed anti-Ty staining was therefore extracellular and flagellar pocket localized (Fig. 1D). Upon methanol permeabilization and anti-Ty immunostaining of fixed T. brucei brucei SRA-Ty cells, intracellular SRA-containing vesicles were observed between the kinetoplast and nucleus (Fig. 1E). Although lysosomal staining was also detectable under permeabilizing conditions, SRA did not colocalize with this marker (Fig. 1E). Quantitation of SRA distribution showed flagellar pocket localization in only 37.9 ± 9.6% of cells, while all anti-Ty-labeled cells showed intracellularly localized SRA (Table 2). These results confirm that SRA traffics to the flagellar pocket but that the steady-state distribution is mainly within nonlysosomal intracellular vesicles.
Table 2.
Distribution of SRA cellular localization
| Treatment and compartment | % of cells in compartmenta |
|---|---|
| Untreated cells | |
| FP/endosomal | 37.9 ± 9.6 |
| Endosomal | 100 |
| Lysosomal | 0 |
| FMK treated | |
| Endosomal only | 19.6 ± 8.0 |
| Lysosomal only | 35.8 ± 16.4 |
| Endosomal/lysosomal | 44.6 ± 8.7 |
Quantitation of cellular distribution of untreated and FMK-treated SRA-expressing cells. Percentages for untreated and for FMK-treated cells are presented as means ± SEM (n = 4 [untreated]; n = 3 [FMK treated]). The total number of untreated cells was 102; the total number of FMK-treated cells was 115.
The initial interaction between TLF-1 and SRA does not occur within the flagellar pocket.
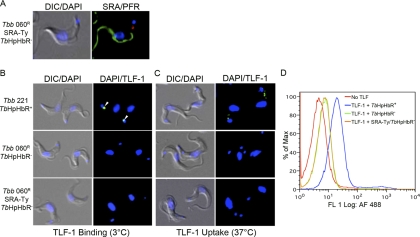
Previous studies demonstrated that SRA binds TLF-1 via apoL-1 (39) and that these proteins colocalize in intracellular vesicles (25). Since SRA traffics to the flagellar pocket, which is also the site of receptor-mediated uptake of TLF-1 by the haptoglobin/hemoglobin receptor TbHpHbR (40), we investigated whether the initial colocalization between SRA and TLF-1 could occur within the flagellar pocket. To examine this possibility, we expressed Ty-tagged SRA in a cell line that does not express TbHpHbR (Fig. 2A; see also Fig. S1C and D in the supplemental material) (20). In these cells, SRA-Ty localized to the flagellar pocket, based on localization studies at 3°C with anti-Ty (Fig. 2A). The SRA-Ty -expressing transfectants, T. brucei brucei 427 060R-SRA-Ty, were also assayed for binding and uptake of Alexa Fluor 488-conjugated TLF-1 by fluorescence microscopy. T. brucei brucei 427-221 cells expressing TbHbHpR exhibited TLF-1 binding and uptake (Fig. 2B and C; see also Fig. S1D and S2 in the supplemental material), while T. brucei brucei 427-060R cells that did not express the receptor, based on the lack of detectable mRNA by reverse transcription-PCR (see Fig. S1D) did not bind or endocytose TLF-1 (Fig. 2B and C). In addition to fluorescence microscopy, 427-221 (TbHpHbR-positive) and 427-060R (TbHpHbR-negative) cell lines were also assayed for Alexa Fluor 488 TLF uptake by flow cytometry. Consistent with the localization studies, T. brucei brucei 427-221 cells bound Alexa Fluor 488 TLF at 37°C. T. brucei brucei 427-060R and 427-060R-SRA-Ty cells had low levels of cell-associated TLF-1, which were likely due to nonspecific cell surface labeling. Despite the localization of TLF-1 and SRA individually at the flagellar pocket, we were unable to detect the colocalization of these two infectivity factors at the flagellar pocket in the TbHpHbR-negative cells. These findings indicate that expression of SRA in the absence of the TbHpHbR is not sufficient to allow for TLF-1 binding at the surface of the trypanosome, suggesting that the proteins independently enter the endocytic pathway.
Fig. 2.
The initial interaction between SRA and TLF does not take place at the flagellar pocket. (A) Binding of Alexa Fluor 594–anti-Ty (SRA) (red) in the flagellar pocket of T. brucei brucei 060R SRA-Ty cells. Anti-PFR staining is shown in green. (B and C) Binding (B) and uptake (C) of Alexa Fluor 488-TLF (green) in T. brucei brucei 221 (TbHpHbR(+)), T. brucei brucei 060R (TbHpHbR−), and T. brucei brucei 060R SRA-Ty (TbHpHbR−) cells. White arrowheads show binding in the flagellar pocket. DIC, differential interference contrast microscopy. (D) Flow cytometry analysis of Alexa Fluor 488-TLF uptake. Strains used included T. brucei brucei 221 (TbHpHbR+), T. brucei brucei 060R (TbHpHbR−), and T. brucei brucei 060R SRA-Ty (SRA-Ty/TbHpHbR−).
SRA is resident within an early endosomal compartment.
Extensive characterization of trafficking vesicles in bloodstream-form T. brucei has defined several classes of endosomes by their associated Rab GTPases, as well as cargo, such as transferrin, for which the endocytic trafficking pathway has been well documented (14, 27). The Rab5a GTPase has been identified as a marker for early endosomes and has been shown to colocalize with transferrin within minutes of receptor-mediated uptake (27, 28). In order to localize the early endosomal compartment, we transfected T. brucei brucei 427-221 and T. brucei brucei 427-221 SRA-Ty cells with HA epitope-tagged Rab5a. Anti-HA Western blotting showed a 33-kDa band in transfectant cell lines that was absent in untransfected cells (Fig. 3A). Using anti-HA immunofluorescence microscopy, we localized HA-tagged Rab5a endosomes distributed between the kinetoplast and nucleus in both cell lines (Fig. 3B and C). This discrete nonlysosomal distribution of HA-tagged Rab5a endosomes is consistent with previous reports on Rab5a localization in T. brucei (28).
Fig. 3.
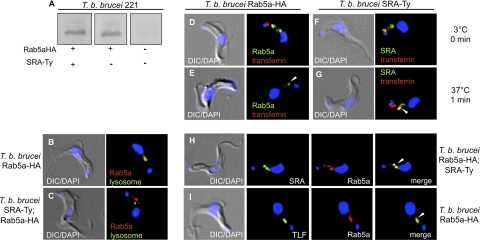
Endosomal localization of SRA. (A to C) Western blot (A) and immunofluorescence staining (B and C) with anti-HA (Rab5a) (red) of T. brucei brucei 221 TbRab5aHA transfectants. Lysosomal (anti-TbCatL) staining is shown in green. (D to G) Binding and uptake studies using T. brucei brucei 221 TbRab5aHA (D and E) and T. brucei brucei 221 SRA-Ty (F and G) transfectants. Live cell binding results of Alexa Fluor 594-transferrin (red) at 3°C, 0 min (D and F), followed by temperature shift to 37°C for 1 min then fixation with 1% paraformaldehyde to halt vesicle trafficking (F and G) are shown. Postfixation, TbRab5aHA was stained with anti-HA (green) (D and E), and SRA was stained with anti-Ty (green) (F and G). (H) Steady-state localization of SRA-Ty and TbRab5aHA. Fixed, permeabilized T. brucei brucei 221 TbRab5aHA;SRA-Ty transfectants were stained with anti-Ty (SRA; green) and anti-HA (Rab5a endosomes; red). (I) Uptake of Alexa Fluor 488-TLF (green) in T. brucei brucei 221 TbRab5aHA transfectants. Rab5a endosomes are shown by the anti-HA (red) staining. The nucleus and kinetoplast were localized by DAPI staining. White arrowheads indicate colocalization.
To validate HA-tagged Rab5a as an early endosomal marker, we carried out live cell binding and uptake studies with Alexa Fluor 594-transferrin, a known cargo of early endosomes. Both T. brucei brucei 427-221 Rab5a-HA and T. brucei brucei 427-221 SRA-Ty transfectant cell lines were allowed to bind Alexa Fluor 594-transferrin at 3°C and then shifted to 37°C to permit resumption of vesicle trafficking. Vesicle trafficking was halted by fixation after 1 min at 37°C, followed by permeabilization and either immunostaining with anti-Ty (SRA-Ty transfectants) or anti-HA (Rab5a-HA transfectants). In T. brucei brucei 427-221 Rab5a-HA transfectants, transferrin bound discretely to the flagellar pocket at 3°C (Fig. 3D). At 1 min postuptake, Alexa Fluor 594-transferrin colocalized intracellularly with Rab5a-HA (Fig. 3E). The same pattern of Alexa Fluor 594-transferrin localization was observed in T. brucei brucei 427-221 SRA-Ty transfectants, with flagellar pocket colocalization of transferrin and SRA at 3°C (Fig. 3F) and colocalization with SRA-containing intracellular vesicles after 1 min at 37°C (Fig. 3G). In addition to endosomally localized Alexa Fluor 594-transferrin, a fraction of the labeled cargo was also visible within the flagellar pocket (Fig. 3G). After fixation and permeabilization, the steady-state localization of SRA by anti-Ty staining revealed the distribution of SRA from the flagellar pocket throughout the early endosomal compartment (Fig. 3F). Finally, anti-Ty and anti-HA staining of T. brucei brucei 427-221 SRA-Ty;Rab5a-HA transfectants showed colocalization of SRA-Ty with Rab5a endosomes, thereby identifying the intracellular SRA-Ty-containing vesicles as early endosomes (Fig. 3H).
Previous studies have presented conflicting observations on the intracellular localization of SRA and TLF-1 (25, 39). Vanhamme et al. (39) found that SRA and TLF-1 colocalized in the lysosome, while Oli et al. (25) reported a nonlysosomal localization. To address this discrepancy, we carried out live cell binding and uptake studies with Alexa Fluor 488-conjugated TLF-1 in T. brucei brucei SRA-Ty;Rab5a-HA transfectants. Cells were incubated with Alexa Fluor 488-TLF at 3°C for binding studies and then shifted to 37°C for 1 min to permit endocytosis and vesicle trafficking. After 1 min at 37°C, Alexa Fluor 488-TLF colocalized with both anti-HA staining (Fig. 3I) and anti-Ty staining (see Fig. 5A), indicating that both TLF-1 and SRA traffic through the early endosomes, the earliest site of observed colocalization of these virulence factors.
Fig. 5.
Localization of SRA to the flagellar pocket is not critical to resistance. (A and B) Uptake of Alexa Fluor 488-TLF (green) at 37°C for 1 min postbinding in SRA-Ty (A) and SRAΔGPI (B) cells. SRA was visualized via anti-Ty staining (red). White arrowheads indicate colocalization. (C) Untransfected T. brucei brucei 221, T. brucei brucei 221 SRA-Ty, and T. brucei brucei 221 SRAΔGPI cells were incubated with increasing concentrations of purified TLF-1. Percent lysis was determined after 2 h at 37°C.
The GPI anchor of SRA is necessary for flagellar pocket, but not endosomal, localization.
The GPI anchor modification has previously been shown to play a role in the trafficking and localization of glycoproteins in T. brucei brucei (32, 38). To investigate the role of the GPI anchor in SRA trafficking, we engineered a Ty-tagged deletion mutant of SRA to be translated without the C-terminal hydrophobic peptide and the GPI anchor addition site, thus preventing posttranslational addition of the GPI anchor (Fig. 4A). Western blot analysis of the mutant T. brucei brucei 427-221 SRAΔGPI and T. brucei brucei 427-221 SRA-Ty cell lysates revealed comparable levels of SRA protein expression; however, SRAΔGPI migrated faster than soluble, GPI-phospholipase C (PLC)-cleaved wild-type SRA-Ty (Fig. 4B). Under the solubilization conditions used here (1% NP-40), we expected wild-type SRA-Ty to be released by GPI-PLC activity. Therefore, neither the mutant SRAΔGPI nor soluble wild-type SRA-Ty is membrane bound (6). The small apparent size of the SRAΔGPI mutant is due to the complete lack of the amino acid anchor addition site, as well as the phosphoethanolamine, glycan, inositol, and diacylglycerol moieties that comprise the GPI anchor. Wild-type SRA-Ty retains all these components except for diacylglycerol following GPI-PLC cleavage. SRAΔGPI therefore lacks the cleaved GPI anchor fragment present on soluble wild-type SRA-Ty, which likely accounts for the 5-kDa difference in migration of the proteins. Deglycosylation of SRAΔGPI by PNGase F treatment led to a shift in migration, indicating that the protein without the GPI anchor traffics through the endoplasmic reticulum (ER), where N-linked glycans are added (see Fig. S1E in the supplemental material).
To examine the cellular localization of SRAΔGPI, we performed 3°C binding studies with Alexa Fluor 594–anti-Ty antibody and found that 100% of SRAΔGPI cells observed had a marked absence of fluorescence in the flagellar pocket region (Fig. 4D), in contrast to cells expressing wild-type SRA-Ty (Fig. 4C). To determine whether SRAΔGPI was being secreted into the culture medium, we conducted anti-Ty immunoprecipitation on medium, but we were unable to detect SRAΔGPI, despite being able to immunoprecipitate SRAΔGPI protein from cell lysates (data not shown). Despite the loss of cell surface localization, intracellular localization of SRAΔGPI showed a similar distribution of SRA-containing vesicles, as was observed with wild-type SRA-Ty (Fig. 4E and F). Consistent with this finding, TLF-1 was found to colocalize with SRA within endosomes in both wild-type SRA (Fig. 5A) and in deletion mutant cells (Fig. 5B), respectively. Furthermore, upon in vitro incubation with TLF-1, untransfected cells were highly sensitive to TLF-1-killing, while transfectants expressing wild-type SRA and SRAΔGPI were equally resistant to TLF-1 toxicity (Fig. 5C).
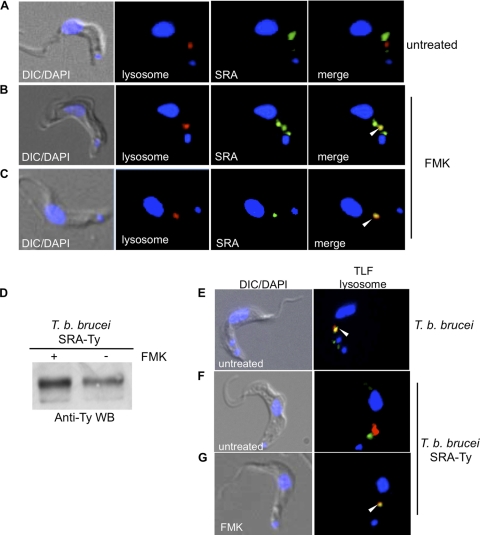
The endosomal localization of SRA that we observed differs from reports by others that SRA is a lysosomal resident protein (39) (Fig. 6A). To address the discrepancy in lysosomal localization of SRA, we treated T. brucei brucei 427-221 SRA-Ty cells with the lysosomal protease inhibitor FMK024, in order to inhibit protein turnover (3). Under these conditions, we were able to observe an accumulation of SRA in the lysosome that was specifically dependent on inhibition of protein degradation by this thiol protease inhibitor (Fig. 6B and C). This lysosomal accumulation was more complete in some cases, with only lysosomal SRA detected in 35.8 ± 16.4% (mean ± SEM) of cells (Fig. 6C; Table 2), while in others, both endosomal and lysosomal SRA was detected by anti-Ty staining (Fig. 6B; Table 2) in 44.6 ± 8.7% of cells. This lysosomal accumulation was also observed by anti-Ty Western blotting of whole-cell SRA-Ty lysates, with an increase (1.4-fold) in SRA in FMK-treated cells compared to untreated cells (Fig. 6D). Thus, while SRA traffics to the trypanosome lysosome, abundance is limited due to rapid degradation, suggesting that the major intracellular pool of SRA is endosomal.
Fig. 6.
SRA is rapidly turned over by lysosomal proteolysis. Immunofluorescence staining of fixed, permeabilized T. brucei brucei 221 SRA-Ty transfectants is shown. (A to C) Cells were untreated (A) or FMK024 treated (B and C) prior to fixation (20 μM; 37°C, 1 h). Anti-Ty (SRA; green) and anti-TbCatL (red) staining showed SRA and lysosomal localization, respectively. Image acquisition was carried out at the same exposure, and images were contrasted to the same extent. (D) Anti-Ty (SRA) Western blot of untreated and FMK024-treated T. brucei brucei 221 SRA-Ty transfectants, showing accumulation of SRA due to protease inhibition. (E to G) Uptake of Alexa Fluor 488-TLF (green) in T. brucei brucei 221 (E) and T. brucei brucei 221 SRA-Ty (F and G) cells. Cells were either untreated (E and F) or treated with FMK024 (20 μM; 37°C, 1 h) (F) during TLF-1 endocytosis. Anti-TbCatL (lysosomal) staining is shown in red. The nucleus and kinetoplast were localized by DAPI staining. White arrowheads indicate colocalization.
In a previous study, we reported that TLF-1 was not observed in the lysosomes of SRA-expressing cells, and thus SRA was thought to alter the trafficking pathway of TLF-1 (25). Here we found that while TLF-1 accumulated in the lysosomes of untreated susceptible T. brucei brucei 427-221 cells (Fig. 6E), a similar accumulation was not observed in SRA-expressing transfectants (Fig. 6F) unless protein turnover was inhibited with FMK024 treatment (Fig. 6G). In T. brucei brucei 427-221 cells, 81.5 ± 3.7% of cells showed TLF-1 localized only to the lysosome, while 74.2 ± 2.5% of SRA-expressing cells showed a similar distribution of TLF-1 upon treatment with FMK024 (Table 3). The protease inhibitor-dependent accumulation of TLF-1 in the lysosome of SRA-expressing cells indicates that TLF-1 may be more rapidly turned over in the presence of SRA, or that the rate of trafficking from endosome to lysosome may be slowed in the presence of SRA. These results suggest that the mechanism of SRA-mediated resistance to TLF-1 may therefore be due to accelerated lysosomal proteolysis of the TLF-1/SRA complex or slowed trafficking between endocytic compartments.
Table 3.
Distribution of TLF-1 cellular localization in FMK-treated cells
| Cell type and compartment | % of cells in compartmenta |
|---|---|
| T. brucei brucei (untransfected) | |
| Endosomal | 4.1 ± 0.5 |
| Lysosomal | 81.5 ± 3.7 |
| Endosomal/lysosomal | 14.4 ± 5.4 |
| T. brucei brucei SRA-Ty | |
| Endosomal | 3.2 ± 0.8 |
| Lysosomal | 74.2 ± 2.5 |
| Endosomal/lysosomal | 22.6 ± 3.1 |
Quantitation of cellular distribution of TLF-1 in FMK-treated untransfected T. brucei brucei cells and T. brucei brucei SRA-Ty cells. Results are presented as means ± SEM (n = 3). The total number of T. brucei brucei cells was 303. The total number of T. brucei brucei SRA-Ty cells was 108.
DISCUSSION
Human infectious T. brucei rhodesiense is able to resist TLF-1 killing through the expression of the VSG-like protein SRA. The molecular basis of resistance remains unclear. However, it is evident that the intracellular colocalization of SRA and TLF-1 is required for neutralization of the trypanolytic activity (39). In an effort to gain insight into the mechanism of human serum resistance, we carried out a thorough characterization of SRA by examination of its cellular trafficking and steady-state localization. In this study, we have for the first time defined the steady-state localization of SRA and its point of initial colocalization with TLF-1. Furthermore, we report that in the absence of protease inhibition, TLF-1 accumulates in the lysosomes of T. brucei brucei cells, but not in SRA-expressing transfectants. This finding suggests a difference in the stability of TLF-1 or a difference in the rate of trafficking of TLF-1 from endosomes to lysosome in resistant SRA-expressing cells compared to sensitive T. brucei brucei cells. Thus, we propose that accelerated turnover of TLF-1 or altered kinetics of trafficking is involved in the mechanism of SRA-mediated resistance.
Bioinformatic analysis of the amino acid sequence of SRA has shown the presence of posttranslational features, such as an N-terminal signal sequence, N-linked glycosylation sites, and a GPI anchor attachment site (5), that are characteristic of cell surface-localized VSG family proteins, such as VSG and the trypanosome transferrin receptor (10, 35, 36). VSGs exist as densely packed homodimers that coat the entire plasma membrane, while transferrin receptor is more discretely localized to the flagellar pocket (22). Given the shared features of the VSG family proteins, it was therefore surprising that SRA exhibited a primarily intracellular localization.
Binding studies with live cells at 3°C and immunofluorescence microscopy of fixed, nonpermeabilized cells showed that a portion of steady-state SRA localized to the cell surface (Fig. 1). The cell surface distribution of SRA was observed solely in the flagellar pocket region of the cell, as delineated by several previously characterized markers. Both the kinetoplast and basal bodies are directly associated with the pocket, while the paraflagellar rod extends along the flagellum of the trypanosome to the point of entry of the flagellum into the pocket (13, 21). The region between the kinetoplast and the posterior end of the paraflagellar rod therefore includes the flagellar pocket. In addition to these cellular markers, we also utilized binding of fluorescently labeled transferrin as a flagellar pocket marker. Immunostaining of SRA in fixed, nonpermeabilized cells revealed that SRA colocalized with receptor-bound transferrin within the flagellar pocket region; thus, we showed conclusively that SRA traffics to the flagellar pocket. Prior to these studies, the apparent lack of SRA in the flagellar pocket was most likely due to the difficulty of visualizing SRA as a consequence of the remarkably high rate of endocytosis that is a hallmark of bloodstream-form T. brucei cell biology (9, 26). Flagellar pocket localization of SRA is therefore transient, as the protein is likely to be rapidly endocytosed almost immediately upon transport to the cell surface.
Since SRA was found to traffic to the flagellar pocket, which is also the site of receptor-mediated endocytosis of TLF-1, we examined the possibility that the initial point of colocalization of SRA and TLF-1 would take place in this specialized region of the plasma membrane. We expressed SRA in a cell line that does not express TbHpHbR, in order to determine whether SRA colocalizes with TLF-1 at the cell surface. Although SRA localized to the flagellar pocket, in the absence of the TLF-1 receptor we did not detect binding or uptake of Alexa Fluor 488 TLF, indicating that SRA is not sufficient for binding or uptake of TLF-1. Based on proposed models of VSG and SRA protein structure (5), SRA appears to be a shorter molecule and may therefore be inaccessible to TLF-1 on the trypanosome cell surface. This unavailability of cell surface SRA may be due to steric hindrance by the larger, more abundant VSGs, which are continually endocytosed via the flagellar pocket.
In a previous study, we reported that the steady-state distribution of SRA within cytoplasmic vesicles localized between the kinetoplast and nucleus (25). Characterization of trafficking vesicles in bloodstream-form T. brucei has defined several classes of endosomes by their associated Rab GTPases and cargo, including the early endosomal Rab5a GTPase (11, 12, 14, 17, 18, 27, 28). Taking advantage of these classifications, we generated a cell line that expressed an epitope-tagged Rab5a as an early endosomal marker. Immunostaining and fluorescence microscopy revealed that SRA colocalized with the early endosomal marker (Fig. 3). Given that the other known VSG family proteins, VSG and TfR, are resident cell surface proteins, SRA therefore has an atypical localization and is the first example of a resident early endosomal VSG family protein in T. brucei. This localization raises important questions about trafficking and retention of GPI-anchored proteins. While both VSG and TfR traffic as homodimers and heterodimers, respectively, no binding partner has been identified for SRA to date. Campillo and Carrington (5) have proposed models that indicated SRA is likely to exist as a dimer. SRA may therefore be retained in the endosomal compartment through protein-protein interactions within these trafficking vesicles. In addition, studies by Wang et al. (40a) have shown that, unlike VSG and TfR, SRA is not released but remains cell associated upon GPI-PLC activation, which suggests that the GPI anchor of SRA may not be susceptible to GPI-PLC activity and may differ from that of VSG and TfR in a way that leads to the retention of the protein in the endosomes.
When Alexa Fluor 488-TLF was incubated with live cells, it localized to SRA-containing Rab5a endosomes. Given that a previous report showed that SRA binds to apoL-I (39), it is likely that SRA interacts with TLF-1 via apoL-I binding within this compartment. Thus, the initial point of interaction between TLF-1 and SRA may occur early on in the trafficking pathway, rather than later in the terminal lysosomal compartment. The shared trafficking pathway via the early endosomes was also observed for Alexa Fluor 594-transferrin, an independent cargo, unrelated to TLF-1 susceptibility. This overlap of independent cargo is not surprising, as both transferrin and TLF-1 are endocytosed by GPI-anchored receptors that localize to the flagellar pocket. The transient flagellar pocket localization and the steady-state, intracellular accumulation of SRA within endosomes may be accounted for by the high rates of endocytosis of proteins associated with the flagellar pocket (9).
In order to determine whether altering SRA trafficking has an effect on trypanosome susceptibility to lysis, we generated a deletion mutant of SRA that lacked the GPI anchor modification (Fig. 4). The loss of the GPI anchor resulted in the disruption of the flagellar pocket localization of SRA, and the protein was no longer detectable in the flagellar pocket. Despite the loss of cell surface localization, the endosomal localization of SRAΔGPI was unaltered. We were also unable to detect SRAΔGPI in cell growth medium, suggesting that the GPI deletion mutant is not secreted. These findings are consistent with studies by Triggs and Bangs (38), as well as by Böhme and Cross (2a). Neither set of investigators detected secretion of GPI-deficient VSG, and they found delayed ER exit to be a consequence of GPI anchor deletion. Some fraction of SRAΔGPI may therefore be ER localized; however, SRAΔGPI also colocalized with endocytosed TLF-1, indicating that at least some SRAΔGPI localizes to the endosomal compartment. This endosomal localization may therefore be the result of direct targeting by an alternative sorting pathway to the endosomes. Cell surface localization of SRA did not appear to be critical to TLF-1 resistance, as SRAΔGPI mutants were resistant to TLF-1-mediated lysis. These data were consistent with the finding that SRA does not facilitate binding or uptake of TLF-1 but mediates human serum resistance via an interaction that begins with endosomal colocalization.
Contrary to previous reports (39), we were not able to localize SRA to the lysosome in the absence of protease inhibitors. However, in cells treated with the thiol protease inhibitor FMK024, we were able to detect an increase in SRA by Western blotting, as well as SRA staining that colocalized with the lysosomal marker. We therefore attribute this localization to accumulation of the protein due to inhibition of the major trypanosome lysosomal protease, T. brucei brucei cathepsin L (4). Since we could only observe SRA in the lysosomal compartment when protein turnover was disrupted, SRA appears to be highly unstable in the lysosomal environment and is therefore likely to be rapidly degraded once it reaches the protease-rich organelle. This susceptibility to rapid lysosomal degradation is consistent with findings with other VSG family proteins: TfR and VSGs (2, 19, 32, 38).
In a previous study, we reported that TLF-1 was not observed in the lysosomal compartment of SRA-expressing cells and SRA was thought to alter the trafficking pathway of TLF-1 (25). The localization of SRA and TLF-1 to the lysosome in FMK024-treated cells clearly shows that altered trafficking to the lysosome in SRA-expressing cells is not the mechanism of resistance. Rather, the apparent loss of lysosomal accumulation of TLF-1 in SRA-expressing cells indicates that the mechanism of resistance may instead be due to accelerated turnover of TLF-1 in the presence of SRA. Alternatively, the rate of TLF-1 trafficking from the endosomes to the lysosome may be slower in the presence of SRA; therefore, TLF-1 accumulation within the lysosome is not observed in the absence of protease inhibition. In the absence of SRA, however, rapid trafficking of TLF-1 into the lysosomal compartment allows for accumulation and subsequent detection. Thus, SRA-mediated resistance may also involve differential rates of trafficking between endosomes and the terminal lysosomal compartment.
The studies reported here allow definitive description of the complete trafficking pathway of SRA from synthesis to degradation. Upon translation of the N-terminal signal sequence, SRA is targeted to the ER, where it is posttranslationally modified (5, 39). The mature protein then moves through the Golgi apparatus for additional processing of N-linked glycans (39) and is then targeted to the flagellar pocket. SRA is only transiently localized at the cell surface before it is rapidly endocytosed into early endosomes, where it initially encounters TLF-1. The interaction between TLF-1 and SRA may occur within the endosomal compartment, or TLF-1 and SRA may traffic independently to the lysosome before binding and subsequently degradation takes place. Neutralization of the trypanosome toxin may be a consequence of accelerated degradation or slowed trafficking from endosomes to lysosomes. Furthermore, the identification of the lysosomal cysteine protease TbCathepsin L as at least one class of proteases involved in turnover implicates the lysosome as the final site of detoxification of TLF-1. While the molecular changes that lead to SRA-mediated inhibition of TLF-1 remain to be elucidated, the findings presented here have brought us closer to resolving the mechanism of inhibition of TLF-1 and human infectivity by T. brucei rhodesiense.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rudo Kieft and John Harrington for critical comments on the manuscript, as well as all the members of the Hajduk laboratory for helpful discussions. We also thank Keri Jamison for her contribution to the cloning of SRA transfectant cell lines. We are indebted to Jay Bangs and Dianne McMahon-Pratt for providing reagents.
This work was supported by the National Institutes of Health (AI39033).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Bangs J. D., Ransom D. M., McDowell M. A., Brouch E. M. 1997. Expression of bloodstream variant surface glycoproteins in procyclic stage Trypanosoma brucei: role of GPI anchors in secretion. EMBO J. 16:4285–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biebinger S., Helfert S., Steverding D., Ansorge I., Clayton C. 2003. Impaired dimerization and trafficking of ESAG6 lacking a glycosyl-phosphatidylinositol anchor. Mol. Biochem. Parasitol. 132:93–96 [DOI] [PubMed] [Google Scholar]
- 2a. Böhme U., Cross G. A. 2002. Mutational analysis of the variant surface glycoprotein GPI-anchor signal sequence in Trypanosoma brucei. J. Cell. Sci. 115:805–816 [DOI] [PubMed] [Google Scholar]
- 3. Caffrey C. R., et al. 2001. Active site mapping, biochemical properties and subcellular localization of rhodesain, the major cysteine protease of Trypanosoma brucei rhodesiense. Mol. Biochem. Parasitol. 118:61–73 [DOI] [PubMed] [Google Scholar]
- 4. Caffrey C. R., Steverding D. 2009. Kinetoplastid papain-like cysteine peptidases. Mol. Biochem. Parasitol. 167:12–19 [DOI] [PubMed] [Google Scholar]
- 5. Campillo N., Carrington M. 2003. The origin of the serum resistance associated (SRA) gene and a model of the structure of the SRA polypeptide from Trypanosoma brucei rhodesiense. Mol. Biochem. Parasitol. 127:79–84 [DOI] [PubMed] [Google Scholar]
- 6. Cardoso de Almeida M. L., Turner M. J. 1983. The membrane form of variant surface glycoproteins of Trypanosoma brucei. Nature 302:349–352 [DOI] [PubMed] [Google Scholar]
- 7. De Greef C., Chimfwembe E., Kihang'a Wabacha J., Bajyana Songa E., Hamers R. 1992. Only the serum-resistant bloodstream forms of Trypanosoma brucei rhodesiense express the serum resistance associated (SRA) protein. Ann. Soc. Belg Med. Trop. 72(Suppl. 1):13–21 [PubMed] [Google Scholar]
- 8. Drain J., Bishop J. R., Hajduk S. L. 2001. Haptoglobin-related protein mediates trypanosome lytic factor binding to trypanosomes. J. Biol. Chem. 276:30254–30260 [DOI] [PubMed] [Google Scholar]
- 9. Engstler M., et al. 2004. Kinetics of endocytosis and recycling of the GPI-anchored variant surface glycoprotein in Trypanosoma brucei. J. Cell Sci. 117:1105–1115 [DOI] [PubMed] [Google Scholar]
- 10. Ferguson M. A., Homans S. W., Dwek R. A., Rademacher T. W. 1988. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239:753–759 [DOI] [PubMed] [Google Scholar]
- 11. Field H., Farjah M., Pal A., Gull K., Field M. C. 1998. Complexity of trypanosomatid endocytosis pathways revealed by Rab4 and Rab5 isoforms in Trypanosoma brucei. J. Biol. Chem. 273:32102–32110 [DOI] [PubMed] [Google Scholar]
- 12. Field M. C., Carrington M. 2004. Intracellular membrane transport systems in Trypanosoma brucei. Traffic 5:905–913 [DOI] [PubMed] [Google Scholar]
- 13. Gadelha C., Wickstead B., de Souza W., Gull K., Cunha-e-Silva N. 2005. Cryptic paraflagellar rod in endosymbiont-containing kinetoplastid protozoa. Eukaryot. Cell 4:516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grunfelder C. G., et al. 2003. Endocytosis of a glycosylphosphatidylinositol-anchored protein via clathrin-coated vesicles, sorting by default in endosomes, and exocytosis via RAB11-positive carriers. Mol. Biol. Cell 14:2029–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hager K. M., et al. 1994. Endocytosis of a cytotoxic human high density lipoprotein results in disruption of acidic intracellular vesicles and subsequent killing of African trypanosomes. J. Cell Biol. 126:155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hajduk S. L., et al. 1989. Lysis of Trypanosoma brucei by a toxic subspecies of human high density lipoprotein. J. Biol. Chem. 264:5210–5217 [PubMed] [Google Scholar]
- 17. Hall B. S., Pal A., Goulding D., Acosta-Serrano A., Field M. C. 2005. Trypanosoma brucei: TbRAB4 regulates membrane recycling and expression of surface proteins in procyclic forms. Exp. Parasitol. 111:160–171 [DOI] [PubMed] [Google Scholar]
- 18. Hall B. S., Pal A., Goulding D., Field M. C. 2004. Rab4 is an essential regulator of lysosomal trafficking in trypanosomes. J. Biol. Chem. 279:45047–45056 [DOI] [PubMed] [Google Scholar]
- 19. Kabiri M., Steverding D. 2000. Studies on the recycling of the transferrin receptor in Trypanosoma brucei using an inducible gene expression system. Eur. J. Biochem. 267:3309–3314 [DOI] [PubMed] [Google Scholar]
- 20. Kieft R., et al. 2010. Mechanism of Trypanosoma brucei gambiense (group 1) resistance to human trypanosome lytic factor. Proc. Natl. Acad. Sci. U. S. A. 107:16137–16141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lacomble S., et al. 2009. Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J. Cell Sci. 122:1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ligtenberg M. J., et al. 1994. Reconstitution of a surface transferrin binding complex in insect form Trypanosoma brucei. EMBO J. 13:2565–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Milner J. D., Hajduk S. L. 1999. Expression and localization of serum resistance associated protein in Trypanosoma brucei rhodesiense. Mol. Biochem. Parasitol. 104:271–283 [DOI] [PubMed] [Google Scholar]
- 24. Molina-Portela M. P., Samanovic M., Raper J. 2008. Distinct roles of apolipoprotein components within the trypanosome lytic factor complex revealed in a novel transgenic mouse model. J. Exp. Med. 205:1721–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oli M. W., Cotlin L. F., Shiflett A. M., Hajduk S. L. 2006. Serum resistance-associated protein blocks lysosomal targeting of trypanosome lytic factor in Trypanosoma brucei. Eukaryot. Cell 5:132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Overath P., Engstler M. 2004. Endocytosis, membrane recycling and sorting of GPI-anchored proteins: Trypanosoma brucei as a model system. Mol. Microbiol. 53:735–744 [DOI] [PubMed] [Google Scholar]
- 27. Pal A., Hall B. S., Jeffries T. R., Field M. C. 2003. Rab5 and Rab11 mediate transferrin and anti-variant surface glycoprotein antibody recycling in Trypanosoma brucei. Biochem. J. 374:443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pal A., Hall B. S., Nesbeth D. N., Field H. I., Field M. C. 2002. Differential endocytic functions of Trypanosoma brucei Rab5 isoforms reveal a glycosylphosphatidylinositol-specific endosomal pathway. J. Biol. Chem. 277:9529–9539 [DOI] [PubMed] [Google Scholar]
- 29. Rifkin M. R. 1978. Trypanosoma brucei: some properties of the cytotoxic reaction induced by normal human serum. Exp. Parasitol. 46:189–206 [DOI] [PubMed] [Google Scholar]
- 30. Rudenko G., Blundell P. A., Taylor M. C., Kieft R., Borst P. 1994. VSG gene expression site control in insect form Trypanosoma brucei. EMBO J. 13:5470–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schell D., et al. 1991. A transferrin-binding protein of Trypanosoma brucei is encoded by one of the genes in the variant surface glycoprotein gene expression site. EMBO J. 10:1061–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwartz K. J., Peck R. F., Tazeh N. N., Bangs J. D. 2005. GPI valence and the fate of secretory membrane proteins in African trypanosomes. J. Cell Sci. 118:5499–5511 [DOI] [PubMed] [Google Scholar]
- 33. Shimamura M., Hager K. M., Hajduk S. L. 2001. The lysosomal targeting and intracellular metabolism of trypanosome lytic factor by Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 115:227–237 [DOI] [PubMed] [Google Scholar]
- 34. Smith A. B., Esko J. D., Hajduk S. L. 1995. Killing of trypanosomes by the human haptoglobin-related protein. Science 268:284–286 [DOI] [PubMed] [Google Scholar]
- 35. Steverding D., Stierhof Y. D., Fuchs H., Tauber R., Overath P. 1995. Transferrin-binding protein complex is the receptor for transferrin uptake in Trypanosoma brucei. J. Cell Biol. 131:1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Strickler J. E., Patton C. L. 1980. Trypanosoma brucei brucei: inhibition of glycosylation of the major variable surface coat glycoprotein by tunicamycin. Proc. Natl. Acad. Sci. U. S. A. 77:1529–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tomlinson S., Muranjan M., Nussenzweig V., Raper J. 1997. Haptoglobin-related protein and apolipoprotein AI are components of the two trypanolytic factors in human serum. Mol. Biochem. Parasitol. 86:117–120 [PubMed] [Google Scholar]
- 38. Triggs V. P., Bangs J. D. 2003. Glycosylphosphatidylinositol-dependent protein trafficking in bloodstream-stage Trypanosoma brucei. Eukaryot. Cell 2:76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vanhamme L., et al. 2003. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422:83–87 [DOI] [PubMed] [Google Scholar]
- 40. Vanhollebeke B., et al. 2008. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science 320:677–681 [DOI] [PubMed] [Google Scholar]
- 40a. Wang J., Böhme U., Cross G. A. 2003. Structural features affecting variant surface glycoprotein expression in Trypanosoma brucei. Mol. Biochem. Parasitol. 128:135–145 [DOI] [PubMed] [Google Scholar]
- 41. Xong H. V., et al. 1998. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell 95:839–846 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.