Abstract
A screening procedure was used to identify cell fusion (hyphal anastomosis) mutants in the Neurospora crassa single gene deletion library. Mutants with alterations in 24 cell fusion genes required for cell fusion between conidial anastomosis tubes (CATs) were identified and characterized. The cell fusion genes identified included 14 genes that are likely to function in signal transduction pathways needed for cell fusion to occur (mik-1, mek-1, mak-1, nrc-1, mek-2, mak-2, rac-1, pp2A, so/ham-1, ham-2, ham-3, ham-5, ham-9, and mob3). The screening experiments also identified four transcription factors that are required for cell fusion (adv-1, ada-3, rco-1, and snf5). Three genes encoding proteins likely to be involved in the process of vesicular trafficking were also identified as needed for cell fusion during the screening (amph-1, ham-10, pkr1). Three of the genes identified by the screening procedure, ham-6, ham-7, and ham-8, encode proteins that might function in mediating the plasma membrane fusion event. Three of the putative signal transduction proteins, three of the transcription factors, the three putative vesicular trafficking proteins, and the three proteins that might function in mediating cell fusion had not been identified previously as required for cell fusion.
INTRODUCTION
The process of cell-to-cell fusion plays a vital role in the life cycles of almost all multicellular organisms. Fertilization, the fusion of an egg and sperm, is a required cell-to-cell fusion event for sexually reproducing organisms. For vertebrates, cell fusion is a critical step in the development of muscle, placenta, and bone and plays an essential role in the formation of multinucleated giant cells in the immune system. Hyphal cell fusion plays an important role in the life cycle of Neurospora crassa and other filamentous fungi (1, 6, 12, 24, 38). During the life cycle of the filamentous fungus Neurospora crassa, cell fusions occur during at least three stages (12, 38). During sexual development, fertilization occurs as a protoperithecium (immature female mating structure) generates a trichogyne (long specialized hyphae) that chemotrophically grows toward a conidium (asexual spore) or hypha of the opposite mating type and undergoes cell fusion with it. During the germination of N. crassa conidia, the cells produce short, specialized thin hyphae called conidial anastomosis tubes (CATs), which mediate cell fusion between the germlings and generate an interconnected hyphal network (38, 41). This process of cell fusion between conidia allows the cells to share resources and may be critical to the establishment of a colony under some environmental conditions. Cell fusions also occur during the growth of a vegetative N. crassa colony. A few millimeters behind the growing edge of the colony, specialized fusion hyphae are formed as branches from the vegetative hyphae. The fusion hyphae grow toward each other in a directed manner and undergo cell fusion to generate an interconnected hyphal network within the colony (18). Cytoplasm and organelles flow freely through the fused fusion hyphae.
Based on morphological criteria, the process of hyphal cell fusion can be roughly divided into three stages (17), beginning with a stage in which the tips of two fusion hyphae grow toward each other. Chemotrophic interactions have been shown to direct the growth from the tips of the two fusion hyphae. Recent work by Fleissner et al. (13) suggests that mitogen-activated protein (MAP) kinase pathways are activated at the tips of the fusion hyphae during this stage of the cell fusion process. Once the two fusion hyphae meet, there is a stage where adhesion occurs and the hyphae cease directed growth. During this adhesion stage, the cell wall between the two hyphal tips is remodeled. This remodeling includes the digestion of the wall between the plasma membranes of the two hyphal tips. It also includes the joining and strengthening of the cell wall peripheral to the future site of membrane fusion in order to protect the hyphae from osmotic lysis (12). The final stage in the process is the fusion of the two plasma membranes, followed by the movement and mixing of the cytoplasm from the two fusion hyphae.
A number of N. crassa cell fusion mutants have been identified and characterized previously (1, 10, 11, 12, 17, 35, 38, 50, 51). These mutants are defective in the process of fusion between fusion hyphae (anastomosis) and the process of CAT formation. Components of the mik-1/mek-1/mak-1 and nrc-1/mek-2/mak-2 MAP kinase pathways are affected in most of these mutants.
We report on the identification and characterization of 12 new cell fusion genes. These genes were identified from a screening of the Neurospora single-gene-deletion library generated by the Neurospora genome project (7). The library is arrayed in 96-well plates and contains deletion mutants for most of the genes encoded in the Neurospora genome. The mutants were identified as being defective in hyphal fusion and CAT formation. The proteins encoded by the cell fusion genes identified include signal transduction pathway components, transcription factors, proteins that function in vesicular trafficking, and proteins that may be involved in mediating fusion between the plasma membranes of the fusing cells.
MATERIALS AND METHODS
Growth of N. crassa.
N. crassa cells were grown in Vogel's sucrose medium and in synthetic crossing medium (SCM) as described by Davis and DeSerres (8). In all of the mating experiments, the cell fusion mutants were used as the male (conidial) partner. Matings and isolation of ascospore progeny were performed as described by Davis and DeSerres (8). N. crassa strains and the deletion mutant library were obtained from the Fungal Genetics Stock Center (FGSC; Kansas City, MO). The his-3 A mutant (FGSC catalog no. 6103) and a his-3 a strain, which we obtained by mating the his-3 A mutant with the wild-type strain (FGSC catalog no. 4200), were used as females in matings with the deletion mutants. Progeny with the deletion mutations and the his-3 mutation were isolated and used in complementation experiments. The formation of CATs was assessed as described by Roca et al. (42). Growth rates were measured by inoculating a spot of conidia onto a petri dish containing Vogel's sucrose agar medium and measuring the linear extension of the colony across the plate as a function of time. The morphological characteristics of each of the mutants were ascertained by growing the mutants on agar plates and slants containing Vogel's sucrose or synthetic crossing medium.
Screening procedures.
The N. crassa deletion library is found in 96-well plates, each well of which contains a conidial suspension of a deletion mutant. Most of the mutants in the library are found as homokaryons, but a significant number of the isolates are provided in the library as heterokaryons due to the essential nature of the genes that are deleted in these isolates. A 3-μl aliquot of conidia was taken for each of the deletion mutants contained in the first 110 plates of the deletion library and was used to inoculate a 3-ml slant of synthetic crossing medium (8). The first 110 plates contain deletion mutants for most, but not all, of the genes identified in the genome. For many of the genes, deletion isolates of both mating types are present in the library. The slants were allowed to grow for 7 days. Each slant was then examined under the dissecting microscope for the presence of protoperithecia on the glass neighboring the growth medium and for the pattern of conidial production. When grown on SCM, wild-type isolates produce large numbers of protoperithecia, while cell fusion mutants either are defective in the formation of protoperithecia or require a much longer time to produce them. Wild-type N. crassa isolates also produce an abundance of conidia from long aerial hyphae produced at the top of the slant, while cell fusion mutants produce short aerial hyphae over the entire slant surface to give a “flat,” carpet-like conidiation pattern (1). During the screening, putative cell fusion mutants were identified by a “flat conidiation” pattern and a defect in the production of protoperithecia. These mutants were then subjected to cosegregation analysis in order to verify that the deletion mutation could be responsible for the mutant phenotype.
Cosegregation experiments.
In characterizing the deletion library, we found that many of the mutants contain mutations in addition to the targeted deletion. Investigators have previously noted a high rate of spontaneous mutations giving rise to mutants with cell fusion phenotypes (22). Thus, as one way to demonstrate that the deletion was responsible for the mutant phenotype, we carried out cosegregation experiments. The deletion mutations were created by replacement of the coding region of the gene being deleted with a hygromycin resistance cassette (7), which provides a selectable marker identifying the location of the deletion mutation. Wild-type strains (FGSC catalog no. 2489 and 4200) were used as females and were mated with the deletion mutants. Individual ascospore progeny were isolated, and their phenotypes were examined by growing the cells on SCM (to assess them for protoperithecium formation), Vogel's sucrose medium (to assess them for the conidiation pattern), and Vogel's sucrose medium containing hygromycin (to assess whether the progeny had the deletion mutation). Cosegregation of the hygromycin resistance phenotype with the conidiation pattern and protoperithecium formation phenotypes was taken as evidence that the deletion was responsible for the mutant phenotype. A lack of cosegregation was taken as definitive evidence that the deletion was not responsible for the mutant phenotype.
Mutant characterization.
Those mutants showing cosegregation of the mutant phenotype and the deletion mutation were further characterized by examining the ability of the mutant cells to participate in intracolonial cell-to-cell fusion events as described by Aldabbous et al. (1). The mutant cells were grown on a cellophane filter placed on top of Vogel's sucrose agar medium. A second layer of cellophane was placed over the colony to create a sandwich, and the cells were allowed to grow between the two layers of cellophane. A 2-mm2 piece of the cellophane sandwich was cut from the growing edge of the colony, placed on a slide with 20 μl of growth medium, and examined with a differential interference contrast (DIC) microscope. In wild-type isolates, the presence of fused hyphae was readily observed by the flow of cytoplasmic organelles through the fused hyphae. Such fusion hyphae were not observed in the cell fusion mutants.
The mutants were also examined for the ability to form CATs by using the procedure described by Roca et al. (42). Conidia were collected, washed, and placed in Vogel's sucrose medium at a titer of 106 cells/ml. The cells were allowed to germinate and grow for 4 h in a Lab-Tek chamber slide (Nalge Nunc Inc., Rochester, NY) and were observed with an inverted DIC microscope. In wild-type isolates, CATs can easily be observed between germinating conidia. In the cell fusion mutants, the germinating conidia fail to form CATs.
Complementation experiments.
Complementation assays were used to verify that the deletion mutation was responsible for the cell fusion mutant phenotype. Complementation experiments were conducted by using the pBM60/pBM61 plasmid system (30) to insert a wild-type copy of the deleted gene into the intergenic region 3′ of the his-3 locus. Wild-type copies of the putative cell fusion genes were amplified using AccuTaq polymerase from Invitrogen (Carlsbad, CA). Primers for the genes were designed to amplify the coding regions, with approximately 1,500 bp of upstream (5′) sequence and 500 bp of downstream (3′) sequence. In most cases, the primers also contained an added restriction enzyme site to allow for the insertion of the amplified DNA into the multicloning sites of pBM60 or pBM61. The choice of pBM60 or pBM61 was made such that the direction of transcription from the amplified gene was the same as the direction of transcription from the his-3 gene so as to minimize the possibility of “antisense” transcripts. The amplified DNAs were cut with restriction enzymes and were ligated into restriction enzyme-digested plasmid pBM60 or pBM61. The ligated DNA was used to transform Escherichia coli, and minipreps were prepared from individual transformants (29). The miniprep DNAs were sequenced, and plasmids containing wild-type copies of the cell fusion genes were chosen for complementation experiments.
Two-week-old conidia from his-3 isolates with deletion mutations were transformed with a pBM60- or pBM61-derived plasmid containing a wild-type copy of the deleted gene (30). The pBM60/pBM61 plasmids contain the 3′ end of the his-3 coding region, followed by a short 3′ untranslated region (UTR) and intergenic sequence, a multicloning region, into which we inserted the wild-type copies of the cell fusion genes, and some additional sequences from the intergenic region. The mutant strains used in the complementation experiments contain a mutation near the 3′ end of the his-3 gene coding region. During transformation, homologous recombination between the plasmid and the genomic sequences generates a functional wild-type his-3 gene and the insertion of the wild-type copy of the cell fusion gene into the intergenic region adjacent to the his-3 locus. The ability of the wild-type copy of the cell fusion gene to restore a wild-type phenotype was taken as proof that the deletion mutation was responsible for the mutant phenotype and that the gene was required for cell fusion.
RIP experiments.
A RIP (repeat induced point mutation) experiment was used to determine if mutations in the ham-10 gene give rise to a cell fusion-defective phenotype. RIP is a phenomenon in which genes that are present in two or more copies in the haploid genome are mutated during the sexual cycle in N. crassa (46). Both copies of the duplicated gene receive multiple C-to-T transition mutations during the RIP process. The RIP experiment was performed by cloning the ham-10 gene into the pBM60 plasmid as described above. A plasmid containing a copy of the ham-10 gene was then used to transform the his-3 A isolate (FGSC catalog no. 6103) to yield a his-3+ transformant containing a transforming copy of the ham-10 gene as well as the endogenous gene copy. Transformants were then mated with the his-3 a isolate to activate the RIP process, and progeny from the mating were characterized. Individual his-3 progeny (progeny lacking the transforming copy of the ham-10 gene) with the phenotype of the deletion mutant were isolated, and the ham-10 gene was sequenced.
RESULTS
Mutant identification.
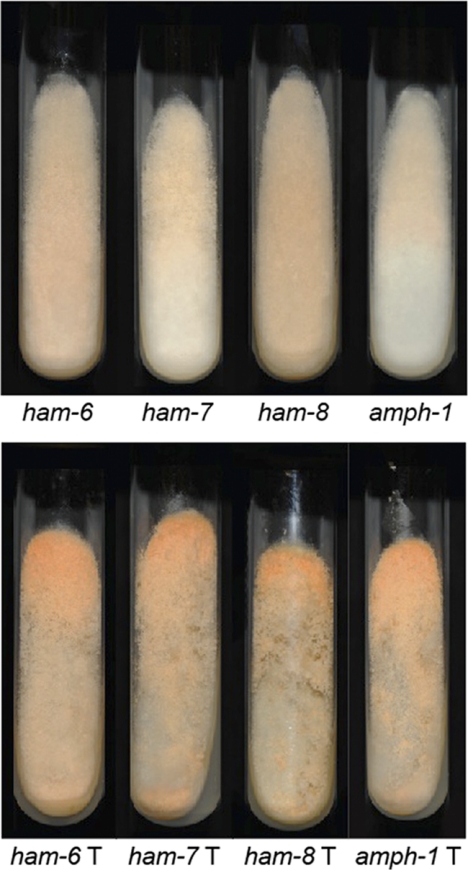
The Neurospora single gene deletion library provides an opportunity to identify the genes required for any phenotype that can be easily identified in a screening assay. We developed a simple screening procedure to help us identify mutants in which the process of cell fusion was affected. Characterization of previously identified cell fusion mutants showed that they share two readily observed phenotypes (1). First, the pattern in which conidia (asexual spores) are produced when the cells are grown on an agar slant is affected in the mutants. Wild-type colonies produce an abundance of aerial hyphae and conidia at the top of the slant, while cell fusion mutants produce short aerial hyphae and conidia as a carpet over the entire surface of the slant. Second, the ability of cell fusion mutants to produce protoperithecia is affected. Some of the cell fusion mutants are unable to produce protoperithecia (mik-1, mek-1, mak-1, nrc-1, mek-2, mak-2, ham-5, rco-1, rcm-1, and pp-1 mutants) (1, 12, 38), while others have been reported to be able to produce protoperithecia (ham-2, ham-3, and ham-4 mutants), but protoperithecium production requires a longer time than for the wild type (47). We used these shared phenotypes in identifying putative cell fusion mutants. All of the mutants in the first 110 plates of the deletion library were examined individually for these phenotypes as described in Materials and Methods. Among the approximately 10,000 deletion strains examined, we found 274 putative cell fusion mutants (see Table S1 in the supplemental material). The library contains many mutants with deletions of genes previously identified as cell fusion genes (rco-1, mik-1, mek-1, mak-1, nrc-1, mek-2, mak-2, so/ham-1, ham-2, ham-3, ham-5, and mob3), and these were found among the mutants we identified in our screening procedure, demonstrating that the screening procedure was effective at identifying cell fusion genes. Figure 1 shows the wild-type conidiation pattern and the flat conidiation pattern seen in the ham-6, ham-7, ham-8, and amph-1 deletion mutants, which were identified as cell fusion mutants in our screening.
Fig. 1.
Slants showing the conidiation pattern phenotypes of cell fusion mutants. Images of the wild-type strain and the ham-6, ham-7, ham-8, and amph-1 deletion mutants grown on Vogel's sucrose medium slants for 72 h are shown.
To determine whether or not the deletion mutations might be responsible for the mutant phenotypes, cosegregation experiments were carried out in which the segregation of mutant phenotypes and the segregation of hygromycin resistance were followed in the progeny from matings between wild-type isolates and each of the 274 deletion mutants. The deletion mutants were created by replacing the deleted gene with a hygromycin resistance cassette, so the segregation of hygromycin resistance was used as a marker for the deletion mutation. We found that hygromycin resistance and the cell fusion-defective phenotype cosegregated in 57 of the putative cell fusion mutants, representing 35 putative cell fusion genes (see Table S1 in the supplemental material). The cosegregation of the mutant phenotype with the hygromycin resistance gene used in creating the deletion mutations suggests that the cell fusion defect might be attributed to the deletion. These mutants were then characterized further. We found that hygromycin resistance and the mutant phenotype did not cosegregate in the remaining 217 mutants, indicating that these mutants have mutations in addition to the deletion mutation and that the mutant phenotype was due to one or more of these additional mutations (see Table S1).
We assayed the 57 putative deletion mutants directly for the presence of fusion hyphae as described by Aldabbous et al. (1). Figure 2 shows the presence of joined fusion hyphae from a wild-type isolate. The flow of cytoplasmic organelles through joined fusion hyphae was easily observed in wild-type cells and was absent from cell fusion mutants. All of the putative cell fusion mutants were defective in the formation of joined fusion hyphae as assessed by this assay.
Fig. 2.
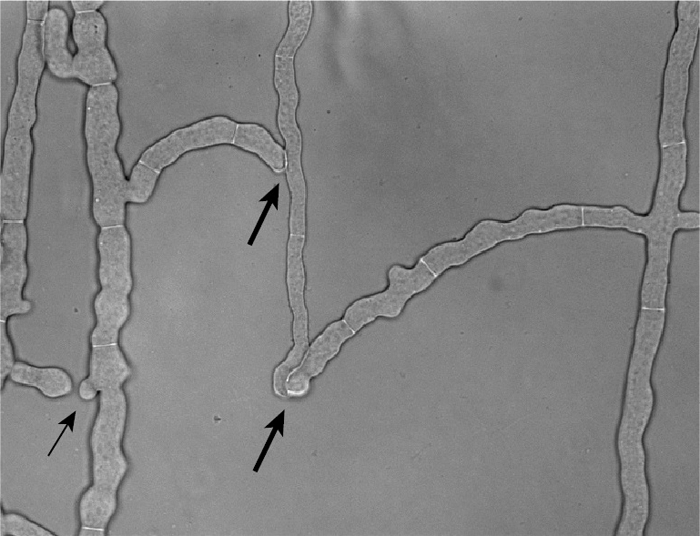
Photograph of the wild-type strain showing fusion hyphae. The wild-type strain was grown between two sheets of cellophane, and a region behind the growing edge of the colony containing fusion hyphae was photographed. The thicker arrows show locations where the fusion hyphae have completed the process of cell fusion. The thinner arrow points to two fusion hyphae that are growing toward each other but have not yet fused.
As a second way to determine directly whether the putative mutants were defective in cell fusion, we ascertained whether or not the mutants could form conidial anastomosis tubes (CATs) (see Fig. S1 in the supplemental material). CATs are formed by germinating conidia and facilitate cell fusion between the germlings to produce a connected network of germlings (42). Most previously identified cell fusion mutants have been found to be incapable of making CATs (rco-1, rcm-1, mik-1, mek-1, mak-1, nrc-1, mek-2, mak-2, ham-1, ham-2, ham-3, ham-4, ham-5, and mob3 mutants) (1, 12, 35, 38). Of our 57 putative cell fusion mutants, we found that 46 mutants (representing 28 genes) were unable to form CATs (Tables 1 to 4). The 11 mutants that were able to form CATs defined 7 genes. These genes are apparently required for intracolonial hyphal cell fusion without being essential for CAT formation. These were NCU00528, NCU01475, NCU01545, NCU02110, NCU04459, NCU08566, and NCU09142 (see Table S1 in the supplemental material). Two of these genes, ham-4 (NCU00528) and nox-1 (NCU02110), had been identified previously as required for hyphal cell fusion (12). Another three of these genes, ro-6 (NCU09142; dynein intermediate chain 2), ro-11 (NCU08566; NudE homolog needed for nuclear distribution), and atg-8 (NCU01545; homolog of the yeast Saccharomyces cerevisiae autophagy protein 8, a subunit of microtubule-associated proteins 1A and 1B), are likely to be needed for the formation and functioning of the cytoskeleton. The phenotypes of the ham-4 (NCU00528), atg-8 (NCU01545), nox-1 (NCU02110), ro-11 (NCU08566), and ro-6 (NCU9142) mutants have been verified by complementation and RIP experiments (see Table S1 in the supplemental material).
Table 1.
Transcription factors needed for cell fusion
| Gene name (locus no.)a | Mutant characteristic |
Verification by complementation exptb | ||
|---|---|---|---|---|
| CAT formation | Growth (mm/h) | Phenotypec | ||
| Wild type | Yes | 2.46 | Wild type | |
| adv-1 (NCU07392)* | No | 0.23 | Fc Pd | No |
| ada-3 (NCU02896)* | No | 0.27 | Fc Pd | No |
| rco-1 (NCU06205) | No | 0.04 | Fc Pd aconidial | Verified by RIP mutation† |
| snf5 (NCU00421)* | No | 0.19 | Fc Pd | No |
Asterisks mark genes newly identified by the screening procedure as required for cell fusion.
A dagger indicates that the gene was identified previously as required for cell fusion.
Fc, flat conidiation phenotype; Pd, delayed or absent protoperithecium development.
Table 4.
Cell fusion genes that might function in mediating membrane fusion
| Gene name (locus no.)a | Mutant characteristic |
Verification by complementation exptb | Predicted function of protein | ||
|---|---|---|---|---|---|
| CAT formation | Growth (mm/h) | Phenotypec | |||
| ham-6 (NCU02767)* | No | 0.92 | Fc Pd | Yes** | Membrane fusion |
| ham-7 (NCU00881)* | No | 0.25 | Fc Pd | Yes** | GPI anchor attachment |
| ham-8 (NCU02811)* | No | 0.83 | Fc Pd | Yes** | Membrane fusion |
Asterisks mark genes newly identified by the screening procedure as required for cell fusion.
Double asterisks mark complementation experiments carried out in conjunction with this study.
Fc, flat conidiation phenotype; Pd, delayed or absent protoperithecium development.
Complementation experiments.
Twelve genes that had been identified previously as required for cell fusion were identified in our screening of the library (Tables 1 to 4). For many of the newly identified mutants, complementation experiments were carried out to demonstrate that a wild-type copy of the gene restored the wild-type phenotype to the mutants. Complementation was taken as definitive proof that the deletion was responsible for conferring the mutant phenotype. PCR-amplified wild-type copies of the genes with 1,500 bp of genomic DNA upstream of the coding region and 500 bp of genomic DNA downstream of the coding region were cloned into a gene-targeting vector and were used to transform the deletion mutants as described in Materials and Methods. A listing of the genes cloned and the PCR primers used in cloning these genes is found in Table S2 in the supplemental material. As examples of these complementation experiments, Fig. 3 shows images of the complementation of the ham-6, ham-7, ham-8, and amph-1 mutants with wild-type copies of these genes as assessed by the conidiation pattern phenotype. The CAT fusion defect and the protoperithecium formation phenotype were also complemented by transformation with the wild-type copy of the gene. During the complementation assays, we found that the wild-type copies of 4 of the genes (NCU00311, NCU02062, NCU03306, and NCU09263), representing 7 of the mutants, did not complement the mutant phenotypes. A flow chart of the protocol used in isolating the mutants is given in Fig. 4, and a listing of all of the putative mutants examined in the protocol is provided in Table S1 in the supplemental material.
Fig. 3.
Complementation of cell fusion gene deletion mutations with wild-type copies of cell fusion genes. (Top) Conidiation patterns of the ham-6, ham-7, ham-8, and amph-1 mutants grown for 72 h on Vogel's sucrose medium. (Bottom) Transformants of these mutants (ham-6T, ham-7T, ham-8T, and amph-1T) in which a wild-type copy of the cell fusion gene complements the mutant conidiation pattern phenotype.
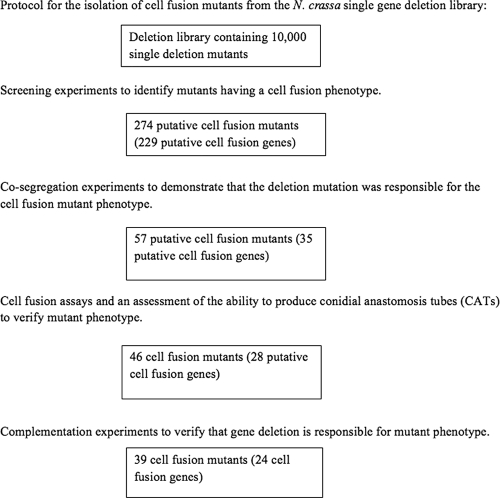
Fig. 4.
Protocol used to identify cell fusion genes. The four steps in the protocol used to identify genes required for cell fusion are shown. The first step involved screening each of the 10,000 mutants in the single gene deletion library to identify those with a cell fusion phenotype. The second step involved mating each of the putative mutants with a wild-type isolate to demonstrate that the mutant phenotype cosegregated with the deletion mutation. The third step involved assessing whether the mutants were able to generate CATs. In the final step, complementation experiments were carried out to demonstrate that the deletion mutation was responsible for the mutant phenotype.
RIP experiments.
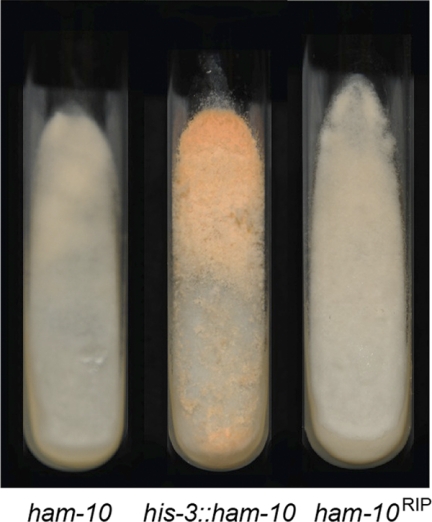
The ham-10 (NCU02833) mutant does not make macroconidia (Table 3), the cell type used in the transformation experiment for complementation analysis, and we were therefore unable to demonstrate by complementation analysis that ham-10 is required for cell fusion. RIP is a phenomenon in which multiple mutations are induced in genes that are found in two or more copies in the N. crassa haploid genome (46). As described in Materials and Methods, we used a RIP experiment to create mutations in the ham-10 gene, and we assessed whether mutations in ham-10 give rise to the mutant phenotype. We found that 12% of the progeny we obtained from mating of a ham-10 transformant (with a transforming copy of ham-10 inserted downstream of the his-3 locus and an endogenous copy of the ham-10 gene) with a his-3 mutant isolate had the cell fusion-defective phenotype characteristic of the ham-10 mutant. An image of the ham-10 deletion mutant and one of the ham-10RIP mutants is found in Fig. 5. We sequenced the ham-10RIP gene from three of the mutant his-3 progeny with the mutant phenotype, and we found that the genes contained 61, 45, and 55 mutations within the coding region. All three mutant genes had stop codon mutations near the beginning of the coding region. We conclude that mutations in the ham-10 gene give rise to a cell fusion-defective phenotype.
Table 3.
Known and putative vesicular trafficking genes required for cell fusion
| Gene name (locus no.)a | Mutant characteristic |
Verification by complementation exptb | ||
|---|---|---|---|---|
| CAT formation | Growth (mm/h) | Phenotypec | ||
| amph-1 (NCU01069)* | No | 0.58 | Fc Pd difficulty in completing conidiation | Yes** |
| ham-10 (NCU02833)* | No | 0.27 | Fc Pd sickly aconidial (microconidia made) | Verified by RIP mutation** |
| pkr1 (NCU00506)* | No | 0.54 | Fc Pd sickly aconidial (microconidia only) | Yes** |
Asterisks mark genes newly identified by the screening procedure as required for cell fusion.
Double asterisks mark complementation and RIP experiments carried out in conjunction with this study.
Fc, flat conidiation phenotype; Pd, delayed or absent protoperithecium development.
Fig. 5.
Mutations in the ham-10 gene give rise to a cell fusion phenotype. To demonstrate that ham-10 is a cell fusion gene, RIP mutants with mutations of ham-10 were generated. Shown are photographs of slants containing the ham-10 deletion mutant, a transformant containing a copy of the ham-10 gene at the his-3 locus (his-3::ham-10), and a ham-10 RIP mutant (ham-10RIP) that was obtained as progeny from the transformant.
Mutant characterization.
Experiments examining the morphological characteristics and linear growth rates of each of the mutants were carried out as described in Materials and Methods. Information about each of the mutants we identified and the gene deletion responsible for the mutant phenotype can be found in Tables 1 through 4. The 24 cell fusion genes we identified are organized into the tables according to their likely functions, but these functions should be considered provisional.
In Table 1, we list 4 transcription factors that we identified as required for cell fusion. Among the transcription factors listed in Table 1, RCO-1 (NCU06205) has been identified previously as necessary for cell fusion (1). RCO-1 and RCM-1 are the N. crassa homologs of the yeast TUP-1 and SSN6 proteins, which in yeast form a heterodimer that functions in regulating cell growth (23, 31, 40, 52). Two of the other transcription factor genes listed in Table 1, ada-3 (NCU002896) and adv-1 (NCU07392), have been identified previously as required for normal sexual and asexual development in N. crassa (7). The Sordaria macrospora PRO1 gene, which has been defined as a major transcription factor regulating female development, is homologous to the adv-1 gene (32). Our data suggest that these genes regulate the synthesis of proteins needed for cell fusion. Mutations in the ada-1 (NCU00499) and ada-2 (NCU02017) transcription factors have been shown to give rise to a defect in protoperithecium formation and conidiation (7), but we found that cell fusion is normal in these mutants. We also identified the snf5 gene (NCU00421), which is homologous to yeast SNF5, as required for cell fusion. In yeast, Snf5p has been characterized as a transcription activator that coordinates the assembly and activity of the Snf/Swi complex in remodeling chromatin structure and affects the expression of a variety of genes, including those regulated by glucose repression (16, 25).
The largest group of cell fusion genes identified in our screening contained 14 genes that are likely to be involved in signal transduction pathways regulating the directed growth of the CATs and fusion hyphae toward fusion partners (Table 2). These genes include 11 genes that had been identified previously as required for cell fusion. The genes in two MAP kinase signal transduction pathways, the mik-1 (NCU02234)/mek-1 (NCU06419)/mak-1 (NCU11376) “cell wall integrity” pathway and the nrc-1 (NCU06182)/mek-2 (NCU04612)/mak-2 (NCU02393) pathway, have been identified previously as necessary for cell fusion (12, 17, 38). The so/ham-1 gene encodes an intracellular protein that associates with and dissociates from the plasma membrane at the tip of a growing CAT and is thought to function in regulating MAK-2 activity (10, 13). The ham-2 gene encodes a 4-transmembrane-domain protein that is a homolog of the Far11p protein from Saccharomyces cerevisiae (51). The ham-3 gene (NCU08741) encodes a homolog of yeast Far8p, which is found in association with Far11p (12, 47). The mob3 gene encodes a homolog of mammalian phocein and has been shown previously to be necessary for cell fusion (28). The ham-5 gene encodes a cytoplasmic protein that has been found to be necessary for cell fusion (1). The pp2A gene (NCU06563) encodes the protein phosphatase 2A protein, which has been demonstrated to modulate a number of different signal transduction pathways, including the MAP kinase pathway, by dephosphorylation of the component kinases (21). As part of our screening, we identified the rac-1 gene (NCU02160), which has also been reported to be part of the Neurospora MAP kinase pathway (45). Based on their known activities, the pp2A and rac-1 genes would have been predicted to be required for cell fusion.
Table 2.
Known and putative signal transduction pathway genes required for cell fusion
| Gene name (locus no.)a | Mutant characteristic |
Verification by complementation exptb | Predicted function of proteinc | ||
|---|---|---|---|---|---|
| CAT formation | Growth (mm/h) | Phenotyped | |||
| mik-1 (NCU02234) | No | 0.33 | Fc Pd sickly microconidial | Yes† | MAPKK kinase |
| mek-1 (NCU06419) | No | 0.08 | Fc Pd sickly | Yes† | MAPK kinase |
| mak-1 (NCU11376) | No | 0.25 | Fc Pd sickly microconidial | Yes† | MAP kinase |
| nrc-1 (NCU06182) | No | 0.21 | Fc Pd sickly | Yes† | MAPKK kinase |
| mek-2 (NCU04612) | No | 1.14 | Fc Pd sickly | Yes† | MAPK kinase |
| mak-2 (NCU02393) | No | 0.21 | Fc Pd sickly | Yes† | MAP kinase |
| rac-1 (NCU02160)* | No | 0.01 | Colonial growth Pd | No | MAPK pathway |
| pp2A (NCU06563)* | No | 0.58 | Fc Pd | No | MAPK pathway |
| so/ham-1 (NCU02784) | No | 0.83 | Fc Pd | Yes† | Signaling activity |
| ham-2 (NCU03727) | No | 0.38 | Fc Pd | Yes† | Signaling pathway |
| ham-3 (NCU08741) | No | 0.17 | Fc late protoperithecia | No† | Signaling pathway |
| ham-5 (NCU01789) | No | 0.83 | Fc Pd | No† | MAPK pathway |
| mob3 (NCU07674) | No | 0.42 | Fc Pd | Yes†** | Signaling pathway |
| ham-9 (NCU07389)* | No | 1.82 | Fc Pd | Yes** | Signaling pathway |
Asterisks mark genes newly identified by the screening procedure as required for cell fusion.
Daggers mark genes previously identified as required for cell fusion. Double asterisks mark complementation experiments carried out in conjunction with this study.
MAPK, MAP kinase; MAPKK, MAPK kinase.
Fc, flat conidiation phenotype; Pd, delayed or absent protoperithecium development.
In keeping with the system of naming cell fusion mutants used by Wilson and Dempsey (50), we have opted to use the ham designation (hyphal anastomosis) for the new cell fusion genes we identified unless a homolog had been identified and characterized in a different system. We identified the ham-9 gene (NCU07389), which encodes a cytoplasmic protein with pleckstrin homology (PH) and sterile alpha motif (SAM) domains, as being required for cell fusion. These domains have been found previously to function in protein-protein interactions (39, 44).
Our screening protocol identified three genes encoding proteins needed for normal vesicular trafficking (Table 3). We hypothesize that the proteins are required for the exocytosis and endocytosis of vesicles at the growing tip of the fusion hypha. Mutations in the amph-1 (NCU01069) (see Fig. 1 and 3) and ham-10 (NCU02833) genes affect CAT formation, and the proteins encoded may be associated with the actin cytoskeleton, which has been shown to be required for CAT formation (43). The amph-1 gene is homologous to the S. cerevisiae RVS161 and RVS167 (amphiphysin) genes, known to be required for cell fusion events in yeast (14, 15). The ham-10 gene encodes a C2 domain-containing protein. C2 domains are thought to interact with membranes and are found in proteins such as synaptotagmin (48). C2 domain-containing proteins are likely to be involved in vesicular trafficking (48). The ham-10 mutant is unable to produce macroconidia, suggesting that the HAM-10 protein is required for conidial development as well as for cell fusion events. We also identified the pkr1 gene (NCU00506) as required for cell fusion. The encoded PKR1 is a 167-amino-acid protein, and homologs are found in other fungi. The yeast homolog, PKR1, has been shown to encode an assembly factor needed for the formation of the V-type ATPase and has been localized to the endoplasmic reticulum (ER) membrane (9). The pkr1 mutant is defective in making macroconidia (but does make microconidia) and has a spreading colonial growth pattern. The pkr1 mutant is also defective in producing protoperithecia (Table 3). Transformation of the mutant with a wild-type copy of pkr1 complements all of the phenotypic characteristics. All of these vesicular trafficking genes were identified as necessary for cell fusion in N. crassa for the first time by the screening experiments.
As shown in Table 4, we identified three genes, ham-6, ham-7, and ham-8 (Fig. 1 and 3), that encode proteins that might mediate the fusion between the plasma membranes at the tips of the fusion hyphae. All three of these genes are highly conserved in the genomes of filamentous fungi but not in other organisms. The ham-6 gene (NCU02767) encodes a small, 145-amino-acid protein with a putative signal peptide. This protein is rather hydrophobic and is the most likely candidate for a fusion protein among the proteins we have identified in our screening. The ham-7 gene (NCU00881) encodes a 230-amino-acid protein with a putative N-terminal signal peptide and a putative glycosylphosphatidylinositol (GPI) anchor attachment signal on its C terminus. HAM-7 is likely to be associated with the plasma membrane and cell wall space. The HAM-7 protein is the only protein we identified that is predicted to have an “extracellular” location. The ham-8 gene (NCU02811) encodes a 527-amino-acid protein with 4 putative transmembrane domains. As a transmembrane protein, it could function in mediating the cell fusion process or could participate in a signal transduction pathway, either as an active participant in the pathway or as an anchor to which pathway elements could bind and become localized to the plasma membrane.
DISCUSSION
The availability of a single gene deletion library greatly facilitates the identification of genes required for growth, morphology, and development in N. crassa. In this report, we used a morphology-based screening protocol to identify 24 genes that are required for hyphal cell fusion. In using the library, we found that it was important to use cosegregation and complementation analyses to verify that the deletion mutation was responsible for the mutant phenotype, since additional mutations exist in many of the mutants. These additional mutations might have arisen during the transformation events used to create the deletion mutations in the mus-51 and mus-52 strains or during the mating of these transformants to generate the single-ascospore isolates found in the library. The mus-51 and mus-52 strains were used in creating the deletion mutations because they facilitate homologous recombination events, but they are defective in elements of DNA damage repair (33). Previous work with N. crassa has shown that cell fusion mutants often arise spontaneously (22), and this phenomenon may have contributed to the large number of cell fusion mutants we obtained in our screening procedure that were not associated with a deletion mutation. Our screening procedure was helpful in identifying easily observed morphological mutants but is not meant to supplant the more detailed and extensive analysis being carried out by the Neurospora genome project. The detailed screening will be helpful in identifying many more mutant phenotypes than the rapid screening we used to identify cell fusion mutants.
The cell fusion genes we identified can be roughly divided into 4 groups, as defined by the known and/or hypothetical functions of the encoded proteins. As given in Table 1, four transcription factor genes (adv-1, ada-3, snf5, and rco-1) were found to be required for cell fusion. Our results suggest that ADV-1 and ADA-3 might be required for the transcription of cell fusion genes and that the lack of cell fusion is likely to be a contributing factor in the observed developmental phenotypes of adv-1 and ada-3 mutants.
The involvement of the MAK-1 and MAK-2 signal transduction pathways in regulating N. crassa growth and development is well established, and the two pathways are known to regulate the formation and growth of CATs (12, 17, 27, 36, 38). We identified all of the kinase elements in the MAK-1 pathway (mik-1, mek-1, and mak-1) and in the MAK-2 pathway (nrc-1, mek-2, and mak-2) among the genes defined by our cell fusion mutants (Table 2). Our screening also identified the rac-1 and pp2A genes, whose protein products have been shown previously to function in MAP kinase pathways (12, 45). In addition to these genes, we also identified the so/ham-1, ham-2, ham-3, mob3, and ham-5 genes, which have previously been shown to be cell fusion genes (1, 10, 12, 28, 50, 51). The elegant work of Fleissner et al. (13) demonstrated that SO/HAM-1 and MAK-2 are recruited to the tips of fusion hyphae in an oscillatory manner. This recruitment occurs in a “ping pong” manner between two fusion hyphae as they grow toward each other. One of the fusion hyphae will have SO/HAM-1 associated with the plasma membrane at the tip of the hypha, and the other hypha will have MAK-2 associated with the plasma membrane at its tip. Within each of the hyphae, the recruitment of SO/HAM-1 oscillates every 6 to 12 min with the recruitment of MAK-2 (13, 37). The SO/HAM-1 protein has been suggested to function in regulating the recruitment of MAK-2 to the hyphal tip. HAM-5 is a homolog of the Podospora anserina IDC1 protein and is thought to function in directing the import of MAP kinases into the nucleus (1, 20).
HAM-2, HAM-3, and MOB3 are likely to be components of a complex that functions as part of a signal transduction pathway during hyphal cell fusion. HAM-2 was identified as a highly conserved putative transmembrane protein that was required for hyphal fusion (51). HAM-2 is a homolog of yeast Far11p, which has been shown to regulate the cell cycle during cell fusion events in mating yeast. The S. macrospora homolog of HAM-2, PRO22, is required for protoperithecium development and has been shown to be localized to the vesicular vacuolar network near the tips of the growing hyphae (5). HAM-3 is a homolog of striatin and Far8p and is a known cell fusion protein (12, 47). In yeast, Far8p is found in association with Far11p as part of a signaling complex. The striatin proteins contain a calmodulin-binding domain, a coiled-coil region, a WD repeat domain, and a caveolin-binding domain and are thought to function in linking signal transduction to vesicular trafficking within neurons (3). In Aspergillus nidulans, striatin has been shown to be needed for sexual development and to be localized to the ER (49). MOB3 is a homolog of phocein, a striatin binding protein found in neurons, where it has been suggested to function with striatin in signal transduction (2). The S. macrospora MOB3 homolog, SmMOB3, has been shown to be required for cell fusion and female development (4). These functions for Far11p, Far8p/striatin, and phocein suggest that the HAM-2, HAM-3, and MOB3 proteins may be part of a signal transduction system that regulates vesicular trafficking during cell fusion.
We identified a new putative signal transduction gene, ham-9, which we tentatively assigned to the signal transduction group of cell fusion genes. The ham-9 gene was assigned to this group because the encoded 869-amino-acid protein has a pleckstrin homology (PH) domain and a sterile alpha motif (SAM) domain. These domains are often found mediating protein-protein interactions between elements of signal transduction pathways. The SAM domain has been identified in the yeast MAP kinase pathway protein Ste11p and more than 60 other proteins, many of which function in signal transduction pathways (44). A conserved tyrosine within the domain is likely to be a site at which SH-2 domain-containing proteins interact with the SAM domain (44). PH domains have been shown to interact with phosphatidylinositols, heterotrimeric G proteins, and protein kinases and could function to localize HAM-9 to the membrane along with a number of other cell fusion proteins (39). They are thought to play a role in targeting signal transduction proteins to the plasma membrane and in signal transduction events (39). Further experimentation will be required to determine how HAM-9 functions during hyphal cell fusion. The ham-9 gene is highly conserved within the filamentous fungi.
We identified three cell fusion genes that we have placed in a vesicular trafficking group. Vesicular trafficking is required for the delivery of those proteins needed for remodeling the cell wall and for plasma membrane fusion to the tips of the hyphae. The amph-1 (NCU01069) gene encodes an amphiphysin protein that is a close homolog of the yeast Rvs161p and Rvs167p proteins and of vertebrate amphiphysin. The protein encoded by amph-1 has a Bin-amphyphysin-Rvs (BAR) domain. BAR domains are able to interact with lipid bilayers, and BAR domain proteins function in the formation of transport and endocytic vesicles (53). The S. cerevisiae RVS161 and RVS167 genes are required for cell fusion between yeast mating cells (15). Rvs161p and Rvs167p function as a dimer in the endocytosis of vesicles from the plasma membranes of mating cells (14, 26). N. crassa AMPH-1 could function in a similar manner in the endocytosis of vesicles from the tips of fusion hyphae.
The ham-10 (NCU02833) gene encodes a 1,422-amino-acid protein with a putative C2 domain. C2 domains are thought to function as calcium-dependent lipid binding domains and are found in proteins involved in vesicular trafficking, exocytosis, or signal transduction (48). HAM-10 could function to mediate intracellular events in response to calcium. The identification of HAM-10, whose activity is likely to be regulated by calcium levels, suggests that calcium signaling could play a role in CAT growth and cell fusion. The role of calcium signaling in exocytosis is well established, and our results suggest that calcium signaling is required for cell fusion. We have placed the pkr1 gene, which encodes a small protein that functions in the assembly of the V-type ATPase (9), in the vesicular trafficking group of genes because acidification of the vesicular elements in the secretory pathway is required for the trafficking of proteins along the pathway. The PKR1, AMPH-1, and HAM-10 proteins are predicted to disrupt different steps in the process of vesicular trafficking, and their identification in our screening protocol highlights the need for normal vesicular trafficking during CAT formation and growth.
The proteins encoded by the ham-6, ham-7, and ham-8 genes have some interesting properties. These three proteins have characteristics that suggest they could function in mediating the process of plasma membrane fusion. All three of these genes are highly conserved in the genomes of the filamentous fungi. The ham-7 gene encodes a 230-amino-acid protein with an N-terminal signal sequence and a C-terminal signal for the addition of a GPI anchor. This suggests that HAM-7 would be found on the extracellular side of the plasma membrane and would be anchored to the membrane through the GPI anchor. Given its probable location, HAM-7 might be involved in interactions between the two fusion hyphae. It could function in the adhesion process, as part of the apparatus that mediates fusion between the two plasma membranes, or as part of a signal transduction pathway. Currently, HAM-7 is the only known cell fusion protein predicted to have an extracellular location. The ham-6 gene encodes a small, 145-amino-acid protein. The TMPRED protein topography prediction program (19) suggests that HAM-6 has three transmembrane domains, but inspection of the amino acid sequence shows that the entire protein is quite hydrophobic. The hydrophobic nature of HAM-6 makes it the most likely candidate for a “fusion protein” that could mediate fusion between the opposing plasma membranes among the cell fusion proteins we have identified in N. crassa. The HAM-6 protein is a homolog of S. macrospora PRO41, which has been shown to be a membrane protein required for protoperithecium production (34). Based on experiments using a green fluorescent protein (GFP)-tagged version of PRO41, Nowrousian et al. (34) suggested that the protein is localized to the ER, but because the GFP tag was cleaved from the chimeric protein in the localization experiment, the location of HAM-6 remains open to question. The ham-8 gene encodes a 597-amino-acid protein with four predicted transmembrane domains. The HAM-8 protein could function in the plasma membrane to facilitate cell fusion, or it could function as part of a signal transduction pathway. A diagram indicating how these different cell fusion proteins might be functioning and their likely cellular locations is given in Fig. 6. Additional experiments will be required to demonstrate clearly how the various proteins interact and function during cell fusion events.
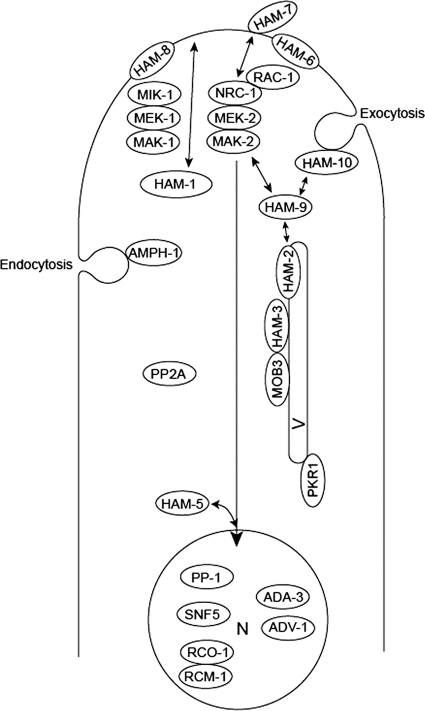
Fig. 6.
Schematic representation of the functions proposed for cell fusion proteins. The transcription factor PP-1 is a target for phosphorylation by MAK-2. PP-1, along with the other transcription factors identified (ADV-1, ADA-3, SNF5, and the dimeric RCO-1/RCM-1), is proposed to reside in the nucleus (N) and to direct the transcription of the cell fusion genes. HAM-5 is proposed to function in directing MAK-2 into the nucleus, where it phosphorylates PP-1 and perhaps some of the other transcription factors. In addition to regulating transcriptional activity, the two MAP kinase signaling pathways (NRC-1/MEK-2/MAK-2 and MIK-1/MEK-1/MAK-1) also regulate CAT formation and growth by controlling vesicular trafficking to and from the hyphal tip. The two MAP kinase pathways are activated by cell-to-cell signaling, and the plasma membrane-associated HAM-6, HAM-7, and HAM-8 proteins might function as receptors for the signaling factor(s) or might be directly involved in fusion between the plasma membranes. As part of the cell-to-cell signaling process, the HAM-1/SO protein and the RAC-1/NRC-1/MEK-2/MAK-2 complex transiently associate with the plasma membrane in an oscillatory “ping pong” manner. The PP2A phosphatase might function in dephosphorylating the MAP kinase pathway proteins. The HAM-2/HAM-3/MOB3 complex is located on an intracellular vesicle (V) and is proposed to regulate vesicular trafficking to the hyphal tip. We propose that the HAM-2/HAM-3/MOB3 complex could receive input from the MAP kinase pathways and that the HAM-9 protein could function to connect the HAM-2/HAM-3/MOB3 complex to the MAP kinase pathways. AMPH-1 is proposed to play roles in mediating the formation of endosomes and to be necessary to maintain a balance between exocytosis and endocytosis of vesicles during CAT growth. The HAM-10 protein may function in regulating the movement of vesicles at the hyphal tip. The PKR1 protein is found in the ER and is necessary for efficient vesicular trafficking in the secretory pathway.
An examination of the proteins we identified as required for cell fusion shows that we failed to identify any proteins that function during the cell wall remodeling that must take place as part of the fusion process. Inspection of the N. crassa genome shows that there are more than 200 genes in the genome that might be functioning during cell wall biogenesis and remodeling (secreted glycosyltransferases, glucanases, chitinases, glucan synthases, and chitin synthases). A likely explanation for the fact that we did not identify cell wall-remodeling proteins in our screening is that several proteins participate in cell wall remodeling and that there is a great deal of functional redundancy among these proteins.
There are three reasons why the list of 24 genes given in Tables 1 through 4 is an incomplete listing of cell fusion genes. First, the 110 plates of the deletion library do not cover the entire Neurospora genome, and there may be some additional cell fusion genes in the portion of the genome not covered by the screening experiments. For example, the rcm-1 (NCU06842) gene is known to be required for cell fusion (1), but the deletion mutant for this gene is not found in the first 110 plates of the library. Second, some of the deletion mutants in the library are found as heterokaryons, isolates that contain both transformed and nontransformed nuclei. These heterokaryons have wild-type morphology, and our screening procedure would miss cell fusion mutants found as heterokaryons in the library. For example, the PP-1 transcription factor (NCU00340) is activated by the MAK-2 signal transduction pathway and has been shown previously to be required for cell fusion (12), but the mutant is found as a heterokaryon in the library and was not identified in our experiments. Third, our screening protocol may have missed some mutants. However, deletion mutants for most of the genes in the Neurospora genome are found in the library, and the fact that we picked up the known available cell fusion genes demonstrates that the screening procedure was effective.
Supplementary Material
ACKNOWLEDGMENTS
Funding for these studies was provided by grants R01GM078589 and 3R01GM078589-04S1 from the National Institutes of Health.
We thank the Neurospora Genome Project for the creation of the single gene deletion library and the Fungal Genetics Stock Center for making the library available. We are grateful to Jason W. Arnold and Jennifer Chinnici for assistance in screening the Neurospora single-gene-deletion library. We thank James Stamos and Alan Siegel for help in manuscript preparation.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Aldabbous M. S., et al. 2010. The ham-5, rcm-1 and rco-1 genes regulate hyphal fusion in Neurospora crassa. Microbiology 156:2621–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baillat G., et al. 2001. Molecular cloning and characterization of Phocein, a protein found from the Golgi complex to dendritic spines. Mol. Biol. Cell 12:663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benoist M., Gaillard S., Castets F. 2006. The striatin family: a new signaling platform in dendritic spines. J. Physiol. Paris 99:146–153 [DOI] [PubMed] [Google Scholar]
- 4. Bernhards Y., Poggeler S. 2011. The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora. Curr. Genet. 57:133–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bloemendal S., et al. 2010. A mutant defective in sexual development produces aseptate ascogonia. Eukaryot. Cell 9:1856–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buller A. H. R. 1933. Researches on fungi, vol. 15. Longman, London, England [Google Scholar]
- 7. Colot H. V., et al. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 103:10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis R. H., DeSerres F. J. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 27:79–143 [Google Scholar]
- 9. Davis-Kaplan S. R., et al. 2006. PKR1 encodes an assembly factor for the yeast V-type ATPase. J. Biol. Chem. 281:32025–32035 [DOI] [PubMed] [Google Scholar]
- 10. Fleissner A., et al. 2005. The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot. Cell 4:920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fleissner A., Glass N. L. 2007. SO, a protein involved in hyphal fusion in Neurospora crassa, localizes to septal plugs. Eukaryot. Cell 6:84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fleissner A., Simonin A. R., Glass N. L. 2008. Cell fusion in the filamentous fungus, Neurospora crassa. Methods Mol. Biol. 475:21–38 [DOI] [PubMed] [Google Scholar]
- 13. Fleissner A., Leeder A. C., Roca M. G., Read N. D., Glass N. L. 2009. Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 106:19387–19392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friesen H., et al. 2006. Characterization of the yeast amphiphysins Rvs161p and Rvs167p reveals roles for the Rvs heterodimer in vivo. Mol. Biol. Cell 17:1306–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gammie A. E., Brizzio V., Rose M. D. 1998. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 9:1395–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geng F., Cao Y., Laurent B. C. 2001. Essential roles of Snf5p in Snf-Swi chromatin remodeling in vivo. Mol. Cell. Biol. 21:4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glass N. L., Jacobson D. J., Shiu P. K. T. 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34:165–186 [DOI] [PubMed] [Google Scholar]
- 18. Hickey P. C., Jacobson D. J., Read N. D., Glass N. L. 2002. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 37:109–119 [DOI] [PubMed] [Google Scholar]
- 19. Hofmann K., Stoffel W. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166 [Google Scholar]
- 20. Jamet-Vierny C., Debuchy R., Prigent M., Silar P. 2007. IDC1, a pezizomycotina-specific gene that belongs to the PaMpk1 MAP kinase transduction cascade of the filamentous fungus Podospora anserina. Fungal Genet. Biol. 44:1219–1230 [DOI] [PubMed] [Google Scholar]
- 21. Janssens V., Goris J. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signaling. Biochem. J. 353:417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kafer E. 1982. Improved backcrossed strains giving consistent map distances. Neurospora Newsl. 29:41–44 [Google Scholar]
- 23. Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68:709–719 [DOI] [PubMed] [Google Scholar]
- 24. Koehler E. 1930. Zur Kenntnis der vegetativen anastomosen der Pilze (II. Mitteilung). Planta 10:495–522 [Google Scholar]
- 25. Laurent B. C., Treitel M. A., Carlson M. 1990. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol. Cell. Biol. 10:5616–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lombardi R., Riezman H. 2001. Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo. J. Biol. Chem. 276:6016–6022 [DOI] [PubMed] [Google Scholar]
- 27. Maerz S., et al. 2008. The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa. Genetics 179:1313–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maerz S., et al. 2009. Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa. Mol. Microbiol. 74:707–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maniatis T., Sambrook J., Fritsch E. F. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Margolin B. S., Freitag M., Selker E. U. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44:34–36 [Google Scholar]
- 31. Marquez J. A., Pascual-Ahuir A., Proft M., Serrano R. 1998. The Ssn6-Tup1 repressor complex of Saccharomyces cerevisiae is involved in the osmotic induction of HOG-dependent and -independent genes. EMBO J. 17:2543–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Masloff S., Poggeler S., Kuck U. 1999. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152:191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ninomiya Y., Suzuki K., Ishii C., Inoue H. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101:12248–12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nowrousian M., et al. 2007. The novel ER membrane protein PRO41 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol. Microbiol. 64:923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pandey A., Roca M. G., Read N. D., Glass N. L. 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3:348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park G., Pan S., Borkovich K. A. 2008. Mitogen-activated protein kinase cascade required for regulation of development and secondary metabolism in Neurospora crassa. Eukaryot. Cell 7:2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Read N. D., Lichius A., Shoji J., Goryachev A. B. 2009. Self-signaling and self-fusion in filamentous fungi. Curr. Opin. Microbiol. 12:608–615 [DOI] [PubMed] [Google Scholar]
- 38. Read N. D., Fleissner A., Roca M. G., Glass N. L. 2010. Hyphal fusion, p. 260–273 In Borkovich K. A., Ebbole D. J. (ed.), Cellular and molecular biology of filamentous fungi. ASM Press, Washington, DC [Google Scholar]
- 39. Rebecchi M. J., Scarlata S. 1998. Pleckstrin homology domains: a common fold with diverse functions. Annu. Rev. Biophys. Biomol. Struct. 27:503–528 [DOI] [PubMed] [Google Scholar]
- 40. Redd M. J., Arnaud M. B., Johnson A. D. 1997. A complex composed of tup1 and ssn6 represses transcription in vitro. J. Biol. Chem. 272:11193–11197 [DOI] [PubMed] [Google Scholar]
- 41. Roca M. G., Read N. D., Wheals A. E. 2005. Conidial anastomosis tubes in filamentous fungi. FEMS Microbiol. Lett. 249:191–198 [DOI] [PubMed] [Google Scholar]
- 42. Roca M. G., Arlt J., Jeffree J. E., Read N. D. 2005. Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot. Cell 4:911–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roca M. G., Kuo H.-C., Lichius A., Freitag M., Read N. D. 2010. Nuclear dynamics, mitosis, and the cytoskeleton during the early stages of colony initiation in Neurospora crassa. Eukaryot. Cell 9:1171–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schultz J., Ponting C. P., Hofmann K., Bork P. 1997. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 6:249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seiler S., Plamann M. 2003. The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14:4352–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Selker E. U. 1999. Gene silencing: repeats that count. Cell 97:157–160 [DOI] [PubMed] [Google Scholar]
- 47. Simonin A. R., Rasmussen C. G., Yang M., Glass N. L. 2010. Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 47:855–868 [DOI] [PubMed] [Google Scholar]
- 48. Sutton R. B., Davletov B. A., Berghuis A. M., Sudhof T. C., Sprang S. R. 1995. Structure of the first C2 domain of synaptotagmin I; a novel Ca2+/phospholipid-binding fold. Cell 80:929–938 [DOI] [PubMed] [Google Scholar]
- 49. Wang C.-L., Shim W.-B., Shaw B. D. 2010. Aspergillus nidulans striatin (StrA) mediates sexual development and localizes to the endoplasmic reticulum. Fungal Genet. Biol. 47:789–799 [DOI] [PubMed] [Google Scholar]
- 50. Wilson J. F., Dempsey J. A. 1999. A hyphal fusion mutant in Neurospora crassa. Fungal Genet. Newsl. 46:31 [Google Scholar]
- 51. Xiang Q., Rasmussen C., Glass N. L. 2002. The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160:169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamashiro C. T., et al. 1996. Characterization of rco-1 of Neurospora crassa, a pleiotropic gene affecting growth and development that encodes a homolog of Tup1 of Saccharomyces cerevisiae. Mol. Cell. Biol. 16:6218–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Youn J.-Y., et al. 2010. Dissecting BAR domain function in the yeast amphiphysin Rvs161 and Rvs167 during endocytosis. Mol. Biol. Cell 21:3054–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.