Abstract
Acetyl coenzyme A (acetyl-CoA) is a crucial metabolite for energy metabolism and biosynthetic pathways and is produced in various cellular compartments with spatial and temporal precision. Our previous study on ATP citrate lyase (ACL) in Gibberella zeae revealed that ACL-dependent acetyl-CoA production is important for histone acetylation, especially in sexual development, but is not involved in lipid synthesis. In this study, we deleted additional acetyl-CoA synthetic genes, the acetyl-CoA synthetases (ACS genes ACS1 and ACS2), to identify alternative acetyl-CoA production mechanisms for ACL. The ACS1 deletion resulted in a defect in sexual development that was mainly due to a reduction in 1-palmitoyl-2-oleoyl-3-linoleoyl-rac-glycerol production, which is required for perithecium development and maturation. Another ACS coding gene, ACS2, has accessorial functions for ACS1 and has compensatory functions for ACL as a nuclear acetyl-CoA producer. This study showed that acetate is readily generated during the entire life cycle of G. zeae and has a pivotal role in fungal metabolism. Because ACSs are components of the pyruvate-acetaldehyde-acetate pathway, this fermentation process might have crucial roles in various physiological processes for filamentous fungi.
INTRODUCTION
The homothallic ascomycete fungus Gibberella zeae (anamorph Fusarium graminearum) is a prominent plant pathogen of major cereal crops, such as wheat, barley, maize, and rice. Fungal disease caused by this fungus leads to yield loss, and the harvested grains are frequently contaminated with mycotoxins that threaten human and animal health (15, 42). This fungus reproduces both sexually and asexually, and ascospores produced by sexual reproduction are believed to play a role in fungal survival and disease initiation. Many studies have been performed to characterize the mechanisms of sexual reproduction (4, 24, 28, 32, 35, 38, 39, 47, 55, 65, 68, 72, 75). Recently, we showed that acetyl coenzyme A (acetyl-CoA) is an important metabolite for sexual development in this fungus (57).
Acetyl-CoA is a crucial metabolite in energy metabolism and the biosynthesis of many cellular components. First, mitochondrial acetyl-CoA supplied from the mitochondrion-associated pyruvate dehydrogenase complex or β-oxidation is used to replenish the mitochondrial tricarboxylic acid (TCA) cycle. Therefore, mitochondrial acetyl-CoA is essential in the production of energy sources for the survival and normal growth of eukaryotic cells (40). However, in some eukaryotes, including the protozoan parasite Trypanosoma brucei and some plants, mitochondrial acetyl-CoA is crucial for lipid biosynthesis through the conversion to acetate for either cytosolic acetyl-CoA generation or direct mitochondrial fatty acid synthesis (20, 50, 51).
Second, peroxisomal acetyl-CoA, which is mainly produced via β-oxidation, enters the glyoxylate cycle for acetyl unit utilization. Plants and several microorganisms have a glyoxylate cycle for the utilization of nonfermentable carbon sources, such as fatty acids, ethanol, and acetate. In consideration of the function of peroxisomes as places for the biosynthesis of lipids and some amino acids, it is also possible to predict that peroxisomal acetyl-CoA takes part in lipid synthesis (14). Moreover, recent studies of fungi have revealed a connection between peroxisomal acetyl-CoA and the early steps of mycotoxin production (11, 31).
Lastly, cytosolic acetyl-CoA is an essential building block for the biosynthesis of fatty acids and numerous secondary metabolites. Acetyl-CoA carboxylase mainly controls fatty acid synthesis via malonyl-CoA production from cytosolic acetyl-CoA. The mevalonate pathway, which is required for the synthesis of various metabolites including sterol, is also created from cytosolic acetyl-CoA through the composition of 3-hydroxy-3-methylglutaryl-CoA by two enzymatic reactions (29, 40). Acetyl-CoA is also used for the acetylation of lysine residues in many proteins, and this posttranslational modification has a key role in protein stability and function (67, 74). Because the nuclear envelop is permeable to acetate, citrate, and even acetyl-CoA, histone acetylation is also strictly regulated by nucleocytosolic acetyl-CoA levels and related genes (48, 57, 63, 70).
The origins of cytosolic acetyl-CoA vary and depend on the living conditions and developmental stages of organisms. In most cases, cytosolic acetyl-CoA is translocated from peroxisomes and mitochondria via specified shuttle systems. Carnitine acetyl-transferases (CATs) are known to transport acetyl groups between peroxisomes or mitochondria and the cytosol and have important roles in several metabolic pathways (6, 59, 62). Carnitine-independent acetyl group movement by peroxisomal thioesterase was also proposed to occur in mammalian cells (41). ATP citrate lyase (ACL)-dependent cytosolic acetyl-CoA generation is the most common pathway in eukaryotes and some prokaryotes (18, 25). Citrate that is exported from the TCA cycle in mitochondria through tricarboxylate carriers is cleaved into oxaloacetate and cytosolic acetyl-CoA by ACL. In the case of Saccharomyces cerevisiae and Candida albicans, however, cytosolic acetyl-CoA is synthesized through the pyruvate-acetaldehyde-acetate pathway, which is mediated by acetyl-CoA synthetases (ACSs) (10, 17, 60, 61, 63, 66). ACSs are conserved in most eukaryotes and are thought to evolve from a mitochondrial origin (34).
Because of the significant roles of acetyl-CoA, most organisms have evolved their own appropriate metabolic processes for the precise temporal (developmental stages) and spatial (organelles) generation of acetyl-CoA. One in silico survey revealed an astounding diversity of metabolic pathways, even within the fungal kingdom, and showed variant metabolic processes across fungi (54). Thus, for basic or applied studies of single species, an in-depth understanding of acetyl-CoA metabolism is important.
A previous study on the functional characterization of ACL in G. zeae revealed that both ACL subunits are essential for ACL function in glucose utilization, development, trichothecene production, and virulence. The ACL genes were not required for de novo lipid synthesis but were required for histone acetylation during sexual development and possibly during asexual growth (57). In this previous work, we had three questions: (i) Which pathways in acetyl-CoA production are required for lipid synthesis? (ii) What enzymes enable ACL mutants to survive and grow to some degree? (iii) What are the functions and functional relationships among the cytosolic acetyl-CoA generating enzymes?
In this study, we studied the two ACS genes (ACS1 and ACS2) as target candidates for addressing the first two questions. Phylogenetic analysis revealed that some fungal species, including G. zeae, have two ACS genes; however, Aspergillus nidulans and Neurospora crassa have only one ACS coding gene each, facA and acu-5, respectively (2, 13, 19, 53). We hypothesized that two ACSs are major acetyl-CoA producers for lipid synthesis. However, since acetate is not a physiological carbon source for G. zeae, we also hypothesized that acetate-producing mechanisms, such as aerobic fermentation, are important for acetyl-CoA production in this fungus.
MATERIALS AND METHODS
Fungal strains and media.
G. zeae wild-type strain GZ3639 (8) and transgenic G. zeae strains derived from this strain were used in this study. All mutant strains used in this study are listed in Table 1. Conidial suspensions of all of the derived or received strains were stored in 20% glycerol at −70°C. Minimal medium containing 5 mM agmatine (MMA) was used for trichothecene production (21). The other media used in this study were made and used according to the Fusarium laboratory manual (42).
Table 1.
G. zeae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| GZ3639 | Wild type | 8 |
| HK22 | Δacs1::gen | This study |
| HK24 | Δacs2::gen | This study |
| HK23 | Δacs1::ACS1-GFP-hyg | This study |
| HK25 | Δacs2::ACS2-GFP-hyg | This study |
| HK31 | Δacs1::gen Δacs2::gen | This study |
| Δmat2 strain | Δmat1-2::GFP-hyg | 36 |
| HK29 | Δmat1-2::GFP-hyg Δacs1::gen | This study |
| HK30 | Δmat1-2::GFP-hyg Δacs2::gen | This study |
| acl2 | Δacl2::gen | 57 |
| HK32 | Δacl2::gen Δacs1::gen | This study |
| HK33 | Δacl2::gen ACS2::hyg-Pzear-ACS2 | This study |
| Δmat1 strain | Δmat1-1::gen | 36 |
| mat1r | Δmat1-1::gen hH1-RFP-gen | 57 |
| HK34 | gen-RFP-SKL | This study |
| HK35 | Δmat1-2::GFP-hyg gen-RFP-SKL | This study |
| HK36 | Δacs1::ACS1-GFP-hyg hH1-RFP-gen | This study |
| HK37 | Δacs2::ACS2-GFP-hyg hH1-RFP-gen | This study |
| HK38 | Δacs1::ACS1-GFP-hyg gen-RFP-SKL | This study |
| HK39 | Δacs2::ACS2-GFP-hyg gen-RFP-SKL | This study |
| Pzear-GzmetE strain | GzmetE::hyg-pzear-GzmetE | 37 |
Nucleic acid manipulation and PCR primers.
Fungal genomic DNA was extracted as previously described (42), and other standard procedures for Southern and Northern hybridization with 32P-labeled probes were performed following standard protocols (52). Total RNA was extracted using an Easy-Spin Total RNA Extraction Kit (Intron Biotech, Seongnam, South Korea) following the manufacturer's instructions. PCR primers used in this study were synthesized by an oligonucleotide synthesis facility (Bionics, Seoul, South Korea) (see Table S1 in the supplemental material).
Fungal transformation for deletion and complementation.
Constructs for targeted gene deletion and green fluorescent protein (GFP)-tagged complementation were generated according to the double-joint (DJ) PCR method (73). For deletion of either the ACS1 or ACS2 genes, a Geneticin resistance cassette (gen) was amplified with primers previously designed (26) and fused with 5′ and 3′ flanking sequences for each targeted region, which was amplified with appropriate primer pairs (see Table S1 in the supplemental material).
To complement each mutant, the DNA fragment carrying the promoter and the open reading frame (ORF) of each gene was fused with green fluorescent protein and the hygromycin resistance cassette (hyg) amplified with pIGPAPA-sGFP F/HYG-F1 primers from the pIGPAPA vector (27). This construct was fused with the 3′ flanking region of each gene as previously described (57). All of the fungal transformations for gene deletion and complementation procedures were followed as previously described (24).
Lipid analysis.
Total lipid extraction and analyses were done according to a previous study (57). In brief, each strain was inoculated on plates containing 20 ml of carrot agar and harvested 5 days after inoculation. Harvested samples were dried in a ventilated hood for 5 days. The dried cultures were ground in a blender, and 1 g of powder was extracted with 8 ml of n-hexane. The extract was evaporated to dryness in a Speed Vac concentrator (Savant Instrument, Inc., Farmingdale, NY), and the total amount of lipid was weighed.
Purification and characterization of POL.
Dried cultures (300 g) were extracted with 1 liter of n-hexane three times and concentrated to dryness. The residual extract was dissolved in 30 ml of n-hexane and was applied at homogeneity by chromatography on a column (1 m by 5 cm [inside diameter]) containing silica gel 60 (70 to 230 mesh; Merck, Germany). The column was then eluted with n-hexane followed by n-hexane-ethyl acetate (10:1, vol/vol). The fractions containing 1-palmitoyl-2-oleoyl-3-linoleoyl-rac-glycerol (POL) were combined, and the purity was confirmed by thin-layer chromatography (57).
The authentic POL was purchased from Sigma-Aldrich Co. (St. Louis, MO). The chemical identification of POL was determined by using spectroscopic analyses. Atmospheric pressure chemical ionization (APCI)-mass spectrum was recorded on a quadrupole mass spectrometer equipped with an APCI source (HP 1100 series LC/MSD; Hewlett-Packard Co., Palo Alto, CA). The vaporizer was operated at 400°C, and the inlet capillary was operated at 350°C. The corona discharge needle was set to 4.0 μA. High-purity nitrogen was used for the sheath and auxiliary gases. Nebulizer pressure and drying gas flow were set to 60 lb/in2 and 4 ml/min, respectively. 1H-NMR (where NMR is nuclear magnetic resonance) spectra were recorded on a Bruker AMX500 (500 MHz) instrument (Bruker Analytische Messtechnik GmbH, Rheinstetten, Germany) in deuterochloroform with tetramethylsilane (TMS) used as an internal standard.
In order to determine the composition of fatty acids of POL, fatty acid methyl esters (FAMEs) were made by acid transmethylation according to a previously described method (12). FAMEs were analyzed by a capillary gas chromatograph-mass spectrophotometer (GC-MS) (Shimadzu GC-MS-QP5050; Shimadzu, Kyoto, Japan). The analytical conditions used were as follows: column, DB-5 fused-silica column (30 m by 0.32 mm [inside diameter], 0.25-μm film thickness (J & W Scientific, Folsom, CA); column temperature, 90°C for 5 min and then increased to 210°C at a rate of 1.5°C/min; injector temperature, 250°C (1 μl of a 10 mg/ml sample was injected).
Sexual crosses and genotyping.
For self-fertilization, mycelia grown on carrot agar for 5 days were gently removed with a glass spreader in the presence of 2.5% of sterilized Tween 60 solution to induce sexual reproduction (42). For the chemical complementation with POL, carrot agar supplemented with 0.5% (vol/vol) purified POL was used. Female strains grown for 5 days on carrot agar were spermatized with 1 ml of conidial suspension of each male strain for out-crosses (36). After sexual induction, all of the cultures were incubated under near-UV light (wavelength, 365 nm; HKiv Import and Export Co., Ltd., Xiamen, China) at 25°C.
To generate the double deletion mutant HK31 (Δacs1 Δacs2), the Δmat2 mutant was out-crossed with Δacs2 to obtain HK30 (Δmat2 Δacs2). HK30 was then out-crossed with HK22 (Δacs1) again to generate HK31 (Δacs1 Δacs2). HK29 (Δmat2 Δacs1) was selected from the Δmat2 × HK22 (Δacs1) out-cross. For double deletion between the ACS genes and ACL2, both HK29 (Δmat2 Δacs1) and HK30 (Δmat2 Δacs2) were fertilized with strain acl2 (Δacl2). Dozens of ascospores were randomly isolated from each out-cross, and the genotype of each progeny was determined by antibiotic resistance and PCR screening using the combination of primers ACS1-5F and Gen-with 5F (see Table S1 in the supplemental material).
Promoter replacement of ACS2 with Pzear in the ACL2 mutant.
To replace the ACS2 promoter with the zearalenone-inducible promoter (Pzear) in the ACL2 deletion mutant, hyg-pzear was amplified from the Pzear-GzmetE strain (where GzmetE is G. zeae metE gene) (37) with HYG-F1 and zear-r2 primers, and the 5′ and 3′ flanking regions of the ACS2 gene were amplified from GZ3639 with primers ACS2-3F/ACS2-5R pzear and ACS2-3F pzear/ACS2-3R pzear, respectively. Three fragments were fused according to the DJ PCR method (73), and the final construct was amplified with primers ACS2-5N/ACS2-3N pzear. For Pzear replacement in the ACL2 mutant, 30 μM ZEA was added to the medium during the regeneration, overlay, and mutant selection processes (37).
Mycelial growth, conidium production, germination, trichothecene analysis, and virulence test.
For mycelium growth, 1 ml of conidium suspension (1 × 105 conidia/ml) was inoculated in 50 ml of minimal medium (MM) supplemented with 2% glucose as the sole carbon source (MMG), MMG supplemented with 40 mM potassium acetate (MMGAc), or MMGAc supplemented with 30 μM ZEA (MMGAc-ZEA) and cultured for 72 h at 25°C on a rotary shaker (150 rpm). Radial growth was measured from mycelia grown for 5 days on MM supplemented with 40 mM potassium acetate as the sole carbon source (MMAc), MM supplemented with 40 mM ethanol as the sole carbon source (MMEtOH), or MM supplemented with 5 mM acetaldehyde as the sole carbon source (MMACD). Conidium production was measured by counting the number of conidia produced after incubating 10 μl of conidial suspension (1 × 105 conidia/ml) in 5 ml of carboxymethyl cellulose (CMC) medium (9) for 72 h at 25°C on a rotary shaker (150 rpm). Trichothecenes (deoxynivalenol and 15-acetyldeoxynivalenol) from MMA were analyzed with a Shimadzu QP-5050 GC-MS with a selected ion monitoring and quantified based on biomasses produced by each strain (57).
The virulence of fungal strains was determined on the wheat cultivar Eunpamil as previously described (57). In brief, 10 μl of conidial suspension (1 × 105 conidia/ml) was injected into a center spikelet of wheat head at midanthesis. Inoculated plants were placed in a greenhouse after 3 days of incubation in a humidity chamber, and spikelets with head blight symptoms were counted after 14 days.
Localization and expression of ACS1-GFP and ACS2-GFP.
To observe the localization of ACS1-GFP and ACS2-GFP with nuclei, the mat1r strain carrying histone H1 fused with red fluorescent protein (RFP) was fertilized with either HK23 (ACS1c) or HK25 (ACS2c). Ascospores carrying ACS1-GFP/hH1-RFP (HK36) and ACS2-GFP/hH1-RFP (HK37) were selected by antibiotic resistance, genotyped by PCR, and observed under a fluorescent microscope. For generation of the HK35 (Δmat2 RFP-SKL) mutant, the gen-EF (gen with elongation factor 1α promoter from Fusarium verticillioides) was amplified from the pSKGEN vector (see Fig. S1 in the supplemental material) with primers Neo-For new and EF Pro-Rev. The gen-EF was then fused to RFP-SKL amplified from the pLC25FgH4Tomloxneolox vector provided by Michael Freitag (Oregon State University, Corvallis, OR) with RFP-For EF/RFP-Rev SKL tail primers. The final round of PCR was conducted with primers Neo-For 5N and RFP-Rev SKL 3N, and the construct was then randomly introduced into the wild-type strain. HK35 (Δmat2 RFP-SKL) was then spermatized with HK23 (ACS1c) or HK25 (ACS2c) to generate HK38 (ACS1-GFP RFP-SKL) or HK39 (ACS2-GFP RFP-SKL). Mitochondria were stained with MitoTracker (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Microscopic observation was performed with a DE/Axio Imager A1 microscope (Carl Zeiss) using the filter set 38HE (excitation, 470/40 nm; emission, 525/50 nm) for GFP and the filter set 15 (excitation, 546/12 nm; emission, 590 nm) for RFP and MitoTracker.
RESULTS
Identification of ACS genes and phylogenetic analysis.
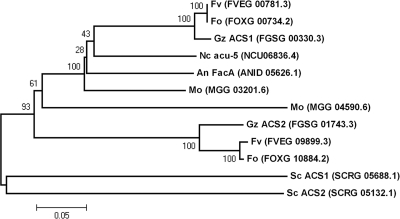
The ACS genes are named differently in many organisms, including fungal species. We identified two ACS genes in G. zeae from the Fusarium Comparative Database (http://www.broadinstitute.org/annotation/genome/fusarium_group/) using both BLASTp and InterProScan (IPR011904 family) (58). The protein sequences of ACS1 (FGSG_00330.3) and ACS2 (FGSG_01743.3) had marked identity (80 and 71%) with the protein sequence of acu-5 of N. crassa, respectively (19).
Phylogenetic relationships among the ACS homologs (IPR011904 family) were estimated using the MEGA program, version 4.0 (64). The phylogenetic tree was constructed using a neighbor-joining algorithm with bootstrap values calculated from 1,000 iterations. The Fusarium species, Magnaporthe oryzae, and S. cerevisiae had two ACSs, while A. nidulans and N. crassa had only one ACS each. One of the ACSs in Fusarium spp. and M. oryzae was grouped with FacA and acu-5, and the other was grouped outside the ACS1 group (Fig. 1).
Fig. 1.
Phylogenetic tree of acu-5/FacA homologs in several fungal species. The alignment was performed with ClustalW, and the MEGA program, version 4.0, was used to perform a 1,000-bootstrap phylogenetic analysis using the neighbor joining method. Fv, F. verticillioides; Fo, F. oxysporum; Gz, G. zeae; Nc, N. crassa; An, A. nidulans; Mo, M. oryzae; and Sc, S. cerevisiae.
Targeted gene deletion and genetic complementation.
To elucidate the functions of ACSs in G. zeae, we deleted ACS1 and ACS2 individually through homologous recombination. Each gene was successfully replaced with the gen. For genetic complementation, each ORF fused with GFP was introduced into a corresponding deletion mutant. All deletion and complementation mutants were confirmed by Southern hybridization (see Fig. S2 in the supplemental material). After 30 discharged ascospores were isolated from an out-cross between HK30 (Δmat2 Δacs2) and HK22 (Δacs1), we obtained three strains carrying a double deletion of ACS1 and ACS2 (HK31).
The ACS1 deletion mutant showed normal conidium production but a defect in mycelial growth on MMAc, MMEtOH, and MMACD. Normal mycelial growth resumed when the colony was transferred to MM supplemented with sucrose (MMS). The ACS2 deletion mutant did not show any defect in growth or conidium production. The double deletion mutant (HK31) had a more severe defect in growth than the ACS1 mutant (HK22) and even showed impairment in growth on MMS and conidium production (Table 2). The virulence of all of the mutants used in this study was similar to that of the wild-type strain (see Fig. S3 in the supplemental material), and trichothecene production of all of the mutants was not significantly different from that of the wild-type strain (P = 0.67) (see Fig. S4).
Table 2.
Radial growth and conidium production in G. zeae strains
| Strain | Radial growth by medium type (mm)a |
Conidiation (no. of conidia/ml)b | |||
|---|---|---|---|---|---|
| MMS | MMAc | MMEtOH | MMACD | ||
| Wild type | 84.0 A | 84.6 A | 69.0 A | 4.0 A | 2.31 × 106 A |
| Δacs1 strain | 83.3 A | 40.0 B | 35.3 B | 2.4 B | 2.34 × 106 A |
| Δacs2 strain | 84.0 A | 84.0 A | 65.0 A | 4.2 A | 2.40 × 106 A |
| Δacs1 Δacs2 strain | 73.3 B | 18.3 C | 10.7 C | 1.0 C | 1.16 × 106 B |
| ACS1c | 84.0 A | 84.6 A | 67.0 A | 4.3 A | 2.31 × 106 A |
| ACS2c | 84.3 A | 85.3 A | 65.3 A | 4.2 A | 2.47 × 106 A |
Radial growth was measured after 5 days of incubation. All data were repeated three times with three replications. Values within a column with different letters are significantly different (P < 0.05) based on a Tukey test. MMS, minimal medium supplemented with 2% of sucrose; MMAc, minimal medium supplemented with 40 mM potassium acetate as the sole carbon source; MMEtOH, minimal medium supplemented with 40 mM ethanol as the sole carbon source; MMACD, minimal medium supplemented with 5 mM acetaldehyde as the sole carbon source.
Conidiation was measured by counting the number of conidia after 3 days of incubation.
Lipid quantification and characterization of POL.
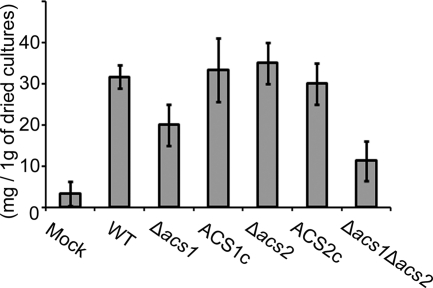
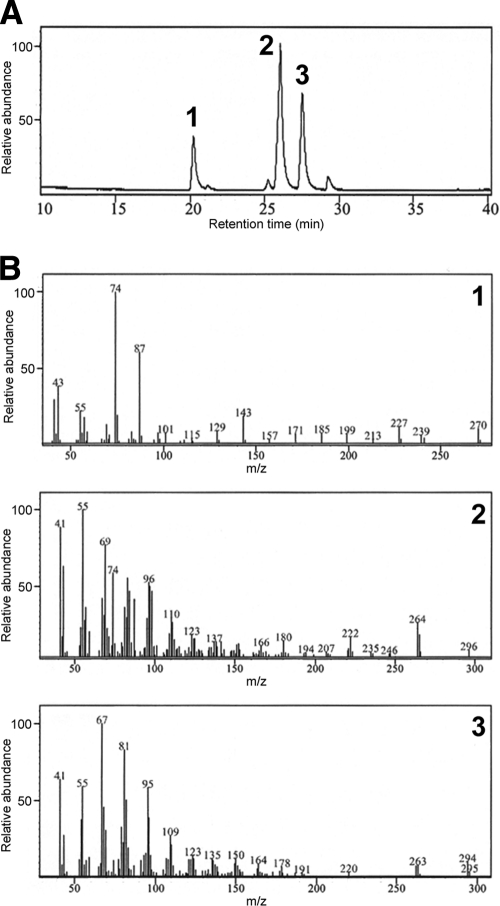
Total lipid production in the ACS1 mutant and double deletion mutant was lower than that of the wild-type strain, and the double deletion mutant produced approximately 50% of the amount of lipid as the ACS1 mutant (Fig. 2). We purified 4.4 g of the major compound in the lipid extract from 300 g of fungal cultures. The compound showed a [M+Na]+ ion at m/z 880 and major fragmentation ions at m/z 854, 604, 578, and 265 by APCI mass spectrum. The 1H-NMR spectrum was identical to that of authentic POL (see Fig. S5 in the supplemental material). In the GC-MS analysis of FAMEs, three peaks appeared in the total ion chromatogram at retention times of 20.1 min, 25.8 min, and 27.4 min, and their mass spectra were identical to those of the methyl esters of palmitic acid, oleic acid, and linoleic acid, respectively (Fig. 3).
Fig. 2.
Total lipid in 1 g of dried carrot agar cultures. WT, G. zeae wild-type strain GZ3639; Δacs1, ACS1 deletion mutant; ACS1c, ACS1- derived strain complemented with ACS1-GFP; Δacs2, ACS2 deletion mutant; ACS2c, ACS2- derived strain complemented with ACS2-GFP; and Δacs1 Δacs2, double deletion mutant of ACS1 and ΔACS2.
Fig. 3.
Total ion chromatogram of the fatty acid methyl esters of POL isolated from G. zeae (A) and mass spectra (B) of methyl esters of palmitic acid (1), oleic acid (2), and linoleic acid (3) with retention times of 20.1 min, 25.8 min, and 27.4 min, respectively.
Fertility test.
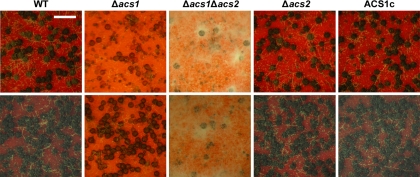
The HK22 (Δacs1) strain produced fewer mature perithecia than the wild-type strain 7 days after sexual induction. HK24 (Δacs2) and complemented strains produced normally melanized mature perithecia containing viable ascospores. The defect in perithecium development was more severe in the double deletion mutant (HK31) (Fig. 4). Perithecium maturation of HK31 (Δacs1 Δacs2) was variable. In five independent sexual induction tests, the HK31 strain did not produce any visible perithecia in two of the trials and produced a few mature perithecia in the remaining three trials. After 14 days, a few perithecia from both the ACS1 and double mutants were matured and normally discharged ascospores. When 0.5% POL was exogenously added to carrot agar, the perithecium maturation of HK22, but not the HK31 (Δacs1 Δacs2) double deletion mutant, was mostly restored. Treatment of the wild-type strain with POL enhanced melanin accumulation and perithecium production (Fig. 4).
Fig. 4.
Self-fertility of G. zeae strains grown on carrot agar. Dissecting microscopic pictures were taken 10 days after sexual induction. Each strain was inoculated on carrot agar without (upper) and with (lower) 0.5% POL. WT, G. zeae wild-type strain GZ3639; Δacs1, ACS1 deletion mutant; Δacs2, ACS2 deletion mutant; Δacs1 Δacs2, double deletion mutant of ACS1 and ACS2; and ACS1c, ACS1- derived strain complemented with ACS1-GFP. Scale bar, 0.5 mm.
Transcriptional analyses of ACSs.
To determine whether deletion of ACS1 or ACS2 affects the expression of the other ACS gene, we performed Northern hybridization in the wild-type strain and each deletion mutant. Transcription profiles of ACS1 and ACS2 were not altered in the ACS2 and ACS1 deletion mutants, respectively, suggesting that transcriptional regulations of the genes are independent of each other (see Fig. S6 in the supplemental material).
Generation of double deletion mutants carrying the ACL2 deletion.
Each HK29 (Δmat2 Δacs1) and HK30 (Δmat2 Δacs2) strain was fertilized with strain acl2 (Δacl2) to generate a double deletion mutant carrying both ACL2 and ACS1 or ACS2. We isolated 60 ascospores from the HK29 × acl2 out-cross and obtained six progenies carrying both ACL2 and ACS1 deletions. From the HK30 × acl2 out-cross, however, we did not obtain any progeny carrying both ACL2 and ACS2 deletions even though we isolated 194 ascospores from two independent out-crosses. In these out-crosses, 19 isolated ascospores (9%) did not germinate. Repeated attempts to generate double deletion mutants of ACS2 and ACL2 by transformation were also unsuccessful.
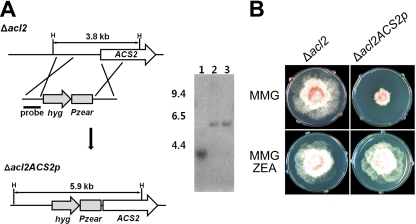
Since we hypothesized that the Δacl2 Δacs2 mutant was lethal, we replaced the promoter of ACS2 with Pzear in the ACL2 deletion mutant using the previously reported strategy (37). The hyg-pzear construct amplified from the Pzear-GzmetE strain was successfully introduced into the promoter of ACS2 in the acl2 strain (Fig. 5A). The promoter-replaced mutant, HK33 (Δacl2 ACS2p), exhibited a severe defect in growth on complete medium, but the defect was restored by exogenous treatment with 30 μM ZEA (Fig. 5B).
Fig. 5.
Replacement of the ACS2 promoter by Pzear in the ACL2 deletion mutant. (A) Strategy for promoter replacement. The promoter of ACS2 was successfully replaced with the zearalenone-inducible promoter (Pzear). H, HindIII; hyg, hygromycin B resistance gene cassette. Lane 1, wild-type strain GZ3639; lanes 2 and 3, promoter-replaced mutant. The sizes of the DNA standards (kb) are indicated on the left of each blot. (B) Growth of G. zeae strains in minimal medium supplemented with 2% glucose (MMG) and MMG supplemented with 30 μM ZEA (MMG-ZEA). When the promoter was successfully replaced, the growth of the Δacl2 ACS2p mutant was severely retarded on minimal medium but was restored by exogenous treatment with ZEA. Δacl2, ACL2 deletion mutant; Δacl2 ACS2p, ACS2 promoter-replaced mutant in the Δacl2 strain.
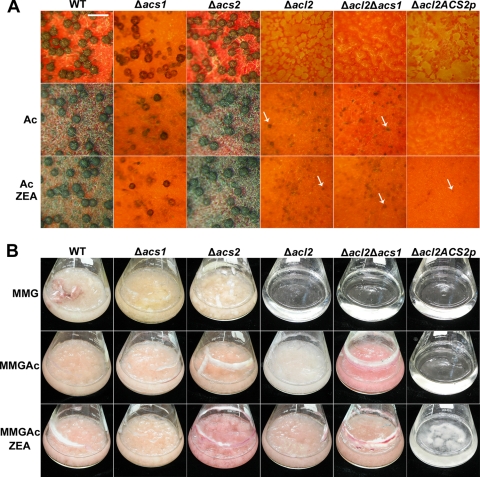
Similar to the acl2 (Δacl2) mutant, HK32 (Δacl2 Δacs1) did not produce any initial structures for perithecia but was somewhat restored by exogenous treatment with 40 mM potassium acetate in carrot agar (Fig. 6A). HK33 (Δacl2 ACS2p) did not produce any initial perithecia even when potassium acetate was supplemented in the medium but produced immature perithecia with ZEA treatment. Similar to the sexual development observations, mycelium growth was somewhat recovered in the acl2 and HK32 mutants when potassium acetate was added, but the HK33 mutant did not grow (Fig. 6B).
Fig. 6.
Chemical complementation by potassium acetate in G. zeae strains. (A) Self-fertility of the G. zeae strains on carrot agar 10 days after sexual induction. Each strain was inoculated on carrot agar (upper), carrot agar supplemented with 40 mM potassium acetate (middle), and carrot agar supplemented with both 40 mM potassium acetate and 30 μM zearalenone (bottom). The white arrows indicate immature perithecia. Scale bar, 0.5 mm. (B) Mycelial growth of G. zeae strains in liquid minimal medium supplemented with 2% of glucose (MMG) 5 days after inoculation. The conidium suspension of each strain was inoculated in MMG (upper), MMG supplemented with 40 mM potassium acetate (middle), and MMG supplemented with both 40 mM potassium acetate and 30 μM ZEA (bottom). WT, G. zeae wild-type strain GZ3639; Δacs1, ACS1 deletion mutant; Δacs2, ACS2 deletion mutant; Δacl2, ACL2 deletion mutant; Δacl2 Δacs1, double deletion mutant of ACL2 and ACS2; Δacl2 ACS2p, ACS2 promoter-replaced mutant in the Δacl2 strain. Ac, exogenous treatment of 40 mM potassium acetate; MMGAc, MMG supplemented with 40 mM potassium acetate; ZEA, exogenous treatment of 30 μM ZEA.
Cellular localization of ACS1-GFP, ACS2-GFP, and ACL2-GFP.
We successfully introduced constructs containing wild-type genes fused with GFP into the corresponding deletion mutants and generated HK36 (ACS1-GFP hH1-RFP), HK37 (ACS2-GFP hH1-RFP), HK38 (ACS1-GFP RFP-SKL), and HK39 (ACS2-GFP RFP-SKL) strains using out-crossing strategies as previously described (see Fig. S2 in the supplemental material) (57).
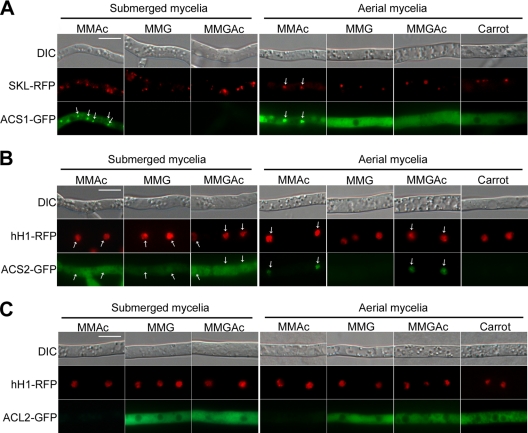
Even though both ACS1 and ACS2 do not contain any subcellular localization signals, they showed different subcellular localization patterns. ACS1-GFP was highly fluorescent in peroxisomes when acetate was used as the sole carbon source in submerged culture but was not visible when MMG and MMGAc liquid cultures were used. In aerial mycelia, ACS1-GFP localized in peroxisomes when MMAc was used but mainly localized to the cytosol when MMG, MMGAc, and carrot agar were used (Fig. 7A). The colocalization study of ACS1-GFP suggested that it did not notably localize in nuclei and mitochondria (see Fig. S7 in the supplemental material). ACS2-GFP was also highly expressed and localized in the cytosol and nuclei of submerged mycelia regardless of the presence of either glucose or acetate (Fig. 7B). In aerial mycelia, ACS2-GFP was not detected in the cytosol, and addition of acetate in solid medium caused ACS2-GFP to mainly localize in nuclei (Fig. 7B). As previously reported, ACL2-GFP localized in the cytosol in hyphae and was highly expressed in mycelia grown in all media except for MMAc (57) (Fig. 7C).
Fig. 7.
Cellular localization of ACS1-GFP (A), ACS2-GFP (B), and ACL2-GFP (C) depending on the culture conditions. Submerged mycelia were harvested for microscopic observation 24 h after conidium inoculation in liquid minimal medium supplemented with potassium acetate (MMAc), glucose (MMG), and both potassium acetate and glucose (MMGAc). Aerial mycelia were collected 3 days after inoculation on solid MMAc, MMG, MMGAc, and carrot agar (Carrot). The middle panels show RFP-SKL and hH1-RFP localized in peroxisomes and nuclei, respectively. All of the white arrows indicate peroxisomal localization (A) and nuclear localization (B and C). DIC, differential interference contrast. Scale bar, 20 μm.
DISCUSSION
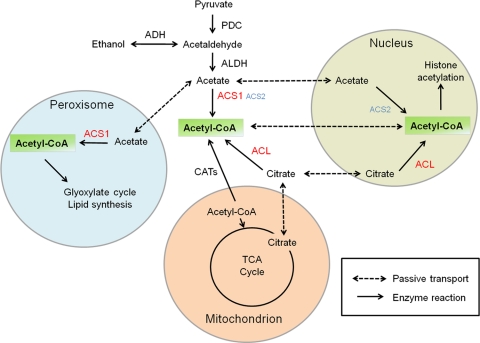
Our current study demonstrated that two ACSs have both overlapping and distinct functions in G. zeae. ACSs are not required for virulence and trichothecene production (see Fig. S3 and S4 in the supplemental material). However, ACS1 is required for the utilization of several nonfermentable carbon sources, suggesting that ACS1 mainly functions in the pyruvate-acetaldehyde-acetate pathway in this fungus (Fig. 8). In addition, ACS1 is required for perithecium maturation. Lipid quantification and complementation with POL suggested that the sexual defect of the deletion mutant is mainly caused by reduced lipid production. Although the ACS2 single mutant did not show any phenotype, ACS2 also had some overlapping roles with ACS1 since the double mutant (Δacs1 Δacs2) showed more severe defects than the ACS1 mutant. Moreover, ACS2 had accessorial functions for ACL that were required for nuclear acetyl-CoA production in G. zeae (Fig. 8).
Fig. 8.
Metabolic pathways for acetyl-CoA generation in G. zeae. The pyruvate-acetaldehyde-acetate pathway has vital roles in acetyl-CoA production. The full name and locus numbers of each enzyme are as follows: PDC, pyruvate decarboxylase (FGSG_09834.3); ACS1, acetyl-CoA synthetase (FGSG_00330.3); ACS2, acetyl-CoA synthetase (FGSG_01743.3); and ACL, ATP-citrate lyase (FGSG_06039.3 and FGSG_12857.3 for each subunit).
ACS1 is required for cytosolic and peroxisomal acetyl-CoA production in G. zeae (Fig. 8). Our lipid analysis and POL complementation test suggested that cytosolic acetyl-CoA produced by ACS1 is crucial for perithecium development (Fig. 2 and Fig. 4). However, since perithecium development of the double deletion mutant was not restored even with POL treatment, ACS-dependent acetyl-CoA production might be required for variable physiological processes in addition to lipid production. Considering that peroxisomes also take part in lipid synthesis, ACS1-derived lipid synthesis might be related to its localization (Fig. 7A) (14). Furthermore, one of the functions of the peroxisome is to detoxify toxic metabolites produced by oxidative reactions (40). Because acetaldehyde, ethanol, and acetate are toxic intermediates, they may need to be detoxified in peroxisomes by ACS1 and proceed to the glyoxylate cycle simultaneously.
POL is a stimulatory compound for perithecium development in G. zeae. Previous studies have reported that G. zeae accumulates an abundance of triacylglycerides for sexual development (23, 57). Because perithecium formation requires enormous energy sources and building blocks, previous researchers hypothesized that triacylglycerides accumulated for storage. We characterized a major compound of triacylglyceride, POL, which enhanced melanin production and perithecium development (Fig. 4).
ACS2 localized to both the cytosol and nuclei and exhibited functions that compensated for ACL since the double deletion of ACL2 and ACS2 was lethal (Fig. 8). The double deletion of the acetyl-CoA synthetase gene facA and the acl gene was also lethal in A. nidulans (29). However, ACS1 is not a compensatory enzyme for the function of ACL even though the roles of ACS1 are similar to those of FacA in A. nidulans, which is also required for acetate utilization (13). The conditional gene expression of ACS2 was created using a ZEA-inducible promoter (37) to generate HK33 (Δacs1 ACS2p) in order to confirm the roles of ACS2 in the ACL mutant. Chemical complementation of the ACL mutant by exogenous treatment with acetate during both sexual and asexual development was repressed only in the HK33 mutant without ZEA treatment (Fig. 6). This result suggests that ACS2 is required for acetate-derived acetyl-CoA production, which is deficient in ACL mutants. Because nuclear acetyl-CoA deficiency was caused by ACL deletion during sexual development and possibly during asexual growth in G. zeae (57), ACS2 seems to specifically participate in nuclear acetyl-CoA production. Nuclear localization of ACS2 in mycelia from both submerged and solid cultures also supported the function of ACS2 as a nuclear acetyl-CoA producer.
Differences in mycelial growth of the ACL mutant in liquid and solid MMG correlated with features of ACSs in catabolic repression. After germination, hyphal elongation of ACL mutants stopped when they were grown in liquid MMG (57). In solid MMG, however, the ACL mutant grew normally, suggesting that some pathways for acetyl-CoA compensation are more functional in solid MMG than liquid medium. Both subunits of ACL were constitutively expressed and localized to the cytosol regardless of the medium, with the exception of growth in MMAc. ACS1 is glucose repressible in G. zeae, similar to other fungal species (16, 53), and the expression level of ACS2 was also reduced in submerged MMG (Fig. 7). On the other hand, ACS1 was highly expressed in the cytosol when the strain was grown on solid MMG, indicating that aerial mycelia were not influenced by glucose repression. Because of the decreased expression of both ACSs in liquid MMG, the ACL knockout mutant may fail to counterbalance acetyl-CoA production but not when the mutant is grown in solid MMG.
The functions of ACSs in G. zeae have some similarities and differences with S. cerevisiae. Similar to ACS1 in G. zeae, ACS1 in yeast is required for growth on several nonfermentable carbon sources, such as acetate and acetaldehyde. ACS1 in yeast, however, is an alternative nuclear acetyl-CoA producer in acetate-utilizing medium and is not required for ethanol utilization, indicating the diverse metabolic roles of ACSs among different organisms (16, 63). As an epigenetic regulator through histone acetylation, yeast ACS2 is required for growth on glucose and was shown to localize primarily to the nuclei, with a minor amount also observed in the cytosol (63, 66). Moreover, a recent study revealed that ACS2 is involved in replicative longevity in yeast (17). Involvement in glucose utilization and histone acetylation, which are lethal features of the double deletion mutant of ACL and ACS2, suggests that ACS2 in G. zeae also has similar functions as ACS2 in yeast (57, 63). However, lipid biogenesis is a process that involves ACS2 in S. cerevisiae but is a function of ACS1 in G. zeae.
One ACS in mammalian cells (AceCS1) also has functions that compensate for ACL (70) but does not have any significant phenotype when silenced (25, 46). The other ACS (AceCS2), however, has been shown to be functional under hypoxic conditions (71). Mammalian ACL, therefore, seems to have more important roles in cell proliferation and lipid biosynthesis than the ACSs (5, 70).
Here, we demonstrated that acetate-derived acetyl-CoA generation by ACSs is important for lipid synthesis and the nuclear acetyl-CoA supply in G. zeae. Since acetate is not a major bioenergetic substrate for G. zeae, acetate should be produced internally by pyruvate decarboxylase (PDC), which is a component of the fermentation pathway. Under anaerobic conditions, fermentation is an alternative pathway for several eukaryotic cells because mitochondria do not have major role in energy metabolism under those conditions (3, 7, 22). Moreover, some obligate aerobic fungi are even known to use the fermentation pathway for survival under hypoxic conditions (43).
In several eukaryotes, the fermentation pathway is also useful for other nonanaerobic conditions. For example, in N. crassa, the transcription of the PDC coding gene was strongly induced by sucrose or glucose, and PDC was hypothesized to be a key postglycolytic enzyme (1). The pdcA (one PDC of A. nidulans) transcript was highly expressed under aerobic conditions in A. nidulans (43). In plants, pyruvate decarboxylase is also required not only for hypoxic growth, such as waterlogged root and seed, but also for survival in an aerobic environment (22, 49). This “aerobic fermentation” produces acetaldehyde from glycolysis-derived pyruvate by PDC and, subsequently, acetate by acetaldehyde dehydrogenase.
An active glyoxylate cycle under aerobic conditions in many fungi also underscores the importance of aerobic fermentation. In fungi, glyoxylate bypass, which comprises isocitrate lyase and malate synthase, was exclusively involved in acetate utilization, indicating its linkage with ACS activity. The glyoxylate cycle has been shown to be crucial for sexual development and fungal virulence (30, 33, 38, 44, 45, 56, 69). A previous study of enzymes related to the glyoxylate cycle in G. zeae revealed that they are highly expressed in aerial mycelia (38). In addition, peroxisomal localization of ACS1 suggests that it may function as an acetyl-CoA producer for the glyoxylate cycle (Fig. 7A).
In conclusion, we had previously characterized ACL subunits that are functional in nucleocytosolic acetyl-CoA generation. ACL-dependent cytosolic acetyl-CoA production is not essential for lipid synthesis but is required for histone acetylation (57). Taken together, these findings suggest that ACS1 and ACS2 are alternative cytosolic and nuclear acetyl-CoA producers, respectively. ACS1 is required for acetate utilization and lipid biosynthesis, and ACS2 has some overlapping functions with both ACS1 and ACL. Moreover, ACS-mediated fermentation seems to have important roles in lipid synthesis in G. zeae. Further studies will focus on the functional characterization of PDC homologs and the functions of the pyruvate-acetaldehyde-acetate pathway in G. zeae.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a National Research Foundation of Korea grant funded by the Korean government (MEST) (2010-0001826) and by a grant (CG 1141) from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Korean Ministry of Education, Science, and Technology.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Alvarez M. E., et al. 1993. The 59-kDa polypeptide constituent of 8-10-nm cytoplasmic filaments in Neurospora crassa is a pyruvate decarboxylase. Gene 130:253–258 [DOI] [PubMed] [Google Scholar]
- 2. Apirion D. 1965. The two-way selection of mutants and revertants in respect of acetate utilization and resistance to fluoro-acetate in Aspergillus nidulans. Genet. Res. 6:317–329 [DOI] [PubMed] [Google Scholar]
- 3. Bakker B. M., et al. 2000. The mitochondrial alcohol dehydrogenase Adh3p is involved in a redox shuttle in Saccharomyces cerevisiae. J. Bacteriol. 182:4730–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baldwin T. K., Urban M., Brown N., Hammond-Kosack K. E. 2010. A role for topoisomerase I in Fusarium graminearum and F. culmorum pathogenesis and sporulation. Mol. Plant Microbe Interact. 23:566–577 [DOI] [PubMed] [Google Scholar]
- 5. Bauer D. E., Hatzivassiliou G., Zhao F., Andreadis C., Thompson C. B. 2005. ATP citrate lyase is an important component of cell growth and transformation. Oncogene 24:6314–6322 [DOI] [PubMed] [Google Scholar]
- 6. Bhambra G. K., Wang Z. Y., Soanes D. M., Wakley G. E., Talbot N. J. 2006. Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol. Microbiol. 61:46–60 [DOI] [PubMed] [Google Scholar]
- 7. Boubekeur S., et al. 1999. A mitochondrial pyruvate dehydrogenase bypass in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274:21044–21048 [DOI] [PubMed] [Google Scholar]
- 8. Bowden R. L., Leslie J. F. 1999. Sexual recombination in Gibberella zeae. Phytopathology 89:182–188 [DOI] [PubMed] [Google Scholar]
- 9. Cappellini R. A., Peterson J. L. 1965. Macroconidium formation in submerged cultures by a non-sporulating strain of Gibberella zeae. Mycologia 57:962–966 [Google Scholar]
- 10. Carman A. J., Vylkova S., Lorenz M. C. 2008. Role of acetyl coenzyme A synthesis and breakdown in alternative carbon source utilization in Candida albicans. Eukaryot. Cell 7:1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chanda A., et al. 2009. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. U. S. A. 106:19533–19538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christie W. W. 1973. Lipid analysis. Isolation, separation, identification and structural analysis of lipids. Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- 13. Connerton I. F., Fincham J. R. S., Sandeman R. A., Hynes M. J. 1990. Comparison and cross-species expression of the acetyl-CoA synthetase genes of the ascomycete fungi, Aspergillus nidulans and Neurospora crassa. Mol. Microbiol. 4:451–460 [DOI] [PubMed] [Google Scholar]
- 14. Cooper G. M., Hausman R. E. 2000. The cell: a molecular approach. ASM Press, Washington, DC. [Google Scholar]
- 15. Desjardins A. E. (ed.) 2006. Fusarium mycotoxins: chemistry, genetics, and biology. APS Press, St. Paul, MN. [Google Scholar]
- 16. De Virgilio C., et al. 1992. Cloning and disruption of a gene required for growth on acetate but not on ethanol: the acetyl-coenzyme A synthetase gene of Saccharomyces cerevisiae. Yeast 8:1043–1051 [DOI] [PubMed] [Google Scholar]
- 17. Falcón A., Chen S., Wood M., Aris J. 2010. Acetyl-coenzyme A synthetase 2 is a nuclear protein required for replicative longevity in Saccharomyces cerevisiae. Mol. Cell. Biochem. 333:99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fatland B. L., et al. 2002. Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiol. 130:740–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flavell R. B., Fincham J. R. S. 1968. Acetate-nonutilizing mutants of Neurospora crassa. I. Mutant isolation, complementation studies, and linkage relationships. J. Bacteriol. 95:1056–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Focke M., et al. 2003. Fatty acid biosynthesis in mitochondria of grasses: malonyl-coenzyme A is generated by a mitochondrial localized acetyl-coenzyme A carboxylase. Plant Physiol. 133:875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gardiner D. M., Kazan K., Manners J. M. 2009. Novel genes of Fusarium graminearum that negatively regulate deoxynivalenol production and virulence. Mol. Plant Microbe Interact. 22:1588–1600 [DOI] [PubMed] [Google Scholar]
- 22. Gass N., et al. 2005. Pyruvate decarboxylase provides growing pollen tubes with a competitive advantage in petunia. Plant Cell 17:2355–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guenther J. C., Hallen-Adams H. E., Bücking H., Shachar-Hill Y., Trail F. 2009. Triacylglyceride metabolism by Fusarium graminearum during colonization and sexual development on wheat. Mol. Plant Microbe Interact. 22:1492–1503 [DOI] [PubMed] [Google Scholar]
- 24. Han Y.-K., Kim M.-D., Lee S.-H., Yun S.-H., Lee Y.-W. 2007. A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol. Microbiol. 63:768–779 [DOI] [PubMed] [Google Scholar]
- 25. Hatzivassiliou G., et al. 2005. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 8:311–321 [DOI] [PubMed] [Google Scholar]
- 26. Hong S.-Y., et al. 2010. Functional analyses of two syntaxin-like SNARE genes, GzSYN1 and GzSYN2, in the ascomycete Gibberella zeae. Fungal Genet. Biol. 47:364–372 [DOI] [PubMed] [Google Scholar]
- 27. Horwitz B. A., et al. 1999. A G protein alpha subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet. Biol. 26:19–32 [DOI] [PubMed] [Google Scholar]
- 28. Hou Z., et al. 2002. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 15:1119–1127 [DOI] [PubMed] [Google Scholar]
- 29. Hynes M. J., Murray S. L. 2010. ATP-citrate lyase is required for the production of cytosolic acetyl-CoA and development in Aspergillus nidulans. Eukaryot. Cell 9:1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Idnurm A., Howlett B. J. 2002. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to Canola (Brassica napus). Eukaryot. Cell 1:719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Imazaki A., et al. 2010. Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen Alternaria alternata. Eukaryot. Cell 9:682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jenczmionka N. J., Maier F. J., Lösch A. P., Schäfer W. 2003. Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase gpmk1. Curr. Genet. 43:87–95 [DOI] [PubMed] [Google Scholar]
- 33. Jennings D. 1995. The physiology of fungal nutrition. Cambridge University Press, London, United Kingdom. [Google Scholar]
- 34. Karan D., David J. R., Capy P. 2001. Molecular evolution of the AMP-forming acetyl-CoA synthetase. Gene 265:95–101 [DOI] [PubMed] [Google Scholar]
- 35. Kim J.-E., et al. 2009. Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr. Genet. 55:449–459 [DOI] [PubMed] [Google Scholar]
- 36. Lee J., Lee T., Lee Y.-W., Yun S.-H., Turgeon B. G. 2003. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol. Microbiol. 50:145–152 [DOI] [PubMed] [Google Scholar]
- 37. Lee J., Son H., Lee S., Park A. R., Lee Y.-W. 2010. Development of a conditional gene expression system using a zearalenone-inducible promoter for the ascomycete fungus Gibberella zeae. Appl. Environ. Microbiol. 76:3089–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee S.-H., Han Y.-K., Yun S.-H., Lee Y.-W. 2009. Roles of the glyoxylate and methylcitrate cycles in sexual development and virulence in the cereal pathogen Gibberella zeae. Eukaryot. Cell 8:1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee S.-H., et al. 2009. GzSNF1 is required for normal sexual and asexual development in the ascomycete Gibberella zeae. Eukaryot. Cell 8:116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lehninger A. L., Nelson D. L., Cox M. M. 1993. Principles of biochemistry. Worth Publishers, New York, NY. [Google Scholar]
- 41. Leighton F., Bergseth S., Rørtveit T., Christiansen E. N., Bremer J. 1989. Free acetate production by rat hepatocytes during peroxisomal fatty acid and dicarboxylic acid oxidation. J. Biol. Chem. 264:10347–10350 [PubMed] [Google Scholar]
- 42. Leslie J. F., Summerell B. A. 2006. The Fusarium laboratory manual. Blackwell Publishing, Ames, IA. [Google Scholar]
- 43. Lockington R. A., Borlace G. N., Kelly J. M. 1997. Pyruvate decarboxylase and anaerobic survival in Aspergillus nidulans. Gene 191:61–67 [DOI] [PubMed] [Google Scholar]
- 44. Lorenz M. C., Fink G. R. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83–86 [DOI] [PubMed] [Google Scholar]
- 45. Lorenz M. C., Fink G. R. 2002. Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryot. Cell 1:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luong A., Hannah V. C., Brown M. S., Goldstein J. L. 2000. Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 275:26458–26466 [DOI] [PubMed] [Google Scholar]
- 47. Min K., et al. 2010. A novel gene, Roa, is required for proper morphogenesis and discharge of ascospores in Gibberella zeae. Eukaryot. Cell 9:1495–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paine P. L., Moore L. C., Horowitz S. B. 1975. Nuclear envelope permeability. Nature 254:109–114 [DOI] [PubMed] [Google Scholar]
- 49. Peschke V. M., Sachs M. M. 1993. Multiple pyruvate decarboxylase genes in maize are induced by hypoxia. Mol. Gen. Genet. 240:206–212 [DOI] [PubMed] [Google Scholar]
- 50. Rivière L., et al. 2009. Acetate produced in the mitochondrion is the essential precursor for lipid biosynthesis in procyclic trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 106:12694–12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rivière L., et al. 2004. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. J. Biol. Chem. 279:45337–45346 [DOI] [PubMed] [Google Scholar]
- 52. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 53. Sandeman R. A., Hynes M. J. 1989. Isolation of the facA (acetyl-coenzyme A synthetase) and acuE (malate synthase) genes of Aspergillus nidulans. Mol. Gen. Genet. 218:87–92 [DOI] [PubMed] [Google Scholar]
- 54. Shen Y.-Q., Burger G. 2009. Plasticity of a key metabolic pathway in fungi. Funct. Integr. Genomics 9:145–151 [DOI] [PubMed] [Google Scholar]
- 55. Shim W.-B., et al. 2006. FSR1 is essential for virulence and female fertility in Fusarium verticillioides and F. graminearum. Mol. Plant Microbe Interact. 19:725–733 [DOI] [PubMed] [Google Scholar]
- 56. Solomon P. S., Lee R. C., Wilson T. J. G., Oliver R. P. 2004. Pathogenicity of Stagonospora nodorum requires malate synthase. Mol. Microbiol. 53:1065–1073 [DOI] [PubMed] [Google Scholar]
- 57. Son H., Lee J., Park A. R., Lee Y.-W. 2011. ATP citrate lyase is required for normal sexual and asexual development in Gibberella zeae. Fungal Genet. Biol. 48:408–417 [DOI] [PubMed] [Google Scholar]
- 58. Starai V. J., Escalante-Semerena J. C. 2004. Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. 61:2020–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stemple C. J., Davis M. A., Hynes M. J. 1998. The facC gene of Aspergillus nidulans encodes an acetate-inducible carnitine acetyltransferase. J. Bacteriol. 180:6242–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Strijbis K., Distel B. 2010. Intracellular acetyl unit transport in fungal carbon metabolism. Eukaryot. Cell 9:1809–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Strijbis K., et al. 2010. Contributions of carnitine acetyltransferases to intracellular acetyl unit transport in Candida albicans. J. Biol. Chem. 285:24335–24346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Strijbis K., et al. 2008. Carnitine-dependent transport of acetyl coenzyme A in Candida albicans is essential for growth on nonfermentable carbon sources and contributes to biofilm formation. Eukaryot. Cell 7:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Takahashi H., McCaffery J. M., Irizarry R. A., Boeke J. D. 2006. Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Mol. Cell 23:207–217 [DOI] [PubMed] [Google Scholar]
- 64. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 65. Urban M., Mott E., Farley T., Hammond-Kosack K. 2003. The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 4:347–359 [DOI] [PubMed] [Google Scholar]
- 66. Van den Berg M. A., Steensma H. Y. 1995. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur. J. Biochem. 231:704–713 [DOI] [PubMed] [Google Scholar]
- 67. Wang Q., et al. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327:1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Y., et al. 2011. A novel transcriptional factor important for pathogenesis and ascosporogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 24:118–128 [DOI] [PubMed] [Google Scholar]
- 69. Wang Z. Y., Thornton C. R., Kershaw M. J., Debao L., Talbot N. J. 2003. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol. Microbiol. 47:1601–1612 [DOI] [PubMed] [Google Scholar]
- 70. Wellen K. E., et al. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324:1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yoshii Y., et al. 2009. Cytosolic acetyl-CoA synthetase affected tumor cell survival under hypoxia: the possible function in tumor acetyl-CoA/acetate metabolism. Cancer Sci. 100:821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yu H.-Y., et al. 2008. Functional analyses of heterotrimeric G protein Gα and Gβ subunits in Gibberella zeae. Microbiology 154:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yu J.-H., et al. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973–981 [DOI] [PubMed] [Google Scholar]
- 74. Zhao S., et al. 2010. Regulation of cellular metabolism by protein lysine acetylation. Science 327:1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhou X., Heyer C., Choi Y.-E., Mehrabi R., Xu J.-R. 2010. The CID1 cyclin C-like gene is important for plant infection in Fusarium graminearum. Fungal Genet. Biol. 47:143–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.