Abstract
The protein kinase Chk1 is required for cell cycle arrest in response to DNA damage. We have found that the 14-3-3 proteins Rad24 and Rad25 physically interact with Chk1 in fission yeast. Association of Chk1 with 14-3-3 proteins is stimulated in response to DNA damage. DNA damage results in phosphorylation of Chk1 and the 14-3-3 proteins bind preferentially to the phosphorylated form. Genetic analysis has independently implicated both Rad24 and Rad25 in the DNA-damage checkpoint pathway. We suggest that DNA damage-dependent association of phosphorylated Chk1 with 14-3-3 proteins mediates an important step along the DNA-damage checkpoint pathway, perhaps by directing Chk1 to a particular substrate or to a particular location within the cell. An additional role for 14-3-3 proteins in the DNA-damage checkpoint has been suggested based on the observation that human Chk1 can phosphorylate Cdc25C in vitro creating a 14-3-3 binding site. Our results suggest that in fission yeast the interaction between the 14-3-3 proteins and Cdc25 does not require Chk1 function and is unaffected by DNA damage, in sharp contrast to the interaction between the 14-3-3 proteins and Chk1.
Keywords: Chk1, 14-3-3 protein, checkpoint, cell cycle, Cdc25, DNA damage
The cellular response to DNA damage includes a transient arrest of cell cycle progression, presumably to allow time for DNA repair (Hartwell and Weinert 1989; Elledge 1996). A number of proteins important for the DNA-damage checkpoint pathway have been identified genetically. Mutants lacking such proteins are unable to arrest the cell cycle in response to DNA damage and die at doses of damaging agents that would be tolerated by wild-type cells (Weinert and Hartwell 1988; Enoch et al. 1992; Rowley et al. 1992; Walworth et al. 1993; Weinert and Hartwell 1993; al-Khodairy et al. 1994; Ford et al. 1994; Weinert et al. 1994). The fission yeast protein kinase Chk1 is essential for cell cycle arrest when DNA is damaged (Walworth et al. 1993; al-Khodairy et al. 1994). Chk1 is phosphorylated in response to damage, and phosphorylation is dependent on the function of several other gene products including Rad3, a fission yeast homolog of the gene ATM, which is defective in the human genetic disorder ataxia telangiectasia (Savitsky et al. 1995; Walworth and Bernards 1996).
Like Chk1, the protein encoded by the fission yeast rad24 gene is important for cell cycle arrest in response to DNA damage (Ford et al. 1994). rad24 encodes a member of the 14-3-3 family of proteins. A mutant allele of rad24 was identified in a genetic screen for checkpoint-defective mutants. A second 14-3-3 family member in Schizosaccharomyces pombe, encoded by the rad25 gene, was found to suppress the rad24 defect when present on a multicopy plasmid (Ford et al. 1994). Mutation of rad25 results in mild sensitivity to DNA-damaging agents and a marginal reduction in the duration of mitotic delay following irradiation (Ford et al. 1994). Simultaneous disruption of rad24 and rad25 is lethal. In mammalian cells, seven isoforms of 14-3-3 proteins have been identified (Aitken 1996). These proteins are thought to play roles on signal transduction pathways, perhaps as scaffolding or adapter proteins that bring together signaling molecules. Although little is known about the biological function of 14-3-3 proteins, the crystal structures of two isoforms have been solved (Liu et al. 1995; Xiao et al. 1995). These structures suggest that 14-3-3 proteins exist as dimers with each monomer containing nine helices that surround a groove. One interpretation of the crystal structure suggests that the groove could accommodate an amphipathic helix (Liu et al. 1995). Consistent with this proposal, the 14-3-3 protein-binding domain on the platelet adhesion receptor, glycoprotein Ib-IX, has been mapped to a serine-rich sequence that could form a helix with serine residues clustered on one side (Du et al. 1996).
A number of additional proteins have been identified that bind to 14-3-3 proteins (Aitken 1996). Association of the protein kinase Raf-1 with 14-3-3 proteins requires a serine residue at amino acid 259 (Michaud et al. 1995). On the basis of this observation, Muslin et al. (1996) defined a consensus motif for 14-3-3 protein binding consisting of RXXp–SXP (where X is any amino acid and p–S is phosphorylated serine). This motif is found in several proteins that have been shown by various techniques to associate with 14-3-3 proteins (Muslin et al. 1996). Nonetheless, it remains to be determined whether all proteins that bind to 14-3-3 proteins contain some form of this motif or whether this motif is necessary for 14-3-3 binding in all of the proteins that do contain it. One example of a protein that does contain the motif and has been shown to bind to 14-3-3 proteins is Cdc25. Cdc25 is a phosphatase that dephosphorylates and activates the cyclin-dependent kinase Cdc2 leading to entry into mitosis. Using Cdc25 as the bait for a two-hybrid protein interaction screen, Conklin and coworkers (1995) identified two 14-3-3 isoforms, β and ε, as Cdc25A- and Cdc25B-binding proteins. Subsequently, Cdc25B and another Cdc25 isoform, Cdc25C, were found to contain the RXXp–SXP consensus sequence for 14-3-3 association. The phosphorylated serine in Cdc25C lies at amino acid position 216. As is the case for Raf-1, phosphorylation of Ser216 within the RXXp–SXP motif is important for association of Cdc25C with 14-3-3 proteins in vitro (Peng et al. 1997).
The regulation of Ser-216 phosphorylation is cell cycle dependent. Ser-216 is phosphorylated during interphase, but unphosphorylated in mitosis. Cdc25C–14-3-3 complexes exist during interphase but are absent in cells in mitosis (Peng et al. 1997). Thus, it has been proposed that dephosphorylation of Ser-216 is coincident with entry into mitosis and further that Ser-216-phosphorylated Cdc25C is kept inactive through its interaction with 14-3-3 proteins (Peng et al. 1997). The fission yeast protein kinase Chk1 as well as the protein kinase encoded by the human homolog of Chk1 are capable of phosphorylating Cdc25C on Ser-216 in an in vitro assay (Peng et al. 1997; Sanchez et al. 1997). These observations led to the proposition that DNA damage might lead to phosphorylation of Cdc25C by Chk1 and subsequent binding to 14-3-3 proteins (Peng et al. 1997; Sanchez et al. 1997; Weinert 1997). According to the model, phosphorylated Cdc25C bound to 14-3-3 proteins would be prevented, by an as-yet-unidentified mechanism, from dephosphorylating Cdc2 and driving cells into mitosis. Although attractive, most aspects of this model remain to be tested. In particular, it has not yet been determined whether phosphorylation of Cdc25C by Chk1 and association of Cdc25C with 14-3-3 proteins occur in response to DNA damage. In the Xenopus system it has been demonstrated that Cdc25 is stoichiometrically bound to 14-3-3 proteins in interphase extracts (Kumagai et al. 1998b). Thus, if Chk1 promotes the association between Cdc25C and 14-3-3 proteins it may do so simply to prevent the dissociation of a Cdc25–14-3-3 complex that is already present in the cell. Recently, it has been demonstrated that depletion of Xenopus Chk1 from egg extracts reduces phosphorylation of Cdc25 by only 20%, which suggests that additional kinases are capable of phosphorylating that site (Kumagai et al. 1998a). Other kinases have been identified that phosphorylate Ser-216 of human Cdc25C, including C-TAK1 (Peng et al. 1998). In addition, the protein kinase Cds1 is capable of phosphorylating fission yeast Cdc25 in vitro on the same sites phosphorylated by Chk1 (Zeng et al. 1998).
The proposition that Chk1 regulates entry into mitosis in response to DNA damage by preventing activation of Cdc25 is consistent with genetic experiments that have been carried out in fission yeast (Furnari et al. 1997; Rhind et al. 1997). Tyrosine-phosphorylated Cdc2 has been shown to persist following DNA damage during the time in which entry into mitosis is delayed (O’Connell et al. 1997; Rhind et al. 1997). This observation is consistent with all models of Cdc2 function: During G2 Cdc2 is tyrosine phosphorylated and it becomes dephosphorylated as cells enter mitosis (Gould and Nurse 1989). Whether maintenance of tyrosine phosphorylation is responsible for cell cycle arrest or a consequence of it is open to question. It is possible that the checkpoint pathway signals arrest through alternative pathways but that eliminating tyrosine phosphorylation of Cdc2 overrides the checkpoint-induced arrest.
Overexpression of Chk1 results in the accumulation of elongated cells in the G2 phase of the cell cycle (Walworth et al. 1993; O’Connell et al. 1997). In these G2-arrested cells, Cdc2 is phosphorylated on tyrosine (O’Connell et al. 1997). Cells lacking the Wee1 protein kinase, which inactivates Cdc2 by phosphorylation of Tyr-15, do not elongate within the same time frame as do wild-type cells (Furnari et al. 1997; O’Connell et al. 1997), suggesting that Chk1-dependent cell cycle arrest could be mediated, at least in part, by Wee1 function. Consistent with this suggestion, recombinant Chk1 can phosphorylate recombinant Wee1 in vitro (O’Connell et al. 1997). However, Wee1 activity is not altered by Chk1 phosphorylation (O’Connell et al. 1997). As noted above, the alternative model, that Chk1 regulates Cdc25 activity, has also been proposed (Furnari et al. 1997; Peng et al. 1997; Sanchez et al. 1997; Weinert 1997). Thus, although it is likely that cell cycle arrest in response to DNA damage is mediated through the regulation of Cdc2 activity, the precise role of Chk1 in this process is not yet clear.
To further define the function of Chk1 we searched for proteins that associate with it by performing a yeast two-hybrid screen using full-length S. pombe Chk1 as the bait. Both fission yeast 14-3-3 proteins, Rad24 and Rad25, interact with Chk1 in this screen. Because mutation of rad24 results in a defective DNA-damage checkpoint (Ford et al. 1994), we pursued the interaction between Chk1 and Rad24/Rad25 in more detail. We find that Chk1 associates with Rad24 and Rad25 in vivo and that the association is stimulated at least 10-fold by DNA-damage. Furthermore, the phosphorylated form of Chk1 preferentially associates with the 14-3-3 proteins. The stimulated association of Chk1 with 14-3-3 proteins is compromised in mutants defective for the DNA-damage checkpoint pathway. These observations suggest that complex formation between Chk1 and 14-3-3 proteins is an important step in the response of fission yeast cells to DNA damage.
Results
Physical interaction of 14-3-3 proteins with Chk1
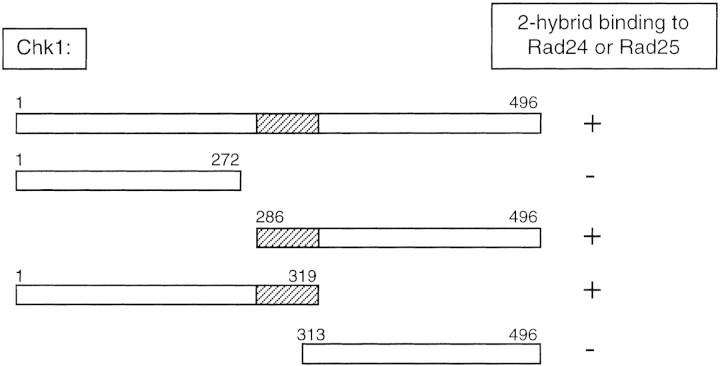
To identify proteins that may physically interact with Chk1, we performed a two-hybrid screen using S. pombe Chk1 as bait. Several interacting proteins were identified including Rad25, a member of the 14-3-3 family of proteins. The second 14-3-3 family member in fission yeast is encoded by the rad24 gene and, when cloned into the appropriate vector (see Materials and Methods), Rad24 also interacts with Chk1 in the two-hybrid assay. The domain of Chk1 necessary for interaction with Rad24 and Rad25 was mapped by constructing amino- and carboxy-terminal deletion mutants of Chk1 and assaying for interaction in the two-hybrid assay (Fig. 1). The catalytic domain of Chk1 lies in the amino-terminal half of the protein ending with amino acid 272 (Walworth et al. 1993). A region of Chk1 within the noncatalytic domain, including amino acids 285–312, appears to be important for interacting with both Rad24 and Rad25 (Fig. 1).
Figure 1.
A domain of Chk1 within amino acids 286–312 is important for interaction with 14-3-3 proteins. Full-length Chk1 (amino acids 1–496) or amino- and carboxy-terminal truncation mutants (indicated by the amino acid number) were expressed as fusions with the DNA-binding domain of Gal4 from the vector pAS2. Rad24 and Rad25 were expressed as fusions with the transactivation domain of Gal4 from the vector pACT. A yeast strain, Y190, was transformed with pAS2–Chk1 vectors and pACT/Rad24 or pACT/Rad25 and assayed for expression of HIS3 and lacZ. (+) The fusion proteins interact allowing expression of the reporter genes; (−) no His+ colonies were obtained.
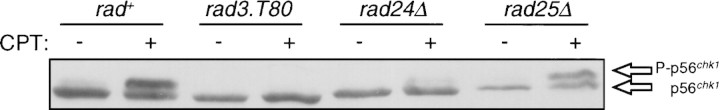
The protein kinase Chk1 becomes phosphorylated when cells are exposed to DNA-damaging agents. Phosphorylation of Chk1 requires the activity of several genetically defined components of the DNA-damage checkpoint including rad1, rad3, rad9, rad17, rad26 (Walworth and Bernards 1996), and hus1 (N.C. Walworth, unpubl.). To test whether rad24 or rad25 function is required for Chk1 phosphorylation we constructed strains in which disruptions of either rad24 or rad25 were combined with an epitope-tagged allele of chk1 (chk1:ep) (Walworth and Bernards 1996). Cells were exposed to the topoisomerase I poison camptothecin (CPT) to generate DNA damage (Wan et al. 1999) and lysates were prepared for Western blot analysis. As shown in Figure 2, the Chk1 (p56chk1) polypeptide becomes phosphorylated following exposure of cells to CPT. Phosphorylation of Chk1 decreases its mobility on SDS-PAGE. As shown in Figure 2, no phosphorylation of Chk1 is seen in a strain with a mutation in the rad3 gene that is defective in the ability to arrest the cell cycle in response to DNA damage (al-Khodairy and Carr 1992; Matsumoto 1997). Phosphorylation of Chk1 appears to be as efficient in a rad25 deletion strain (rad25Δ) as it is in a wild-type (rad+) background consistent with the observation that the rad25Δ strain has an essentially intact checkpoint. In a rad24 deletion strain (rad24Δ), a low level of Chk1 phosphorylation is detected. This could reflect the fact that the rad24Δ mutant is not completely defective in the DNA damage checkpoint. Cells lacking rad24 function are sensitive to DNA damage generated by UV light or γ radiation but not as sensitive as cells lacking chk1 (Ford et al. 1994). Disruption of chk1 results in the complete absence of G2 delay following irradiation whereas disruption of rad24 results in a partial delay (Ford et al. 1994). The partial delay accomplished by a rad24Δ mutant after DNA damage may be caused by Rad25 function as increasing the copy number of rad25 can complement the radiation sensitivity of rad24 (Ford et al. 1994). Similarly, it is possible that the low level of Chk1 phosphorylation in a rad24Δ mutant is caused by Rad25 function.
Figure 2.
Dependence of Chk1 phosphorylation on components of the DNA-damage checkpoint pathway. Wild-type (NW223), rad3.T80 (NW324), rad24Δ (NW250), and rad25Δ (NW278) cells, in which genomic chk1+ was replaced by chk1:ep (Walworth and Bernards 1996), were grown to mid-log phase, after which portions of cell culture were incubated with 40 μm CPT for 2 hr to generate DNA damage. Lysates were prepared and analyzed by immunoblot with antibody to the HA epitope (12CA5).
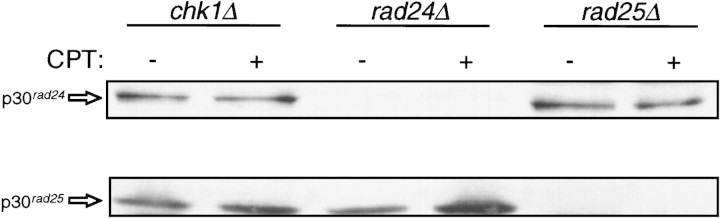
The interaction between the fission yeast 14-3-3 proteins and Chk1 was tested in vitro. GST, GST–Rad24, and GST–Rad25 were purified from Escherichia coli and incubated with a fission yeast lysate in which epitope-tagged Chk1 was overexpressed. Glutathione–Sepharose (GSH–Sepharose) was added to the lysate to precipitate the GST–14-3-3 proteins along with any associated proteins. Western blot analysis of the precipitates demonstrated that Chk1 could bind to GST–Rad24 or GST–Rad25 but not to GST alone (data not shown). We have been unable to produce full-length Chk1 as a fusion protein with GST in E. coli. Therefore, we expressed GST–Chk1 in S. pombe from a regulatable promoter (nmt1) that is repressed by thiamine (Maundrell 1993). Under repressing conditions, sufficient GST–Chk1 is produced to complement the UV sensitivity of a chk1 deletion strain (data not shown) suggesting that GST–Chk1 is a functional protein. Cells were grown for 12 hr in the absence of thiamine to derepress the nmt1 promoter and then CPT was added for 2 hr to generate DNA damage. Lysates were made and GSH–Sepharose added to precipitate GST-Chk1 and associated proteins. Western blot analysis of the precipitates demonstrated that both Rad24 and Rad25 associate with GST–Chk1 in S. pombe (Fig. 3).
Figure 3.
Fission yeast 14-3-3 proteins bind to GST–Chk1 in vivo. Strains with disruptions of chk1 (NW291), rad24 (NW296), or rad25 (NW300) were transformed by pREP1–GST–Chk1. Cells in early log phase were washed free of thiamine and incubated for 12 hr in the absence of thiamine to induce expression of GST–Chk1. Cells were treated for 2 hr with 40 μm CPT and lysates prepared as described in Materials and Methods. GSH–Sepharose was added to precipitate GST–Chk1 and associated proteins. Precipitates were separated by SDS-PAGE and probed by immunoblot with antibodies to Rad24 (UMDNJ-21) and Rad25 (UMDNJ-18) followed by detection with ECL. Antibodies to Rad24 and Rad25 show little cross-reactivity by Western blot, as no signal is seen in the GST–Chk1 pull-down from a rad24Δ strain with the anti-Rad24 antibody. Similarly, little signal is seen in the GST–Chk1 pull-down from a rad25Δ strain with the anti-Rad25 antibody.
Association between 14-3-3 proteins and Chk1 is stimulated by DNA damage
To confirm the interaction between Chk1 and the 14-3-3 proteins in vivo, we placed GST–Rad24 and GST–Rad25 under the control of the nmt1 promoter for expression in S. pombe. Expression of GST–Rad24 or GST–Rad25 from the pREP1 plasmid with a wild-type nmt1 promoter allows high-level expression of the proteins when cells are grown in the absence of thiamine. In the presence of thiamine, a low level of expression is achieved. Low-level expression of either GST–Rad24 or GST–Rad25 complements the UV sensitivity of the rad24Δ strain indicating that GST–Rad24 and GST–Rad25 are functional proteins (data not shown).
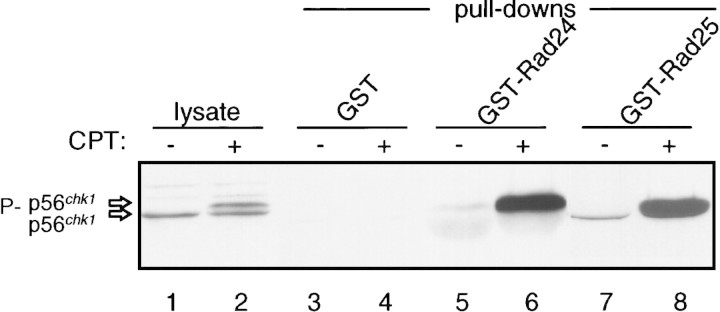
To determine whether Chk1 associates with 14-3-3 proteins in S. pombe, GST–14-3-3 precipitates were assayed for the presence of Chk1 by Western blot analysis. As shown in Figure 4, Chk1 associates with both Rad24 and Rad25. Strikingly, the association of Chk1 with either Rad24 or Rad25 is stimulated when cells are exposed to CPT to generate DNA damage. Quantitation suggests that the amount of Chk1 bound to either GST–Rad24 or GST–Rad25 is increased ∼10-fold by DNA damage.
Figure 4.
Association of Chk1 with 14-3-3 proteins is stimulated by DNA damage. Wild-type cells (NW223) were transformed by pREP1–GST, pREP1–GST–Rad24, or pREP1–GST–Rad25. Expression of the GST proteins was induced by incubating cells in PMA media without thiamine for 12 hr. CPT was then added to 40 μm for an additional 2 hr. Lysates were prepared and GST proteins precipitated with GSH–Sepharose. Precipitates were washed and separated by SDS-PAGE, followed by immunoblot analysis with mAb12CA5 for the detection of 3× HA-tagged Chk1 (Chk1:ep). (Lysate) Lanes contain lysate from cells expressing GST prior to addition of GSH–Sepharose. The shadow band in the GST–Rad24 precipitates is caused by distortion of the Chk1 protein by the presence of a large amount of GST–Rad24 protein on the gel, which has the same mobility as Chk1. Immunoblot analysis of serial dilutions of these samples indicates that binding of Chk1 to GST–Rad24 or GST–Rad25 is stimulated ∼10 fold after DNA damage.
14-3-3 proteins preferentially associate with phosphorylated Chk1
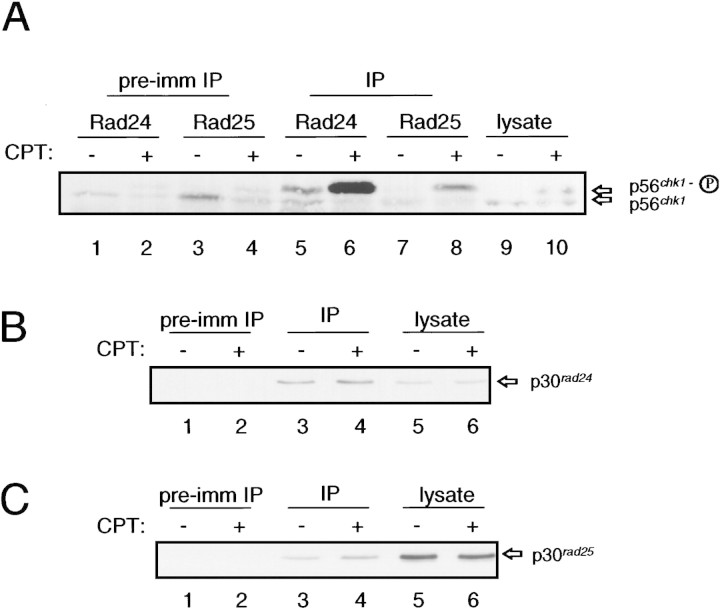
We wished to determine whether the increased association of Chk1 with the 14-3-3 proteins was caused by association of the phosphorylated form of Chk1. GST–Rad24 and GST–Rad25 migrate on SDS-PAGE with approximately the same mobility as Chk1 preventing the resolution of the two forms of Chk1 in lanes that also contain the GST–14-3-3 proteins (see Fig. 4). Chk1 can be detected, however, in immunoprecipitates of Rad24 or Rad25 from S. pombe cells (Fig. 5). Furthermore, both the phosphorylated and unphosphorylated forms of Chk1 can be readily resolved in this assay. As shown in Figure 5A, it is apparent that the 10-fold increase in the amount of Chk1 that is associated with the 14-3-3 proteins is due primarily, if not exclusively, to association of the phosphorylated form. Control experiments demonstrate that the abundance of Rad24 and Rad25 was unchanged by DNA damage and that equal amounts of Rad24 and Rad25 were immunoprecipitated from CPT-treated and untreated cells (Fig. 5B,C). The coimmunoprecipitation experiment also demonstrates that phosphorylated Chk1 is present in cells that have not been exposed to agents that damage DNA. As seen in Figure 5A, lane 5, the Rad24 antiserum precipitates phosphorylated Chk1 from cells that have not been treated with CPT. On occasion low levels of phosphorylated Chk1 can be detected by Western blot analysis of lysates prepared from wild-type cells that have not been exposed to a DNA damaging agent (N.C. Walworth, unpubl.). It is possible that Rad24 acts as an affinity reagent for isolating phosphorylated Chk1 and the small amount of phosphorylated Chk1 that may be present in undamaged cells is concentrated in the Rad24 immunoprecipitate.
Figure 5.
Increased association of Chk1 with 14-3-3 proteins is specific to the phosphorylated form of Chk1. (A) Wild-type cells (NW223) were grown to mid-log phase and then treated with 40 μm CPT for 2 hr. Lysates were prepared and immunoprecipitated with Rad24 or Rad25 antiserum or the corresponding preimmune sera. Immunoprecipitates were resolved on 7.5% SDS-PAGE and subjected to immunoblotting with mAb12CA5. Some background detection of Chk1 is apparent in precipitates with the preimmune sera (lanes 1–4). The background level of the phosphorylated and unphosphorylated forms of Chk1 in these control precipitates reflects the abundance of each in the lysate. Samples of lysate prior to immunoprecipitation were included on the gel (lanes 9,10) to provide markers for the phosphorylated and unphosphorylated forms of Chk1. (B,C) The abundance of 14-3-3 proteins in lysates and in immunoprecipitates is unaffected by DNA damage. The samples shown in A were diluted, electrophoresed on 12% gels, and immunoblotted with antibody to Rad24 (B) and Rad25 (C). In B the samples from lanes 1,2,5,6,9, and 10 of A correspond to lanes 1–6. In C the samples from lanes 3,4,7,8,9, and 10 of A correspond to lanes 1–6.
DNA damage-stimulated association is dependent on the checkpoint pathway
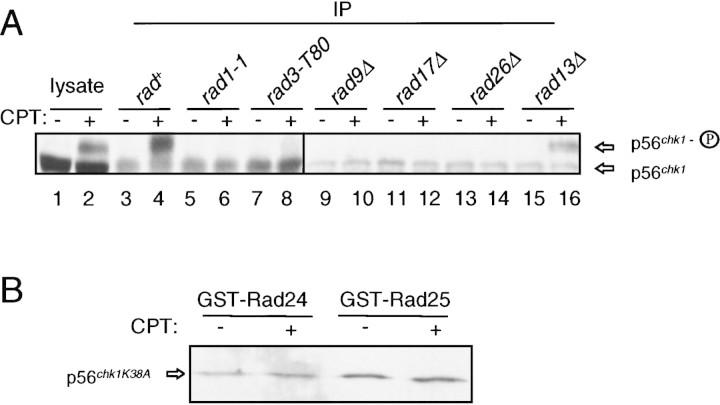
Because phosphorylation of Chk1 in response to DNA damage is dependent on several components of the checkpoint pathway, we predicted that the increased association of Chk1 with 14-3-3 proteins would also be dependent on these proteins. To test this possibility, Rad24 was immunoprecipitated from checkpoint-defective strains harboring mutations in rad1, rad3, rad9, rad17, and rad26. As a control, a DNA repair-defective mutant (rad13Δ) with an intact G2 checkpoint was examined. Cells were grown to mid-log phase and treated with CPT for 2 hr. Lysates were prepared and Rad24 was immunoprecipitated. Immunoprecipitates were examined for the presence of Chk1 by Western blot analysis. As shown in Figure 6A, there is no change in the amount of Chk1 that coimmunoprecipitates with Rad24 in the checkpoint-defective strains (lanes 5–14). Identical results were seen when Rad25 was immunoprecipitated from these lysates (data not shown).
Figure 6.
Increased association of Chk1 with 14-3-3 proteins is checkpoint dependent. (A) Rad24 was immunoprecipitated with antibody UMDNJ-21 from strains with chk1:ep and mutations in the checkpoint genes rad1-1 (NW238), rad3.T80 (NW324), rad9Δ (NW247), rad17Δ (NW249), rad26Δ (NW251). Wild-type cells (NW223) and a strain defective in DNA repair but with an intact G2 checkpoint (rad13Δ, NW248) were included as controls. CPT treatment and immunoprecipitation of Rad24 were carried out as described in the legend to Fig. 5. Chk1 was detected by immunoblot analysis with mAb12CA5. Immunoprecipitates are shown in lanes 3–16. Samples of lysate from wild-type cells (lanes 1,2) are included as controls to demonstrate the mobilities and relative abundance of the two forms of Chk1. (B) Association of Chk1K38A with 14-3-3 proteins is not increased after DNA damage. A strain with integrated chk1K38A:ep that shows no phosphorylation of Chk1K38A after DNA damage was transformed by pREP1–GST–Rad24 or pREP1–GST–Rad25. Expression of GST-14-3-3 proteins, CPT treatment, and precipitation of GST proteins was carried out as described in the legend to Fig. 4 and in Materials and Methods. Chk1K38A was detected by immunoblot with mAb12CA5. No Chk1K38A precipitates with GST alone (data not shown).
We also tested whether a mutant allele of Chk1 that is unable to function on the checkpoint pathway could associate with 14-3-3 proteins. Mutation of a conserved lysine residue (lysine 38 to alanine, K38A) in the catalytic domain of Chk1 compromises its ability to function in the DNA-damage checkpoint pathway (Walworth and Bernards 1996). When the Chk1K38A allele is integrated as the sole copy of Chk1 in the genome (NW307), the cells are as sensitive to UV light or CPT treatment as a chk1Δ disruption strain (Wan et al. 1999). The Chk1K38A protein is not phosphorylated in response to DNA damage (Walworth and Bernards 1996). The NW307 strain was transformed with the plasmids that express GST–Rad24 and GST–Rad25 and assayed for the association of Chk1K38A with the 14-3-3 proteins. As shown in Figure 6B, although the Chk1K38A protein does associate with each of the 14-3-3 proteins, the association is not stimulated by DNA damage.
Association of Rad24 and Rad25 with Cdc25 does not require Chk1 function
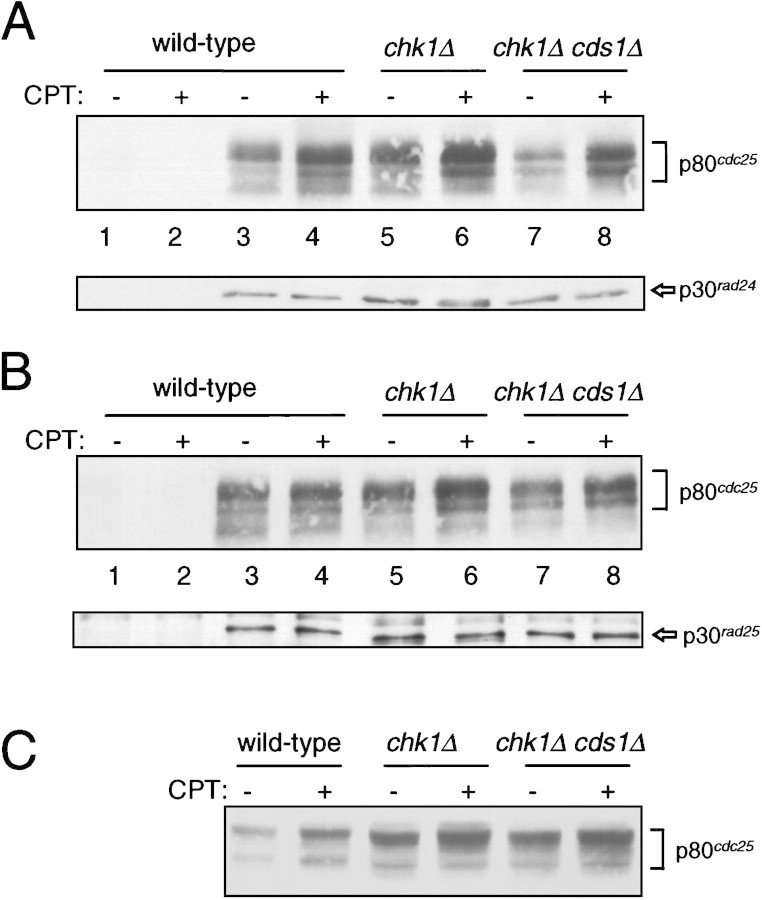
Recently it has been demonstrated that Xenopus and human Cdc25 associate with 14-3-3 proteins. To test whether fission yeast Cdc25 binds to fission yeast 14-3-3 proteins in vivo, we immunoprecipitated the 14-3-3 proteins from S. pombe strains with an epitope-tagged allele of Cdc25 integrated into the chromosome (Nefsky and Beach 1996). Following exposure of cells to CPT, lysates were prepared and immunoprecipitates with Rad24 or Rad25 antisera were isolated and analyzed by Western blot analysis for the presence of Cdc25 protein. As shown in Figure 7, A and B (lanes 3,4), Cdc25 associates with Rad24 and Rad25, respectively. Consistent with experiments performed in a Xenopus in vitro system (Kumagai et al. 1998b), no dramatic alteration in the amount of Cdc25 associated with the 14-3-3 proteins is seen in vivo following DNA damage. Apparent increases in the amount of Cdc25 coprecipitated with 14-3-3 proteins from CPT-treated cells reflects an increase in the amount of Cdc25 that is extracted from the cells as assayed by Western blot analysis of the total cell lysates (Fig. 7C). Quantitative Western blot analysis of serial dilutions of both lysate samples and immunoprecipitates indicates that the difference in abundance of Cdc25 in samples prepared from CPT-treated and untreated cells is no more than twofold (data not shown).
Figure 7.
Association of Cdc25 with 14-3-3 proteins before and after DNA damage is not dependent on Chk1 or Cds1. Strains with integrated cdc25:3HA were exposed to 40 μm CPT for 2 hr. Lysates were prepared and subjected to immunoprecipitation with antibody to Rad24 (UMDNJ-55) and Rad25 (UMDNJ-56). (A) Rad24 immunoprecipitates were separated by SDS-PAGE and immunoblotted with antibody to detect Cdc25 (12CA5, top) and Rad24 (UMDNJ-55, bottom). The samples in lanes 1 and 2 are precipitates of wild-type cells with preimmune sera. The samples in lanes 3–8 are precipitates with Rad24 antisera (UMDNJ-55). (Lanes 3,4 Wild-type cells (BN71); (lanes 5,6) chk1Δ cells (NW291); (lanes 7,8) chk1Δ cds1Δ cells (NW373). (B) Rad25 immunoprecipitates were separated by SDS-PAGE and immunoblotted with antibody to detect Cdc25 (12CA5, top) and Rad25 (UMDNJ-55, bottom). The samples in lanes 1 and 2 are precipitates of wild-type cells with preimmune sera. The samples in lanes 3–8 are precipitates with Rad25 antisera (UMDNJ-56). (Lanes 3,4) Wild-type cells (BN71); (lanes 5,6) chk1Δ cells (NW291); (lanes 7,8) chk1Δ cds1Δ cells (NW373). (C) Samples of the lysates used for immunoprecipitation in A and B were immunoblotted for Cdc25 protein with mAb12CA5. The amount of Cdc25 in the lysates is increased no more than twofold by DNA damage (estimated by Western blot analysis of serial dilutions of the samples from wild-type cells).
We next evaluated the association between Cdc25 and the 14-3-3 proteins for dependence on Chk1 function. In a strain lacking Chk1, Cdc25 coprecipitates with Rad24 (Fig. 7A, lanes 5,6) and Rad25 (Fig. 7B, lanes 5,6) both before and after exposure to CPT. It has recently been reported that the fission yeast protein kinase Cds1 phosphorylates fission yeast Cdc25 on the same sites that are phosphorylated by Chk1 (Zeng et al. 1998). The sites phosphorylated by Chk1 and Cds1 have been suggested to be important for Cdc25 binding to 14-3-3 proteins (Zeng et al. 1998). Therefore, we tested whether Cdc25–14-3-3 association occurs in cells lacking both Chk1 and Cds1. As shown in Figure 7 (lanes 7,8), Cdc25–14-3-3 association does not require either Chk1 or Cds1 function either before or after DNA damage.
Discussion
The DNA-damage checkpoint pathway is required to prevent entry into mitosis in the presence of DNA damage. We have demonstrated that the protein kinase Chk1 associates with the fission yeast 14-3-3 proteins, Rad24 and Rad25. This association is stimulated 10-fold by DNA damage (Fig. 4), and it is the phosphorylated form of Chk1 generated in response to DNA damage that preferentially binds to the 14-3-3 proteins (Fig. 5). Whereas the amount of DNA damage generated in the experiments shown here results in phosphorylation of only 30%–50% of Chk1, the increased association of Chk1 to the 14-3-3 proteins is attributable almost exclusively to binding of the phosphorylated form. To our knowledge, this is only the second example of an association between a target protein and a 14-3-3 protein that changes in response to activation of a signal transduction pathway. The other example of such regulation is the increased association between the protooncogene Cbl and 14-3-3τ in T lymphocytes (Liu et al. 1996) following T-cell activation by anti-CD3. In contrast, Raf1 association with 14-3-3τ remains constant in these cells (Liu et al. 1996).
Whereas association of Chk1 with 14-3-3 proteins is stimulated by DNA damage, association of 14-3-3 proteins with GST–Chk1 does not appear to change in cells treated with CPT (Fig. 3). We believe that this result is attributable to the fact that GST–Chk1 is overexpressed in this experiment in order to recover sufficient Chk1 for the assay. We and others have observed that overexpression of Chk1 leads to a cell cycle delay in the absence of DNA damage (Walworth et al. 1993; Ford et al. 1994; Furnari et al. 1997; O’Connell et al. 1997). When Chk1 is overexpressed in cells it is not possible to detect a mobility shift of Chk1 following DNA damage (N.C. Walworth, unpubl.). Thus, in the cells used for this experiment, it is possible that the DNA-damage checkpoint pathway leading to phosphorylation of Chk1 is not activated because the cells are already experiencing a mitotic delay. Alternatively, because GST–Chk1 is overexpressed, it is possible that the pool of 14-3-3 proteins available for binding to Chk1 has been depleted.
Phosphorylation of Chk1 and its stimulated association with 14-3-3 proteins requires an intact DNA-damage checkpoint pathway. In strains with mutations in the genetically defined components of the checkpoint pathway, rad1, rad3, rad9, rad17, rad26, and hus1, Chk1 is not phosphorylated in response to DNA damage. The association between Chk1 and Rad24 is not stimulated in these strains consistent with the suggestion that increased complex formation is dependent on an intact checkpoint signaling pathway (Fig. 6A). Mutation of the conserved lysine residue in the Chk1 catalytic domain (K38A) compromises its ability to function in the DNA damage checkpoint (Walworth and Bernards 1996). This mutant protein is not phosphorylated in response to DNA damage and its association with 14-3-3 proteins following DNA damage is not stimulated (Fig. 6B).
Immunoprecipitation experiments clearly demonstrate that it is the phosphorylated form of Chk1 that associates with 14-3-3 proteins following DNA damage (Fig. 5). It has been proposed that binding partners for 14-3-3 proteins contain a sequence motif surrounding a phosphorylated serine residue (Muslin et al. 1996). Several proteins that have been shown to bind to 14-3-3 proteins contain the RXXp–SXP motif and, in some cases, mutation of the serine residue to an alanine has been shown to compromise binding to 14-3-3 proteins (Peng et al. 1997). Although phosphorylation of Chk1 correlates with its ability to associate with 14-3-3 proteins, Chk1 does not contain a motif that matches the RXXp–SXP motif. Other motifs have been suggested to be important for binding of target proteins to 14-3-3 proteins including a serine-rich patch on one side of an α-helix (Du et al. 1996) or an amphipathic α-helix (Xiao et al. 1995). We are currently trying to define the 14-3-3 protein binding site on Chk1 in more detail.
Recent evidence from in vitro experiments suggests that human Chk1 is capable of phosphorylating the tyrosine phosphatase Cdc25C on Ser-216. Phosphorylation of Ser-216 is necessary for association of Cdc25C with 14-3-3 protein family members (Peng et al. 1997). An attractive model was proposed to explain the events that govern cell cycle arrest in response to DNA damage (Furnari et al. 1997; Peng et al. 1997; Sanchez et al. 1997; Weinert 1997). The model proposes that in response to DNA damage, Chk1 becomes phosphorylated in a Rad3- or ATM-dependent manner. Chk1 phosphorylates Cdc25 to create a binding site for association of a 14-3-3 protein. When bound to 14-3-3 protein, Cdc25 is prevented from dephosphorylating and thus activating Cdc2. With the exception of Rad3-dependent Chk1 phosphorylation (Walworth and Bernards 1996), none of the subsequent steps of the model have been shown to occur in response to DNA damage. Our data suggest that this model requires revision. The association of Cdc25 with 14-3-3 protein in fission yeast does not require Chk1 function. On DNA damage, the apparent twofold increase in the binding of Cdc25 to 14-3-3 proteins reflects an increase in the total amount of Cdc25 present in the lysate. Thus, the increase in Cdc25 concentration could represent, for example, a change in extractability of Cdc25 or stabilization of the protein following DNA damage. Regardless, this behavior of Cdc25 is not Chk1 dependent. Our result is consistent with observations made with a Xenopus in vitro system demonstrating that 95% of Cdc25 is bound to 14-3-3 protein in interphase extracts and that no change in the association of Cdc25 with 14-3-3 proteins is seen following activation of a checkpoint pathway (Kumagai et al. 1998b). It has been suggested, therefore, that Chk1 may simply maintain phosphorylation of Cdc25 that is normally present during interphase, allowing Cdc25 to remain bound to 14-3-3 protein in an inactive state at a time when the cell might otherwise progress into mitosis (Kumagai et al. 1998b). Our data suggest, however, that association of Cdc25 to 14-3-3 protein is not sufficient to prevent entry into mitosis. Cells with a disrupted allele of Chk1 clearly do not arrest the cell cycle in response to DNA damage (al-Khodairy et al. 1994), yet the Cdc25–14-3-3 complex is present in these cells just as it is in wild-type cells both before and after DNA damage (Fig. 7). Because the Cdc25–14-3-3 complex exists in cells that have no Chk1, phosphorylation of Cdc25 in the absence of DNA damage must be carried out by another protein kinase. Consistent with our observations it has been demonstrated recently that depletion of Xenopus Chk1 from egg extracts reduces phosphorylation of Xenopus Cdc25 in an in vitro assay by no more than 20% (Kumagai et al. 1998a).
Recently it has been demonstrated that the protein kinase Cds1, like Chk1, can phosphorylate fission yeast Cdc25 in vitro on sites that influence 14-3-3 protein binding (Zeng et al. 1998). It is not known, however, whether either kinase phosphorylates those sites in vivo (Zeng et al. 1998). Our results indicate that, in cells lacking both Cds1 and Chk1, Cdc25 still associates with 14-3-3 proteins both before and after DNA damage (Fig. 7). Furthermore, cells expressing a mutant allele of Cdc25 (Cdc25–S3) lacking the 3 phosphorylation sites still show a significant checkpoint arrest when treated with hydroxyurea (Zeng et al. 1998). Even in strains expressing the Cdc25–S3 mutant that additionally have the wee1 or mik1 genes deleted, more than 60% of the cells still arrest the cell cycle (Zeng et al. 1998). Finally, fission yeast completely lacking Cdc25 function, but kept alive by a semidominant allele of cdc2 (cdc2-3w cdc25Δ), are not sensitive to DNA damage generated by UV light (al-Khodairy and Carr 1992). In contrast, cells lacking Chk1 or Rad24 function are sensitive to UV light (Walworth et al. 1993; al-Khodairy et al. 1994). Thus, it is highly unlikely that phosphorylation of Cdc25 by Chk1 and/or Cds1 is sufficient to mediate the checkpoint arrest that leads to survival after DNA damage.
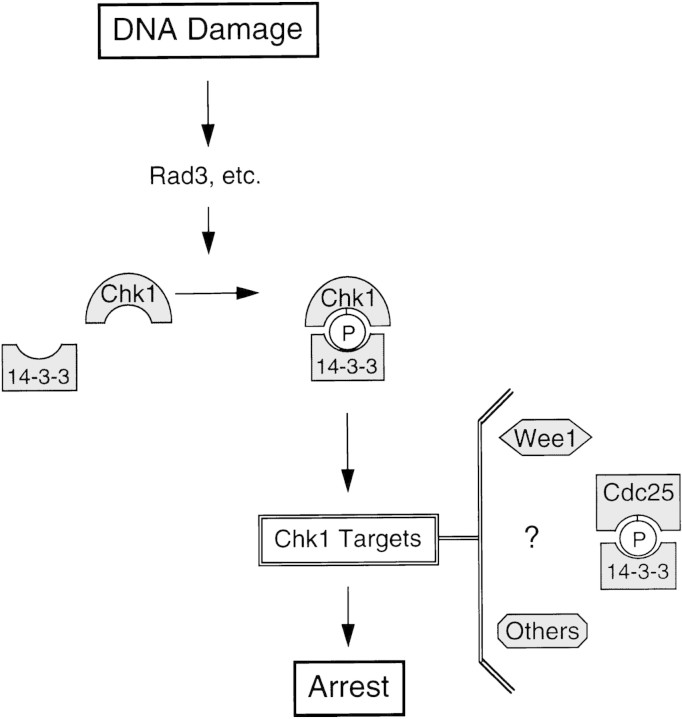
We hypothesize that association of 14-3-3 proteins with phosphorylated Chk1 following DNA damage plays a critical role in the checkpoint response (Fig. 8). Whereas it is possible that this interaction mediates the targeting of Chk1 to Cdc25, it is clear that phosphorylation of Cdc25 by Chk1 (or by Chk1 and Cds1 acting redundantly) to create a 14-3-3 binding site is not sufficient to explain how cell cycle arrest is achieved. Wee1 has also been suggested to be a target of Chk1 as overproduction of Chk1 in fission yeast, like DNA damage, leads to Wee1 phosphorylation (O’Connell et al. 1997). However, despite the fact that Chk1 can phosphorylate Wee1 in vitro, this phosphorylation event has no effect on the catalytic activity of Wee1 (O’Connell et al. 1997). We anticipate that there will be additional targets of Chk1 that mediate DNA damage-dependent cell cycle arrest before mitosis (Fig. 8).
Figure 8.
Model of the DNA-damage checkpoint pathway. In response to DNA damage, Chk1 becomes phosphorylated in a manner that is dependent on the activity of Rad3 and other checkpoint Rad proteins. The association of Chk1 with 14-3-3 proteins is stimulated by phosphorylation of Chk1. Phosphorylated Chk1 communicates with target proteins to propagate the signal for arrest. Wee1 and Cdc25 have been suggested as potential in vivo targets of Chk1; however, it is likely that other targets exist as well (see text for discussion).
Although the association between Chk1 and 14-3-3 proteins is increased 10-fold, the significance of this association is not yet clear. The association could target Chk1 to a relevant substrate or to a particular subcellular location. It is also possible that binding of Chk1 to 14-3-3 alters Chk1 catalytic activity. Clearly, however, the association of Chk1 with 14-3-3 proteins correlates with phosphorylation of Chk1, an event that appears to be necessary for Chk1 function when DNA is damaged (Walworth and Bernards 1996). Therefore, it is likely that stimulated Chk1–14-3-3 association is an integral step in the DNA-damage checkpoint pathway. To prove that the association is essential for the function of the pathway it will be necessary to identify a mutant Chk1 protein that is phosphorylated in response to DNA damage but fails to bind to 14-3-3 proteins: Such a mutant allele would be expected to be checkpoint defective. An alternative approach would be to identify mutant alleles of Rad24 or Rad25 that are specifically defective in binding to Chk1 and assess their functionality in the checkpoint response. These approaches and screens for additional Chk1 substrates are under way.
Materials and methods
Yeast strains, media, and growth conditions
Yeast strains used in these studies are listed in Table 1. S. pombe strains were transformed using the LiAc transformation procedure as described (Moreno et al. 1991). S. pombe strains were grown in pombe minimal (PM) medium with the appropriate supplements at 75 μg/ml in the presence or absence of 20 μm thiamine or in rich yeast extract (YE) medium as indicated. Standard recipes were used for YE (Moreno et al. 1991), and minimal medium was prepared using Edinburgh minimal medium (EMM) from Bio-101 according to the manufacturer’s instructions. Cells were grown at 30°C. Strains were constructed by random spore analysis. The identity of the chk1::ura4 cds1::ura4 double mutant was confirmed by comparing the ability of the cells to arrest in the presence of hydroxyurea (HU) relative to either single mutant. After 4 hr in the presence of 12 mm HU, the septation index of either single mutant was <5% whereas the double mutant was 68%.
Table 1.
Yeast strains
| Strain
|
Genotype
|
|---|---|
| NW158 | h+, chkl∷ura4, ura4–D18, leu1–32, ade6–216 |
| NW223 | h+, chkl:ep, leu1–32, ade6–216 |
| NW238 | h+, chkl:ep, rad1–1, leu1–32, ade6–216 |
| NW247 | h+, chkl:ep, rad9∷ura4, ura4–D18, leu1–32, ade6–216 |
| NW248 | h+, chkl:ep, rad13∷ura4, ura4–D18, leu1–32, ade6–216 |
| NW249 | h+, chkl:ep, rad17∷ura4, ura4–D18, leu1–32, ade6–216 |
| NW250 | h+, chkl:ep, rad24∷ura4, ura4–D18, leu1–32, ade6–216 |
| NW251 | h+, chkl:ep, rad26∷ura4, ura4–D18, leu1–32, ade6–216 |
| NW278 | h+, chkl:ep, rad25∷ura4, ura4–D18, leu1–32, ade6–216 |
| NW291 | h+, chkl∷ura4, ura4–D18, leu1–32, cdc25(HA)3 |
| NW296 | h+, rad24∷ura4, ura4–D18, leu1–32, cdc25(HA)3 |
| NW300 | h+, rad25∷ura4, ura4–D18, leu1–32, cdc25(HA)3 |
| NW307 NW324 NW373 | h+, chklK38A:ep, leu1–32, ade–216chk1:ep, rad3T.80, leu1–32, ade6–216chk1∷ura4, cds1∷ura4, ura4–D18, leu1–32, ade6–216, cdc25(HA)3 |
| BN71 | h+, cdc25(HA)3, ura4–D18, leu1–32, ade6–216 |
Two-hybrid screen
Plasmids, strains, and the S. pombe cDNA library for the two-hybrid screen were generously provided by Dr. Stephen J. Elledge (Baylor College of Medicine, Houston, TX). Chk1 was cloned into the pAS1–CYH2 vector using an NdeI site engineered at the initiation codon of Chk1 (see below) and by filling in an XhoI site at the 3′ end of the cDNA. This fragment was cloned into the vector cut with NdeI and SmaI to generate pAS–Chk1. The reporter strain Y190 was used for the two-hybrid screen (Bai and Elledge 1997). Transformation of Y190 with pAS–Chk1 did not produce His+ colonies. Cotransformation of Y190 with pAS–Chk1 and the S. pombe cDNA library in the pACT vector gave rise to 77 colonies that could grown in the absence of histidine and the presence of 3-aminotriazole (3-AT) at 25 mm. These colonies were assayed for the production of β-galactosidase on nitrocellulose filters as described (Bai and Elledge 1997). False positives were eliminated by crossing Y190 strains that had been forced to lose the pAS–Chk1 plasmid but retained the pACT–library plasmids to Y187 tester strains carrying GAL4 DNA-binding domain fusion protein vectors that expressed lamin, p53, SNF1, and CDK2. After retesting, nine positive clones were identified. Of these, one encodes Rad25, three encode a putative 40S ribosomal subunit protein, one encodes a DEAD-box protein (Dep1) to be described elsewhere (H.-Y. Liu and N.C. Walworth, in prep.), one encodes a putative homolog of ATP citrate–lyase, and three others encode novel proteins. The cDNA for Rad24 was amplified by PCR from the pACT cDNA library using the oligonucleotides listed below and cloned into the pACT vector. Interaction with Chk1 was tested by cotransforming pAS–Chk1 (or truncation mutants, described below) and pACT–Rad24 into Y190 and testing for His prototrophy and β-galactosidase production.
PCR and construction of plasmids for two-hybrid assays
Amplification of DNA fragments by PCR was done with Vent DNA polymerase (New England Biolabs) according to the manufacturer’s instructions.
An NdeI site was introduced into the chk1 cDNA at the start codon to facilitate cloning into the two-hybrid bait vector (pAS1–CYH2). A fragment of the 5′ end of chk1 was amplified using an oligonucleotide that introduced an NdeI site at the initiation codon. The oligonucleotide for the 5′ end was 5′CCCTTTACACCAACATATGGCTCA. The NdeI site is underlined. The oligonucleotide for the 3′ end was 5′GTCATAAAGCTTATCTATAATGG, which includes base changes that would mutagenize chk1 to include a HindIII site. However, this change is not relevant to the experiments described here. Following amplification and purification, the fragment was digested with NdeI and AatII for cloning back into the chk1 cDNA in pBluescript to create pBS/Chk1–NdeI. The chk1 sequence includes an NdeI site 47 base pairs upstream from the ATG. Thus, by digesting the cDNA with NdeI and introducing the fragment with NdeI at the start site, 47 base pairs of 5′ untranslated region are deleted. The fragment was sequenced to ensure that no other mutations were introduced.
The rad24 cDNA was amplified from the S. pombe cDNA library in pACT1 using 5′CGCACATTTTGGATCCTTTGGTAAAA and 5′CGGGATCCACAGAGCTATCTATGCGTCGCC as primers. BamHI sites are underlined. The fragment was digested with BamHI and cloned into the BamHI site of pACT2 (Bai and Elledge 1997) and pREP1 (Maundrell 1993).
Oligonucleotides for cloning of truncated Chk1 alleles are listed below. Fragments of chk1 were amplified by PCR such that an NdeI site (underlined) was introduced at the 5′ end for fusion in frame with the GAL4 activation domain in pAS1–CYH2. For the carboxy-terminal truncations, the amplified fragments were digested with AatII (within chk1) and BamHI (engineered in the oligonucleotides, underlined below) and cloned into pAS–Chk1. Stop codons (bold) were included in the 3′ primers for amplification of the amino-terminal truncations. For the amino-terminal truncations, the fragment amplified includes the stop codon and some 3′-untranslated region. Fragments were digested with NdeI (engineered in the oligonucleotides) and AvrII (a site within the 3′ end of Chk1) and cloned into pAS–Chk1. Primers were amino-terminal 1, 5′-CCCTTTACACCAACATATGGCTCA; carboxy-terminal 272, 5′-GGCGGATCCTTAAAGCCATGGGTGCTGTAC; carboxy-terminal 319, 5′CGCGGATCCTTAACCGCTATCGTTTTGAGATGC; amino-terminal 286, 5′-TAGTCTATCATATGGCCGATCCTGTAGCATTGGC; amino-terminal 313, 5′-GGATTACTCATATGGCATCTCAAAACGATAGC; carboxy-terminal 496, 5′-GATATTTGCTCGAGAAAGCG. Primer combinations for the following truncation constructs were pAS–chk1(1–272) amino-terminal 1 and carboxy-terminal 272; pAS–chk1 (1–319) amino-terminal 1 and carboxy-terminal 319; pAS–chk1 (286–496) amino-terminal 286 and carboxy-terminal 496; pAS–chk1 (313–496) amino-terminal 313 and carboxy-terminal 496.
Construction of plasmids for GST fusion proteins
For expression in E. coli, rad24 and rad25 cDNAs were cloned into the vector pGEX-1 (Smith and Johnson 1988). The BamHI fragment of rad24 was moved from pREP1/rad24 into pGEX-1 resulting in the incorporation of three additional amino acids between GST and Rad24. The BamHI fragment of rad25 was moved from pACT/rad25 into pGEX-1 using the BamHI site in the polylinker of pACT1 and a BamHI site 3′ of the stop codon in rad25. This construction results in the incorporation of 11 additional amino acids between GST and Rad25.
For expression as GST fusion proteins in S. pombe, Rad24 and Rad25 were first cloned into pREP1 as BamHI fragments. For Rad24, the BamHI sites were introduced during amplification by PCR as described above. For Rad25, the BamHI site in the polylinker of the pACT vector and a BamHI site 3′ of the stop codon were used for cloning. A PstI–NdeI fragment from pREP1–KZwis1 (a generous gift of Dr. Paul Russell, Scripps Research Institute, La Jolla, CA) containing the nmt1 promoter and GST coding sequence was cloned into pREP1/rad24 and pREP1/rad25 resulting in fusion of GST with either Rad24 or Rad25. Therefore, included in these constructs downstream of GST is a thrombin cutting site and a stretch of glycine residues plus the 3 or 11 additional amino acids encoded by the 5′-untranslated regions of rad24 and rad25, respectively. pREP1/GST was generated by amplifying GST from pGEX-2T (Smith and Johnson 1988) using 5′CGGGATCCTACATATGTCCCCTATACTAGG and 5′GCGGATCCTCATTTTGGAGGATGGTCG. The BamHI sites are underlined. The carboxy-terminal oligonucleotide includes an in-frame stop codon. The BamHI fragment from the amplified product was cloned into pREP1.
Expression of GST-14-3-3 proteins in E. coli and antibody production
Plasmids were transformed into E. coli strain BL21. Cells were grown at 37°C and induced to express GST, GST–Rad24, or GST–Rad25 by addition of 4 μm IPTG for 2 hr at 37°C. Lysates were prepared in PBS by sonication followed by centrifugation at 13,000 rpm in an SS-34 rotor for 10 min. Lysates were loaded onto a GSH–sepharose (Pharmacia) column, washed with PBS, and eluted with 50 mm glutathione. GST–Rad24 and GST–Rad25 were provided to Cocalico Biologicals Inc. (Reamstown, PA) for the generation of antibodies in rabbits. The anti-Rad24 antibodies are UMDNJ-21 and UMDNJ-55 and the anti-Rad25 antibodies are UMDNJ-18 and UMDNJ-56.
Expression of GST fusion proteins in S. pombe and camptothecin treatment
Cells were grown in the presence of 20 μm thiamine to repress the nmt1 promoter until expression was desired. To induce expression, cells were grown to early log phase, washed with PMA lacking thiamine, and resuspended in PMA. After 12 hr of induction, CPT was added to 40 μm and incubation continued for 2 hr. CPT lactone was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment, National Cancer Institute, National Institutes of Health (NIH). CPT was dissolved in DMSO to 10 mm and aliquots were stored at −20°C.
Preparation of lysates and precipitation of GST-containing proteins
Cell lysates were prepared based on the method described by Ansari et al. (1999). After cells were harvested, they were washed with cold ddH2O once, then again with HMKGD buffer (10 mm HEPES at pH 7.9, 5 mm MgCl2, 150 mm KCl, 10% glycerol, 0.5 mm DTT). Cells were resuspended in HMKGD buffer and frozen in liquid nitrogen. Frozen cells were lysed in liquid nitrogen by grinding with a pestle and mortar. After gentle stirring for 30 min at 4°C, lysates were centrifuged at 18,000 rpm in an SS-34 rotor for 30 min. Supernatant from this spin was then subjected to another 1 hr centrifugation at 37,000 rpm in a Ti70 rotor. The supernatant from the second spin was used as ‘lysate’ for all experiments. Aliquots of lysate were stored at −80°C. Chk1, Cdc25, Rad24, Rad25, and GST or GST fusions all remained soluble in the lysates.
To precipitate GST and GST fusion proteins, 50 μl of glutathione (GSH)–Sepharose was added to 1 ml of lysate containing 1.25 mg of protein. Incubation was carried out at 4°C with continuous rocking of the tube. GSH–Sepharose was pelleted by centrifugation at 3000 rpm in a microcentrifuge for 3 min. The beads were washed four times with 10 ml of HMKGD buffer and were resuspended in 15 μl of 2× Laemmli sample buffer for running on SDS-PAGE.
Immunoprecipitation of 14-3-3 proteins and immunoblotting
Lysates were prepared as described above, thawed on ice, and centrifuged at 14,000 rpm for 5 min in a microcentrifuge. Prior to the addition of antibody, the lysates were cleared by incubation with 15 μl of protein A–Sepharose (Pharmacia) for 15 min. The Sepharose beads were removed by centrifugation. To 1 ml of cleared lysate containing 10 mg of protein, 150 μl of Rad24 or Rad25 antibody or preimmune serum was added and incubated for 1 hr at 4°C. The lysate and antibody mixture was spun for 5 min at 14,000 rpm in a microcentrifuge to remove any protein aggregates. Protein A–Sepharose was added (50 μl) and incubated for 1 hr at 4°C to precipitate the immune complexes. The immunoprecipitates were washed four times with 1 ml of HMKGD buffer by centrifugation at 3000 rpm in a microcentrifuge. Precipitates were resuspended in 2× Laemmli sample buffer and resolved on SDS-PAGE.
For immunoblot analysis, proteins were transferred to nitrocellulose (Schleicher & Schuell, BA83). Blocking of the membranes and all antibody incubations and washes were done with 1% milk and 0.05% Tween 20 in PBS. mAb12CA5 was used at a 1:1000 dilution, and antibodies to Rad24 and Rad25 were used at a 1:10,000 dilution. Peroxidase-conjugated secondary antibodies were obtained from Boehringer-Mannheim (goat anti-mouse) and Jackson Immunoresearch (goat anti-rabbit) and used at 1:2500 and 1:50,000, respectively. Chemiluminescent detection of the peroxidase-conjugated secondary antibodies was done with the luminol (Renaissance) detection reagent from New England Nuclear Life Sciences Products. Filters were exposed to Kodak BioMax film for 2 min to 1 hr.
Acknowledgments
We are grateful to members of our laboratory for their contributions to this work. In particular, we thank Hsing-Yin Liu for generating pAS–Chk1 and initiating the two-hybrid screen, Lian Duan for the pREP1/GST–Chk1 construct, Shanhong Wan for constructing the chk1K38A:ep strain (NW307), and Jinfeng Liu for assistance in construction of Chk1 truncations. We thank Dr. Stephen J. Elledge for generously providing plasmids, libraries, and strains for the two-hybrid screen. We thank Dr. Antony M. Carr for the rad24::ura4, rad25::ura4, and cds1::ura4 mutants; Dr. Tomohiro Matsumoto for the rad3.T80 mutant; and Dr. Paul Russell for providing pREP1–KZwis1 for generating GST fusion protein expression vectors for S. pombe. We are indebted to Dr. Brad Nefsky for many helpful suggestions and discussions and for critically reading the manuscript. This research is supported by a grant from the NIH. (RO1-GM53194). N.C.W. gratefully acknowledges the UMDNJ Foundation and the Cancer Institute of New Jersey for support of this work in its early stages. N.C.W is the recipient of a Sinsheimer Scholar Award and a Junior Faculty Research Award from the American Cancer Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL walworna@umdnj.edu; FAX (732) 235-4073.
References
- Aitken A. 14-3-3 and its possible role in co-ordinating multiple signaling pathways. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJF, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoints and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari, A., T.-H. Cheng, and M. Gartenberg. 1999. Isolation of selected chromatin fragments from yeast by site-specific recombination in vivo. Methods Enzymol. 17: (in press). [DOI] [PubMed]
- Bai C, Elledge SJ. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1997;283:141–156. doi: 10.1016/s0076-6879(97)83013-3. [DOI] [PubMed] [Google Scholar]
- Conklin DA, Galaktionov K, Beach D. 14-3-3 proteins associate with cdc25 phosphatases. Proc Natl Acad Sci. 1995;92:7892–7896. doi: 10.1073/pnas.92.17.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Fox JE, Pei S. Identification of a binding sequence for the 14-3-3 protein within the cytoplasmic domain of the adhesion receptor, platelet glycoprotein Iba. J Biol Chem. 1996;271:7362–7367. doi: 10.1074/jbc.271.13.7362. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: Preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Enoch T, Carr AM, Nurse P. Fission yeast genes involved in coupling mitosis to the completion of DNA replication. Genes & Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Ford JC, al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Carr AM. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science. 1994;265:533–535. doi: 10.1126/science.8036497. [DOI] [PubMed] [Google Scholar]
- Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by Chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. JCell Biol. 1998a;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Yakowec PS, Dunphy WG. 14-3-3 proteins act as negative regulators of the mitotic inducer Cdc25 in Xenopus egg extracts. Mol Biol Cell. 1998b;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Bienkowska J, Petosa C, Collier RJ, Fu H, Liddington R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- Liu Y-C, Elly C, Yoshida H, Bonnefoy-Berard N, Altman A. Activation-modulated association of 14-3-3 proteins with Cbl in T cells. J Biol Chem. 1996;271:14591–14595. doi: 10.1074/jbc.271.24.14591. [DOI] [PubMed] [Google Scholar]
- Matsumoto T. A fission yeast homolog of CDC20/p55CDC/Fizzy is required for recovery from DNA damage and genetically interacts with p34cdc2. Mol Cell Biol. 1997;17:742–750. doi: 10.1128/mcb.17.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Michaud NR, Fabian JR, Mathes KD, Morrison DK. 14-3-3 is not essential for Raf-1 function: Identification of Raf-1 proteins that are biologically activated in a 14-3-3- and Ras-independent manner. Mol Cell Biol. 1995;15:3390–3397. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Nefsky B, Beach D. Pub1 acts as an E6-AP-like protein ubiquitin ligase in the degradation of cdc25. EMBO J. 1996;15:1301–1312. [PMC free article] [PubMed] [Google Scholar]
- O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu A, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: Regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ, III, Wu Z, Stephenson MT, Piwnica-Worms H. C-TAK1 protein kinase phosphorylates human Cdc25C on 216 and promotes 14-3-3 protein binding. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes & Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- Rowley R, Subramani S, Young PG. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: Linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single Ataxia Telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- Wan, S., H. Capasso, and N.C. Walworth. 1999. The topoisomerase I poison camptothecin generates a Chk1-dependent DNA-damage checkpoint signal in fission yeast. Yeast (in press). [DOI] [PubMed]
- Weinert T. A DNA damage checkpoint meets the cell cycle engine. Science. 1997;277:1450–1451. doi: 10.1126/science.277.5331.1450. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- ————— Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Xiao B, Smerdon SJ, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ. Structure of a 14-3-3 protein and implications for coordination of multiple signaling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]