Abstract
Leptospira interrogans is the causative agent of leptospirosis, which is an emerging zoonotic disease. Resistance to stress conditions is largely uncharacterized for this bacterium. We therefore decided to analyze a clpB mutant that we obtained by random transposon mutagenesis. The mutant did not produce any of the two isoforms of ClpB. The clpB mutant exhibited growth defects at 30° and 37°C and in poor nutrient medium and showed increased susceptibility to oxidative stress, whereas the genetically complemented strain was restored in ClpB expression and in vitro wild-type growth. We also showed that the clpB mutant was attenuated in virulence in an animal model of acute leptospirosis. Our findings demonstrate that ClpB is involved in the general stress response. The chaperone is also necessary, either directly or indirectly, for the virulence of the pathogen L. interrogans.
INTRODUCTION
Spirochetes belonging to the pathogenic Leptospira spp. are the causative agents of leptospirosis, which is an emerging disease with more than 500,000 annual severe cases worldwide (1). Leptospires colonize renal tubules of animal reservoirs and are excreted into the environment, where the bacteria can survive for weeks or even months in moist soil and water (37). Leptospirosis is usually transmitted to humans by contact of abraded skin or mucous membranes with water that is contaminated with the urine of infected animals. Leptospires then disseminate into the host, causing febrile illnesses and severe disease manifestations that can lead to death (16).
Like many other pathogenic bacteria, leptospires are subjected to environmental stresses during infection. Temperature constitutes an important signal in Leptospira spp., leading to alterations in gene transcription (20, 21, 32) and protein expression (3, 21). Stressful conditions may result in damage to intracellular proteins. Proteome analysis of L. interrogans has revealed that stress and heat shock proteins, including the chaperones DnaK, GrpE, and GroEL as well as the Hsp100/Clp chaperone ClpB, are upregulated at 37°C in comparison to 30°C, which is the optimal temperature for Leptospira in vitro growth (21).
The Clp chaperones and proteases are highly conserved across prokaryotes and are found in eukaryotes. Clp ATPases are divided into two main classes (35). Members of class I, which includes ClpA, ClpB, ClpC, ClpD, ClpE, and ClpL, contain two ATP nucleotide-binding domains or AAA+ (ATPases associated with various cellular activities) domains that have characteristic Walker A and Walker B nucleotide-binding motifs. Proteins of class II, which include ClpX and ClpY (HslU), are smaller and contain only one AAA+ domain. Most of these Clp ATPases (except ClpB) are associated with a proteolytic subunit (ClpP or ClpQ/HslV). These chaperones and proteases function mainly in the disaggregation, unfolding, and degradation of native as well as misfolded proteins. They have been shown to be involved in general stress responses in various bacteria.
In Escherichia coli, ClpB is a chaperone required for resistance to high temperature that assists DnaK, DnaJ, and GrpE in repairing damaged proteins (13, 36). A clpB mutant was discovered during a systematic search for insertional mutants obtained by random transposon mutagenesis in the pathogen Leptospira interrogans (24). In previous studies, clpB mutants of Salmonella enterica serovar Typhimurium, Mycoplasma pneumoniae, Porphyromonas gingivalis, Listeria monocytogenes, and Francisella tularensis have been shown to be relatively avirulent compared with their parental strains (5, 6, 15, 22, 38). In the present study, inactivation of the clpB gene of L. interrogans resulted in a mutant impaired in in vitro growth under stress conditions and exhibiting attenuation in virulence. Complementation with the full-length wild-type gene restored, at least partially, the stress survival and virulence phenotype to the clpB mutant, therefore demonstrating that ClpB is essential for survival under stress conditions and during in vivo infection. The only other described example of functional complementation in L. interrogans was obtained for the loa22 mutant (34). This study should help us to understand the role of ClpB in leptospiral pathogenesis.
MATERIALS AND METHODS
Leptospiral strains and culture conditions.
The pathogens L. interrogans serogroup Canicola strain Kito and L. interrogans serovar Manilae strain L495 were used in this study. Leptospira organisms were cultivated in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (9, 14) and on 1% agar-EMJH plates at 30°C. When appropriate, spectinomycin or kanamycin was added to the culture medium at 50 μg ml−1.
Identification of clpB mutants and construction of complemented strains.
Random insertion mutagenesis was carried out with L. interrogans serogroup Canicola strain Kito, L. interrogans serovar Manilae strain L495, and a kanamycin-resistant Himar1 transposon as previously described (24). Among the kanamycin transformants, we identified mutants with an insertion in clpB (LA1879) at positions 1852876 and 1853989 in the large chromosomes (nucleotide positions based on the genome of L. interrogans serovar Lai strain 56601) of strains L495 and Kito, respectively.
For complementation, clpB was amplified with primers ClpBasc1 and ClpBasc2 (Table 1) from L. interrogans strain Kito, and the amplified product, which was flanked by AscI restriction sites, was cloned into pCR2.1 (Invitrogen), thereby generating pCRclpB. The plasmid construct was verified by nucleotide sequencing. A spectinomycin resistance cassette was amplified with primers SpcSmA and SpcSmB and cloned into SmaI-digested pCRclpB. The resulting plasmid was then digested with AscI, thereby generating a 4.4-kb DNA fragment carrying the wild-type clpB gene and the spectinomycin resistance cassette. The DNA fragment was inserted into the AscI restriction site of the transposon carried by the conjugative plasmid pCJTKS1 (30). The plasmid was introduced into clpB mutants by conjugation as previously described (30). After 4 to 6 weeks of incubation, spectinomycin-resistant transformants were inoculated into liquid medium, and after DNA extraction, the transposon insertion site was identified by semirandom PCR (24). We selected one mutant, KS2 from strain Kito, exhibiting the additional transposon in an intergenic region between LA1293 and LA1294 of the large chromosome of L. interrogans. Two other mutants, KS1 and KS3, had the transposon inserted into LA2656 and LA0140. Another complemented strain, exhibiting the transposon in LA4340, was also obtained from the L495 clpB mutant strain (serovar Manilae).
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′)a | Reference |
|---|---|---|
| RTclb1 | AACCAAAGTGATCGGACAGG | This study |
| RTclb2 | ATACATGGAGGCTCATTCCG | This study |
| CLPBA | CGAGTGTAATCTTATTTTTTA | This study |
| 998-1 | CCGAAAAATTGGGAAATCCG | This study |
| ClpBascI | TTGGCGCGCCCGGGTGCAAAACATACTTTCAACC | This study |
| ClpBasc2 | TTGGCGCGCCTAAAAAATAAGATTACACTCG | This study |
| SpcSmA | TCCCCCGGGGGCCCGAGCTTCAAGGAAGA | This study |
| SpcSmB | TCCCCCGGGGGAAAGTAAGCACCTGTTATTGC | This study |
| KS2R | GCACTTTCTGCTCCATTCAA | This study |
| KS2F | CGGCTTTCCTTCTAACAAAGG | This study |
| RTFLAB1 | GAGAGAAACACCGAAGACGG | 20 |
| RTFLAB2 | TGAATAGCAAGAACCCGGAT | 20 |
| dnaK1 | CGATTACGAACGCGGTAAAT | This study |
| dnaK2 | TTGGGGAGTGAATTCTCCTG | This study |
| hsp1 | GATTAAACCTCTGGGCGACA | This study |
| hsp2 | GGATGAGTTTCCCGTCTTCA | This study |
| LA | GGCGGCGCGTCTTAAACATG | 23 |
| LB | TTCCCCCCATTGAGCAAGATT | 23 |
| rt rpob1 | ATGGAGCGGAACGTGTAGTC | This study |
| rt rpob2 | CTTCGTTCGTTCCATGTCCT | This study |
AscI restriction sites are underlined.
Genomic DNAs were extracted from pellets of 20-ml cultures by using a QIAamp DNA blood minikit (Qiagen, Inc., Valencia, CA). Confirmation of genotypes was performed by using PCR with primer pairs 998-1-RTclb2 and KS2R-KS2F (Table 1), which target the flanking sequences of the insertion site of the transposon, and Southern blots of EcoRI-digested DNA were probed for hybridization with the kanamycin resistance cassette.
Western blotting.
Expression of ClpB was verified by Western blotting. Polyclonal rabbit antiserum was prepared against L. interrogans ClpB (domain 158-334) by Proteogenix (Oberhausbergen, France). Protein concentrations of whole-cell lysates were determined by the Bradford protein assay (Coomassie Plus protein assay; Pierce). Ten micrograms of crude protein extract was separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were then transferred to Immobilon polyvinylidene fluoride (PVDF) membranes and probed with anti-E. coli ClpB (7) and L. interrogans ClpB antibodies at dilutions of 1:5,000 and 1:1,000, respectively. ImpL63 antiserum diluted 1:10,000 (a generous gift from Dave Haake) was also used as a loading control. The secondary antibody was anti-rabbit peroxidase, used at a dilution of 1:150,000. Blots were developed with a chemiluminescence kit (SuperSignal West Pico chemiluminescent substrate; Thermo Scientific, IL) according to the manufacturer's instructions.
Growth with limited nutrient concentrations and oxidative stress assay.
To test for growth of Leptospira strains with limited nutrient concentrations, exponential-phase cultures of L. interrogans strains were inoculated (0.1 ml in 9 ml) in EMJH and EMJH diluted 1:2 in sterile water, and cultures were grown at 30°C under agitation. Cells were also grown in EMJH supplemented with 10% rabbit serum or M199 medium (PAN Biotech) with agitation at 37°C.
For disk diffusion assays, bacterial cells (approximately 106 bacteria) were spread onto EMJH plates, and 6-mm- or 12-mm-diameter filter disks containing 20 and 50 mM hydrogen peroxide and 10 and 20 mM butyl peroxide were placed in the center of plates. The plates were incubated for 15 days at 30°C, and the diameters of the zones of inhibition created around the disks were measured. Plate assays were repeated at least three times, and similar results were obtained each time.
Heat shock assay.
For heat shock assays, cultures were grown with agitation at 30°C until mid-exponential phase (optical density at 420 nm [OD420], 0.2 to 0.3) and then transferred to 30 or 42°C for 1, 2, 4, or 6 h. Cells were then harvested for RNA extraction. One milliliter of TRIzol (Invitrogen) was added to a pellet from a 2-ml culture of leptospires, and RNA was purified and reverse transcribed as previously described (2). The expression of flaB, rpoB, clpB, dnaK, and hsp10 was determined with the primer pairs indicated in Table 1. The PCRs were performed and analyzed with a CFX96 real-time PCR detection system (Bio-Rad). The fold change in the amount of target gene was normalized to the amount of 16S cDNA obtained with primers LA and LB (Table 1) and relative to the expression at time zero at 30°C by using the comparative threshold cycle (CT) method [also called the 2(−ΔΔCT) method] (19).
Animal experiments.
L. interrogans strains were tested for virulence in the gerbil model of acute leptospirosis. Groups of 28-day-old gerbils (Charles River Laboratories) were inoculated intraperitoneally with 108 leptospires. Animals were monitored daily for clinical signs of leptospirosis (i.e., prostration, jaundice, etc.) and for survival for up to 21 days postinfection. Protocols for animal experiments were prepared according to the guidelines of the Animal Care and Use Committees of the Institut Pasteur.
Statistical analysis.
Data were graphed and analyzed using GraphPad Prism 5.0c (GraphPad Software, San Diego, CA). Susceptibility to oxidative stress was statistically analyzed by one-way analysis of variance. Survival curves were compared using the log rank (Mantel Cox test) analysis. P values of <0.05 were considered to be significant.
RESULTS
Genome analysis of clp genes in Leptospira spp.
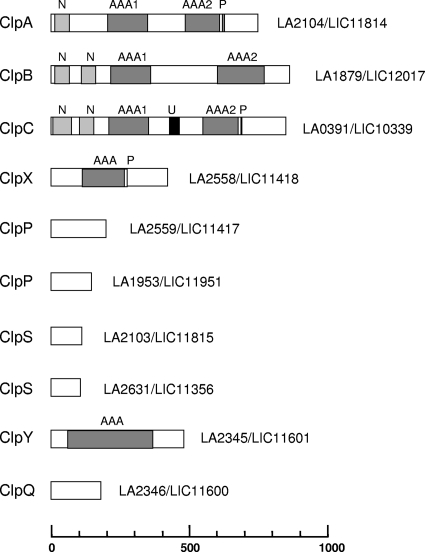
The complete genome sequences of strains belonging to two pathogenic species, L. interrogans and Leptospira borgpetersenii, and one saprophytic species, Leptospira biflexa, were recently determined (4, 28, 31, 33). The genomes of the sequenced strains of Leptospira spp. possess clpA, clpB, clpX, clpP, clpS, clpY, and clpQ. Leptospira spp. also possess clpC, which is usually present in Gram-positive but absent in Gram-negative bacteria (Fig. 1). It has to be noted that clpC is also present in the spiro- chetes Treponema pallidum (TP0549) and Borrelia burgdorferi (BB0834). The genes of the Clp system are conserved in saprophytic and pathogenic Leptospira species and constitute part of the core genes of the genus Leptospira (31). ClpA, ClpB, ClpC, ClpX, and ClpY are proteins belonging to the Clp/Hsp100 family that carry domains typical of the AAA+ superfamily of ATPases. Amino acid sequence analysis predicted the presence of two well-conserved AAA domains in L. interrogans ClpA, ClpB, and ClpC and one AAA domain in ClpX and ClpY/HslU (Fig. 1; see Fig. S1 in the supplemental material). The Clp ATPase proteins ClpA, ClpC, and ClpX possess a ClpP recognition binding motif (also called a P-loop motif), (L/I/V)G(F/L), that allows association with the ClpP peptidase, resulting in a Clp proteolytic complex (see Fig. S1). ClpS is an adaptor protein binding to the ClpA N-terminal domain and altering the substrate specificity of ClpA. In Leptospira spp., clpA and clpS are adjacent to each other on the chromosome; another putative clpS gene was found elsewhere in the chromosome. No genes for adaptor proteins that target E. coli ClpX (SspB, RssB, and UmuD) have been identified thus far in the Leptospira genomes. Although the sequences and functions of ClpA and ClpC are quite similar, ClpC can be distinguished from ClpA by a relatively longer spacer region between the two ATP-binding domains (Fig. 1; see Fig. S1). In addition, a UVR domain resembling the UVRB/UVRC interaction domain was identified in several ClpC proteins (12). The ClpB ATPases do not associate with ClpP and function as chaperones. Finally, ClpQ interacts with the ClpY chaperone, forming the ClpYQ (also known as HslUV) protease (17).
Fig. 1.
Genome analysis of Clp chaperones and proteases in Leptospira spp. ClpA, ClpB, ClpC, ClpX, and ClpY belong to the family of Clp ATPases. The ATP domains of the AAA+ superfamily of ATPases are indicated (AAA). The P-loop domain (P), involved in the interaction with the ClpP peptidase, the N-terminal protein binding domain (N), and a domain resembling the UVRB/UVRC interaction domain (U) are also indicated. The genes in L. interrogans serovar Lai strain 56601 and L. interrogans serovar Copenhageni strain Fiocruz L1-130 are defined by their LA and LIC numbers, respectively.
ClpB expression in L. interrogans.
The clpB gene of L. interrogans encodes a protein of 860 amino acids which exhibits 95% and 62% similarity with orthologs in the pathogen L. borgpetersenii and the saprophyte L. biflexa, respectively. Among the pathogenic strains L. interrogans serogroup Canicola strain Kito, L. interrogans serovar Copenhageni strain Fiocruz L1-130, L. interrogans serovar Lai strain 56601, and L. interrogans serovar Manilae strain L495, the complete nucleotide sequence of clpB, including the promoter region (300 bp upstream of the start codon), exhibits 99% identity at the nucleotide level (data not shown).
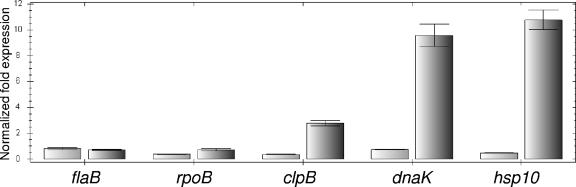
In order to characterize ClpB properties in Leptospira spp., we assessed the effects of heat shock on the transcription of clpB in L. interrogans serogroup Canicola strain Kito (Fig. 2). Transcription of the clpB gene increased during a temperature shift from 30°C to 42°C, reaching a 3-fold increase after 2 h of exposure to 42°C. Transcriptional responses of the L. interrogans dnaK and hsp10 genes to heat shock conditions were also measured by reverse transcription-quantitative PCR (RT-qPCR) and showed elevated expression of both genes. In contrast, the expression of the rpoB and flaB genes, which were included as controls to ensure reliability of the comparative CT analysis, was unchanged (Fig. 2).
Fig. 2.
Relative gene expression of L. interrogans clpB, dnaK, and hsp10. Exponential-phase cultures of L. interrogans were incubated at 30°C (left columns) and 42°C (right columns) for 2 h. Transcripts of the selected genes were measured by RT-qPCR, and the data were analyzed using the comparative critical threshold (ΔΔCT) method. The error bars indicate the standard deviations for three replicates.
Construction of L. interrogans mutants.
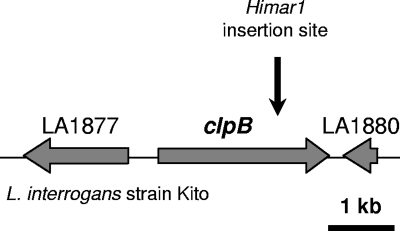
We previously obtained about 1,000 random mutants with characterized transposon insertion points in L. interrogans pathogenic strains (24). We pursued this work in the laboratory, and we recently identified one transposon mutant exhibiting an insertion in clpB (LA1879) (Fig. 3). The insertion of a single transposon in the mutant strain was confirmed by Southern blotting using a transposon-specific probe (data not shown).
Fig. 3.
Genetic locus of clpB in L. interrogans. The insertion site of Himar1 in the chromosome of the clpB mutant in strain Kito is indicated.
Because there is no replicative plasmid vector available for pathogenic Leptospira (16), we also generated a clpB-complemented mutant by insertion of the transposon carrying clpB and a spectinomycin resistance cassette. We identified the transposon insertion sites in several transformants and selected one strain, KS2, exhibiting the transposon in an intergenic region. PCR and Southern blotting confirmed the presence of the wild-type clpB gene in the wild-type and complemented strains (data not shown).
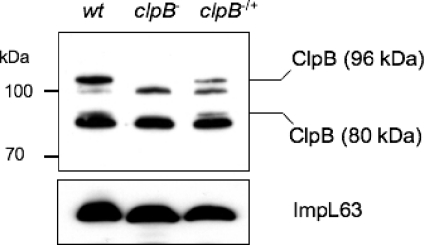
Western blots of whole-cell lysates of the L. interrogans wild-type strain, obtained by using antibodies raised against L. interrogans ClpB, showed several bands (Fig. 4). The use of E. coli ClpB antiserum showed similar results (data not shown). The L. interrogans ClpB protein (96 kDa) shares >60% similarity with L. interrogans ClpA (83 kDa) and ClpC (95 kDa), which are also members of the Hsp100/Clp family. This may explain the presence of more than one ClpB-reactive band. Interestingly, E. coli was found to use two transcriptional start sites to express two forms of ClpB, a 93-kDa and a 79-kDa protein (36). The L. interrogans ClpB protein could also be synthesized in two forms. Nucleotide sequence analysis of the L. interrogans clpB gene revealed a classical ribosome binding site (RBS) immediately upstream of at least three putative alternative start codons (ATG or TTG). These potential internal translation initiation sites should result in proteins with calculated molecular masses of 94, 89, and 80 kDa (instead of the 96-kDa mass for the full-length ClpB protein). Alignment of amino acid sequences showed that the alternative translational start of the 80-kDa polypeptide of L. interrogans ClpB coincides with amino acid 149 of E. coli ClpB, which is the start codon of the smaller form of E. coli ClpB (79 kDa) (36). Gene inactivation of clpB resulted in the disappearance of two bands (Fig. 4). Western blot analysis of the complemented strain with the ClpB antiserum detected two reactive bands similar to those for the wild-type strain (Fig. 4), further confirming the identity of the 96-kDa polypeptide and another smaller polypeptide (80 kDa) as ClpB.
Fig. 4.
ClpB in L. interrogans strains. Western analysis was performed with whole-cell extracts of wild-type (wt), clpB mutant (clpB−), and complemented clpB mutant (clpB−/+) strains grown at 30°C, using an antibody to L. interrogans ClpB. Data shown are from a single experiment and are representative of at least three experiments performed with similar results. ImpL63 antibody was used as a loading control in Western blots.
In vitro phenotype.
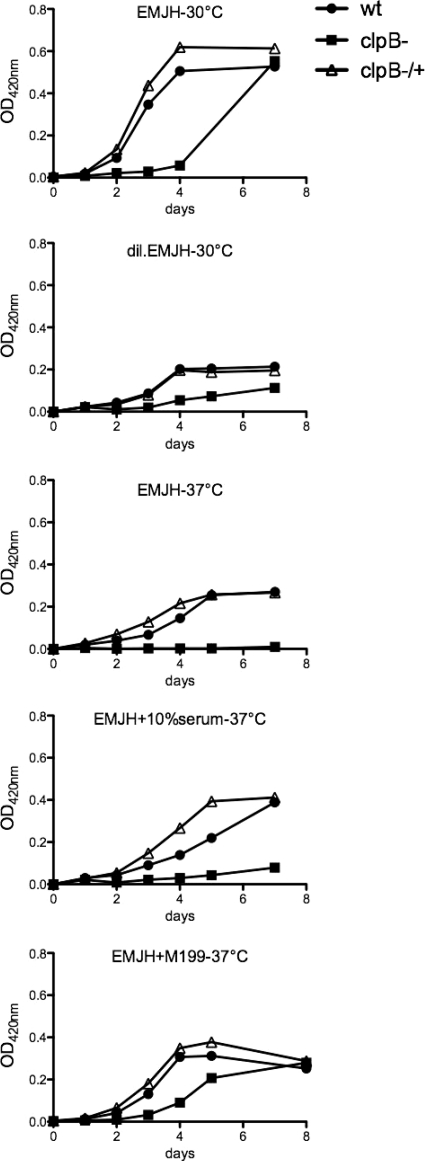
The L. interrogans clpB mutant presented a similar motility and cell morphology to those of the wild-type strain (data not shown). However, analysis of the growth curves of the clpB mutant under normal growth conditions (i.e., aerobic conditions at 30°C) indicated that the lag phase of the growth curve was 48 h longer than that for the wild-type and complemented strains (Fig. 5). The clpB mutant did reach the maximum growth yield of the wild-type strain. The wild-type and complemented strains had similar cell growth kinetics in EMJH medium at 30°C. The capacity of the clpB mutant to grow under a variety of stress conditions was also examined. No significant differences in growth between the complemented and parental strains were observed when cells were grown at 30°C with limited nutrient concentrations (diluted EMJH). Under similar conditions, the clpB mutant had greatly reduced growth (Fig. 5). Pathogenic Leptospira strains usually do not grow well in artificial medium at 37°C (10). In our hands, the Kito strain showed greatly reduced growth in EMJH, even with supplemented serum or tissue culture medium, at 37°C in comparison to that for cultures at 30°C. Under these conditions, the growth rate of the clpB mutant was dramatically reduced, sometimes with no evidence of multiplication, compared to those of the wild-type and complemented strains (Fig. 5).
Fig. 5.
Growth kinetics of wild-type, clpB mutant, and complemented clpB mutant strains of L. interrogans. Strains were grown in EMJH, EMJH supplemented with 10% rabbit serum, EMJH supplemented with 10% M199 medium, and diluted EMJH medium, and cultures were incubated at 30°C or 37°C with shaking. Growth was monitored by measuring the OD420.
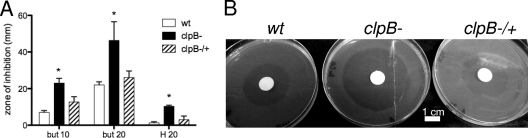
To examine whether ClpB contributes to oxidative stress resistance, disk diffusion assays were carried out with hydrogen peroxide and butyl peroxide. The zone of inhibition for the clpB mutant was significantly larger than the zone of inhibition for the wild type, indicating that the clpB mutant is more susceptible to oxidative stress (Fig. 6). Additionally, complementation of the clpB mutant restored resistance to the wild-type level (Fig. 6). These results indicate that ClpB plays a major role in resistance to peroxides.
Fig. 6.
Effects of hydrogen peroxide and butyl peroxide on L. interrogans strains. (A) Disk diffusion assays with 10 mM and 20 mM butyl peroxide (but) and 20 mM hydrogen peroxide (H), using the wild-type (wt), clpB mutant (clpB−), and complemented clpB mutant (clpB−/+) strains. Bars show the diameters of the zones of inhibition in millimeters. The means and standard deviations for three independent experiments are shown, with statistical significance indicated by asterisks (P < 0.05). (B) Plate assay with 20 mM butyl peroxide.
Together, our results show that both in vitro growth deficiencies and the lower resistance to heat and oxidative stresses in the mutant are due to inactivation of clpB and can be complemented by a transposon-carried clpB copy.
In vivo phenotype.
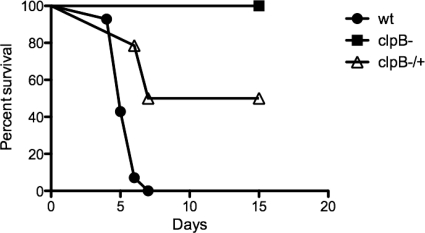
To determine if ClpB is important for the ability of L. interrogans to cause disease, gerbils were infected with 108 organisms (three independent experiments with groups of 4 to 6 animals) of either the wild-type, clpB mutant, or complemented clpB mutant version of L. interrogans strain Kito. The clpB mutant demonstrated a complete loss of virulence, with no clinical signs of leptospirosis, during the 21-day follow-up period, while the wild-type strain caused death in 4 to 7 days after challenge (Fig. 7). The survival curve for the mutant was statistically different from the curve generated for gerbils infected with the wild type (P < 0.0001). Necropsy evaluation of gerbils infected with the clpB mutant did not show macroscopic lesions associated with leptospirosis (data not shown). The mutant phenotype could be complemented by reintroduction of clpB, with the log rank test between the mutant and complemented strains giving a P value of 0.0027. The complemented strain caused death in 50% of the inoculated animals (Fig. 7). Our results showed that the lack of virulence of the clpB mutant could be complemented partially in vivo by expressing clpB from the Himar1-derived transposon, which is stably integrated into the chromosome of L. interrogans without antibiotic selection.
Fig. 7.
Cumulative survival rates for gerbils infected with wild-type, clpB mutant, and complemented clpB mutant strains of L. interrogans. The survival curves (n = 14 animals) show the results for three independent experiments.
DISCUSSION
The Clp system is involved in the general stress response as well as the virulence of many pathogenic bacteria. Given the variety of environmental stresses that L. interrogans may encounter during its life cycle, it is likely that the Clp proteins play a role in stress survival and virulence.
Genome sequencing of Leptospira strains has enabled the identification of putative molecular chaperones and proteases. These include the ClpXP chaperone-protease, which is the most ubiquitous of the Clp proteases; ClpYQ, which is found in most proteobacteria and in certain Gram-positive bacteria; and ClpAP and its adaptor, ClpS, which are found in the Gram-negative proteobacteria. Leptospira spp. possess a double-membrane structure, including an outer membrane which contains lipopolysaccharide (LPS), and are therefore related to Gram-negative bacteria. In contrast to Gram-negative bacteria, which possess ClpA but not ClpC, Leptospira spp. were found to harbor both ClpA and ClpC, which are orthologs associated with ClpP and showing similar functions in other bacteria. Finally, like other organisms, including eukaryotes, Leptospira spp. possess a ClpB-encoding gene. Immunoblotting and sequence analysis indicated that two forms of ClpB may be produced in L. interrogans, like the case in E. coli (36). The biological role of the smaller ClpB form, which does not contain the substrate-interacting N-terminal domain, is not well understood. In E. coli, the two isoforms form hetero-oligomers that optimize the chaperone activity by synergistic cooperation (27).
A previous microarray study showed that L. interrogans clpB or LA1879/LIC12017 (misannotated clpA), together with other stress and heat shock genes, was upregulated 3.5-fold at 37°C in comparison to 30°C (20). This was also shown at the protein level (21). We confirmed that clpB is upregulated during heat stress. ClpB was also found to be downregulated in response to serum (29) and under iron-depleted conditions (11).
Only a few L. interrogans mutants showed attenuation in virulence in the animal model of leptospirosis. These avirulent mutants were disrupted in genes of the LPS biosynthesis locus (25) and in genes encoding a heme oxygenase (26), a lipoprotein, Loa22, of unknown function (34), or the flagellar motor switch protein FliY (18). However, only the attenuation of the loa22 mutant was confirmed by complementation studies (34).
In this study, we isolated a mutant with an insertion in clpB in L. interrogans strain Kito. Our results suggest that ClpB plays an important role in in vitro growth and in responses to oxidative and temperature stresses in L. interrogans. Transformation of the clpB mutant with a construct to express wild-type ClpB restored the wild-type phenotype in vitro, confirming that all of the phenotypes described here for the clpB mutant were indeed due to the loss of function of this gene.
The clpB mutant of L. interrogans strain Kito demonstrated a loss of virulence. The attenuation of lethality in infected gerbils could be a consequence of downregulation of virulence factors, in vivo growth deficiency, or increased susceptibility to stress conditions. The attenuation of lethality was only partially restored in gerbils. The incomplete complementation of the clpB mutant may have been due to poor in vivo expression of clpB from the integrated Himar1 transposon. Integration of the wild-type clpB gene into a chromosomal locus other than the original one may not allow optimal expression or regulation of clpB in complemented strains, as suggested by the level of expression of ClpB-reactive bands in Western blot analysis (Fig. 4). The only other example of complementation of an avirulent mutant of pathogenic Leptospira spp. also showed partial restoration of virulence, with the complemented strain being lethal in 57 to 80% of infected animals (34).
Interestingly, we recently identified another clpB mutant in the pathogen L. interrogans serovar Manilae strain L495 among random transposon mutants (24). Inactivation of clpB was therefore obtained in two distinct L. interrogans serogroups, i.e., serogroups Canicola (strain Kito) and Pyrogenes (strain L495). The L495 clpB strain exhibited growth defects at 37°C and at 30°C in poor nutrient medium, but not at 30°C in regular EMJH medium, in comparison to the parental and complemented strains. The susceptibility to oxidative stress of the clpB mutant was significantly higher than the susceptibility of both the wild-type and complemented cells (see Fig. S2 in the supplemental material). However, the clpB mutant of strain L495 did not show decreased virulence compared to the wild-type strain in the animal model of leptospirosis (see Fig. S2). The reason for this difference of phenotypes between the clpB mutants of strains Kito and L495 is unclear. The clpB gene was not found to be duplicated in the genome of the L. interrogans clpB mutant (B. Adler, unpublished data). Differences in the regulation and/or expression of clpB may exist among clinical strains. Gene inactivation of clpB in strain L495 may also result in compensatory mechanisms, such as increased activity levels of Clp proteases or other chaperone machineries. The virulence of pathogens is partly due to their ability to survive in the host by escaping natural defense mechanisms. ClpB has been shown to be a key determinant in tolerance to oxidative stress conditions. Other mechanisms of resistance to oxidative stress, such as enzymatic degradation of peroxide, may also vary among pathogenic Leptospira strains (8), allowing persistence of virulence in the L495 clpB strain.
In conclusion, we demonstrated that ClpB, which is induced by heat stress, is required for growth in poor nutrient media and for resistance to oxidative and heat stresses. ClpB also appears to play a major role in virulence, as assessed using the Kito mutant. We believe that our results will provide insights for understanding the mechanisms of tolerance of pathogenic Leptospira spp. to stress conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Adler and G. Murray (Monash University) for supplying the Manilae strains. We are thankful to M. Zolkiewski (Kansas State University) for the generous gift of E. coli ClpB antiserum and to P. Béguin (Institut Pasteur) for the preparation of recombinant L. interrogans ClpB. We thank N. Benaroudj for critical reading of the manuscript and for analysis of Clp sequences. We also thank G. Santos and P. Ristow (Gonçalo Moniz Research Center) for animal experiments.
This work was supported by the Institut Pasteur, by National Institutes of Health grants (R01 AI052473, U01 AI0088752, and D43 TW000919), by the French Ministry of Research (ANR-08-MIE-018), and by the doctoral program from the Région Île-de-France (DIM Malinf).
This work is part of the doctoral thesis of K. Lourdault.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 5 July 2011.
REFERENCES
- 1. Abela-Ridder B., Sikkema R., Hartskeerl R. A. 2010. Estimating the burden of human leptospirosis. Int. J. Antimicrob. Agents 36:S5–S7 [DOI] [PubMed] [Google Scholar]
- 2. Aviat F., Slamti L., Cerqueira G. M., Lourdault K., Picardeau M. 2010. Expanding the genetic toolbox for Leptospira species by generation of fluorescent bacteria. Appl. Environ. Microbiol. 76:8135–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beck M., et al. 2009. Visual proteomics of the human pathogen Leptospira interrogans. Nat. Methods 6:817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bulach D. M., et al. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capestany C. A., Tribble G. D., Maeda K., Demuth D. R., Lamont R. J. 2008. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J. Bacteriol. 190:1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chastanet A., Derre I., Nair S., Msadek T. 2004. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 186:1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chow I. T., Barnett M. E., Zolkiewski M., Baneyx F. 2005. The N-terminal domain of Escherichia coli ClpB enhances chaperone function. FEBS Lett. 579:4242–4248 [DOI] [PubMed] [Google Scholar]
- 8. Corin R. E., Boggs E., Cox C. D. 1978. Enzymatic degradation of H2O2 by Leptospira. Infect. Immun. 22:672–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellinghausen H. C., McCullough W. G. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am. J. Vet. Res. 26:45–51 [PubMed] [Google Scholar]
- 10. Ellinghausen H. C. J. 1973. Growth temperatures, virulence, survival, and nutrition of leptospires. J. Med. Microbiol. 6:487–497 [DOI] [PubMed] [Google Scholar]
- 11. Eshghi A., Cullen P. A., Cowen L., Zuerner R. L., Cameron C. E. 2009. Global proteome analysis of Leptospira interrogans. J. Proteome Res. 8:4564–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frees D., Savijoki K., Varmanen P., Ingmer H. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63:1285–1295 [DOI] [PubMed] [Google Scholar]
- 13. Goloubinoff P., Mogk A., Zvi A. P., Tomoyasu T., Bukau B. 1999. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. U. S. A. 96:13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson R. C., Harris V. G. 1967. Differentiation of pathogenic and saprophytic leptospires. J. Bacteriol. 94:27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kannan T. R., Musatovova O., Gowda P., Baseman J. B. 2008. Characterization of a unique ClpB protein of Mycoplasma pneumoniae and its impact on growth. Infect. Immun. 76:5082–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ko A. I., Goarant C., Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kress W., Maglica Z., Weber-Ban E. 2009. Clp chaperone-proteases: structure and function. Res. Microbiol. 160:618–628 [DOI] [PubMed] [Google Scholar]
- 18. Liao S., et al. 2009. Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol. 9:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) methods. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 20. Lo M., et al. 2006. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 74:848–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lo M., Cordwell S. J., Bulach D. M., Adler B. 2009. Comparative transcriptional and translational analysis of leptospiral outer membrane protein expression in response to temperature. PLoS Negl. Trop. Dis. 3:e560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meibom K. L., et al. 2008. The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol. Microbiol. 67:1384–1401 [DOI] [PubMed] [Google Scholar]
- 23. Merien F., Amouriaux P., Perolat P., Baranton G., Saint Girons I. 1992. PCR for detection of Leptospira spp. in clinical samples. J. Clin. Microbiol. 30:2219–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murray G. L., et al. 2009. Genome-wide transposon mutagenesis in pathogenic Leptospira spp. Infect. Immun. 77:810–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murray G. L., et al. 2010. Mutations affecting Leptospira interrogans lipopolysaccharide attenuate virulence. Mol. Microbiol. 78:701–709 [DOI] [PubMed] [Google Scholar]
- 26. Murray G. L., et al. 2009. Leptospira interrogans requires heme oxygenase for disease pathogenesis. Microbes Infect. 11:311–314 [DOI] [PubMed] [Google Scholar]
- 27. Nagy M., et al. 2010. Synergistic cooperation between two ClpB isoforms in aggregate reactivation. J. Mol. Biol. 396:697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nascimento A. L., et al. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patarakul K., Lo M., Adler B. 2010. Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum. BMC Microbiol. 10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Picardeau M. 2008. Conjugative transfer between Escherichia coli and Leptospira spp. as a new genetic tool. Appl. Environ. Microbiol. 74:319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Picardeau M., et al. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qin J. H., et al. 2006. Genome-wide transcriptional analysis of temperature shift in L. interrogans serovar Lai strain 5660. BMC Microbiol. 6:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ren S., et al. 2003. Unique and physiological and pathogenic features of Leptospira interrogans revealed by whole genome sequencing. Nature 422:888–893 [DOI] [PubMed] [Google Scholar]
- 34. Ristow P., et al. 2007. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 3:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schirmer E. C., Glover J. R., Singer M. A., Lindquist S. 1996. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21:289–296 [PubMed] [Google Scholar]
- 36. Squires C. L., Pedersen S., Ross B. M., Squires C. 1991. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 173:4254–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trueba G., Zapata S., Madrid K., Cullen P., Haake D. 2004. Cell aggregation: a mechanism of pathogenic Leptospira to survive in fresh water. Int. Microbiol. 7:35–40 [PubMed] [Google Scholar]
- 38. Turner A. K., Lovell M. A., Hulme S. D., Zhang-Barber L., Barrow P. A. 1998. Identification of Salmonella Typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immun. 66:2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.