Abstract
After transmission by an infected tick, the Lyme disease spirochete, Borrelia burgdorferi sensu lato, colonizes the mammalian skin and may disseminate systemically. The three major species of Lyme disease spirochete—B. burgdorferi sensu stricto, B. garinii, and B. afzelii—are associated with different chronic disease manifestations. Colonization is likely promoted by the ability to bind to target tissues, and Lyme disease spirochetes utilize multiple adhesive molecules to interact with diverse mammalian components. The allelic variable surface lipoprotein decorin binding protein A (DbpA) promotes bacterial binding to the proteoglycan decorin and to the glycosaminoglycan (GAG) dermatan sulfate. To assess allelic variation of DbpA in GAG-, decorin-, and cell-binding activities, we expressed dbpA alleles derived from diverse Lyme disease spirochetes in B. burgdorferi strain B314, a noninfectious and nonadherent strain that lacks dbpA. Each DbpA allele conferred upon B. burgdorferi strain B314 the ability to bind to cultured kidney epithelial (but not glial or endothelial) cells, as well as to purified decorin and dermatan sulfate. Nevertheless, allelic variation of DbpA was associated with dramatic differences in substrate binding activity. In most cases, decorin and dermatan sulfate binding correlated well, but DbpA of B. afzelii strain VS461 promoted differential binding to decorin and dermatan sulfate, indicating that the two activities are separable. DbpA from a clone of B. burgdorferi strain N40 that can cause disseminated infection in mice displayed relatively low adhesive activity, indicating that robust DbpA-mediated adhesive activity is not required for spread in the mammalian host.
INTRODUCTION
Lyme disease spirochetes of the genus Borrelia are transmitted to humans from a bite by an infected Ixodes tick. The first stage of Lyme disease is local infection of the skin, typically giving rise to a characteristic rash termed erythema migrans. In the absence of antibiotic therapy, some strains may disseminate from the skin, via the blood, to multiple secondary sites including the joints, heart, and brain, resulting in the varied clinical manifestations of Lyme disease such as arthritis, carditis, and neuroborreliosis (for a review, see reference 49).
At least seven Borrelia species are associated with Lyme disease (11, 20, 45, 47, 49) and are collectively referred to as B. burgdorferi sensu lato. The three most clinically important Lyme disease spirochetes are B. burgdorferi sensu stricto, B. garinii, and B. afzelii. B. burgdorferi sensu stricto, referred to here simply as B. burgdorferi, is the most prevalent Lyme disease spirochete in the United States. In Europe all three species are associated with Lyme disease, with B. garinii and B. afzelii being the more prevalent (49). The three species are each capable of causing long-term infection in humans but are associated with different chronic manifestations: B. burgdorferi with Lyme arthritis, B. garinii with neuroborreliosis, and B. afzelii with the chronic skin lesion acrodermatitis (55). In addition, within a species, there is apparent strain-to-strain variation in the ability to cause disseminated infection (54, 59). B. burgdorferi strains, typed on the basis of polymorphisms in the rRNA operon or the highly polymorphic ospC gene, have been shown to vary in their association with bloodstream infection in humans (14, 56, 58). Although OspC, a surface lipoprotein, is required for experimental infection of the mouse (22, 37), allelic variation of OspC cannot fully account for observed differences in the capacity to disseminate within the mammalian host by various B. burgdorferi strains (1). Thus, other virulence factors likely contribute to the apparent differences in tissue colonization and clinical manifestations exhibited by diverse Lyme disease spirochetes.
Attachment to host tissues is thought to be a critical step in the ability of many pathogens to disseminate within the mammalian host. B. burgdorferi attaches to a wide variety of cells in vitro, including epithelial, endothelial, and glial cells, platelets, and lymphocytes (12, 13, 18, 19, 51, 52). In addition, multiple host cell and extracellular matrix molecules are recognized by B. burgdorferi, such as integrins (10), fibronectin (6, 21, 43), laminin (7, 53), collagen (61), and proteoglycans (24, 30, 33). Proteoglycans consist of a protein core covalently linked to one or more glycosaminoglycan (GAG) chains, which are long linear repeating disaccharides (for a review, see reference 34). GAGs are typically quite heterogeneous in size and charge and vary structurally between tissues and host species (42). Different GAGs can be segregated into classes based on epimerization of the glycan chain, the location and degree of sulfation, and sensitivity to enzymatic cleavage by different lyases.
We previously showed that diverse spirochetes recognize distinct classes of GAGs, and differences in GAG-binding specificity can lead to differences in the types of mammalian cells recognized (40). For example, B. burgdorferi strain N40, which recognizes heparan sulfate and dermatan sulfate GAGs, binds efficiently to glial and endothelial cells in vitro, while B. afzelii strain VS461, which predominantly recognizes dermatan sulfate, binds selectively to glial cells (40). These findings suggest that strain-specific differences in GAG recognition by Lyme disease spirochetes might influence the specificity of host cell attachment and thus tissue colonization during infection.
B. burgdorferi encodes multiple GAG binding proteins, including the highly related Dbp (decorin binding protein) A and DbpB, which comprise a single operon and were first identified on the basis of their ability to bind decorin, a chondroitin/dermatan sulfate proteoglycan that “decorates” collagen fibers in mammalian tissues (23–25). DbpA has been shown to be expressed by diverse B. burgdorferi sensu lato strains during murine infection (3, 25, 26) and, on the basis of ubiquitous seroreactivity among Lyme patients, is likely to be expressed during human Lyme disease as well (for a review, see reference 57). DbpA and DbpB, when expressed in a noninfectious and otherwise nonadherent B. burgdorferi strain, were shown to be sufficient to promote spirochetal attachment to isolated dermatan sulfate GAGs as well (16). The dbpA (and presumably dbpB) gene is not expressed in the unfed tick, but transcription is induced upon tick feeding (28). In addition, DbpA and DbpB are required for maximal infectivity and dissemination after intradermal inoculation of either immunodeficient or immunocompetent mice (3, 45, 54). These findings are consistent with the hypothesis that spirochetal binding to extracellular matrix mediated by DbpA and DbpB promotes colonization of the mammalian host.
DbpA and DbpB are related proteins that each bind to dermatan sulfate but differ in their GAG and mammalian cell type binding specificities (16). In addition, whereas DbpB is highly conserved among Lyme disease spirochete strains, DbpA is highly polymorphic (44), and recombinant derivatives of DbpA allelic variants differ in decorin binding (41). In the present study, we found that allelic variation of DbpA is associated with dramatic differences in the ability of this adhesin to promote bacterial attachment to decorin, GAGs, and mammalian cells.
MATERIALS AND METHODS
Bacterial strains and cell lines.
The high-passage B. burgdorferi strain B314 (46) was a generous gift from Tom Schwan (Rocky Mountain Labs, Hamilton, MT). B. burgdorferi strains N40 clone D10/E9 (N40D10/E9), B356, B31, and 297, B. garinii strain PBr, and B. afzelii strain VS461 have been described previously (10, 17). B314 and its derivatives were cultured at 33°C in BSKII complete medium (2), supplemented with 200 μg of kanamycin/ml where appropriate. For a list of bacterial strains used in the present study, see Table 1 . 293 (human kidney epithelial) cells, C6 (rat glioma) cells, and EA-Hy926 (human umbilical vein endothelial) cells were cultured as described previously (40).
Table 1.
Bacterial strains used in this study
| Strain | Species | Origin and/or notesa | Source or reference |
|---|---|---|---|
| N40 (clone D10/E9) | B. burgdorferi | Tick, United States | 9 |
| B356 | B. burgdorferi | Skin | 54 |
| B31 | B. burgdorferi | Tick, Switzerland | 2, 17 |
| 297 | B. burgdorferi | Human, CSF, United States | 31 |
| PBr | B. garinii | Human, CSF, Germany | 9 |
| VS461 | B. afzelii | Tick, Switzerland | 35 |
| B314/pJF21 | B. burgdorferi | B314 clone JF5 (harboring vector control) | 15 |
| B314/DbpAN40 | B. burgdorferi | B314 clone VB2-7 | This study |
| B314/DbpAB356 | B. burgdorferi | B314 clone JF12 | This study |
| B314/DbpAB31 | B. burgdorferi | B314 clone YL1 | This study |
| B314/DbpA297 | B. burgdorferi | B314 clone JF11 | This study |
| B314/DbpAPBr | B. burgdorferi | B314 clone JF8 | This study |
| B314/DbpAVS461 | B. burgdorferi | B314 clone VB1-7 | This study |
CSF, cerebrospinal fluid.
Plasmids and cloning.
For initial sequence analysis, dbpA from (i) B. burgdorferi strains N40D10/E9, B356, B31, and 297, (ii) B. afzelii strain VS461, and (iii) B. garinii strain PBr were amplified by PCR from genomic DNA using primers and reaction conditions described previously (44). Briefly, the forward primer, 10F4 5′-GTGGTTAAGGAAAAACAAAA-3′, is homologous to a sequence in the highly conserved dbpB gene (44), and the reverse primer, 5R1 5′-CCAAATAACATCAAAAAGGA-3′, is homologous to a sequence downstream of dbpA. PCR with these primers resulted in the generation of amplicons ∼1 kb in size, which were inserted into pCR-XL-TOPO vector (Invitrogen) and then sequenced. Nucleotide sequence analysis showed that dbpA cloned from strains B31, 297, PBr, and VS461 were identical to previously published sequences (44, 60). DbpA from strain N40D10/E9 was found to be 95% identical to DbpA from an independent clone of N40 (44) and is, for clarity, referred to as DbpAN40-D10/E9 in the present study.
For expression in B314, the entire coding sequence of each dbpA allele was amplified from genomic DNA by PCR using the primers listed in Table 2. These amplicons were then cloned into a modified shuttle vector, pJF21 (15), using engineered SalI and BamHI sites at the 5′ and 3′ ends, respectively, and sequenced with the M13F and M13R primers. For production of recombinant polyhistidine-tagged proteins, dbpA genes from strains B31, N40D10/E9, PBr, and VS461 without their lipoprotein signal sequences were amplified by PCR using the primers described in Table 2 and cloned into pET15b (Novagen, Madison, WI).
Table 2.
Primers used in this study
| Purpose | Primer | Sequence (5′–3′)a | Nucleotide positionb |
|---|---|---|---|
| B314/pDbpAN40-D10/E9 | AN40F | ACGCGTCGACATGAATAAATATCAAAAAACTTTC | 1–24 |
| AN40R | CGCGGATCCTTAGTTATTTTTGCATTTTTCATCAGT | 559–582 | |
| B314/pDbpAB356 | A356F | ACGCGTCGACATGAATAAATATCAAAAAACTTTC | 1–24 |
| A356R | CGCGGATCCTTAGTTATTTTTGCATTTTTCATC | 562–585 | |
| B314/pDbpAB31 | AB31F | CGGTCGACATGATTAAATGTAAT | 1–15 |
| AB31R | CGGGATCCTTAGTTATTTTTGCA | 562–576 | |
| B314/pDbpA297 | A297F | ACGCGTCGACATGATTAAATGTAATAATAAAACT | 1–24 |
| A97R | CGCGGATCCTTACGATTTAGCAGTGCTGTCTTC | 541–564 | |
| B314/pDbpAPBr | APBrF | ACGCGTCGACATGATTAAATATAATAAAATATTG | 1–24 |
| APBrR | CGCGGATCCTTATGTAGTAGTAGCAGTTTTGGC | 535–558 | |
| B314/pDbpAVS461 | AVS461F | ACGCGTCGACATGATTAAATATAATAAAATTATA | 1–24 |
| AVS461R | CGGGATCCTTATTTTTGATTTTTAGTTTGTTTTTCTTTAATGTTTTCC | 487–510 | |
| BL21/pHis-DbpAB31 | B31 HisDbpAF | GCGGATCCGGACTAACAGGAGCAACA | 76–93 |
| B31 HisDbpAR | CGCTCGAGTTAGTTATTTTTGCATTT | 559–576 | |
| BL21/pHis-DbpAPBr | PBrHisDbpAF | GCGGATCCGGCTTAACAGGAGAAACT | 64–81 |
| PBrHisDbpAR | CGCTCGAGTTATGTAGTAGTAGCAGT | 541–558 | |
| BL21/pHis-DbpAVS461 | 5VS461DBPA | GGAATTCCATATGAGTTTAACAGGAAAAGCTAGATTGGAA | 64–90 |
| AVS461R | CGCGGATCCTTATTTTTGATTTTTAGTTTGTTTTTCTTTAATGTTTTCC | 487–510 |
Underlining denotes the restriction sites.
The nucleotide position is given relative to the first nucleotide of the initiation codon. Note that to generate recombinant DbpA proteins, the open reading frame of only the mature proteins (i.e., those lacking a signal sequence) was amplified.
Purification of human decorin.
Recombinant human decorin, a generous gift from David Mann (MedImmune, Inc.), was purified from stably transfected Chinese hamster ovary cells as described previously (29).
Generation of recombinant proteins and antisera.
Plasmids encoding recombinant His-tagged DbpA proteins were transformed into Escherichia coli strain BL21, and protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Bacteria were lysed, and proteins were purified by nickel affinity chromatography as described in the manufacturer's instructions (Novagen). The homogeneity of recombinant proteins was confirmed by SDS-PAGE and Coomassie blue staining. Five-week-old BALB/c mice were immunized with 100 μg of His-DbpAB31, His-DbpAN40-D10/E9, His-DbpAPBr, or His-DbpAVS461 in complete Freund adjuvant. The animals were boosted twice with 100 μg of the same proteins in incomplete Freund adjuvant at 2-week intervals, and antisera were collected from blood after terminal cardiac puncture.
Attachment of immobilized recombinant DbpA proteins to biotinylated glycosaminoglycans.
Dermatan sulfate (Calbiochem), heparin, and chondroitin-6-sulfate (Sigma) were biotinylated using EZ-Link Biocytin Hydrazide (Pierce) and then dialyzed in phosphate-buffered saline (PBS) using a Slide-A-Lyzer cassette (Pierce) with 10,000-kDa molecular mass cutoff. Microtiter plates (96 well; Linbro) were coated overnight at 4°C with 1 μg of His-tagged recombinant DbpA in PBS. The next day, proteins were removed, and the wells were washed twice with PBS and then blocked with 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature. The blocking buffer was removed, and 50 μl of each biotinylated GAG, diluted to a final concentration of 100 μg of 1% BSA/ml in PBS, was added to the wells, followed by incubation for 2 h. After a washing step, bound GAGs were detected by enzyme-linked immunosorbent assay (ELISA) using horseradish peroxidase-tagged anti-biotin antibody, followed by TMB substrate.
Transformation of B. burgdorferi B314.
Electrocompetent B314 spirochetes were prepared and transformed as described previously (36). Briefly, 100-ml portions of mid-log-phase cultured spirochetes were harvested and washed twice in electroporation solution (EPS; 15% [vol/vol] glycerol, 0.27 M sucrose) and resuspended in 100 μl of EPS. Then, 30 to 40 μg of plasmid DNA was added to the suspension, and the mixtures were electroporated at 2,200 V in a 0.2-cm cuvette and cultured in BSKII complete medium at 33°C for 24 h. The transformation mixture was added to 1.7% analytical-grade agarose (Invitrogen) and plated onto a 1.5× BSKII/agarose bottom layer in a sterile tissue culture dish (100 mm × 20 mm; Corning) in the presence of kanamycin (200 μg/ml), and the plates were incubated at 34°C in a 2% CO2 atmosphere for 2 weeks. Colonies were picked and cultured at 33°C to mid-log-phase density in BSKII complete medium containing kanamycin (200 μg/ml).
Proteinase K treatment, SDS-PAGE, and Western blotting.
To detect DbpA proteins, lysates from 5 × 107 spirochetes were separated by SDS–15% PAGE. DbpA and FlaB were identified by immunoblotting with polyclonal antibodies against the appropriate DbpA (diluted 1:5,000) or a monoclonal antibody, CB1 (a gift from J. Benach, Stony Brook University, Stony Brook, NY), against FlaB (diluted 1:500), respectively. Surface localization of DbpA proteins in recombinant B314 strains was determined as previously described (43). Briefly, 5 × 107 spirochetes were centrifuged, and the pellets were washed twice in PBS plus 0.2% BSA. After the final wash, pellets were gently lifted with 5 mM MgCl2 in PBS supplemented with 4 mg of proteinase K (Sigma)/ml or buffer only and incubated at room temperature for 30 min. To inactivate the proteinase K, 150 μg of phenylmethylsulfonyl fluoride (PMSF) was added to each pellet. Pellets were washed twice with PBS plus 0.2% BSA, lysed, and separated by SDS–15% PAGE. The DbpA and FlaB proteins were identified by immunoblotting as described above.
Triton X-114 fractionation.
Membrane fractions of B314 expressing DbpA were prepared as described previously (5). Briefly, Borrelia cultures were grown to log phase as determined by counting spirochetes by dark-field microscopy, and 109 spirochetes were harvested and washed twice with 0.2% BSA in PBS. Spirochetes were then resuspended in 2% Triton X-114 and allowed to incubate overnight with rocking at 4°C. The following day, insoluble material was removed by centrifugation, and the supernatants were phase separated as follows. The supernatants were warmed to 37°C for 15 min and then centrifuged at 13,000 rpm for 15 min to separate the aqueous and detergent fractions. The aqueous phase was discarded, and the detergent phase was washed three times by adding cold PBS to the original volume and rewarming and recentrifuging the samples as described above in the phase separation step. After the final wash, the proteins were precipitated by adding 9 volumes of cold 100% ethanol, incubated overnight at −20°C, and recovered by centrifugation. After a washing with 90% ethanol, the proteins were resuspended in PBS. The protein concentration was determined by a BCA assay (Pierce). Then, 15 μg of total protein from the outer membrane (detergent) fraction was separated on a 15% SDS-PAGE gel and stained with Coomassie blue.
Attachment of spirochetes to mammalian cells.
Radiolabeled bacteria were prepared by growing spirochetes at 33°C in BSKII complete medium supplemented with 60 μCi of [35S]methionine/ml. When the cultures achieved mid-log phase (approximately 5 × 107/ml), bacteria were harvested at 10,000 × g, and the pellets were washed twice with 0.2% BSA in PBS. Labeled spirochetes were then stored as aliquots at −80°C in BSK-H (Sigma) containing 20% glycerol.
One day before each assay, mammalian cells were lifted with 0.5% trypsin–0.5 mM EDTA (Invitrogen) and plated in 96-well break-apart microtiter plates (Nunc) which were previously UV sterilized and coated with MBP-Inv497, a maltose-binding protein fusion containing the cell-binding domain of the invasin protein from Yersinia pseudotuberculosis (32). Frozen aliquots of radiolabeled B. burgdorferi were thawed and resuspended at 108cells/ml in BSK-H, followed by incubation at room temperature for 2 h. Prior to the addition of radiolabeled spirochetes, the cell monolayers were washed twice with PBS. Radiolabeled spirochetes were then diluted 1:3 into GHS buffer (10 mM glucose, 10 mM HEPES, 50 mM NaCl [pH 7.0]) and added to quadruplicate wells at 106 spirochetes/well. To enhance spirochete-cell contact, the plates were centrifuged at 1,000 rpm for 5 min and then rocked at room temperature for 1 h. Unbound spirochetes were removed by washing wells four times with 0.2% BSA in PBS. The plates were then air-dried, and the percentage of bound bacteria in each well was determined by liquid scintillation. Each strain was tested for cell binding in three to five independent experiments.
Enzymatic removal of specific classes of GAGs.
Monolayers were incubated for 2 h with 0.5 U of heparinase I, heparitinase, or chondroitinase ABC (Sigma)/ml at 37°C in RPMI supplemented with 1% BSA, 10−2 U of aprotinin/ml, and 165 μg of PMSF/ml. After the monolayers were washed with PBS, radiolabeled spirochetes were added to the pretreated monolayers as described above.
Inhibition of binding with exogenous GAGs.
Radiolabeled spirochetes were prepared as described above and incubated for 30 min at room temperature in BSK-H supplemented with between 80 ng and 6.25 mg of the GAGs/ml. After incubation, the spirochetes were diluted 1:3 in GHS buffer before addition to the cell monolayers.
Attachment of radiolabeled bacteria to purified GAGs and decorin.
Prior to each assay, wells from Nunc 96-well break-apart microtiter plates were coated with either a titration of purified human decorin (1.25 to 0.156 μg/ml) or dermatan sulfate (2.5 to 0.625 mg/ml) in PBS at 4°C overnight. Wells were washed three times with 0.05% Tween 20 in PBS. The wells were then blocked with 1% BSA in PBS for 1 h at room temperature. After removal of the blocking buffer, radiolabeled B. burgdorferi were added to the wells as described above.
RESULTS
Ectopic expression of diverse DbpA alleles on the surface of a nonadherent B. burgdorferi strain.
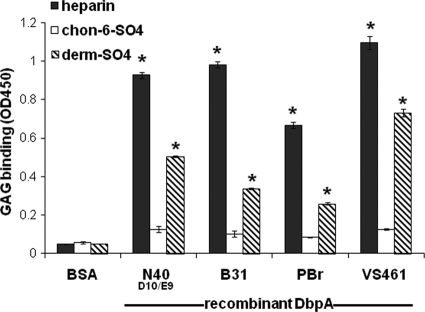
We previously showed that recombinant DbpA from B. burgdorferi strain N40 clone D10/E9 (N40D10/E9) bound to purified GAGs (16, 31). To determine whether GAG binding activity is a common property of diverse DbpA alleles, we measured GAG binding by recombinant DbpA from strain N40D10/E9 and three additional Lyme disease spirochetes. B. burgdorferi strain B31 is the type strain for this organism and is highly infectious in the mouse model (17), B. garinii strain PBr displays high levels of the hemagglutination activity that is associated with GAG binding (33), and strain VS461 is a representative of B. afzelii (35). DbpA from strain N40D10/E9 was found to be 95% identical to DbpA from an independent clone of N40 (44) and for clarity is referred to here as DbpAN40-D10/E9. Immobilized recombinant His-tagged DbpA alleles were tested for their ability to bind to biotinylated GAGs. DbpA from each strain bound to heparin, as well as to dermatan sulfate, but not to a control GAG, chondroitin-6-sulfate (Fig. 1). Biotinylated heparin and dermatan sulfate did not bind to BSA-coated wells in this experiment (Fig. 1) or to a control His-tagged protein in previously described studies (38).
Fig. 1.
Recombinant DbpA proteins bind to biotinylated GAGs. Microtiter wells were mock coated (with PBS) or coated with His-tagged DbpAN40,DbpAB31, DbpAPBr, or DbpAVS461 and probed with biotinylated heparin, dermatan sulfate, or chondroitin-6-sulfate (see Materials and Methods). After a washing step, bound GAGs were detected by ELISA, as indicated by the absorbance at 450 nm. Each bar represents the mean of four independent determinations ± the standard errors. Asterisks indicate that GAG bound to DbpA-coated wells significantly (P < 0.05 [Student t test]) better than to mock-coated wells.
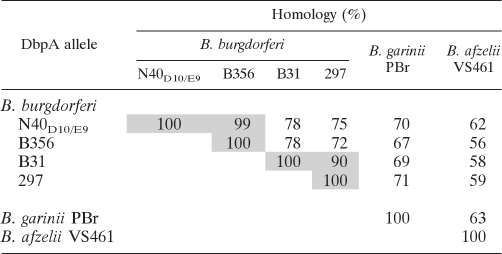
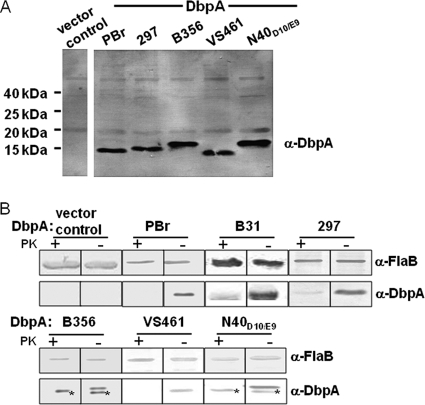
Allelic differences of DbpA could result in differences in binding activity. Although the initial analysis of recombinant DbpA proteins revealed potential differences in GAG binding (Fig. 1), given that it is not clear how faithfully the binding activities of recombinant DbpA reflect its activities when expressed on the spirochetal outer membrane, we tested the adhesive function of DbpA alleles on the surface of an otherwise nonadherent B. burgdorferi strain (16, 46). B. burgdorferi strain B314 is a high-passage strain that lacks all discernible linear plasmids, including lp54 (46), which encodes the dbpBA operon, and appears to be incapable of binding to GAGs or cultured mammalian cells. Alleles of dbpA were cloned into the expression shuttle vector pJF21 downstream of the ospC promoter (16, 50), which is highly active in strain B314 and is predicted to facilitate high-level transcription (46). Recombinant plasmids were transformed into B. burgdorferi strain B314. In addition to the alleles of B. garinii PBr, B. afzelii VS461, and B. burgdorferi N40D10/E9 and B31, we also analyzed dbpA from B. burgdorferi 297, a commonly studied strain, and from B. burgdorferi B356, a skin biopsy isolate that is incapable of causing persistent disseminated infection in mice (54). The set of six DbpA alleles share as little as 56% identify (Table 3). To confirm that these DbpA alleles were expressed in strain B314, bacterial outer membrane proteins were isolated by Triton X-114 extraction and immunoblotted using mouse anti-DbpA antisera. As expected, each recombinant strain harboring a recombinant plasmid encoding dbpA was found to produce DbpA, whereas B314 harboring the pJF21 vector control did not (Fig. 2A). Proteinase K digestion of intact spirochetes resulted in complete (e.g., Fig. 2B, PBr) or near-complete (e.g., Fig. 2B, B31) loss of reactivity to DbpA antisera in immunoblot analysis, indicating that all DbpA alleles were largely exported to the surface of strain B314. As predicted, the periplasmic protein FlaB remained intact, indicating that the integrity of the outer membrane during proteinase K digestion was maintained (Fig. 2B).
Table 3.
Homology of DbpA alleles used in this studya
A pairwise comparison of amino acid identities between DbpA alleles is shown. Values for the two groups of alleles that share ≥90% homology are shaded.
Fig. 2.
DbpA alleles from diverse Lyme disease spirochetes are expressed on the surface of a nonadherent B. burgdorferi strain. (A) Triton X-114 extractions of B. burgdorferi strain B314 expressing the indicated DbpA allele were subjected to SDS-15% PAGE and immunoblotted with a mixture of antisera raised against DbpAB31, DbpAN40-D10/E9, DbpAPBr, and DbpAVS461. (B) Spirochetes expressing DbpA protein were digested with proteinase K (PK+) or incubated in PBS (PK−). Lysates from 5 × 107 treated bacteria were separated by SDS–15% PAGE, and DbpA and FlaB proteins were identified by Western blotting with an individual antiserum against DbpAN40-D10/E9, DbpAB31, DbpAPBr, or DbpAVS461. The proteinase K sensitivity experiments were performed several times and Fig. 2 is representative of such studies. Proteinase K-resistant species in lysates of B314/pDbpAB356 and B314/pDbpAN40-D10/E9, indicated by asterisks, were consistently observed and may represent intracellular nonlipidated DbpA protein. Flagellin, a periplasmic protein, was immunoblotted as a control for the retention of outer membrane integrity. B. burgdorferi strain B314 expressing DbpAB31 was analyzed in a separate experiment.
DbpA displays allelic variation in the recognition of dermatan sulfate on the surface of cultured epithelial cells.
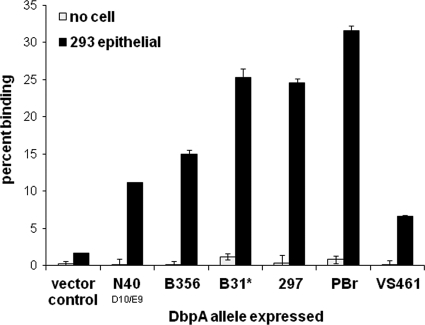
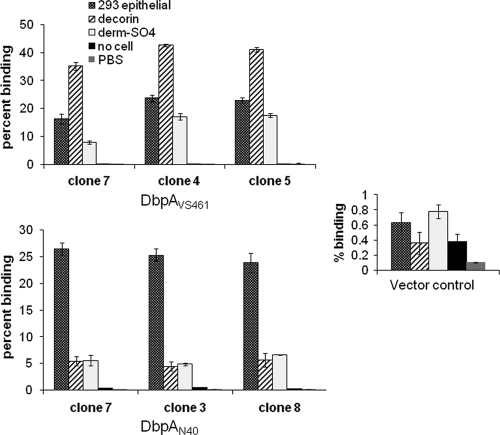
DbpAN40-D10/E9 was previously shown to promote attachment to cultured epithelial cells but not to endothelial nor glial cells (16). Therefore, recombinant B314 strains expressing different DbpA alleles were tested for gain-of-function in attachment to 293 epithelial, EA-Hy926 endothelial, and C6 glial cells. Similar to our previous analysis of DbpAN40-D10/E9, all DbpA alleles promoted spirochete attachment to cultured epithelial cells (Fig. 3) but not glial or endothelial cells (data not shown). DbpAPBr promoted levels of epithelial cell attachment significantly (P < 0.05) higher than other DbpA alleles (Fig. 3). DbpA297 and DbpAB31 mediated spirochetal binding less efficiently than DbpAPBr, but more efficiently than DbpAN40-D10/E9, DbpAB356, and DbpAVS461 (Fig. 3). Complementation of the nonadherent strain B314 gave reproducible results across multiple assays, and independently derived clones expressing the same allele displayed similar binding to 293 epithelial cells, purified decorin, and dermatan sulfate (Fig. 4). These results suggested that the binding phenotype measured in this way reflects intrinsic properties of the allele rather than the particular clone analyzed.
Fig. 3.
DbpA-mediated binding to cultured mammalian cells is cell type specific and variable among alleles. Radiolabeled B. burgdorferi strain B314 spirochetes expressing the indicated DbpA allele were added to wells containing 293 epithelial cells or to empty wells (media). Each bar represents the mean (± the standard error) of four independent determinations. B. burgdorferi strain B314 expressing DbpAB31 was analyzed in a separate experiment in which the binding by B314 strains expressing DbpAPBr or no DbpA (vector control), analyzed in parallel as controls, bound with an efficiency similar to that in the depicted experiment.
Fig. 4.
Independently derived clones expressing the same allele of DbpA display similar in vitro binding phenotypes. Radiolabeled B. burgdorferi strain B314 spirochetes expressing either DbpAVS461 (top panel), DbpAN40 (bottom panel), or vector control (inset panel) were added to wells containing 293 epithelial cells or to wells containing either no cells (media) or decorin, dermatan sulfate, or PBS (mock coated). Each bar represents the mean (± the standard error) of four independent determinations.
We previously showed that spirochetal binding to 293 cells mediated by DbpAN40-D10/E9 required dermatan sulfate (16). To determine whether GAGs promoted binding of the recombinant B314 strains to 293 cells, monolayers were digested with specific lyases to remove different classes of GAGs from the mammalian cell surface. Consistent with this, the removal of dermatan or chondroitin sulfates with chondroitinase ABC virtually eliminated attachment promoted by all DbpA alleles, whereas the removal of heparin or heparan sulfates with heparinase or heparitinase had no significant effect (Table 4). In addition, exogenous dermatan sulfate, but not heparin or chondroitin-6-sulfate, significantly impaired cell binding by recombinant B314 expressing DbpA from strains B356, 297, or PBr (Table 5), suggesting that cell attachment was associated with GAG binding activity.
Table 4.
Removal of chondroitin sulfate GAGs from the surfaces of epithelial cells diminishes binding by spirochetes expressing diverse DbpA alleles
| Plasmid | Mean binding (%) ± SEa |
|||
|---|---|---|---|---|
| Mock digestion | Heparinase | Heparatinase | ChonABC | |
| Vector control | 2.9 ± 0.2 | NA | NA | NA |
| pDbpAN40-D10/E9 | 10.3 ± 0.7 | 8.3 ± 1.3 | 10.2 ± 0.9 | 0.6 ± 0.2* |
| pDbpAB356 | 17.0 ± 2.4 | 17.6 ± 0.5 | 23.6 ± 2.8 | 0.2 ± 0.1* |
| pDbpA297 | 33.7 ± 3.3 | 34.8 ± 3.3 | 42.3 ± 2.3 | 2.0 ± 0.6* |
| pDbpAPBr | 42.3 ± 1.3 | 38.4 ± 3.0 | 42.6 ± 0.6 | 1.1 ± 1.0* |
| pDbpAVS461 | 6.6 ± 0.4 | 4.6 ± 0.8 | 5.5 ± 0.3 | 0.5 ± 0.2* |
Binding of radiolabeled transformants to 293 epithelial monolayers was determined after pretreatment of cells with the indicated lyase, as described previously (15, 16). ChonABC, chondroitinase ABC. Each value represents the mean of four independent determinations ± SE. For all strains, <2% of bacteria bound to identically treated wells without mammalian cells (data not shown). *, Significant (P < 0.05) difference in binding to mock-treated versus lyase-treated monolayers as determined by Student t test analysis. NA, not applicable.
Table 5.
Dermatan sulfate inhibits cell binding by B. burgdorferi strain B314 expressing DbpA alleles from diverse Lyme disease spirochetes
| Plasmid | Mean binding (%) ± SEa |
|||
|---|---|---|---|---|
| No inhibitor | Heparin | C6S | DS | |
| Vector control | 2.9 ± 0.5 | NA | NA | NA |
| pDbpAB356 | 15.6 ± 1.9 | 22.6 ± 0.7 | 18.4 ± 2.1 | 3.8 ± 0.3* |
| pDbpA297 | 42.5 ± 2.8 | 33.0 ± 2.2 | 47.6 ± 3.5 | 26.9 ± 1.5* |
| pDbpAPBr | 57.5 ± 4.2 | 59.1 ± 1.9 | 58.8 ± 3.7 | 32.9 ± 4.3* |
Binding of radiolabeled transformants to 293 epithelial monolayers was determined after pretreatment of bacteria with the indicated soluble GAG, as described previously (15, 16). DS, dermatan sulfate; C6S, chondroitin-6-sulfate. Each point represents the mean of four independent determinations ± SE. For all strains, <2% of bacteria bound to identically treated wells without mammalian cells (data not shown). *, Significant (P < 0.05) difference in binding to mock-treated versus GAG-treated bacteria as determined by Student t test analysis. NA, not applicable.
DbpA displays allelic variation in the ability to promote spirochetal attachment to purified decorin or dermatan sulfate.
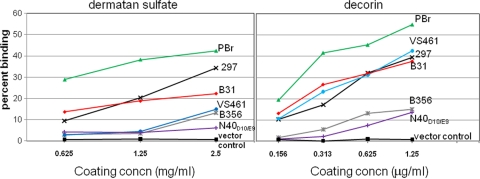
We found previously that DbpAN40-D10/E9 was able to confer binding to purified dermatan sulfate to strain B314 (16). To determine whether the variation in the ability of different DbpA alleles is associated with differences in their ability to promote binding to purified GAGs, we tested for the binding of the recombinant B314 strains to microtiter wells coated with increasing concentrations of dermatan sulfate. Of the DbpA-expressing strains, B314 expressing DbpAPBr bound significantly (P < 0.05) better to dermatan sulfate than all of the other B314 strains at each of the concentrations tested (Fig. 5, left panel). DbpA297 and DbpAB31 promoted binding somewhat less efficiently than DbpAPBr but significantly more efficiently than DbpAVS461, DbpAB356, or DbpAN40-D10/E9 (Fig. 5, left panel). Binding mediated by the latter three alleles, while not robust, was nevertheless considerably (and significantly) above the background levels observed for strain B314 harboring the vector control. Thus, variation in the ability of the DbpA alleles to promote binding to purified dermatan sulfate correlated with their ability to promote binding to 293 cells (compare Fig. 3 and the left panel of Fig. 5), suggesting that 293 cell attachment reflects dermatan sulfate binding activity.
Fig. 5.
Decorin- and GAG-binding levels vary among DbpA alleles. Radiolabeled B314 spirochetes expressing the indicated DbpA allele were added to wells coated with increasing concentrations of decorin or dermatan sulfate, and the percentage of bacteria stably bound was determined. Analysis of B314 expressing DbpAB31 was performed in a separate experiment in which the binding by B314 strains expressing DbpAPBr or no DbpA, analyzed in parallel to the controls, bound with an efficiency similar to that in the depicted experiment. Each point represents the mean of four independent determinations, with the standard error omitted for clarity. At a concentration of 1.25 mg of dermatan sulfate/ml, DbpAPBr bound to this substrate at a significantly higher percentage (P < 0.05) than both DbpA297 and DbpAB31, and the latter two strains bound at a significantly higher percentage than DbpAB356, DbpAN40, and DbpAVS461. At a concentration of 1.25 μg of decorin/ml, DbpAPBr bound at a significantly higher percentage (P < 0.05) than DbpA297,DbpAB31, and DbpAVS461, and the latter three bound at a significantly higher percentage (P < 0.05) than DbpAB356 and DbpAN40.
DbpA was originally identified by its ability to bind to decorin, a dermatan sulfate proteoglycan (23). To test whether allelic variation of DbpA in binding to decorin correlates with binding to dermatan sulfate and 293 cells, the B314 recombinants expressing different DbpA alleles were similarly tested for the ability to bind to increasing concentrations of immobilized decorin. Again, DbpAPBr promoted decorin binding significantly better than did other alleles, and DbpA297 and DbpAB31 promoted attachment to decorin better than DbpAB356 or DbpAN40-D10/E9 (Fig. 5, right panel). Notably, DbpAVS461 promoted relatively efficient binding to decorin, in spite of promoting only low level binding to dermatan sulfate and epithelial cells (compare to Fig. 5, left and right panels, and Fig. 3). These results indicate that DbpA-mediated bacterial attachment to decorin and dermatan sulfate are distinguishable, with binding to purified dermatan sulfate correlating better with 293 epithelial cell binding.
The C terminus of DbpAVS461 is required for spirochetal attachment to mammalian cells and to purified decorin or dermatan sulfate.
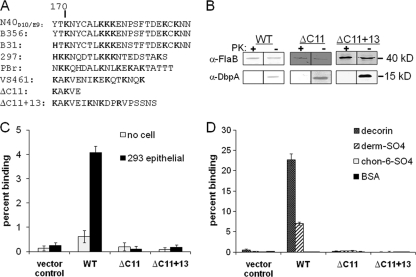
Basic amino acids are often critical for the ability of GAG-binding proteins to bind their substrate (27), and three lysine residues of DbpA297—K82, K163, and K170—were previously implicated in decorin binding (8, 41). The sequences flanking K82 of DbpAB31 or DbpAN40-D10/E9, which differed significantly in their ability to promote binding to epithelial cells, dermatan sulfate and decorin (Fig. 3 and 5), are nearly identical (44), suggesting that other segments of the proteins may contribute to GAG, decorin, and cell binding. A comparison of DbpA sequences shows that C terminal to K170, all alleles contain a lysine-rich segment of amino acids (Fig. 6A). Among the alleles from different Lyme disease species, this region is not highly conserved in specific amino acid sequence, but its basic nature could promote binding to GAGs and/or decorin. In fact, analysis of a fortuitous PCR-generated deletion-substitution mutant of DbpAVS461 that lacks this segment did not bind to decorin, dermatan sulfate, or 293 epithelial cells (Fig. 6C and D, ΔC11+13). The defect in binding was apparently due to the loss of the 11-residue highly basic C-terminal DbpA sequence rather than the acquisition of 13 residues of exogenous sequence because simple deletion of the DbpAVS461 C-terminal 11 amino acids also eliminated the ability of DbpAVS461 to promote bacterial attachment (Fig. 6, ΔC11). The defect in binding by either mutant was not due to alteration in surface localization of DbpA (Fig. 6B), suggesting that the extreme C-terminal region of this DbpA allele is critical for decorin, dermatan sulfate, and 293 cell binding.
Fig. 6.
The C-terminal region of DbpAVS461 is required for binding to GAGs and mammalian cells. (A) The C-terminal amino acid sequences of DbpA proteins analyzed in the present study. (B) Strain B314 expressing wild- type (WT) or mutant DbpAVS461 were subjected to proteinase K (PK+) or mock (PK−) digestion and immunoblotted for DbpA or flagellin, as described for Fig. 1. Radiolabeled B314 expressing the vector control or the indicated DbpAVS461 derivative was added to wells with 293 epithelial cells or no cells (C), or dermatan sulfate, decorin, chondroitin-6-sulfate, or BSA (D). The percentage of bacteria bound was determined by scintillation counting. Each bar represents the mean (± the standard errors) of four independent determinations.
DISCUSSION
The bacterial factors that contribute to the ability of some Lyme disease spirochete strains to cause disseminated infection are not fully characterized, and may include surface proteins that mediate attachment to mammalian cells or extracellular matrix (ECM) in target tissues. The Lyme disease spirochete exhibits strain-specific variation in GAG-binding specificity that corresponds to variation in the mammalian cell types that are recognized in vitro (40). B. burgdorferi encodes multiple GAG-binding proteins, including DbpA, DbpB (16), Bgp (39), and BBK32 (15), and the expression pattern of these adhesins might vary with strain. In addition, B. burgdorferi may encode allelic-variable GAG-binding adhesins that exhibit differences in GAG recognition. DbpA has long been known to be highly variable among Lyme disease spirochetes (44) and could contribute to such strain-specific variation. In the present study, we analyzed DbpA alleles from each of the three major species of Lyme disease spirochete, as well as representatives of B. burgdorferi sensu stricto that were associated with different abilities to disseminate in the mammalian host.
After confirming that different recombinant DbpA alleles possessed GAG-binding activity, we utilized an ospC promoter-based vector to ectopically express different DbpA alleles on the surface of the high-passage, noninfectious strain B. burgdorferi B314. This strain lacks the dbpA-encoding Lp54, as well as any other linear plasmid (46; unpublished observations), and is not known to adhere to any cell line (16). The outer membrane of B. burgdorferi strain B314 lacks many lipoproteins (46) and undoubtedly differs from that typical of infectious B. burgdorferi strains. Nevertheless, this strain provides one means to analyze the ability of an adhesin to promote spirochetal binding in the absence of confounding alternate binding pathways. In addition, a preliminary survey of Lyme disease strains corresponding to several of the DbpA alleles analyzed in the present study indicates that B. garinii strain PBr, which we show here encodes a DbpA allele that mediates efficient spirochetal attachment to 293 cells, also binds with high efficiency to this cell line (N. Parveen and D. Robbins, unpublished observations).
For each DbpA allele, ectopic expression conferred the ability to attach to epithelial (but not endothelial or glial) cells and to purified dermatan sulfate. In all cases, DbpA-mediated mammalian cell attachment was eliminated upon enzymatic removal of this class of GAGs, indicating that diverse DbpA alleles are capable of promoting epithelial cell attachment through the recognition of dermatan sulfate GAGs.
Each DbpA-expressing B314 derivative also bound to decorin significantly above background levels. Interestingly, upon a recent survey of allelic variants of DbpA (and DbpB), J. Salo and coworkers found that several alleles, including all B. afzelii DbpA alleles, were devoid of detectable decorin-binding activity (47a). The apparent discrepancy in the two studies could be due to technical differences or to the analysis of different alleles, given that the sets of DbpA alleles analyzed in the two studies were nonoverlapping. Clarification of the reason(s) behind the different findings will require further investigation.
In the present study, although we detected common binding activities for each DbpA allele, we also observed allelic differences in the level of adhesiveness. For example, DbpAPBr promoted >4-fold more efficient epithelial cell attachment than DbpAVS461 and >20-fold more efficient dermatan sulfate binding. These differences were reproducible and were a property of the allele expressed because multiple clones of a given allele were associated with similar binding activities. The differences in binding could not be attributed to the levels of expression of different alleles. First, no dramatic differences in expression levels were detected upon immunoblotting of outer membrane preparations with pooled antisera that collectively recognize the entire collection of DbpA alleles. Second, to the degree that SDS-PAGE and Coomassie blue staining revealed minor potential differences in DbpA levels (data not shown), we found no correlation between expression level and substrate binding. For example, DbpAPBr and DbpAVS461, which as noted above differed dramatically in cell and GAG binding, were expressed at roughly equivalent levels.
As predicted, highly homologous alleles displayed similar binding activities. For example, the dermatan sulfate, decorin and 293 cell binding profiles of DbpAN40-D10/E9 and DbpAB356, which are 99% identical in sequence, were virtually indistinguishable. Similarly, DbpA297 and DbpAB31, which are 90% identical, also mediated roughly equivalent levels of cell, decorin, and dermatan sulfate binding. Consistent with the previously documented sequence diversity of DbpA alleles, we observed considerable intraspecies variation in adhesive activities: DbpA alleles derived from B. burgdorferi sensu stricto that varied considerably in sequence, e.g., DbpA297 and DbpAN40-D10/E9, also varied in attachment activities. Interestingly, given that strain N40D10/E9 causes disseminated infection in mice (9), the observation that DbpAN40-D10/E9 promoted levels of adhesiveness lower than most other alleles indicates that robust DbpA-mediated in vitro adhesiveness is not a prerequisite for disseminated infection. Related to this, DbpAN40-D10/E9 and DbpAB356 displayed similar binding activities in spite of the fact that B. burgdorferi N40D10/E9 and B356 differ considerably in their ability to disseminate in mice (9, 54).
Decorin contains a chondroitin or dermatan sulfate GAG chain that is critical for recognition by DbpA (24) and, for five of the six DbpA alleles analyzed here, we observed a strong correlation between decorin and dermatan sulfate binding. However, one allele, DbpAVS461, promoted relatively high-level spirochetal attachment to decorin but low-level binding to purified dermatan sulfate. Therefore, although the dermatan sulfate GAG chain of decorin is critical for recognition by DbpA (24), the dermatan sulfate and decorin binding properties are not identical. Consistent with this observation, the protein core of decorin was previously shown to be required for detectable binding by DbpA in a radiometric assay (24). We found here that DbpAVS461 displayed a relatively poor activity in promoting 293 cell attachment, suggesting that, at least in this instance, cell attachment correlates more closely with dermatan sulfate binding than decorin binding. Definitive determination of the relative contribution of decorin and dermatan sulfate binding in cell attachment mediated by DbpA is complicated by the close structural relationship of these ligands and awaits further study.
Previous work showed that the lysine residues K163 and K170 in the C terminus of DbpA297, along with the centrally located lysine residue K82, were required for binding to decorin (8, 41). The sequence of the DbpA C terminus is highly divergent, and in particular the DbpAVS461 C terminus is quite dissimilar from that of DbpA297 (44). Nevertheless, the C termini of all of the alleles tested are highly basic and thus might contribute to GAG binding. We found that the 11-residue C-terminal segment of VS461 just C-terminal to K170 is required for the ability of DbpAVS461 to promote spirochetal binding to cells, decorin, and dermatan sulfate. This and other relatively variable regions of DbpA might contribute to the allelic diversity of attachment activity.
B. burgdorferi mutants that lack DbpA demonstrate a colonization defect in mice (4, 48). In the present study, we showed that the adhesive activity of DbpA is subject to considerable allelic variation. A detailed understanding of the molecular basis of this variation, combined with experimental infection by isogenic infectious strains that express different alleles of DbpA, may provide insight into the adhesive activities of this protein that contribute to infectivity and colonization in the mammalian host.
ACKNOWLEDGMENTS
We thank Jenifer Coburn and Mark Hanson for valuable technical advice. Jon Goguen, Linden Hu, Brian Akerley, and Victor Boyartchuk provided critical review of the manuscript. We also thank Ira Schwartz for helpful discussion and for providing B. burgdorferi strain B356. We are also extremely grateful to Nancy Ulbrandt and David Mann (MedImmune, Inc., Gaithersburg, MD), without whom the experiments with recombinant human decorin would not have been possible. We thank Jemiina Salo and Jukka Hytönen for the communication of unpublished results.
This study was supported by National Institutes of Health grants R01AI37601 and R01AI093104.
The authors declare they have no competing financial interests.
Footnotes
Published ahead of print on 27 June 2011.
REFERENCES
- 1. Alghaferi M. Y., et al. 2005. Borrelia burgdorferi ospC heterogeneity among human and murine isolates from a defined region of northern Maryland and southern Pennsylvania: lack of correlation with invasive and noninvasive genotypes. J. Clin. Microbiol. 43:1879–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbour A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521–525 [PMC free article] [PubMed] [Google Scholar]
- 3. Barthold S. W., Hodzic E., Tunev S., Feng S. 2006. Antibody-mediated disease remission in the mouse model of Lyme borreliosis. Infect. Immun. 74:4817–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blevins J., Hagman K., Norgard M. 2008. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brandt M. E., Riley B. S., Radolf J. D., Norgard M. V. 1990. Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect. Immun. 58:983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brissette C. A., et al. 2010. The borrelial fibronectin-binding protein RevA is an early antigen of human Lyme disease. Clin. Vaccine Immunol. 17:274–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brissette C. A., Verma A., Bowman A., Cooley A. E., Stevenson B. 2009. The Borrelia burgdorferi outer-surface protein ErpX binds mammalian laminin. Microbiology 155:863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown E. L., Guo B. P., O'Neal P., Höök M. 1999. Adherence of Borrelia burgdorferi. J. Biol. Chem. 274:26272–26278 [DOI] [PubMed] [Google Scholar]
- 9. Coburn J., Barthold S. W., Leong J. M. 1994. Diverse Lyme disease spirochetes bind integrin αIbβ3 on human platelets. Infect. Immun. 62:5559–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coburn J., Leong J. M., Erban J. K. 1993. Integrin αIbβ3 mediates binding of the Lyme disease agent Borrelia burgdorferi to human platelets. Proc. Natl. Acad. Sci. U. S. A. 90:7059–7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collares-Pereira M., et al. 2004. First isolation of Borrelia lusitaniae from a human patient. J. Clin. Microbiol. 42:1316–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Comstock L. E., Thomas D. D. 1989. Penetration of endothelial cell monolayers by Borrelia burgdorferi. Infect. Immun. 57:1626–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dorward D. W., Fischer E. R., Brooks D. M. 1997. Invasion and cytopathic killing of human lymphocytes by spirochetes causing Lyme disease. Clin. Infect. Dis. 25(Suppl. 1):S2–S8 [DOI] [PubMed] [Google Scholar]
- 14. Dykhuizen D. E., et al. 2008. The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am. J. Trop. Med. Hyg. 78:806–810 [PMC free article] [PubMed] [Google Scholar]
- 15. Fischer J. R., LeBlanc K. T., Leong J. M. 2006. Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect. Immun. 74:435–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fischer J. R., Parveen N., Magoun L., Leong J. M. 2003. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. U. S. A. 100:7307–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fraser C. M., et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 18. Galbe J. L., Guy E., Zapatero J. M., Peerschke E. I., Benach J. L. 1993. Vascular clearance of Borrelia burgdorferi in rats. Microb. Pathol. 14:187–201 [DOI] [PubMed] [Google Scholar]
- 19. Garcia-Monco J. C., Fernandez-Villar B., Benach J. L. 1989. Adherence of the Lyme disease spirochete to glial cells and cells of glial origin. J. Infect. Dis. 160:497–506 [DOI] [PubMed] [Google Scholar]
- 20. Gern L. 2008. Borrelia burgdorferi sensu lato, the agent of Lyme borreliosis: life in the wilds. Parasite 15:244–247 [DOI] [PubMed] [Google Scholar]
- 21. Grab D. J., Givens C., Kennedy R. 1998. Fibronectin-binding activity in Borrelia burgdorferi. Biochim. Biophys. Acta 1407:135–145 [DOI] [PubMed] [Google Scholar]
- 22. Grimm D., et al. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. U. S. A. 101:3142–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo B. P., Brown E. L., Dorward D. W., Rosenberg L. C., Höök M. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30:711–723 [DOI] [PubMed] [Google Scholar]
- 24. Guo B. P., Norris S. J., Rosenberg L. C., Höök M. 1995. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect. Immun. 63:3467–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hagman K. E., et al. 1998. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect. Immun. 66:2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanson M. S., Patel N. K., Cassatt D. R., Ulbrandt N. D. 2000. Evidence for vaccine synergy between Borrelia burgdorferi decorin binding protein A and outer surface protein A in the mouse model of Lyme borreliosis. Infect. Immun. 68:6457–6460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hileman R. E., Fromm J. R., Weiler J. M., Linhardt R. J. 1998. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays 20:156–167 [DOI] [PubMed] [Google Scholar]
- 28. Hodzic E., Feng S., Freet K. J., Borjesson D. L., Barthold S. W. 2002. Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect. Immun. 70:3382–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McBain A. L., Mann D. M. 2001. Purification of recombinant human decorin and its subdomains, p. 221–229. In Walker J. M. (ed.), Proteoglycan protocols, vol. 171 Humana Press, Inc., Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 30. Isaacs R. D. 1994. Borrelia burgdorferi bind to epithelial cell proteoglycans. J. Clin. Invest. 93:809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leong J. M., Moitoso de Vargas L., Isberg R. R. 1992. Binding of cultured mammalian cells to immobilized bacteria. Infect. Immun. 60:683–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leong J. M., Morrissey P. E., Marra A., Isberg R. R. 1995. An aspartate residue of the Yersinia pseudotuberculosis invasin protein that is critical for integrin binding. EMBO J. 14:422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leong J. M., Morrissey P. E., Ortega-Barria E., Pereira M. E., Coburn J. 1995. Hemagglutination and proteoglycan binding by the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malavaki C., Mizumoto S., Karamanos N., Sugahara K. 2008. Recent advances in the structural study of functional chondroitin sulfate and dermatan sulfate in health and disease. Connect. Tissue Res. 49:133–139 [DOI] [PubMed] [Google Scholar]
- 35. Marconi R. T., Konkel M. E., Garon C. F. 1993. Variability of osp genes and gene products among species of Lyme disease spirochetes. Infect. Immun. 61:2611–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Samuels D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi, p. 253–259. In Walker J. M. (ed.), Electroporation protocols for microorganisms, vol. 47 Humana Press, Inc., Totowa, NJ: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pal U., et al. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Invest. 113:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parveen N., Caimano M. J., Radolf J. D., Leong J. M. 2003. Adaptation of the Lyme disease spirochaete to the mammalian host environment results in enhanced glycosaminoglycan and host cell binding. Mol. Microbiol. 47:1433–1444 [DOI] [PubMed] [Google Scholar]
- 39. Parveen N., Leong J. M. 2000. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:1220–1234 [DOI] [PubMed] [Google Scholar]
- 40. Parveen N., Robbins D., Leong J. M. 1999. Strain variation in glycosaminoglycan recognition influences cell-type-specific binding by Lyme disease spirochetes. Infect. Immun. 67:1743–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pikas D. S., et al. 2003. Decorin-binding sites in the adhesin DbpA from Borrelia burgdorferi. J. Biol. Chem. 278:30920–30926 [DOI] [PubMed] [Google Scholar]
- 42. Población C. A., Michelacci Y. M. 1986. Structural differences of dermatan sulfates from different origins. Carbohydr. Res. 147:87–100 [DOI] [PubMed] [Google Scholar]
- 43. Probert W. S., Johnson B. J. B. 1998. Identification of a 47-kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 30:1003–1015 [DOI] [PubMed] [Google Scholar]
- 44. Roberts W. C., Mullikin B. A., Lathigra R., Hanson M. S. 1998. Molecular analysis of sequence heterogeneity among genes encoding decorin binding proteins A and B of Borrelia burgdorferi sensu lato. Infect. Immun. 66:5275–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rudenko N., et al. 2009. Molecular detection of Borrelia bissettii DNA in serum samples from patients in the Czech Republic with suspected borreliosis. FEMS Microbiol. Lett. 292:274–281 [DOI] [PubMed] [Google Scholar]
- 46. Sadziene A., Wilske B., Ferdows M. S., Barbour A. G. 1993. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun. 61:2192–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saito K., et al. 2007. Borrelia valaisiana infection in a Japanese man associated with traveling to foreign countries. Am. J. Trop. Med. Hyg. 77:1124–1127 [PubMed] [Google Scholar]
- 47a. Salo J., Loimaranta V., Lahdenne P., Viljanen M. K., Hytonen J. 2011. Decorin binding by DbpA and B of Borrelia garinii, Borrelia afzelil, and Borrelia burdorferi sensu stricto. J. Infect. Dis. 204:65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shi Y., Xu Q., McShan K., Liang F. T. 2008. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect. Immun. 76:1239–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steere A. C., Coburn J., Glickstein L. 2004. The emergence of Lyme disease. J. Clin. Invest. 113:1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stewart P. E., Thalken R., Bono J. L., Rosa P. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714–721 [DOI] [PubMed] [Google Scholar]
- 51. Szczepanski A., Furie M. B., Benach J. L., Lane B. P., Fleit H. B. 1990. Interaction between Borrelia burgdorferi and endothelium in vitro. J. Clin. Invest. 85:1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thomas D. D., Comstock L. E. 1989. Interaction of Lyme disease spirochetes with cultured eucaryotic cells. Infect. Immun. 57:1324–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Verma A., Brissette C. A., Bowman A., Stevenson B. 2009. Borrelia burgdorferi BmpA is a laminin-binding protein. Infect. Immun. 77:4940–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang G., et al. 2001. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect. Immun. 69:4303–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang G., van Dam A. P., Schwartz I., Dankert J. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang I.-N., et al. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wilske B. 2005. Epidemiology and diagnosis of Lyme borreliosis. Ann. Med. 37:568–579 [DOI] [PubMed] [Google Scholar]
- 58. Wormser G. P., et al. 2008. Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J. Infect. Dis. 198:1358–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wormser G. P., et al. 1999. Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J. Infect. Dis. 180:720–725 [DOI] [PubMed] [Google Scholar]
- 60. Wywial E., et al. 2009. Fast, adaptive evolution at a bacterial host-resistance locus: the PFam54 gene array in Borrelia burgdorferi. Gene 445:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zambrano M. C., Beklemisheva A. A., Bryksin A. V., Newman S. A., Cabello F. C. 2004. Borrelia burgdorferi binds to, invades, and colonizes native type I collagen lattices. Infect. Immun. 72:3138–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]