Abstract
Chlamydia trachomatisis an obligate intracellular bacterium that is dependent on its host cell for nucleotides. Chlamydiaimports ribonucleotide triphosphates (NTPs) but not deoxyribonucleotide triphosphates (dNTPs) and instead uses ribonucleotide reductase to convert imported ribonucleotides into deoxyribonucleotides for DNA synthesis. The genes encoding ribonucleotide reductase have been recently shown to be negatively controlled by a conserved regulator called NrdR. In this study, we provide direct evidence that Escherichia coliNrdR is a transcriptional repressor and that C. trachomatisCT406 encodes its chlamydial ortholog. We showed that CT406 binds specifically to two NrdR boxes upstream of the nrdABoperon in C. trachomatis. Using an in vitrotranscription assay, we confirmed that these NrdR boxes function as an operator since they were necessary and sufficient for CT406-mediated repression. We validated our in vitrofindings with reporter studies in E. colishowing that both E. coliNrdR and CT406 repressed transcription from the E. colinrdHand C. trachomatisnrdABpromoters in vivo. This in vivorepression was reversed by hydroxyurea treatment. Since hydroxyurea inhibits ribonucleotide reductase and reduces intracellular deoxyribonucleotide levels, these results suggest that NrdR activity is modulated by a deoxyribonucleotide corepressor.
INTRODUCTION
Chlamydia trachomatisis a Gram-negative obligate intracellular parasite and a significant human pathogen. It is the causative agent of the most frequently reported bacterial sexually transmitted disease in the United States (5). C. trachomatisalso causes trachoma, the world's most common form of infectious blindness (23). Within an infected cell, this organism exists as a metabolically active form called the reticulate body (RB), which divides by binary fission. RBs reside at all times within a membrane-bound cytoplasmic inclusion and acquire lipids, proteins, and other nutrients from the host cell (27). Chlamydiaspp. are unusual among bacteria in having a second morphological form called the elementary body (EB), which is specialized for extracellular survival and the ability to infect a new host cell to initiate another round of intracellular infection.
Chlamydiahas been so successful in adapting to life inside a eukaryotic cell that it is now completely dependent on its host cell for survival and replication. Over the course of its evolution, Chlamydiahas reduced its genome to approximately 1,000 genes (22, 33), compared to about 5,000 in Escherichia coli(2), dispensing with many genes whose functions can be replaced by the host cell. For example, C. trachomatisdoes not encode enzymes involved in the biosynthesis of amino acids such as aspartate and tyrosine and has only a partial biosynthetic pathway for tryptophan (33). Chlamydiae also depend on the host cell for nucleotides, which are the essential building blocks of RNA and DNA (18). While C. trachomatiscan synthesize CTP, it is auxotrophic for ATP, GTP, and UTP, which it imports from the host cell (39, 40). Chlamydiae do not import deoxyribonucleotide triphosphates (dNTPs), however, and instead use the enzyme ribonucleotide reductase to convert imported ribonucleotides into deoxyribonucleotides (38).
Ribonucleotide reductase is conserved from prokaryotes to eukaryotes and is responsible for production of all four dNDPs from their respective NDPs (17). E. coliexpresses three different ribonucleotide reductases from the nrdAB, nrdDG, and nrdHIEF(nrdH) operons (2). In contrast, all Chlamydiaspp. express a single class Ia ribonucleotide reductase encoded by the nrdABoperon (26). In bacteria, expression of the ribonucleotide reductase genes can be positively or negatively regulated at the transcriptional level. For example, in E. coli, the DNA replication initiation factor DnaA represses nrdABwhen bound to ATP (9). Fis and IciA, on the other hand, are factors that activate nrdABtranscription (12, 14).
The expression of ribonucleotide reductase genes is also regulated by a transcription factor called NrdR. NrdR was first described in Streptomyces coelicolor(3), and its cognate operator, the NrdR box, was predicted from a comparative bioinformatics analysis of 63 bacterial genomes (24). A consensus NrdR box sequence has been proposed (acaCwAtATaTwGtgt), and tandem NrdR boxes have been identified upstream of genes encoding ribonucleotide reductase and factors involved in DNA maintenance and replication (24). There is genetic evidence in several bacterial species that NrdR negatively regulates nrdtranscription in vivo(3, 10, 19, 21, 41). NrdR has also been shown to bind to NrdR boxes (10, 11, 41), although it has not been directly shown to repress transcription in vitro. NrdR contains an ATP cone for nucleotide binding and a zinc ribbon for DNA binding (24), and binding to nucleotides and zinc has been demonstrated (10). These observations have led to speculation that NrdR may regulate intracellular pools of nucleotides in response to a nucleotide or metal ion, but a cofactor requirement has not been shown.
A putative NrdR ortholog has been identified in all Chlamydiaspp. (24), but there have been no functional studies to validate these predictions. In C. trachomatis, the candidate NrdR ortholog is encoded by CT406, which has 45% amino acid similarity to E. coliNrdR (E. D. R. Case and M. Tan, unpublished observation). In addition, tandem NrdR boxes have been predicted upstream of the nrdABoperon, which is the only predicted target in Chlamydiaspp. (24). To examine whether CT406 is the chlamydial ortholog of NrdR, we performed functional studies to determine if CT406 is able to bind the predicted NrdR boxes and repress transcription of nrdABand if repression is regulated by intracellular nucleotide levels.
MATERIALS AND METHODS
Extraction of chlamydial total RNA.
To prepare chlamydial RNA, eight T150 flasks of subconfluent mouse L929 fibroblast monolayers were infected with C. trachomatisserovar D strain UW-3/Cx at a multiplicity of infection (MOI) of 20. At 24 h postinfection, the chlamydial RBs were harvested as previously described (35). Total RNA was extracted using RNA STAT-60 according to the manufacturer's instructions. The purified RNA was treated with RQ1 DNase (Roche) to remove any contaminating genomic DNA.
Mapping of nrdABtranscription start site by 5′ RACE.
Ten micrograms of chlamydial total RNA was used to synthesize nrdAcDNA by using 2 pmol of the gene-specific primer T653 and AMV reverse transcriptase (Promega). The sequences of all primers and DNA oligonucleotides used in this study are presented in Table 1. Following reverse transcription, the RNA was hydrolyzed and the cDNA was purified using the QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions. The 5′ RACE protocol was adapted from the 5′ RACE system for rapid amplification of cDNA ends, version 2.0 (Invitrogen). Briefly, a poly(dC) tail was added to the 5′ end of the purified cDNA by 1 U terminal deoxynucleotidyl transferase (Promega). Five microliters of tailed cDNA was amplified using primers T602 and T694. The resulting PCR product was digested with EcoRI (Promega) and ligated into pGEM-7Z (f)+ cloning vector (Promega). The DNA sequences of this and other cloned inserts used in this study were verified.
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Oligonucleotide sequence (5′–3′) | Applicationa |
|---|---|---|
| T653 | TCTGCGAGTGTCTCGAAAAG | nrdAB5′ RACE primer |
| T602 | CGCGAATTCCTCTTCTAGATGGGIIGGGIIGGGIIG | 5′ RACE adapter primer |
| T694 | TTCTAAAGCCTGAAAAATACGG | nrdABnested 5′ RACE primer |
| T1434 | ATGCTGTGCCCGTTCTGCAA | CT406 5′ cloning primer |
| T1435 | GGTCTGTTTTTCACCGTCCGG | CT406 3′ cloning primer |
| T1441 | ATGCATTGCCCATTCTGTTTCGC | nrdR5′ cloning primer |
| T1442 | GTCCTCCAGGCGCGCGAT | nrdR3′ cloning primer |
| T1137 | AATTCACGTTGCTAGCTTCTATATATGGTATACAAGAGCCGAGACGTTCACAATATCTTGGGTTTTTAGGGGC | nrdABtandem NrdR box probe fwd |
| T1138 | GATCCGCCCCTAAAAACCCAAGATATTGTGAACGTCTCGGCTCTTGTATACCATATATAGAAGCTAGCAACGTG | nrdABtandem NrdR box probe rev comp |
| T1326 | AATTCCTAGATTTTCCTTTTGGTTGAGGATATAAATTCATTCT | ytgABCRDprobe fwd |
| T1327 | GATCCAGAATGAATTTATATCCTCAACCAAAAGGAAAATCTAG | ytgABCRDprobe rev comp |
| T1350b | GCGCCTTTGTTGTATCGTCAGTTCAGGGTAAAATAGATTTCCGTTAACGTGTAGGCTGGAGCTGCTTC | nrdR5′ deletion primer |
| T1351 | TCCTCGTTGCGCCAGCTTTAGCGCCCGCGCCATGTAATACTCGTCCTGCATATGAATATCCTCCTTAGTTCC | nrdR3′ deletion primer |
| T1490 | CAGCAGAGGCGGATAAAAGT | luxA5′ cloning primer |
| T1513 | GATCAGAAGAACGCTTTGAATTCATAAAATTG | luxA3′ cloning primer |
| T1491 | TCAGAGGTTTTCACCGTCATC | luxB5′ cloning primer |
| T1512 | CAATTTTATGAATTCAAAGCGTTCTTCTGATC | luxB3′ cloning primer |
| T1521 | ATCGGTACCGGTTTCTTTTTGCCTGGTGA | nrdHIEFreporter 5′ cloning primer |
| T1522 | CGTCTCGAGTTGATACAGAGATACTAGAT | nrdHIEFreporter 3′ cloning primer |
| T1523 | CGAGGTACCGCTTAAAAGTTTGCTCTCGA | nrdABreporter 5′ cloning primer |
| T1524 | TCGCTCGAGAGCCCCTAAAAACCCAAGAT | nrdABreporter 3′ cloning primer |
| T1599 | TTCCTTTTGGTTGAGGATATAAATTCATTCTGTTAAAAGTATCTTTACAAT | ytgABCRDreporter insert fwd |
| T1600 | ATTGTAAAGATACTTTTAACAGAATGAATTTATATCCTCAACCAAAAGGAA | ytgABCRDreporter insert rev comp |
| T1527 | GGTGAATTCCTAGCACCTTCTATATATGGTATCTAAAATTCTTGACCATCACAATATCTTGGGTTATAATGACACCAACTT | dnaKpromoter + nrdABNrdR boxes fwd |
| T1528 | AAGTTGGTGTCATTATAACCCAAGATATTGTGATGGTCAAGAATTTTAGATACCATATATAGAAGGTGCTAGGAATTCACC | dnaKpromoter + nrdABNrdR boxes rev comp |
| T1575 | GAGGAATTCCTAGCACTTTGCTATATATTGTGTTAAAATTCTTGACCCAACTACATCTAGTACTATAATGACACCAACTT | dnaKpromoter + nrdHIEFNrdR boxes fwd |
| T1576 | AAGTTGGTGTCATTATAGTACTAGATGTAGTTGTGGTCAAGAATTTTAACACAATATATAGCAAAGTGCTAGGAATTCCTC | dnaKpromoter + nrdHIEFNrdR boxes rev comp |
| T1689 | AATTCACGTTGCTAGCATGTTGTTAAGCATTACAAGAGCCGAGACGTTCACAATATCTTGGGTTTTTAGGGGC | nrdABupstream NrdR box probe fwd |
| T1690 | GATCCGCCCCTAAAAACCCAAGATATTGTGAACGTCTCGGCTCTTGTAATGCTTAACAACATGCTAGCAACGT | nrdABupstream NrdR box probe rev comp |
| T1693 | AATTCACGTTGCTAGCTTCTATATATGGTATACAAGAGCCGAGACGTACTCTTAATGTACCGTTTTTAGGGGC | nrdABdownstream NrdR box probe fwd |
| T1694 | GATCCGCCCCTAAAAACGGTACATTAAGAGTACGTCTCGGCTCTTGTATACCATATATAGAAGCTAGCAACGT | nrdABdownstream NrdR box probe rev comp |
fwd, forward; rev, reverse; comp, complement.
Construction of protein expression plasmids.
A codon-optimized version of CT406 from Chlamydia trachomatisserovar D was generated (Verdezyne, Carlsbad, CA) for the optimal expression in E. coli(13). Codon-optimized CT406 was designed by changing the codon bias of the CT406 open reading frame to better match that of E. coliwithout changing its translated amino acid sequence. The codon-optimized open reading frame was amplified from the manufacturer-supplied plasmid by PCR using primers T1434 and T1453. E. colinrdRwas amplified from E. coliK-12-MG1655 genomic DNA by PCR using primers T1441 and T1442. The CT406 and E. colinrdRDNA inserts were cloned into the expression vector pBAD-ITO (generous gift of G. W. Hatfield, University of California, Irvine) at blunted NdeI and HindIII sites to generate pMT1565 and pMT1566, respectively. Both recombinant proteins contained C-terminal His6and myctags.
Overexpression and purification of CT406.
An overnight culture of pMT1565 transformed into E. coliTOP10 (Invitrogen) was diluted 1:100 in LB broth containing 100 mg/ml ampicillin. After growth at 37°C to an optical density at 600 nm (OD600) of ∼0.5, CT406 expression was induced with 0.002% (wt/vol) l-arabinose followed by overnight incubation at 16°C. Cells were harvested by centrifugation at 2,000 × gfor 10 min at 4°C, and the cell pellet was stored at −80°C. To purify recombinant CT406 protein, thawed cells were resuspended in protein purification buffer (20 mM ethanolamine [pH 10], 300 mM NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml pepstatin A, and 100 μg/ml lysozyme) and disrupted by sonication. The lysate was cleared by centrifugation at 20,000 × gfor 30 min at 4°C and incubated with a 1-ml bed volume of equilibrated Talon metal affinity resin (Clontech) at 4°C for 1 h with agitation. The resin was collected by centrifugation and washed 3 times with 20 volumes of protein purification buffer at 4°C for 20 min with agitation. Protein was eluted with 5 volumes of protein purification buffer containing 50 mM imidazole. The eluate was pooled and concentrated to 1 ml using a Vivaspin 6 column (GE Healthcare) and then dialyzed against protein storage buffer. For protein stocks used in electrophoretic mobility shift assay (EMSA) experiments, storage buffer contained 20 mM ethanolamine (pH 10), 10 mM MgCl2, 10 mM 2-mercaptoethanol, 100 mM NaCl, and 30% (wt/vol) glycerol. For in vitrotranscription experiments, a low-salt storage buffer was used (10 mM Tris [pH 8], 10 mM MgCl2, 10 mM 2-mercaptoethanol, and 30% [wt/vol] glycerol). Aliquots of rCT406 were stored at −80°C. Protein concentration was determined using the Bio-Rad protein assay.
Production of polyclonal antibodies.
Recombinant CT406 was gel purified by SDS-PAGE and used to generate rabbit polyclonal antibodies (Harlan Bioproducts for Science). Antibodies were then affinity purified using a protein A-agarose column according to the manufacturer's instructions (Bio-Rad).
Construction and labeling of gel shift assay probes.
The following DNA probes were generated by annealing cDNA oligonucleotides: the nrdABpromoter region with predicted tandem NrdR boxes from C. trachomatisserovar D (T1137 and T1138); the nrdABupstream NrdR box only (T1689 and T1690); the nrdABdownstream NrdR box only (T1693 and T1694); and a nonspecific oligonucleotide probe containing sequences upstream of the C. trachomatisytgoperon (T1326 and T1327). Each annealed oligonucleotide probe contained a 5′ EcoRI overhang and a 3′ BamHI overhang for fill-in labeling with [γ-32P]dATP as previously described (43).
DNA gel shift assay.
Electrophoretic mobility shift assay (EMSA) reactions were performed by incubating a range of concentrations of recombinant CT406 with 0.5 nM labeled DNA probe in 1× binding buffer (40 mM Tris-HCl [pH 8.0], 4 mM MgCl2, 70 mM KCl, 125 μM EDTA, 100 μM dithiothreitol [DTT], 4% [wt/vol] glycerol). Some reactions also contained anti-mycantibodies. Reaction mixtures were incubated at room temperature for 30 min and then loaded onto a 6% polyacrylamide gel and electrophoresed at 150 V at 4°C in 0.5× Tris-borate buffer. Following electrophoresis, gels were dried, exposed to a phosphorimager plate, and visualized as previously described (43).
Generation of E. colinrdRnull mutant.
A strain of E. colilacking the nrdRgene was generated using the method of Datsenko and Wanner (6). The entire nrdRlocus was deleted from the region immediately downstream of the transcription start site to its stop codon. Primers T1350b and T1351, which contain homologous sequences that flank the nrdRgene locus and leave the nrdRpromoter intact, were used to amplify the kanamycin resistance cassette from pKD4 (6). The PCR product was purified by agarose gel electrophoresis with the QIAquick gel extraction kit (Qiagen) and electroporated into E. coliBW25113/pKD46 (6). Following selection for kanamycin resistance, homologous recombination of the Kanrcassette was verified by PCR. The kanamycin resistance marker was then transduced into E. coliK-12-MG1655 by P1vir(37). Transductants were selected on LB medium containing kanamycin and verified by colony PCR. A positive transductant was then electroporated with pCP20 for removal of the kanamycin resistance gene by FLP recombinase (6) to generate E. coliK-12-MG1655ΔnrdR. Removal of Kanrwas confirmed by PCR and sequencing of the target locus, which matched the pKD4 FLP scar sequence described previously (6). We verified that the loss of nrdRdid not produce a significant phenotype during normal growth, which is in agreement with published studies (3, 41), and we confirmed that transcription of the downstream gene ribDwas unaffected by reverse transcription (RT)-PCR (data not shown). Strains and plasmids used in this study are shown in Table 2.
Table 2.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| Chlamydia trachomatisserovar D | Trachoma type D strain UW-3/Cx | ATCC |
| Escherichia coliK-12-MG1655 | F−λ−ilvG−rfb-50 rph-1 | ATCC |
| Escherichia coliBW25113 | Δ(araD-araB)567ΔlacZ4787(::rrnB-3) λ−rph-1Δ(rhaD-rhaB)568 hsdR514 | Coli Genetic Stock Center (6) |
| Escherichia coliΔnrdR | F−λ−ilvG−rfb-50 rph-1 ΔnrdR | This study |
| Escherichia coliTOP10 | F−mcrAΔ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139Δ(ara-leu)7697 galE15 galK16 rpsL(Strr) endA1λ− | Invitrogen |
| Escherichia coliXL1-Blue | endA1 gyrA96(Nalr) thi-1 recA1 relA1 lac glnV44F′[::Tn10 proAB+lacIqΔ(lacZ)M15]hsdR17(rK−mK+) | Stratagene |
| Plasmids | ||
| pGEM-7Z(f)+ | Ampicillin-resistant cloning vector | Promega |
| pBAD-ITO | Ampicillin-resistant, arabinose-inducible expression vector | Gift of G. W. Hatfield, UC Irvine |
| pKD4 | kanRtemplate plasmid for gene disruption | Coli Genetic Stock Center (6) |
| pKD46 | Lambda Red recombinase expression plasmid | Coli Genetic Stock Center (6) |
| pCP20 | Temperature-sensitive yeast FLP recombinase expression plasmid | Coli Genetic Stock Center (6) |
| pACYC177 | Kanamycin-resistant cloning vector | ATCC |
| pQF110 | Vibrio harveyiluxAB/phoAreporter plasmid | ATCC (25) |
| pMT1125 | Ampicillin-resistant promoterless transcription template | Wilson and Tan (43) |
| pMT1161 | dnaKpromoter in pMT1125 | Schaumburg and Tan (30) |
| pMT1565 | Codon-optimized CT406 in pBAD-ITO | This study |
| pMT1566 | E. colinrdRin pBAD-ITO | This study |
| pMT1580 | Vibrio harveyiluxABreporter gene in pACYC177 | This study |
| pMT1581 | E. colinrdHpromoter in pMT1580 | This study |
| pMT1582 | C. trachomatisnrdApromoter in pMT1580 | This study |
| pMT1587 | dnaKpromoter with NrdR boxes from C. trachomatisnrdAin pMT1125 | This study |
| pMT1608 | dnaKpromoter with NrdR boxes from E. colinrdHin pMT1125 | This study |
| pMT1616 | C. trachomatisytgpromoter in pMT1580 | This study |
Construction of luciferase reporter plasmids.
The luciferase reporter vector pMT1580 was constructed by cloning the luxABgenes from Vibrio harveyiinto the pACYC177 cloning vector (ATCC). First, the luxAgene was amplified by PCR using PfuDNA polymerase with primers T1490 and T1513 from the pQF110 template (25). The insert was ligated into pACYC177 at a blunted BamHI site. Clones were selected on LB medium containing ampicillin, and the correct insert was verified by PCR. This luxAplasmid was purified from E. coliXL1-Blue by using the Nucleobond AX kit (Macherey-Nagel) according to the manufacturer's instructions.
The luxBgene was also amplified by PCR using primers T1491 and T1512 from pQF110. This insert was digested with EcoRI at its 5′ end and ligated into EcoRI and blunted AatII sites of the luxAplasmid. After verifying the DNA sequence of luxAB, the ampicillin resistance cassette on the vector backbone was disrupted by digestion with ApaLI and AhdI, followed by blunting and religation to produce pMT1580. The disruption of Amprwas confirmed by lack of growth on LB medium containing ampicillin.
The following reporter plasmids were constructed from PCR-amplified promoter inserts digested with KpnI and cloned upstream of luxABat KpnI and blunted XbaI sites in pMT1580: pMT1581 containing the nrdHpromoter amplified with T1521 and T1522 from E. coliK-12-MG1655 genomic DNA; pMT1582 containing the nrdABpromoter region amplified with T1523 and T1524 from C. trachomatisserovar D genomic DNA (7). The pMT1616 reporter containing the C. trachomatis ytgoperon promoter region was constructed by annealing complementary oligonucleotides T1599 and T1600 and ligated between blunted XbaI sites in pMT1580.
Luciferase reporter assays.
Each luciferase reporter construct was cotransformed with either empty pBAD-ITO vector, pMT1565, or pMT1566 into E. coliK-12-MG1655ΔnrdRand selected on LB medium containing ampicillin and kanamycin. An overnight culture was diluted 1:100 in LB medium containing ampicillin, kanamycin, and 0.2% l-arabinose and incubated at 37°C. Hydroxyurea (15 mM or 100 mM) was added to some cultures once an OD600of ∼0.2 was reached. After growth to an OD600of ∼0.8, a sample of the culture was diluted to an OD600of 0.1 in culture medium. Twenty-five microliters of the diluted sample was added to 75 μl of luciferase reaction buffer (50 mM sodium phosphate buffer [pH 7], 50 mM 2-mercaptoethanol, 2% [wt/vol] bovine serum albumin [BSA], and n-decanal [1:2,000 final dilution]). The reactions were incubated at room temperature for 10 min in the dark, and then light production was measured over 10 s on a Sirius luminometer V3.1 (Berthold Detection Systems) and reported as relative light units (RLU) per second. All measurements were performed in triplicate, and the data presented are the means of three independent experiments. Statistical analysis of the results by one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparison test was performed using GraphPad Prism v5.0 software. Expression of pMT1565 and pMT1566 was confirmed by SDS-PAGE and Western blot analysis.
Construction of in vitrotranscription templates.
Two synthetic promoters were generated by addition of an NrdR box to the C. muridarum(also known as C. trachomatisMoPn) dnaKpromoter. The insert for pMT1587 (NrdR boxes from C. trachomatisserovar D nrdAB) was generated by annealing single-stranded oligonucleotides T1527 and T1528, followed by digestion with EcoRI (Promega) and phosphorylation with T4 polynucleotide kinase (New England BioLabs). Similarly, the insert for pMT1608 (NrdR boxes from E. colinrdH) was generated by annealing oligonucleotides T1575 and T1576. The inserts were ligated into pMT1125 digested with EcoRI and EcoRV (43). Construction of the wild-type dnaKpromoter template (pMT1161) was described previously (30).
Purification of chlamydial RNA polymerase.
RNA polymerase holoenzyme was biochemically purified from C. trachomatisserovar L2 reticulate bodies at 21 h postinfection as previously described (36).
In vitrotranscription assay.
Transcription of chlamydial promoter templates was performed as described previously (36), with several modifications. DNA template (1 μl [5 nM]) was incubated in the presence of varying concentrations of purified recombinant CT406 protein at room temperature for 15 min in 9 μl of protein storage buffer. Following preincubation of protein and the DNA template, the transcription reaction mixture (1 μl rRNAsin [Promega], 0.5 μl chlamydial RNA polymerase, 400 μM ATP, 400 μM UTP, 100 μM mGTP, and 0.8 μl [32P]CTP) was added to a final volume of 20 μl. The reaction mixtures were incubated at 37°C for 15 min, and the reactions were stopped with the addition of an equal volume of stop buffer (36), followed by incubation for 5 min at 65°C. The transcripts were resolved by electrophoresis with visualization and quantification as previously described (43).
RESULTS
The nrdABpromoter overlaps predicted NrdR boxes.
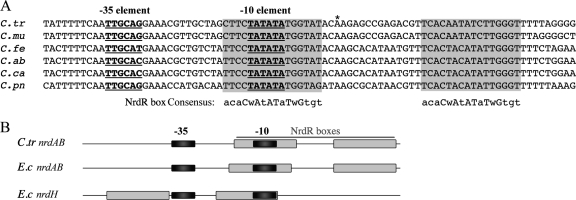
To study the regulation of the ribonucleotide reductase genes in Chlamydia, we first identified the promoter of the nrdABoperon. We mapped the transcription start site for C. trachomatisnrdABby using 5′ RACE and predicted a candidate promoter immediately upstream (Fig. 1A) that has sequence similarity to the preferred C. trachomatisσ66promoter (29, 34, 36). Using an in vitrotranscription assay, we verified that this promoter is transcribed by chlamydial σ66RNA polymerase (data not shown). Intriguingly, the nrdABpromoter is in the immediate vicinity of tandem NrdR boxes that are conserved in all Chlamydiaspp. (24). The upstream NrdR box overlaps the −10 promoter element that we predicted, and the second NrdR box is located just downstream of the transcription start site (Fig. 1A). This overlapping arrangement of the promoter and NrdR boxes is conserved among Chlamydiaspp. (Fig. 1A) and is similar to the E. colinrdABpromoter (Fig. 1B) (41). The close proximity of the tandem NrdR boxes and the nrdABpromoter supports a model for the repression of the nucleotide reductase genes in Chlamydiaby an NrdR ortholog.
Fig. 1.
Relative position of the C. trachomatisnrdABpromoter and predicted NrdR boxes. (A) Sequence alignment of the nrdABpromoter regions from C. trachomatis(C.tr), C. muridarum(C.mu), C. felis(C.fe), C. abortus(C.ab), C. caviae(C.ca), and C. pneumoniae(C.pn). The transcription start site mapped in C. trachomatisserovar D is marked by an asterisk (*), and the −35 and −10 promoter elements are underlined and shown in bold. Predicted NrdR boxes are highlighted in gray and aligned with the bacterial NrdR box consensus sequence (24). (B) Diagram showing position of NrdR boxes (gray rectangles) relative to the −35 and −10 elements (black rectangles) in the nrdABpromoters from C. trachomatis, E. coli, and the E. colinrdHpromoter.
CT406 binds upstream of nrdAB.
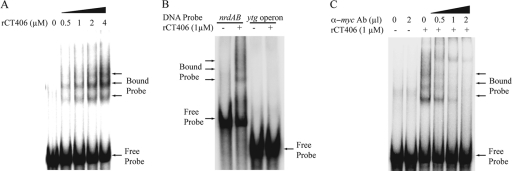
To test whether CT406 is the chlamydial NrdR ortholog, we first examined whether it can bind the NrdR boxes of the nrdABoperon. In an electrophoretic mobility shift assay (EMSA), myc-tagged recombinant CT406 produced a concentration-dependent gel shift with a 78-bp oligonucleotide DNA probe containing the tandem NrdR boxes in their native location relative to the nrdABpromoter (Fig. 2A). We detected at least three shifted bands, indicating the formation of multiple higher-order rCT406-DNA complexes, which have been described for NrdR orthologs in E. coliand S. coelicolor(10, 11, 41). Binding was sequence specific, since rCT406 did not produce a gel shift with a probe containing the C. trachomatisytgpromoter, which does not contain a recognizable NrdR box (Fig. 2B). We verified that the observed gel shift is due to rCT406 by showing that antibodies to the myctag disrupted the gel shift in a concentration-dependent manner (Fig. 2C).
Fig. 2.
Gel shift assays showing that rCT406 binds to the C. trachomatisnrdABpromoter region. (A) Concentration-dependent binding of rCT406, with the gel migration positions of free and bound probe as indicated. (B) rCT406 bound to the nrdABpromoter region but did not produce a gel shift with a probe containing the upstream sequence of the ytgoperon. (C) Disruption of rCT406-DNA binding by increasing amounts of monoclonal anti-myc(α-myc) antibody directed against the myc-tagged rCT406.
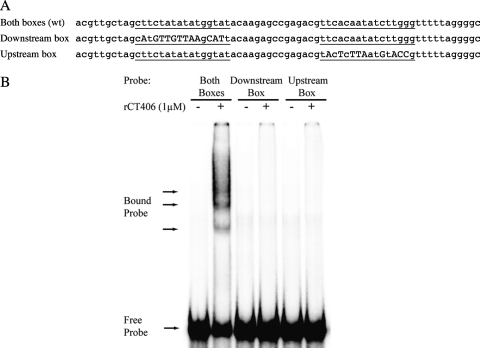
To examine whether both NrdR boxes are necessary for rCT406 binding, we repeated the EMSA studies with DNA probes in which either box was disrupted by sequence substitutions at the most conserved positions within the chlamydial NrdR boxes (Fig. 3A) (24). We observed no gel shift when we tested mutant nrdABDNA probes containing only an intact downstream or upstream NrdR box (Fig. 3B). These results indicate that the tandem NrdR boxes are necessary for binding of CT406 to the C. trachomatisnrdABpromoter and together make up the functional operator.
Fig. 3.
rCT406 requires both NrdR boxes for binding. (A) DNA sequence of the wild-type (wt) C. trachomatisnrdABprobe containing tandem NrdR boxes (underlined) and two mutant probes each containing only one intact NrdR box. For the “downstream box” probe, transversions (marked in uppercase letters) were introduced at 12 positions in the upstream NrdR box. Similarly, for the “upstream box” probe, transversions were introduced at 9 positions in the downstream NrdR box. (B) rCT406 (1 μM) produced a gel shift with the wild-type probe containing tandem NrdR boxes but not the mutant probes containing only one NrdR box each.
CT406 represses transcription in vitro.
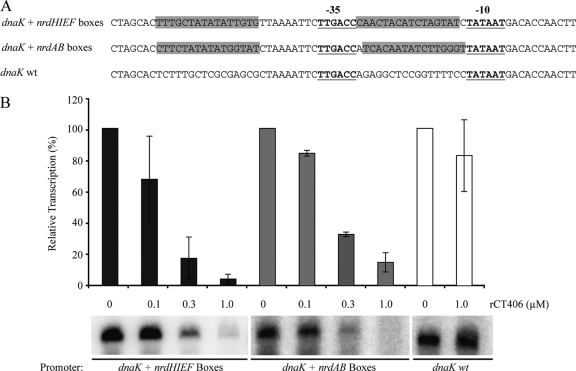
We used a chlamydial in vitrotranscription assay to test if rCT406 is a functional repressor. Our assay utilizes a G-less cassette transcription template, which allows transcription of a specific promoter in the absence of GTP to limit background transcription initiated elsewhere on the plasmid template (35). However, we could not test the native C. trachomatisnrdABpromoter under these conditions because the NrdR box downstream of the transcription start site encodes G residues (Fig. 1A). We were also unable to test the native nrdABpromoter with a run-off transcription assay because the nrdABpromoter was only weakly transcribed from a linear template (data not shown). As a solution, we tested NrdR boxes in the context of a well-defined chlamydial promoter. We placed tandem NrdR boxes upstream and downstream of the −35 element of the dnaKpromoter to preserve the spacing between the two boxes without altering the promoter sequence (Fig. 4A). rCT406 caused a concentration-dependent decrease in transcription of a dnaKpromoter containing tandem NrdR boxes from either E. colinrdHor C. trachomatisnrdAB(Fig. 4B). At the highest concentration of rCT406 tested, there was 96% repression with the nrdHNrdR boxes and 85% repression with the nrdABNrdR boxes (Fig. 4B). In contrast, rCT406 did not cause a significant change in transcription from the wild-type dnaKpromoter (Fig. 4B). These results indicate that CT406 can function as a transcriptional repressor and that its activity is dependent on the presence of the NrdR boxes in the vicinity of the target promoter.
Fig. 4.
rCT406 repressed transcription in vitro. (A) Sequence alignment of transcription templates containing the wild-type dnaKpromoter and mutant forms of the dnaKpromoter engineered to contain tandem NrdR boxes from either E. colinrdHor C. trachomatisnrdAB. The sequences of the inserted NrdR boxes are highlighted in gray. (B) In vitrotranscription of these three transcription templates in the presence of rCT406. For each template, the amount of transcription in the absence of rCT406 was defined as 100% and used to normalize transcript levels obtained with increasing concentrations of rCT406. The values shown are the means of three independent experiments, and the error bars represent standard deviations.
CT406 represses transcription in vivo.
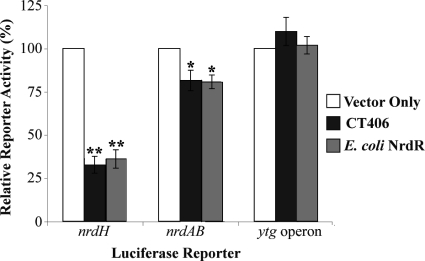
Since there is no experimental genetic system for Chlamydia, we used a luciferase reporter assay in E. colito validate these in vitroresults and to test for repression of the native chlamydial nrdABpromoter. We constructed reporter plasmids by cloning the promoter sequences of E. colinrdH, C. trachomatisnrdAB, or the C. trachomatisytgoperon upstream of Vibrio harveyiluxAB(25). To avoid confounding effects from endogenous E. coliNrdR, we performed these experiments in a strain in which we had deleted the nrdRlocus. Expression of recombinant CT406 reduced the activity of the nrdHreporter by approximately 70% (Fig. 5). We measured a similar level of repression when we expressed E. coliNrdR from a plasmid in this nrdRdeletion strain. We observed more modest repression of the nrdABreporter with expression of either CT406 or E. coliNrdR, although the effect was statistically significant. This differential regulation of promoters is consistent with studies in E. coliin which NrdR caused less repression of the nrdABpromoter compared to the nrdHpromoter (41). In a control experiment, neither CT406 nor E. coliNrdR repressed expression from the control ytgreporter, which does not contain a predicted NrdR box. These results provide additional experimental support for CT406 as a transcriptional repressor of ribonucleotide reductase genes.
Fig. 5.
rCT406 repressed transcription in vivo. A reporter plasmid containing either the promoter of E. colinrdH, that of C. trachomatisnrdAB, or the C. trachomatis ytgoperon was transformed into E. coliΔnrdR. For each reporter, the luciferase activity from cells cotransformed with an empty expression vector (white bars; marked as “vector only”) was defined as 100% and used to normalize the reporter activity measured when either rCT406 (dark gray bars) or E. coliNrdR (light gray bars) was expressed from a plasmid. The values shown are the means of three independent experiments, and the error bars represent standard deviations. Asterisks indicate that the values reported are significantly different from the vector-only control as determined by one-way ANOVA followed by Dunnett's multiple comparison test, with reported Pvalues of <0.0001 (**) and 0.0019 (*).
CT406- and E. coliNrdR-mediated repression is reversed by hydroxyurea treatment.
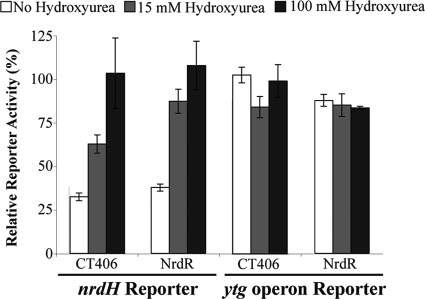
To examine whether NrdR-mediated repression is regulated by a nucleotide cofactor, we treated the E. coliΔnrdRstrain used in the reporter assay with hydroxyurea to inhibit ribonucleotide reductase and lower intracellular dNTP levels (31). When the NrdR deletion strain expressed CT406 from a plasmid, repression of the E. colinrdHreporter was partially reversed by treatment with 15 mM hydroxyurea and completely reversed at 100 mM hydroxyurea (Fig. 6). We observed similar results with control experiments in which the ΔnrdRstrain was complemented with E. coliNrdR. For these experiments, we verified that expression levels of CT406 and E. coliNrdR were adequate by Western blot (data not shown). This hydroxyurea-mediated derepression was specific for promoters regulated by CT406 and NrdR, since we observed no effect on ytgreporter activity at either hydroxyurea concentration. These data indicate that repression by NrdR and CT406 is sensitive to hydroxyurea, which is consistent with recent findings in Salmonella(21). Since hydroxyurea reduces deoxyribonucleotide levels by inhibiting the conversion of NDPs to dNDPs (31), our results support a model in which transcriptional repression by CT406 and E. coliNrdR is modulated by deoxyribonucleotides.
Fig. 6.
Effect of hydroxyurea on NrdR- and CT406-mediated repression. The luciferase activity from E. coliΔnrdRcells cotransformed with an E. colinrdHreporter and an empty expression vector was defined as 100% and used to normalize the reporter activity measured when either E. coliNrdR or C. trachomatisCT406 was expressed. Reporter activity was measured in the absence (white bars) or presence of 15 mM (light gray bars) or 100 mM (dark gray bars) of hydroxyurea. Parallel experiments were performed with a control reporter containing the promoter of the C. trachomatisytgoperon. The values shown are the means of three independent experiments, and the error bars represent standard deviations.
DISCUSSION
In this study, we provide direct functional evidence that E. coliNrdR is a transcriptional repressor and that CT406 is the ortholog of NrdR in Chlamydia. We showed that rCT406 binds upstream of the nrdABoperon, which encodes the only copy of ribonucleotide reductase in the genomes of C. trachomatisand other Chlamydiaspp. (26, 33). Our data suggest that the tandem NrdR boxes upstream of C. trachomatisnrdABmake up the functional operator since rCT406 required both boxes for binding in vitro. This result differs from what is seen for E. coli, in which only the downstream NrdR box of nrdABwas necessary for binding by NrdR (41). In an in vitrotranscription assay, rCT406 repressed transcription in a manner that was dependent on the NrdR boxes. We validated these results with an in vivoluciferase reporter assay in which both CT406 and E. coliNrdR repressed the E. colinrdHpromoter, and to a lesser extent the C. trachomatisnrdABpromoter. We also showed that NrdR-dependent repression of nrdHwas reversed by hydroxyurea treatment, suggesting that NrdR and CT406 regulate transcription in response to intracellular nucleotide levels.
The modest repression of the C. trachomatisnrdABpromoter by CT406 and E. coliNrdR is consistent with prior studies that showed that NrdR does not repress all its target promoters to the same degree (10, 41). These promoter-specific effects have been proposed to be due to either the location of the operator relative to the promoter or the operator sequence (41). While we found that there was less repression from the C. trachomatisnrdABpromoter than the E. colinrdHpromoter, the NrdR boxes of either promoter were sufficient to produce a high level of repression by CT406 in the context of the dnaKpromoter (Fig. 4B). This result indicates that the difference in the two operators is not due to the operator sequence, per se. Instead, there is a correlation between the greater repression of the E. colinrdHpromoter, which has two NrdR boxes that overlap the promoter elements and the lower repression of the nrdABpromoters from C. trachomatisand E. coli, which are overlapped by only one of their two NrdR boxes (Fig. 1B) (10, 41). This location of the NrdR boxes relative to the nrdABpromoters may lead to less steric hindrance of RNA polymerase and consequently less transcriptional repression (41).
There has been speculation that the activity of NrdR is regulated by a nucleotide cofactor. There are many examples where the activity of a transcription factor that controls the expression of a metabolic enzyme is regulated by a cofactor. For instance, the chlamydial aporepressor TrpR requires tryptophan as a corepressor in order to bind its cognate operator and repress transcription of the tryptophan biosynthetic genes (1, 4). While both E. coliNrdR and CT406 have the potential to bind ATP and dATP via the conserved ATP cone domain (10, 11), a role for a nucleotide cofactor has not been demonstrated with functional evidence. For example, the addition of nucleotide triphosphates (NTPs) or dNTPs did not affect the binding of E. coliNrdR to its operator in EMSA experiments (41). In our in vitrostudies, CT406 operator binding was similarly insensitive to NTPs and dNTPs (data not shown), and CT406 was sufficient to repress transcription in vitrowithout the addition of exogenous dNTPs. Recombinant NrdR purified from bacteria, however, has been shown to contain ATP and dATP (10, 41), which may explain why a nucleotide cofactor requirement has not been demonstrated with in vitrostudies. We were unable to directly assess whether our CT406 preparation contains bound nucleotides because we were unable to purify large enough quantities of this recombinant protein.
Our in vivofinding that hydroxyurea treatment reversed repression by both E. coliNrdR and CT406 provides evidence that the activities of these repressors can be modulated by intracellular nucleotide levels. Hydroxyurea decreases intracellular dNTP pools by inhibiting the activity of class I ribonucleotide reductases (31). We propose that deoxyribonucleotide is required as a corepressor for NrdR-mediated repression and that its depletion by hydroxyurea treatment caused the observed derepression. In contrast, it is unlikely that ribonucleotide acts as an inducer of ribonucleotide reductase genes, because CT406 was able to repress even when a molar excess of ribonucleotides was present for the transcription reaction (Fig. 4B). A deoxyribonucleotide corepressor would allow NrdR to sense high intracellular dNTP levels and respond by repressing ribonucleotide reductase expression. This corepressor model would also provide a mechanism to explain the observation that hydroxyurea treatment induces the transcriptional expression of ribonucleotide reductase (8, 15, 16, 32, 42).
Work with other bacteria has shown that the ribonucleotide reductase genes can be transcriptionally regulated by additional factors, but we do not know if these mechanisms are operative in Chlamydia. In E. coli, nrdABtranscription is also controlled by DnaA, Fis, and IciA (9, 12, 14). Chlamydiadoes not encode obvious orthologs of Fis and IciA, but two DnaA genes have been predicted in the C. trachomatisgenome (33). DnaA is best known as a DNA replication initiation factor, but when it is bound to ATP, it represses the transcription of nrdABin E. coliby binding to an upstream operator (9, 20). Interestingly, we have identified an A/T-rich sequence immediately upstream of the C. trachomatisnrdAB−35 element (Fig. 1A), which resembles a DnaA box for binding by DnaA (28).
In summary, this study provides evidence that CT406 is an ortholog of NrdR and that its regulatory target is the nrdABribonucleotide reductase operon. CT406 is likely to be a key regulator in Chlamydia, since ribonucleotide reductase is the only means that chlamydiae have to produce deoxyribonucleotides by conversion from imported ribonucleotides (38). Furthermore, by regulating this critical conversion step, chlamydial NrdR has the potential to control the balance between ribonucleotides for transcription and energy production, and deoxyribonucleotides for DNA replication.
ACKNOWLEDGMENTS
We thank Aishwarya Ramaswamy and Kirsty Salmon for technical support in constructing the E. coliNrdR deletion strain, and Christopher Rosario, Allan Chen, Eric Cheng, Kirsten Johnson, and Jennifer Lee for critical reading of the manuscript and helpful advice. We also thank Mikhail Gelfand for his help in initiating the project.
This work was supported by a grant from the NIH(AI 44198). M.T. was supported by an NIH Independent Scientist Award(AI 057563).
Footnotes
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Akers J. C., Tan M. 2006. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J. Bacteriol. 188:4236–4243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coliK-12. Science 277:1453–1474 [DOI] [PubMed] [Google Scholar]
- 3. Borovok I., et al. 2004. Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol. Microbiol. 54:1022–1035 [DOI] [PubMed] [Google Scholar]
- 4. Carlson J. H., Wood H., Roshick C., Caldwell H. D., McClarty G. 2006. In vivo and in vitro studies of Chlamydia trachomatis TrpR:DNA interactions. Mol. Microbiol. 59:1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. CDC 2009. Sexually transmitted disease surveillance, 2008. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Washington, DC [Google Scholar]
- 6. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Engel J. N., Ganem D. 1987. Chlamydial rRNA operons: gene organization and identification of putative tandem promoters. J. Bacteriol. 169:5678–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Filpula D., Fuchs J. A. 1977. Regulation of ribonucleoside diphosphate reductase synthesis in Escherichia coli: increased enzyme synthesis as a result of inhibition of DNA synthesis. J. Bacteriol. 130:107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gon S., et al. 2006. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 25:1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grinberg I., et al. 2006. The StreptomycesNrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons. J. Bacteriol. 188:7635–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grinberg I., et al. 2009. Functional analysis of the Streptomyces coelicolorNrdR ATP-cone domain: role in nucleotide binding, oligomerization, and DNA interactions. J. Bacteriol. 191:1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han J. S., Kwon H. S., Yim J. B., Hwang D. S. 1998. Effect of IciA protein on the expression of the nrd gene encoding ribonucleoside diphosphate reductase in E. coli. Mol. Gen. Genet. 259:610–614 [DOI] [PubMed] [Google Scholar]
- 13. Hatfield G. W., Roth D. A. 2007. Optimizing scaleup yield for protein production: Computationally Optimized DNA Assembly (CODA) and Translation Engineering. Biotechnol. Annu. Rev. 13:27–42 [DOI] [PubMed] [Google Scholar]
- 14. Jacobson B. A., Fuchs J. A. 1998. Multiple cis-acting sites positively regulate Escherichia coli nrd expression. Mol. Microbiol. 28:1315–1322 [DOI] [PubMed] [Google Scholar]
- 15. Jordan A., Aragall E., Gibert I., Barbe J. 1996. Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Mol. Microbiol. 19:777–790 [DOI] [PubMed] [Google Scholar]
- 16. Jordan A., Gibert I., Barbe J. 1995. Two different operons for the same function: comparison of the Salmonella typhimurium nrdAB and nrdEF genes. Gene 167:75–79 [DOI] [PubMed] [Google Scholar]
- 17. Kolberg M., Strand K. R., Graff P., Andersson K. K. 2004. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta 1699:1–34 [DOI] [PubMed] [Google Scholar]
- 18. McClarty G., Tipples G. 1991. In situ studies on incorporation of nucleic acid precursors into Chlamydia trachomatisDNA. J. Bacteriol. 173:4922–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mowa M. B., Warner D. F., Kaplan G., Kana B. D., Mizrahi V. 2009. Function and regulation of class I ribonucleotide reductase-encoding genes in mycobacteria. J. Bacteriol. 191:985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Olliver A., Saggioro C., Herrick J., Sclavi B. 2010. DnaA-ATP acts as a molecular switch to control levels of ribonucleotide reductase expression in Escherichia coli. Mol. Microbiol. 76:1555–1571 [DOI] [PubMed] [Google Scholar]
- 21. Panosa A., Roca I., Gibert I. 2010. Ribonucleotide reductases of Salmonella typhimurium: transcriptional regulation and differential role in pathogenesis. PLoS One 5:e11328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Read T. D., et al. 2000. Genome sequences of Chlamydia trachomatisMoPn and Chlamydia pneumoniaeAR39. Nucleic Acids Res. 28:1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Resnikoff S., et al. 2004. Global data on visual impairment in the year 2002. Bull. World Health Organ. 82:844–851 [PMC free article] [PubMed] [Google Scholar]
- 24. Rodionov D. A., Gelfand M. S. 2005. Identification of a bacterial regulatory system for ribonucleotide reductases by phylogenetic profiling. Trends Genet. 21:385–389 [DOI] [PubMed] [Google Scholar]
- 25. Ronald S. L., Kropinski A. M., Farinha M. A. 1990. Construction of broad-host-range vectors for the selection of divergent promoters. Gene 90:145–148 [DOI] [PubMed] [Google Scholar]
- 26. Roshick C., Iliffe-Lee E. R., McClarty G. 2000. Cloning and characterization of ribonucleotide reductase from Chlamydia trachomatis. J. Biol. Chem. 275:38111–38119 [DOI] [PubMed] [Google Scholar]
- 27. Saka H. A., Valdivia R. H. 2010. Acquisition of nutrients by Chlamydiae: unique challenges of living in an intracellular compartment. Curr. Opin. Microbiol. 13:4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schaper S., Messer W. 1995. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 270:17622–17626 [DOI] [PubMed] [Google Scholar]
- 29. Schaumburg C. S., Tan M. 2003. Mutational analysis of the Chlamydia trachomatisdnaKpromoter defines the optimal −35 promoter element. Nucleic Acids Res. 31:551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schaumburg C. S., Tan M. 2000. A positive cis-acting DNA element is required for high level transcription in Chlamydia. J. Bacteriol. 182:5167–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sinha N. K., Snustad D. P. 1972. Mechanism of inhibition of DNA synthesis in Escherichia coliby hydroxyurea. J. Bacteriol. 112:1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sitjes J., Ysern P., Barbe J., Llagostera M. 1992. Induction of ribonucleoside diphosphate reductase gene transcription by chemicals in Escherichia coli. Mutagenesis 7:47–49 [DOI] [PubMed] [Google Scholar]
- 33. Stephens R. S., et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759 [DOI] [PubMed] [Google Scholar]
- 34. Tan M. 2006. Regulation of gene expression. In Bavoil P., Wyrick P.(ed.), Chlamydia: genomics, pathogenesis and implications for control. Horizon Bioscience, Wymondham, United Kingdom [Google Scholar]
- 35. Tan M., Engel J. N. 1996. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatisrRNA P1 promoter. J. Bacteriol. 178:6975–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tan M., Gaal T., Gourse R. L., Engel J. N. 1998. Mutational analysis of the Chlamydia trachomatisrRNA P1 promoter defines four regions important for transcription in vitro. J. Bacteriol. 180:2359–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomason L. C., Costantino N., Court D. L. 2007. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 1:1.17. [DOI] [PubMed] [Google Scholar]
- 38. Tipples G., McClarty G. 1991. Isolation and initial characterization of a series of Chlamydia trachomatisisolates selected for hydroxyurea resistance by a stepwise procedure. J. Bacteriol. 173:4932–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tipples G., McClarty G. 1993. The obligate intracellular bacterium Chlamydia trachomatis is auxotrophic for three of the four ribonucleoside triphosphates. Mol. Microbiol. 8:1105–1114 [DOI] [PubMed] [Google Scholar]
- 40. Tjaden J., et al. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 181:1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Torrents E., et al. 2007. NrdR controls differential expression of the Escherichia coliribonucleotide reductase genes. J. Bacteriol. 189:5012–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Torrents E., Roca I., Gibert I. 2003. Corynebacterium ammoniagenes class Ib ribonucleotide reductase: transcriptional regulation of an atypical genomic organization in the nrd cluster. Microbiology 149:1011–1020 [DOI] [PubMed] [Google Scholar]
- 43. Wilson A. C., Tan M. 2002. Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J. Bacteriol. 184:6566–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]