Abstract
Crenarchaea, such as Sulfolobus acidocaldariusand Sulfolobus tokodaii, produce antimicrobial proteins called sulfolobicins. These antimicrobial proteins inhibit the growth of closely related species. Here we report the identification of the sulfolobicin-encoding genes in S. acidocaldarius. The active sulfolobicin comprises two proteins that are equipped with a classical signal sequence. These proteins are secreted by the cells and found to be membrane vesicle associated. Gene inactivation studies demonstrate that both proteins are required for the bacteriostatic antimicrobial activity. Sulfolobicins constitute a novel class of antimicrobial proteins without detectable homology to any other protein.
INTRODUCTION
A large variety of ribosomally synthesized polypeptides that inhibit the growth of microorganisms have been described. Many of these polypeptides are cationic amphiphilic peptides comprising ≤50 amino acids, and they are also called bacteriocins (11). Some peptides are posttranslationally modified, such as the lanthionine-containing nisin produced by Lactococcus lactis(26). Bacteriocins are a highly diverse group of molecules that also include large proteins that are active against bacteria that are closely related to the producer strain. These peptides are usually secreted by specific mechanisms, i.e., either by the general secretion (Sec) system or by ABC-type exporter proteins. Some bacteriocins are released through cell lysis. The genes involved in bacteriocin production, secretion, and immunity are often plasmid encoded and/or contained in an operon. Under laboratory and natural conditions, bacteriocins have been shown to offer a competitive growth advantage to the producer strain (21). Some groups of bacteriocins, such as colicins, are highly polymorphic proteins, whereas bacteriocins produced by Gram-positive lactic acid bacteria form a much more homogeneous group of smaller cationic peptides (6).
Next to bacteria and eukaryotes, archaea constitute the third domain of life. Culture-independent surveys have shown them to be present in virtually every natural habitat (5). Archaea produce so-called archaeocins, but until now only a few of these antimicrobial (poly)peptides have been described (14, 16, 17). Halocins are archaeocins that are produced by halophilic euryarchaea (13). Some halocins, such as halocin H4, have a rather narrow activity spectrum, whereas halocin A4, for instance, is a broad-spectrum antimicrobial agent that is also active against thermoacidophilic crenarchaea (10, 16). Halocins comprise a wide range of polypeptides with molecular masses from about 4 kDa up to 35 kDa. Some originate from larger precursor proteins that are processed to maturate to the active polypeptide. For example, halocin C8 is produced as a 283-amino-acid-long precursor protein by Halobacteriumthat after processing results in an N-terminal 207-amino-acid-long immunity protein HalI and a C-terminus-derived antimicrobial halocin of 76 amino acid residues (24). Archaeocins appear to be inactive against bacteria. Conversely, the bacteriocin nisin is effective against certain archaea (16). Currently, there is insufficient data to determine whether the archaeocins are evolutionarily related to bacteriocins (21).
Members of the genus Sulfolobusgrow at temperatures between 65 and 85°C and pH values ranging from 2 to 4. The genomes of four Sulfolobusspecies, i.e., Sulfolobus solfataricus, Sulfolobus acidocaldarius, Sulfolobus tokodaii, and Sulfolobus islandicus, have been sequenced, and these four species are the most intensively studied thermoacidophilic crenarchaea (3, 12, 20, 23). Some strains of S. islandicusproduce an antimicrobial protein with a molecular mass of about 20 kDa. This protein was named sulfolobicin, and its activity was found to be associated with the cytoplasmic membrane and with small membrane vesicles found in the medium (17). The identity of sulfolobicin has, however, remained obscure. Here we report on the identification of the sulfolobicin-encoding genes in Sulfolobus acidocaldarius. Our data suggest that the active entity comprises two proteins that are both needed for antimicrobial activity against closely related Sulfolobusspecies.
MATERIALS AND METHODS
Strains and growth conditions.
All Sulfolobusstrains used in this study are listed in Table 1and grown at pH 3.2 in Brock medium (2), supplemented with 0.2% (wt/vol) tryptone and when necessary 0.2% (wt/vol) N-Z-Amine, 20 μg/ml uracil, and 50 mg/ml 5-fluoroorotic acid (5-FOA). S. islandicusstrains HEN2/2 and Rey15A were kindly provided by D. Prangishvili (17) and Q. She (4), respectively. Precultures (50 ml) were grown at 78°C until the late log phase, and then 3 ml of each preculture was transferred into 400 ml of fresh medium. Growth was continued until the stationary phase using 3-liter flasks that were shaken at 160 rpm at 78°C. For the isolation of Sulfolobus tokodaiimembrane vesicles and medium proteins, cultures of 800 ml were grown in 4-liter flasks.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coliDH5α | F−endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupGφ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK−mK+) λ− | Invitrogen |
| Archaeal strains | ||
| S. acidocaldariusMR31 | Wild type, 18 bp deleted in pyrE | 18 |
| S. acidocaldariusMW001 | Wild type, 324 bp deleted in pyrE | M. Wagner and S. V. Albers, unpublished results |
| S. islandicusRey15A | Wild type | 4 |
| S. islandicusHEN2/2 | Wild type | 17 |
| S. solfataricusP2 (DSM1617) | Wild type | DSMZ |
| S. solfataricusP1 (DSM1616) | Wild type | DSMZ |
| S. tokodaiiDSM16993 | Wild type | DSMZ |
| S. acidocaldarius | ||
| ΔsulABmutant | MW001, ΔSaci_1271 Saci_1272, 324 bp deleted in pyrE | This study |
| ΔsulAmutant | MW001, ΔSaci_1271, 324 bp deleted in pyrE | This study |
| ΔsulBmutant | MW001, ΔSaci_1272, 324 bp deleted in pyrE | This study |
| SulAB strain | MW001, Saci_1271/Saci_1272, 324 bp deleted in pyrE | This study |
| Plasmids | ||
| pΔ2pyrEF | pBluescript-based vector with S. solfataricuspyrEFgene | 25 |
| pΔsulAB | pΔ2pyrEF carrying 923 bp of Saci_1272downstream region and 658 bp of Saci_1271upstream region | This study |
| pSVA406 | pGEM-T Easy-based vector containing the S. solfataricuspyrEFgene | Wagner and Albers, unpublished |
| pSVA427 | pSVA406 carrying 688 bp and 727 bp of the down- and upstream regions of Saci_1271, respectively | This study |
| pSVA428 | pSVA406 carrying 607 bp and 658 bp of the down- and upstream regions of Saci_1272, respectively | This study |
| pCMal | pRN1-based shuttle vector with S. solfataricuspyrEFgene | 1 |
| pC1271/72 | pCMal containing 2.1-kb fragment containing 347 and 348 bp of the downstream and upstream regions of Saci_1272and Saci_1271, respectively | This study |
Isolation of secreted proteins and membrane vesicles.
Membrane vesicles and the culture supernatant fraction were obtained as described previously (7). Stationary grown cultures of 800 ml were cooled down on ice for 20 min, followed by a low spin centrifugation (10 min, 12,000 × g, 4°C) to remove intact cells. Subsequently, the membrane vesicles were collected from the supernatant by ultracentrifugation (45 min, 125,000 × g, 4°C). Membrane pellets were resuspended into 1.5-ml portions of residual supernatant and centrifuged for 30 s at 16,000 × gto remove remaining cell debris and aggregates followed by a second ultracentrifugation step (45 min, 376,000 × g, 4°C). The vesicles were resuspended in 50 to 200 μl demineralized water and stored at −20°C. The supernatant of the first ultracentrifugation step (spent medium fraction) was passed through a 0.45-μm filter and concentrated down to 2 to 6 ml in a 200-ml stirred cell using an YM10 ultrafiltration membrane (10-kDa cutoff filter; Amicon).
Antimicrobial overlay assay.
A lawn of Sulfolobusreporter cells in 0.1% (wt/vol) tryptone Brock medium pH 3.0 to 3.5 was spread on gelrite plates containing 0.1% (wt/vol) tryptone and 20 μg/ml uracil as described previously (22). Isolated membrane vesicles (1 to 2 μl), cells, or (concentrated) soluble medium proteins were spotted onto the lawn after it had solidified. The zones of growth inhibition were recorded after 2 days of growth at 78°C.
For the in-gel activity assay, ∼25 μg of membrane vesicle protein was heated to 90°C for 3 min in sample buffer, loaded on a 12% SDS-polyacrylamide gel, and subjected to electrophoresis at room temperature for 1.5 h. The gel was fixed for 10 min in 40% methanol–10% acetic acid after which the gel was stained for 1 h in 0.12% Coomassie brilliant blue G250, 10% phosphoric acid, 10% (wt/vol) ammonium sulfate, and 20% methanol. Destaining was done for 10 min in demineralized water, followed by a 10-min wash in 40% methanol–10% acetic acid. Finally, the gel was washed twice for 10 min with demineralized water. Next, the gels were sliced, and the slices were embedded in a solidifying lawn of Sulfolobus solfataricusstrain P1 reporter cells.
For protease treatment, about 0.5 μl of isolated S. tokodaiimembrane vesicle suspension was added to 19.5 μl of 20 mM Tris (pH 7.0) buffer with or without protease K (25 μg/ml). After 1 h at 37°C, the protease K was heat inactivated for 10 min at 80°C after which 1 μl was spotted onto a lawn of S. solfataricusstrain P1. For a control, 1 μl of the protease K-containing buffer was spotted onto a lawn of S. solfataricusstrain P1.
Liquid culture growth inhibition.
The sulfolobicin-sensitive S. solfataricusP2 strain was grown in 25 ml of Brock medium supplemented with tryptone until the culture reached an optical density at 600 nm (OD600) of 0.09. S. acidocaldariusMW001 and ΔsulAB(control) strains were grown in Brock medium supplemented with tryptone and uracil. In the late log phase (OD600of 0.8), cells were removed by centrifugation (40 min at 10,000 rpm), and the supernatant fractions were added to the S. solfataricusP2 cultures at a ratio of 2:1, 1:1, and 0.6:1. Growth was monitored during the next 4 days. After 24 and 48 h, samples of the cultures were plated in serial dilutions on medium containing tryptone to determine the viability of the S. solfataricuscells.
Construction of a sulAand sulBdisruption strain.
All plasmids used in this study are listed in Table 1. To inactivate the Saci_1271and Saci_1272genes named sulAand sulB, genomic DNA was isolated with a QuickPick SML genomic DNA (gDNA) kit (Bio-Nobile, Turku, Finland) from the S. acidocaldariusΔpyrEFstrain MR31 (19). A fragment of 923 bp containing a portion of the upstream region and of sulBwas amplified using the forward primer ForSulB, which contained a SacII restriction site, and the reverse primer RevSulB, which contained a PstI restriction site (all primer sequences are shown in Table S1 in the supplemental material). A 658-bp fragment containing a part of the downstream region and of sulAwas amplified using the forward primer ForSulA, which contains a KpnI restriction site, and a reverse primer RevSulA, which contains an XhoI restriction site. The amplified up- and downstream regions were cloned in the pΔ2pyrEF vector (25). S. acidocaldariusstrain MW001 (M. Wagner and S. V. Albers, unpublished results) was transformed with the resulting plasmid, and the sulABdisruption mutant was isolated as described previously (8).
The individual sulAand sulBdeletion mutants were isolated by means of a similar procedure (see Fig. S1 in the supplemental material). The upstream and downstream flanking regions of sulAwere amplified using primers 997 and 998 and primers 999 and 1051. The sites for restriction enzymes BamHI and NcoI were incorporated into primers 997 and 1051, respectively. These two fragments were used as templates by overlapping PCR using primers 997 and 1051. This resulted in a 1,418-bp product that was digested by BamHI/NcoI and ligated into vector pSVA406, yielding plasmid pSVA427. pSVA406 is a pGEM-T Easy-based vector containing the S. solfataricus pyrEFcassette for selection in S. acidocaldarius(Wagner and Albers, unpublished). Likewise, the up- and downstream flanking regions of sulBwere obtained by PCR using primer pair 993 and 994 and primer pair 995 and 996. The BamHI and NcoI restriction sites were incorporated into primers 993 and 996, respectively. Overlap PCR using these two fragments as templates was performed using primers 993 and 996, resulting in a 1,260-bp product that was digested by BamHI/NcoI and ligated into vector pSVA406, yielding plasmid pSVA428. All constructs were confirmed by sequencing. pSVA427 and pSVA428 were transformed into S. acidocaldariusstrain MW001, yielding the individual ΔsulAandΔsulBmutants, respectively. The individual strains were checked by PCR for the presence of the sulAand sulBgenes (see Fig. S1B and S1C in the supplemental material).
Gene complementation.
A fragment of 2,182 kb containing 347 and 348 bp of the downstream and upstream regions of sulABwere amplified from S. acidocaldariusgenomic DNA using the 1271for and 1272rev primers with the EagI and NcoI restriction sites, respectively (see Table S1 in the supplemental material). The amplified fragment was cloned into vector pCMal (1) using the restriction enzymes EagI and NcoI, yielding pC1271/72. Transformation of pC1271/1772 was performed as described previously (8).
qPCR analysis.
The expression of sulAand sulBwas analyzed by real-time quantitative PCR (qPCR). Total RNA of the indicated strains was isolated from cells grown in Brock medium supplemented with tryptone in the presence or absence of uracil for the times indicated in the figure legends. Total RNA isolation was with TRIzol (Invitrogen), and additional DNase treatment using the Turbo DNA-free kit (Ambion). The concentration of total RNA was measured with a NanoDrop ND-1000. The primer sets used are listed in Table S2 in the supplemental material. All primers were designed so that fragments of 300 bp were synthesized. cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad) in a volume of 10 μl. A negative reverse transcriptase (RT) control was used to determine the gDNA contamination in isolated total RNA. The expression levels were analyzed in three replicate samples with a MiniOpticon system (Bio-Rad) using the Bio-Rad CFX manager software, with which the threshold cycle (CT) values were determined automatically by regression. The SensiMix SYBR mix (Bioline) was used as a master mix for qPCRs using primers at 0.4 μM each. The following thermocycler conditions were used: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 57°C for 30 s, and 72°C for 30 s. Subsequently, a melting curve was generated to determine the specificity of the qPCRs.
In-gel trypsin digestion and matrix-assisted laser desorption ionization–tandem time of flight mass spectrometry (MALDI-TOF/TOF MS).
Gel slices from SDS-polyacrylamide gels were washed with 200 μl water, destained twice with 200 μl of 50% acetonitrile (ACN) in 50 mM NH4HCO3, and subsequently dehydrated in 100% ACN. Destained gel slices were then incubated for 1 h at 55°C in 100 μl of 50 mM NH4HCO3and 10 mM dithiothreitol (DTT), followed by 30-min incubation with 100 μl of 55 mM iodoacetamide at room temperature. Gel pieces were dehydrated in 100% ACN and air dried. Next, 20-μl portions of 10 ng/μl sequencing-grade trypsin (Promega, Leiden, The Netherlands) or porcine elastase (USB Corporation, Cleveland, OH) in 40 mM NH4HCO3were added to the dried gel pieces and incubated overnight at 37°C. The overlay solution was collected, and peptides were subsequently extracted with 20 μl of 1% trifluoroacetic acid (TFA), followed by extractions with 20 μl of 50% ACN in 0.5% TFA and 20 μl of 100% ACN. Fractions of the extracted peptides were pooled, vacuum dried, and dissolved in 20 μl of 0.1% TFA.
For nano-liquid chromatography (nLC) coupled to MALDI MS (nLC-MALDI MS), peptide mixtures were separated on a C18capillary column (C18PepMap 300 column; 75 μm by 150 mm; 3-μm particle size; LC-Packing) mounted on an Ultimate 3000 nanoflow liquid chromatography system (LC-Packing, Amsterdam, The Netherlands). Solutions of 0.05% TFA and 80% ACN in 0.05% TFA were used as mobile phase A and B, respectively. A gradient from 4 to 40% mobile phase B was used for 60 min with a flow rate of 300 nl·min−1. Column effluent was mixed 1:4 (vol/vol) with a matrix solution containing 2.3 mg/ml α-cyano-4-hydroxycinnamic acid (LaserBio Labs, Sophia-Antipolis, France), whereupon 12-second fractions were spotted onto a blank MALDI target with a Probot system (LC Packings, Amsterdam, The Netherlands). Mass spectrometry analysis was carried out with a MALDI-TOF/TOF 4800 proteomics analyzer (Applied Biosystems, Foster City, CA) in the m/zrange from 800 to 4,000. Data acquisition was performed in positive ion mode. Peptides with a signal-to-noise (S/N) level above 80 were selected for tandem MS (MS/MS) fragmentation. Peak lists of the acquired MS/MS spectra were generated, using default settings and an S/Nthreshold of 10.
Protein identification was carried out using the software ProteinPilot 2.0 (Applied Biosystems, Foster City, CA). Searches against the Sulfolobussequence database, combined with reversed entries for all protein sequences were performed. Protein identification was based on at least 2 peptides of 8 amino acids or longer and independently identified with a probability higher than 95%.
RESULTS
Antimicrobial activity of Sulfolobusspecies.
To test the ability of Sulfolobusspecies to produce compounds that inhibit the growth of closely related species as reported by Prangishvili et al. (17), four different species, i.e., S. solfataricusP1 and P2, S. acidocaldarius, S. tokodaii, and S. islandicusHEN2/2 were spread on gelrite plates to create an even lawn of growth. On these lawns, cells of different Sulfolobusspecies were spotted. S. acidocaldariusand S. islandicusproduce a clear zone of growth inhibition around their colonies when spotted onto lawns of S. solfataricusstrains P1 and P2, but no inhibition was observed against S. tokodaii(Table 2). S. solfataricusstrains P2 and P1 showed no significant activity against any of the reporter cells tested. S. tokodaiiformed no colonies except on its own lawn, which suggests that all other strains in the lawn produce a strong activity against S. tokodaiiwhich precluded the definition of its antimicrobial activities in this cell-based assay.
Table 2.
Sulfolobicin activity and susceptibility of various Sulfolobusspeciesa
| Fraction and species | Antimicrobial activitybon the following reporter cells: |
||||
|---|---|---|---|---|---|
| S. solfataricusP1 | S. solfataricusP2 | S. acidocaldarius | S. tokodaii | S. islandicus | |
| Cells | |||||
| S. acidocaldarius | + | + | − | − | − |
| S. tokodaii | −† | −† | − | − | − |
| S. islandicus | + | + | − | − | − |
| Cleared medium supernatant | |||||
| S. acidocaldarius | + | + | − | − | − |
| S. tokodaii | + | + | − | − | − |
| S. islandicus | − | − | − | − | − |
| Membrane vesicles | |||||
| S. acidocaldarius | + | +/− | − | − | − |
| S. tokodaii | + | + | − | − | +/− |
| S. islandicus | + | +/− | − | − | − |
The strains used were S. acidocaldariusstrain DSM639, S. tokodaiistrain DSM16993, S. solfataricusP2 strain DSM1617, and P1 strain DSM1616; and S. islandicusstrain HEN2/2.
Symbols: +, clear halo formation; +/−, weak halo formation; −, no effect; †, the producer strain did not grow.
Next, liquid cultures were analyzed for the presence of a growth-inhibiting compound. To this end, the various Sulfolobusspecies were grown in liquid culture until stationary phase. Spent medium was subjected to ultracentrifugation to yield a membrane vesicle fraction and a soluble protein fraction. Small aliquots of the membrane vesicle fraction were spotted onto lawns of different Sulfolobusspecies and examined for growth inhibition. The S. islandicusmembrane vesicle fraction was active against S. solfataricusstrain P1 but only weakly active against strain P2, while no activity was observed against the other Sulfolobusspecies, confirming a previous report (17). Interestingly, the S. tokodaiimembrane vesicle fraction was active against S. solfataricusstrains P1 and P2 and S. islandicusbut not against S. acidocaldarius. The S. acidocaldariusmembrane vesicles were active against S. solfataricusP1. Finally, S. solfataricusP1 and P2 membrane vesicle showed no activity against any of the other Sulfolobusspecies tested.
Treatment of the S. tokodaiimembrane vesicle fraction with proteinase K resulted in an almost complete loss of the activity against S. solfataricusstrain P1. On the other hand, in the presence of trypsin, the activity against S. solfataricusstrain P1 was retained, which suggests that a trypsin-resistant protein(s) causes the growth inhibition. The S. tokodaiivesicles did not inhibit growth when spotted onto a lawn of Escherichia colior Bacillus subtilis(data not shown). The membrane vesicle-cleared soluble medium protein fraction of S. tokodaii(and S. acidocaldarius) also produced a growth inhibition zone on lawns of S. solfataricusstrains P1 and P2. When the S. tokodaiimedium fraction was concentrated up to ∼60-fold by ultrafiltration using a 10-kDa-cutoff filter, the activity was retained in the filtrate, suggesting that the active component has a molecular mass of >10 kDa. Moreover, semiquantitative comparison of the growth inhibition halos suggests that the majority of the activity in S. tokodaiiis membrane vesicle associated (more than 80%). These data demonstrate that membrane vesicles of S. tokodaiiand S. acidocaldariusare equipped with antimicrobial activity against closely related species.
Identification of the sulfolobicin proteins.
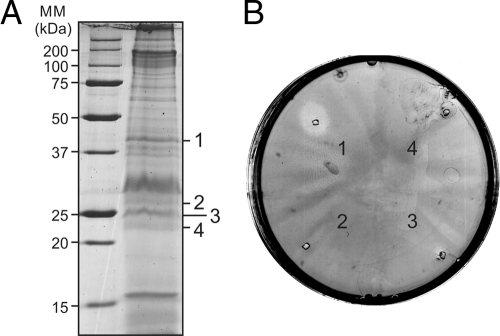
The S. islandicussulfolobicin retains its activity even under the harsh denaturing conditions of SDS-PAGE (17). To determine whether the S. tokodaiisulfolobicin behaves in a similar manner, membrane vesicles were subjected to SDS-PAGE, and the lanes were sliced in thin pieces that were tested for growth inhibition on a lawn of S. solfataricusP1. Strong growth-inhibiting activity was found in a slice corresponding to proteins with an apparent mass of about 42 kDa (Fig. 1). Thinner slices were generated, and the slice with the highest activity was subjected to in-gel digestion with trypsin and subsequently LC-MS. This led to the identification of 7 proteins (see Tables S3 and S4 in the supplemental material). When elastase was used for cleavage, an additional 8 proteins were identified.
Fig. 1.
Antimicrobial activity of proteins isolated from membrane vesicles derived from Sulfolobus tokodaiicells. (A) S. tokodaiivesicles were loaded on an SDS-polyacrylamide gel, and proteins were separated. Protein bands were excised from the gel. The protein bands were numbered 1 to 4 and are shown to the right of the gel. The positions of molecular mass markers (MM) (in kilodaltons) are shown to the left of the gel. (B) The protein bands excised from the gel were loaded on plates containing a lawn of Sulfolobus solfataricusstrain P1 reporter cells. The active slice (band 1) was analyzed by LC-MS.
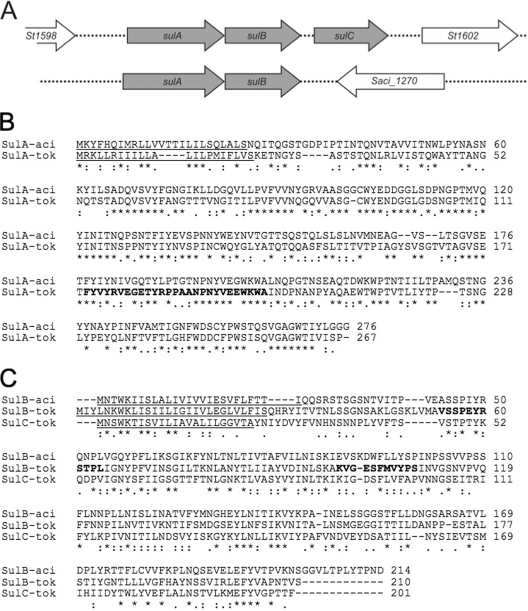
In order to select possible candidates from the above mass spectrometry data for gene inactivation analysis, we assumed that the putative sulfolobicin must be a protein(s) with unknown function containing a signal sequence for secretion and be present in both S. tokodaiiand S. acidocaldarius. The most likely candidates were the hypothetical protein St1599/St1600, St1072, and St1616 that are all predicted to contain an N-terminal signal sequence by PRED-SIGNAL (see Table S3 in the supplemental material). Since the antimicrobial activity was insensitive to trypsin, we also rationalized that the proteins was not identified by mass spectrometry in the trypsin-treated samples. Therefore, the St1599/St1600 and St1616 proteins appear to be the most likely candidates. The latter protein is relatively large, 417 amino acids, with homologs also present in the other Sulfolobusspecies. On the other hand, the St1599and St1600genes colocalize in the genome of S. tokodaiiand encode proteins of 276 and 210 amino acids, respectively, with molecular masses of 29,418 and 22,764 Da, respectively (Fig. 2A). S. tokodaiicontains a homologue of St1600, i.e., St1601, which is located directly adjacent to St1600in the genome (Fig. 2A). Importantly, the S. acidocaldariusSaci_1272 and Saci_1271 proteins are highly homologous to St1600 and St1599, respectively (Fig. 2B and C). We could not identify homologs in the publically available genome sequences of different S. islandicusstrains, except for S. islandicusREY15A that contains an open reading frame (ORF) (SiRe_1467) that encodes a 176-amino-acid-long polypeptide highly homologous to the St1599 and Saci_1271 proteins. This ORF appears to be a C-terminal truncated version of St1599, while a homolog of St1600 or Saci_1272 protein is missing. In this respect, S. islandicusREY15A showed no antimicrobial activity (data not shown). No further homologs were found in the protein database, suggesting that these are unique secretory proteins of S. tokodaiiand S. acidocaldarius.
Fig. 2.
Genomic organization and amino acid sequences of the sulfolobicin genes. (A) Genomic organization of the sulfolobicin genes of Sulfolobus tokodaii(top line) and Sulfolobus acidocaldarius(bottom line). The S. tokodaii sulCgene appears to be a duplication of sulBbut is absent from the S. acidocaldariusgenome. (B and C) ClustalW2 alignment of SulA (B) and SulBC (C) proteins from S. acidocaldariusDSM 639 (-aci) and S. tokodaiistrain 7 (-tok). Predicted signal peptides are underlined, and peptides identified by LC-MS are depicted in bold type. Gaps introduced to maximize alignment are indicated by dashes. Amino acids that are identical in the different sequences are indicated by an asterisk below the sequence alignment. For S. tokodaii, the sulAgene is also called St1599, the sulBgene is also called St1600, and the sulCgene is also called St1601. For S. acidocaldarius, the sulAgene is also called Saci_1271, and the sulBgene is also called Saci_1272.
Inactivation of the S. acidocaldariussulfolobicin genes.
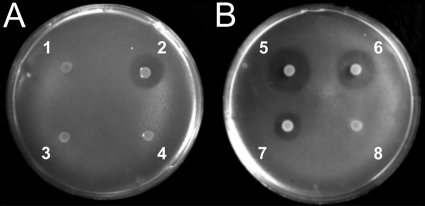
Since there is an effective gene disruption method for S. acidocaldarius, we focused on the inactivation of the Saci_1271and Saci_1272genes that we call sulAand sulB, respectively, in the remainder of this article. In the created deletion mutant, the entire sequences coding for the Saci_1272 and Saci_1271 proteins were removed. A colony of the mutant with the double-disruption ΔsulABtogether with the parental strain S. acidocaldariusMW001 was spotted onto a lawn of S. solfataricusP2 reporter cells. The halo of growth inhibition that is visible around the colony of S. acidocaldariusMW001 completely disappeared in the disruption mutant (Fig. 3A). To test whether the disruption mutant had become sensitive to sulfolobicin, S. acidocaldariusMW001 cells were spotted onto a lawn of the disruption mutant, but no halo was observed. To verify that the loss in antimicrobial activity is indeed due to the loss of the sulABgenes, the disruption mutant was transformed with a vector containing the sulABgenes for complementation. The isolated transformants were spotted onto a plate with a lawn of S. solfataricusP2. All transformants showed the restoration of the halos with sizes slightly larger than those of the parental strain S. acidocaldariusMW001 (Fig. 3B). Quantitative PCR (qPCR) analysis showed a higher transcriptional level of sulAand sulBin the complemented strain compared to S. acidocaldariusMW001 (see Fig. S2A in the supplemental material). Summarizing, these data suggest that the sulABgenes encode the sulfolobicin active antimicrobial compound.
Fig. 3.
Overlay assay with colonies of S. acidocaldariusMW001 grown on a lawn of S. solfataricusstrain P2. The S. acidocaldariusMW001 strains used are indicated by the following numbers on the plates: 1, ΔsulABmutant; 2, wild type; 3, ΔsulBmutant; 4, ΔsulAmutant; 5 and 6, ΔsulABmutant complemented with sulAB[MW001 (sulAB)]; 7, wild type; and 8, ΔsulABmutant.
Sulfolobicin activity requires both sulAand sulB.
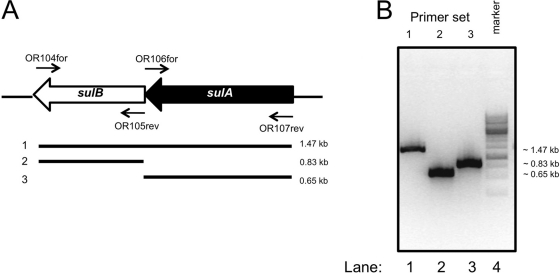
Next, we addressed the question whether sulAand sulBare both required for antimicrobial activity or whether the individual genes are sufficient. The sulAand sulBgenes are arranged in an operon. They are cotranscribed as a single transcript as demonstrated by PCR analysis using the cDNA derived from S. acidocaldariusMW001 strain as a template (Fig. 4). This suggests that these proteins also function together. To test this hypothesis further, single-deletion strains of sulAand sulBof S. acidocaldariusMW001 were isolated. In the individual deletion strains, the loss of the respective sulBand sulAgenes was demonstrated by PCR (see Fig. S1B and S1C in the supplemental material). Deletion of sulBresulted in a reduction of the expression of sulAas examined by qPCR, whereas deletion of the more distal gene of the transcript, sulB, had no effect on the expression of sulA(see Fig. S2B in the supplemental material). The polar effect is likely due to a reduction in mRNA stability, but the expression levels are still sufficiently high to allow for the detection of activity. The single-deletion mutants were spotted onto plates with a lawn of S. solfataricusP2. With both deletion mutants, a complete loss of antimicrobial activity was observed, which suggests that sulAand sulBare both required for antimicrobial activity.
Fig. 4.
The sulAand sulBgenes are coexpressed from a single transcript. (A) Positions of the primers used to detect cDNA fragments and the expected PCR fragments. (B) cDNA was synthesized from RNA isolated from S. acidocaldariusMW001 cells grown to late exponential phase (OD600of ∼0.7). The indicated primer sets were used to detect the presence of PCR products. When reverse transcriptase was left out of the cDNA synthesis reaction, no PCR products were detected.
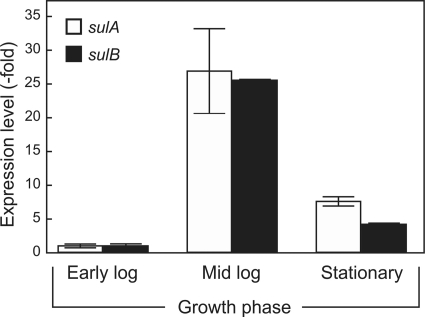
Mixing of the supernatants of the two individual deletion strains did not result in the recovery of antimicrobial activity, which suggests that both proteins need to be expressed together. Possibly, the individual expressed proteins are unstable in the supernatant, and expression of both genes concomitantly is needed to assemble the active sulfolobicin. To determine when the sulABgenes are expressed during growth, RNA was isolated from S. acidocaldariusMW001 at different growth stages, and expression was determined by qPCR. The highest level of expression occurred in the mid-exponential phase and dropped to lower levels in the stationary phase (Fig. 5). Cogrowth of S. acidocaldariustogether with S. solfataricusP2 did not result in an altered expression of the sulAand sulBgenes (data not shown).
Fig. 5.
Expression levels of the sulABgenes from S. acidocaldariusMW001 during the indicated growth stages. Early log, mid log, and stationary growth phase correspond to OD600s of 0.1, 0.5, and 1.5, respectively. Expression was determined by real-time quantitative PCR and normalized relative to the expression of sulAand sulBin the early log phase with the secYgene as a reference.
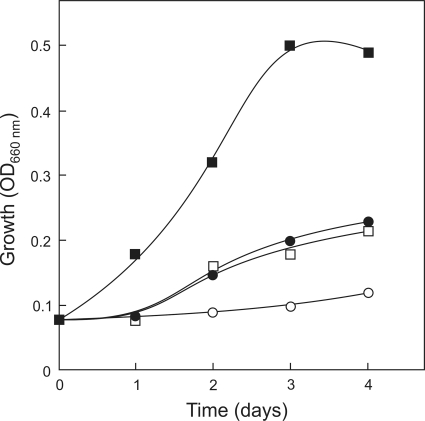
When sulfolobicin-containing spent medium from S. acidocaldariusMR31 was added to a growing culture of S. solfataricusP2, growth was arrested in a dose-dependent manner (Fig. 6) Growth was not inhibited when spent medium of the S. acidocaldariusΔsulABstrain was used instead. Plating of the S. solfataricuscells showed a dramatic loss in the viable cell count when the cells were exposed to spent medium containing sulfolobicin (Table 3). The cells were also stained with a LIVE/DEAD cell viability stain. Remarkably, the addition of sulfolobicin did not result in cell death (Fig. 6) but only in growth arrest. Negative stain electron microscopy investigation of the sulfolobicin-treated S. solfataricuscells did not reveal any morphology changes (data not shown). These data suggest that sulfolobicin is bacteriostatic.
Fig. 6.
Effect of sulfolobicin produced by S. acidocaldariusMW001 on the growth of S. solfataricusP2 in liquid culture. Supernatant from medium in which wild-type S. acidocaldarius(0 to 50 ml) and ΔsulABcells (0 to 50 ml) was grown (spent medium) was mixed with a growing culture of S. solfataricusP2 (25 ml). Next, growth was monitored over time by measuring the OD660. Symbols: ▪, 50 ml of ΔsulABmutant supernatant (control); ○, 50 ml of wild-type strain MW001 supernatant; □, 25 ml (each) of supernatant from the ΔsulABmutant and MW001 strain; •, 15 ml of supernatant from strain MW001 and 35 ml of supernatant from ΔsulABmutant.
Table 3.
Effect of sulfolobicin-containing spent medium on the viability of S. solfataricusP2a
| Spent medium (ml) from the following cells: |
Fresh medium (ml) | Viability of S. solfataricusP2 (CFU/ml) after the following incubation time: |
||
|---|---|---|---|---|
| ΔsulABmutant | Wild-type | 24 h | 48 h | |
| 50 | 0 | 25 | 140 × 105± 10 × 105 | 150 × 105± 10 × 105 |
| 35 | 15 | 25 | 2.3 × 105± 0.3 × 105 | 2.1 × 105± 0.5 × 105 |
| 25 | 25 | 25 | 0.2 × 105± 0.1 × 105 | 0.3 × 105± 0.1 × 105 |
| 0 | 50 | 25 | 0 | 0 |
Spent medium from S. acidocaldariusMW001 wild-type and ΔsulABcells was mixed with fresh Brock medium and incubated for the indicated time with S. solfataricusP2, and then the number of viable cellswas determined as described in Materials and Methods.
DISCUSSION
Here we reported for the first time the identification of the antimicrobial proteins produced by Sulfolobusspecies that for long have been known as sulfolobicins. The presence of sulfolobicin proteins SulA and SulB from S. tokodaiiin the slices of an SDS-polyacrylamide gel of the medium supernatant correlated with a high antimicrobial activity. However, firm evidence that SulA and SulB indeed specify the sulfolobicin activity was provided by the genetic inactivation of the sulABgenes in S. acidocaldarius. The sulAand sulBgenes of S. acidocaldariusare organized in an operon and cotranscribed. A similar operon structure is present in S. tokodaiithat specifies a putative third sulfolobicin gene that encodes a protein that is highly homologous to SulB. These proteins do not share any sequence homology with any other protein known so far and therefore represent an entirely novel class of antimicrobial proteins.
A unique feature of sulfolobicins is their thermostability: these proteins are produced and remain active at 78°C (17), which is one of the highest temperatures reported for any antimicrobial polypeptide so far (15). Sulfolobicins are stable proteins as demonstrated by their resistance to SDS treatment and exposure to low and high pH (3 to 10.7). Furthermore, long-term storage up to 17 months at 4°C, without the addition of preservatives, as well as treatment with trypsin, did not affect antimicrobial activity. Importantly, the active sulfolobicin relates to a pair of proteins that seem to comigrate during SDS-PAGE, as activity could be extracted from slices from the SDS-polyacrylamide gel corresponding to the 42-kDa molecular mass region, whereas the individual masses of SulA and SulB are only 22 kDa. This suggests that SulA and SulB form a stable, likely stoichiometric, complex that resists the electrophoresis conditions.
Halocins of euryarchaea have been described in some detail, and in some cases, the structural and immunity genes have also been identified (16, 24). These halocins are not homologous to sulfolobicin. The production of halocins by halophilic euryarchaea is a common process, although their precise ecological role is elusive (13). The release of antimicrobial polypeptides targeted at related species therefore seems to be a common feature in archaea, although not all Sulfolobusstrains are equipped with this activity as will be discussed below. Interestingly, euryarchaeal halocins are active against S. acidocaldarius, and it has been proposed that the molecular target of halocins is conserved in the euryarchaea and crenarchaea (10). Archaeocins appear not to be active against bacteria. Indeed, the S. tokodaiisulfolobicins were ineffective against E. coliand B. subtilis. At this time, the exact molecular target of sulfolobicins is unknown and the target still need to be purified and its function further analyzed. In this respect, it would also be of interest to examine the relation between the mode of action of sulfolobicins and other antimicrobial proteins.
S. islandicusHEN2/2 used in our study also produced an activity against S. solfataricusP1 and P2. We could, however, not find sulfolobicin homologs in any of the sequenced S. islandicusstrains, but the genome sequence of the strain used in this study has not been determined. S. islandicusREY15A appears to encode a C-terminally truncated version of SulA, but this must be an inactive form, as the strain showed no antimicrobial activity. We noted that the sulAgenes of S. tokodaiiand S. islandicusare situated directly adjacent to genetic mobile elements. In bacteria, many bacteriocins are highly mobile as they are plasmid encoded (21), and thus the possibility exists that this is also the case for archaeocins. It should be stressed that out of 420 tested S. islandicusstrains, only about 10% seemed to produce sulfolobicin activity (17). This implies that this trait is rather rare among the S. islandicusisolates.
For S. islandicus(17) and S. tokodaii(this study), most of the sulfolobicin activity in the medium fraction was found to be associated with membrane vesicles. We noted, however, that the activity could be effectively extracted from these membrane vesicles by alkaline carbonate treatment, suggesting a peripheral association (unpublished data). Perhaps the membrane vesicle association is a mechanism for delivery. Membrane association is not a strict requirement for activity, as the sulfolobicin extracted by alkaline carbonate treatment was also active. In bacteria, membrane vesicle-associated toxins are known to be highly protease resistant, which probably increases the toxin's lifetime (9). We cannot exclude the possibility that other proteins contribute to the sulfolobicin activity, as this will require the overexpression and purification of the proteins. Future studies should also address the mechanism by which sulfolobicins act on closely related species and the mechanisms that provide resistance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Netherlands Proteomics Centre (NPC). S.-V.A received a VIDI grant from the Netherlands Science Organization (NWO)and intramural funds from the Max Planck Society.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Berkner S., Wlodkowski A., Albers S. V., Lipps G. 2010. Inducible and constitutive promoters for genetic systems in Sulfolobus acidocaldarius. Extremophiles 14:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brock T. D., Brock K. M., Belly R. T., Weiss R. L. 1972. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84:54–68 [DOI] [PubMed] [Google Scholar]
- 3. Chen L., et al. 2005. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 187:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Contursi P., et al. 2006. Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles 10:615–627 [DOI] [PubMed] [Google Scholar]
- 5. DeLong E. F., Pace N. R. 2001. Environmental diversity of bacteria and archaea. Syst. Biol. 50:470–478 [PubMed] [Google Scholar]
- 6. Dykes G. A., Hastings J. W. 1997. Selection and fitness in bacteriocin-producing bacteria. Proc. Biol. Sci. 264:683–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ellen A. F., et al. 2009. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 13:67–79 [DOI] [PubMed] [Google Scholar]
- 8. Ellen A. F., Albers S. V., Driessen A. J. 2010. Comparative study of the extracellular proteome of Sulfolobus species reveals limited secretion. Extremophiles 14:87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellis T. N., Leiman S. A., Kuehn M. J. 2010. Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect. Immun. 78:3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haseltine C., et al. 2001. Secreted euryarchaeal microhalocins kill hyperthermophilic crenarchaea. J. Bacteriol. 183:287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hilpert K., Fjell C. D., Cherkasov A. 2008. Short linear cationic antimicrobial peptides: screening, optimizing, and prediction. Methods Mol. Biol. 494:127–159 [DOI] [PubMed] [Google Scholar]
- 12. Kawarabayasi Y., et al. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8:123–140 [DOI] [PubMed] [Google Scholar]
- 13. Kis-Papo T., Oren A. 2000. Halocins: are they involved in the competition between halobacteria in saltern ponds? Extremophiles 4:35–41 [DOI] [PubMed] [Google Scholar]
- 14. Li Y., Xiang H., Liu J., Zhou M., Tan H. 2003. Purification and biological characterization of halocin C8, a novel peptide antibiotic from Halobacterium strain AS7092. Extremophiles 7:401–407 [DOI] [PubMed] [Google Scholar]
- 15. Martirani L., Varcamonti M., Naclerio G., De Felice M. 2002. Purification and partial characterization of bacillocin 490, a novel bacteriocin produced by a thermophilic strain of Bacillus licheniformis. Microb. Cell Fact. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Connor E. M., Shand R. F. 2002. Halocins and sulfolobicins: the emerging story of archaeal protein and peptide antibiotics. J. Ind. Microbiol. Biotechnol. 28:23–31 [DOI] [PubMed] [Google Scholar]
- 17. Prangishvili D., et al. 2000. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J. Bacteriol. 182:2985–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reilly M. S., Grogan D. W. 2001. Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J. Bacteriol. 183:2943–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reilly M. S., Grogan D. W. 2002. Biological effects of DNA damage in the hyperthermophilic archaeon Sulfolobus acidocaldarius. FEMS Microbiol. Lett. 208:29–34 [DOI] [PubMed] [Google Scholar]
- 20. Reno M. L., Held N. L., Fields C. J., Burke P. V., Whitaker R. J. 2009. Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl. Acad. Sci. U. S. A. 106:8605–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Riley M. A. 1998. Molecular mechanisms of bacteriocin evolution. Annu. Rev. Genet. 32:255–278 [DOI] [PubMed] [Google Scholar]
- 22. Schleper C., Kubo K., Zillig W. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. U. S. A. 89:7645–7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. She Q., et al. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U. S. A. 98:7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun C., et al. 2005. A single gene directs both production and immunity of halocin C8 in a haloarchaeal strain AS7092. Mol. Microbiol. 57:537–549 [DOI] [PubMed] [Google Scholar]
- 25. Wagner M., et al. 2009. Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem. Soc. Trans. 37:97–101 [DOI] [PubMed] [Google Scholar]
- 26. Willey J. M., A. van der Donk W. 2007. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61:477–501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.