Abstract
Most biological nitrogen (N2) fixation results from the activity of a molybdenum-dependent nitrogenase, a complex iron-sulfur enzyme found associated with a diversity of bacteria and some methanogenic archaea. Azotobacter vinelandii, an obligate aerobe, fixes nitrogen via the oxygen-sensitive Mo nitrogenase but is also able to fix nitrogen through the activities of genetically distinct alternative forms of nitrogenase designated the Vnf and Anf systems when Mo is limiting. The Vnf system appears to replace Mo with V, and the Anf system is thought to contain Fe as the only transition metal within the respective active site metallocofactors. Prior genetic analyses suggest that a number of nif-encoded components are involved in the Vnf and Anf systems. Genome-wide transcription profiling of A. vinelandiicultured under nitrogen-fixing conditions under various metal amendments (e.g., Mo or V) revealed the discrete complement of genes associated with each nitrogenase system and the extent of cross talk between the systems. In addition, changes in transcript levels of genes not directly involved in N2fixation provided insight into the integration of central metabolic processes and the oxygen-sensitive process of N2fixation in this obligate aerobe. The results underscored significant differences between Mo-dependent and Mo-independent diazotrophic growth that highlight the significant advantages of diazotrophic growth in the presence of Mo.
INTRODUCTION
Biological nitrogen fixation, the reduction of dinitrogen (N2) to ammonia, is an essential reaction in the global nitrogen (N) cycle. Biological N2fixation accounts for roughly two-thirds of the fixed N produced on Earth and is catalyzed by the nitrogenase complex (54). Homologs of nitrogenase have been identified in a diversity of bacteria and in several lineages of methanogenic archaea (8, 53). Most present-day biological N2fixation is catalyzed by molybdenum (Mo) nitrogenase (encoded by nifHDK), an oxygen-sensitive metalloenzyme complex composed of the Fe protein (product of nifH) and the MoFe protein (product of nifDK) (9). The Fe protein is a homodimer bridged by an intersubunit [4Fe-4S] cluster that serves as the obligate electron donor to the MoFe protein (23). The MoFe protein is an α2β2heterotetramer that houses the P clusters, [8Fe-7S] clusters that shuttle electrons to the FeMo cofactors (FeMo-co). The FeMo-co are [Mo-7Fe-9S-homocitrate] clusters where substrate reduction occurs (44). Two genetically distinct “alternative” forms of nitrogenase have been identified in a subset of diazotrophs that also encode nif(8, 53, 61). The nitrogenase encoded by the vnfHDKgenes contains vanadium in place of molybdenum in the active-site cofactor, whereas the nitrogenase encoded by the anfHDKgenes appears to contain Fe as the only metal constituent of its active-site cofactor (11, 24). In some diazotrophs, the expression and activity of the alternative forms are regulated by the availability of Mo or V when fixed N is also limiting (34, 35); in contrast, at least one diazotroph synthesizes the alternatives in the absence of fixed N when the Mo-dependent nitrogenase is not functional, regardless of trace metal availability (48). Biochemical analysis indicates that the N2reduction rates for the Mo nitrogenase are much higher than the rates observed for the alternative forms (19). In addition, a larger proportion of the electron flux during nitrogen reduction is directed toward proton reduction by the alternative forms of nitrogenase than toward proton reduction by Mo nitrogenase (20). Although the alternative nitrogenases are genetically distinct, they are mechanistically similar to Mo nitrogenase. Because neither the Vnf nor the Anf system contains a complete suite of genes necessary for the maturation of the corresponding catalytic partners, it is likely that significant cross talk occurs either between or among the different systems during the assembly and regulation of these different enzyme complexes (36, 41).
Azotobacter vinelandiiis an obligately aerobic member of the class Gammaproteobacteriathat is common in terrestrial soils sampled from across the world (42) and is known to produce both alternative forms of nitrogenase in addition to Nif (32, 34, 35). Owing to its genetic tractability, A. vinelandiihas emerged as the principal model organism for exploring the nitrogenase mechanism, the assembly of complex metalloclusters, and the regulatory features that coordinate the differential expression of the large number of genes associated with N2fixation (43). The recently completed A. vinelandiigenome sequence (59), in combination with the results of extensive biochemical and genetic characterization (49, 54, 58), has implicated at least 82 gene products in the formation and regulation of the three forms of nitrogenase (17). In addition to the energetic demands associated with maintaining these 82 genes in A. vinelandii, the nitrogenase reaction is energetically demanding, requiring 16 molecules of ATP and 8 electrons for the reduction of a single molecule of N2. These high metabolic costs, in conjunction with the demands for fixed N in growing cells and the requirement to protect the nitrogenase system from oxidative inactivation, presumably imposed a strong selective pressure to efficiently integrate N2fixation with other cellular metabolic processes.
In the present study, we examined patterns of global gene expression in A. vinelandiiwhen it is cultured under N-replete conditions (fixed-N source is readily available) or when cells were grown under N2-fixing conditions (fixed-N source is limiting) in several experimental iterations that individually favored the expression of the nif, vnf, or anfsystem. Mo-dependent diazotrophy was maintained by growing cells in medium lacking fixed N and amended with excess molybdate (Mo-dependent diazotrophy). Diazotrophic growth using the V-dependent nitrogenase was achieved by growth in medium lacking fixed N and molybdate and amended with excess vanadate (V-dependent diazotrophy). Diazotrophic growth using the Fe-only nitrogenase was achieved by growth in medium lacking fixed N, molybdate, and vanadate (heterometal-independent diazotrophy). Because alternative nitrogenase-dependent diazotrophic growth of A. vinelandiioccurs only in the absence of even trace levels of Mo, reverse transcriptase PCR (RT-PCR) was used to confirm the presence of transcripts of the appropriate alternative nitrogenase structural genes under the given cultivation conditions prior to transcriptome analysis. High-throughput cDNA sequencing revealed distinct patterns of transcript levels in cells cultivated under the various growth conditions examined. These results have broad implications for the physiology of the diazotrophic growth of A. vinelandiiand the evolution of nitrogenase.
MATERIALS AND METHODS
Strains and growth conditions.
A. vinelandiiDJ was cultured at 30°C with rotary shaking (200 rpm) in modified liquid Burk medium (6). Medium designed to favor growth under N-replete conditions was amended with 25 mM filter-sterilized ammonium acetate (NH4OAc). For alternative nitrogenase-dependent diazotrophic growth, Mo-deficient medium was prepared as previously described and NH4OAc was not added (6). For V-dependent diazotrophy, V2O5was added to N- and Mo-deficient medium to a final concentration of 1 μM. Cells were grown in triplicate under each condition, and nitrogenase activity was monitored by acetylene reduction (25). Expression of genes encoding each type of nitrogenase was confirmed by RT-PCR using the AccessQuick RT-PCR system (Promega Corp., Madison, WI) according to the manufacturer's protocol. The targets and primers used for RT-PCR are listed in Table S10 in the supplemental material.
Preparation of total RNA, cDNA library constructions, and SOLiD sequencing.
After subculturing for at least three transfers under the specified growth conditions, cells from 30-ml cultures at an optical density at 600 nm of 0.7 were harvested by centrifugation at 10,000 × gat 4°C for 10 min. Cell pellets were flash-frozen in liquid N2and stored at −80°C until further processed. Total RNA was prepared from cell pellets from each of the four conditions as previously described (45). Total RNA (0.5 μg) from each of the four cell growth conditions was submitted to the Genomics Core Facility at The Pennsylvania State University (University Park, PA) for cDNA library construction and SOLiD sequencing (45).
Data processing and analysis.
Raw sequencing reads were mapped against the A. vinelandiigenome as previously described (45). Reads that mapped to more than one region of the genome (2 to 5% of the total) could not be unambiguously mapped and were excluded from subsequent analyses (see Table S7 in the supplemental material). Summary information for sequences obtained by SOLiD is provided in Table S7 in the supplemental material. Statistical analyses were performed using DESeq(2) (version 1.2.0) from the Bioconductor R statistical packages (http://www.r-project.org/contributors.html) (22). Transcript level differences with adjusted Pvalues of <0.01 (generally, genes with more than 20 transcripts detected) and also a fold change ratio of 2.0 or greater (see reference 45) were considered to be significant (see Fig. 3). Previous comparisons of biological replicates have shown that 2-fold changes in transcript levels are significant and that much smaller changes are probably significant for highly expressed genes (45). The 15 genes most highly transcribed under each condition are summarized in Table S8 in the supplemental material.
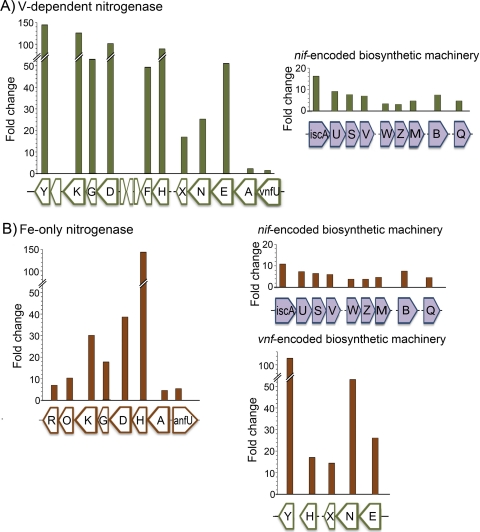
Fig. 3.
Differential expression of proteins necessary for Mo-independent nitrogen fixation. (A) Ratios of transcript levels (fold changes) of genes encoding structural proteins, assembly machinery, and regulatory proteins during V-dependent diazotrophy (Vnf, green) to transcript levels in fixed-N replete (control) cultures. Transcript levels of nifgenes observed to undergo large changes for V-dependent diazotrophy (Vnf, green) compared to transcript levels of the fixed-N replete (control) cultures are also indicated. (B) Ratios of transcript levels (fold changes) of genes encoding structural proteins, assembly machinery, and regulatory proteins for heterometal-independent diazotrophy (Anf, rust) to transcript levels in fixed-N replete (control) cultures. Transcript levels of nifand vnfgenes observed to undergo large changes during heterometal-independent diazotrophy (Anf, rust) compared to transcript levels in fixed-N replete (control) cultures are also shown. Each gene name is indicated by a letter(s). Full gene names, descriptions, and locus tags are given in Table S3 in the supplemental material.
Quantitative RT-PCR (qRT-PCR).
To confirm the results of SOLiD sequencing, 14 genes were chosen for qRT-PCR analyses that spanned the range of coverage depth from the four transcriptomes (see Table S9 in the supplemental material). qRT-PCR was performed using the PowerSYBR Green RNA-to-CT1-StepKit from Invitrogen (Carlsbad, CA) according to the manufacturer's protocol, and reaction products were assayed on a RotorGene-Q real-time PCR detection system from Qiagen (Valencia, CA). Reactions were performed in triplicate with 25 ng of total RNA quantified as described above, with 200 nM forward and reverse primers (see Table S10 in the supplemental material) in a final reaction volume of 20 μl. Control reaction mixtures contained either no RT or no template RNA. Results of the qRT-PCR assays are summarized in Fig. S1 in the supplemental material.
RESULTS AND DISCUSSION
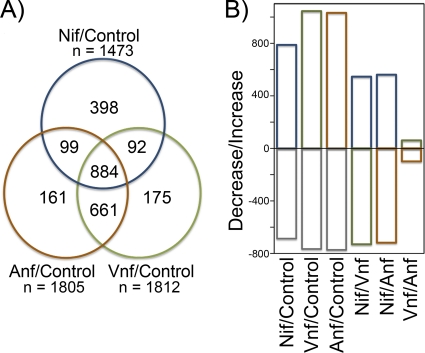
Transcript levels for nearly 30% of the A. vinelandiigenes changed more than 2-fold during diazotrophic growth relative to those in the fixed-N replete control. This included 884 genes for which the changes in expression were shared for all of the diazotrophic growth conditions tested (Mo-dependent, V-dependent, or heterometal-independent diazotrophy) compared to expression in fixed-N replete control cultures (Fig. 1A). Mo-dependent diazotrophic growth resulted in the differential expression of 398 genes (at least a 2-fold change compared to expression in fixed-N replete control cultures) that did not undergo a similar change in abundance under conditions of V-dependent or heterometal-independent diazotrophy (Fig. 1A). In addition to the 884 genes that changed under all nitrogen-fixing conditions, V-dependent or heterometal-independent diazotrophic growth resulted in an additional set of 661 common differentially expressed genes relative to the fixed-N replete control that were not observed to be differentially expressed (at least a 2-fold change relative to the control) under Mo nitrogenase-dependent diazotrophic growth. However, the number of genes uniquely affected by either V-dependent or heterometal-independent diazotrophy was much smaller than for Mo-dependent growth: 161 or 175, respectively (Fig. 1A).
Fig. 1.
Global response of the A. vinelandiitranscriptome during diazotrophic growth. (A) Venn diagram comparing the total number of mRNA species undergoing at least a 2.0-fold change in transcript level under Mo-dependent diazotrophy (Nif, blue circle), under V-dependent diazotrophy (Vnf, green circle), and under heterometal-independent diazotrophy (Anf, rust circle) compared to a fixed-N replete (control) culture. Values in overlapping circles indicate the numbers of mRNA species that undergo a change in transcript level under both or all three conditions. nis the total number of mRNA species undergoing a change in transcript level in each culture. (B) Bar graph of the total number of mRNA species that undergo a larger-than-2-fold increase or decrease in transcript abundance as a result of metal availability and nitrogen limitation. Nif, Mo-dependent diazotrophy; Vnf, V-dependent diazotrophy; Anf, heterometal-independent diazotrophy; Control, N-replete culture.
The largest changes in transcript abundance were observed during growth requiring expression of the alternative nitrogenases compared to Mo-dependent diazotrophy, when transcript abundance increased for over 1,000 genes and more than 700 decreased (by at least 2-fold) compared to the fixed-N replete control (Fig. 1B). Mo-dependent diazotrophic growth resulted in a smaller change in the transcriptome: transcripts for 788 genes increased and 685 decreased more than 2-fold (Fig. 1B). Interestingly, only 160 genes were differentially expressed as a result of diazotrophic growth dependent on the alternative nitrogenases when V-dependent diazotrophic growth was compared to heterometal-independent diazotrophic growth (Fig. 1B). Collectively, the results indicated that the patterns of gene expression under conditions of V-dependent or heterometal-independent diazotrophic growth are very similar to one another but distinctly different from the patterns of gene expression under Mo-dependent diazotrophic growth.
Expression of nifgenes under Mo-dependent nitrogen-fixing conditions compared to nondiazotrophic growth.
The genetic complexity of nitrogenase assembly and the regulation of nifgene expression of nitrogenase have been extensively characterized using the model diazotroph A. vinelandii(43). Previous specific gene deletion (28, 29, 33, 36) and Northern blot hybridization analyses (30, 33) provided key insights into the genes involved in biological N2fixation by A. vinelandii. The genes encoding Mo nitrogenase and the machinery to assemble the enzyme are colocalized in a major nifcluster and a secondary cluster that contains a few key biosynthetic and regulatory genes (28, 33) (Fig. 2A). Transcription of nifgenes is activated by a σ54-dependent activator, NifA, and a number of nif-specific promoter sequences have been identified within the two nifclusters (15, 28).
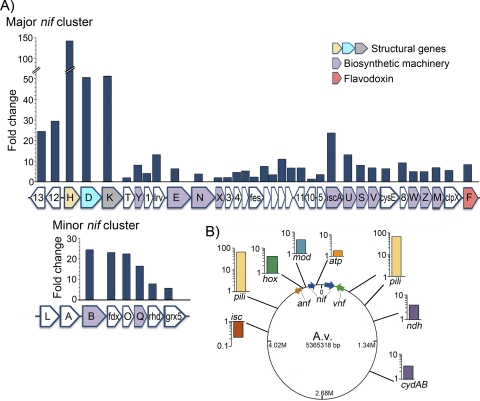
Fig. 2.
Differential expression of proteins necessary for Mo-dependent diazotrophy. (A) The ratio of transcript levels (fold change) of genes encoding structural proteins, assembly machinery, and regulatory proteins for Mo-dependent diazotrophy (Nif, blue) are compared to transcript levels in fixed-N replete (control) cultures. Transcripts mapping to the regulatory genes nifAand nifLwere detected at very low levels in both Mo-dependent diazotrophic and fixed-N replete cultures and, as a result, are not included here (see Table S1 in the supplemental material). (B) The ratio of transcript levels of genes or groups of genes encoding proteins important for the global response of A. vinelandii(A.v.) during Mo-dependent diazotrophic growth compared to the N-replete (control) culture, presented on a log scale. The genomic locations of these genes are indicated, as well as the genomic locations of the gene clusters encoding the Mo-dependent nitrogenase (nif, blue arrows) and the alternative nitrogenase (vnf[green arrow] and anf[rust arrow]). isc, ISC; pili, pilus machinery; hox, membrane-bound [NiFe] hydrogenase; mod, molybdate transport; atp, ATP synthase; ndh, NADH-dependent oxidoreductase; cydAB, cytochrome bdoxidase I. Each gene name is indicated by a letter(s). Full gene names, descriptions, and locus tags are given in Tables S1 and S2 in the supplemental material.
Under N-fixing conditions, nitrogenase components make up nearly 10% of the total cellular proteins, the majority of which are structural proteins (14). The catalytic components necessary for FeMo-co biosynthesis are presumably necessary in much smaller amounts. A large increase in the transcript levels of the structural genes nifH(Avin01380), nifD(Avin01390), and nifK(Avin01400) was observed under Mo-dependent diazotrophy compared to those in the fixed-N replete (control) culture (Fig. 2A; also see Table S1 in the supplemental material). The transcript levels of nifHincreased much more (143-fold) than those of nifDand nifK(∼54-fold) (Fig. 2A). This observation is consistent with previous analyses and supports the observation that nitrogenase activity is optimal under conditions of high Fe protein to MoFe protein ratios (13, 14). Interestingly, the abundances of transcripts of nonessential nifgenes (28) downstream of nifKincreased by only 2- to 14-fold (Fig. 2A). The different abundances of these apparently cotranscribed genes suggested that these gene-specific regions of the transcripts have different processing and segmental stabilities or that their synthesis may be regulated by transcriptional attenuation or processing.
The operon adjacent to the nifHDKstructural genes contains nifE(Avin01450), nifN(Avin01470), and nifX(Avin01480), encoding the scaffold protein necessary for FeMo-co biosynthesis (NifEN) and a carrier protein (NifX) (54) (Fig. 2A; also see Table S1 in the supplemental material). The Shethna protein (FeSII, encoded by fesII[Avin01520]), which is involved in protecting the Fe protein from O2damage (46), is also encoded within this operon. Transcripts mapping to these genes increased 2- to 6-fold under Mo-dependent nitrogen fixation conditions (Fig. 2A).
The majority of the remaining genes within the major nifcluster encode proteins important for FeMo-co and P cluster biosynthesis and maturation of nitrogenase (54). Transcripts mapping to the genes iscAnif(Avin01610) and nifUSV(Avin01620 to Avin01640) increased (∼6- to 24-fold) under N2-fixing conditions (Fig. 2A; also see Table S1 in the supplemental material). NifU, a scaffold protein, and NifS, a cysteine desulfurase, are similar to proteins of the Isc and Suf housekeeping systems for [Fe-S] cluster biosynthesis and facilitate the assembly of Nif-specific metal clusters (38). NifU and NifS are required for the maturation of nitrogenase in A. vinelandiiand, despite the similarity to IscU and IscS and evidence that some cross talk may exist (18), are unable to supplant the function of the ISC (iron-sulfur cluster) system (39). NifV is a homocitrate synthase and is required for synthesis of homocitrate for FeMo-co in A. vinelandii(66). Previous biochemical and genetic analyses indicate that these gene products are necessary for optimal diazotrophic growth in A. vinelandii(29). orf8(Avin01660) and nifZW(Avin01670 and Avin01680) transcript levels, also important for Mo-dependent diazotrophic growth (29), increased ∼5- to 9-fold (Fig. 2A). Increases of ∼6- to 8-fold were observed for transcripts mapping to nifM(Avin01690), the product of which is necessary for Fe protein maturation (26), and clpXnif(Avin01700), which is cotranscribed (Fig. 2A), as well as the adjacent, independently transcribed gene nifF(Avin01710), which encodes a flavodoxin (4) (Fig. 2A). Transcripts mapping to genes encoding several hypothetical proteins located within the major nifoperon also increased 3- to 11-fold (Fig. 2A; also see Table S1 in the supplemental material). However, previous gene deletion studies indicated that these products are not required for diazotrophic growth (28, 29).
The minor nifcluster contains two operons that include the genes encoding the nif-regulatory proteins NifL (Avin50990) and NifA (Avin5100) and maturation proteins encoded by nifB(Avin51010) and nifQ(Avin51040), which are involved in FeMo-co biosynthesis. The genes nifB, fdx, nifO, nifQ, rhd, and grx5(Avin51010 to Avin51060) are cotranscribed. A large increase (∼12- to 18-fold) in the number of transcripts mapping to nifB, fdx, nifO, and nifQwas detected, and a smaller increase (4- to 6-fold) in those for rhdand grx5was noted (Fig. 2A). NifB is a radical S-adenosylmethionine (SAM) protein and a key enzyme required for FeMo-co biosynthesis. NifQ (Avin51040) has been implicated in Mo acquisition and FeMo-co biosynthesis (54). The nifLand nifAregulatory genes are located upstream of nifBand are under the control of a separate promoter. As noted above, the σ54-dependent activator NifA is required for transcriptional activation of Mo nitrogenase genes; NifA activity is controlled by its regulatory partner protein NifL in response to excess oxygen and fixed N, ensuring that Mo-dependent nitrogen fixation occurs only under appropriate physiological conditions (16). Low nifLand nifAtranscript levels were detected under both conditions (see Table S1 in the supplemental material).
Differences in global patterns of gene expression implicated during Mo-dependent diazotrophic growth compared to nondiazotrophic growth.
Fixed N is essential for the biosynthesis of cellular components and growth; however, when fixed N is limiting, biological N2fixation also imposes a high demand for ATP and reducing power on cells. These overlapping and competing requirements were expected to cause significant differences in the patterns of gene expression in the presence and absence of fixed N. Other than the major nifgene cluster, the largest increases (60- to >200-fold) in transcript abundance under Mo-dependent diazotrophic growth occurred for genes that encode type IV pili, namely, pilG, pilE, pilA, pilN, and pilM(Avin03180, Avin11830, Avin12104, Avin45290, and Avin45300, respectively) (Fig. 2B; also see Table S2 in the supplemental material). Type IV pili are involved in a number of processes, including cell motility, DNA uptake, sensing, attachment, and aggregation (55); the latter could be a previously unrecognized O2protection mechanism in A. vinelandii.
Several strategies of O2protection for nitrogenase have been noted, including respiratory protection by active consumption of oxygen (50), spatial decoupling, for which N2fixation occurs in differentiated anoxic heterocysts (21), and temporal separation by free-living cyanobacteria that fix nitrogen at night, when oxygenic photosynthesis cannot occur (62). When carbon is not limiting, A. vinelandiiemploys a respiratory protection mechanism to limit levels of cytoplasmic O2, effectively protecting oxygen-sensitive enzymes such as nitrogenase from inactivation (50). The genome of A. vinelandiiencodes four NADH:ubiquinone oxidoreductases and five terminal oxidases. Transcripts mapping to the genes coding for cytochrome bdoxidase I (CydAB I; Avin19880 and Avin19890) and the type II NADH-dependent oxidoreductase Ndh (Avin12000) increased 3- to 4-fold in nitrogen-fixing cells compared to those in the control (Fig. 2B; also see Table S2 in the supplemental material). These results are consistent with previous studies indicating a role for both CydAB I and Ndh in the diazotrophic growth of A. vinelandii, especially at high O2concentrations (5, 50). Small increases in transcript abundance for two other terminal oxidases, cytochrome cbb3oxidase (Cco; Avin19940 to Avin20010) and cytochrome coxidase (Cox; Avin11170 and Avin11180), were also observed during diazotrophic growth (see Table S2 in the supplemental material).
The increased levels of transcripts for terminal oxidases and respiratory enzymes could be coordinately regulated to meet the energy demands for nitrogen fixation. Transcripts mapping to both ATP synthases (Avin19670 to Avin19750 and Avin52150 to Avin52230) also increased (∼1- to 5-fold) under N2-fixing conditions (Fig. 2B; also see Table S2 in the supplemental material). Transcript levels also increased ∼5-fold for the well-characterized, membrane-bound [NiFe] hydrogenase (hoxVTRQOLMZGK[Avin50500 to Avin50590]) (Fig. 2B) (57), which presumably recycles the hydrogen produced as a by-product of nitrogenase-catalyzed N2reduction, thereby capturing reducing equivalents that would otherwise be lost.
In addition to nif-specific genes, whose products are necessary for the initial steps in targeting Fe and S for assembly of metalloclusters associated with the maturation of nitrogenase (nifUand nifS), the genome of A. vinelandiialso includes a suite of genes that encode proteins required for the maturation of [Fe-S] cluster-containing proteins related to other metabolic functions. The more general components of the machinery responsible for maturation of “housekeeping” [Fe-S] proteins have been designated ISC components and include iscR, iscU, iscS, iscA, hscBhscA, fdx, and iscX(Avin40340 to Avin40410) (38). Among the ISC proteins, IscS is paralogous to NifS (38% identity) and IscU is similar in sequence (49% identity) to the N-terminal domain of NifU. Biochemical studies have established that NifS and IscS have cysteine desulfurase activity associated with the mobilization of S for [Fe-S] cluster assembly and that both IscU and the N-terminal domain of NifU can serve as invitroand invivoscaffolds for [Fe-S] cluster formation (18, 60). Genetic and physiological studies have also established that there is some functional cross talk between the ISC and Nif systems for [Fe-S] cluster formation (18). This indicates that the Nif counterparts function primarily to satisfy the increased demand for the mobilization and targeting of Fe and S for [Fe-S] cluster formation associated with N2fixation. However, NifU and NifS cannot entirely replace the housekeeping [Fe-S] biogenesis supported by ISC in A. vinelandii(18, 39). The present work provides further evidence for functional cross talk between the ISC and Nif systems because stimulation of NifU and NifS expression under N2-fixing conditions is accompanied by a decrease in the expression of the ISC system (Fig. 2B; also see Table S2 in the supplemental material). This observation also extends to the expression of other housekeeping components associated with [Fe-S] protein maturation, including ErpA (Avin46030) and NfuA (Avin28760). Interestingly, ErpA has primary structural similarity to the IscA-like protein contained within the major nifcluster and NfuA has primary structural similarity to the C-terminal domain of NifU. Although the specific function of these proteins in A. vinelandiihas not yet been clearly established, current evidence indicates that they could function as [Fe-S] cluster assembly scaffolds or as intermediate carriers of [Fe-S] clusters, a role that has been demonstrated for NfuA in Synechococcussp. strain PCC 7002 (31). The observed ability of the Nif-specific [Fe-S] maturation components to replace the analogous housekeeping functions partially (18) may reflect the lower level of expression of the ISC components when cells are cultured under N2-fixing conditions. However, expression of housekeeping [Fe-S] cluster biosynthetic proteins is subject to feedback regulation through the action of IscR, an [Fe-S] cluster-containing transcription regulator. It is also possible that the respiratory protection mechanism of nitrogen-fixing cells also protects other [Fe-S] proteins from oxygen damage, thus lowering the demand for [Fe-S] protein maturation.
A. vinelandiiutilizes an efficient molybdate transport system, encoded by the modgenes (Avin50650 to Avin50690), to provide the Mo necessary for Mo-containing enzymes, including nitrogenase (47). The A. vinelandiigenome contains genes encoding a high-affinity transport system (Avin50650 to Avin50690) located adjacent to the minor nifcluster and preceded by a putative σ54-dependent promoter. Transcripts mapping to this molybdate transport system increased ∼4-fold during N limitation (Fig. 2B; also see Table S2 in the supplemental material). In addition, smaller increases in transcript abundance (∼1- to 5-fold) were also observed for the two other Mo transport systems (Avin01280 to Avin01300 and Avin50700 to 50730) (see Table S2 in the supplemental material).
Transcription profiles in cells expressing alternative nitrogenases.
The alternative forms of nitrogenase were first discovered through the observation that strains of A. vinelandiiin which nifstructural genes had been deleted were still capable of diazotrophic growth in Mo-deficient, fixed-N-free medium (7). Subsequent genetic and DNA sequence analysis has revealed that distinct alternative (V and Fe-only) nitrogenases are encoded by nifparalogs within the vnfand anfclusters which are not colocalized with either of the nifgene clusters (Fig. 2A and 3) (59). The expression of the alternative systems in A. vinelandiiis regulated by the availability of Mo (30). Transcripts of the vnfand anfstructural genes have been observed by Northern blot hybridization in N-limited A. vinelandiicultures in the absence of Mo with added V (27) or in the absence of both Mo and V (51).
The alternative forms of nitrogenase are present only in the genomes of organisms that also harbor the nifsystem (8, 53, 61), and although the structural genes are distinct, the gene operons encoding the alternative enzymes do not include paralogs of all of the genes for the biosynthetic machinery necessary for the assembly and maturation of Mo-dependent nitrogenase. In A. vinelandii, limited biochemical evidence has suggested that VnfH can play a role analogous to that of NifH in FeMo-co maturation (10); however, diazotrophic growth is not supported in mutants containing deletions of each of the respective structural genes. Moreover, mutational analyses have indicated that some components of the nif-encoded biosynthetic machinery, such as nifU, nifS, nifV, nifM, and nifB, are required for nitrogen fixation by cells utilizing either of the alternative enzymes in A. vinelandii(36, 41) and a role for the NifEN scaffold protein has also been observed (65).
The transcriptome of cultures relying on V-dependent nitrogenase for diazotrophic growth revealed a large increase (68- to 137-fold) in the vnfHFand vnfDGKgenes, compared to the levels in fixed-N replete (control) cultures (Fig. 3A). The large difference in transcript abundance observed for nifHversus nifDK, which are cotranscribed from a single promoter, was not observed for vnfH, vnfD, and vnfK, which are located in separate operons (3). A small increase (∼2 fold) was observed for nifHtranscripts, but given the high vnfHtranscript abundance, it is unclear what the potential role of nifH-encoded Fe protein might be during V-dependent diazotrophy. These genes encode an Fe protein specific to the V-dependent system (encoded by vnfH) and the VFe protein (encoded by vnfDGK) analogous to the MoFe protein which houses the active-site metalloclusters, the FeV cofactors (FeV-co). The vnfgene cluster in A. vinelandiialso encodes a putative FeV-co assembly scaffold, VnfEN (Avin02770 and Avin02750), paralogous to the NifEN scaffold protein necessary for FeMo-co biosynthesis. Mutant strains of A. vinelandiiwith knockouts in vnfENcan still grow diazotrophically under Mo-limiting but V-replete diazotrophic growth conditions (65). However, transcriptome analysis revealed a large increase (26- to 56-fold) in the expression of vnfEand vnfNtranscripts (Fig. 3) and a very small increase in the expression of nifENtranscripts (∼1- to 2.5-fold; see Table S3 in the supplemental material) during V-dependent diazotrophy. The large increase in vnfENtranscript levels supports the limited biochemical evidence that suggested that VnfEN, and not NifEN, is the preferred scaffold for FeV-co maturation (65). The vnfoperon also encodes VnfX (Avin02740) and VnfY (Avin02570) (Fig. 2A), paralogs of NifX and NifY. VnfY is required for optimal V-dependent diazotrophy (55), and large increases in vnfXand vnfYtranscript levels (15- and 117-fold) were observed during V-dependent diazotrophy (Fig. 3A). Interestingly, this is markedly different from the nifX(Avin01480) and nifYtranscript levels observed under Mo-dependent diazotrophy, in which much smaller increases (<8-fold) were observed (see Tables S1 and S3 in the supplemental material). This implies that perhaps VnfX and VnfY have a role in FeV-co biosynthesis more significant than that of NifX and NifY in FeMo-co biosynthesis.
Transcripts mapping to the nif-encoded cluster biosynthesis machinery implicated in playing a role in the assembly of alternative cofactors such as nifUSV, nifM, and nifBalso increased (5- to 10-fold), consistent with what was observed from previous deletion mutant analysis (36), indicating that these biosynthetic genes are also involved in V-dependent diazotrophy. Interestingly, an increase in transcript abundance of nifQ, which is necessary for Mo acquisition for FeMo-co, was also observed, and this may reflect the pressure to acquire Mo to utilize the Mo-dependent nitrogenase preferentially or may be simply the result of the transcriptional coupling of nifQwith nifB. Our analysis thus implicated the involvement of nif-encoded maturation proteins in V-dependent diazotrophy in A. vinelandii.
The Fe-only nitrogenase (AnfHDGK; Avin49000 to Avin48970) responsible for heterometal-independent diazotrophy is encoded by the anfgene cluster. This cluster includes the anfHDGKstructural genes encoding an additional Fe protein, AnfH, specific to the Fe-only system and the genes encoding the Fe-only version of the substrate reduction component, the FeFe protein (AnfDGK), where the FeFe cofactors reside. In addition, this cluster includes anfA(Avin49020), a regulatory gene, and anfO(Avin48960) and anfR(Avin48950) (Fig. 3B). Transcripts mapping to the anfHDGKstructural genes increased 18- to 144-fold in cultures dependent on the Fe-only nitrogenase for diazotrophic growth compared to the levels in the fixed-N replete cultures (Fig. 3B). The differences in transcript abundance of anfHversus anfDand anfK, which are presumably cotranscribed, mirrors what is observed for the nifstructural genes, where the increase in nifHtranscript abundance was much larger than the observed increases in nifDand nifKtranscript abundance. Large increases (14- to 117-fold) in the vnf-encoded H, E, N, X, and Ytranscripts were observed during heterometal-independent diazotrophy (Fig. 3B). This observation suggested that these vnf-encoded products are preferred over their nif-encoded counterparts for maturation of the Fe-only nitrogenase. Therefore, both the V-dependent and Fe-only nitrogenases apparently utilize the VnfEN scaffold protein for cofactor assembly but the nif-encoded accessory proteins NifU, NifS, NifM, NifV, and NifB for maturation (41). Importantly, vnfENin A. vinelandiiis derived from nifEN, suggesting that the preferential utilization of the VnfEN scaffold over the NifEN scaffold in alternative systems is a recent evolutionary feature (8). In addition, VnfH has been demonstrated to be necessary for transcription of anfHDGK; however, in vnfHmutants, the presence of NifH is detected but lower growth rates of heterometal-independent diazotrophically grown cells are observed (37) An increase in vnfHtranscripts (∼17-fold) and nifHtranscripts (∼2.5-fold) was observed during heterometal-independent diazotrophy (see Table S3 in the supplemental material). Together, these results may suggest that Vnf preceded and is evolutionarily ancestral to Anf, which is consistent with evidence reported previously (8, 53).
Electron transport for Mo-, V-, and Fe-only nitrogenase-dependent diazotrophic growth.
The energy and electron transport requirements differ for the three forms of nitrogenase, and both the nifand vnfsystems encode specific flavodoxins, i.e., those encoded by the nifF(Avin01710) and vnfF(Avin02650) genes. Although nifFis not required for Mo-dependent diazotrophy (4), it is not known if vnfFis required for V-dependent or heterometal-independent diazotrophy. Both vnfFand nifFtranscripts increased under Mo-independent diazotrophic conditions, and the increases in vnfF(47- to 102-fold) were much larger than the increase in nifF(∼4-fold) (Fig. 3). During Mo-dependent diazotrophy, transcripts mapping to nifFincreased 8-fold.
Three clusters of genes, rnf1(Rnf1, Avin50930 to Avin50980), rnf2(Rnf2, Avin19220 to Avin19270), and fix(Avin10510 to Avin10550), encode electron transport systems that are thought to provide reducing equivalents to nitrogenase in A. vinelandii(40, 56). The rnfgenes were originally identified and characterized in Rhodobactercapsulatus, and they encode a membrane-bound complex involved in electron transport to nitrogenase (56). The expression of the genes encoding the Rnf1 complex is NifA dependent, while the genes encoding Rnf2 are not regulated as a result of N availability (see Table S4 in the supplemental material) (12). In A. vinelandii, the rnfgenes are necessary for full Mo-dependent nitrogenase activity and have been suggested to be regulated by NifA, potentially by modulation of the redox state of NifL (12). In addition, Rnf1 proteins are required for accumulation of the [4Fe-4S] cluster in the nifH-encoded Fe protein (12). The fixoperon, which is preceded by a putative σ54-dependent promoter (R.D., unpublished data), encodes five proteins, FixP, -A, -B, -C, and -X, that form an electron transfer complex that seems to be required to support nitrogen fixation in some diazotrophs (40). Both rnf1and fixPABCXtranscript levels increased markedly during diazotrophic growth; however, during V-dependent or heterometal-independent diazotrophy, fixPABCXoperon transcript levels were higher than rnf1operon transcript levels (∼231-fold versus ∼18-fold) (see Table S4 in the supplemental material). In contrast, the increases in transcripts from the rnf1operon and the fixPABCXoperon were similar in magnitude during Mo-dependent diazotrophy (∼40-fold). As expected, levels of transcripts from the rnf2operon, which is not NifA regulated, were relatively unchanged.
Regulation of gene expression during diazotrophic growth.
The σ54-dependent activator proteins VnfA and AnfA are required for expression of V-dependent nitrogenase and the Fe-only nitrogenase, respectively (52), but unlike the activity of NifA, their activity is not subject to regulation by a NifL-like protein. As observed for nifA, the vnfAand anfAtranscripts were detectable at low levels under all of the growth conditions tested here; however, their transcript levels increased under V-dependent and heterometal-independent diazotrophic growth (Fig. 3) and decreased in the presence of Mo (see Table S5 in the supplemental material). This pattern of regulation of vnfAand anfAexpression has been observed previously, and the Mo-responsive transcriptional regulator ModE has been shown to be at least partially involved in Mo-dependent repression of anfA(52). Transcriptional regulation of vnfAand anfAis likely to be responsible for the absence of the VFe- and Fe-only nitrogenases when molybdenum is available, because transcription of the genes encoding these alternative enzymes cannot be activated in the absence of VnfA or AnfA, respectively (64). The A. vinelandiigenome contains an additional gene homologous to nifA, nifA2(Avin26490), and two additional vnfAhomologs, vnfA2(Avin33440) and vnfA3(Avin47100) (59). Interestingly, the pattern of regulation of vnfA2and vnfA3expression was similar to that of vnfAand anfAexpression, as transcripts for both increased under Mo-depleted diazotrophic conditions compared with those in cultures grown with molybdate (see Table S5 in the supplemental material). Although the precise role of these activator paralogs is unclear, the sequence similarity of their DNA binding determinants to those of the cognate NifA and VnfA activators (59) suggests that they may be able to provide additional regulatory input.
Differences in global patterns of gene expression implicated during Mo-independent nitrogen-fixing growth.
The complexity of the nitrogenase enzyme and the absolute requirement for fixed N to meet cellular growth might be expected to result in a similar global response to fixed-N limitation, regardless of metal availability, but as noted above, the patterns of gene expression for Mo-dependent diazotrophy are significantly different from those in cells grown under Mo-independent diazotrophic conditions. The alternative forms of nitrogenase are markedly less efficient than the Mo nitrogenase (19), and coupled with the demand for fixed N levels necessary to maintain cell growth, this inefficiency would presumably result in significant differences in the transcriptional response of V-dependent or heterometal-independent diazotrophy from that in N-replete control cultures. Key differences are observed in the A. vinelandiitranscriptome in response to N-limiting conditions and the availability of Mo. Most notably, transcripts for hutU(Avin26180), the gene encoding urocanate hydratase, increased dramatically (30- to 40-fold) under diazotrophic conditions in the absence of Mo (see Table S5 in the supplemental material). Urocanate hydratase catalyzes a key step in the degradation of histidine (63). The increased hutUtranscript levels presumably reflect the low catalytic efficiency of the alternative forms and imply that cells seek to supplement the fixed N provided by the alternative forms by liberating fixed N during protein turnover by His degradation to maintain cell viability until Mo or fixed N becomes available. This increase in enzymes capable of accessing fixed N was also noted in the transcriptome analysis of Rhodopseudomonas palustrisunder diazotrophic conditions requiring expression of the alternative nitrogenases (48). Transcript levels of other hutgenes in the same operon, hutG(Avin26150), hutI(Avin26160), and the gene for a hypothetical protein (Avin26170), and just downstream, hutH(Avin26290) and hutF(Avin26300), were also observed to increase (∼5- to 40-fold) under these conditions (see Table S5 in the supplemental material). Interestingly, the large increase in transcripts for the formation of pili was not observed during Mo-independent, diazotrophic growth. Also notable was an increase in the levels of transcripts for the genes (Avin04360 to Avin04410) encoding an uncharacterized soluble [NiFe] hydrogenase, which increased ∼10-fold during V-dependent diazotrophy and ∼20-fold during heterometal-independent diazotrophy (see Table S5 in the supplemental material). The increase in transcripts may reflect the need to recycle the larger amount of hydrogen produced by the activity of the alternative enzymes to counteract the catalytic inefficiency. The alternative nitrogenases have been shown to produce more hydrogen during dinitrogen reduction, which presumably provides additional pressure to avoid the loss of needed reducing equivalents (19). Finally, transcripts mapping to a number of genes annotated as hypothetical proteins were also observed to undergo large changes as a result of Mo-independent diazotrophy. For example, Avin02580, Avin16830, Avin47120, Avin46150, and Avin10510 transcript levels increased and Avin22850, Avin09340, Avin49410, and Avin47230 transcript levels decreased under these conditions (see Table S6 in the supplemental material).
Although our analysis indicates similar patterns of gene expression when cultures are grown either under conditions of V-dependent diazotrophy and heterometal-independent diazotrophy, several key differences were noted. Importantly, transcripts mapping to several genes near the vnfoperon increased dramatically (53- to 100-fold change) during V-dependent diazotrophy (see Table S6 in the supplemental material). These genes, Avin02540, Avin02550, and Avin02560, encode proteins that are homologous to the inner membrane transport protein and substrate binding protein of an ABC transporter, which is annotated as a phosphonate transport system. Due to the proximity of these genes to the vnfgene cluster and due to the large increase in transcripts mapping to these genes during V-dependent diazotrophy, we hypothesize that these gene products are responsible for vanadate uptake.
Like Mo-dependent diazotrophy, both V-dependent and heterometal-independent diazotrophic growth resulted in an increased abundance of transcripts for CydAB I (∼3- to 5-fold), Ndh (∼3-fold), and both ATP synthases (see Table S5 in the supplemental material) compared to that in N-replete control cultures. Likewise, similar increases in levels of transcripts mapping to the hox-encoded, membrane-bound [NiFe] hydrogenase and the Mod transport systems were also observed in cultures expressing the alternative nitrogenases. Interestingly, a decrease in the ISC machinery was also noted, further supporting the hypothesis that iscgene expression levels observed under normal growth conditions are sufficient to provide [Fe-S] clusters for general cell metabolism (see Table S5 in the supplemental material). A significant decrease in transcript abundance was noted for some genes regardless of metal availability during diazotrophic growth (see Table S6 in the supplemental material). The differential expression of these proteins presumably reflects changes in the transcriptome of A. vinelandiiin response to fixed-N limitation; however, most of these transcripts mapped to proteins without defined functions (for example, those of Avin09340, Avin22850, Avin47230, and Avin49410).
Evolutionary implications.
The results presented herein have fundamental implications for our understanding of nifevolution. Several phylogenetic studies have suggested that the alternative nitrogenases are evolutionary ancestors of Mo nitrogenase (8, 53, 61). This is considered to be consistent with early Earth history, because the bioavailability of Mo and perhaps V would have been very low prior to the advent of oxygenic photosynthesis and subsequent oxygenation of Earth's atmosphere (1). Interestingly, however, the V-dependent and Fe-only nitrogenases in extant organisms have not yet been observed in a background devoid of the Mo nitrogenase (8, 53). In this study, we clearly show that FeMo-co biosynthetic genes clustered with and required for the nifsystem are expressed as presumably essential components of alternative systems, extending mutant and biochemical analyses that have suggested this functional cross talk (36, 41) and in line with previous genetic experiments which indicated that the alternative nitrogenases are not functional in the absence of these nif-encoded gene products. The available information regarding alternative nitrogenase genomic occurrence, the results of previous genetic studies, and the transcription profiling results described here collectively make it difficult to consider the alternative nitrogenases as ancestral forms of the Mo nitrogenase. Instead, a simpler hypothesis is that the alternative nitrogenases evolved from Mo nitrogenase to expand the fundamental niche of diazotrophic populations under conditions where Mo may, in fact, be limiting.
Conclusion.
The carefully controlled transcriptional analysis presented here has clearly defined the suite of genes associated with each of the three nitrogenase systems in A. vinelandii. In addition, these results have uncovered several interesting surprises regarding the evolutionary relationships between Mo nitrogenase and the alternative nitrogenases, which can be examined further through combined genetic, biochemical, and evolutionary analyses. The results of the global expression under N2-fixing conditions suggest that Mo limitation, whether in the presence or in the absence of V, results in marked differences in gene expression in A. vinelandii. These results also support the hypothesis that the lower catalytic efficiencies of the V-dependent and Fe-only nitrogenases impose significant physiological constraints upon A. vinelandiiwhen it is grown under N2-fixing conditions in the absence of Mo. These studies will help to refine our understanding of the complex metabolic and regulatory interactions that control N2fixation in this important model organism.
Supplementary Material
Acknowledgements
This work was supported by NASA Astrobiology Institutegrant NNA08C-N85Ato J.W.P. D.A.B. acknowledges support form NASA Astrobiology: Exobiology and Evolutionary Biologyaward NNX09AM87G, and work in the laboratory of D.R.D. is supported by NSFMCB-071770. T.L.H. was supported by an NSF-Integrated Graduate Educational Research and Training fellowshipgrant, and E.S.B. was supported by a fellowship from the NASA Astrobiology Institute postdoctoral program.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Anbar A. D., et al. 2007. A whiff of oxygen before the great oxidation event? Science 317:1903–1906 [DOI] [PubMed] [Google Scholar]
- 2. Anders S., Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bageshwar U. K., Raina R., Choudhury N. R., Das H. K. 1998. Analysis of upstream activation of the vnfHpromoter of Azotobacter vinelandii. Can. J. Microbiol. 44:405–415 [PubMed] [Google Scholar]
- 4. Bennett L. T., Jacobson M. R., Dean D. R. 1988. Isolation, sequencing, and mutagenesis of the nifFgene encoding flavodoxin from Azotobacter vinelandii. J. Biol. Chem. 263:1364–1369 [PubMed] [Google Scholar]
- 5. Bertsova Y. V., Bogachev A. V., Skulachev V. P. 2001. Noncoupled NADH:ubiquinone oxidoreductase of Azotobacter vinelandiiis required for diazotrophic growth at high oxygen concentrations. J. Bacteriol. 183:6869–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bishop P. E., Jarlenski D. M., Hetherington D. R. 1982. Expression of an alternative nitrogen fixation system in Azotobacter vinelandii. J. Bacteriol. 150:1244–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bishop P. E., et al. 1986. Nitrogen fixation by Azotobacter vinelandiistrains having deletions in structural genes for nitrogenase. Science 232:92–94 [DOI] [PubMed] [Google Scholar]
- 8. Boyd E. S., et al. 2011. A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology 9(3):221–232 [DOI] [PubMed] [Google Scholar]
- 9. Burgess B. K., Lowe D. J. 1996. Mechanism of molybdenum nitrogenase. Chem. Rev. 96:2983–3012 [DOI] [PubMed] [Google Scholar]
- 10. Chatterjee R., Allen R. M., Ludden P. W., Shah V. K. 1997. In vitro synthesis of the iron-molybdenum cofactor and maturation of the nif-encoded apodinitrogenase. Effect of substitution of VNFH for NIFH. J. Biol. Chem. 272:21604–21608 [DOI] [PubMed] [Google Scholar]
- 11. Chisnell J. R., Premakumar R., Bishop P. E. 1988. Purification of a second alternative nitrogenase from a nifHDKdeletion strain of Azotobacter vinelandii. J. Bacteriol. 170:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Curatti L., Brown C. S., Ludden P. W., Rubio L. M. 2005. Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. Proc. Natl. Acad. Sci. U. S. A. 102:6291–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dilworth M. J., Subramanian D., Munson T. O., Burris R. H. 1965. The ATP requirement for nitrogen fixation in cell-free extracts of Clostridium pasteurianum. Biochim. Biophys. Acta 99:486–503 [DOI] [PubMed] [Google Scholar]
- 14. Dingler C., Kuhla J., Wassink H., Oelze J. 1988. Levels and activities of nitrogenase proteins in Azotobacter vinelandiigrown at different dissolved oxygen concentrations. J. Bacteriol. 170:2148–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dixon R. 1998. The oxygen-responsive NIFL-NIFA complex: a novel two-component regulatory system controlling nitrogenase synthesis in γ-proteobacteria. Arch. Microbiol. 169:371–380 [DOI] [PubMed] [Google Scholar]
- 16. Dixon R., Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621–631 [DOI] [PubMed] [Google Scholar]
- 17. Dos Santos P. C., Dean D. R. 2011. Coordination and fine-tuning of nitrogen fixation in Azotobacter vinelandii. Mol. Microbiol. 79:1132–1135 [DOI] [PubMed] [Google Scholar]
- 18. Dos Santos P. C., Johnson D. C., Ragle B. E., Unciuleac M. C., Dean D. R. 2007. Controlled expression of nifand isciron-sulfur protein maturation components reveals target specificity and limited functional replacement between the two systems. J. Bacteriol. 189:2854–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eady R. R. 1996. Structure-function relationships of alternative nitrogenases. Chem. Rev. 96:3013–3030 [DOI] [PubMed] [Google Scholar]
- 20. Eady R. R., Robson R. L., Richardson T. H., Miller R. W., Hawkins M. 1987. The vanadium nitrogenase of Azotobacterchroococcum. Purification and properties of the VFe protein. Biochem. J. 244:197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fay P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56:340–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gentleman R. C., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Georgiadis M. M., et al. 1992. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257:1653–1659 [DOI] [PubMed] [Google Scholar]
- 24. Hales B. J., Case E. E., Morningstar J. E., Dzeda M. F., Mauterer L. A. 1986. Isolation of a new vanadium-containing nitrogenase from Azotobacter vinelandii. Biochemistry 25:7251–7255 [DOI] [PubMed] [Google Scholar]
- 25. Hardy R. W., Holsten R. D., Jackson E. K., Burns R. C. 1968. The acetylene-ethylene assay for N(2) fixation: laboratory and field evaluation. Plant Physiol. 43:1185–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Howard K. S., et al. 1986. KlebsiellapneumoniaenifMgene product is required for stabilization and activation of nitrogenase iron protein in Escherichia coli. J. Biol. Chem. 261:772–778 [PubMed] [Google Scholar]
- 27. Jacobitz S., Bishop P. E. 1992. Regulation of nitrogenase-2 in Azotobacter vinelandiiby ammonium, molybdenum, and vanadium. J. Bacteriol. 174:3884–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jacobson M. R., et al. 1989. Physical and genetic map of the major nifgene cluster from Azotobacter vinelandii. J. Bacteriol. 171:1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jacobson M. R., et al. 1989. Biochemical and genetic analysis of the nifUSVWZMcluster from Azotobacter vinelandii. Mol. Gen. Genet. 219:49–57 [DOI] [PubMed] [Google Scholar]
- 30. Jacobson M. R., Premakumar R., Bishop P. E. 1986. Transcriptional regulation of nitrogen fixation by molybdenum in Azotobacter vinelandii. J. Bacteriol. 167:480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin Z., et al. 2008. Biogenesis of iron-sulfur clusters in photosystem I: holo-NfuA from the cyanobacterium Synechococcussp. PCC 7002 rapidly and efficiently transfers [4Fe-4S] clusters to apo-PsaC invitro. J. Biol. Chem. 283:28426–28435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joerger R. D., Bishop P. E. 1988. Bacterial alternative nitrogen fixation systems. Crit. Rev. Microbiol. 16:1–14 [DOI] [PubMed] [Google Scholar]
- 33. Joerger R. D., Bishop P. E. 1988. Nucleotide sequence and genetic analysis of the nifB-nifQregion from Azotobacter vinelandii. J. Bacteriol. 170:1475–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joerger R. D., Jacobson M. R., Premakumar R., Wolfinger E. D., Bishop P. E. 1989. Nucleotide sequence and mutational analysis of the structural genes (anfHDGK) for the second alternative nitrogenase from Azotobacter vinelandii. J. Bacteriol. 171:1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joerger R. D., et al. 1990. Nucleotide sequences and mutational analysis of the structural genes for nitrogenase 2 of Azotobacter vinelandii. J. Bacteriol. 172:3400–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Joerger R. D., Premakumar R., Bishop P. E. 1986. Tn5-induced mutants of Azotobacter vinelandiiaffected in nitrogen fixation under Mo-deficient and Mo-sufficient conditions. J. Bacteriol. 168:673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Joerger R. D., Wolfinger E. D., Bishop P. E. 1991. The gene encoding dinitrogenase reductase 2 is required for expression of the second alternative nitrogenase from Azotobacter vinelandii. J. Bacteriol. 173:4440–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247–281 [DOI] [PubMed] [Google Scholar]
- 39. Johnson D. C., Dos Santos P. C., Dean D. R. 2005. NifU and NifS are required for the maturation of nitrogenase and cannot replace the function of isc-gene products in Azotobacter vinelandii. Biochem. Soc. Trans. 33:90–93 [DOI] [PubMed] [Google Scholar]
- 40. Kaminski P. A., et al. 1988. Characterization of the fixABCregion of Azorhizobium caulinodansORS571 and identification of a new nitrogen fixation gene. Mol. Gen. Genet. 214:496–502 [DOI] [PubMed] [Google Scholar]
- 41. Kennedy C., Dean D. 1992. The nifU, nifSand nifVgene products are required for activity of all three nitrogenases of Azotobacter vinelandii. Mol. Gen. Genet. 231:494–498 [DOI] [PubMed] [Google Scholar]
- 42. Kennedy C., Rudnick P., MacDonald T., Melton T. 2005. Genus Azotobacter, p. 384–401In Garrity G. M.(ed.), Bergey's manual of systematic bacteriology, vol. 2, part B Spring Verlag, New York, NY [Google Scholar]
- 43. Kennedy C., Toukdarian A. 1987. Genetics of azotobacters: applications to nitrogen fixation and related aspects of metabolism. Annu. Rev. Microbiol. 41:227–258 [DOI] [PubMed] [Google Scholar]
- 44. Kim J., Rees D. C. 1992. Structural models for the metal centers in the nitrogenase molybdenum-iron protein. Science 257:1677–1682 [DOI] [PubMed] [Google Scholar]
- 45. Ludwig M., Bryant D. A. 2011. Transcription profiling of the model cyanobacterium Synechococcussp. strain PCC 7002 by Next-Gen (SOLiDTM) sequencing of cDNA. Front. Microb. Physiol. Metab. 2:41.doi:10.3389/fmicb.2011.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maier R. J., Moshiri F. 2000. Role of the Azotobacter vinelandiinitrogenase-protective shethna protein in preventing oxygen-mediated cell death. J. Bacteriol. 182:3854–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mouncey N. J., Mitchenall L. A., Pau R. N. 1995. Mutational analysis of genes of the modlocus involved in molybdenum transport, homeostasis, and processing in Azotobacter vinelandii. J. Bacteriol. 177:5294–5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oda Y., et al. 2005. Functional genomic analysis of three nitrogenase isozymes in the photosynthetic bacterium Rhodopseudomonas palustris. J. Bacteriol. 187:7784–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peters J. W., Fisher K., Dean D. R. 1995. Nitrogenase structure and function: a biochemical-genetic perspective. Annu. Rev. Microbiol. 49:335–366 [DOI] [PubMed] [Google Scholar]
- 50. Poole R. K., Hill S. 1997. Respiratory protection of nitrogenase activity in Azotobacter vinelandii—roles of the terminal oxidases. Biosci. Rep. 17:303–317 [DOI] [PubMed] [Google Scholar]
- 51. Premakumar R., Jacobson M. R., Loveless T. M., Bishop P. E. 1992. Characterization of transcripts expressed from nitrogenase-3 structural genes of Azotobacter vinelandii. Can. J. Microbiol. 38:929–936 [DOI] [PubMed] [Google Scholar]
- 52. Premakumar R., Pau R. N., Mitchenall L. A., Easo M., Bishop P. E. 1998. Regulation of the transcriptional activators AnfA and VnfA by metals and ammonium in Azotobacter vinelandii. FEMS Microbiol. Lett. 164:63–68 [DOI] [PubMed] [Google Scholar]
- 53. Raymond J., Siefert J. L., Staples C. R., Blankenship R. E. 2004. The natural history of nitrogen fixation. Mol. Biol. Evol. 21:541–554 [DOI] [PubMed] [Google Scholar]
- 54. Rubio L. M., Ludden P. W. 2008. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62:93–111 [DOI] [PubMed] [Google Scholar]
- 55. Rüttimann-Johnson C., Rubio L. M., Dean D. R., Ludden P. W. 2003. VnfY is required for full activity of the vanadium-containing dinitrogenase in Azotobacter vinelandii. J. Bacteriol. 185:2383–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schmehl M., et al. 1993. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241:602–615 [DOI] [PubMed] [Google Scholar]
- 57. Seefeldt L. C., Arp D. J. 1986. Purification to homogeneity of Azotobacter vinelandiihydrogenase: a nickel and iron containing αβ dimer. Biochimie 68:25–34 [DOI] [PubMed] [Google Scholar]
- 58. Seefeldt L. C., Hoffman B. M., Dean D. R. 2009. Mechanism of Mo-dependent nitrogenase. Annu. Rev. Biochem. 78:701–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Setubal J. C., et al. 2009. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J. Bacteriol. 191:4534–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smith A. D., et al. 2005. NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry 44:12955–12969 [DOI] [PubMed] [Google Scholar]
- 61. Soboh B., Boyd E. S., Zhao D., Peters J. W., Rubio L. M. 2010. Substrate specificity and evolutionary implications of a NifDK enzyme carrying NifB-co at its active site. FEBS Lett. 584:1487–1492 [DOI] [PubMed] [Google Scholar]
- 62. Stal L. J., Krumbien W. E. 1985. Nitrogenase activity in the non-heterocystous cyanobacterium Oscillatoriasp. grown under alternating light-dark cycles. Arch. Microbiol. 143:67–71 [Google Scholar]
- 63. Tabor H., Mehler A. H., Hayaishi O., White J. 1952. Urocanic acid as an intermediate in the enzymatic conversion of histidine to glutamic and formic acids. J. Biol. Chem. 196:121–128 [PubMed] [Google Scholar]
- 64. Walmsley J., Toukdarian A., Kennedy C. 1994. The role of regulatory genes nifA, vnfA, anfA, nfrX, ntrC, and rpoNin expression of genes encoding the three nitrogenases of Azotobacter vinelandii. Arch. Microbiol. 162:422–429 [DOI] [PubMed] [Google Scholar]
- 65. Wolfinger E. D., Bishop P. E. 1991. Nucleotide sequence and mutational analysis of the vnfENXregion of Azotobacter vinelandii. J. Bacteriol. 173:7565–7572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zheng L., White R. H., Dean D. R. 1997. Purification of the Azotobacter vinelandiinifV-encoded homocitrate synthase. J. Bacteriol. 179:5963–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.