Abstract
This study investigated a metabolic network (MN) from Mycobacterium vanbaaleniiPYR-1 for polycyclic aromatic hydrocarbons (PAHs) from the perspective of structure, behavior, and evolution, in which multilayer omics data are integrated. Initially, we utilized a high-throughput proteomic analysis to assess the protein expression response of M. vanbaaleniiPYR-1 to seven different aromatic compounds. A total of 3,431 proteins (57.38% of the genome-predicted proteins) were identified, which included 160 proteins that seemed to be involved in the degradation of aromatic hydrocarbons. Based on the proteomic data and the previous metabolic, biochemical, physiological, and genomic information, we reconstructed an experiment-based system-level PAH-MN. The structure of PAH-MN, with 183 metabolic compounds and 224 chemical reactions, has a typical scale-free nature. The behavior and evolution of the PAH-MN reveals a hierarchical modularity with funnel effects in structure/function and intimate association with evolutionary modules of the functional modules, which are the ring cleavage process (RCP), side chain process (SCP), and central aromatic process (CAP). The 189 commonly upregulated proteins in all aromatic hydrocarbon treatments provide insights into the global adaptation to facilitate the PAH metabolism. Taken together, the findings of our study provide the hierarchical viewpoint from genes/proteins/metabolites to the network via functional modules of the PAH-MN equipped with the engineering-driven approaches of modularization and rationalization, which may expand our understanding of the metabolic potential of M. vanbaaleniiPYR-1 for bioremediation applications.
INTRODUCTION
With the 2010 Deepwater Horizon BP oil spill in the Gulf of Mexico (http://www.epa.gov/BPSpill), concerns have been raised regarding the effect of polycyclic aromatic hydrocarbons (PAHs) on the environment. PAHs are a diverse class of organic compounds with two or more fused benzene rings (4). These compounds are highly hydrophobic and not easily bioavailable to microorganisms for degradation, and they pose a significant toxicological risk to human and environmental health (4). Microbial activities represent one of the primary processes by which PAHs are eliminated from the environment (4).
Mycobacterium vanbaaleniiPYR-1, originally isolated from oil-contaminated estuarine sediment, was the first bacterium reported to degrade high-molecular-weight (HMW) PAHs with four or more fused benzene rings (10, 18). Since it has the ability to mineralize or degrade various kinds of PAHs, such as phenanthrene, anthracene, fluoranthene, pyrene, benzo[a]pyrene, benz[a]anthracene, and 7,12-dimethylbenz[a]anthracene (10, 11, 15–17, 24, 34–38), strain PYR-1 has been studied extensively as a prototype organism to elucidate pathways (19) and has been used to remediate PAH-contaminated soils (31). Recently, with the completion of the genome sequence of M. vanbaaleniiPYR-1, efforts to elucidate the molecular background for the metabolism of PAHs have been initiated (21, 22, 26).
Recently, a global biodegradation network has been reconstructed from public resources independently of the microbial hosts (40). In this study, we conducted research on the metabolism of PAHs by M. vanbaaleniiPYR-1 from the standpoint of a metabolic network (MN). We used high-throughput one-dimensional (1D) gel electrophoresis coupled to nanoflow-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) approaches to yield large-scale proteome data for the response of strains PYR-1 to seven aromatic hydrocarbons, phthalate, fluorene, acenaphthylene, anthracene, phenanthrene, pyrene, and benzo[a]pyrene, which well represent characteristics of aromatic hydrocarbons degraded by strain PYR-1 in terms of their structure and metabolism. The whole-cell proteome results then were combined with previous metabolic, biochemical, physiological, and genomic information (20–22, 29) to reconstruct an experimental evidence-driven PAH-MN for M. vanbaaleniiPYR-1. We analyzed the network from the viewpoints of structure, behavior, and evolution, which provided a system-wide perspective on the biodegradation of PAHs and insights on how M. vanbaaleniiPYR-1 satisfies the demands for unusual metabolic capability toward HMW PAHs.
MATERIALS AND METHODS
Bacterial strain, chemicals, and culture conditions.
M. vanbaaleniiPYR-1 (DSM 7251) was used in this study. Phthalate, fluorene, acenaphthylene, anthracene, phenanthrene, pyrene, and benzo[a]pyrene were purchased from Sigma-Aldrich (St. Louis, MO). A 100-ml aliquot of the initial culture was grown for a week in 7H9 Middlebrook broth. The culture was washed, resuspended, and transferred into 250-ml flasks containing 50 ml of phosphate-based minimal (PBM) medium (23) supplemented with 0.5% sorbitol and further incubated with shaking at 200 rpm for 5 days at 28°C. Aromatic hydrocarbons were dissolved in dimethyl sulfoxide (DMSO) and added to the flasks (final concentration, 25 μM). The cells were induced two more times by repeated additions of aromatic hydrocarbons at 24-h intervals. The control culture had the same incubation conditions, except that only DMSO was added.
SDS-PAGE and in-gel digestion.
After 6 h of the third induction, the cells were harvested by centrifugation. Protein extracts were prepared by homogenization using glass beads as described previously (22). Twenty μg of each sample was resolved on a 1.5-mm-thick, 4 to 12% Bis-Tris SDS gel (Invitrogen, Carlsbad, CA), and the separation was duplicated. The SDS-PAGE gels were visualized by staining with SimplyBlue (Invitrogen). Each lane was cut into 20 bands, and the duplicate bands then were combined. The bands were destained overnight in 25 mM NH4HCO3–50% acetonitrile. After destaining, the gel bands were digested with 10 ng/μl of trypsin (Promega, Madison, WI) overnight at 37°C, and the peptides were extracted by 70% acetonitrile and 5% formic acid with sonication. The extracted peptides were lyophilized to dryness.
Nano-LC-MS/MS analysis.
The peptides extracted from each gel band were analyzed by reversed-phase nano-LC-MS/MS (RP nano-LC-MS/MS). RP nano-LC-MS/MS was performed using an Agilent 1200 nano-LC system (Agilent Technologies) coupled on line to a linear ion trap mass spectrometer (LTQ XL; Thermo Electron). Reverse-phase separation was performed using a fused silica capillary column (75-μm inner diameter, 360-μm outer diameter, 9 cm long; Polymicro Technologies, Phoenix, AZ) that was slurry packed with Jupiter 5-μm, 300-Å-pore-size C18silica-bonded stationary phase (Phenomenex, Torrance, CA). Each sample was dissolved in 20 μl of 0.1% formic acid, and 5 μl was injected into the RP column. During sample loading, the column was held for 30 min with 98% of solvent A (0.1% [vol/vol] formic acid in water). Peptides were eluted at a flow rate of 0.25 μl/min using a step linear gradient of 2 to 42% solvent B (0.1% [vol/vol] formic acid in acetonitrile) for 40 min and 42 to 98% solvent B for 10 min, followed by 98% solvent B for 5 min. The mass spectrometer was operated in a data-dependent mode, in which each full MS scan was followed by seven MS/MS scans, where the seven most abundant peptide molecular ions were dynamically selected from the prior MS scan for collision-induced dissociation (CID) using a normalized collision energy of 35%.
Protein identification and quantitation.
The raw MS/MS data were searched using SEQUEST, running under BioWorks (Rev. 3.3.1 SP1) (Thermo Electron, San Jose, CA), against the M. vanbaaleniiPYR-1 protein database with the addition of a PhtAc protein sequence to identify peptides. A peptide mass tolerance of 2 Da and fragment ion tolerance of 1 Da were allowed with tryptic specificity allowing two missed cleavages. SEQUEST criteria were a cross-correlation score of ≥1.7 for [M+H]1+ions, ≥2.5 for [M + 2H]2+ions, and ≥3.2 for [M + 3H]3+ions (P≤ 0.01 for the identification of fully tryptic peptides). The false-positive rate in peptide identification was evaluated to be less than 1% by a SEQUEST search of the database comprising forward and reverse protein sequences. The peak areas of identified peptides were calculated using BioWorks' PepQuan module (Thermo Electron, San Jose, CA), by which peak areas were integrated from their extracted ion chromatograms (XIC) using a minimum intensity threshold of 50,000 counts, mass tolerance of 1.5 Da, and smoothing point of 5. Each peptide area was normalized to the total area of all peptides identified from the control sample or each treatment and expressed as parts per million (ppm). The areas of unique peptides from the same protein were summed to represent the abundance of that protein.
Bioinformatic and statistical analysis.
Some of the proteins identified in the proteome were further determined with respect to function using a BlastP search. Subcellular localizations of identified proteins were predicted using PSORTb ver. 2.0.4 (http://psort.org). The log Pvalues, which measure the water solubility of the compounds, were calculated by MarvinSketch (version 4.1.8). Relative protein expression levels against the control were normalized by using the following logarithm base 2 form: log2(treatment expression/control expression). If the expression in the controls was not detectable but was measurable in treatments, we arbitrarily set the expression level to 5; on the other hand, if the expression level in treatment is not detectable, we set it to −5. For the hierarchical clustering analysis with the average linkage method, the dissimilarity measurement between proteins was calculated as 1 − r, where ris the pairwise Pearson correlation of protein expression profiles. Afterwards, the number of clusters (c) was determined by a combined assessment with external criteria R2, pseudo-F, and pseudo-T2statistics, which all are based on between- and within-cluster sums of squares. Here, R2is a measure of the quality of a partition. In search of an optimal number of clusters, we look for local peaks of the pseudo-Fcombined with a small value of the pseudo-T2statistic and a larger pseudo-T2value for the next fusion. R2= 1 − (SSwithin)/(SStotal) = (SSbetween/SStotal) is related to the proportion of the total variation explained by clusters, where SSwithinstands for the within-cluster sum of squares, SSbetweenstands for the between-cluster sum of squares, and SStotalstands for the total sum of squares. Pseudo-F= [SSbetween/(c− 1)]/[SStotal/(n− c)] mimics the conventional Fstatistic, where nis the total number of observations. Unlike the other two criteria, pseudo-T2= [(SSwithin_t− SSwithin_r− SSwithin_s)(nr+ ns− 2)]/(SSwithin_r+ SSwithin_s) monitors every step of fusion more directly, where rand sdenote two merged old clusters while trepresents the merged cluster. The pseudo-Fand pseudo-T2statistics that performed the best out of 30 methods for estimating the number of clusters in the simulation study by Cooper and Milligan (5) and Milligan and Cooper (33) served as criteria for determining the number of clusters. Based on the clustering analysis, the proteins identified as Mvan numbers were classified by the clusters of orthologous groups (COG) functions. The methods were implemented in R (version 2.4.1; downloadable from http://cran.r-project.org).
RESULTS
Proteome analysis.
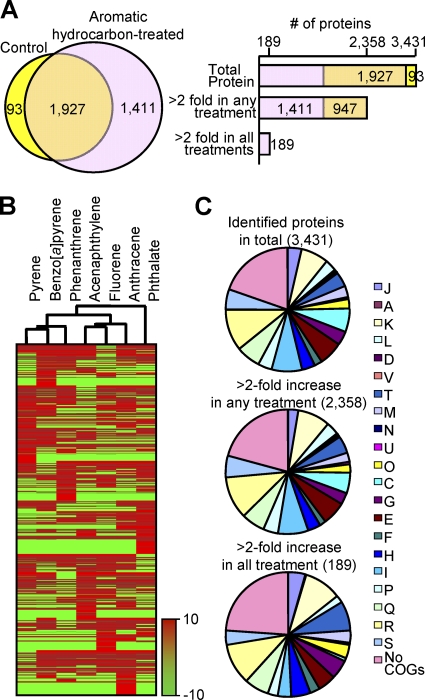
We identified between 2,119 and 2,380 proteins, which added up to 3,431 proteins in total (57.38% of the 5,979 genome-predicted proteins) from both control samples and aromatic hydrocarbon-treated samples treated with phthalate, fluorene, acenaphthylene, anthracene, phenanthrene, pyrene, and benzo[a]pyrene (Table 1; also see Table S1 in the supplemental material). Briefly, as shown in Fig. 1A, 1,927 proteins were shared among the control and seven treatments, with 93 and 1,411 proteins identified only in control and PAH-treated samples, respectively. The comparative analysis of protein expression profiles showed that 2,358 proteins were upregulated more than 2-fold in abundance under at least a single set of growth conditions, and among them, 189 proteins commonly were more than 2-fold more abundant in all aromatic hydrocarbon treatments (see Table S2 in the supplemental material). An overall comparison of protein expression profiles by cluster analysis is shown in Fig. 1B. Figure 1C shows the groupings of these 3,431 proteins according to the COG categories (45) (Table 1). Generally, most of the COGs showed similar percentages of detection in the proteome compared to those of protein-coding genes predicted from the genome of M. vanbaaleniiPYR-1. Identified proteins also were grouped by COG categories based on the culture conditions (see Fig. S1 in the supplemental material).
Table 1.
Summary of identified proteins grouped by COGs
| COG |
No. of proteins predicted from PYR-1 genome | % of genome | No. of identified proteinsa | % of observed proteome | % identified in proteome | No. of proteins showing >2-fold increase in any treatmentb | % of proteins showing >2-fold increase in any treatment | No. of proteins showing >2-fold increase in all treatmentsb | % of proteins showing >2-fold increase in all treatments | |
|---|---|---|---|---|---|---|---|---|---|---|
| Category | Function | |||||||||
| J | Translation, ribosomal structure, and biogenesis | 173 | 2.89 | 134 | 3.91 | 77.46 | 72 | 3.05 | 9 | 4.76 |
| A | RNA processing and modification | 1 | 0.02 | 1 | 0.03 | 100.00 | 1 | 0.04 | 1 | 0.53 |
| K | Transcription | 514 | 8.60 | 280 | 8.16 | 54.47 | 219 | 9.29 | 20 | 10.58 |
| L | Replication, recombination, and repair | 280 | 4.68 | 111 | 3.24 | 39.64 | 85 | 3.60 | 4 | 2.12 |
| D | Cell cycle control, cell division | 32 | 0.54 | 24 | 0.70 | 75.00 | 14 | 0.59 | 0 | 0.00 |
| V | Defense mechanisms | 54 | 0.90 | 20 | 0.58 | 37.04 | 14 | 0.59 | 0 | 0.00 |
| T | Signal transduction mechanisms | 236 | 3.95 | 144 | 4.20 | 61.02 | 113 | 4.79 | 15 | 7.94 |
| M | Cell wall/membrane/envelope biogenesis | 173 | 2.89 | 107 | 3.12 | 61.85 | 72 | 3.05 | 7 | 3.70 |
| N | Cell motility | 9 | 0.15 | 1 | 0.03 | 11.11 | 0 | 0.00 | 0 | 0.00 |
| U | Intracellular trafficking, secretion,, and vesicular transport | 41 | 0.69 | 22 | 0.64 | 53.66 | 11 | 0.47 | 1 | 0.53 |
| O | Posttranslational modification, protein turnover, chaperones | 148 | 2.48 | 95 | 2.77 | 64.19 | 55 | 2.33 | 7 | 3.70 |
| C | Energy production and conversion | 368 | 6.15 | 228 | 6.65 | 61.96 | 142 | 6.02 | 2 | 1.06 |
| G | Carbohydrate transport and metabolism | 238 | 3.98 | 140 | 4.08 | 58.82 | 78 | 3.31 | 10 | 5.29 |
| E | Amino acid transport and metabolism | 421 | 7.04 | 243 | 7.08 | 57.72 | 158 | 6.70 | 10 | 5.29 |
| F | Nucleotide transport and metabolism | 97 | 1.62 | 70 | 2.04 | 72.16 | 45 | 1.91 | 5 | 2.65 |
| H | Coenzyme transport and metabolism | 157 | 2.63 | 130 | 3.79 | 82.80 | 87 | 3.69 | 10 | 5.29 |
| I | Lipid transport and metabolism | 499 | 8.35 | 304 | 8.86 | 60.92 | 206 | 8.74 | 8 | 4.23 |
| P | Inorganic ion transport and metabolism | 311 | 5.20 | 133 | 3.88 | 42.77 | 96 | 4.07 | 5 | 2.65 |
| Q | Secondary metabolite biosynthesis, transport, and catabolism | 527 | 8.81 | 245 | 7.14 | 46.49 | 164 | 6.96 | 13 | 6.88 |
| R | General function prediction only | 841 | 14.07 | 401 | 11.69 | 47.68 | 289 | 12.26 | 22 | 11.64 |
| S | Function unknown | 344 | 5.75 | 200 | 5.83 | 58.14 | 140 | 5.94 | 8 | 4.23 |
| Not in COGs | Unclassified proteins | 1,506 | 25.19 | 751 | 21.89 | 49.87 | 546 | 23.16 | 49 | 25.93 |
| Total | 5,979 | 3,431 | 57.38 | 2,358 | 189 | |||||
Proteins were identified by proteomic analysis in this study.
M. vanbaaleniiPYR-1 was treated with seven aromatic hydrocarbons, which include phthalate, fluorene, acenaphthylene, anthracene, phenanthrene, pyrene, and benzo[a]pyrene.
Fig. 1.
Summary of proteins identified in this proteome study. (A) Venn diagram showing numbers of proteins identified in control samples and aromatic hydrocarbon-treated samples treated with phthalate, fluorene, acenaphthylene, anthracene, phenanthrene, pyrene, and benzo[a]pyrene. The bar graph shows a comparative analysis of protein expression profiles (explained in detail in the text). (B) Heat map of proteomic data sets. Cluster analysis shows similarities of protein expression between samples. (C) Pie chart showing observed proteins based on COG functions. COG category descriptions are provided in Table S1 in the supplemental material.
Out of ∼200 genes involved in the degradation of aromatic hydrocarbons, which were identified previously in the 6.4-Mb genome of strain PYR-1 (21), about 160 genes in this proteome study were expressed as proteins. Among them, 10 ring-hydroxylating oxygenases (RHOs), 20 cytochrome P450 monooxygenases (CYPs), and 10 other monooxygenases were variably expressed, depending on the structural differences of the aromatic substrates. These enzymes were thought to be involved mostly in the initial step of aromatic hydrocarbon oxidation. M. vanbaaleniiPYR-1 incubated with pyrene upregulated at least six RHOs, including NidAB (Mvan_0488/0487) and PhtAaAb (Mvan_0463/0464), whereas only one RHO, PhtAaAb, was upregulated when incubated with phthalate. These observations clearly indicated that the higher the molecular weight and size of the substrate, the more initial oxidation reactions are required and the more RHOs, of three different types (1, 26), are expressed. Interestingly, the phthalate dioxygenase PhtAaAb was identified in all aromatic hydrocarbon-induced conditions except for that of benzo[a]pyrene. The electron transport chain (ETC) of PhtAc and PhtAd (Mvan_0467) was identified in all culture conditions, including the control.
Although many of their physiological roles are not known, the involvement of CYPs in the initial oxidation of PAHs has been demonstrated in M. vanbaaleniiPYR-1 (12). The expression of 20 CYPs out of 50 gene copies found in the genome of strain PYR-1, together with the expression of epoxide hydrolases (Mvan_0521/4998), which are required to form trans-dihydrodiols (12), further supported the possibility of alternative reactions for the oxidation of aromatic hydrocarbons. They include two CYPs, CYP151 (Mvan_1848) and CYP150 (Mvan_4141), whose functions in oxidizing PAHs were experimentally proposed (2). Another CYP, Mvan_2007, was identified as upregulated in all induction conditions; its function should be further characterized. Several types of monooxygenases, including flavin-containing monooxygenases, also were detected and could function in a wide variety of biological processes.
In contrast, a relatively limited number of cis-dihydrodiol dehydrogenases and ring cleavage dioxygenases were identified. Among five dihydrodiol dehydrogenase genes in the genome of M. vanbaaleniiPYR-1, the genes, Mvan_0466 and Mvan_0544 were detected as proteins. However, since the specific activity of PhtB (Mvan_0466) is for 3,4-dihydroxy-3,4-dihydrophthalate (44), the dehydrogenase Mvan_0544 is believed to cover most dihydrodiol dehydrogenation reactions in strain PYR-1. This protein was upregulated by all aromatic hydrocarbons except for phthalate and benzo[a]pyrene. M. vanbaaleniiPYR-1 is able to support both intra- and extradiol enzymatic ring cleavage of catechol derivatives (22, 29). Of the four identified ring cleavage dioxygenases, PhdI (Mvan_0468) and PcaG (Mvan_0560 and Mvan_0561) were shown previously to be involved in the ring cleavage of 1-hydroxy-2-naphthoate and protocatechuate, respectively (22). The other two ring cleavage dioxygenases, Mvan_0470 and Mvan_0545, appear to catalyze the fission of catechols. These observations suggest that the dihydrodiol dehydrogenases and ring cleavage dioxygenases have extremely broad substrate specificities. Alterations in the abundance of a number of other important PAH metabolic enzymes also were observed, such as hydrolases, hydratase-aldolases, decarboxylases, alcohol (or aldehyde)-dehydrogenases, and enzymes involved in the β-ketoadipate pathway (see Tables S1 and S3 in the supplemental material).
Structure of the PAH-MN.
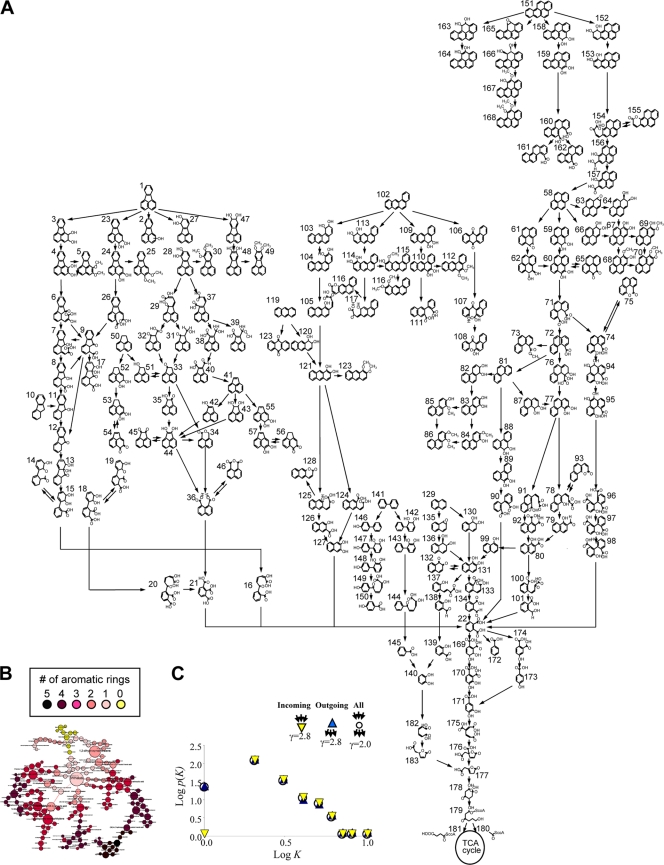
(i) Reconstruction of PAH-MN.A PAH-MN was reconstructed based on the PAH metabolites of M. vanbaaleniiPYR-1 (Fig. 2A and B; see Fig. S2 and Table S3 in the supplemental material). An initial set of 183 PAH metabolites identified from the metabolism of 10 aromatic hydrocarbons, which are biphenyl, naphthalene, acenaphthylene, acenaphthene, anthracene, phenanthrene, pyrene, fluoranthene, benz[a]anthracene, and benzo[a]pyrene, by strain PYR-1 (15, 22, 24, 29, 34–37) was mapped to produce a rudimentary metabolite-centric network with 183 nodes and 224 edges.
Fig. 2.
PAH-MN in M. vanbaaleniiPYR-1 (183 nodes and 224 edges). (A) The reconstructed PAH-MN of M. vanbaaleniiPYR-1. Names of PAHs and their metabolites are given in Table S3 in the supplemental material. (B) Scale-free PAH-MN showing the highlighted larger-sized hubs. The color and size of the nodes indicate benzene ring number and connectivity, respectively. For example, the main hub node, phthalate, which shows high connectivity with an in-degree of 7 and out-degree of 3, was shown to be the biggest yellow node in size and color. (C) Log-log plots showing the number of compounds versus connectivity. Connections involved either as a substrate or as a product are counted. The exponents of the power law distribution (γ) are shown. k, connectivity; P(k), number of compounds. Cytoscape (http://www.cytoscape.org/) was used for network analysis and visualization.
(ii) A metabolite-centric view.Many of the complex networks that occur in nature share global structural features. The analysis of the PAH-MN suggests it has scale-free properties. The log-log plots of probability distribution P(k) versus the interactions kshow a linear correlation, convincingly indicating that the degree of distribution follows a power law, P(k) ≈ k−γ, with γin or out= 2.8, which is similar to that of many other metabolic networks (Fig. 2C). The power law connectivity of PAH-MN implies that a few nodes dominate the overall connectivity. PAH-MN has a network diameter of 24, the largest distance between two substrates, with an average shortest path length of 7.2, which is similar to that of other metabolic networks (13, 40). As shown in Fig. 2A (also see Fig. S2A in the supplemental material), PAH-MN had an apparent funnel-like structure in which peripheral pathways converge to a central aromatic pathway via a common intermediate, phthalate. The pyrene and fluoranthene subnetworks are linked together via phthalate, which also connected with the benz[a]anthracene and anthracene branches. The degree of analysis of all nodes of PAH-MN gave a top rank to phthalate of 10 degrees (see Fig. S2B in the supplemental material), with a 2.4 degree average. Phthalate shows high connectivity, with an in-degree of 7 and out-degree of 3, and a low clustering coefficient, a typical property of hub nodes in the network. Phthalate is a common metabolic intermediate during phenanthrene, pyrene, and fluoranthene metabolism in strain PYR-1 (17, 22, 43, 44).
Behavior of the PAH-MN.
(i) Functional modules.Cellular networks are known to be organized into functionally separable modules (9, 41). To better understand the modularity and organization principle of PAH-MN (30), we identified functional modules by rational decomposition based on the functions of network components (42).
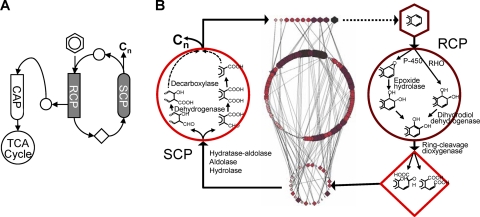
The 224 reactions of the PAH-MN were grouped into three functional categories based on the input/output compounds and the mode of reaction in the following order of occurrence: (i) oxidoreduction of the aromatic substrates to produce catechols and ring cleavage products; (ii) hydrolysis of the α-keto side chain of the ring-cleaved compounds to form a biological precursor (pyruvate) and metabolites with an aldehyde group, which should be removed through oxidation/decarboxylation in the following reaction; and (iii) CoA transfer and subsequent degradative thiolase reactions to form acetyl-CoA and succinyl-CoA. These categories were defined as three functional processes: ring cleavage process (RCP), side chain process (SCP), and central aromatic process (CAP), respectively (Fig. 3and Table 2). The repeating pattern of the chemical properties during the degradation process further supports the functional modules (Fig. 4). It also indicates that the PAH-MN follows the common metabolic logic, with the activation of the thermodynamically stable benzene rings, ring cleavage, side chain removal, and production of biological metabolic precursors.
Fig. 3.
(A and B) General scheme of PAH metabolism in M. vanbaaleniiPYR-1. Hexagons, circles, and rhombuses indicate starting PAH, intermediates, and ring cleavage metabolites, respectively. In RCP, aromatic hydrocarbons are mono-oxygenated with the aid of epoxide hydrolase or dioxygenated to dihydrodiols, which are ring cleaved via the corresponding catechols. The output of the RCP, ring cleavage compounds (rhombus), then go through SCP, which converts them to metabolic intermediates (circle or hexagon), which are ready to enter another round of RCP or CAP and produce active biological precursors. SCP shows various numbers of steps, from one to more than five, in the functional order of (i) the hydroxylation of side chains of the ring cleavage compounds, (ii) the oxidation of the hydroxylation-generated aldehyde group to a carboxylic acid, and (iii) the decarboxylation of the carboxyl group from the aromatic nucleus. The functional linearity for the rearrangement of metabolites to enter another round of RCP or CAP results in the relatively low in- and out-degrees of the metabolic compounds. Because more benzene rings may still remain, RCP and SCP reiterate until the pathways produce an intermediate to enter CAP. The number of repetitions depends on the number of benzene rings of the HMW PAHs. Diverse PAHs are transformed into the common metabolic intermediate protocatechuate, which then is funneled into CAP. The CAP consists of a series of reactions for the conversion of protocatechuate to small aliphatic compounds, which can directly enter central metabolism. In M. vanbaaleniiPYR-1, the β-ketoadipate pathway functions for the completion of the pathway into the TCA cycle intermediates. Please refer to articles by Kim et al. (21) and Kweon et al. (29) for more information about the genes and enzymes in detail.
Table 2.
Characteristics of functional processes for the degradation of HMW PAHs
| Parameter | Characteristics of: |
||
|---|---|---|---|
| RCP | SCP | CAP | |
| Input | (HMW) PAHs | Ring cleavage compounds | Protocatechuate |
| Output | Ring cleavage compounds | Pyruvate | Acetyl-CoA/succinyl-CoA |
| Metabolites acceptable to RCP | |||
| Metabolite centric | Relatively highly branched pathways | Linear or less branched pathways | Partial RCP and SCP |
| High-degree metabolites | Low-degree metabolites | Linear pathway | |
| Gradual increase in MW | Gradual decrease in MW | ||
| Enzyme centric | O2dependent | Productive | Productive |
| Non-productive metabolites | Constitutively/loosely regulated | Constitutive expression | |
| Tightly regulated | Generally broad substrate specificities | Entering TCA cycle | |
| Diverse substrate specificity | Preparative for RCP or CAP | Belong to EC 1-5 | |
| Pathway determining | Belong to EC 1-5 | ||
| Preparative for SCP | |||
| Mostly belong to EC 1 | |||
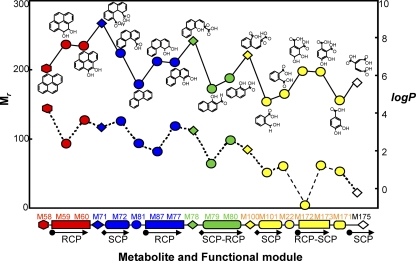
Fig. 4.
Relationships between nodes (chemical compounds) and their molecular weights (Mr) and water solubility (log P) on the network. Modularity does not always guarantee clear-cut subnetworks linked in well-defined ways, but there is a high degree of overlap and cross-talk between modules (8). As degradation proceeds, metabolites repeatedly move up and down in molecular weight and hydrophobicity, which is in accordance with the functional modules in the degradation process. This repeating pattern of the chemical properties over the degradation process strongly supports the concept of functional modules in the PAH-MN of M. vanbaaleniiPYR-1. Hexagons, circles, and rhombuses indicate starting PAH, intermediates, and ring cleavage metabolites, respectively. The color denotes the number of aromatic rings: red, 4; blue, 3; green, 2; yellow, 1; white, 0. Numbers with M represent PAHs and their metabolites, which can be found in Fig. 2and Table S4 in the supplemental material. The log Pvalues for the water solubility of the compounds were calculated by MarvinSketch version 4.1.8.
(ii) An enzyme-centric view.To obtain an enzymatic viewpoint based on the functional modules, we correlated metabolites with enzymes via the chemical reactions based on protein expression data. Encouragingly, the expression profiles of the PAH-MN-related enzymes are fully compatible with the PAH-MN. As shown in Fig. 5, the profiles of pyrene and phenanthrene, whose pathways mostly overlap, are clustered together, while the profile of phthalate is broadly grouped with that of the control.
Fig. 5.
Heat map of the proteins involved in the metabolism of aromatic hydrocarbons in M. vanbaaleniiPYR-1. Profiles of protein expression showed a close relationship with functional modules. We correlated three functional modules with the concept of peripheral pathways and central pathways. Clearly, the heat map shows that the regulation of enzyme expression involved in RCP is much tighter that those of SCP and CAP, which additionally supports the idea of a system with three functional modules.
Generally, the functional distribution and expression of the enzymes are not homogeneous in the PAH-MN. All of the enzymes responsible for RCP belong to the group EC 1 (oxidoreductases), with the exception of epoxide hydrolase, which is in EC 3 (hydrolases), whereas SCP and CAP seem to consist of functionally diverse groups of enzymes from EC 1 to EC 5 (Fig. 3and 5). The β-ketoadipate–succinyl-coenzyme A (CoA) transferase (EC 2.8.3.6) performs the penultimate step in CAP, the conversion of β-ketoadipate to β-ketoadipyl-CoA. Overall, the functional distributions and modularity of enzymes satisfied a typical O2-dependent catabolic strategy.
A set of enzymes, including RHOs, CYPs, epoxide hydrolases, cis-dihydrodiol dehydrogenases, and ring cleavage dioxygenases, are responsible for RCP in M. vanbaaleniiPYR-1. Although the process is nonproductive, preparing metabolites only for entering SCP or CAP, it is a crucial process that determines substrate range and pathways (28). As shown in Fig. 2and 5, the oxygenation steps in PAH-MN required more enzymes to completely handle HMW PAHs, since the same suite of enzymes could not serve the isofunctional reactions for substrates with different sizes and architecture with full activity. Actually, the initial enzymes, such as RHOs and CYPs for RCP, showed relatively high genetic and functional redundancy and more complex regulation. Interestingly, there is a clear correlation between the number of aromatic benzene rings and the number of expressed RHOs, whereas CYPs showed no such relationship with the substrates. This finding clearly shows that RHOs are the main enzymes for initial O2activation and further points to how M. vanbaaleniiPYR-1 regulates RHO expression to regiospecifically control the cis-dihydrodiol products with the best conversion rate for the preferential metabolic route of each aromatic substrate. In detail, NidAB (Mvan_0488) is induced only by pyrene and phenanthrene. NidAB regiospecifically oxidizes pyrene to only one metabolite, pyrene cis-4,5-dihydrodiol (28). Since the route through pyrene C-4,5 dioxygenation is a productive pathway, channeled into the tricarboxylic acid (TCA) cycle, but the other route, C-1,2 dioxygenation, forms O-methylated derivatives as dead-end products, the expression and regiospecificity of NidAB with respect to pyrene degradation are of great advantage to M. vanbaaleniiPYR-1. On the other hand, cis-dihydrodiol dehydrogenases and ring cleavage dioxygenases, for the second and third steps of RCP, show low genetic and functional redundancy, indicating their extremely broad substrate specificity.
Functionally diverse enzymes, with numerical abundance and relatively relaxed substrate specificities, are involved in the SCP (Fig. 5; also see Table S4 in the supplemental material). Copies of genes for the beginning steps of SCP, such as hydratase-aldolase and hydrolase, are abundant, and their level of expression is high, enabling the SCP to accept an enormous range of ring cleavage compounds. However, the decarboxylase (Mvan_0543) is an exception to this observation. The genome of strain PYR-1 has only one gene copy, from which it can be inferred that this one enzyme functions for all of the decarboxylation reactions in PAH-MN. All of the enzymes belonging to SCP, including the decarboxylase, appeared to be expressed constitutively, which suggests that their regulation is more relaxed than that of others which belong to the RCP.
Phthalate is the main hub node, showing an in-degree of 7 and out-degree of 3, respectively. All three outgoing routes are channeled into protocatechuate, which has only one out-degree, to β-carboxy-cis,cis-muconate. Therefore, the β-ketoadipate pathway, which has been reported for HMW PAH metabolism, inevitably functions for CAP (22, 29). Through the β-ketoadipate pathway, which consists of reactions of partial RCP and SCP, protocatechuate is converted to TCA cycle intermediates. The proteome shows a coordinated expression of CAP genes arranged as pcaHGBLIJ(21), enhancing the linearity of the metabolic route. The paralogs of β-ketoadipyl-CoA thiolase (pcaF), catalyzing the last step of the pathway, transforming β-ketoadipyl-CoA to succinyl-CoA and acetyl-CoA, also were identified in the proteome (Fig. 5; see Table S4 in the supplemental material).
Overall, the enzyme-centric analysis demonstrates that PAH-MN is organized into three functional modules in a hierarchical modularity. In the structure/function of the functional modules, which operate on inputs that satisfy some constraints, RCP and SCP show an apparent funnel shape, while CAP shows a cylinder shape with the same input and output diameters. Therefore, the hierarchical functional modularity of PAH-MN is in a funnel-shaped structure, which is consistent with its metabolite-centric scale-free characteristics. As exemplified in pyrene metabolism, the coordination of the regulation, substrate specificity, and product regiospecificity of the components at the level of module and network suggests that M. vanbaaleniiPYR-1 has a channel management with an apparent preferred route(s) or pathway(s) for each substrate. However, enzyme-based analysis showed that PAH-MN probably was less tolerant of error(s) than expected from the metabolite-centric topological robustness of the PAH-MN.
Global adaptive response of M. vanbaaleniiPYR-1.
We further analyzed 189 proteins whose expression was commonly upregulated by the presence of aromatic hydrocarbons. Although it only represents 3.2% of the annotated PYR-1 genes (5,979 open reading frames) and 5.5% of the observed proteome, the agreement of upregulation among seven independent measurements enhances the confidence in the expression of changes of these proteins upon treatment with aromatic hydrocarbons, which significantly contributed to the elucidation of the adaptation mechanisms in the context of PAH-MN.
A classification of the 189 proteins based on the annotated COGs is listed in Table S2 in the supplemental material. The percentages of most of the COGs were similar in the whole proteome (Fig. 1C), which shows that the treatment of aromatic hydrocarbons influenced not only their metabolism but also the global response of the cellular process. Results of these 189 protein location predictions also showed that that the largest number of proteins belongs to the cytoplasm, but that those located in the cell membrane or cell wall also were found. Among 189 proteins, we associated 105 proteins with the metabolism of aromatic hydrocarbons, which include 26 proteins related to the signal transduction/transcriptional regulation of COGs (see Table S5 in the supplemental material). For the remaining 84 proteins, 4 were catabolic enzymes, which we mentioned with other enzymes in the pathways of aromatic hydrocarbon degradation. Functions of the other 80 proteins were not clear.
We identified the upregulation of two ABC transporter-related proteins (Mvan_2845/5577) and a membrane translocase-related protein (Mvan_6075) which could affect the uptake of aromatic substrates or other substrates. Permease-associated proteins (Mvan_2277/2474) also were found when PYR-1 is cultivated with aromatic hydrocarbons except for phthalate. Thus far, no transport systems for the entry of hydrophobic substrates into cells have been identified. We also identified a considerable number of proteins (15 proteins) commonly upregulated which have predicted cellular functions associated with cell wall/membrane organization and biosynthesis. Adaptation mechanisms have been well observed with strains of environmental microorganisms in response to the poor aqueous solubility of aromatic hydrocarbon substrates, which include PAH bioavailability-enhancing mechanisms (14) such as the modification of the cell wall mycolic acid composition (48), production of biosurfactants (7), and alteration of membrane fluidity and permeability by changing membrane lipid composition (47).
Defense or detoxification-related proteins also have been identified in the proteome, which include several methyltransferases listed as being related to cell wall biogenesis and secondary metabolites. The toxicity of aromatic hydrocarbons is well established as a result of the production of aldehyde- and hydroxyl-quinone intermediates (24, 46). M. vanbaaleniihas been shown to have strategies to neutralize the potential toxic effect of catechols and o-quinones, which have been exemplified by catechol O-methyltransferase and o-quinone reductases (23, 25). We have identified a putative GCN5-related N-acetyltransferase (Mvan_3906) which is known to modify toxic phenolic compounds in Bradyrhizobium japonicum(39).
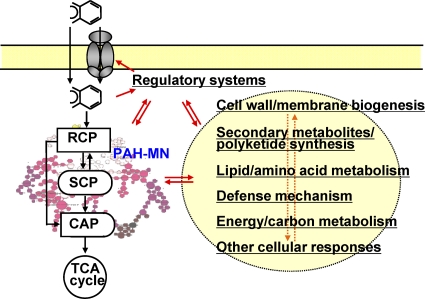
Based on these commonly upregulated proteins, we obtained a picture depicting basic cellular response processes influenced by aromatic hydrocarbons (Fig. 6). Once aromatic hydrocarbons enter into the cell, regulatory processes affect not only the PAH-MN but also various other cellular responses related to cell wall/envelope biosynthesis, membrane transporters, the production of secondary metabolites, signal transduction/transcriptional regulation, defense, or detoxification-related proteins. All of these responses are interconnected and affect each other and likely are involved in the enhancement of the efficiency of aromatic hydrocarbon degradation.
Fig. 6.
Predicted cellular processes influenced by the growth of M. vanbaaleniiPYR-1 with aromatic hydrocarbons based on the commonly upregulated proteins.
At least 33 proteins annotated as hypothetical or whose functions were not categorized in COGs were found to be upregulated under aromatic hydrocarbon incubation states. Some of these proteins, encoded by Mvan_1099, Mvan_3829, and Mvan_5946 genes, increased 7-, 50-, and 19-fold, respectively, suggesting that they have an important role in the adaptive response to aromatic hydrocarbons.
Evolution of the PAH-MN.
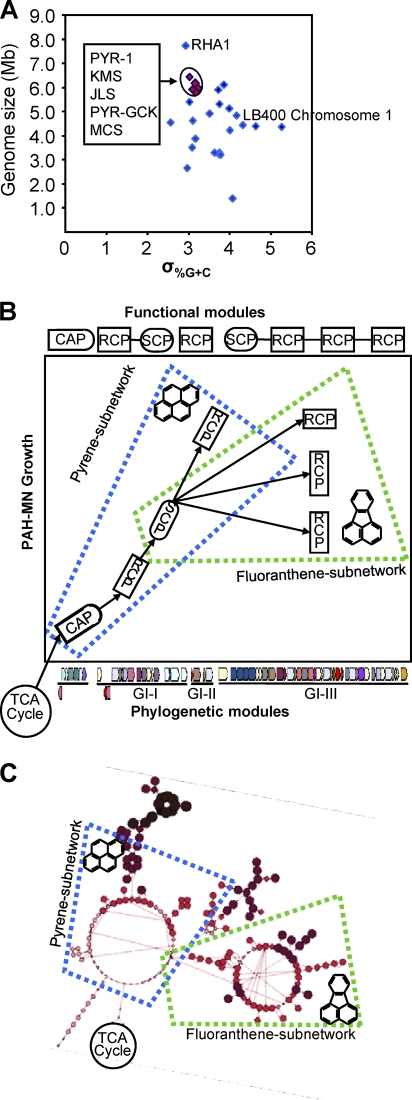
Phylogenetic network modules are evolutionarily conserved functional units in the metabolic network (49). To trace a probable pathway for network evolution, we investigated the structures of evolutionary modules and their relationships to functional modules, which would explain PAH-MN building principles. As suggested by McLeod et al. (32), we analyzed the horizontal gene transfer (HGT)-acquired catabolic genomic islands (GIs) and the standard deviations (σ%G+C) to determine their contributions to the evolution of PAH metabolism and to analyze the degree to which recent HGT shaped a genome. We analyzed 23 aromatic hydrocarbon-degrading bacteria whose genomes are available, including four mycobacteria closely related to M. vanbaaleniiPYR-1 (Fig. 7A; also see Table S6 in the supplemental material). Interestingly, all five PAH-degrading mycobacteria, isolated from geographically diverse locations, shared the conserved ≈150-kb catabolic gene cluster with slightly different genetic configurations, with the value of σ%G+Cranging from 2.99 to 3.17, indicating that they occasionally have acquired the gene cluster by one ancient recent HGT event from a common ancestor rather than by vertical descent, promoting rapid pathway evolution that likely confers immediate selective advantages to the recipients.
Fig. 7.
Evolution of PAH-MN. (A) Genome size versus σ%G+Cfor 24 replicons from 23 aromatic hydrocarbon-degrading bacteria (http://www.pathogenomics.sfu.ca/islandpath/update/IPindex.pl). The M. vanbaaleniiPYR-1 replicon and those of the four PAH-degrading mycobacteria show relatively low σ%G+Cvalues, predicting that they have undergone relatively little HGT. Notably, the σ%G+Cof M. vanbaaleniiPYR-1 is similar to that of the Gram-positive polychlorinated bisphenyl (PCB) degrader Rhodococcus jostiiRHA1 but is smaller than that of another PCB degrader, Burkholderia xenovoransLB400. The 23 bacteria are listed in Table S6 in the supplemental material. (B and C) The relationships between phylogenetic and functional modules and the growth of PAH-MN. The genomic island prediction was analyzed using IslandViewer (http://www.pathogenomics.sfu.ca/islandviewer/query.php).
The further analysis of the conserved region suggests that the catabolic genes have been acquired from separate HGT events from neighbors with close physiological, biochemical, and phylogenetic relationships. We found three GIs in the conserved region, which contains ≈57% (85 kb) of an ≈150-kb catabolic gene cluster, that showed high sequence similarity to the catabolic genes of nocardioform actinomycetes, such as Terrabacterand Nocardioides(Fig. 7B and C). The first GI (GI-I) contains the genes functioning in RCP and SCP. It includes the type V RHO system, phtAaAb(phthalate dioxygenase). GI-II contains the type V RHO gene, nidAB, that is involved in RCP (26). GI-III contains three oxygenases and other enzymes involved in SCP. Two of these oxygenases are type V, and the other belongs to type X (26). Interestingly, however, genes/operons for CAP were not found in the catabolic regions of the GIs, strongly suggesting that the CAP operon was not acquired through recent HGT at about the same time as the three GIs. Its acquisition probably precedes the recruitment of the three GIs, which function mainly for the RCP and SCP. Further supporting the hypothesis of separate HGT events is the existence of plasmids harboring GI-II or GI-III in Mycobacteriumsp. strains MCS and KMS. The functional association of phylogenetic modules opens the possibility of hierarchical modularity evolution (41) in which PAH-MN seems initially to have been elevated from CAP to RCP via SCP. The CAP gene cluster is in a phylogenetically conserved operon structure, suggesting an ancient origin, whereas the ones for the RCP, with an atypical mosaic structure, appear to be young. All of these observations together suggest that initially a common ancestor progressively recruited the phylogenetic modules in a way that functions coordinately, promoting rapid pathway evolution, which enables the mycobacteria to overcome evolutionary pressure to enhance the interactions among the functional modules. In spite of dynamic gene reorganization, the catabolic gene cluster seemed to resist the gene-scrambling events.
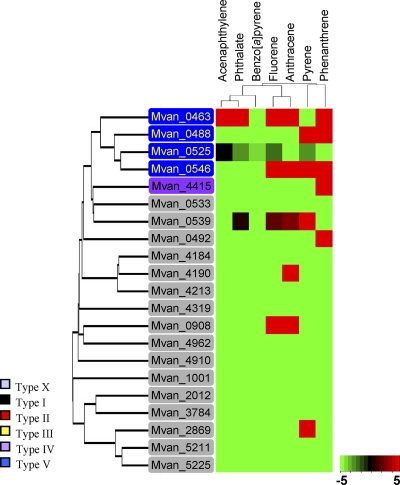
Although such an evolutionary scenario has advantages, the demands for the practical contribution of the phylogenetic modules to the metabolic capacity of the host require more sophisticated functional associations at the level of proteins or modules. Such functional association generally leads to a strong genomic association and evolutionary cohesion, resulting in coexpression or genetic linkage (3). In this respect, of particular interest are the RHO enzymes, due to their extremely low evolutionary cohesiveness and significant numerical imbalance between oxygenases and ETC (21, 26). In M. vanbaaleniiPYR-1, 21 genes encoding oxygenase components were identified, whereas only one copy of the type V ETC genes (phtAcAd) was found. Interestingly, the genes are under relaxed regulation, and PhtAcAd functions with diverse oxygenase components (28). Moreover, as shown in Fig. 8, among 21 oxygenases the 5 oxygenases expressed belong to types V and IV, all of which might be functionally compatible with PhtAcAd. The recruitment of a [3Fe-4S]-type ferredoxin (28) would be functionally and evolutionarily beneficial to M. vanbaaleniiPYR-1.
Fig. 8.
Classification of 21 RHOs of M. vanbaaleniiPYR-1 and their protein expression patterns under PAH treatments. The expressed 10 RHOs were classified as type IV (Mvan_4415), type V (Mvan_0463/0488/0525/0546), and type X (Mvan_0492/0539/0908/2869/4190) based on Kweon's classification (1, 26).
Enzymes in the phylogenetic modules GI-I and GI-II cannot support all of the degradation steps from pyrene to protocatechuate (22) due to the absence of two enzymes, decarboxylase (Mvan_0543) and dihydrodiol dehydrogenase (Mvan_0544). The genes for the enzymes, which show very low genetic and functional redundancy, are found on GI-III, indicating that all enzymes of the three GIs should be together for the complete pyrene subnetwork. In addition, the GI-III RHO enzymes NidA3B3, Mvan_0539/0540, and Mvan_0546/0547, which might function in the lateral or angular dioxygenation of fluoranthene, 9-fluorenone, acenaphthylene, and naphthalene, are essential for the fluoranthene subnetwork. The oxygenases NidA3B3 and Mvan_0539/0540 are upregulated by fluoranthene but not by pyrene, and the enzyme NidA3B3 dioxygenates fluoranthene regiospecifically to fluoranthene cis-2,3-dihydrodiol with the highest conversion rate (29). Therefore, interaction and complementation among the three GIs are necessary for the complete pyrene and fluoranthene subnetworks. The recruitment of metabolic capability toward pyrene and fluoranthene probably has co-occurred, indicating that the two HMW PAHs are the preferential choices of M. vanbaaleniiPYR-1.
DISCUSSION
Since its isolation in 1986, M. vanbaaleniiPYR-1 (10, 18) has been extensively studied with respect to the bacterial metabolism of HMW PAHs (19, 27). The reconstruction of PAH-MN was essentially an effort to organize the dispersed data into an integrated resource for the multiscale mining of chemical and biological data. The proteome was important not only for analyzing cell behavior during aromatic hydrocarbon metabolism but also for bridging the gap between the genome of strain PYR-1 and metabolites with a holistic perspective. The conversion from gene redundancy (genomic view) to functional redundancy (integrated view of proteome and genome) was crucial for a rational decomposition into the functional modules and enzyme-centric interpretation of the PAH-MN. Experimentally founded system-level interpretation of PAH-MN led to new and deepened insight into M. vanbaaleniiPYR-1's metabolic activities for growth despite the extremely low bioavailability of HMW PAHs.
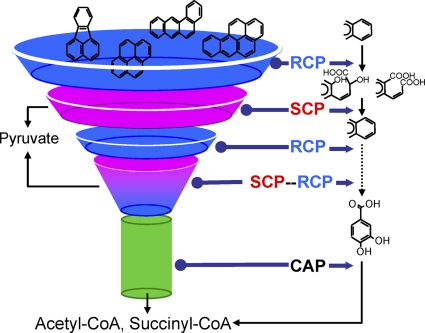
The scale-free architectural features of the PAH-MN are shared to a large degree with other biological networks. The funnel-like topology of MN is intimately related to its behavior and evolution, in which many peripheral pathways converge to a widely conserved central aromatic pathway, the β-ketoadipate pathway. The CAP module for the central aromatic pathway is more conserved in evolution and function, whereas the SCP and RCP modules appeared later in evolution, being able to accomplish their functions with relatively diverse specificity. The structures and functions of the RCP and SCP modules apparently are funnel shaped with a wide conical mouth and a narrow stem, whereas CAP shows almost the same input and output diameters in its function (Fig. 9). The CAP enzymes are relatively loosely regulated and functionally shared, while the function of RCP enzymes is substrate dependent, and expression is tightly regulated (Fig. 7B and C). The enzymes for SCP are dispersed in modularity and redundant in function. The integrated view of structure, behavior, and evolution of PAH-MN suggests how M. vanbaaleniiPYR-1 can benefit from the funnel effect (Fig. 9). The funnel effect and its practical benefits include the following: (i) enhancing input diversity with the controlled production of limited outputs, which concentrates the flux of intermediates to central metabolism; (ii) increasing the connectivity between functional modules, which decreases the epimetabolome (6); (iii) allowing more coordinated regulation; and (iv) enhancing the linearity of metabolic pathways, which reduces metabolite dissipation and ensures a more efficient metabolic flow. Overall, such channel management to maximize the benefits of the funnel effect at the levels of the functional module and network may expand and optimize the catabolic potential of an ≈150-kb catabolic gene cluster of M. vanbaaleniiPYR-1 to exceptional metabolic diversity, including HMW PAHs with the accepted efficiency. The link of new modules, such as an RCP module, to the existing modules in the PAH-MN has merit to expand economically the metabolic capability without affecting the ability of other modules or even causing the malfunction of the whole system.
Fig. 9.
Funnel effect in the structure/function of the functional modules RCP, SCP, and CAP in the PAH-MN.
In conclusion, this study introduces a channel management model of M. vanbaaleniiPYR-1 for global PAH metabolism, along with technical endeavors for mounting and integrating omics data. The hierarchical viewpoint of genes/proteins/metabolites of the network via functional modules of PAH-MN, equipped with the engineering-driven approaches of modularization and rationalization, may guide how we use biodegradation knowledge for practical applications via sound descriptive and predictive modeling.
Supplementary Material
ACKNOWLEDGMENTS
We thank John B. Sutherland and Steven L. Foley for the critical review of the manuscript and Jeff A. Runnells for his assistance with graphics.
This work was supported in part by an appointment to the Postgraduate Research Fellowship Program(D.K.) at the National Center for Toxicological Research, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration. The views presented in this article do not necessarily reflect those of the U.S. FDA.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Baek S., et al. 2009. ClassRHO: a platform for classification of bacterial rieske non-heme iron ring-hydroxylating oxygenases. J. Microbiol. Methods 76:307–309 [DOI] [PubMed] [Google Scholar]
- 2. Brezna B., et al. 2006. Molecular characterization of cytochrome P450 genes in the polycyclic aromatic hydrocarbon degrading Mycobacterium vanbaaleniiPYR-1. Appl. Microbiol. Biotechnol. 71:522–532 [DOI] [PubMed] [Google Scholar]
- 3. Campillos M., von Mering C., Jensen L. J., Bork P. 2006. Identification and analysis of evolutionarily cohesive functional modules in protein networks. Genome Res. 16:374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cerniglia C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 3:351–368 [Google Scholar]
- 5. Cooper M. C., Milligan G. W. 1984. The effect of error on determining the number of cluster. College Administrative Science Working Paper Series. The Ohio State University, Columbus, OH.
- 6. de Lorenzo V. 2008. Systems biology approaches to bioremediation. Curr. Opin. Biotechnol. 19:579–589 [DOI] [PubMed] [Google Scholar]
- 7. Desai J. D., Banat I. M. 1997. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61:47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Han J. D., et al. 2004. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430:88–93 [DOI] [PubMed] [Google Scholar]
- 9. Hartwell L. H., Hopfield J. J., Leibler S., Murray A. W. 1999. From molecular to modular cell biology. Nature 402:C47–C52 [DOI] [PubMed] [Google Scholar]
- 10. Heitkamp M. A., Cerniglia C. E. 1988. Mineralization of polycyclic aromatic hydrocarbons by a bacterium isolated from sediment below an oil field. Appl. Environ. Microbiol. 54:1612–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heitkamp M. A., Cerniglia C. E. 1989. Polycyclic aromatic hydrocarbon degradation by a Mycobacteriumsp. in microcosms containing sediment and water from a pristine ecosystem. Appl. Environ. Microbiol. 55:1968–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heitkamp M. A., Freeman J. P., Miller D. W., Cerniglia C. E. 1988. Pyrene degradation by a Mycobacteriumsp.: identification of ring oxidation and ring fission products. Appl. Environ. Microbiol. 54:2556–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeong H., Tombor B., Albert R., Oltvai Z. N., Barabasi A. L. 2000. The large-scale organization of metabolic networks. Nature 407:651–654 [DOI] [PubMed] [Google Scholar]
- 14. Johnsen A. R., Wick L. Y., Harms H. 2005. Principles of microbial PAH-degradation in soil. Environ. Pollut. 133:71–84 [DOI] [PubMed] [Google Scholar]
- 15. Kelley I., Freeman J. P., Cerniglia C. E. 1990. Identification of metabolites from degradation of naphthalene by a Mycobacteriumsp. Biodegradation 1:283–290 [DOI] [PubMed] [Google Scholar]
- 16. Kelley I., Freeman J. P., Evans F. E., Cerniglia C. E. 1991. Identification of a carboxylic acid metabolite from the catabolism of fluoranthene by a Mycobacteriumsp. Appl. Environ. Microbiol. 57:636–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kelley I., Freeman J. P., Evans F. E., Cerniglia C. E. 1993. Identification of metabolites from the degradation of fluoranthene by Mycobacteriumsp. strain PYR-1. Appl. Environ. Microbiol. 59:800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khan A. A., Kim S. J., Paine D. D., Cerniglia C. E. 2002. Classification of a polycyclic aromatic hydrocarbon-metabolizing bacterium, Mycobacterium sp. strain PYR-1, as Mycobacterium vanbaaleniisp. nov. Int. J. Syst. Evol. Microbiol. 52:1997–2002 [DOI] [PubMed] [Google Scholar]
- 19. Kim S. J., Kweon O., Cerniglia C. E. 2010. Degradation of polycyclic aromatic hydrocarbons by Mycobacteriumstrains, p. 1865–1880 In Timmis K. N. (ed.), Handbook of hydrocarbon and lipid microbiology, vol. 3 Springer, Braunschweig, Germany [Google Scholar]
- 20. Kim S. J., Kweon O., Cerniglia C. E. 2009. Proteomic applications to elucidate bacterial aromatic hydrocarbon metabolic pathways. Curr. Opin. Microbiol. 12:301–309 [DOI] [PubMed] [Google Scholar]
- 21. Kim S. J., Kweon O., Jones R. C., Edmondson R. D., Cerniglia C. E. 2008. Genomic analysis of polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaaleniiPYR-1. Biodegradation 19:859–881 [DOI] [PubMed] [Google Scholar]
- 22. Kim S. J., et al. 2007. Complete and integrated pyrene degradation pathway in Mycobacterium vanbaaleniiPYR-1 based on systems biology. J. Bacteriol. 189:464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim Y. H., Engesser K. H., Cerniglia C. E. 2003. Two polycyclic aromatic hydrocarbon o-quinone reductases from a pyrene-degrading Mycobacterium. Arch. Biochem. Biophys. 416:209–217 [DOI] [PubMed] [Google Scholar]
- 24. Kim Y. H., Freeman J. P., Moody J. D., Engesser K. H., Cerniglia C. E. 2005. Effects of pH on the degradation of phenanthrene and pyrene by Mycobacterium vanbaaleniiPYR-1. Appl. Microbiol. Biotechnol. 67:275–285 [DOI] [PubMed] [Google Scholar]
- 25. Kim Y. H., Moody J. D., Freeman J. P., Engesser K. H., Cerniglia C. E. 2004. Evidence for the existence of PAH-quinone reductase and catechol-O-methyltransferase in Mycobacterium vanbaaleniiPYR-1. J. Ind. Microbiol. Biotechnol. 31:507–516 [DOI] [PubMed] [Google Scholar]
- 26. Kweon O., et al. 2008. A new classification system for bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. BMC Biochem. 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kweon O., Kim S. J., Cerniglia C. E. 2010. Genomic view of mycobacterial high molecular weight polycyclic aromatic hydrocarbon degradation, p. 1165–1178 In Timmis K. N. (ed.), Handbook of hydrocarbon and lipid microbiology, vol. 2 Springer, Braunschweig, Germany [Google Scholar]
- 28. Kweon O., et al. 2010. Substrate specificity and structural characteristics of the novel Rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaaleniiPYR-1. mBio 1:e135–e110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kweon O., et al. 2007. A polyomic approach to elucidate the fluoranthene degradative pathway in Mycobacterium vanbaaleniiPYR-1. J. Bacteriol. 189:4635–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma H. W., Zhao X. M., Yuan Y. J., Zeng A. P. 2004. Decomposition of metabolic network into functional modules based on the global connectivity structure of reaction graph. Bioinformatics 20:1870–1876 [DOI] [PubMed] [Google Scholar]
- 31. MacLeod C. T., Daugulis A. J. 2003. Biodegradation of polycyclic aromatic hydrocarbons in a two-phase partitioning bioreactor in the presence of a bioavailable solvent. Appl. Microbiol. Biotechnol. 62:291–296 [DOI] [PubMed] [Google Scholar]
- 32. McLeod M. P., et al. 2006. The complete genome of Rhodococcussp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Milligan G. W., Cooper M. C. 1985. An examination of procedures for determining the number of clusters in a data set. Psychometrika 50:159–179 [Google Scholar]
- 34. Moody J. D., Doerge D. R., Freeman J. P., Cerniglia C. E. 2002. Degradation of biphenyl by Mycobacteriumsp. strain PYR-1. Appl. Microbiol. Biotechnol. 58:364–369 [DOI] [PubMed] [Google Scholar]
- 35. Moody J. D., Freeman J. P., Cerniglia C. E. 2005. Degradation of benz[a]anthracene by Mycobacterium vanbaaleniistrain PYR-1. Biodegradation 16:513–526 [DOI] [PubMed] [Google Scholar]
- 36. Moody J. D., Freeman J. P., Doerge D. R., Cerniglia C. E. 2001. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacteriumsp. strain PYR-1. Appl. Environ. Microbiol. 67:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moody J. D., Freeman J. P., Fu P. P., Cerniglia C. E. 2004. Degradation of benzo[a]pyrene by Mycobacterium vanbaaleniiPYR-1. Appl. Environ. Microbiol. 70:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moody J. D., Fu P. P., Freeman J. P., Cerniglia C. E. 2003. Regio- and stereoselective metabolism of 7,12-dimethylbenz[a]anthracene by Mycobacterium vanbaaleniiPYR-1. Appl. Environ. Microbiol. 69:3924–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nienaber A., Huber A., Gottfert M., Hennecke H., Fischer H. M. 2000. Three new NifA-regulated genes in the Bradyrhizobium japonicumsymbiotic gene region discovered by competitive DNA-RNA hybridization. J. Bacteriol. 182:1472–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pazos F., Valencia A., De Lorenzo V. 2003. The organization of the microbial biodegradation network from a systems-biology perspective. EMBO Rep. 4:994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ravasz E., Somera A. L., Mongru D. A., Oltvai Z. N., Barabasi A. L. 2002. Hierarchical organization of modularity in metabolic networks. Science 297:1551–1555 [DOI] [PubMed] [Google Scholar]
- 42. Rives A. W., Galitski T. 2003. Modular organization of cellular networks. Proc. Natl. Acad. Sci. U. S. A. 100:1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stingley R. L., Brezna B., Khan A. A., Cerniglia C. E. 2004. Novel organization of genes in a phthalate degradation operon of Mycobacterium vanbaaleniiPYR-1. Microbiology 150:3749–3761 [DOI] [PubMed] [Google Scholar]
- 44. Stingley R. L., Khan A. A., Cerniglia C. E. 2004. Molecular characterization of a phenanthrene degradation pathway in Mycobacterium vanbaaleniiPYR-1. Biochem. Biophys. Res. Commun. 322:133–146 [DOI] [PubMed] [Google Scholar]
- 45. Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vasiliou V., Pappa A., Petersen D. R. 2000. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem. Biol. Interact. 129:1–19 [DOI] [PubMed] [Google Scholar]
- 47. Wick L. Y., Pelz O., Bernasconi S. M., Andersen N., Harms H. 2003. Influence of the growth substrate on ester-linked phospho- and glycolipid fatty acids of PAH-degrading Mycobacteriumsp. LB501T. Environ. Microbiol. 5:672–680 [DOI] [PubMed] [Google Scholar]
- 48. Wick L. Y., Wattiau P., Harms H. 2002. Influence of the growth substrate on the mycolic acid profiles of mycobacteria. Environ. Microbiol. 4:612–616 [DOI] [PubMed] [Google Scholar]
- 49. Yamada T., Kanehisa M., Goto S. 2006. Extraction of phylogenetic network modules from the metabolic network. BMC Bioinformatics 7:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.