Abstract
The Gram-negative intracellular pathogen Legionella pneumophilareplicates in a membrane-bound compartment known as the Legionella-containing vacuole (LCV), into which it abundantly releases its chaperonin, HtpB. To determine whether HtpB remains within the LCV or reaches the host cell cytoplasm, we infected U937 human macrophages and CHO cells with L. pneumophilaexpressing a translocation reporter consisting of the Bordetella pertussisadenylate cyclase fused to HtpB. These infections led to increased cyclic AMP levels, suggesting that HtpB reaches the host cell cytoplasm. To identify potential functions of cytoplasmic HtpB, we expressed it in the yeast Saccharomyces cerevisiae, where HtpB induced pseudohyphal growth. A yeast-two-hybrid screen showed that HtpB interacted with S-adenosylmethionine decarboxylase (SAMDC), an essential yeast enzyme (encoded by SPE2) that is required for polyamine biosynthesis. Increasing the copy number of SPE2induced pseudohyphal growth in S. cerevisiae; thus, we speculated that (i) HtpB induces pseudohyphal growth by activating polyamine synthesis and (ii) L. pneumophilamay require exogenous polyamines for growth. A pharmacological inhibitor of SAMDC significantly reduced L. pneumophilareplication in L929 mouse cells and U937 macrophages, whereas exogenously added polyamines moderately favored intracellular growth, confirming that polyamines and host SAMDC activity promote L. pneumophilaproliferation. Bioinformatic analysis revealed that most known enzymes required for polyamine biosynthesis in bacteria (including SAMDC) are absent in L. pneumophila, further suggesting a need for exogenous polyamines. We hypothesize that HtpB may function to ensure a supply of polyamines in host cells, which are required for the optimal intracellular growth of L. pneumophila.
INTRODUCTION
Chaperonins constitute a family of highly conserved proteins found in all prokaryotic and eukaryotic organisms (34). Their primary role is to facilitate the folding of nascent and stress-denatured proteins into their functional native states in an ATP-dependent manner (54). Group I chaperonins, referred to as Hsp60, Cpn60, or GroEL, are prokaryotic proteins found in bacteria and in eukaryotic organelles such as mitochondria and chloroplasts (34). Group II chaperonins, also known as CCT or TCP-1, are found in the eukaryotic cytosol and in the archaea (34). Structural and functional studies of Escherichia coliGroEL have established the role of group I chaperonins as intracellular mediators of protein folding (7, 94). GroEL is an essential protein in E. coli(23) whose intracellular level increases substantially in response to defined stressful stimuli (55, 85). The protein-folding paradigm of group I chaperonins has changed with accumulating reports of surface- and membrane-associated chaperonins that perform other diverse functions. For instance, the extracytoplasmically localized chaperonins of Haemophilus ducreyi(25), Helicobacter pylori(9, 92), Borrelia burgdorferi(77), and Clostridium difficile(37) have been implicated in adhesion and/or cell invasion. It has also been shown that some surface-exposed bacterial chaperonins have the capacity to interact with mammalian cell surface receptors to initiate signaling events that result in cytokine production (71). Moreover, the functional flexibility of group I chaperonins is demonstrated by the role of Mycobacterium lepraechaperonin as a protease (69), Enterobacter aerogenesGroEL as an insect toxin (93), and E. coliGroEL as a lipochaperonin (83).
Legionella pneumophila, a Gram-negative intracellular amoebal pathogen, is also an opportunistic human pathogen that replicates in mononuclear leukocytes (41) and causes Legionnaires' disease in susceptible individuals (59, 91). The L. pneumophila60-kDa chaperonin, encoded by the htpBgene (14, 39), is expressed at high levels under steady-state conditions, with an only 2-fold increase in expression following heat shock (53). This is in sharp contrast to the normally low levels of expression of GroEL in E. coliand the ∼20-fold increase in expression upon heat shock (39, 53). We have been unable to delete htpBfrom the L. pneumophilagenome (16), suggesting that it is an essential gene. Therefore, our HtpB studies are based on the use of functional protein tests.
HtpB expression is upregulated in the presence of L929 cells and monocytes, even prior to Legionellainternalization, and a high level of expression is maintained throughout intracellular infections (24), leading to accumulation of HtpB in the lumen of the Legionella-containing vacuole (LCV), as observed in L929 cells, monocytes, and HeLa cells (24, 28, 40). More than 40% of the cell-associated HtpB epitopes detectable by immunogold labeling are membrane associated, periplasmic, or cell surface localized in L. pneumophila(28), and we have previously established that surface-localized HtpB acts as an adhesion and invasion factor in HeLa cells (30). Furthermore, microbeads coated with purified HtpB (but not uncoated beads or beads coated with control proteins) were sufficient to attract mitochondria, modestly delay fusion with lysosomes, and transiently modify the organization of actin microfilaments when taken up by human macrophage and Chinese hamster ovary (CHO) cell lines (16), thus mimicking the early trafficking of LCVs. Although HtpB could function by signaling across the cell and LCV membranes after binding to host cell surface receptors, it is also possible that HtpB reaches the cytoplasm of infected cells and interacts with cytoplasmic targets.
In this study, we determined that HtpB is not confined to the lumen of the LCV but reaches the host cell cytosol. To identify potential functions of cytoplasmic HtpB, we expressed it in the genetically tractable eukaryote Saccharomyces cerevisiaeand found that it induces pseudohyphal growth (PHG) and interacts with S-adenosylmethionine decarboxylase (SAMDC), an enzyme required for biosynthesis of the polyamines spermidine and spermine in eukaryotic cells. Pharmacological inhibition of SAMDC activity significantly reduced the intracellular multiplication of L. pneumophilain mammalian cell lines. Moreover, the addition of exogenous polyamines moderately enhanced the intracellular growth of L. pneumophila, collectively suggesting that host polyamine biosynthesis and elevated levels of polyamines are important for the optimal intracellular growth of L. pneumophila.
MATERIALS AND METHODS
Culture conditions for bacteria and S. cerevisiae.
The bacterial and yeast strains used in this study are listed in Table 1. L. pneumophilastrains were grown at 37°C on buffered charcoal yeast extract agar (BCYE) (61), or at 37°C with agitation (200 rpm in a New Brunswick C25KC shaker incubator) in buffered yeast extract (BYE) broth. BCYE and BYE broth were supplemented with streptomycin (100 μg/ml) and, for growth of Lp02, thymidine (100 μg/ml). The defined medium (DM) of Pine et al. (68) was also used, but without choline. Medium ingredients were from Sigma-Aldrich (St. Louis, MO), MP Biomedicals (Santa Ana, CA [previously ICN Biomedicals]), Fisher Scientific (Fair Lawn, NJ), or BDH (Toronto, Ontario, Canada). E. colistrains were grown at 37°C on Luria-Bertani (LB) agar (74) or at 37°C with agitation (as described above) in LB broth. Bacterial transformants harboring the plasmids listed in Table 1were grown in culture medium containing the appropriate antibiotics, i.e., ampicillin (100 μg/ml) for E. coli, and kanamycin (50 μg/ml) and/or chloramphenicol (5 μg/ml) for L. pneumophila. All bacterial strains were stored at −70°C in nutrient broth containing 10% dimethyl sulfoxide.
Table 1.
Microbial strains and plasmids used in this study
| Strain or plasmid | Selection marker(s) | Characteristic(s) | Source/referencea |
|---|---|---|---|
| L. pneumophila | |||
| JR32 | Smr | Salt-sensitive isolate of AM511 (AM511: Philadelphia-1, serogroup 1, streptomycin resistant, restriction deficient, modification positive) | H. Shuman/73 |
| Lp02 | Smr | Philadelphia-1, serogroup 1, salt sensitive, restriction deficient, thymidine auxotroph | R. Isberg/6 |
| E. coli | |||
| DH5α | F−Φ80 ΔlacZΔM15 Δ(lacZYA-argF)U169 supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Clontech, Mountain View, CA/—c | |
| JM109/pSH16 | JM109 carries pSH16 for expression of HtpA and HtpB | P. Hoffman/39 | |
| S. cerevisiae | |||
| W303-1b | MATα leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 | G. Johnston/2 | |
| Y153 | MATagal4 gal80 his3 trp-902 ade2-101 ura3-52 leu2-3,112 URA3::GAL-lacZ LYS2::Gal-HIS3 | S. Elledge/21 | |
| 21R | MATaade1 leu2-3,112 ura3-52 | J. Hopper/44 | |
| Bacterial plasmids | |||
| pAC2 | Ampr, Cmr | pMMB207C with the cyaA-htpBchimera | This study |
| pAC17 | Ampr, Cmr | pMMB207C with the htpB-cyaAchimera | This study |
| pBluescript II KS | Ampr | High-copy-number plasmid used as a general cloning vector in E. coli | Stratagene, La Jolla, CA/— |
| pBS::htpB-1 | Ampr | pBluescript with modified htpBcontaining an added Kozak sequence and the first LEU codon optimized for CHO expression (TTA CTG) | This studyb |
| pBS::htpB-2 | Ampr | pBluescript with promoterless htpBlacking its stop codon, in which htpBis flanked by KpnI and XbaI sites | This study |
| pBS::htpB-3 | Ampr | pBluescript with promoterless htpB, in which htpBis flanked by XbaI and SphI sites | This study |
| pBS::htpB-4 | Ampr | pBlueScript with promoterless htpB, in which htpBis flanked by XbaI and SalI sites | This study |
| pBS::htpB-5 | Ampr | pBlueScript with promoterless htpB, in which htpBis flanked by EcoRI and SalI sites | This study |
| p2CyaA | Ampr, Cmr | pMMB207C with cyaA-cyaA | This study |
| pJC158 | Ampr, Cmr | pMMB207C derivative carrying the lepA-cyaAfusion | H. Shuman/12 |
| pJC203 | Ampr, Cmr | pMMB207C derivative carrying the cyaAgene | H. Shuman/12 |
| pMMB207C | Ampr, Cmr | RSF1010 (IncQ lacIqPtacoriT) derivative with ΔmobAand the PtacIPTG-inducible promoter | H. Shuman /12 |
| pSH16 | Ampr | pUC19 derivative carrying an EcoRI fragment from the L. pneumophilachromosome containing the htpABoperon | P. Hoffman/39 |
| pTrc99a::groELS | Ampr | pTrc99a derivative carrying the groELSoperon from E. coli | P. Sigler/— |
| Yeast shuttle vectors | |||
| pEMBLyex4 | Ampr, Ura+, Leu+ | High-copy-number plasmid used as a yeast expression vector controlled by the galactose-inducible GAL1-CYC1promoter | G. Johnston/22 |
| pEMBLyex4::groEL | Ampr, Ura+, Leu+ | pEMBLyex4 carrying the E. coli groELgene | This study |
| pEMBLyex4::htpB | Ampr, Ura+, Leu+ | pEMBLyex4 carrying the L. pneumophilahtpBgene optimized for expression in S. cerevisiae | This study |
| pEMBLyex4::HSP60 | Ampr, Ura+, Leu+ | pEMBLyex4 carrying the S. cerevisiaewild-type HSP60gene | This study |
| pEMBLyex4::hsp60Δ1-72 | Ampr, Ura+, Leu+ | pEMBLyex4 carrying the S. cerevisiaehsp60gene lacking the N-terminal region encoding the mitochondrial targeting sequence | This study |
| pGAD-C1 | Ampr, Leu+ | Yeast two-hybrid plasmid encoding the GAL4 trans-activating domain controlled by a modified S. cerevisiaePADH1promoter. | P. James/43 |
| pGAD-C1::DNAx, pGAD-C2::DNAx, pGAD-C3::DNAx | Ampr, Leu+ | pGAD-C1, -C2, and -C3 carrying highly representative genomic libraries from S. cerevisiaestrain YM706 in reading frames 1, 2, and 3, respectively | P. James/43 |
| pGBD-C1 | Ampr, Trp+ | Yeast two-hybrid plasmid encoding the GAL4DNA binding domain controlled by a modified PADH1promoter | P. James/43 |
| pGBD-C1::htpB | Ampr, Trp+ | pGBD-C1 carrying the GAL4DBD::htpBgene fusion | This study |
| pLM86 | Ampr, His+ | pPP389::htpBin which URA3was replaced with HIS3 | This study |
| pLM87 | Ampr, Trp+ | pPP389::htpBin which LEU2was replaced with TRP1 | This study |
| pPP389 | Ampr, Ura+, Leu+ | pEMBLyex4 in which the leu2-dgene (encoding a low-activity enzyme) was replaced with a wild-type LEU2allele | P. Poon/— |
| pPP389::HSP60 | Ampr, Ura+, Leu+ | pPP389 carrying the S. cerevisiaewild-type HSP60gene | This study |
| pPP389::hsp60Δ1-72 | Ampr, Ura+, Leu+ | pPP389 carrying the S. cerevisiaehsp60gene lacking the N-terminal region encoding the mitochondrial targeting sequence | This study |
| pPP389::htpB | Ampr, Ura+, Leu+ | pPP389 carrying the htpBgene | This study |
| pPP389::SPE2 | Ampr, Ura+, Leu+ | pPP389 carrying the S. cerevisiaeSPE2gene | This study |
| pSE1111 | Ampr, Trp+ | pGAD-C1 encoding the yeast Snfl protein fused to the GAL4 DNA binding domain | S. Elledge/21 |
| pSPE2.3 | Ampr, Ura+ | Derivative of high-copy-number plasmid YEp352 carrying the S. cerevisiaeSPE2gene | D. Balasundaram/4 |
David Allan, Dalhousie University; David Balasundaram, NIH Center for Scientific Review; Gerald Johnston, Dalhousie University; Howard Shuman, Columbia University College of Physicians and Surgeons; H.-U. Mösch, Philipps University; Joseph Vogel, Washington University School of Medicine; Paul Hoffman, University of Virginia; Paul Sigler, Harvard University; Jim Hopper, The Ohio State University; Pak Poon, Dalhousie University; Stephen Elledge, Baylor College of Medicine; Philip James, University of Wisconsin; Ralph Isberg, Tufts University Medical School.
The plasmid construction method used is described in Materials and Methods.
—, no reference available.
S. cerevisiaestrains were cultured at 30°C on YEP-dextrose agar, on YEP-galactose agar, or in YM-1 broth (35) containing the appropriate supplements (all at a concentration of 10 μg/ml) to compensate for strain auxotrophies. Sugars added to yeast culture medium were used at 2% (wt/vol). Synthetic complete or synthetic defined (SD) medium (35) was used to select and grow plasmid-carrying prototrophs. SD medium containing kanamycin (40 μg/ml) was used to select and grow kanamycin-resistant yeast transformants. Yeast strains were stored at −70°C in YM-1 broth containing 10% glycerol.
Culture of mammalian cell lines and amoebae.
Human U937 cells (a gift from Andrew Issekutz, Dalhousie University) were routinely cultured in suspension in RPMI 1640 medium (Gibco-Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT), 2 mM l-glutamine (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin and incubated at 37°C in 5% CO2. Before infection, U937 cells were induced to differentiate into adherent, macrophage-like cells with fresh medium containing 60 ng/ml phorbol myristate 13-acetate (PMA; Sigma). PMA-activated U937 cells were washed three times with RPMI 1640 medium and transferred to 24-well plates (Falcon-BD Biosciences Canada, Mississauga, Ontario, Canada) at approximately 1 × 105/well. Mouse L929 cells (ATCC clone CCL1) were routinely grown at 37°C in 5% CO2in minimal essential medium (MEM; Gibco) supplemented with 5% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B (Gibco). CHO-AA8 Tet-Off cells (Clontech-BD, Palo Alto, CA) carrying htpBwithin the integrated plasmid pTRE2hyg, referred to as CHO-htpBcells (16), were used in the initial translocation assays (below). CHO-htpBcells were grown and maintained at 37°C in 5% CO2in αMEM (Gibco) containing 10 ng/ml doxycycline (Gibco) to repress HtpB expression (16). Before infection, L929 or CHO-htpBcells were allowed to attach to 24-well plates at ∼1 × 105/well. Acanthamoeba castellanii(obtained from David Spencer, Dalhousie University) was grown to confluence in Neff's medium in 25-cm2cell culture flasks at room temperature. Trophozoites were gently scraped off the surface of one flask before encystation had begun, harvested by centrifugation at 700 × gfor 10 min, and then washed in phosphate-buffered saline (PBS).
Transformation and general genetic methods.
Lithium acetate-treated yeast cells were transformed according to a standard protocol (31). Glycerol-treated electrocompetent E. coliDH5α cells were transformed by electroporation with plasmid DNA (10 to 80 ng) using a Gene Pulser (Bio-Rad Laboratories Inc.), incubated in LB broth at 37°C for 1 h, and then plated on LB agar with 100 μg/ml ampicillin. L. pneumophilaLp02 and JR32 were transformed by electroporation as detailed by Viswanathan and Cianciotto (86).
Plasmid purification from E. coliDH5α was performed with the Spin Miniprep or Midiprep kit (Qiagen, Mississauga, Ontario, Canada). Isolation of DNA fragments from agarose gels was carried out with a QIAquick gel purification kit (Qiagen). PCR primers (Table 2) were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). PCR amplifications were performed on a TPersonal thermal cycler (Biometra, Göttingen, Germany) using TaqDNA polymerase (MBI Fermentas) or Platinum PfxDNA polymerase (Invitrogen Canada Inc., Burlington, Ontario, Canada).
Table 2.
PCR primers used in this study
| Primer | Sequence (5′ to 3′) | Restriction sitea |
|---|---|---|
| PH16-F | GGTACCATGATAATGGCTAAAGAATTACGTTTTGGT | KpnI |
| PH16-R | TCTAGACATCATTCCGCCCATGCCACCCAT | XbaI |
| htpB-Forward | GCCATTGCTCAAGTTGGAACTAT | None |
| htpB-Reverse | GCGTTGAAACCGTAGTTGTCTTT | None |
| CyaA-F | CCGGGGTACCATGCAGCAATCGCATCAGGCT | KpnI |
| CyaA-R | GCCTCTAGACGATCCCACCCCATCAAGGCT | XbaI |
| PBShtpB-F | TCTAGAATGATAATGGCTAAAGAATTACGTTTTGGT | XbaI |
| PBShtpB-R | GCATGCTTATTACATCATTCCGCCCATGCCACCCAT | SphI |
| GAL4htpB-F | GGAATTCCGTTTTGGTGATGAC | EcoRI |
| GAL4htpB-R | GCGTCGACTATTGGATAACCGGGAG | SalI |
| groELpEMBL-F | CGCGAGCTCATGGCAGCTAAAGACGT | SacI |
| pTrc99a-R | TCAGAAAGCTTCTGCGTTC | HindIII |
| mtHSP60-F | CGCGAGCTCATGTTGAGATCATCCGT | SacI |
| mtHSP60-R | CGGGATCCATCATACCTGGCATTCCT | BamHI |
| mthsp60Δ1-72-F | CGCGAGCTCATGAAAGAATTGAAATTCGGT | SacI |
| SPE2-F | CCCGGATCCATGACTGTCACCATAAAAGAAT | BamHI |
| SPE2-R | AATTCTGCAGTATTTTCTTCTGCAATTTC | PstII |
Restriction sites are shown as underlined boldface letters.
Construction of plasmids. (i)Translocation assay constructs.
N- and C-terminal htpBfusions to cyaA(encoding the calmodulin-dependent Bordetella pertussisadenylate cyclase subunit) were constructed in pJC158 (12). htpBwas amplified by PCR using primers PH16-F and PH16-R (Table 2) on template plasmid pSH16 and T/A cloned (74) into the EcoRV site of pBluescript to create pBS::htpB-2. A KpnI-XbaI fragment from pBS::htpB-2, containing the open reading frame of htpB, was used to replace the KpnI-XbaI lepAfragment in pJC158 to generate pAC17, which encodes adenylate cyclase fused to the C terminus of HtpB (htpB:cyaA). cyaAwas amplified by PCR using primers CyaA-F and CyaA-R on template plasmid pJC203 (12) and used to replace the KpnI-XbaI lepAfragment of pJC158, generating p2CyaA, which carries two in-tandem copies of cyaA. htpBwas amplified by PCR using primers PBShtpB-F and PBS::htpB-R on template plasmid pSH16 and T/A cloned into the EcoRV site of pBluescript to create pBS::htpB-3. A XbaI-SphI fragment from pBShtpB-3 carrying htpBin the correct orientation replaced the XbaI-SphI fragment in p2CyaA, containing one of the two copies of cyaA, to generate pAC2, which encodes an in-frame adenylate cyclase fusion to the N terminus of HtpB (cyaA:htpB).
(ii) Constructs for protein expression in yeast.
A 1,889-bp DraI fragment from pSH16, encoding the promoterless htpBgene, was cloned into the EcoRV site of pBluescript to generate pBS::htp-4. A SalI-XbaI fragment from pBS::htp-4, containing htpB, was cloned into the same sites of pEMBLyex4 to generate pEMBLyex4::htpB. Expression of htpBin yeast was optimized by removal of the GC-rich region of the pBluescript multiple cloning site (between the GAL1-CYChybrid promoter of pEMBLyex4 and the start of the htpBgene) by SacI/SmaI digestion, treatment with Klenow, and blunt-end ligation. A 1,900-bp XhoI-SalI fragment from pEMBLyex4::htpB, containing the htpBgene, was cloned into pPP389 to generate pPP389::htpB. Plasmid pPP389 is a derivative of pEMBLyex4 carrying a fully functional LEU2allele. In medium lacking leucine, pPP389 is maintained at a low copy number, in contrast to the high copy number at which pEMBLyex4 is maintained. The E. colichaperonin gene, groEL, was amplified by PCR using primers groELpEMBL-F and pTrc99a-R on template plasmid pTRC99a::groELSand cloned into the SacI-HindIII sites of pEMBLyex4 to generate pEMBLyex4::groEL. The yeast mitochondrial HSP60gene, with or without its encoded mitochondrial targeting sequence, was amplified by PCR using, respectively, primer pairs mtHSP60-F and mtHSP60-R or mthsp60Δ1-72-F and mtHSP60-R on template genomic DNA of S. cerevisiaestrain 21R. The corresponding amplification products were cloned into the SacI-BamHI sites of pEMBLyex4 and pPP389 to generate plasmids pEMBLyex4::HSP60, pEMBLyex4::hsp60Δ1-72, pPP389::HSP60, and pPP389::hsp60Δ1-72. SPE2, encoding yeast SAMDC, was amplified by PCR using primers SPE2-F and SPE2-R on template genomic DNA of S. cerevisiaestrain 21R and ligated into the PstI and BamHI sites of pPP389 to create pPP389::SPE2.
(iii) Yeast two-hybrid bait.
htpBwas PCR amplified using primers GAL4htpB-F and GAL4htpB-R on template plasmid pSH16 and cloned into the EcoRV site of pBluescript to create pBS::htpB-5. An EcoRI-SalI fragment from pBS::htpB-5, carrying htpBin the correct orientation, was cloned into the same sites of pGBD-C1 to generate pGBD-C1::htpB, encoding the GAL4DNA binding domain fused with HtpB at the N terminus (GAL4DBD-HtpB).
All constructed plasmids were verified by DNA sequence analysis carried out by DalGen (Dalhousie University, Halifax, Nova Scotia, Canada).
Translocation assays.
L. pneumophilastrains Lp02 and JR32 carrying plasmid pAC17, pAC2, pJC203, pJC158, or pMMB207C were grown to mid-exponential phase at 37°C in BYE broth containing 5 μg/ml chloramphenicol and treated with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for at least 2 h to induce expression of the plasmid-encoded proteins. Bacteria were then pelleted and resuspended in αMEM with 5% FBS, 5 μg/ml chloramphenicol, and 1 mM IPTG before infection of CHO-htpBcells at a bacterium-to-cell ratio of ∼600:1. Doxycycline (10 ng/ml) was maintained throughout the assays with CHO-htpBcells to repress the expression of HtpB (16). For infection of U937-derived macrophages, bacteria were resuspended in RPMI 1640 medium with 10% FBS, 1% glutamine, 5 μg/ml chloramphenicol, and 1 mM IPTG and inoculated at a bacterium-to-cell ratio of ∼100:1. Centrifugation at 500 × gfor 15 min was used to promote contact between host cells and bacteria. Cells were then incubated for 90 min (CHO-htpBcells) or 30 min (U937 macrophages) at 37°C in 5% CO2to allow internalization and intracellular establishment of legionellae. Cyclic AMP (cAMP) was extracted and measured using the enzyme immunoassay Biotrak kit, following the protocol suggested by the manufacturer (Amersham, Pharmacia). Femtomoles of cAMP/internalized bacterium were then calculated, for which a gentamicin protection assay was performed in triplicate to quantify the number of internalized bacteria. Briefly, after the 30- or 90-min infection period and 3 washes with warm PBS, monolayers of infected L929 or U937 cells were treated for 1 h with αMEM or RPMI 1640 medium containing 5% FBS and 100 μg/ml gentamicin. The monolayers were then washed 3 times with warm PBS and lysed in double-deionized water (ddH2O) to determine, by dilution plating, the number of intracellular bacteria (reported as CFU per well) that survived the gentamicin treatment. Dilution plating was performed using ddH2O as the diluent and BCYE agar with the appropriate supplements. Numbers of CFU/well were determined after plate incubation for at least 3 days at 37°C.
The functionality and expression of the CyaA-HtpB fusion, as well as the lack of effect of any ectopic HtpB on cAMP levels, were confirmed in CHO-htpBcell lysates by quantifying cAMP levels after mixing with whole-cell lysates of strain JR32 carrying pAC2 or pAC17. Lysates were produced by sonication as follows. After IPTG induction for 2 h, strain JR32 cells in late exponential phase were harvested and suspended to ∼109/ml in αMEM with 5% FBS, 1 mM IPTG, and 86 μg/ml protease inhibitor cocktail (Sigma). CHO-htpBcells grown to confluence were trypsinized from one 25-cm2flask (∼4 × 105cells) and suspended in 7 ml of αMEM with 5% FBS, 1 mM IPTG, and 74 μg/ml protease inhibitor cocktail. Bacteria were sonicated (Vibra-Cell; Sonics & Materials, Inc.) in 10 cycles of a 1-min pulse followed by a 3-min incubation on ice, whereas CHO cells were sonicated for 3 cycles. Bacterial lysates (1 ml) were incubated with 0.8 ml of CHO cell lysate for 20 min at 37°C in a 24-well plate. Proteins were precipitated from the mixture with HCl added to a final concentration of 50 mM, followed by neutralization with 0.5 M NaOH. cAMP was extracted and quantified as described above.
Pseudohypha formation and invasive growth.
Yeast cells grown at 30°C with agitation (150 rpm) in YEP-dextrose or SD medium with 2% dextrose, including appropriate selection, were harvested in exponential phase. Cells (∼107) were washed in water; diluted to 10−3, 10−4, 10−5, or 10−6in inducing medium; spotted in duplicate 100-μl drops on solid inducing medium; and incubated at 30°C in a humid chamber. Cell elongation and unipolar budding were scored 15 to 20 h after inoculation (while the inoculum drops were still wet) by light microscopy using the 40× objective of a Nikon DIAPHOT-TMD inverted microscope. To test for invasive growth, plates were incubated for 5 days, surface washed with a stream of ddH2O, and observed as described above. Photographs were captured with a Nikon 2000 camera using 35-mm Fuji film or digitally with a Pro series monochrome camera and Image-Pro 4.0 software (Media Cybernetics, Inc., Bethesda, MD).
Yeast two-hybrid (Y2H) screen.
Plasmid pGBD-C1::htpBwas transformed into strain Y153, and expression of the GAL4DBD::htpBgene fusion was confirmed by immunoblotting (see below) using monoclonal antibody (MAb) GW2X4B8B2H6 (36), directed against HtpB. Strain Y153 harboring pGBD-C1::htpBwas then transformed with a yeast genomic DNA plasmid library consisting of a combination of pGAD-C1::DNAx, pGAD-C2::DNAx, and pGAD-C3::DNAx constructs and plated on medium lacking leucine and tryptophan (to select transformants carrying the bait and prey plasmids), as well as histidine (to select for positive protein interactions via activation of the HIS3reporter gene). To minimize the growth of false positives, 30 mM 3-aminotriazole (Sigma) was used (43). A β-galactosidase filter assay (17) was performed to identify clones expressing interacting proteins that led to the activation of the lacZreporter gene. The library plasmids from positive clones were isolated and transformed into E. coliDH5α for plasmid amplification and subsequent DNA sequencing. The BLAST-P algorithm (1) was used to identify yeast proteins encoded by the positive Y2H library plasmids.
Protein electrophoresis, immunoblotting, and densitometry.
Bacterial cell pellets from 1-ml suspensions with an optical density at 620 nm (OD620) of 1.0 were solubilized in 100 μl of sample buffer, and 10 μl per lane was subjected to SDS-PAGE in a 12% (wt/vol) acrylamide vertical slab minigel. For yeast samples, 108pelleted cells (800 × gfor 5 min) were resuspended in 200 μl of sample buffer containing the α-yeast protease inhibitor cocktail (Sigma) and mechanically broken by adding ∼100 μl of acid-washed and baked glass beads (BT-5 high-impact beads, 40 to 50 μm in diameter; Supply America Company Inc., Norfolk, VA) and vortex mixing at 4°C for 15 min. Samples were then boiled for 5 min, unbroken cells and cell wall debris were pelleted at 15,000 × g, and 10 μl of the supernatant per lane was subjected to SDS-PAGE. For immunoblotting (84), proteins resolved by SDS-PAGE were transferred onto nitrocellulose membranes using a Bio-Rad electrotransfer apparatus and then immunostained with the appropriate MAb (GW2X4B8B2H6 [36] for HtpB, mtHsp60 MAb [Stressgen] for yeast mitochondrial Hsp60p, or GroEL MAb [Stressgen] for the E. colichaperonin) or polyclonal antibody (PAb) (in-house HtpB-specific rabbit serum [16]). All MAbs were diluted 1:1,000 and the PAb was diluted 1:5,000 in 0.1 M Tris-buffered saline pH 8 (TBS) containing 0.1% (wt/vol) bovine serum albumin (BSA; Sigma Chemical Co.). Secondary antibodies were alkaline phosphatase conjugates of either anti-mouse or anti-rabbit IgG (Cedarlane Laboratories Ltd.) diluted 1:5,000 in TBS containing 0.1% (wt/vol) BSA. Densitometry of immunostained proteins was done as follows. To first visualize transferred proteins, nitrocellulose membranes were stained with a 0.2% (wt/vol) solution of Ponceau-S (Allied Chemical Co., New York, NY) prepared in 3% (wt/vol) trichloroacetic acid–3% (wt/vol) sulfosalicylic acid and a reference digital image was acquired. Membranes were then immunostained and analyzed using the “single-band analysis” function of the GelPro 2.0 software (Media Cybernetics, Inc.). OD values were corrected for differences in loading and electrotransfer efficiency using the OD data of three well-defined protein bands from the Ponceau-S-stained reference image.
Blot overlay assays.
HtpB and GroEL purified from E. coli(16) and BSA (New England BioLabs) were subjected to SDS-PAGE (at 5 and 10 μg/lane) and electrotransferred to nitrocellulose membranes as described above or spotted (in 10-μg aliquots) onto a nitrocellulose membrane using a 96-well Bio-Dot SF microfiltration apparatus (Bio-Rad). Lysates of A. castellanii, L929 cells, and undifferentiated U937 cells were prepared in radioimmunoprecipitation assay (RIPA) buffer containing a protease inhibitor cocktail for mammalian cells plus 1 mM phenylmethylsulfonyl fluoride (all from Sigma Chemicals). The protein concentration of lysates was determined by the DC Protein Assay (Bio-Rad), and 60 μg of total protein/lane was subjected to SDS-PAGE and electrotransfer to nitrocellulose membranes as described above. For nonreducing conditions, 2-mercaptoethanol was not included in the sample buffer. Nitrocellulose membranes were then stained with Ponceau-S red to obtain a reference image for densitometry, as described above, before being blocked with SuperBlock buffer (Pierce/ThermoFisher) and overlaid as follows. (i) Membranes with purified proteins subjected to SDS-PAGE were overlaid with the L929 cell lysate (at 40 μg total protein per ml of RIPA buffer). (ii) The dot blot assay membrane was cut into three sections that were overlaid with RIPA buffer, the L929 cell lysate (40 μg/ml), or the U937 cell lysate (40 μg/ml). (iii) Membranes with the cell lysates separated in reducing or nonreducing SDS-PAGE gels were cut into three sections that were overlaid with purified HtpB, purified GroEL, or BSA (all prepared at 10 μg/ml in TBS). Overlays were rocked for 2 h at 4°C and then washed 3 times with TBS. Immunostaining of chaperonins was performed as described above. For SAMDC immunostaining, we used a chicken anti-SAMDC primary PAb, a peroxidase-conjugated rabbit anti-chicken secondary antibody, and development with the CN/DAB reagent (all from Pierce/ThermoFisher). Immunostained membranes were analyzed with the “single-band analysis” or the “dot blot analysis” function of the GelPro 2.0 software (Media Cybernetics, Inc.) using the Ponceau-S-stained reference image to calculate OD ratios.
Effects of pharmacological inhibitors of polyamine synthesis on the intracellular growth of L. pneumophila.
Methylglyoxal bis(guanylhydrazone) (MGBG) and α-difluoromethylornithine (DFMO; MP Biomedicals), at concentrations of up to 100 μM in MEM (for L929 cells) or RPMI 1640 medium (for U937 macrophages), were prepared immediately before use. L. pneumophilastrain JR32 grown for 2 to 3 days on BCYE was harvested and suspended in MEM or RPMI 1640 medium (containing the different concentrations of MGBG or DFMO) to ∼107bacteria/ml and added in triplicate to L929 or U937 cells in 24-well plates to a bacterium-to-cell ratio of 100:1. Plates were centrifuged at 500 × gfor 10 min to promote bacterial contact and incubated at 37°C in 5% CO2for 90 min. Monolayers were then washed 3 times with PBS to remove free bacteria. Cells were then either lysed in ddH2O to determine the number of CFU/well (set as the time zero value) or replenished with FBS-free MEM or RPMI 1640 medium containing the corresponding concentrations of MGBG or DFMO and then lysed in ddH2O to determine the number of CFU/well at 24 and/or 48 h postinfection, as described above for translocation assays.
Effect of MGBG or DFMO on cell viability.
The vital stain trypan blue was used to determine the effects of MGBG and DFMO on host cell viability. L929 or U937cells were plated in a 24-well plate (BD, Franklin Lakes, NJ) in 1 ml MEM or RPMI, respectively, at 105/well. After cells attached and spread, the medium was removed and 1 ml of fresh medium containing 100 μM MGBG or DFMO was added to 12 wells (test) and 1 ml of fresh medium without drugs was added to the remaining 12 wells (control). Cells were then incubated at 37°C in 5% CO2for up to 48 h. At 0, 24, and 48 h, the supernatants (1 ml) from 3 control and test wells (containing floating cells) were collected in separate microcentrifuge tubes. Adherent cells were then detached using 500 μl of 0.25% trypsin-EDTA in PBS (Invitrogen) per well for 10 min and added to the previously collected supernatant. Equal volumes of sample and 0.4% trypan blue (Invitrogen) were mixed, and cells were counted using a hemocytometer (Improved Neubauer chamber; Hausser Scientific, Horsham, PA). The percentage of blue (nonviable) cells was calculated over a total count of 1,000 cells per sample.
Effects of exogenous polyamines on L. pneumophilagrowth.
BYE broth or DM containing cadaverine, putrescine, spermine, or spermidine (MP Biomedicals) was inoculated as follows. BYE broth was inoculated to an initial OD620of 0.2 with a culture of strain JR32 grown in BYE broth to an OD620of 2 to 3. DM was inoculated to an initial OD620of 0.09 with a culture of strain JR32 grown in polyamine-free DM to an OD620of 0.18. The inoculated media were dispensed into a flat-bottom 96-well plate (Falcon Plastics-BD) at 16 wells/condition and 250 μl/well. Cultures were incubated at 37°C in a humid box. Static growth was monitored by measuring OD620in a Benchmark Plus multiwell plate reader (Bio-Rad Canada, Mississauga, Ontario, Canada). Aerated 50-ml cultures of BYE broth or DM with or without polyamines were prepared in 100-ml Erlenmeyer flasks inoculated as described above and incubated at 37°C with agitation (200 rpm in a New Brunswick C25KC shaker incubator).
L929 cells were prepared for infection and processed to obtain CFU/well counts as described above under “Effects of pharmacological inhibitors of polyamine synthesis on the intracellular growth of L. pneumophila,” with the following changes. The FBS-free MEM used to add the bacterial inoculum and maintain the polyamine-treated cells during the infection period was supplemented with spermine and spermidine (both from MP Biomedicals and used at a final concentration of 100 μM). For wells infected with L. pneumophilapretreated with polyamines, the bacterial inoculum was prepared from strain JR32 grown to stationary phase in BYE broth supplemented with spermine and spermidine (both at a final concentration of 100 μM). Harvested bacteria were washed to ensure that no additional spermidine or spermine was present in the inoculum.
Bioinformatic analysis of polyamine biosynthetic enzymes.
DNA sequences of genes encoding known polyamine biosynthetic enzymes in E. coliand Vibrio cholerae(11, 52, 79, 81, 82) were obtained from GenBank at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov). To identify gene sequences similar to the sequences obtained, the nucleotide BLAST tool (NCBI website) was used to compare the sequences obtained from E. coliand V. choleraeagainst the four L. pneumophilagenome sequences available from NCBI (Philadelphia-1, Lens, Paris, and Corby), as well as the genome sequences of Chlamydia trachomatis, Coxiella burnetii, Francisella tularensis, Listeria monocytogenes, Salmonella enterica, Shigella flexneri, pathogenic E. coli, and Yersinia enterocolitica. The search was optimized for the “somewhat similar sequences (blastn)” option.
Statistical analysis.
Statistical significance was assessed using the Student ttest or the one- or two-way analysis of variance (ANOVA) test using Minitab software version 15.1.30.0 (Minitab Inc., State College, PA). For multiple comparisons, one-way ANOVA and a Bonferroni posttest were used (http://graphpad.com/quickcalcs/posttest1.cfm).
RESULTS
HtpB reaches the cytosol of L. pneumophila-infected cells.
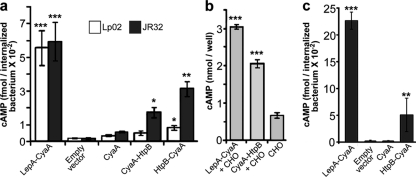
Immunoelectron microscopy data previously suggested that HtpB is exposed on the cytoplasmic face of LCVs (15, 24). To determine whether HtpB indeed reaches the cytoplasm of infected cells, we used the CyaA reporter system, which has been used before to establish the translocation of L. pneumophilatype IV secretion effectors (8, 12, 20, 58). Initially, we used CHO-htpBcells as part of a series of functional studies aimed at assessing the responses of mammalian cells to HtpB (16). CHO-htpBcells not expressing ectopic HtpB and infected with strain Lp02 or JR32 expressing the HtpB-CyaA fusion showed a significant increase in cAMP levels (P< 0.05, Fig. 1a) compared to those of host cells infected with L. pneumophilaharboring the empty vector control or the CyaA-only construct. Strain Lp02 was much less efficient than strain JR32 at transferring the HtpB fusion proteins but as efficient as strain JR32 in the translocation of the known Dot/Icm substrate LepA (Fig. 1a). Cells infected with L. pneumophilaexpressing the CyaA-HtpB fusion showed lower cAMP levels than those infected with L. pneumophilaexpressing HtpB-CyaA (Fig. 1b), suggesting that the N terminus of HtpB either is important for translocation or interferes with CyaA activity. To rule out the latter, we showed that a large increase in cAMP was detected after mixing a host cell lysate with a lysate from strain JR32 expressing the CyaA-HtpB fusion (Fig. 1b). This increase was comparable to that induced by the lysate of JR32 expressing the LepA-CyaA fusion, suggesting that the lower level of cAMP produced by CHO-htpBcells infected with L. pneumophilaexpressing CyaA-HtpB was not due to a defect in CyaA activity or to low expression of the fusion protein. That HtpB reaches the cytoplasm of infected cells was then confirmed in U937-derived macrophages (Fig. 1c). Overall, the levels of cAMP in macrophages were higher than those in CHO-htpBcells.
Fig. 1.
HtpB reaches the cytosol of L. pneumophila-infected CHO-htpBcells. (a) cAMP levels in CHO-htpBcells infected for 90 min with L. pneumophilastrain Lp02 or JR32 expressing the CyaA-HtpB or HtpB-CyaA fusion protein. L. pneumophilastrains carrying the empty vector pMMB207C and strains expressing only CyaA served as negative translocation controls, whereas strains expressing the LepA-CyaA fusion served as positive translocation controls. (b) cAMP levels measured after mixing CHO-htpBwhole-cell lysates with lysates from JR32 expressing the LepA-CyaA fusion (positive control) or the CyaA-HtpB fusion. The lysate of CHO-htpBcells alone served as a negative control and indicated that even if some ectopic HtpB was produced by these cells, it did not result in increased cAMP levels. (c) cAMP levels in U937-derived macrophages infected for 30 min with L. pneumophilastrain JR32 expressing the HtpB-CyaA or the LepA-CyaA fusion protein. JR32 cells carrying the CyaA-only construct or the empty vector served as negative translocation controls. Means and standard deviations were obtained from triplicate samples (n= 3). The results shown are representative of two independent experiments. *, P< 0.05; **, P< 0.01; ***, P< 0.001.
Inducible expression of HtpB in S. cerevisiae.
Knowing that HtpB reaches the cytoplasm of infected cells, we set out to investigate potential functions for cytoplasmic HtpB in the genetically tractable eukaryote S. cerevisiae. Expression of ectopic HtpB in yeast cells grown in YEP-galactose (inducing medium) was confirmed by immunoblotting (Fig. 2a). In YEP-galactose liquid medium, no growth differences between two HtpB-expressing clones and a clone carrying the empty vector control were detected (compare Fig. 2b and c). However, the expression of HtpB caused an approximately 2-fold reduction in colony numbers in relation to those of yeast cells grown on dextrose (Fig. 2b, inset) or yeast cells carrying the empty plasmid pEMBLyex4 (Fig. 2c, inset), suggesting that solid medium imposed a slight growth restriction on yeast cells expressing HtpB. Expression of HtpB in liquid cultures (not shown) coincided with the resumption of cell division 10 h after yeast cell transfer from YEP-dextrose into YEP-galactose (Fig. 2c).
Fig. 2.
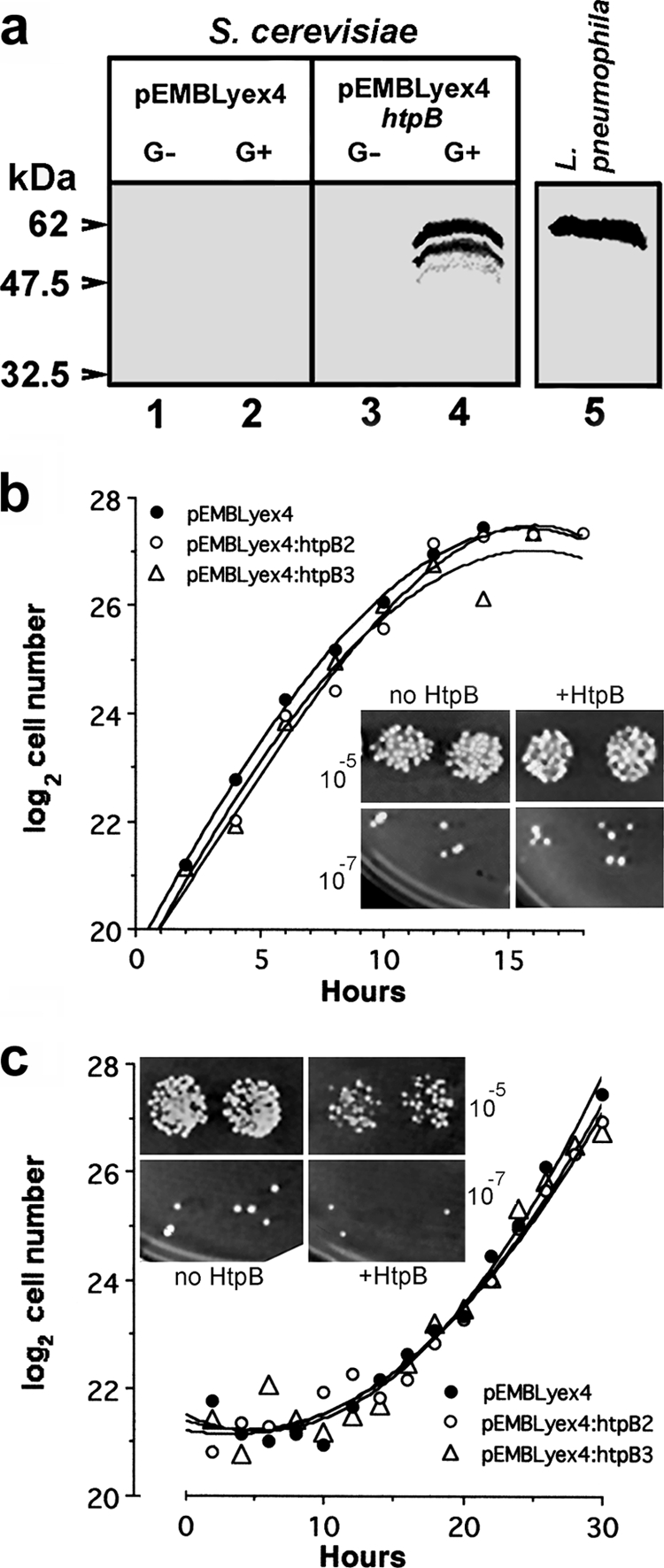
Expression of the L. pneumophilachaperonin, HtpB, in S. cerevisiae. (a) Immunoblotting of whole-cell lysates from yeast strain W303-1b bearing the vector pEMBLyex4 (lanes 1 and 2) or the galactose-inducible construct pEMBLyex4::htpB(lanes 3 and 4) and grown in noninducing medium with dextrose (G−) or inducing medium with galactose (G+) as a carbon source. The blot was probed with an HtpB-specific MAb. Full-length ectopic HtpB expressed in yeast (lane 4) and HtpB from L. pneumophila(lane 5) migrated to similar positions, but ectopic HtpB showed degradation products. The positions and molecular masses (kDa) of prestained marker proteins are indicated. (b and c) Growth curves of yeast strain W303-1b carrying pEMBLyex4 (one clone) or pEMBLyex4::htpB(two clones designated pEMBLyex4::htpB2 and pEMBLyex4::htpB3) in YEP-dextrose (b) or YEP-galactose (c) at 30°C. Insets show growth at 30°C of serially diluted suspensions spotted in duplicate (10−5and 10−7dilutions are shown) on solid medium containing dextrose (b) or galactose (c). The inoculum for the spots contained equivalent numbers of yeast cells carrying either pEMBLyex4 (no HtpB) or pEMBLyex4::htpB(+HtpB). The experiments shown in panels a and c were repeated several times as part of subsequent experiments requiring the growth of yeast strain W303-1b carrying pEMBLyex4 or pEMBLyex4::htpB.
HtpB stimulates S. cerevisiaeto form pseudohyphae.
In nature, S. cerevisiaecells form pseudohyphae when nitrogen becomes limiting (reviewed in reference 27). Haploid yeast strains may also differentiate and become invasive under glucose limitation (49, 50). On nitrogen-replete solid medium containing galactose, cells of haploid yeast strain W303-1b expressing HtpB from plasmid pEMBLyex4::htpBelongated and budded in a unipolar direction (Fig. 3a), and after 5 days at 30°C, they produced some agar-invasive filamentous colonies (Fig. 3c). Yeast cells carrying the empty vector, pEMBLyex4, remained ovoid when grown on the same medium (Fig. 3b) and were not agar invasive (Fig. 3d). The same pseudohyphal phenotype (as shown in Fig. 3a and c) was observed in cells of strain W303-1b harboring the construct pPP389::htpB. To assess the HtpB-mediated phenotype in a glucose-replete medium, HtpB was expressed from pGBD-C1::htpB, a construct that drives expression of the GAL4DBD-HtpB chimera using a modified alcohol dehydrogenase promoter (PADH1). Yeast strain W303-1b carrying pGBD-C1::htpBalso formed pseudohyphae 15 to 20 h after inoculation onto solid SD medium containing glucose (data not shown). Therefore, induction of PHG by HtpB occurs in nitrogen- and glucose-replete medium and is not dependent on the plasmid vector used.
Fig. 3.
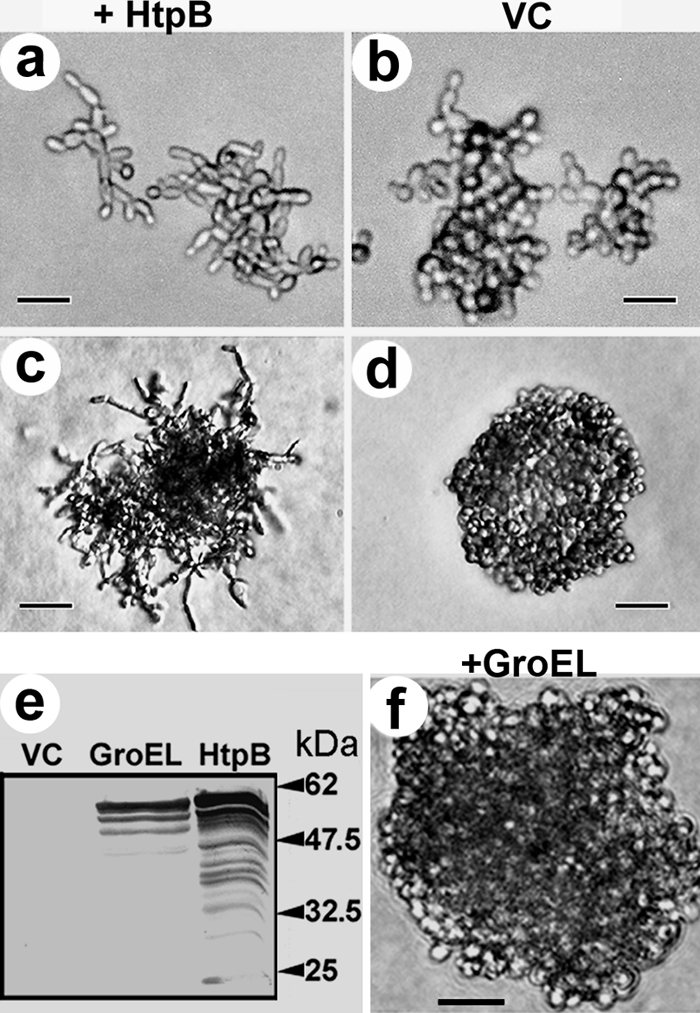
HtpB, but not GroEL, induces S. cerevisiaeto form pseudohyphae that invade solid medium. Shown are cells within microcolonies (a and b) and small colonies (c, d, and f) of yeast strain W303-1b grown on solid medium containing galactose. Cells carry the galactose-inducible construct pEMBLyex4::htpBfor expression of HtpB (a and c, +HtpB), the vector control (VC) pEMBLyex4 (b and d), or the galactose-inducible construct pEMBLyex4::groEL(f, +GroEL). The filamentous colony shown in panel c had penetrated the agar. (e) Immunoblotting of whole-cell lysates of yeast strain W303-1b grown in inducing medium containing galactose immunostained with an HtpB-specific PAb (cross-reactive with GroEL). Arrows and numbers indicate the positions and masses (in kilodaltons) of prestained marker proteins. Size bars represent 9 μm (a and b), 25 μm (c and d), and 21.5 μm (f). The experiment showing the ability of HtpB to induce PHG was repeated several times as part of subsequent experiments where clones carrying pEMBLyex4::htpBor pPP389::htpBwere used as positive controls for PHG.
Other type I chaperonins do not stimulate S. cerevisiaeto form pseudohyphae.
GroEL, the E. colichaperonin (accession no. AAC77103; NCBI Entrez Protein), shows 75.5% protein sequence identity with HtpB (accession no. AAA25299; NCBI Entrez Protein), according to the Lalign program (63). We set out to determine if the amino acid differences between GroEL and HtpB would result in functional differences in relation to pseudohypha formation in yeast. We found that yeast strain W303-1b expressing E. coliGroEL from pEMBLyex4::groEL(Fig. 3e) did not form pseudohyphae (Fig. 3f).
The S. cerevisiaemitochondrial chaperonin (Hsp60p) (systematic name, YLR259C, SGD), shows 54.5% protein sequence identity with HtpB, according to the Lalign program (63). Unlike HtpB, Hsp60p has a positively charged motif at its N terminus that is necessary for import into yeast mitochondria. A complete HSP60construct and a construct lacking the mitochondrial import motif (hsp60Δ1-72) were expressed in strain W303-1b to produce either full-length Hsp60p or truncated Hsp60Δ1-24p, as confirmed by immunoblotting (data not shown). In contrast to HtpB-expressing cells, W303-1b cells expressing either Hsp60p or Hsp60Δ1-24p did not form pseudohyphae (data not shown). Collectively, these data indicate that the ability to alter yeast cell morphology does not represent a general characteristic of group I chaperonins.
SAMDC interacts with HtpB in S. cerevisiae.
We hypothesized that there is a specific interaction between HtpB and a yeast protein involved in PHG. To test this hypothesis, we conducted a Y2H screen using as bait the Gal4DBD-HtpB chimera, which is functional for PHG induction (data not shown). Circa 8 × 108yeast clones carrying yeast genomic library plasmids were screened, and two strongly positive clones were identified. DNA sequence determination revealed that these two clones carried independent fusions of the GAL4 trans-activating domain with SPE2, the S. cerevisiaegene that encodes SAMDC. In yeast, SAMDC is a conserved and essential enzyme required for aerobic growth (3–5, 10) and for synthesis of the polyamines spermidine and spermine (reviewed in references 18, 81, and 82).
Overexpression of SAMDC induces PHG in yeast.
Knowing that a transient increase in both the level of polyamines and the activity of polyamine synthetic enzymes characterizes the yeast-to-hypha transition in many fungal species (33, 38, 42, 57, 76), we hypothesized that overexpression of SAMDC could result in PHG in S. cerevisiae. To test this hypothesis, we increased the SPE2copy number in strain W303-1b as a means to enhance SAMDC activity. It has been noted elsewhere that when SPE2is carried on a yeast high-copy-number plasmid, the activity of SAMDC in S. cerevisiaeincreases ∼50-fold in relation to that of S. cerevisiaethat does not bear the plasmid (46). The presence of the low-copy-number plasmid pPP389::SPE2in strain W303-1b induced PHG (Fig. 4). Thus, overexpression of either HtpB or its interacting protein, SAMDC, induces the same phenotype in S. cerevisiae.
Fig. 4.
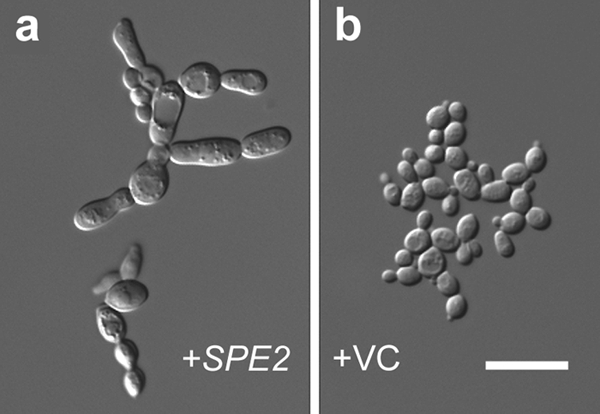
Overexpression of SAMDC induces pseudohyphal growth. (a and b) Microcolonies of yeast strain W303-1b grown on SD medium containing galactose. Yeast cells carried the galactose-inducible construct pPP389::SPE2(a, +SPE2) or the vector control pPP389 (b, +VC). The microcolony shown in panel a had penetrated the agar. The size bar represents 12 μm and applies to both panels. One of two experiments with similar results is shown.
HtpB interacts with mammalian SAMDC.
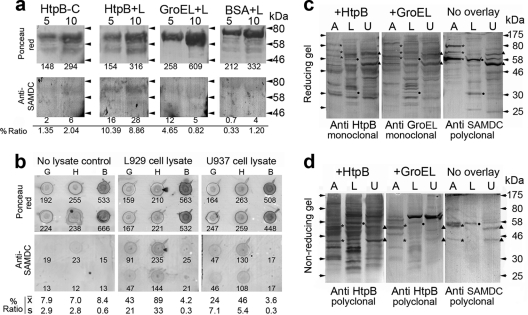
To establish whether HtpB is capable of interacting with the SAMDC of cells that serve as hosts for L. pneumophila, a series of overlay immunoassays were conducted. Initially, in a far-Western assay, HtpB was able to capture SAMDC from a lysate of L929 cells (Fig. 5a). The anti-SAMDC immunostaining signal obtained in the HtpB lanes exposed to the L929 lysate overlay was the only one clearly above the background, with 10 and 9% signal ratios for the 5- and 10-μg aliquots, respectively (Fig. 5a). Next, we performed a dot blot overlay assay to determine, in a more quantitative format, whether HtpB would capture SAMDC from an L929 or an U937 cell lysate (Fig. 5b). In this format, HtpB captured 2-fold more SAMDC than GroEL and between 12- and 20-fold more SAMDC than BSA. It was also shown that HtpB and GroEL interacted twice as well with murine SAMDC (from L929 cells) as with human SAMDC (from U937 cells). Finally, far-Western assays (reversed in relation to those shown in panel a) were performed that included amoeba lysates. Here, the ability of host cell proteins, separated by SDS-PAGE under reducing and nonreducing conditions, to bind HtpB and GroEL was assessed (Fig. 5c and d). The main protein bands recognized by the anti-SAMDC PAb were able to capture HtpB and, with some exceptions, GroEL. The anti-HtpB PAb (panel d) is cross-reactive with GroEL. HtpB was captured by many host cell proteins, particularly in the nonreducing gel (Fig. 5d), suggesting that it might have other cytoplasmic targets in host cells that were not identified in our Y2H assay.
Fig. 5.
HtpB interacts with mammalian SAMDC. (a) Far-Western blot assay with 5 or 10 μg of immobilized purified proteins (HtpB, GroEL, and BSA) overlaid with a lysate of L929 cells (L). The negative control for the SAMDC immunostaining signal was prepared with purified HtpB exposed to buffer instead of lysate (HtpB-C). The small number under each lane is the densitometry reading for the upper band representing full-size chaperonin or BSA. Differences in loading and blotting efficiency were controlled for by calculating the percent ratio of the immunostaining signal over the Ponceau-S staining signal (values under the anti-SAMDC membranes/values under the Ponceau red membranes × 100). The horizontal arrowheads indicate the positions of marker proteins run between samples, and their molecular masses are indicated on the right in kDa. (b) Dot blot assay showing 10 μg of immobilized GroEL (G), HtpB (H), or BSA (B) overlaid with L929 or U937 cell lysate. The control for the negative SAMDC immunostaining signal was prepared with immobilized proteins overlaid with buffer (no-lysate control). The percent ratio of the immunostaining signal over the Ponceau-S staining signal was calculated as described for panel a and is shown as the mean (x̄) and standard deviation (s) (n= 2). Shown are reverse far-Western assays of immobilized host cell proteins separated by SDS-PAGE under reducing (c) or nonreducing (d) conditions overlaid with purified HtpB and GroEL and immunostained with the indicated antichaperonin antibodies. The no-overlay membranes show the protein bands recognized by the anti-SAMDC antibody, whose positions are indicated by asterisks for amoebae (A), black dots for L929 cells (L), and black triangles for U937 cells (U). Arrowheads indicate the positions of marker proteins whose masses are indicated on the right in kDa.
SAMDC activity promotes L. pneumophilareplication in mammalian cells.
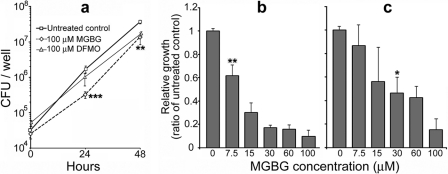
Having determined the ability of HtpB to interact with SAMDC from mammalian cells and amoebae, we next hypothesized that L. pneumophilacould benefit from the activity of SAMDC in its host cells. To determine whether L. pneumophilarequires the activity of host SAMDC during intracellular growth in mammalian cells, we infected cells treated with two inhibitors of polyamine biosynthesis. MGBG acts as a specific, competitive, and irreversible inhibitor of SAMDC (19, 88, 90), and DFMO is an irreversible inhibitor of ornithine decarboxylase (56, 70), an enzyme required for the synthesis of putrescine, a precursor of spermidine. We initially found that treatment of mouse L929 cells with MGBG significantly inhibited L. pneumophilaintracellular replication at 24 and 48 h postinfection, whereas DFMO inhibition was significant only at 48 h postinfection (Fig. 6a). The pharmacological inhibition of SAMDC with MGBG was dose dependent in both L929 cells (Fig. 6b) and U937 macrophages (Fig. 6c), suggesting that host SAMDC, but not necessarily host ornithine decarboxylase, needs to remain fully active to support the optimal early replication of L. pneumophilain mammalian cells.
Fig. 6.
Pharmacological inhibition of polyamine biosynthetic enzymes decreases L. pneumophilareplication in mammalian cells. (a) Intracellular growth of L. pneumophilastrain JR32 in L929 cells treated with MGBG or DFMO. (b and c) Dose-dependent inhibition by MGBG in L929 cells (b) or in U937-derived macrophages (c). Results are shown as the mean ± one standard deviation of the growth ratio of treated to untreated cells for 3 independent experiments (n= 3), each run in triplicate. The statistical significance of differences from the corresponding untreated control was calculated by the two-way ANOVA test. *, P< 0.05; **, P< 0.01; ***, P< 0.001. In panels b and c, statistical significance is shown only above the concentration of inhibitor that first imparted a significant growth difference in relation to the untreated control.
At a concentration of 100 μM, neither MGBG nor DFMO was significantly toxic to L929 cells or U937 macrophages at 24 or 48 h, as determined by trypan blue staining (data not shown). In addition, neither MGBG nor DFMO at a final concentration of 100 μM showed a direct inhibitory effect on L. pneumophilareplication in BYE broth (data not shown). These toxicity results suggest that the inhibition of L. pneumophilagrowth in L929 and U937 cells was not due to a drug-mediated killing of infected cells or to a direct effect of MGBG or DFMO on L. pneumophila.
Addition of exogenous polyamines enhances the growth of L. pneumophilain vitro.
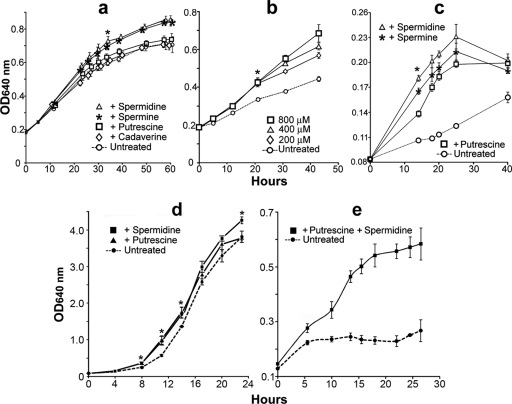
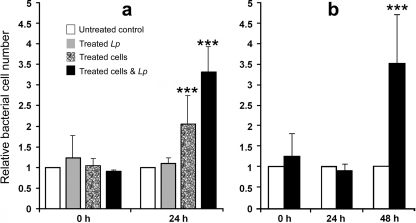
Supplementation of BYE broth with polyamines modestly enhanced the growth of strain JR32 in vitro(Fig. 7a and d), and supplementation with spermidine did so in a dose-dependent manner (Fig. 7b). The beneficial effect of exogenous polyamines was obvious in choline-free DM (Fig. 7c and e), where spermidine was confirmed to be the most beneficial polyamine for L. pneumophilagrowth in vitro. The treatment of L929 cells with exogenous spermidine and spermine (added at the time of infection) caused a modest but significant (2-fold) increase in the intracellular growth of L. pneumophilaat 24 h postinfection relative to that in untreated cells (Fig. 8a). Although not beneficial on its own, pregrowth of L. pneumophilain BYE broth with polyamines, in combination with polyamine treatment of L929 cells, resulted in a significant (>3-fold) increase relative to that in untreated L929 cells infected with strain JR32 cells pregrown in BYE broth without polyamines (Fig. 8a). This suggested that exposure of L. pneumophilato polyamines either contributed to a higher net concentration of these polyamines in intracellular legionellae or predisposed these legionellae to better use the excess polyamines added to host cells (potentially by preinducing uptake or utilization pathways). The >3-fold moderate effect of the combined treatment of host cells and legionellae with exogenous polyamines was confirmed in U937 macrophages (Fig. 8b). In this case, the added polyamines afforded the completion of ∼2 additional cell divisions between 24 and 48 h postinfection, a period for which no legionella growth occurred in the untreated controls (Fig. 8b).
Fig. 7.
Exogenous polyamines enhance the growth of L. pneumophilain vitro. Panels a to c represent experiments performed with strain JR32 in 96-well plates (250 μl/well) incubated without agitation, and panels d and e represent experiments performed with strain JR32 in shaken 50-ml liquid cultures. (a) BYE broth was individually supplemented with the indicated polyamines (100 μM). (b) Dose-response growth experiment with spermidine in BYE broth. (c) Growth in the absence or presence of polyamines (100 μM) in choline-free DM. (d) Growth in BYE broth or BYE broth supplemented with 100 μM spermidine or putrescine. (e) Growth in choline-free DM or DM supplemented with 100 μM putrescine and spermidine. Panels a to c show the mean values ± one standard deviation (n= 16 replicates) from one of three experiments showing similar results. Panels d and e show the mean values ± one standard deviation of three independent cultures (n= 3). The statistical significance of differences from the corresponding untreated control at each time point was calculated by the Student ttest (*, P< 0.05). For panels a to c, the differences between treated and untreated cultures were statistically significant at and after the time point marked with the asterisk, except for the putrescine- and cadaverine-treated cultures in panel a. For panel d, only the time points marked with an asterisk showed significant growth differences between spermidine-treated samples and the untreated control. For panel e, the differences between treated and untreated cultures were statistically significant at all time points except time zero.
Fig. 8.
Treatment of L. pneumophilaand/or host cells with exogenous polyamines enhances bacterial intracellular growth. Treated L. pneumophilastrain JR32 (Lp) was pregrown in BYE broth with spermidine and spermine, and L929 cells or U937-derived macrophages (cells) received medium with spermidine and spermine at the time of infection. Results are shown as relative numbers of intracellular bacteria (calculated as the number of CFU/well of the treated test samples divided by the number of CFU/ml of the untreated control) and represent the mean plus one standard deviation for 3 independent experiments (n= 3), each run in triplicate. The CFU values for the untreated controls (shown as white bars set at a relative value of 1) are as follows: panel a, 1.5 × 105± 0.8 × 105at 0 h and 9.9 × 106± 0.1 × 106at 24 h; panel b, 0.8 × 105± 0.4 × 105at 0 h, 3.8 × 106± 0.5 × 106at 24 h, and 3.4 × 106± 1.9 × 106at 48 h (n= 3). Statistically significant differences in L. pneumophilareplication in relation to that of the untreated control are indicated by *** (P< 0.001).
The L. pneumophilagenome does not encode most of the known prokaryotic polyamine biosynthetic enzymes.
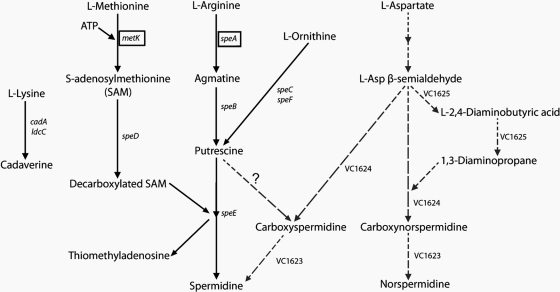
To understand why L. pneumophilabenefits from host SAMDC activity and/or exogenous polyamines, we assessed in silicothe metabolic capacity of L. pneumophilato synthesize polyamines. In a comparative analysis against known conserved prokaryotic pathways of polyamine biosynthesis (11, 52, 79, 81, 82), we found that most of the enzymes required for polyamine biosynthesis in E. colior Vibrio cholerae(Fig. 9) are not encoded by the L. pneumophilagenome. The only genes encoding polyamine biosynthetic enzymes in L. pneumophilawere metK(methionine adenosyltransferase) and speA(arginine decarboxylase) (Fig. 9), suggesting that L. pneumophilacannot synthesize all polyamines. Although our results cannot rule out the possibility that the genome of L. pneumophilaencodes polyamine biosynthetic enzymes bearing no sequence similarity to other known bacterial enzymes, a dependence on exogenous polyamines was strongly supported by our results with liquid cultures (Fig. 7) and L929 cells and U937-derived macrophages (Fig. 8).
Fig. 9.
Integrated polyamine biosynthetic pathways in E. coliand V. cholerae. The V. choleraepathway is marked by the broken-line arrows. Enzymes are represented by the names of the genes that encode them. For V. cholerae, genes encoding biosynthetic enzymes are indicated by number. In alphabetical order, they are cadA(inducible lysine decarboxylase), ldcC(constitutive lysine decarboxylase), metK(methionine adenosyltransferase), speA(arginine decarboxylase), speB(agmatine ureohydrolase), speC(constitutive ornithine decarboxylase), speD(SAMDC), speE(spermidine synthase), speF(inducible ornithine decarboxylase), VC1623 (carboxynorspermidine decarboxylase), VC1624 (carboxynorspermidine dehydrogenase), and VC1625 (a large fusion protein comprising the two enzymes diaminobutyrate aminotransferase and diaminobutyrate decarboxylase). The genes that have homologs in the L. pneumophilagenome are boxed.
The genomes of the intracellular bacterial pathogens Chlamydia trachomatis, Coxiella burnetii, Francisella tularensis, and Listeria monocytogenesalso lacked several of the polyamine biosynthetic enzymes shown in Fig. 9(data not shown), indicating that reduced polyamine biosynthetic capacity is not specific to L. pneumophila. However, all of the intracellular Enterobacteriaceaeanalyzed possessed the complete polyamine biosynthetic pathway of E. coli(data not shown).
DISCUSSION
HtpB reaches the cytoplasm of L. pneumophila-infected cells, as indicated here by the CyaA reporter assay. This result implies that, in addition to its external role as an invasion factor in nonphagocytic cells (30), HtpB could play internal roles as a cytoplasmic effector, as previously suggested by the HtpB-mediated alteration of actin filaments in CHO-htpBcells (16). Using S. cerevisiaeas a model eukaryote, we have now shown that HtpB induces PHG and interacts with the cytoplasmic enzyme SAMDC. This interaction, together with the observation that increased SAMDC activity also induces PHG, suggested a potential link between HtpB function and polyamines and allowed us to speculate that L. pneumophilamight use HtpB to manipulate polyamine levels in host cells and achieve optimal intracellular growth.
Based on our current knowledge, we propose the following physiological model to explain how the known HtpB functions are linked to L. pneumophilapathogenesis. (i) Surface-exposed HtpB, which increases in abundance in the presence of L929 cells and monocytes (24), as well as during differentiation of L. pneumophilainto mature infectious forms (29), serves as a ligand for cell surface receptors. As a ligand, HtpB then mediates attachment to macrophages and other cell types (30, 72) and invasion of human nonphagocytic (HeLa) cells (30). (ii) Intracellular L. pneumophilaabundantly releases HtpB into the lumen of the LCV (24, 28, 40) by an ill-defined mechanism (see below). (iii) In experiments with phagocytosed HtpB-coated beads in CHO cells and macrophages, we showed that HtpB signals across the phagosomal membrane to attract mitochondria and transiently alters the actin cytoskeleton of host cells (16). (iv) HtpB from the LCV lumen reaches the host cell cytoplasm (this study), where it could interact with SAMDC (this study) and putatively increase the intracellular pool of polyamines.
The mechanism by which HtpB is secreted by L. pneumophilaor reaches the host cell cytoplasm is as yet undefined. Previous unpublished data from our lab suggest that the presence of surface-exposed HtpB is not dependent on type II secretion or the TAT pathway, but type IV secretion avirulent mutants show reduced levels of surface-exposed HtpB in relation to those of their virulent parental strains. Surprisingly, the type IV secretion mutants showed increased levels of periplasmic HtpB in relation to those of their parent strains (unpublished results), suggesting that release of HtpB by L. pneumophilais a multistep process that involves passage through the periplasm. We have speculated that once HtpB reaches the periplasm of legionellae (by a yet-to-be-determined mechanism), it could then pass through outer membrane channels (e.g., those of the type IV secretion apparatus) in response to environmental changes. In relation to the HtpB that is abundantly released into the LCV by L. pneumophila(28, 40), the low level of cAMP detected in cells infected with L. pneumophilacarrying the C-terminal HtpB-CyaA fusion (relative to the cAMP levels attained during infections with L. pneumophilacarrying the LepA-CyaA fusion) suggests that only small amounts of HtpB reach the host cell cytoplasm or that fusion to CyaA artificially reduces the efficiency of HtpB passage into the host cell cytoplasm. The amount of HtpB that reached the host cell cytoplasm was particularly small in cells infected with L. pneumophilastrain Lp02. This is reminiscent of the situation proposed for L. pneumophila's flagellin, which also reaches the host cell cytoplasm in small amounts, perhaps passing through the pores formed in the LCV membrane by the Dot/Icm system (89), without being a type IV secretion substrate per se. It is also possible that the free HtpB present in the lumen of the LCV could cross the LCV membrane in a manner similar to that proposed for the cell-penetrating peptides that directly interact with lipid bilayers and alter their structure (95). Alternatively, HtpB contained in outer membrane vesicles (26) could reach the host cell cytoplasm upon the fusion of these vesicles with the LCV membrane. The accidental delivery of HtpB into the host cell cytoplasm (due to bacterial cell lysis and LCV rupture) is unlikely, mainly because it was controlled for by the very low levels of cAMP observed after infection with L. pneumophilacarrying only the CyaA construct and by the inability of L. pneumophilacarrying the N-terminal CyaA-HtpB fusion to efficiently induce increased amounts of cAMP (Fig. 1).
Since none of the aforementioned possible transport processes for HtpB involve direct translocation of HtpB, its transfer across the LCV membrane would differ fundamentally from the active Dot/Icm-dependent translocation of effectors such as LepA (12). The parent strain of Lp02 (the thymidine prototroph Lp01) lacks two conjugation systems (the Lvh and tra1type IV secretion systems) present in other L. pneumophilastrains, whereas strain JR32 carries the Lvh system (75). Strains Lp01 and JR32 also differ in the abilities to infect mice and grow in amoebae and monocyte-derived macrophages, the latter being more efficient (75). The presence of multiple type IV secretion systems in L. pneumophilastrain JR32 might, in part, explain why cells infected with this strain had more cytoplasmic HtpB-CyaA and CyaA-HtpB, particularly if passage of HtpB fusion proteins through type IV system-mediated pores is the mechanism at work. The fact that no difference between strains Lp02 and JR32 in the translocation of LepA-CyaA was observed suggests that these strains have similarly functional Dot/Icm systems and that the differences in HtpB translocation are not likely Dot/Icm related. Thus, we presume that HtpB is not a substrate of the Dot/Icm system, but experimental proof of this notion awaits future resolution.
In spite of the high amino acid sequence similarity that exists between bacterial chaperonins, we have recently shown that HtpB is capable of performing unique roles that are not shared by the E. coliprotein GroEL (16). Microbeads coated with purified HtpB, but not GroEL, attracted mitochondria and transiently modified the organization of actin microfilaments in mammalian cells, mimicking the trafficking of LCVs (16). We demonstrated here that the ability of HtpB to activate PHG in S. cerevisiaeis not shared by GroEL or by the S. cerevisiaemitochondrial chaperonin (Hsp60p) expressed in the yeast cytosol. Unique functions of bacterial chaperonins have been previously reported, which can be attributed to the effect of a few amino acids. Toxic GroEL from endosymbiotic E. aerogenes(comprising 545 amino acids) differs from nontoxic E. coliGroEL (comprising 548 amino acids) by 11 amino acids, of which only 4 are critical for toxicity. When nontoxic E. coliGroEL was engineered at the four critical residues to resemble E. aerogenesGroEL it, too, became a potent insect toxin (93). In the case of the Hsp65 chaperonin of M. leprae, which acts as a protease, only three amino acids (Thr-375, Lys-409, and Ser-502) formed the threonine catalytic group responsible for protease activity (69). Differences in the signaling abilities of bacterial chaperonins, accompanied by a variety of downstream consequences (reviewed in reference 71), have also been previously reported. For instance, the 60-kDa chaperonins of Actinobacillus actinomycetemcomitansand E. coliwere extremely active stimulators of bone resorption in a mouse model, whereas the chaperonins of Mycobacterium tuberculosisand M. lepraeshowed no such activity (48). Because HtpB and E. coliGroEL differ in 137 amino acids scattered throughout the proteins, it is not easily discernible which of these residues confer on HtpB its unique ability to interact with SAMDC and trigger PHG in yeast.
The term “polyamines” describes a group of polycationic compounds present in all cells. Polyamines are similarly important in prokaryotes and eukaryotes, being crucial for normal cell growth, DNA and protein synthesis, and eukaryotic cell differentiation, proliferation, and signaling (18, 65). By definition, these compounds have more than one amino group and are commonly synthesized using amino acids as precursors (18, 65). Based on the fact that a strong correlation exists between elevated polyamine levels and fungal filamentation (33, 38, 42, 57, 76), it seems reasonable to surmise that the activation of PHG by HtpB, via its interaction with SAMDC, is mediated by increased concentrations of intracellular polyamines in S. cerevisiae. In this view, it is predicted that yeast cells expressing HtpB would have elevated levels of spermidine and spermine. SAMDC is a rate-limiting enzyme key in the biosynthesis of polyamines, which is tightly regulated in eukaryotic cells (67). Like other rate-limiting enzymes, SAMDC has a short half-life, its basal activity is low, and it is rapidly induced by different stimuli (64, 66, 67). Furthermore, SAMDC is synthesized as an inactive proenzyme that, in response to different stimuli, undergoes an intramolecular cleavage reaction to form the active enzyme (46, 64, 66). Thus, it is possible that one or more of the processes that affect the activity of SAMDC could be modulated upon interaction with HtpB. For instance, the chaperonin activity of HtpB could extend the half-life of SAMDC, protecting it from early degradation, or the presence of HtpB could increase the rate of proenzyme cleavage.
The physiological model advanced at the beginning of this section predicts that HtpB modulates the intracellular pool of polyamines in host cells. These polyamines could then be transported into the LCV and directly used by L. pneumophilato grow optimally (nutritional role) and/or change the physiology of the host cell to favor L. pneumophilaproliferation (intracellular signaling role). The nutritional role of host polyamines is supported by the in vitroenhancement of L. pneumophilagrowth in liquid medium supplemented with polyamines (Fig. 8) and by the limited ability of L. pneumophilato synthesize polyamines, predicted from our bioinformatic analysis (Fig. 9). However, an alteration of host cell physiology in favor of intracellular L. pneumophila(independent of a merely nutritional effect) cannot be ruled out by our results. For instance, the lack of inhibition by DFMO of L. pneumophilareplication at 24 h postinfection (Fig. 6a) could be interpreted as meaning that SAMDC activity, rather than increased concentrations of spermidine and spermine, is necessary for the optimal growth of L. pneumophila. This is made all the more plausible by the knowledge that inhibition of ornithine decarboxylase by DFMO is known to increase SAMDC activity (66). However, DFMO might simply have a late effect on the polyamine pool of host cells due to its slow uptake (78), its inability to effectively reduce spermine levels in treated cells (32), or the expected long period required to exhaust the levels of putrescine, and consequently spermidine, in host cells.
It should be noted here that the overall biological effect of polyamines on the growth of L. pneumophila in vitroand in cultured cells was modestly enhancing. In all of the instances tested here, L. pneumophilawas able to grow in the absence of added polyamines and/or in the presence of pharmacological inhibitors of polyamine biosynthesis. Nonetheless, optimal growth of L. pneumophilawas achieved only when polyamines were plentiful and host cell SAMDC activity was uninhibited. Thus, while not essential in principle, polyamines still could play an important role in enhancing the growth of L. pneumophiladuring its intricate interactions with host cells and ultimately tilt the outcome of the infection in favor of this pathogen.
The role of polyamines in the growth and virulence of human pathogens has recently attracted increased attention (reviewed in reference 79). H. pyloridecreases macrophage survival by modulating the activity of host ornithine decarboxylase (13). Similarly, Pneumocystis jiroveciiupregulates polyamine biosynthesis, which is, in turn, thought to induce apoptosis of alveolar macrophages (51). Other reports have shown that the deletion of some spegenes (Fig. 9) in several bacterial human pathogens affects biofilm formation. For instance, the inactivation of either speAor speCreduced the attachment of Yersinia pestisto a solid surface (62) and deletion of speABin Proteus mirabiliseliminates swarming (80). In V. cholerae, deletion of genes involved in polyamine metabolism (52) or transport (45) severely reduced its ability to form biofilms. In V. cholerae(47) and E. coli(60), polyamines are important for siderophore production and iron acquisition. Finally, a Streptococcus pneumoniaepotDmutant (unable to effectively transport exogenous polyamines) showed significant attenuation in murine virulence models (87). Because polyamines appear to have multiple functions in bacterial human pathogens, studies of the role of polyamines in L. pneumophilapathogenesis are warranted to uncover new mechanisms used by this intracellular bacterium to survive and proliferate within host cells. Here we have shown that increased levels of exogenous polyamines, as well as host SAMDC activity, have an enhancing effect on the intracellular growth of L. pneumophilaand proposed a potential role for the HtpB chaperonin in this process.
ACKNOWLEDGMENTS
Kaitlyn Carson, Andrew Caddell, and Hany Abdelhady are gratefully acknowledged for their help with statistical analyses, dilution plating for CFU counts, and U937 cell activation, respectively. We thank Howard Shuman for generously providing plasmids, antibodies against LepA, and L. pneumophilastrains, as well as Paul Hoffman for antibodies. We acknowledge the following individuals for providing plasmids and/or strains needed for our work: David Allan, David Balasundaram, Gerald Johnston, H.-U. Mösch, Jim Hopper, Joseph Vogel, Pak Poon, Paul Sigler, Philip James, Ralph Isberg, and Stephen Elledge.
We thank the reviewers of our manuscript for the many valuable comments that helped to improve it.
This work was funded by independent grants from the Natural Sciences and Engineering Research Councilof Canada to R.A.G. and L.E.M.
Footnotes
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Archambault J., Drebot M. A., Stone J. C., Friesen J. D. 1992. Isolation and phenotypic analysis of conditional-lethal, linker-insertion mutations in the gene encoding the largest subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Gen. Genet. 232:408–414 [DOI] [PubMed] [Google Scholar]
- 3. Balasundaram D., Dinman J. D., Tabor C. W., Tabor H. 1994. SPE1and SPE2: two essential genes in the biosynthesis of polyamines that modulate +1 ribosomal frameshifting in Saccharomyces cerevisiae. J. Bacteriol. 176:7126–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balasundaram D., Tabor C. W., Tabor H. 1991. Spermidine or spermine is essential for the aerobic growth of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 88:5872–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balasundaram D., Tabor C. W., Tabor H. 1993. Oxygen toxicity in a polyamine-depleted spe2Δ mutant of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 90:4693–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berger K. H., Isberg R. R. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7–19 [DOI] [PubMed] [Google Scholar]
- 7. Bochkareva E. S., Lissin N. M., Girshovich A. S. 1988. Transient association of newly synthesized unfolded proteins with the heat-shock GroEL protein. Nature 336:254–257 [DOI] [PubMed] [Google Scholar]
- 8. Campodonico E. M., Chesnel L., Roy C. R. 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophilaDot/Icm system. Mol. Microbiol. 56:918–933 [DOI] [PubMed] [Google Scholar]
- 9. Cao P., McClain M. S., Forsyth M. H., Cover T. L. 1998. Extracellular release of antigenic proteins by Helicobacter pylori. Infect. Immun. 66:2984–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chattopadhyay M. K., Tabor C. W., Tabor H. 2002. Absolute requirement of spermidine for growth and cell cycle progression of fission yeast (Schizosaccharomyces pombe). Proc. Natl. Acad. Sci. U. S. A. 99:10330–10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chattopadhyay M. K., Tabor C. W., Tabor H. 2009. Polyamines are not required for aerobic growth of Escherichia coli: preparation of a strain with deletions in all of the genes for polyamine biosynthesis. J. Bacteriol. 191:5549–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J., et al. 2004. Legionellaeffectors that promote nonlytic release from protozoa. Science 303:1358–1361 [DOI] [PubMed] [Google Scholar]
- 13. Cheng Y., et al. 2005. Helicobacter pylori-induced macrophage apoptosis requires activation of ornithine decarboxylase by c-Myc. J. Biol. Chem. 280:22492–22496 [DOI] [PubMed] [Google Scholar]
- 14. Chien M., et al. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968 [DOI] [PubMed] [Google Scholar]
- 15. Chong A. 2007. Characterization of the virulence-related roles of the Legionella pneumophilachaperonin, HtpB, in mammalian cells. Ph.D. dissertation. Dalhousie University, Halifax, Nova Scotia, Canada.
- 16. Chong A., Lima C. A., Allan D. S., Nasrallah G. K., Garduño R. A. 2009. The purified and recombinant Legionella pneumophilachaperonin alters mitochondrial trafficking and microfilament organization. Infect. Immun. 77:4724–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clontech 1999. MATCHMAKER GAL4 two-hybrid system and libraries user manual. Clontech, Palo Alto, CA.
- 18. Cohen S. (ed.). 1998. A guide to the polyamines. Oxford University Press, New York, NY [Google Scholar]
- 19. Corti A., Dave C., Williams-Ashman H. G., Mihich E., Schenone A. 1974. Specific inhibition of the enzymic decarboxylation of S-adenosylmethionine by methylglyoxal bis(guanylhydrazone) and related substances. Biochem. J. 139:351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Felipe K. S., et al. 2005. Evidence for acquisition of Legionellatype IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 187:7716–7726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Durfee T., et al. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555–569 [DOI] [PubMed] [Google Scholar]
- 22. Erhart E., Hollenberg C. P. 1983. The presence of a defective LEU2gene on 2 mu DNA recombinant plasmids of Saccharomyces cerevisiaeis responsible for curing and high copy number. J. Bacteriol. 156:625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fayet O., Ziegelhoffer T., Georgopoulos C. 1989. The groESand groELheat shock gene products of Escherichia coliare essential for bacterial growth at all temperatures. J. Bacteriol. 171:1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fernandez R. C., Logan S. M., Lee S. H., Hoffman P. S. 1996. Elevated levels of Legionella pneumophilastress protein Hsp60 early in infection of human monocytes and L929 cells correlate with virulence. Infect. Immun. 64:1968–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frisk A., Ison C. A., Lagergard T. 1998. GroEL heat shock protein of Haemophilus ducreyi: association with cell surface and capacity to bind to eukaryotic cells. Infect. Immun. 66:1252–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galka F., et al. 2008. Proteomic characterization of the whole secretome of Legionella pneumophilaand functional analysis of outer membrane vesicles. Infect. Immun. 76:1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gancedo J. M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:107–123 [DOI] [PubMed] [Google Scholar]
- 28. Garduño R. A., Faulkner G., Trevors M. A., Vats N., Hoffman P. S. 1998. Immunolocalization of Hsp60 in Legionella pneumophila. J. Bacteriol. 180:505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garduño R. A., Garduño E., Hiltz M., Hoffman P. S. 2002. Intracellular growth of Legionella pneumophilagives rise to a differentiated form dissimilar to stationary-phase forms. Infect. Immun. 70:6273–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garduño R. A., Garduño E., Hoffman P. S. 1998. Surface-associated Hsp60 chaperonin of Legionella pneumophilamediates invasion in a HeLa cell model. Infect. Immun. 66:4602–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gietz D., St. Jean A., Woods R. A., Schiestl R. H. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grove J., Fozard J. R., Mamont P. S. 1981. Assay of alpha-difluoromethylornithine in body fluids and tissues by automatic amino-acid analysis. J. Chromatogr. 223:409–416 [DOI] [PubMed] [Google Scholar]
- 33. Guevara-Olvera L., Xoconostle-Cazares B., Ruiz-Herrera J. 1997. Cloning and disruption of the ornithine decarboxylase gene of Ustilago maydis: evidence for a role of polyamines in its dimorphic transition. Microbiology 143:2237–2245 [DOI] [PubMed] [Google Scholar]
- 34. Gupta R. S. 1995. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol. Microbiol. 15:1–11 [DOI] [PubMed] [Google Scholar]
- 35. Guthrie C., Fink (ed.) G. R. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1–863 [PubMed] [Google Scholar]
- 36. Helsel L. O., Bibb W. F., Butler C. A., Hoffman P. S., McKinney R. M. 1988. Recognition of a genus-wide antigen of Legionellaby a monoclonal-antibody. Curr. Microbiol. 16:201–208 [Google Scholar]
- 37. Hennequin C., et al. 2001. GroEL (Hsp60) of Clostridium difficileis involved in cell adherence. Microbiology 147:87–96 [DOI] [PubMed] [Google Scholar]
- 38. Herrero A. B., et al. 1999. Control of filament formation in Candida albicansby polyamine levels. Infect. Immun. 67:4870–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoffman P. S., Butler C. A., Quinn F. D. 1989. Cloning and temperature-dependent expression in Escherichia coliof a Legionella pneumophilagene coding for a genus-common 60-kilodalton antigen. Infect. Immun. 57:1731–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoffman P. S., Houston L., Butler C. A. 1990. Legionella pneumophila htpABheat shock operon: nucleotide sequence and expression of the 60-kilodalton antigen in L. pneumophila-infected HeLa cells. Infect. Immun. 58:3380–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horwitz M. A., Silverstein S. C. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Invest. 66:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Inderlied C. B., Cihlar R. L., Sypherd P. S. 1980. Regulation of ornithine decarboxylase during morphogenesis of Mucor racemosus. J. Bacteriol. 141:699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. James P., Halladay J., Craig E. A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnston S. A., Hopper J. E. 1982. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc. Natl. Acad. Sci. U. S. A. 79:6971–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Karatan E., Duncan T. R., Watnick P. I. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio choleraebiofilm formation by norspermidine. J. Bacteriol. 187:7434–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kashiwagi K., Taneja S. K., Liu T. Y., Tabor C. W., Tabor H. 1990. Spermidine biosynthesis in Saccharomyces cerevisiae. Biosynthesis and processing of a proenzyme form of S-adenosylmethionine decarboxylase. J. Biol. Chem. 265:22321–22328 [PubMed] [Google Scholar]
- 47. Keating T. A., Marshall C. G., Walsh C. T. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513–15521 [DOI] [PubMed] [Google Scholar]
- 48. Kirby A. C., et al. 1995. The potent bone-resorbing mediator of Actinobacillus actinomycetemcomitansis homologous to the molecular chaperone GroEL. J. Clin. Invest. 96:1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuchin S., Vyas V. K., Carlson M. 2003. Role of the yeast Snf1 protein kinase in invasive growth. Biochem. Soc. Trans. 31:175–177 [DOI] [PubMed] [Google Scholar]
- 50. Kuchin S., Vyas V. K., Carlson M. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lasbury M. E., et al. 2007. Polyamine-mediated apoptosis of alveolar macrophages during Pneumocystispneumonia. J. Biol. Chem. 282:11009–11020 [DOI] [PubMed] [Google Scholar]
- 52. Lee J., et al. 2009. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J. Biol. Chem. 284:9899–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lema M. W., Brown A., Butler C. A., Hoffman P. S. 1988. Heat-shock response in Legionella pneumophila. Can. J. Microbiol. 34:1148–1153 [DOI] [PubMed] [Google Scholar]
- 54. Lin Z., Rye H. S. 2006. GroEL-mediated protein folding: making the impossible, possible. Crit. Rev. Biochem. Mol. Biol. 41:211–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lund P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93–140 [DOI] [PubMed] [Google Scholar]
- 56. Mamont P. S., Duchesne M. C., Grove J., Bey P. 1978. Anti-proliferative properties of dl-alpha-difluoromethyl ornithine in cultured cells. A consequence of the irreversible inhibition of ornithine decarboxylase. Biochem. Biophys. Res. Commun. 81:58–66 [DOI] [PubMed] [Google Scholar]
- 57. Martinez J. P., Lopez-Ribot J. L., Gil M. L., Sentandreu R., Ruiz-Herrera J. 1990. Inhibition of the dimorphic transition of Candida albicansby the ornithine decarboxylase inhibitor 1,4-diaminobutanone: alterations in the glycoprotein composition of the cell wall. J. Gen. Microbiol. 136:1937–1943 [DOI] [PubMed] [Google Scholar]
- 58. Nagai H., et al. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the LegionellaRalF protein to host cells. Proc. Natl. Acad. Sci. U. S. A. 102:826–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Newton H. J., Ang D. K. Y., van Driel I. R., Hartland E. L. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 23:274–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pan Y. H., et al. 2006. The critical roles of polyamines in regulating ColE7 production and restricting ColE7 uptake of the colicin-producing Escherichia coli. J. Biol. Chem. 281:13083–13091 [DOI] [PubMed] [Google Scholar]
- 61. Pasculle A. W., et al. 1980. Pittsburgh pneumonia agent: direct isolation from human lung tissue. J. Infect. Dis. 141:727–732 [DOI] [PubMed] [Google Scholar]
- 62. Patel C. N., et al. 2006. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 188:2355–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pearson W. R. 1995. Comparison of methods for searching protein sequence databases. Protein Sci. 4:1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pegg A. E. 2009. S-Adenosylmethionine decarboxylase. Essays Biochem. 46:25–45 [DOI] [PubMed] [Google Scholar]
- 65. Pegg A. E. 1986. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem. J. 234:249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pegg A. E., et al. 1988. Regulation of mammalian S-adenosylmethionine decarboxylase. Adv. Enzyme Regul. 27:43–55 [DOI] [PubMed] [Google Scholar]
- 67. Pegg A. E., Xiong H., Feith D. J., Shantz L. M. 1998. S-Adenosylmethionine decarboxylase: structure, function and regulation by polyamines. Biochem. Soc. Trans. 26:580–586 [DOI] [PubMed] [Google Scholar]
- 68. Pine L., Franzus M. J., Malcolm G. B. 1986. Guanine is a growth factor for Legionellaspecies. J. Clin. Microbiol. 23:163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Portaro F. C., et al. 2002. The Mycobacterium lepraehsp65 displays proteolytic activity. Mutagenesis studies indicate that the M. lepraehsp65 proteolytic activity is catalytically related to the HslVU protease. Biochemistry 41:7400–7406 [DOI] [PubMed] [Google Scholar]
- 70. Poulin R., Lu L., Ackermann B., Bey P., Pegg A. E. 1992. Mechanism of the irreversible inactivation of mouse ornithine decarboxylase by alpha-difluoromethylornithine. Characterization of sequences at the inhibitor and coenzyme binding sites. J. Biol. Chem. 267:150–158 [PubMed] [Google Scholar]
- 71. Ranford J. C., Coates A. R., Henderson B. 2000. Chaperonins are cell-signalling proteins: the unfolding biology of molecular chaperones. Expert Rev. Mol. Med. 2(8):1–17 [DOI] [PubMed] [Google Scholar]
- 72. Retzlaff C., et al. 1996. Legionella pneumophilaheat-shock protein-induced increase of interleukin-1 beta mRNA involves protein kinase C signalling in macrophages. Immunology 89(2):281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sadosky A. B., Wiater L. A., Shuman H. A. 1993. Identification of Legionella pneumophilagenes required for growth within and killing of human macrophages. Infect. Immun. 61:5361–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 75. Samrakandi M. M., Cirillo S. L. G., Ridenour D. A., Bermudez L. E., Cirillo J. D. 2002. Genetic and phenotypic differences between Legionella pneumophilastrains. J. Clin. Microbiol. 40:1352–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. San-Blas G., San-Blas F., Sorais F., Moreno B., Ruiz-Herrera J. 1996. Polyamines in growth and dimorphism of Paracoccidioides brasiliensis. Arch. Microbiol. 166:411–413 [DOI] [PubMed] [Google Scholar]
- 77. Scopio A., Johnson P., Laquerre A., Nelson D. R. 1994. Subcellular localization and chaperone activities of Borrelia burgdorferiHsp60 and Hsp70. J. Bacteriol. 176:6449–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seiler N., Delcros J. G., Moulinoux J. P. 1996. Polyamine transport in mammalian cells. An update. Int. J. Biochem. Cell Biol. 28:843–861 [DOI] [PubMed] [Google Scholar]
- 79. Shah P., Swiatlo E. 2008. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 68:4–16 [DOI] [PubMed] [Google Scholar]
- 80. Sturgill G., Rather P. N. 2004. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 51:437–446 [DOI] [PubMed] [Google Scholar]
- 81. Tabor C. W., Tabor H. 1984. Polyamines. Annu. Rev. Biochem. 53:749–790 [DOI] [PubMed] [Google Scholar]
- 82. Tabor C. W., Tabor H. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Török Z., et al. 1997. Evidence for a lipochaperonin: association of active protein-folding GroESL oligomers with lipids can stabilize membranes under heat shock conditions. Proc. Natl. Acad. Sci. U. S. A. 94:2192–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Towbin H., Staehelin T., Gordon J. 1992. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Biotechnology 24:145–149 [PubMed] [Google Scholar]
- 85. VanBogelen R. A., Acton M. A., Neidhardt F. C. 1987. Induction of the heat shock regulon does not produce thermotolerance in Escherichia coli. Genes Dev. 1:525–531 [DOI] [PubMed] [Google Scholar]
- 86. Viswanathan V. K., Cianciotto N. P. 2000. Electroporation of Legionellaspecies, p. 203–211 In Eynard N., Teissie J. (ed.), Electro-transformation of bacteria. Springer Verlag, Heidelberg, Germany [Google Scholar]
- 87. Ware D., Jiang Y., Lin W., Swiatlo E. 2006. Involvement of potDin Streptococcus pneumoniaepolyamine transport and pathogenesis. Infect. Immun. 74:352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Warrell R. P., Jr., Burchenal J. H. 1983. Methylglyoxal-bis(guanylhydrazone) (Methyl-GAG): current status and future prospects. J. Clin. Oncol. 1:52–65 [DOI] [PubMed] [Google Scholar]
- 89. Whitfield N. N., Byrne B. G., Swanson M. S. 2010. Mouse macrophages are permissive to motile Legionellaspecies that fail to trigger pyroptosis. Infect. Immun. 78:423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Williams-Ashman H. G., Schenone A. 1972. Methyl glyoxal bis(guanylhydrazone) as a potent inhibitor of mammalian and yeast S-adenosyl methionine decarboxylases. Biochem. Biophys. Res. Commun. 46:288–295 [DOI] [PubMed] [Google Scholar]
- 91. Winn W. C., Jr 1988. Legionnaires disease: historical perspective. Clin. Microbiol. Rev. 1:60–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yamaguchi H., Osaki T., Kurihara N., Taguchi H., Kamiya S. 1998. The role of heat shock protein 60 (HSP60) of Helicobacter pyloriin adhesion of H. pylorito human gastric epithelial cell. Kansenshogaku Zasshi 72:487–492 [DOI] [PubMed] [Google Scholar]
- 93. Yoshida N., et al. 2001. Protein function. Chaperonin turned insect toxin. Nature 411:44. [DOI] [PubMed] [Google Scholar]
- 94. Zeilstra-Ryalls J., Fayet O., Georgopoulos C. 1991. The universally conserved GroE (Hsp60) chaperonins. Annu. Rev. Microbiol. 45:301–325 [DOI] [PubMed] [Google Scholar]
- 95. Zorko M., Langel ü. 2005. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv. Drug Deliv. Rev. 57:529–545 [DOI] [PubMed] [Google Scholar]