Abstract
Enteropathogenic Escherichia coli(EPEC) requires the tnaA-encoded enzyme tryptophanase and its substrate tryptophan to synthesize diffusible exotoxins that kill the nematode Caenorhabditis elegans. Here, we demonstrate that the RNA-binding protein CsrA and the tryptophan permease TnaB coregulate tryptophanase activity, through mutually exclusive pathways, to stimulate toxin-mediated paralysis and killing of C. elegans.
TEXT
Enteropathogenic Escherichia coli(EPEC) belongs to the attaching and effacing (A/E) family of pathogens, the other members of which include enterohemorrhagic Escherichia coli(EHEC) and Citrobacter rodentium(8, 17, 46, 56, 58, 64, 72). Upon infection, A/E pathogens bind to intestinal epithelia and destroy the cellular microvilli in their vicinity (8, 17, 58). Subsequently, the bacteria recruit several host factors that cooperate to promote the biogenesis of actin-filled membranous protrusions, termed “pedestals,” beneath adherent bacteria (17, 25, 46, 49, 58, 64). Pedestal formation is accompanied by severe diarrhea, which results in significant morbidity and mortality worldwide (17, 34, 74).
Penetrance of the A/E pathomorphology requires the pathogenicity island (PAI), locus of enterocyte effacement (LEE) that encodes for the regulators, structural components of a type III secretion system (T3SS), and several of its secreted effector molecules (8, 18, 20, 25, 28, 46, 47, 58, 60, 73, 91). The LEE1-encoded master regulator (Ler) orchestrates the coordinated transcription from the other LEE operons to promote morphogenesis of the T3SS that forms a continuous conduit between the bacterial and the host cytoplasm (5, 15, 25, 28, 31, 47, 58, 60, 78). Subsequently, effectors, including the translocated intimin receptor (Tir), are trafficked into the host (18, 46, 47). Tir integrates into the host plasma membrane, where it serves as a receptor for its ligand, the adhesin intimin, located on the outer bacterial membrane (47). Tir-intimin interactions initiate a signal transduction cascade that leads to actin polymerization and pedestals (10, 41, 45, 47, 83).
A significant obstacle in elucidating the pathobiology of EPEC infections is that this bacterium is a human pathogen that neither colonizes nor causes disease in mice (62). Over the past decade, the capacity of bacterial pathogens, including EPEC, to kill the nematode Caenorhabditis eleganshas been utilized to identify virulence determinants in the bacteria that may be relevant to pathogenesis in mammalian systems (61, 75). Its small size, rapid generation time, large brood size, amenability to genetic manipulation, and high degree of homology to humans and other mammals make C. elegansa useful experimental system with which to study bacterial toxins or infection (1, 61, 75, 76).
The morbidity and mortality in C. eleganscaused by noxious microbes can be classified into two broad categories on the basis of whether the pathogen makes contact with the worm (1, 61, 75, 76). Contact-dependent killing usually involves the detrimental colonization of the worm in the form of a biofilm (e.g., Yersinia pestis) (21, 84), an invasive infection (e.g., Streptomyces albireticuli) (67), or accumulation within the intestine (e.g., EPEC) (59). The death of the nematode, as a consequence of colonization, typically occurs over several days and is referred to as “slow killing” (75). In contrast, contact-independent killing is mediated through structurally and functionally unrelated exotoxins that are secreted by diverse pathogens, including EPEC (2, 3), Pseudomonas aeruginosa(32, 57), and Burkholderia cenocepacia(51), among others, that lead to toxicity in the nematode (75). Intoxication of the worms is a relatively rapid pathophysiological process occurring over a period of hours and is referred to as “fast killing” (75). The utility of C. elegansas a surrogate host for mimicking bacterial infections has been repetitively substantiated by numerous studies in which novel virulence factors that were identified employing worm-based screens were subsequently shown to modulate virulence in mammalian systems (2, 33, 57, 75, 85). In reciprocal studies, virulence factors originally implicated in mammalian and plant pathogenesis were demonstrated to coregulate pathogenesis in worms (75, 76, 86).
EPEC is capable of killing C. elegansby contact-dependent and -independent means (2, 59). On minimal nematode growth medium (NGM), EPEC kills C. elegansover a period of several days by colonizing its intestinal tract (59). However, no virulence factors that contribute to the contact-dependent killing of the worm have thus far been discovered (59). Moreover, none of the virulence determinants previously implicated in mammalian pathogenesis were necessary for nematocidal activity (59). In contrast, on nutritionally rich medium (Luria-Bertani [LB] or E. colidirect agar [ECD]) supplemented with tryptophan, EPEC synthesizes diffusible exotoxins that lead to rapid paralysis and subsequent death of the nematode within a few hours (2, 3). Exotoxin-induced lethality requires the bacterial enzyme tryptophanase. Subsequently, it was shown that tryptophanase regulates the LEE in both EPEC and EHEC and consequently influences pedestal formation and mammalian pathogenesis (2, 42). However, other than tryptophanase, the EPEC-C. eleganspathosystem has not been exploited to identify additional virulence determinants that may contribute to morbidity in mammals.
In a previous study, we reported that the RNA-binding protein, CsrA, is necessary for EPEC to form pedestals on mammalian cells (7, 8). CsrA and its ortholog, RsmA, recognize AGGA/ANGGA tracts in the 5′-untranslated leader segments of transcripts and modulate mRNA stability and/or translation (7, 8, 26, 68). The relaxed sequence specificity of CsrA/RsmA enables this posttranscriptional regulator to modulate a panoply of physiological traits, such as carbon homeostasis (4, 27, 68–70), peptide uptake (27), biofilm formation (44, 88), motility (7, 14, 52, 90, 92), quorum sensing (19, 55), colicin biosynthesis (93), and virulence (7, 13, 19, 30, 40, 48, 55).
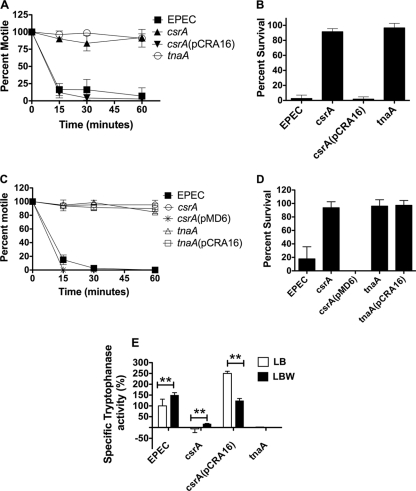
Here, we have evaluated the role of CsrA in the toxin-mediated killing of C. elegans. Bioassays employing worms were conducted on LB agar plates containing or lacking tryptophan essentially as described previously, with the modification that tryptophan was added to a final concentration of 1 mg/ml, and 200 μl of the overnight inoculum was seeded onto plates (2). Disruption of csrAabolished the ability of EPEC to paralyze (Fig. 1A) and kill (Fig. 1B) C. elegans. The csrAmutant regained its pathogenicity when complemented in transwith the plasmid pCRA16 that expresses csrAunder its native promoters (Fig. 1A and B) (Table 1) (89).
Fig. 1.
(A to E) csrAregulates tryptophanase activity to promote toxin-mediated killing of C. elegansby EPEC. (A and B) Young adult worms were exposed to confluent lawns of EPEC, csrAmutant, csrAmutant complemented with a functional csrAallele expressed from a multicopy plasmid [csrA(pCRA16)], and tnaAmutant and monitored for paralysis (A) and killing (B) on LB agar supplemented with tryptophan (LBW). Worms were considered paralyzed if they failed to traverse an entire body length on prodding. Worm mortality was assayed by transferring the pathogen-exposed worms onto NGM plates containing nonpathogenic E. coliOP50 and assaying for motility 24 h later. Error bars indicate standard deviations of results from at least three independent experiments, with each employing at least two biological replicates. A one-way analysis of variance (ANOVA) was used to assess statistical significance. A Pvalue cutoff of <0.05 was considered statistically significant. The calculated Pvalues for both the paralysis and killing assays were <0.02. (C and D) Paralysis (C) and killing (D) of the nematodes were assayed in the presence of EPEC, csrAmutant, csrAmutant overexpressing tnaA[csrA(pMD6)], tnaAmutant, and tnaAmutant overexpressing csrA[tnaA(pCRA16)] essentially as described above. A one-way ANOVA was used to assess statistical significance. The calculated Pvalues for both the paralysis and killing assays were <0.02. (E) Tryptophanase activity was measured from lysates of bacteria cultivated on agar plates. The rate of hydrolysis of SOPC, a chromogenic tryptophan analogue, to ONTP was measured as described previously (2). Error bars indicate standard deviations of results from at least two independent experiments, each with at least three replicates. The unpaired Student ttest was employed to assay for statistical significance between the indicated samples. A Pvalue cutoff of <0.01 was considered statistically significant. **, P< 0.01.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype/phenotype | Reference or source |
|---|---|---|
| Strains | ||
| EPEC | Prototypical EPEC 2348/69 serotype O127:H6 | Jim Kaper |
| EPEC csrA | EPEC 2348/69 ΩcsrA::cat/Cmr | 7 |
| EPEC tnaA | EPEC 2348/69 ΔtnaA::cat/Cmr | 2 |
| EPEC csrA(pCRA16) | EPEC 2348/69 ΩcsrA::cattransformed with the plasmid pCRA16/CmrTcr | This study |
| EPEC csrA(pMD6) | EPEC 2348/69 ΩcsrA::cattransformed with the plasmid pMD6/CmrApr | This study |
| EPEC tnaA(pCRA16) | EPEC 2348/69 ΔtnaA::cattransformed with the plasmid pCRA16/CmrTcr | This study |
| EPEC tnaB | EPEC 2348/69 ΩtnaB::Tn5-kan/Kmr | This study |
| EPEC tnaB(ptnaB) | EPEC 2348/69 ΩtnaB::Tn5-kantransformed with the plasmid ptnaB/KmrApr | This study |
| EPEC mtr | EPEC 2348/69 Δmtr::cat/Cmr | This study |
| EPEC aroP | EPEC 2348/69 ΔaroP::cat/Cmr | This study |
| Plasmids | ||
| pKD3 | pANTSγ-(FRT-cat-FRT) R6KγoriV/AprCmr | 22 |
| pCRA16 | pBR322-Ωbla::(PcsrA-csrA+K-12)/Tcr | 82 |
| pMD6 | pBR322-(PtnaCAB-tnaCAK-12)/Apr | 23 |
| ptnaB | An EcoRI-PstI-restricted amplicon containing the tnaBORF from EPEC 2348/69 cloned downstream of the ParaBADpromoter of the identically restricted plasmid, pBAD24/Apr | This study |
The observation that disruption of csrAgenocopies the effect of deleting tnaA(Fig. 1) suggested that the two genes might constitute components of the same regulatory pathway. In E. coli, tnaAis the central gene within a tricistronic operon that includes the upstream regulatory gene tnaCand the downstream structural gene tnaB(23, 24, 36). tnaCencodes a cis-acting regulatory peptide that governs the expression of tnaAand tnaBin response to tryptophan accumulation (37, 80). tnaAencodes for the catabolic enzyme tryptophanase, which catalyzes the hydrolysis of tryptophan into indole, pyruvate, and ammonia, whereas tnaBspecifies a low-affinity tryptophan permease that facilitates the import of tryptophan into the bacterium (23, 54, 71, 77, 94). To elucidate the regulatory hierarchy of csrAand tnaA, each gene was expressed from a multicopy plasmid in the mutant background of the other. Whereas multicopy expression of csrAfailed to restore virulence to the tnaAmutant, overexpression of tnaA, from the medium-copy-number plasmid pMD6 (Table 1), suppressed the attenuated phenotype of the csrAmutant and restored its ability to paralyze (Fig. 1C) and kill (Fig. 1D) C. elegans. The observation that increased expression of tnaAcircumvents the requirement for a functional csrAallele raised the possibility that tnaAmight act downstream of CsrA in a putative regulatory pathway. To test this possibility, we assayed tryptophanase activity by measuring the hydrolysis of the chromogenic tryptophan analogue S-O-nitrophenyl-l-cysteine (SOPC) to O-nitrothiophenolate (ONTP) in bacterial lysates that had been precultivated on agar plates containing or lacking tryptophan, essentially as described previously (2). Accordingly, tryptophanase activity was dramatically reduced in the csrAmutant (Fig. 1E). Moreover, this effect occurred independently of the addition of exogenous tryptophan (Fig. 1E). Collectively, these results suggest that the inability of the csrAmutant to paralyze and kill the nematode results from reduced tryptophanase activity and that tnaAacts distally to csrA.
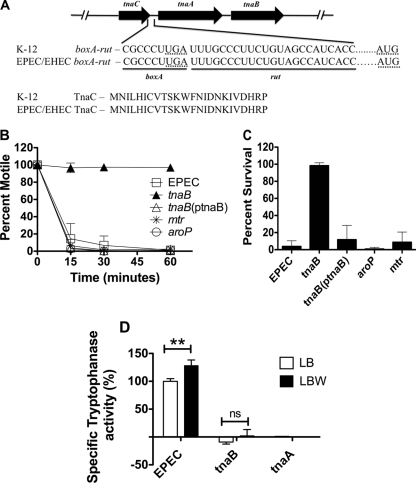
In E. coli, the tnaCABoperon is subject to transcriptional as well as posttranscriptional control (9, 11, 12, 16, 36, 81). The nascent leader peptide, TnaC, while translocating through the exit tunnel of the ribosome, transduces conformational alterations in the ribosome to generate a stereospecific l-tryptophan-binding site near the peptidyltransferase center (79). Bound tryptophan promotes ribosomal stalling, which in turn masks the boxA-rutriboelement of the transcriptional terminator Rho that overlaps the C terminus as well as the segment immediately downstream of the tnaCopen reading frame (ORF) (35, 37, 80). Consequently, Rho does not bind to the transcript and the stalled RNA polymerase is not offloaded and continues to transcribe the downstream genes tnaAand tnaB(35–38). Thus, tryptophan posttranscriptionally induces the expression from the tnaCABoperon in E. coli(79). The primary structure of the TnaC leader peptide as well as the nucleotide sequence of the boxA-rutsite within the tnaCABoperon of EPEC and EHEC are identical to that of E. coliK-12, suggesting that tryptophan-mediated stimulation of the tnaoperon is likely conserved (Fig. 2A). Consistent with this bioinformatic observation, a modest but reproducible increase in tryptophanase activity was observed upon addition of tryptophan to LB medium (Fig. 1E and 2D). LB medium is naturally replete with tryptophan in the form of tryptone, and thus its presence likely masks the actual induction in tryptophanase activity by exogenously added tryptophan.
Fig. 2.
(A to D) TnaB but not AroP or Mtr imports tryptophan into EPEC to stimulate toxin-dependent killing of C. elegans. (A) Comparative analysis of the primary structure of the boxA-rutriboelement and the TnaC leader peptide of the tnaCABoperon in E. coliK-12 and EPEC/EHEC. The translation termination codon of tnaCand the translation initiation codon of tnaAare indicated by dashed underlines. The intersite distance between the terminal nucleotide of rutand the translational initiation nucleotide of tnaAis 197 bases. (B and C) Young adult worms were exposed to EPEC and its congenic mutant derivatives, the tnaB, mtr, and aroPmutants and the tnaBcomplemented strain [tnaB(ptnaB)], and assayed for paralysis (B) and killing (C). Error bars indicate the standard deviations of results from at least three independent experiments, with each using at least two replicates. A one-way ANOVA was used to assess statistical significance. A Pvalue cutoff of <0.05 was considered statistically significant. The calculated Pvalues for both the paralysis and killing assays were <0.02. (D) Specific tryptophanase activity was assayed in the tnaBmutants as described above. The unpaired Student ttest was employed to assay for statistical significance between the indicated samples. A Pvalue cutoff of <0.01 was considered statistically significant. **, P< 0.01; ns, no statistically significant difference.
Because uptake of tryptophan is necessary for killing of C. elegans, we reasoned that tryptophan importers might also be necessary for toxin production. In E. coli, three permeases, tnaB, aroP, and mtr, are responsible for importing tryptophan into the bacterium (94). Orthologs of all the three transporters are present in EPEC (data not shown). Using lambda red-mediated recombineering, we substituted mtrand aroPwith a catcassette as described previously (7, 22, 63) and evaluated the roles of each of the permeases in toxin production and pathogenesis in C. elegans. Inactivation of mtror aroPdid not compromise the ability of EPEC to paralyze or kill C. elegans(Fig. 2B and C). In contrast, inactivation of tnaBwas sufficient to completely abolish EPEC-induced paralysis and killing of C. elegans(Fig. 2B and C). The tnaBmutant regained its pathogenicity when complemented with a functional tnaBallele that was expressed under a heterologous promoter from the low-copy-number plasmid ptnaB (Fig. 2B and C) (Tables 1and 2). The attenuated phenotype of the tnaBmutant correlated with reduced tryptophanase activity (Fig. 2D). Moreover, the tryptophan-mediated induction of tnaAwas no longer evident when tnaBwas inactivated (Fig. 2D). Taken together, these results suggest that on LB agar, TnaB is the primary permease responsible for importing tryptophan into the bacterium, which subsequently induces tnaA. Besides inducing tnaA, tryptophan is also one of the natural substrates of tryptophanase (65, 66). Because overexpression of tnaAin LB medium, without added tryptophan, is insufficient for worm killing, tryptophan must play an important role as a tryptophanase substrate and as a precursor for exotoxin synthesis. Interestingly, inactivation of csrAdoes not disrupt the tryptophan-mediated stimulation of tryptophanase (Fig. 1E), suggesting that the import of the inducer remains unhindered in the csrAmutant. This corroborates the observation that overexpression of tnaA, without tnaB, is sufficient to restore virulence to the csrAmutant when cultivated on LB agar supplemented with tryptophan (LBW) (Fig. 1C and D). Curiously, tryptophan repressed tryptophanase activity when csrAwas overexpressed (Fig. 1E). Biochemical studies with tryptophanase from E. colisuggest that the degradative product of tryptophan, indole, exerts a dose-dependent, feedback inhibitory effect on the enzymatic activity by competing with its substrates for the catalytic site (39). Moreover, derivatives of indole have also been demonstrated to silence the expression of tnaA(53). Thus, the observed phenotype likely stems from the repressive effect of elevated indole levels on the expression and/or activity of tryptophanase. In summary, our results suggest that CsrA and TnaB exert their effects via parallel pathways that converge at the level of regulation of tnaAto synthesize exotoxins that enable EPEC to paralyze and kill C. elegans(Fig. 3).
Table 2.
Oligonucleotides used in this study
| Primer | Sequenceb |
|---|---|
| 5′-aroP-P2-Wanner-EPEC | CCGCCACATACAGCTTATCGCGCTGGGAGGCGCGATAGGGACAGGCATATGAATATCCTCCTTA |
| 3′-aroP-P1-Wanner-EPEC | TACCTAACACGATCAGCCATACCGGGATCAGGTATACCGAAATCGGTGTAGGCTGGAGCTGCTTC |
| 5′-mtr-P2-Wanner-EPEC | TTATCGGCGGCACCATTATTGGCGCAGGGATGTTTTCTCTGCCAGCATATGAATATCCTCCTTA |
| 3′-mtr-P1-Wanner-EPEC | CATTGTGTAGGCAGCAGAAATGTCGGATAAGGCACCGCTGATTACGTGTAGGCTGGAGCTGCTTC |
| 5′-tnaB-EcoRI-pBAD24 | gcggccGAATTCCCTCTAAAGGTGGCATCATGACTG |
| 3′-tnaB-PstI-pBAD24 | gcggccCTGCAGAAAGCGGGACATGGGCTAAAG |
| c1a | TTATACGCAAGGCGACAAGG |
| c2a | GATCTTCCGTCACAGGTAGG |
See reference 22.
Underlined sequences indicate restriction sites. Lowercase indicates additional nucleotides that facilitate cleavage of the PCR product by the restriction enzyme.
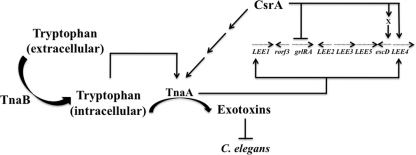
Fig. 3.
Model for the role of CsrA and TnaB in the regulation of tnaAand toxin production. CsrA positively regulates tryptophanase activity independently of the tryptophan permease TnaB. TnaB is the primary importer of tryptophan when EPEC is cultivated on LB medium. Imported tryptophan stimulates the expression of tnaA, as is evident by elevated tryptophanase activity. In turn, tryptophanase catabolizes tryptophan to synthesize exotoxins, which paralyze and kill C. elegans. csrAand tnaAalso regulate the LEEin A/E pathogens. Activating and repressive circuits are depicted as thin lines with arrowheads and blunt ends, respectively. Arrows with dashed lines represent the LEE. Curved arrows indicate catalytic reactions.
Herein, we provide evidence that the dual metabolic and virulence regulator CsrA, previously shown to regulate the virulence of EPEC in mammals (7), also contributes to pathogenicity in nematodes. Our results also suggest that toxin-based bioassays employing C. eleganscan be effectively utilized to identify novel virulence factors of A/E pathogens with relevance to mammalian pathogenesis. Future experiments utilizing a saturated transposon-mutagenized library will provide invaluable insight into evolutionarily conserved virulence determinants of EPEC. Moreover, using worm killing as a readout, we were able to determine the metabolic requirement of the different tryptophan importers in the nematocidal activity of EPEC. Thus, it may be possible to adapt the toxin-based assay to study alternative metabolic pathways and design screens to identify virulence factors for other pathogens. For instance, the murine A/E pathogen C. rodentiumlacks tnaA. However, the closely related enzyme tyrosine phenol lyase (tpl) is present in the genus Citrobacter(29, 43). Both the enzymes utilize the same cofactors and display remarkable conservation of key residues (6). Tpl enzymatically cleaves tyrosine to yield phenol, pyruvate, and ammonia. Because phenolic compounds are nematotoxic (50, 87), substitution of tryptophan with tyrosine in the medium may facilitate evaluation of the toxicity of C. rodentiumtoward C. elegansand identifying virulence factors that may also induce pathology in mammals.
Acknowledgments
We thank Charles Yanofsky and Tony Romeo for their generous gifts of the plasmids pMD6 and pCRA16, respectively.
This work was supported by NIHgrants R01DK074731-01and R01-A1056067-01to DK.
S.B. is the recipient of the National Science Foundation award no. 0450303, subaward no. I-66-606-63, to Emory University.
Footnotes
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Aballay A., Ausubel F. M. 2002. Caenorhabditis elegansas a host for the study of host-pathogen interactions. Curr. Opin. Microbiol. 5:97–101 [DOI] [PubMed] [Google Scholar]
- 2. Anyanful A., et al. 2005. Paralysis and killing of Caenorhabditis elegansby enteropathogenic Escherichia colirequires the bacterial tryptophanase gene. Mol. Microbiol. 57:988–1007 [DOI] [PubMed] [Google Scholar]
- 3. Anyanful A., Easley K. A., Benian G. M., Kalman D. 2009. Conditioning protects C. elegansfrom lethal effects of enteropathogenic E. coliby activating genes that regulate lifespan and innate immunity. Cell Host Microbe 5:450–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baker C. S., Morozov I., Suzuki K., Romeo T., Babitzke P. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgCin Escherichia coli. Mol. Microbiol. 44:1599–1610 [DOI] [PubMed] [Google Scholar]
- 5. Barba J., et al. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187:7918–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barbolina M. V., Phillips R. S., Gollnick P. D., Faleev N. G., Demidkina T. V. 2000. Citrobacter freundiityrosine phenol-lyase: the role of asparagine 185 in modulating enzyme function through stabilization of a quinonoid intermediate. Protein Eng. 13:207–215 [DOI] [PubMed] [Google Scholar]
- 7. Bhatt S., et al. 2009. The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect. Immun. 77:3552–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhatt S., Romeo T., Kalman D. 2011. Honing the message: post-transcriptional and post-translational control in attaching and effacing pathogens. Trends Microbiol. 19:217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blankenhorn D., Phillips J., Slonczewski J. L. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia colirevealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bommarius B., et al. 2007. Enteropathogenic Escherichia coliTir is an SH2/3 ligand that recruits and activates tyrosine kinases required for pedestal formation. Mol. Microbiol. 63:1748–1768 [DOI] [PubMed] [Google Scholar]
- 11. Bordi C., Theraulaz L., Mejean V., Jourlin-Castelli C. 2003. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol. Microbiol. 48:211–223 [DOI] [PubMed] [Google Scholar]
- 12. Botsford J. L., DeMoss R. D. 1971. Catabolite repression of tryptophanase in Escherichia coli. J. Bacteriol. 105:303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brencic A., Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosaRsmA. Mol. Microbiol. 72:612–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burrowes E., Baysse C., Adams C., O'Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosaPAO1, as revealed by transcriptome analysis. Microbiology 152:405–418 [DOI] [PubMed] [Google Scholar]
- 15. Bustamante V. H., Santana F. J., Calva E., Puente J. L. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664–678 [DOI] [PubMed] [Google Scholar]
- 16. Chant E. L., Summers D. K. 2007. Indole signalling contributes to the stable maintenance of Escherichia colimulticopy plasmids. Mol. Microbiol. 63:35–43 [DOI] [PubMed] [Google Scholar]
- 17. Chen H. D., Frankel G. 2005. Enteropathogenic Escherichia coli: unraveling pathogenesis. FEMS Microbiol. Rev. 29:83–98 [DOI] [PubMed] [Google Scholar]
- 18. Croxen M. A., Finlay B. B. 2010. Molecular mechanisms of Escherichia colipathogenicity. Nat. Rev. Microbiol. 8:26–38 [DOI] [PubMed] [Google Scholar]
- 19. Cui Y., Chatterjee A., Liu Y., Dumenyo C. K., Chatterjee A. K. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovorasubsp. carotovorathat controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwiniaspp. J. Bacteriol. 177:5108–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daniell S. J., et al. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3:865–871 [DOI] [PubMed] [Google Scholar]
- 21. Darby C., Hsu J. W., Ghori N., Falkow S. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243–244 [DOI] [PubMed] [Google Scholar]
- 22. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coliK-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deeley M. C., Yanofsky C. 1981. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coliK-12. J. Bacteriol. 147:787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deeley M. C., Yanofsky C. 1982. Transcription initiation at the tryptophanase promoter of Escherichia coliK-12. J. Bacteriol. 151:942–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deng W., et al. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dubey A. K., Baker C. S., Romeo T., Babitzke P. 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11:1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dubey A. K., et al. 2003. CsrA regulates translation of the Escherichia colicarbon starvation gene, cstA, by blocking ribosome access to the cstAtranscript. J. Bacteriol. 185:4450–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elliott S. J., et al. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Faleev N. G., et al. 1988. Tyrosine phenol-lyase from Citrobacter intermedius. Factors controlling substrate specificity. Eur. J. Biochem. 177:395–401 [DOI] [PubMed] [Google Scholar]
- 30. Forsbach-Birk V., McNealy T., Chunwei S., Lynch D., Marre R. 2004. Reduced expression of the global regulator protein CsrA in Legionella pneumophilaaffects virulence-associated regulators and growth in Acanthamoeba castellanii. Int. J. Med. Microbiol. 294:15–25 [DOI] [PubMed] [Google Scholar]
- 31. Friedberg D., Umanski T., Fang Y., Rosenshine I. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941–952 [DOI] [PubMed] [Google Scholar]
- 32. Gallagher L. A., Manoil C. 2001. Pseudomonas aeruginosaPAO1 kills Caenorhabditis elegansby cyanide poisoning. J. Bacteriol. 183:6207–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garsin D. A., et al. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U. S. A. 98:10892–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gomes T. A., et al. 1991. Enteropathogens associated with acute diarrheal disease in urban infants in Sao Paulo, Brazil. J. Infect. Dis. 164:331–337 [DOI] [PubMed] [Google Scholar]
- 35. Gong F., Ito K., Nakamura Y., Yanofsky C. 2001. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro). Proc. Natl. Acad. Sci. U. S. A. 98:8997–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gong F., Yanofsky C. 2002. Instruction of translating ribosome by nascent peptide. Science 297:1864–1867 [DOI] [PubMed] [Google Scholar]
- 37. Gong F., Yanofsky C. 2001. Reproducing tnaoperon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 276:1974–1983 [DOI] [PubMed] [Google Scholar]
- 38. Gong F., Yanofsky C. 2003. Rho's role in transcription attenuation in the tnaoperon of E. coli. Methods Enzymol. 371:383–391 [DOI] [PubMed] [Google Scholar]
- 39. Gooder H., Happold F. C. 1954. The tryptophanase-tryptophan reaction; the nature of the enzyme-coenzyme-substrate complex. Biochem. J. 57:369–374 [PMC free article] [PubMed] [Google Scholar]
- 40. Goodman A. L., et al. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745–754 [DOI] [PubMed] [Google Scholar]
- 41. Gruenheid S., et al. 2001. Enteropathogenic E. coliTir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3:856–859 [DOI] [PubMed] [Google Scholar]
- 42. Hirakawa H., Kodama T., Takumi-Kobayashi A., Honda T., Yamaguchi A. 2009. Secreted indole serves as a signal for expression of type III secretion system translocators in enterohaemorrhagic Escherichia coliO157:H7. Microbiology 155:541–550 [DOI] [PubMed] [Google Scholar]
- 43. Iwamori S., Yoshino S., Ishiwata K., Makiguchi N. 1991. Structure of tyrosine phenol-lyase genes from Citrobacter freundiiand structural comparison with tryptophanase from Escherichia coli. J. Ferment. Bioeng. 72:147–151 [Google Scholar]
- 44. Jackson D. W., et al. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kalman D., et al. 1999. Enteropathogenic E. coliacts through WASP and Arp2/3 complex to form actin pedestals. Nat. Cell Biol. 1:389–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaper J. B., Nataro J. P., Mobley H. L. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 47. Kenny B., et al. 1997. Enteropathogenic E. coli(EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520 [DOI] [PubMed] [Google Scholar]
- 48. Kerrinnes T., et al. 2008. CsrA and CsrB are required for the post-transcriptional control of the virulence-associated effector protein AvrA of Salmonella enterica. Int. J. Med. Microbiol. 299:333–341 [DOI] [PubMed] [Google Scholar]
- 49. Knutton S., Baldwin T., Williams P. H., McNeish A. S. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kohra S., Kuwahara K., Takao Y., Ishibashi Y. 2002. Effect of bisphenol A on the feeding behavior of Caenorhabditis elegans. J. Health Sci. 48:93–95 [Google Scholar]
- 51. Kothe M., et al. 2003. Killing of Caenorhabditis elegansby Burkholderia cepaciais controlled by the cepquorum-sensing system. Cell. Microbiol. 5:343–351 [DOI] [PubMed] [Google Scholar]
- 52. Lawhon S. D., et al. 2003. Global regulation by CsrA in SalmonellaTyphimurium. Mol. Microbiol. 48:1633–1645 [DOI] [PubMed] [Google Scholar]
- 53. Lee J., Bansal T., Jayaraman A., Bentley W. E., Wood T. K. 2007. Enterohemorrhagic Escherichia colibiofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl. Environ. Microbiol. 73:4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee M., Phillips R. S. 1995. The mechanism of Escherichia colitryptophan indole-lyase: substituent effects on steady-state and pre-steady-state kinetic parameters for aryl-substituted tryptophan derivatives. Bioorg. Med. Chem. 3:195–205 [DOI] [PubMed] [Google Scholar]
- 55. Lenz D. H., Miller M. B., Zhu J., Kulkarni R. V., Bassler B. L. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 58:1186–1202 [DOI] [PubMed] [Google Scholar]
- 56. Luperchio S. A., et al. 2000. Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentiumand mouse-pathogenic Escherichia coli. J. Clin. Microbiol. 38:4343–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mahajan-Miklos S., Tan M.-W., Rahme L. G., Ausubel F. M. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis eleganspathogenesis model. Cell 96:47–56 [DOI] [PubMed] [Google Scholar]
- 58. Mellies J. L., Barron A. M., Carmona A. M. 2007. Enteropathogenic and enterohemorrhagic Escherichia colivirulence gene regulation. Infect. Immun. 75:4199–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mellies J. L., Barron A. M., Haack K. R., Korson A. S., Oldridge D. A. 2006. The global regulator Ler is necessary for enteropathogenic Escherichia colicolonization of Caenorhabditis elegans. Infect. Immun. 74:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mellies J. L., Elliott S. J., Sperandio V., Donnenberg M. S., Kaper J. B. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296–306 [DOI] [PubMed] [Google Scholar]
- 61. Mellies J. L., Lawrence-Pine E. R. 2010. Interkingdom signaling between pathogenic bacteria and Caenorhabditis elegans. Trends Microbiol. 18:448–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mundy R., Girard F., FitzGerald A. J., Frankel G. 2006. Comparison of colonization dynamics and pathology of mice infected with enteropathogenic Escherichia coli, enterohaemorrhagic E. coliand Citrobacter rodentium. FEMS Microbiol. Lett. 265:126–132 [DOI] [PubMed] [Google Scholar]
- 63. Murphy K. C., Campellone K. G. 2003. Lambda red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nataro J. P., Kaper J. B. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Newton W. A., Morino Y., Snell E. E. 1965. Properties of crystalline tryptophanase. J. Biol. Chem. 240:1211–1218 [PubMed] [Google Scholar]
- 66. Newton W. A., Snell E. E. 1964. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc. Natl. Acad. Sci. U. S. A. 51:382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Park J. O., El-Tarabily K. A., Ghisalberti E. L., Sivasithamparam K. 2002. Pathogenesis of Streptoverticillium albireticulion Caenorhabditis elegansand its antagonism to soil-borne fungal pathogens. Lett. Appl. Microbiol. 35:361–365 [DOI] [PubMed] [Google Scholar]
- 68. Romeo T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321–1330 [DOI] [PubMed] [Google Scholar]
- 69. Romeo T., Gong M., Liu M. Y., Brun-Zinkernagel A. M. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia colithat affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175:4744–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sabnis N. A., Yang H., Romeo T. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia colivia the gene csrA. J. Biol. Chem. 270:29096–29104 [DOI] [PubMed] [Google Scholar]
- 71. Sarsero J. P., Wookey P. J., Gollnick P., Yanofsky C., Pittard A. J. 1991. A new family of integral membrane proteins involved in transport of aromatic amino acids in Escherichia coli. J. Bacteriol. 173:3231–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schauer D. B., Falkow S. 1993. Attaching and effacing locus of a Citrobacter freundiibiotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sekiya K., et al. 2001. Supermolecular structure of the enteropathogenic Escherichia colitype III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U. S. A. 98:11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Senerwa D., et al. 1989. Enteropathogenic Escherichia coliserotype O111:HNT isolated from preterm neonates in Nairobi, Kenya. J. Clin. Microbiol. 27:1307–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sifri C. D., Begun J., Ausubel F. M. 2005. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13:119–127 [DOI] [PubMed] [Google Scholar]
- 76. Sifri C. D., Begun J., Ausubel F. M., Calderwood S. B. 2003. Caenorhabditis elegansas a model host for Staphylococcus aureuspathogenesis. Infect. Immun. 71:2208–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Snell E. E. 1975. Tryptophanase: structure, catalytic activities, and mechanism of action. Adv. Enzymol. Relat. Areas Mol. Biol. 42:287–333 [DOI] [PubMed] [Google Scholar]
- 78. Sperandio V., et al. 2000. Activation of enteropathogenic Escherichia coli(EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781–793 [DOI] [PubMed] [Google Scholar]
- 79. Stewart V. 2008. The ribosome: a metabolite-responsive transcription regulator. J. Bacteriol. 190:4787–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stewart V., Yanofsky C. 1985. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coliK-12. J. Bacteriol. 164:731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Stewart V., Yanofsky C. 1986. Role of leader peptide synthesis in tryptophanase operon expression in Escherichia coliK-12. J. Bacteriol. 167:383–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Suzuki K., et al. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184:5130–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Swimm A., et al. 2004. Enteropathogenic Escherichia coliuse redundant tyrosine kinases to form actin pedestals. Mol. Biol. Cell 15:3520–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tan L., Darby C. 2004. A movable surface: formation of Yersiniasp. biofilms on motile Caenorhabditis elegans. J. Bacteriol. 186:5087–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tan M.-W., Rahme L. G., Sternberg J. A., Tompkins R. G., Ausubel F. M. 1999. Pseudomonas aeruginosakilling of Caenorhabditis elegansused to identify P. aeruginosavirulence factors. Proc. Natl. Acad. Sci. U. S. A. 96:2408–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tan M.-W., Mahajan-Miklos S., Ausubel F. M. 1999. Killing of Caenorhabditis elegansby Pseudomonas aeruginosaused to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 96:715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tominaga N., Kohra S., Iguchi T., Arizono K. 2003. A multi-generation sublethal assay of phenols using the nematode Caenorhabditis elegans. J. Health Sci. 49:459–463 [Google Scholar]
- 88. Wang X., et al. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56:1648–1663 [DOI] [PubMed] [Google Scholar]
- 89. Wang X., Preston J. F., III, Romeo T. 2004. The pgaABCDlocus of Escherichia colipromotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wei B. L., et al. 2001. Positive regulation of motility and flhDCexpression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245–256 [DOI] [PubMed] [Google Scholar]
- 91. Wilson R. K., Shaw R. K., Daniell S., Knutton S., Frankel G. 2001. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell. Microbiol. 3:753–762 [DOI] [PubMed] [Google Scholar]
- 92. Yakhnin H., et al. 2007. CsrA of Bacillus subtilisregulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol. Microbiol. 64:1605–1620 [DOI] [PubMed] [Google Scholar]
- 93. Yang T. Y., Sung Y. M., Lei G. S., Romeo T., Chak K. F. 2010. Posttranscriptional repression of the celgene of the ColE7 operon by the RNA-binding protein CsrA of Escherichia coli. Nucleic Acids Res. 38:3936–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yanofsky C., Horn V., Gollnick P. 1991. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J. Bacteriol. 173:6009–6017 [DOI] [PMC free article] [PubMed] [Google Scholar]