Abstract
Although reversible phosphorylation on tyrosine residues regulates the activity of many eukaryotic proteins, there are few examples of this type of regulation in bacteria. We have identified the first essential tyrosine phosphatase homolog in a bacterium, Caulobacter crescentusCtpA. ctpAmutants with altered active-site residues are nonviable, and depletion of CtpA yields chains of cells with blebbed outer membranes, linked by unresolved peptidoglycan. CtpA overexpression reduces cell curvature in a manner similar to deleting the intermediate filament protein crescentin, but it does not disrupt crescentin localization or membrane attachment. Although it has no obvious signal sequence or transmembrane-spanning domains, CtpA associates with the Caulobacterinner membrane. Immunolocalization experiments suggest that CtpA accumulates at the division site during the last quarter of the cell cycle. We propose that CtpA dephosphorylates one or more proteins involved in peptidoglycan biosynthesis or remodeling, which in turn affect cell separation, cell envelope integrity, and vibrioid morphology.
INTRODUCTION
Tyrosine phosphorylation is used extensively in eukaryotes to convey signaling information and regulate protein activity. In bacteria, however, few processes are known to be regulated in this manner (23). In many Gram-positive and Gram-negative bacteria, tyrosine phosphorylation regulates the synthesis of capsular and extracellular polysaccharides (collectively referred to as EPS) (12). In both Escherichia coliand Bacillus subtilis, the heat shock response is regulated in part by tyrosine phosphorylation of pathway components (38, 39). Single-stranded DNA-binding proteins are also phosphorylated on tyrosine residues in several species (50), and B. subtiliscells lacking the tyrosine kinase PtkA are impaired in the control of DNA replication (57). Tyrosine-phosphorylated proteins have been identified in various bacteria using proteomics (46, 47) or affinity purification with anti-phosphotyrosine antibodies (50), but these studies have not revealed the enzymes involved or the biological roles of the modifications.
The relative lack of information on tyrosine phosphorylation in bacteria is partly due to the fact that bacterial genomes rarely encode homologs of eukaryotic-type tyrosine kinase proteins (27). Instead, most bacterial tyrosine kinases have sequences and structures similar to P-loop ATPases containing Walker A and B motifs (23, 33, 43, 55). In contrast, both eukaryotic and prokaryotic genomes contain the same families of tyrosine phosphatase genes (4). Most prevalent are the low-molecular-weight tyrosine phosphatases (LMW-PTP), followed by the classical (PTP) and dual-specificity (DSP) tyrosine phosphatases.
The first bacterial tyrosine phosphatase to be characterized was the PTP YopH from Yersinia pseudotuberculosis(26), a plasmid-encoded virulence factor. YopH is transported by a type III secretion system directly into the cytoplasm of host cells (56, 63), where it dephosphorylates host proteins (5). Similar tyrosine phosphatases that are translocated into the eukaryotic host to enhance virulence have been found in other animal and plant pathogens (6, 36, 40, 66, 74). The first tyrosine phosphatase to be identified in the genome of a nonpathogenic bacterium was IphP, from the photosynthetic species Nostoc communeUTEX 584 (59). IphP is a dual-specificity phosphatase, containing the active-site residues of traditional PTPs but capable of dephosphorylating both phosphoserine and phosphotyrosine residues on artificial substrates. Although IphP's activity toward small-molecule substrates has been investigated in detail (30), and at least one tyrosine-phosphorylated protein is present in N. commune(59), the cellular function of IphP remains unknown.
Because of their impact on virulence, EPS production systems are the best-studied bacterial examples of regulation by tyrosine phosphorylation. Group 1 and 4 capsular polysaccharides and the EPS colanic acid are synthesized by multiprotein complexes spanning the inner and outer membranes (77). Homologs of Wzc, a bifunctional protein, are essential for both systems. The periplasmic domain and flanking transmembrane helices of Wzc are thought to function in polysaccharide polymerization and/or export, while the cytoplasmic domain is a tyrosine kinase that phosphorylates itself and other enzymes involved in EPS and capsule biosynthesis (15, 52, 75). A corresponding LMW-PTP, Wzb, dephosphorylates Wzc, as well as other substrates (8, 24, 50, 52). Group 1 capsule production is compromised when either the kinase or the phosphatase is missing (76, 79). wzcphospho-acceptor site mutants synthesize colanic acid, but the molecular mass of the polymer is different than in wild-type cells (54). In contrast, a wzcactive-site mutant produces ∼10 times less colanic acid than wild-type cells (54), suggesting that Wzc-mediated phosphorylation of other substrates is critical for colanic acid production. The exact role of reversible tyrosine phosphorylation in these systems is still unclear, but one model suggests that cycling of the kinase between the phosphorylated and dephosphorylated states is mechanistically coupled to EPS polymerization or export.
Caulobacter crescentusis a Gram-negative alphaproteobacterium that has been the subject of extensive research on cell cycle regulation and cell polarity (7). Each Caulobactercell division is asymmetric, yielding a flagellated swarmer cell that cannot initiate chromosome replication, and a cell with a polar stalk that immediately begins a new round of chromosome replication and division. When the swarmer progeny differentiates into a stalked cell, it initiates DNA replication and reenters the division cycle (31, 68). Daughter cells divide by invaginating the inner membrane, peptidoglycan (PG) and outer membrane, which closely follow each other in time and space (34). However, the swarmer and stalked cell cytoplasms become compartmentalized several minutes before the daughter cells are fully separated (14, 35).
Caulobacteris also a leading model system in studies of bacterial morphogenesis (48). The rod shape of Caulobacterrequires the actin homolog MreB (19), and the vibrioid shape requires the intermediate filament homolog crescentin, encoded by creS(3). Crescentin forms a filamentous structure on the concave surface of the cytoplasmic membrane and generates curvature in a growth-dependent manner (3, 10). MreB helps crescentin to maintain its association with the cell membrane (11). Crescentin filaments are believed to promote cell curvature by exerting an imbalanced force on the cell that decreases the rate of insertion of PG monomers into the adjacent cell wall and increases the rate of insertion into the opposite, convex wall (10).
We show here that the tyrosine phosphatase homolog CtpA, encoded by CC0306, is necessary for viability in Caulobacter. To our knowledge, CtpA is the first example of an essential tyrosine phosphatase in a bacterium. Before causing cell death, depletion of CtpA yields short chains of cells with ragged outer membranes. Overexpression of CtpA reduces cell curvature, though not as severely as a creSdeletion. In CtpA-overexpressing cells, crescentin remains in a filamentous structure associated with the cytoplasmic membrane, yet its ability to generate curvature is apparently reduced. Although it has no obvious signal sequence or transmembrane-spanning domains, CtpA is located in the Caulobacterinner membrane, and it accumulates at the midcell during the last half of the cell cycle. Based on mutant phenotypes, we propose that one function of CtpA is to regulate PG biosynthesis or remodeling via dephosphorylation of tyrosine residues on target proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used here are listed in Table S1 in the supplemental material. All experiments were performed using derivatives of Caulobacter crescentusstrain CB15N (18) grown to mid-exponential phase. Plasmids were mobilized from E. colito C. crescentusby conjugation using E. colistrain S17-1, and generalized transduction was performed by using ΦCr30 (17). The sequences of the primers used for amplification or sequence modification are available upon request. CB15N strains were grown in peptone-yeast extract medium (PYE) (17) at 30°C. Where indicated, strains were grown in PYE plus 0.1% d-glucose (PYED) or 0.3% xylose (PYEX). Solid and liquid PYE media were supplemented with sucrose and antibiotics as described previously (62). E. colistrains were grown in Luria broth at 37°C, and solid and liquid media were supplemented with antibiotics as described previously (59).
To delete sspB, E. colistrain S17-1 was used to mobilize pDL6 into KR684, and single colonies were selected on PYE-spectinomycin (Spec)-nalidixic acid (Nal). These colonies were grown overnight in PYE-Spec and plated on PYE-Spec-sucrose. Sucrose-resistant colonies were screened for resistance to Spec and sensitivity to kanamycin (Kan) to exclude strains that were sucrose resistant due to mutations in the sacBgene. Strain KR1499 is comparable to the sspBdeletion strain previously described (44).
For the ctpAdeletion plasmid pSK189, we amplified 1-kb arms of homology flanking ctpAusing PCR, ligated them into pNPTS138, and inserted a tetracycline resistance cassette between the arms.
To create a ctpAdepletion plasmid, we used PCR to generate an NdeI site at the 5′ end and a BamHI site at the 3′ end of ctpA. This fragment was digested and ligated into pLB13, replacing the divJfragment. The resultant plasmid contained a xylose-inducible promoter (xylX) followed by the ctpAcoding sequence fused to a C-terminal 3×FLAG tag and an ssrAdegradation sequence (37). The entire insert was moved to pJS14 to create pAB6. In plasmids pAB11 and pAB12, site-directed mutagenesis was used to alter amino acids in the CtpA active site, yielding the changes R129A and C123A, respectively.
To create strain KR2423, where the only copy of ctpAis regulated by the xylXpromoter, pAB6 and pSK189 were mobilized into KR1499. Single colonies were selected on PYE-Kan-Nal and were grown overnight in PYEX-chloramphenicol (Chlor). Cells were plated on PYEX-Chlor-oxytetracycline (Oxy)-sucrose, and sucrose-resistant colonies were screened for resistance to Oxy and Chlor and sensitivity to Kan to exclude strains that were sucrose resistant due to mutations in the sacBgene. To determine whether the catalytic residues were essential for the function of CtpA, the same procedure was performed using the plasmids pAB11 or pAB12 in place of pAB6.
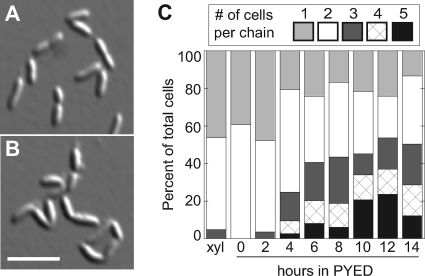
In KR2423, CtpA-3×FLAG-SsrA is expressed from a xylose-inducible promoter on a high-copy-number plasmid, and both ctpAand sspBare deleted from the chromosome. We first attempted to deplete CtpA using a low-copy vector with ctpA::3×FLAG driven by the xylXpromoter. However, after several hours or overnight growth in PYED, CtpA-3×FLAG levels remained undiminished. We then added the ssrAtag to the C terminus of CtpA-3×FLAG to speed its degradation. Unfortunately, the xylX::ctpA::3×FLAG::ssrAgene did not support viability even when expressed in xylose on a high-copy-number plasmid. Reasoning that the ssrAtag was preventing the cells from accumulating sufficient CtpA, we then deleted sspB, which encodes an adaptor that increases the degradation rate of ssrA-tagged substrates (45, 44). In this strain background, xylX::ctpA::3×FLAG::ssrAsupported viability in PYEX and was reduced to ∼10% of starting levels after growth on PYED (Fig. 1D).
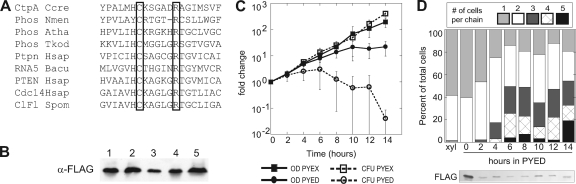
Fig. 1.
(A) Alignment of the active site of Caulobacter crescentusCtpA with its closest homologs for which structural or functional data are available. For references and the full alignment, see the supplemental material. Phos Nmen, nonclassical eukaryote-like phosphatase from Neisseria meningitidis; Phos Atha, putative phosphoprotein phosphatase from Arabidopsis thaliana; Phos Tkod, protein tyrosine/serine phosphatase from Thermococcus kodakarensis; Ptpn Hsap, tyrosine phosphatase Ptpn22 from Homo sapiens; RNA5 Bacu, RNA 5′-phosphatase from Baculovirus; Pten Hsap, PTEN phosphoinositide phosphatase from Homo sapiens; Cdc14 Hsap, proline-directed phosphatase Cdc14 from Homo sapiens; ClF1 Spom, protein phosphatase Clp1/Flp1 from Schizosaccharomyces pombe. Conserved active-site cysteine and arginine residues are boxed. (B) Whole-cell lysates were analyzed by SDS-PAGE and α-FLAG Western blotting. All strains are ΔsspB. Strains in lanes 1, 3, and 5 have pSK189 integrated by single homologous recombination. Lane 1, pAB6; lanes 2 and 3, pAB6-C123A; lanes 4 and 5, pAB6-R129A. (C) Growth curve showing turbidity (OD660) and viability (CFU/ml) during growth of KR2423 (ΔsspBΔctpA+ pAB6) in PYEX or PYED. (D) Graphical representation of the percentage of cell bodies existing as a chain of ncells during the growth of KR2423 in PYED. The number of chains counted in each sample: xylose = 362, 0 h = 163, 2 h = 207, 4 h = 161, 6 h = 173, 8 h = 172, 10 h = 166, 12 h = 111, and 14 h = 110. Below the graph is a Western blot showing the levels of CtpA-3×FLAG after the indicated number of hours in PYED.
To overexpress ctpA::3×FLAG, we used PCR to create an NdeI site at the 5′end and a BamHI site at the3′ end ctpA. We amplified 1 kb of the xylXpromoter, adding a 5′ EcoRV site and an NdeI site overlapping the start codon. We ligated the xylXpromoter and ctpAfragments to an EcoRV-BamHI-digested pSK84 to create pSK166. The EcoRV fragment from pSK166 was moved to pJS14 to create pES133. To create a ctpA::3×FLAG fusion regulated by the native ctpApromoter, the HindIII-SphI fragment from pES133 was ligated to the SphI-EcoRI fragment from pSK77 to create pES137. The 3×FLAG tag adds the amino acid residues DYKDHDGDYKDHDIDYKDDDDK to the C terminus of CtpA.
Preparation of thin sections for transmission electron microscopy.
Strain KR2423 was grown in PYEX overnight to mid-exponential phase and then washed and resuspended either in the same medium or in PYED and allowed to grow for 12 h (cells in PYEX were back-diluted to keep them in exponential phase). Cultures were fixed with 2% glutaraldehyde with shaking for 5 min. Cells were pelleted and resuspended in 1 ml of 2% glutaraldehyde in 1 M cacodylate buffer and shaken at room temperature for 30 min. Cells were washed twice with 1 ml of cacodylate buffer, followed by 10 min of agitation at room temperature. Next, the cells were resuspended in 2% osmium tetroxide and shaken in the dark for 1 h. The cells were washed twice with 1 ml of cacodylate buffer, followed by 5 min of shaking at room temperature. The cells were then resuspended in 2% low-melt agarose, and the suspension was allowed to solidify. The extra agarose was trimmed, and the remaining fragment was washed twice with double-distilled H2O (ddH2O) and stained with 1 ml of 1% uranyl acetate with agitation for 30 min. The fragment was washed twice with water and then dehydrated in successive washes of 35, 50, 75, 80, 95, and 100% ethanol in increasingly cold temperatures from 4°C to −50°C. The fragment was washed twice in acetone and then in successively higher concentrations of Epon resin for 1 h each, and finally in 100% resin overnight. Accelerant was added to 12 ml of resin, and pieces of the cell pellet were allowed to harden in molds for 2 days. Thin sections of 17 nm were sliced by using a Reichart Microtome Ultracut and placed onto glow-discharged 100-mesh grids coated with carbon and Formvar. Grids were stained with 2% uranyl acetate, washed with methanol, stained with lead citrate, and rinsed with water before viewing.
Isolation of sacculi.
Cultures (500 ml) of KR2423 were grown in either PYEX or PYED medium for 12 h to an optical density at 600 nm (OD660) of ∼0.4, and the cells were chilled on ice for 10 min before being harvested by centrifugation at 9,000 rpm for 10 min at 4°C. Cells were washed once in ddH2O and resuspended in 15 ml of ddH2O. The cell suspension was added dropwise to 15 ml of boiling 8% sodium dodecyl sulfate (SDS). The solution was boiled with stirring for 30 min and allowed to stand at room temperature overnight. SDS was removed in four washes with ddH2O by spinning at 26,000 rpm in a Beckman ultracentrifuge rotor type 60Ti. Cells were resuspended in 1 ml of 10 mM Tris-HCl (pH 7) and digested with 100 μg of alpha-amylase (MP Biomedicals) at 37°C for 2.5 h. Predigested pronase (Roche Diagnostics) was added to a final concentration of 20 μg/ml, and the samples were incubated at 60°C for 90 min. Samples were added to an equal volume of 8% SDS and boiled for 15 min. SDS was removed with four washes in ddH2O as described above. Sacculi were applied to glow-discharged 400-mesh grids coated with carbon and Formvar. Grids were washed three times in water, stained with 1% uranyl acetate for 1 min, and washed once in water before examination.
Transmission electron microscopy.
Samples were imaged using a FEI Tecnai 12 transmission electron microscope operated at 120 kV.
Immunofluorescence microscopy.
Swarmer cells of strain KR2828 were harvested, released into PYE, and allowed to proceed synchronously through the cell cycle as described previously (60). Samples from the synchronized culture of KR2828 were withdrawn every 15 min and fixed and stained as described below. Samples from a mixed culture of strain KR2829 were examined in the same manner. Cells were fixed in 2.5% formaldehyde–30 mM sodium phosphate (pH 7.4) for 15 min at room temperature, followed by 1 h on ice. The cells were washed three times with phosphate-buffered saline (PBS; 140 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4) and resuspended in GTE (50 mM glucose, 10 mM EDTA, 20 mM Tris-HCl [pH 7.5]). Cells were adhered to poly-l-lysine-coated glass slides, treated with 10 μg of lysozyme/ml for 2 min, and dried completely. The slides were immersed in PBS–2% bovine serum albumin before incubation with mouse α-FLAG antibodies (Sigma) in the same buffer for 1 h. Slides were washed three times with PBS and incubated with Alexa Fluor 488-labeled goat anti-mouse secondary antibodies (Invitrogen) for 1 h. The cells were then washed three times in PBS and covered with 80% glycerol containing 2 μg of propidium iodide (PI)/ml to stain the nucleic acids. Images were acquired using a Nikon Eclipse 80i microscope with a PlanApo 100×/NA 1.40 objective lens and a Cascade 512B camera (Roper Scientific), driven by Metavue software (Universal Imaging). Alexa Fluor 488 and PI were imaged by using Chroma filter sets 41001 and 41004, respectively.
Fluorescence microscopy.
The cells were immobilized on agarose pads (1% [wt/vol] agarose in distilled water). Enhanced green fluorescent protein (EGFP) was imaged by using a Chroma filter set 41001 and the equipment described above.
Western blotting.
Each lane received the same number of cell equivalents, normalized by OD660. Samples were boiled in 1× SDS loading buffer, separated by SDS-PAGE, and transferred to Immobilon-P (Millipore). The membrane was dried and then blocked in 5% nonfat milk in TBS (150 mM NaCl, 50 mM Tris-HCl [pH 7.4]) for 30 min. Primary antisera were incubated 2 h at room temperature in TBS–5% milk at the following dilutions: anti-CtrA, 1:10,000 (14); anti-McpA, 1:50,000 (1); anti-CreS, 1:10,000 (3); anti-RsaF, 1:5000 (69); and anti-FLAG, 1:10,000 (Sigma). Horseradish peroxidase-conjugated anti-rabbit secondary antibodies (Fisher Scientific) were incubated 1 h at room temperature in TBS–5% milk at a dilution of 1:10,000. The membranes were washed using four 5-min washes with TBS–0.5% Tween 20 after primary and secondary antibody incubations. Chemiluminescent signals were visualized with Western Lightning (Perkin-Elmer).
Cell fractionation.
Whole-cell lysates from 100 ml of exponential-phase culture were separated into soluble and membrane fractions and into inner and outer membrane fractions by sucrose density gradient centrifugation as previously described (64).
RESULTS
Identification of tyrosine phosphatase homologs.
We searched the genome of Caulobacter crescentusCB15 for sequences similar to known protein tyrosine phosphatases. As previously noted (4), Caulobactercontains two genes predicted to encode LMW-PTPs, CC2368and CC2344. These proteins resemble E. coliWzb (75) and Acinetobacter johnsoniiPtp (24), tyrosine phosphatases that regulate EPS biosynthesis, with strongest similarity in the N-terminal region containing the active site. However, the Caulobactergenes are not in operons with other EPS biosynthetic genes, and recognizable tyrosine kinase homologs are not found nearby (53). By searching for proteins similar to the DSP Cdc14, which governs mitotic exit in yeast and higher eukaryotes (25), we also identified CC0306, which encodes a predicted tyrosine phosphatase in a single-gene operon. The amino acid similarity between CC0306and biochemically characterized phosphatases is chiefly limited to the active-site motif, HCX5R, which in PTPs and DSPs is located nearer the C terminus than in LMW-PTPs (Fig. 1A and see Fig. S1 in the supplemental material). A glycine residue is often present in the active-site motif, since this sequence forms a flexible loop between a beta strand and an alpha helix in three-dimensional structures of tyrosine phosphatases (2). A Neisseria meningitidisprotein that contains the active-site motif but has limited overall homology to PTPs was found to have an X-ray crystal structure very similar to classical PTPs, including positioning of the active-site residues (41).
We attempted to replace the CC0306, CC2344, and CC2368coding sequences with antibiotic resistance cassettes (see Materials and Methods). ΔCC2344and ΔCC2368mutants were readily obtained, and for each mutant, the growth rate, morphology, and motility were indistinguishable from the wild-type parent (data not shown). In contrast, we could not delete the chromosomal copy of CC0306unless we provided a second copy of the gene on a plasmid, indicating that CC0306is essential for Caulobacterviability (Table 1). To determine whether the predicted active-site residues of CC0306are essential for the protein's function, we made site-directed mutations in CC0306encoding the amino acid substitution C123A or R129A. In characterized tyrosine phosphatases, the conserved arginine residue helps to position the phosphate group of the substrate, and the side chain of the cysteine forms a covalent intermediate with the phosphate during catalysis (25). Each variant was expressed at a level similar to the wild-type protein (Fig. 1B), but neither protein supported Caulobacterviability in the absence of wild-type CC0306(Table 1). These results suggest that the essential function of CC0306 is related to phosphatase activity, and we henceforth refer to CC0306 as CtpA (Caulobactertyrosine phosphatase A).
Table 1.
The wild-type sequence of ctpAis required for viabilitya
| Covering plasmid | % Colonies with ctpAdeletion (no. of colonies) |
|---|---|
| pAB6 (ctpA) | 22 (159) |
| pAB12 (ctpA-C123A) | 0 (100) |
| pAB11 (ctpA-R129A) | 0 (100) |
| pJS14 (empty) | 0 (100) |
The percentages of colonies expressing the indicated plasmid-borne ctpAalleles in which the chromosomal ctpAgene was deleted are indicated. pAB6 is pJS14-xylX::ctpA::3×FLAG::ssrA.pAB11 and pAB12 were generated by site-directed mutagenesis of pAB6. pSK189 (KanrsacB ctpA::tetAR) was integrated into the Caulobacterchromosome at the ctpAlocus in strains harboring the indicated covering plasmids. First integrants were plated on PYE-Oxy-sucrose to counterselect the sacBmarker. Sucrose-resistant colonies were screened for sensitivity to kanamycin to identify those in which ctpAhad been replaced by ctpA::tetAR.
We attempted to demonstrate CtpA phosphatase activity by measuring its ability to hydrolyze the small molecule substrate p-nitrophenylphosphate (pNPP) in vitro(see the supplemental methods in the supplemental material). pNPP is a generic substrate hydrolyzed by all classes of tyrosine phosphatase (28). Surprisingly, purified CtpA failed to hydrolyze pNPP, while the purified LMW-PTP CC2344 exhibited robust phosphate ester hydrolysis (data not shown). We found no combination of divalent cations, pH, or temperature that stimulated CtpA activity in vitro.
Depletion of CtpA causes outer membrane blebbing and inhibits cell separation.
To examine the terminal phenotype of cells lacking CtpA, we constructed a strain (KR2423) in which expression of an epitope-tagged CtpA protein is driven by the xylose-inducible xylXpromoter (49; see also Materials and Methods). CtpA was depleted to ∼10% of starting levels 2 h after the cells were shifted to medium lacking xylose and stayed at this level for the remainder of the experiment (Fig. 1D, lower panel). At 14 h after CtpA depletion, the number of viable cells/ml decreased at least 10-fold compared to the starting number of cells in the culture (Fig. 1C).
After 8 h of CtpA depletion, differential interference contrast (DIC) microscopy revealed chains of cells that had not separated, as well as cells with ragged outlines (Fig. 2E). These features were never observed when the same strain was maintained in xylose-containing medium (Fig. 2A). DAPI staining revealed that each compartment in a chain contained DNA (data not shown).
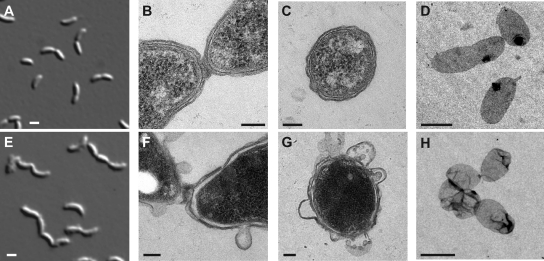
Fig. 2.
(A to D) KR2423 (ΔsspBΔctpA+ pAB6) grown in PYEX. (E to H) KR2423 grown in PYED. (A and E) DIC images at t= 8 h. (B and F) TEM images of predivisional cell junctions at t= 12 h. (C and G) TEM images of transverse sections at t= 12 h. (D and H) TEM images of sacculi at t= 12 h. Scale bars: A, E, D, and H, 1 μm; and B, C, F, and G, 100 nm.
We performed transmission electron microscopy (TEM) on fixed sections of Caulobacterto examine cell junctions and cell walls. In CtpA-depleted samples, we often observed junctions in which the two cytoplasmic membranes of the daughters were separated, and the PG and outer membrane were invaginated, but a mass of material remained connecting the two daughters (Fig. 2F and see Fig. S2E to G in the supplemental material). More than 90% of cell junctions observed in CtpA-depleted cells were similarly joined by disordered material outside the cytoplasmic membrane (n= 16), but no cell junctions in CtpA-replete cells had this appearance (n= 6; Fig. 2B and see Fig. S2D in the supplemental material). The ragged edges of CtpA-depleted cells observed in DIC microscopy corresponded to blebbed or detached sections of outer membrane (Fig. 2F to G and see Fig. S2E to G in the supplemental material).
Because the material connecting the cell chains was of indeterminate origin, we isolated PG sacculi from CtpA-replete and CtpA-depleted cultures and examined them by whole-mount TEM. Sacculi from PYEX cultures retained the shapes of individual Caulobactercells, including stalks and invaginations (Fig. 2D and see Fig. S2A in the supplemental material). In repeated isolations, sacculi from CtpA-depleted cultures were found in contiguous groups joined by darker-staining PG (Fig. 2H and see Fig. S2B and C in the supplemental material), indicating that chains of CtpA-depleted cells were connected by unresolved PG. Sacculi from CtpA-depleted cells also appeared wrinkled or misshapen (Fig. 2H and see Fig. S2B and C in the supplemental material). Sacculi with this morphology were previously observed when E. coliwas starved for diaminopimelic acid, a dibasic amino acid which mediates cross-links between the peptide side chains of adjacent glycan strands in PG (9). By analogy, CtpA depletion may render the sacculi less rigid and more susceptible to mechanical deformation.
Overexpression of CtpA affects cell shape downstream of crescentin or in an independent pathway.
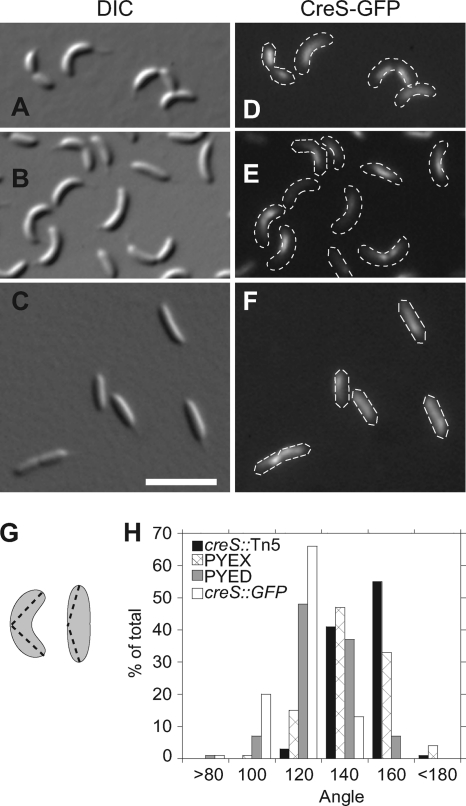
The balance between tyrosine kinase and tyrosine phosphatase activities is essential for proper EPS biosynthesis (8, 76, 79). Reasoning that excess CtpA may also have detrimental effects on Caulobacter, we overexpressed CtpA from the xylXpromoter on a high-copy-number plasmid in CJW815, a strain that expresses both crescentin and crescentin-GFP (CreS-GFP) (3). Cells in PYEX grew at the same rate as those in PYED and were approximately the same length (data not shown), but those with excess CtpA appeared straighter than uninduced cells (compare Fig. 3B and C). We quantified the change in curvature by measuring the angle formed between the endpoints of each cell and its most convex point using Image J (Fig. 3G) (61). Although the distributions overlap, the mean angles described by the cell populations are significantly different (P< 0.010). CJW815 cells averaged 107°. The angles for CJW815 cells with pxylX::ctpAgrown in glucose averaged 118°, while the same cells grown in xylose to overexpress CtpA averaged 134°. Cells lacking functional crescentin (creS::Tn5) lose their vibrioid morphology (3). The average angle described by creS::Tn5cells was 141°, and the distribution of angles resembled that of cells overexpressing CtpA (Fig. 3H).
Fig. 3.
(A and D) CJW815 (creS, creS::GFP). (B and E) KR2616 (CJW815 + JS14-xylX::ctpA) grown in PYED. (C and F) KR2616 grown in PYEX. (G) Diagram of method used to measure cell curvature using ImageJ (61). (H) Cell angle distributions of the strains CJW761 (creS::Tn5), CJW815, and KR2616 grown for 20 h in either PYEX or PYED.
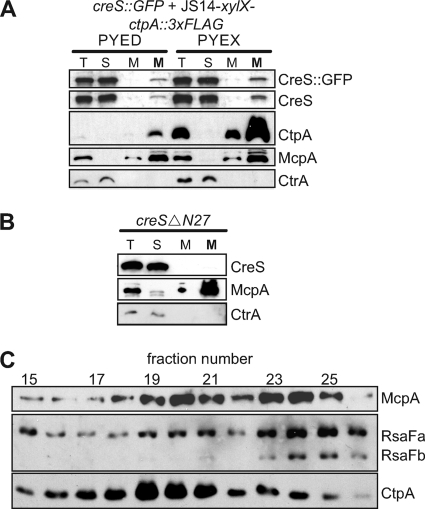
To generate the vibrioid morphology of Caulobacter, crescentin must associate with the cytoplasmic membrane via its N terminus and form a filamentous structure along the inner curvature of the cell (3, 10). Overexpression of CtpA might reduce curvature by acting directly on crescentin, altering its levels, polymerization, or membrane association. However, we found that the amounts of crescentin and CreS-GFP were unchanged in cells overexpressing CtpA (see Fig. 5A), and crescentin-GFP remained in a filamentous structure (Fig. 3D to F).
Fig. 5.
(A) Western blot showing cellular distributions of CreS-GFP, CreS, CtpA-3×FLAG, McpA, and CtrA in KR2570 (creS, creS::GFP+ JS14-xylX::ctpA::3×FLAG) grown in PYED or PYEX. (B) Western blot showing cellular distributions of CreSΔN27-tc, McpA, and CtrA in CJW1537 (ΔcreS+ MR20-xylX::creSΔN27::tc). Normalized volumes of total cell lysate (T), the soluble fraction (S), and the membrane fraction (M) were analyzed by SDS-PAGE and Western blotting. Ten times the normalized volume of the membrane fraction (M) was included for visualization of membrane-associated crescentin. (C) Fractions from sucrose density gradient centrifugation of cell membranes were analyzed by SDS-PAGE and Western blot against the inner membrane protein McpA, the outer membrane proteins RsaFaand RsaFb, and CtpA-3×FLAG. Fraction density increases from left to right.
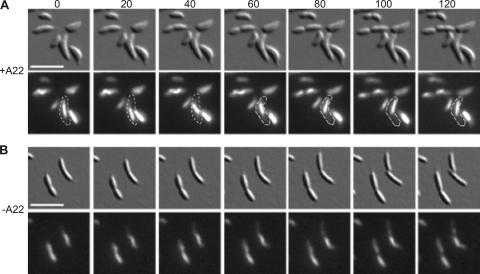
When Caulobactercells are treated with the drug A22 to depolymerize MreB filaments (20), crescentin structures become loose in the cytoplasm, rather than staying attached to the cell membrane (11). Under these conditions, shifting or coiling of GFP-labeled crescentin filaments can be observed during time-lapse fluorescence microscopy. In CtpA-overexpressing cells, CreS-GFP structures (103/103 observed) remained stationary over a period of 120 min, except for changes due to growth and slight cell movements (Fig. 4B). In contrast, when CtpA-overexpressing cells were treated with A22, CreS-GFP structures (71/75 observed) changed position within the cell at least once (Fig. 4A). Thus, CtpA overexpression does not alter the amount, gross level of polymerization, or MreB-dependent localization of crescentin. However, we cannot rule out subtle changes in crescentin polymerization that cannot be detected by fluorescence microscopy.
Fig. 4.
Time-lapse images of KR2616 (creS, creS::GFP+ JS14-xylX::ctpA) grown in PYEX with (A) or without (B) A22. DIC (top) and CreS-GFP fluorescence images (bottom) were taken at 20-min intervals. All cells were grown overnight in PYEX to induce excess CtpA. Cells in A were also treated with 50 μM A22 for 3 h before imaging. Scale bar, 5 μm. Outlines highlight a cell in which the crescentin structure was mobile.
We performed cell fractionation experiments to confirm that crescentin was still associated with the cytoplasmic membrane when CtpA was overexpressed. In cells containing either normal or increased amounts of CtpA, a small amount of crescentin remained in the membrane fraction, along with the inner membrane chemoreceptor McpA (Fig. 5A), a finding consistent with previous studies of wild-type Caulobacter(10). As a control, we fractionated cells expressing only CreSΔN27-tc, which polymerizes but lacks the N-terminal segment that mediates association with the cell membrane (10). CreSΔN27-tc was located exclusively in the cytosolic fraction, along with the soluble protein CtrA (Fig. 5B), indicating that crescentin found in the membrane fraction of CtpA-overexpressing cells does not reflect sedimentation of free cytosolic polymers.
Cell chaining during CtpA depletion is independent of crescentin.
When crescentin is expressed without other Caulobacterproteins, it generates curvature in rod-shaped E. coli(10). In these cells, cytokinesis is impaired, possibly because E. colidoes not contain the machinery necessary to divide crescentin filaments. We reasoned that depletion of CtpA could produce cell chains by preventing the division of crescentin filaments. We transduced the creS::Tn5allele from CJW763 into KR2423 to observe the phenotype when CtpA is depleted from cells lacking crescentin. Even in the absence of crescentin, chains of up to five cell bodies were formed during CtpA depletion (Fig. 6). Thus, the chaining phenotype is not due to an inability of CtpA-depleted cells to sever crescentin filaments.
Fig. 6.
DIC images of KR2612 (ΔcreS ΔsspB ΔctpA+ pAB6) grown in PYEX (A) or PYED (B) for 8 h. Scale bar, 5 μm. (C) Graphical representation of the percentage of cell bodies existing as a chain of ncells during growth of KR2612 in PYED. Number of chains counted in each sample: xylose = 183, 0 h = 207, 2 h = 190, 4 h = 324, 6 h = 297, 8 h = 203, 10 h = 239, 12 h = 176, and 14 h = 188.
CtpA is associated with the cytoplasmic membrane and dynamically localizes to the division plane.
CtpA itself is located in the Caulobactermembrane fraction (Fig. 5A). This result was surprising, since CtpA lacks a signal sequence and transmembrane-spanning domains, as determined by TMHMM2 and SignalP (65). To further localize CtpA, we analyzed Caulobactermembranes by sucrose density centrifugation (64). CtpA was enriched in lighter fractions, along with McpA, whereas the outer membrane proteins RsaFaand RsaFb(72) were enriched in denser fractions (Fig. 5C).
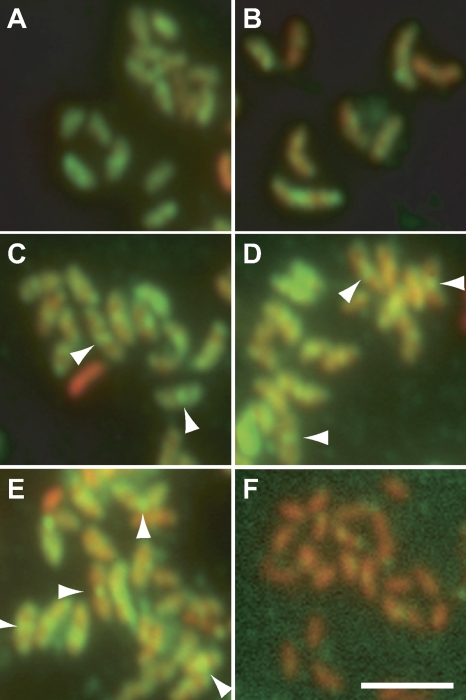
CtpA fused to a fluorescent protein at either the N terminus or the C terminus failed to complement a ctpAdeletion (data not shown), so we used immunofluorescence microscopy to localize the CtpA-3×FLAG protein in strains lacking the wild-type ctpAgene. When expressed from the high-copy-number plasmid pJS14 under its own promoter, CtpA-3×FLAG supported normal growth and cell curvature (data not shown). Swarmer cells of this strain (KR2828) were isolated and allowed to proceed synchronously through the cell cycle. At intervals, samples were fixed and stained with α-FLAG antibodies, as well as PI, to detect nucleic acids. In Caulobacter, the nucleoid is diffuse and fills the cell, so the PI signal corresponds to the extent of each cell. During the first half of the division cycle (Fig. 7A and B), CtpA-3×FLAG was present throughout the cell. At later time points (Fig. 7C to E), brighter foci of CtpA-3×FLAG also appeared at the division plane (Fig. 7C to E, arrowheads). ΔctpAcells expressing untagged CtpA from pJS14 (KR2829), which were processed in parallel contained PI signal, but very little α-FLAG signal (Fig. 7F).
Fig. 7.
Localization of CtpA-3×FLAG during the cell cycle. (A to E) KR2828 (ΔctpA+ pJS14-ctpA::3×FLAG) swarmer cells were isolated, released into PYE medium, and allowed to proceed synchronously through the cell cycle. At the indicated times, samples were withdrawn, fixed, and stained with α-FLAG antibodies, followed by Alexa Fluor 488-labeled secondary antibodies and propidium iodide. α-FLAG images, showing CtpA-3×FLAG, and propidium iodide images, showing nucleic acids, were overlaid. (A) t= 30 min; (B) t= 60 min; (C) t= 90 min; (D) t= 105 min; (E) t= 120 min. (F) Cells from a mixed culture of KR2829 (ΔctpA+ pJS14-ctpA) were fixed, stained, and imaged as described above. Green, CtpA-3×FLAG; red, propidium iodide. Arrowheads indicate midcell accumulations of CtpA-3×FLAG. Scale bar, 5 μm.
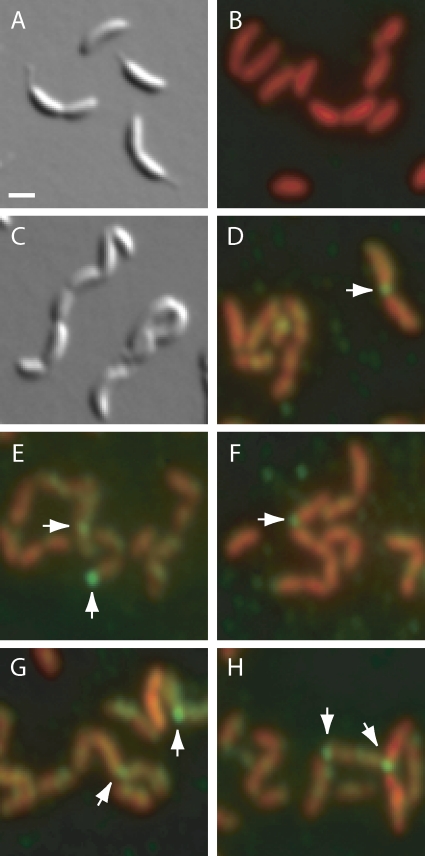
Because CtpA-3×FLAG was expressed from a high-copy-number plasmid in KR2828, it is possible that some of the α-FLAG staining represents mislocalized protein. We therefore localized CtpA-3×FLAG that was expressed from the low-copy-number plasmid pMR10 in the ΔctpAstrain as well. Untagged CtpA expressed from pMR10 complemented the ctpAdeletion, yielding unchained cells with smooth edges (KR2944, Fig. 8A). These cells were stained with PI, but showed almost no α-FLAG signal (Fig. 8B). CtpA-3×FLAG expressed from pMR10 did not fully complement the ctpAdeletion, and isolation of swarmer cells was not possible. This strain (KR3031) grew slowly (data not shown), and cell chains were visible using DIC microscopy (Fig. 8C). Because this phenotype resembles CtpA depletion, we suspect that CtpA-3×FLAG is not fully functional compared to the untagged protein. CtpA-3×FLAG was concentrated at cell junctions or poles (arrows, Fig. 8D to H) and was also present in fainter, randomly located patches.
Fig. 8.
Localization of CtpA-3×FLAG expressed from a low-copy-number plasmid. (A and B) KR2944 (ΔctpA+pMR10-ctpA). (C to H) KR3031 (ΔctpA+ pMR10-ctpA::3×FLAG). (A and C) DIC images of live cells mounted on PYE–1% agarose pads. (B, D to H) Cells were fixed and stained with α-FLAG antibodies, Alexa Fluor 488-labeled secondary antibodies and propidium iodide as in Fig. 7. Arrows indicate foci of CtpA-3×FLAG at cell junctions or poles. Scale bar, 1 μm.
Although neither of these experiments is ideal, they both revealed CtpA-3×FLAG located at the cell division plane. In cells with excess fusion protein, which could be synchronized (Fig. 7), the midcell localization was dynamic, appearing in the last quarter of the cell cycle. When CtpA-3×FLAG was expressed from a low-copy vector, the gross cell morphology was compromised, but the fusion protein was still present at cell junctions (Fig. 8). Dynamic localization of CtpA-3×FLAG to the division plane is consistent with its proposed role in resolving septal PG.
DISCUSSION
Both depletion and overexpression of CtpA cause phenotypes associated with alterations in PG synthesis or remodeling. Cells lacking CtpA have unresolved septal PG, and CtpA-3×FLAG accumulates at the division plane during the last half of the cell cycle. Conversely, cells with excess CtpA exhibit less curvature than their wild-type counterparts, a property conferred by the PG sacculus. In addition, CtpA depletion leads to ragged, blebbed outer membranes, indicating that CtpA is necessary, directly or indirectly, for outer membrane integrity.
Chains of cells with well-separated cytoplasmic compartments are generated when the E. coliPG amidases AmiA, AmiB, and AmiC are deleted (29, 73). However, these cell chains are viable, so we predict that chaining per seis not the cause of death for Caulobactercells lacking CtpA. Instead, loss of outer membrane integrity may be the lethal event. Chains of Caulobactercells with irregularly spaced division sites are also formed by mutants lacking the LytM-domain PG hydrolase DipM (22, 51, 58). Unlike the chains formed during CtpA depletion, ΔdipMchains contain large compartments up to several cell lengths, suggesting that the ΔdipMmutant is also defective in cytoplasmic compartmentalization (58). It remains to be determined whether DipM and CtpA function in the same pathway during cell division.
The Tol-Pal system, conserved in Gram-negative bacteria, links the inner and outer membranes via protein-protein interactions in the periplasm (21). Proteins in this system are essential for Caulobacterviability (16, 80), and mutants depleted of TolA, TolB, or Pal develop prominent outer membrane blebs (80), reminiscent of CtpA-depleted cells. CtpA may regulate the Tol-Pal system or other protein targets directly involved in outer membrane integrity. Alternatively, the outer membrane could be affected indirectly, via changes in the PG layer, as seen in Caulobactermutants lacking DipM (22). Caulobactercells recovering from treatment with the MreB-depolymerizing drug A22 also suggest that changes in PG could cause outer membrane alterations; these cells undergo extensive PG remodeling and develop prominent outer membrane blebs (69).
CtpA overexpression causes cell straightening, even though crescentin filaments appear to be intact and associated with the cell membrane. The Caulobacterenzyme CTP synthase (CtpS) forms filaments and affects curvature via crescentin (32). Like CtpA, overexpression of CtpS causes cell straightening; however, in these cells the crescentin structure is reduced to a single focus. CtpS depletion causes excessive cell curvature, but only when the strain contains crescentin. These results indicate that CtpS regulates vibrioid morphology via crescentin, in contrast to CtpA.
Crescentin is thought to generate curvature by exerting an asymmetric force along one side of a rod-shaped cell, such that the insertion of new PG monomers is kinetically hindered on the same side of the cell and favored on the opposite side (10, 78). In this model, CtpA overexpression could render one or more PG synthetic enzymes unresponsive to the effects of crescentin, causing unbiased addition of PG monomers to the cell wall. A recent modeling study also showed that growth-dependent straightening of curved rods is dependent on the processivity of glycan strand synthesis, with increased processivity yielding straighter cells (67). CtpA overexpression could therefore increase the processivity of glycan strand synthesis, straightening the cells.
CtpA-3×FLAG is present in patches throughout the cell but accumulates at division sites late in the cell cycle. When expressed from a low-copy-number plasmid, CtpA-3×FLAG is also visible in polar foci (Fig. 8). Transient polar localization may be a genuine property of CtpA that is obscured when we express the fusion protein from a high-copy-number plasmid (Fig. 7). Conversely, the polar foci may be an artifact of overall changes in cell morphology when CtpA activity is insufficient for proper cell division. Perhaps CtpA is normally released from the division site around the time of cell separation but is inappropriately retained when division is compromised. Future studies with antibodies directed against the native CtpA protein are necessary to obtain a better understanding of CtpA localization.
Although CtpA exhibited no phosphohydrolyase activity toward pNPP in vitro, the conserved active-site cysteine and arginine residues were required for Caulobacterviability. CtpA consists of a single PTP domain, while tyrosine phosphatases in eukaryotes often contain additional regulatory domains (71). CtpA may have low affinity for this artificial substrate, or its activity may depend on another Caulobacterprotein that was not present in vitro. In particular, the protein(s) that recruit CtpA to the cytoplasmic membrane and to the midcell may stimulate its activity. This model is consistent with PTP regulation in eukaryotic cells, where individual cytosolic PTPs are targeted to various subdomains, such as the cytoplasmic face of the endoplasmic reticulum, cell-cell adhesion complexes, or the nucleus to act on specific substrates (13). Because the known tyrosine kinases and phosphatases act within the cytoplasm or nucleus (13, 70), we predict that CtpA is associated with the inner surface of the cytoplasmic membrane. The active-site mutations argue that CtpA phosphatase activity is necessary in vivo, but we cannot rule out the possibility that CtpA functions exclusively via protein-protein interactions. We are currently isolating CtpA-interacting proteins to identify its membrane anchor and potential targets.
An essential tyrosine phosphatase in Caulobacterimplies the existence of a corresponding tyrosine kinase. Many tyrosine kinases found in bacteria differ from their eukaryotic counterparts in protein sequence and structure, forming a group called the BY-kinases (42). By searching for Caulobacterproteins with homology to known BY-kinases, we have identified candidates whose functions we are assessing genetically and biochemically.
Supplementary Material
ACKNOWLEDGMENTS
We thank Reena Zalpuri of the Electron Microscope Lab at the University of California, Berkeley, for assistance with sample preparation. We are grateful to Senthil Annamalai and Donna Lee for DNA constructs and to Matthew Cabeen and Christine Jacobs-Wagner for bacterial strains, antibodies, and valuable advice. We thank the members of the Ryan lab for helpful discussions and critical reading of the manuscript.
This study was supported by the UC Berkeley Committee on Researchand by NSFgrant MCB0543801.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Alley M. R., Maddock J. R., Shapiro L. 1992. Polar localization of a bacterial chemoreceptor. Genes Dev. 6:825–836 [DOI] [PubMed] [Google Scholar]
- 2. Andersen J. N., et al. 2001. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 21:7117–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ausmees N., Kuhn J. R., Jacobs-Wagner C. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705–713 [DOI] [PubMed] [Google Scholar]
- 4. Bhaduri A., Sowdhamini R. 2005. Genome-wide survey of prokaryotic O-protein phosphatases. J. Mol. Biol. 352:736–752 [DOI] [PubMed] [Google Scholar]
- 5. Bliska J. B., Guan K. L., Dixon J. E., Falkow S. 1991. Tyrosine phosphate hydrolysis of host proteins by an essential Yersiniavirulence determinant. Proc. Natl. Acad. Sci. U. S. A. 88:1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bretz J. R., et al. 2003. A translocated protein tyrosine phosphatase of Pseudomonas syringaepv. tomato DC3000 modulates plant defense response to infection. Mol. Microbiol. 49:389–400 [DOI] [PubMed] [Google Scholar]
- 7. Brown P. J., Hardy G. G., Trimble M. J., Brun Y. V. 2009. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv. Microb. Physiol. 54:1–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bugert P., Geider K. 1997. Characterization of the amsI gene product as a low-molecular-weight acid phosphatase controlling exopolysaccharide synthesis of Erwinia amylovora. FEBS Lett. 400:252–256 [DOI] [PubMed] [Google Scholar]
- 9. Burman L. G., Reichler J., Park J. T. 1983. Evidence for multisite growth of Escherichia colimurein involving concomitant endopeptidase and transpeptidase activities. J. Bacteriol. 156:386–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cabeen M. T., et al. 2009. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 28:1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Charbon G., Cabeen M. T., Jacobs-Wagner C. 2009. Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 23:1131–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cozzone A., Grangeasse C., Doublet P., Duclos B. 2004. Protein phosphorylation on tyrosine in bacteria. Arch. Microbiol. 181:171–181 [DOI] [PubMed] [Google Scholar]
- 13. den Hertog J., Ostman A., Bohmer F.-D. 2008. Protein tyrosine phosphatases: regulatory mechanisms. FEBS J. 275:831–847 [DOI] [PubMed] [Google Scholar]
- 14. Domian I. J., Quon K. C., Shapiro L. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415–424 [DOI] [PubMed] [Google Scholar]
- 15. Duclos B., Grangeasse C., Vaganay E., Riberty M., Cozzone A. J. 1996. Autophosphorylation of a bacterial protein at tyrosine. J. Mol. Biol. 259:891–895 [DOI] [PubMed] [Google Scholar]
- 16. Eisenbeis S., Lohmiller S., Valedebenito M., Leicht S., Braun V. 2008. NagA-dependent uptake of N-acetyl-glucosamine and N-acetyl-chitin oligosaccharides across the outer membrane of Caulobacter crescentus. J. Bacteriol. 190:5230–5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372–384 [DOI] [PubMed] [Google Scholar]
- 18. Evinger M., Agabian N. 1977. Envelope-associated nucleoid from Caulobacter crescentusstalked and swarmer cells. J. Bacteriol. 132:294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Figge R. M., Divakaruni A. V., Gober J. W. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321–1332 [DOI] [PubMed] [Google Scholar]
- 20. Gitai Z., Dye N., Shapiro L. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 101:8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Godlewska R., Wisniewska K., Pietras Z., Jagusztyn-Krynicka E. K. 2009. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol. Lett. 298:1–11 [DOI] [PubMed] [Google Scholar]
- 22. Goley E. D., Comolli L. R., Fero K. E., Downing K. H., Shapiro L. 2010. DipM links peptidoglycan remodeling to outer membrane organization in Caulobacter. Mol. Microbiol. 177:56–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grangeasse C., Cozzone A. J., Deutscher J., Mijakovic I. 2007. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem. Sci. 32:86–94 [DOI] [PubMed] [Google Scholar]
- 24. Grangeasse C., et al. 1998. Functional characterization of the low-molecular-mass phosphotyrosine-protein phosphatase of Acinetobacter johnsonii. J. Mol. Biol. 278:339–347 [DOI] [PubMed] [Google Scholar]
- 25. Gray C. H., Good V. M., Tonks N. K., Barford D. 2003. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J. 22:3524–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guan K. L., Dixon J. E. 1990. Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science 249:553–556 [DOI] [PubMed] [Google Scholar]
- 27. Hanks S. K., Hunter T. 1995. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576–596 [PubMed] [Google Scholar]
- 28. Hardie G.(ed.),. 1999. Protein phosphorylation: a practical approach, second ed. Oxford University Press, Oxford, England [Google Scholar]
- 29. Heidrich C., et al. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167–178 [DOI] [PubMed] [Google Scholar]
- 30. Howell L. D., Griffiths C., Slade L. W., Potts M., Kennelly P. J. 1996. Substrate specificity of IphP, a cyanobacterial dual-specificity protein phosphatase with MAP kinase phosphatase activity. Biochemistry 35:7566–7572 [DOI] [PubMed] [Google Scholar]
- 31. Iba H., Fukuda A., Okada Y. 1977. Chromosome replication in Caulobacter crescentusgrowing in a nutrient broth. J. Bacteriol. 129:1192–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ingerson-Mahar M., Briegel A., Werner J. N., Jensen G. J., Gitai Z. 2010. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 12:739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jadeau F., et al. 2008. Identification of the idiosyncratic bacterial protein tyrosine kinase (BY-kinase) family signature. Bioinformatics 24:2427–2430 [DOI] [PubMed] [Google Scholar]
- 34. Judd E. M., et al. 2005. Distinct constrictive processes, separated in time and space, divide Caulobacterinner and outer membranes. J. Bacteriol. 187:6874–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Judd E. M., Ryan K. R., Moerner W. E., Shapiro L., McAdams H. H. 2003. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 100:8235–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaniga K., Uralil J., Bliska J. B., Galan J. E. 1996. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol. Microbiol. 21:633–641 [DOI] [PubMed] [Google Scholar]
- 37. Keiler K. C., Shapiro L., Williams K. P. 2000. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 97:7778–7783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kirstein J., Zuhlke D., Gerth U., Turgay K., Hecker M. 2005. A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in Bacillus subtilis. EMBO J. 24:3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klein G., Dartigalongue C., Raina S. 2003. Phosphorylation-mediated regulation of heat shock response in Escherichia coli. Mol. Microbiol. 48:269–285 [DOI] [PubMed] [Google Scholar]
- 40. Koul A., et al. 2000. Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J. Bacteriol. 182:5425–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krishna S. S., et al. 2007. Crystal structure of NMA1982 from Neisseria meningitidisat 1.5 Å resolution provides a structural scaffold for nonclassical, eukaryotic-like phosphatases. Proteins 69:415–421 [DOI] [PubMed] [Google Scholar]
- 42. Lee D. C., Jia Z. 2009. Emerging structural insights into bacterial tyrosine kinases. Trends Biochem. Sci. 34:351–357 [DOI] [PubMed] [Google Scholar]
- 43. Lee D. C., Zheng J., She Y. M., Jia Z. 2008. Structure of Escherichia colityrosine kinase Etk reveals a novel activation mechanism. EMBO J. 27:1758–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lessner F. H., Venters B. J., Keiler K. C. 2007. Proteolytic adaptor for transfer-mRNA-tagged proteins from alpha-proteobacteria. J. Bacteriol. 189:272–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Levchenko I., Seidel M., Sauer R. T., Baker T. A. 2000. A specificity-enhancing factor for the ClpXP degradation machine. Science 289:2354–2356 [DOI] [PubMed] [Google Scholar]
- 46. Macek B., et al. 2008. Phosphoproteome analysis of Escherichia colireveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell Proteomics 7:299–307 [DOI] [PubMed] [Google Scholar]
- 47. Macek B., et al. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell Proteomics 6:697–707 [DOI] [PubMed] [Google Scholar]
- 48. Margolin W. 2009. Sculpting the bacterial cell. Curr. Biol. 19:R812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meisenzahl A. C., Shapiro L., Jenal U. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mijakovic I., et al. 2003. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 22:4709–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moll A., Schlimpert S., Briegel A., Jensen G. J., Thanbichler M. 2010. DipM, a new factor required for peptidoglycan remodeling during cell division in Caulobacter crescentus. Mol. Microbiol. 77:90–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morona J. K., Paton J. C., Miller D. C., Morona R. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431–1442 [DOI] [PubMed] [Google Scholar]
- 53. Nierman W. C., et al. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 98:4136–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Obadia B., et al. 2007. Influence of tyrosine-kinase Wzc activity on colanic acid production in Escherichia coliK-12 cells. J. Mol. Biol. 367:42–53 [DOI] [PubMed] [Google Scholar]
- 55. Olivares-Illana V., et al. 2008. Structural basis for the regulation mechanism of the tyrosine kinase CapB from Staphylococcus aureus. PLoS Biol. 6:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Persson C., et al. 1995. Cell-surface-bound Yersiniatranslocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol. Microbiol. 18:1351–1352 [DOI] [PubMed] [Google Scholar]
- 57. Petranovic D., et al. 2007. Bacillus subtilisstrain deficient for the protein-tyrosine kinase PtkA exhibits impaired DNA replication. Mol. Microbiol. 63:1797–1805 [DOI] [PubMed] [Google Scholar]
- 58. Poggio S., Takacs C. N., Vollmer W., Jacobs-Wagner C. 2010. A protein critical for cell constriction in the Gram-negative bacterium Caulobacter crescentuslocalizes at the division site through its peptidoglycan-binding LysM domains. Mol. Microbiol. 177:74–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Potts M., et al. 1993. A protein-tyrosine/serine phosphatase encoded by the genome of the cyanobacterium Nostoc communeUTEX 584. J. Biol. Chem. 268:7632–7635 [PubMed] [Google Scholar]
- 60. Quon K. C., Marczynski G. T., Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93 [DOI] [PubMed] [Google Scholar]
- 61. Rasband W. J. 1997-2009, posting date ImageJ. National Institutes of Health, Bethesda, MD: [Online.] [Google Scholar]
- 62. Reisinger S. J., Huntwork S., Viollier P. H., Ryan K. R. 2007. DivL performs critical cell cycle functions in Caulobacter crescentusindependent of kinase activity. J. Bacteriol. 189:8308–8320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rosqvist R., Magnusson K. E., Wolf-Watz H. 1994. Target cell contact triggers expression and polarized transfer of YersiniaYopE cytotoxin into mammalian cells. EMBO J. 13:964–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ryan K. R., Taylor J. A., Bowers L. M. 2010. The BAM complex subunit BamE (SmpA) is required for membrane integrity, stalk growth and normal levels of outer membrane β-barrel proteins in Caulobacter crescentus. Microbiology 156:742–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schultz J., Milpetz F., Bork P., Ponting C. P. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Singh R., et al. 2003. Disruption of mptpB impairs the ability of Mycobacterium tuberculosisto survive in guinea pigs. Mol. Microbiol. 50:751–762 [DOI] [PubMed] [Google Scholar]
- 67. Sliusarenko O., Cabeen M. T., Wolgemuth C. W., Jacobs-Wagner C., Emonet T. 2010. Processivity of peptidoglycan synthesis provides a built-in mechanism for the robustness of straight-rod cell morphology. Proc. Natl. Acad. Sci. U. S. A. 107:10086–10091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stove J. L., Stanier R. Y. 1962. Cellular differentiation in stalked bacteria. Nature 196:1189–1192 [Google Scholar]
- 69. Takacs C. N., et al. 2010. MreB drives de novorod morphogenesis in Caulobacter crescentusvia remodeling of the cell wall. J. Bacteriol. 192:1671–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tiganis T. 2007. Protein tyrosine phosphatase function: the substrate perspective. Biochem. J. 402:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tonks N. K. 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell. Biol. 7:833–846 [DOI] [PubMed] [Google Scholar]
- 72. Toporowski M. C., Nomellini J. F., Awram P., Smit J. 2004. Two outer membrane proteins are required for maximal type I secretion of the Caulobacter crescentusS-layer protein. J. Bacteriol. 186:8000–8009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Uehara T., Parzych K. R., Dihn T., Bernhardt T. G. 2010. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29:1412–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Underwood W., Zhang S., He S. Y. 2007. The Pseudomonas syringaetype III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J. 52:658–672 [DOI] [PubMed] [Google Scholar]
- 75. Vincent C., et al. 1999. Cells of Escherichia colicontain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J. Bacteriol. 181:3472–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vincent C., et al. 2000. Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J. Mol. Biol. 304:311–321 [DOI] [PubMed] [Google Scholar]
- 77. Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39–68 [DOI] [PubMed] [Google Scholar]
- 78. Wolgemuth C. W., et al. 2005. How to make a spiral bacterium. Phys. Biol. 2:189–199 [DOI] [PubMed] [Google Scholar]
- 79. Wugeditsch T., et al. 2001. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J. Biol. Chem. 276:2361–2371 [DOI] [PubMed] [Google Scholar]
- 80. Yeh Y.-C., Comolli L. R., Downing K. H., Shapiro L., McAdams H. H. 2010. The CaulobacterTol-Pal system is essential for outer membrane integrity and the positioning of a polar localization factor. J. Bacteriol. 192:4847–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.