Abstract
UvrD is an SF1 family helicase involved in DNA repair that is widely conserved in bacteria. Mycobacterium tuberculosishas two annotated UvrD homologues; here we investigate the role of UvrD2. The uvrD2gene at its native locus could be knocked out only in the presence of a second copy of the gene, demonstrating that uvrD2is essential. Analysis of the putative protein domain structure of UvrD2 shows a distinctive domain architecture, with an extended C terminus containing an HRDC domain normally found in SF2 family helicases and a linking domain carrying a tetracysteine motif. Truncated constructs lacking the C-terminal domains of UvrD2 were able to compensate for the loss of the chromosomal copy, showing that these C-terminal domains are not essential. Although UvrD2 is a functional helicase, a mutant form of the protein lacking helicase activity was able to permit deletion of uvrD2at its native locus. However, a mutant protein unable to hydrolyze ATP or translocate along DNA was not able to compensate for lack of the wild-type protein. Therefore, we concluded that the essential role played by UvrD2 is unlikely to involve its DNA unwinding activity and is more likely to involve DNA translocation and, possibly, protein displacement.
INTRODUCTION
Tuberculosis, caused by Mycobacterium tuberculosis, causes more deaths than almost any other infectious agent. It is estimated that one-third of the world population is latently infected with M. tuberculosis(38), with most individuals carrying the organism without showing disease symptoms. During infection, M. tuberculosisbacteria reside within host macrophages, where they are likely to encounter reactive oxygen and nitrogen species (19, 34) which can damage macromolecules, including DNA. Therefore, the ability to repair damaged DNA and to maintain chromosomal integrity will play a role in the persistence of M. tuberculosisin host tissues, and presumably in the maintenance of a reservoir of future infection. Indeed, M. tuberculosisstrains carrying mutations in genes involved in DNA repair have been shown to be attenuated for infection in a nonhuman primate model of infection (8), and an M. tuberculosisstrain deficient in nucleotide excision repair (NER) shows reduced pathogenicity in mice (5). NER is a method of DNA repair by which a single-stranded oligonucleotide surrounding a base carrying a bulky adduct can be removed, which allows DNA polymerase to fill in the gap created, thereby replacing the damaged DNA.
In Escherichia coli, in addition to its role in NER, UvrD appears to play roles in recombination (17), replication (9, 14, 18), plasmid rolling-circle replication (2), and the mismatch repair pathway (16), an alternative method of DNA repair commonly found in prokaryotes but absent in M. tuberculosis(36). Thus, UvrD homologues appear to play roles in a number of distinct pathways, and many of these roles appear to involve the ability to turn over protein-DNA complexes (11), which may be independent of helicase activity (1, 15).
M. tuberculosisand other mycobacterial species possess two homologues of UvrD, annotated UvrD1 and UvrD2 (3). Previously, we have shown that UvrD1 of M. tuberculosisis a DNA helicase with 3′-5′ polarity and with an unwinding preference for nicked DNA resembling an NER intermediate and for substrates resembling stalled replication forks (4). Furthermore, an M. tuberculosisuvrD1mutant strain exhibited increased sensitivity to DNA damaging agents commonly processed by NER (J. Houghton, C. Güthlein, B. Springer, E. C. Böttger, and E. O. Davis, unpublished data), similar to a Mycobacterium smegmatisuvrD1mutant strain (10). Thus, it would appear that it is UvrD1 that functions in NER in M. tuberculosis. Interestingly, M. tuberculosisUvrD1 also suppresses DNA strand-exchange reactions catalyzed by RecA (28), and M. smegmatisUvrD1 appears to play a role in regulating recombination (10), suggesting that mycobacterial UvrD1 has further roles outside NER.
UvrD2 has an unusual protein domain structure, with an N-terminal UvrD domain (Pfam00580) typical of superfamily I helicases linked to a C-terminal HRDC domain (Pfam00570) typical of superfamily II helicases, with an intervening domain carrying a tetracysteine motif (31). In the case of Mycobacterium smegmatisUvrD2, the C-terminal HRDC domain is not required for in vitroDNA helicase activity (31), while the tetracysteine domain (but not the tetracysteine motif) is required for DNA unwinding. Surprisingly, it was found that Ku, a DNA-binding protein which plays a role in the nonhomologous end-joining (NHEJ) pathway, could restore helicase activity to the truncated protein lacking the tetracysteine and HRDC domains. The unusual domain structure of UvrD2 suggests that it may have a distinct biological function from that of UvrD1, which is more similar to typical UvrD-like helicases.
In contrast to uvrD1, uvrD2has been shown to be essential in M. smegmatis(31). Both uvrD1and uvrD2have been predicted to be essential in M. tuberculosisby a global transposon mutagenesis approach (27); nevertheless, we have been able to isolate a mutant strain in which uvrD1is inactivated (our unpublished data). In this study, we demonstrate directly that uvrD2is essential in M. tuberculosisand investigate which properties of the encoded protein are required for its essential function.
MATERIALS AND METHODS
Bacterial strains and media.
The strains and plasmids used in this study are listed in Table 1. E. colistrain DH5α (Invitrogen) was used for all plasmid construction and site-directed mutagenesis (25). A streptomycin-resistant mutant strain of M. tuberculosisH37Rv, 1424, was used as the wild-type M. tuberculosisstrain. E. coliwas grown in Luria-Bertani (LB) broth or on LB agar plates, while M. tuberculosiswas grown in modified Dubos medium (Difco) supplemented with 4% albumin and 0.2% (wt/vol) glycerol or on Difco Middlebrook 7H11 agar (Becton Dickinson) plates supplemented with 4% albumin and 0.5% (wt/vol) glycerol. E. coliliquid cultures were grown at 37°C with shaking at 225 rpm, and M. tuberculosiswas grown at 37°C in a rolling incubator at 2 rpm. All procedures with live M. tuberculosiswere carried out under Advisory Committee on Dangerous Pathogens (ACDP) containment level 3 conditions. Where appropriate, 50 μg kanamycin ml−1and 20 μg gentamicin ml−1(for E. coli) or 25 μg kanamycin ml−1, 15 μg gentamicin ml−1, 50 μg hygromycin ml−1, and 100 μg streptomycin ml−1(for mycobacteria) were added to the media.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| M. tuberculosisstrain | ||
| 1424 | M. tuberculosisH37Rv derivative; Strr | 6 |
| E. colistrains | ||
| DH5α | Invitrogen | |
| BL21(DE3) | Invitrogen | |
| Plasmids | ||
| pBS-Int | pBluescript carrying the L5 integrase gene | 32 |
| pKP186 | Cloning vector carrying L5 attPsite; Kanr | 24 |
| pKP203 | Cloning vector carrying L5 attPsite; Gmr | 24 |
| pAW24 | uvrD2in pKP186 | This study |
| pAW26 | uvrD2in pKP203 | This study |
| pAW27 | uvrD21-600in pKP203 | This study |
| pAW28 | uvrD21-640in pKP203 | This study |
| pAW29 | uvrD2(Q266R) in pKP203 | This study |
| pAW30 | uvrD2(E508A) in pKP203 | This study |
| pMCS5-rpsL-hyg | rpsL-containing allele-exchange vector | 35 |
| pUVRD2-targ | pMCS5-rpsL-hyg containing PCR-amplified 5′ (1,316 bp) and 3′ (1,225 bp) regions surrounding uvrD2 | This study |
| pET-28 | Expression vector, contains histidine tag | Novagen |
| pET28-UvrD2 | uvrD2with C-terminal histidine tag and T7 promoter | This study |
| pET28-Q266R | uvrD2(Q266R) with C-terminal histidine tag | This study |
| pET28-E508A | uvrD2(E508A) with C-terminal histidine tag | This study |
Protein purification.
The uvrD2coding sequence was cloned into pET28 to give a C-terminally histidine-tagged construct. Site-directed mutagenesis of this construct was used to generate Q266R and E508A mutations. The constructs were transformed into E. coliBL21(DE3). Cultures were grown at 37°C until the optical density at 600 nm (OD600) reached 0.5 and then were chilled on ice for 30 min. The expression of the recombinant proteins was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside, followed by incubation at 18°C with constant shaking for 18 h. Cells were collected by centrifugation and stored at −80°C. Thawed bacteria were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.5, 0.5 M NaCl, 10% glycerol, EDTA-free protease inhibitor cocktail [Roche]). Lysozyme was added to a final concentration of 1 mg ml−1, along with 1 U ml−1DNase. Cells were disrupted by sonication, and the clarified lysate was applied to a Ni2+-agarose column (GE Healthcare) in buffer A (50 mM Tris-HCl, pH 8.5, 0.5 M NaCl, 7.5 mM imidazole). The column was washed with buffer A containing 20 mM imidazole and eluted with a gradient of 20 to 500 mM imidazole in buffer A. Eluted fractions containing recombinant UvrD2 were further purified by gel filtration using a Superdex S200 column (GE Healthcare) equilibrated with buffer B (50 mM Tris-HCl, pH 8.5, 400 mM NaCl, 2 mM EDTA, and 5 mM dithiothreitol [DTT]). Fractions containing UvrD2 were pooled and dialyzed against buffer B containing 25% glycerol. Protein concentrations were measured using a bovine serum albumin (BSA) protein assay kit (Thermo Scientific) according to the manufacturer's instructions.
ATPase assay.
The ability of M. tuberculosisUvrD2 to hydrolyze ATP was measured by linking ATP hydrolysis to NADH oxidation and measuring it spectrophotometrically as described previously (4), with the modification of using 10 to 500 μM UvrD2. Briefly, reaction mixtures (200 μl) were incubated at 37°C for 15 min in 50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 10% glycerol in the presence of 0.05 to 10 μM single-stranded DNA (ssDNA) (23 nucleotides [nt]), 2 mM ATP, 0.2 mM NADH, and 2 mM phosphoenolpyruvate, along with 4.7 U lactate dehydrogenase and 5.3 U pyruvate kinase (Sigma-Aldrich). NADH oxidation was measured by the loss of absorbance at 340 nm. The steady-state kinetic parameters for ATP hydrolysis were calculated by plotting the initial rates of hydrolysis at different substrate concentrations against the substrate concentrations and fitting the data to typical Michaelis-Menten equations, using Prism software (GraphPad). To determine the constant KDNA, defined as the concentration of DNA that supports the half-maximal rate of catalysis, reactions were performed in the presence of 1 mM ATP and various concentrations of DNA. The data are reported as averages for at least three independent experiments.
Helicase assay.
A fluorometric assay was used to observe unwinding of a partial duplex DNA substrate. An oligonucleotide complex was designed to include 22 complementary nucleotides with a 3′ 23-bp single-stranded overhang and was labeled at the end of the duplex away from the single-stranded region with adjacent 5′-ROX and 3′-BHQ-2 groups. In a duplex, the fluorescence of the ROX dye is quenched by the BHQ-2 moiety. Upon unwinding of the duplex DNA, the separation of the fluorophore from the quencher increases fluorescence. Labeled oligonucleotides were mixed at a 1:1.5 molar ratio (fluorophore to quencher) to minimize free ROX, heated to 90°C, and allowed to cool to room temperature for 2 h. Reaction mixtures (30 μl) contained 100 nM DNA substrate and 500 nM UvrD2 in helicase buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 10% glycerol). To prevent reannealing of the substrate, a 500 nM capture oligonucleotide corresponding to the 22 bp of the fluorophore oligonucleotide complementary to the quencher strand was added. The reaction mix was equilibrated to 37°C, and the reaction was initiated by addition of 5 mM ATP. DNA unwinding was monitored by measuring ROX fluorescence on a Gemini XPS microplate spectrofluorometer (Molecular Devices), with excitation at 584 nm and emission measured at 612 nm. Reported initial unwinding rates are averages for at least 4 independent experiments.
Streptavidin displacement assay.
In order to measure DNA translocation, the ability of helicase proteins to displace proteins from DNA was used (23). A 30-mer oligonucleotide with a 5′ biotin label was phosphorylated by incubation with [γ-32P]ATP and T4 polynucleotide kinase according to the manufacturer's instructions; the unincorporated nucleotides were removed by passage through a G25 gel filtration spin column (GE Healthcare). A 10 nM radiolabeled oligonucleotide was incubated with 300 nM streptavidin in the presence of 1 mM ATP in helicase buffer for 15 min to allow the streptavidin to bind to the DNA. Free biotin (6 μM) was added to act as a trap for displaced and nonannealed streptavidin, and reactions in 10-μl mixtures were started by the addition of 200 nM UvrD2 or a mutant derivative. The reaction mixtures were incubated for 15 min at 37°C, and the reactions were quenched by the addition of 10 μl stop solution (1 M NaCl, 200 mM EDTA, pH 8.0, 40% glycerol, 0.3% bromophenol blue) containing 50 nM nonbiotinylated oligonucleotide to bind to the protein and prevent band shifting. Control reactions lacking protein were performed and showed no dissociation of streptavidin from the biotinylated oligonucleotide. The reaction products were analyzed by electrophoresis through a 10-cm 15% polyacrylamide gel in 89 mM Tris-borate, 2 mM EDTA. The gels were dried under vacuum and exposed to a phosphorimager screen. Radiolabeled bands were visualized using a Storm860 phosphorimager and processed using ImageQuant software (Molecular Dynamics).
Plasmid construction.
A targeting construct for M. tuberculosisUvrD2 (Rv3198c) was constructed by amplifying the flanking genomic regions by PCR. The 5′ region was amplified from M. tuberculosisgenomic DNA by a PCR using primers 7f and 7r (Table 2), incorporating NdeI and BglII sites, respectively, and the 3′ region was amplified with primers 8f and 8r, introducing BglII and NsiI sites, respectively. The PCR products were cloned sequentially into the suicide vector pMCS5-rpsL-hyg to generate the final targeting construct pUVRD2-targ, which was verified by sequencing. Upon recombination, the construct generates a 1,484-bp deletion in uvrD2, which creates a truncated gene product.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| D2delchkF | GCATGACAACGCACGATTC |
| D2delchkR | CGAGATGCTGCGCCAATACG |
| Q266R_F | CGTCGGCGACGCCAACCGGACCATCTACTCGTTTACCG |
| Q266R_R | CGGTAAACGAGTAGATGGTCCGGTTGGCGTCGCCGACG |
| E508A_F | CACGCCGCCAAGGGACTGGCATGGGACGCGGTGTTCCTGG |
| E508A_R | CCAGGAACACCGCGTCCCATGCCAGTCCCTTGGCGGCGTG |
| D2southF | TGGACCGGTGTGCGTGCTGG |
| D2southR | ACTTGCTGTCCAGCAGCTGC |
| UvrD2int3 | TGCTTGCCGAGCTACGCCG |
| pKP186R | TCGCCACCTCTGACTTGAGC |
| pKP203R | CTACGTCGACATCGATAAGC |
| 7F | GGAATTCCATATGGAACTGGCATGACAACGCACG |
| 7R | GGAAGATCTCACGGCGAACTTGCTGTCCAG |
| 8F | GGAAGATCTGACGTGTGCAGCCGACG |
| 8R | TGCATGCATCCAGGTCGAGGTCTTCC |
| Hel_5_FLO | ROX-CATGGGCACCTAGGAGATCTCACGTACCCGTGGATCCTCTAGAGT |
| Hel_3_BHQ-2 | TGAGATCTCCTAGGTGCCCATG-BHQ-2 |
| Capture | CATGGGCACCTAGGAGATCTCA |
| 5′ Bio | Ct(B)GTCTATAGCGTGCAGTCACTTGAGCAT |
In oligonucleotides used for site-directed mutagenesis, base changes relative to the wild type are highlighted in gray. Restriction sites used for cloning are underlined.
t(B), biotinylated nucleotide.
For complementation, a 2.5-kb PCR product containing uvrD2and its promoter region was cloned into pKP186 (24) to generate plasmid pAW24. pKP186 is an integrase-negative derivative of the integrating vector pMV306 (33) and confers kanamycin resistance. The pKP186 plasmid and derivatives were cotransformed along with the plasmid pBS-int, carrying the integrase gene necessary to achieve integration of the plasmid into the chromosome (32). pBS-int lacks a mycobacterial origin of replication and is therefore lost from the bacterium. The plasmid pKP186 integrates into the L5 attBchromosomal site as a single copy by site-specific recombination (13).
For complement switching, pKP203, a gentamicin resistance-conferring derivative of pKP186, was used. PCR products spanning the uvrD2promoter and full-length gene and truncated copies covering residues 1 to 600 and 1 to 640 were cloned into pKP203 to give plasmids pAW26, pAW27, and pAW28, respectively. To introduce point mutations, site-directed mutagenesis was carried out on plasmid pAW26, using a Stratagene QuikChange XL kit and the oligonucleotides indicated in Table 2. The resulting plasmids, pAW29 [uvrD2(Q266R)] and pAW30 [uvrD2(E508A)], were sequenced to confirm that the point mutations had been introduced and that no other bases had been changed in the gene sequence.
Genetic procedures.
Plasmids were introduced into M. tuberculosisby electroporation as described previously (37). Single-crossover transformants (SCOs) were selected on plates containing hygromycin. Double-crossover transformants (DCOs) were isolated by streaking cells onto plates lacking antibiotics, followed by selection on medium containing streptomycin. Colonies were screened for hygromycin sensitivity and then by PCR using primers D2delchkF and D2delchkR (Table 2). We created a merodiploid strain by electroporating the uvrD2SCO with pAW24 and selecting for kanamycin/hygromycin resistance. As a control, the empty vector pKP186 was electroporated into the uvrD2SCO strain. DCOs were isolated and screened as described above. DCOs were verified by Southern analysis using an ECL direct nucleic acid labeling system (Amersham), using a PCR probe located in the 5′-flanking region of the deletion.
Complement switching was carried out by transforming the complemented uvrD2deletion strain with plasmids pAW26, pAW27, pAW28, pAW29, and pAW30. Colonies were selected for gentamicin resistance and then screened for kanamycin sensitivity. A PCR-based screen was then used to confirm gene replacement, using an internal uvrD2primer (uvrD2int3) and vector-specific primers (pKP186R and pKP203R).
RESULTS
UvrD2 is essential in M. tuberculosis.
Initially, we were interested in assessing the role of UvrD2 in DNA damage repair in M. tuberculosis, particularly in relation to whether there is an overlap in function between UvrD1 and UvrD2. Although both uvrD1and uvrD2had been predicted to be essential (27), we had been able to isolate a mutant in uvrD1, so we attempted to remove uvrD2by allelic exchange.
After transformation of the targeting plasmid into M. tuberculosis1424, Hygrcolonies were checked by PCR and Southern blot analysis to confirm that they resulted from a single crossover at the uvrD2locus. Growth of a single-crossover mutant in nonselective medium should allow a second recombination between the uvrD2flanking region carried in the chromosome and the region carried on the integrated targeting construct, pUvrD2-targ. Selection for resistance to streptomycin would select for a second recombination event and the resulting excision of the targeting plasmid carrying the rpsL+gene (26). Depending on where the second recombination event occurred, this would leave either a wild-type or deletion genotype. We used a PCR-based screen to differentiate between wild-type and deletion genotypes, for which we designed PCR primers that lay outside the region used for the construction of the targeting plasmid. These primers would generate a 4-kb PCR product from the wild-type gene and a 2.6-kb PCR product from a uvrD2deletion, while a single crossover would yield a 10-kb PCR product which we would not expect to amplify under the conditions used, allowing clear assignation of genotype. After selection for the second crossover, we found that 64/64 colonies tested carried a wild-type copy of the gene. This result suggested that uvrD2might be essential and that deletion of the gene was lethal.
To confirm this, we constructed a merodiploid strain by taking a strain carrying a single-crossover mutant of the uvrD2targeting construct and integrating a copy of pAW24 (which carries an intact copy of uvrD2) into the L5 phage attachment site. As a control, we also integrated an empty copy of pKP186, the vector from which pAW24 was derived. Plating of exponential-phase cultures of these strains onto plates containing streptomycin selected for second crossover events. Colonies isolated from either the merodiploid strain or the strain carrying the empty vector were then screened by PCR to check for the genotype. When pKP186 was integrated, 100% (28/28 colonies) of the colonies had a wild-type genotype, while when pAW24 was integrated, 44% (15/34 colonies) of the colonies had a uvrD2deletion (Fig. 1). Deletion of the chromosomal copy of uvrD2was confirmed by Southern blot analysis. These results demonstrate that uvrD2is an essential gene in M. tuberculosis, which correlates with observations in M. smegmatis.
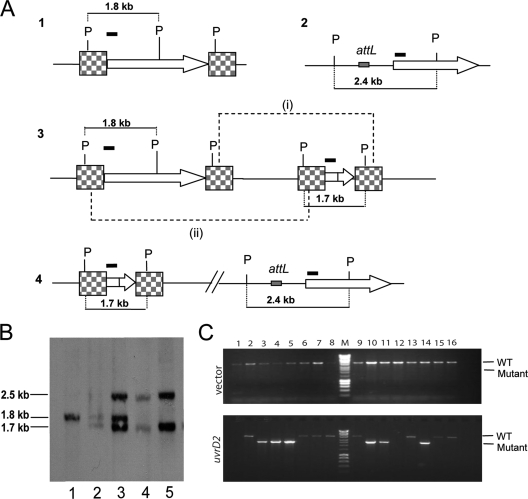
Fig. 1.
Construction of uvrD2mutant strains. (A) Schematic showing the chromosomal locus of uvrD2(1), the complementing copy of uvrD2integrated into the phage attachment site (2), and the single-crossover integrated pUvrD2-targ construct (3). Checkered boxes represent the flanking regions cloned into pUvrD2-targ. A second recombination event between the flanking regions could lead to either (i) the restoration of the wild-type locus or (ii) uvrD2deletion (4). P, PstI sites. (B) Chromosomal DNAs from the wild-type (lane 1), uvrD2single-crossover (lane 2), and uvrD2single-crossover merodiploid (lane 3) strains and from two ΔuvrD2complemented strains (lanes 4 and 5) were extracted, digested with PstI, and analyzed by Southern blotting with a 32P-labeled probe corresponding to the first 300 bp of uvrD2(shown by a black bar in panel A). (C) PCR screen showing that deletion of the chromosomal copy of uvrD2can be achieved only when a second copy of uvrD2is present. Upon integration of the empty vector pKP186, all double-crossover colonies isolated had a wild-type copy of uvrD2, shown by the PCR product of ∼4 kb (upper panel). In the presence of a second copy of uvrD2carried on pAW24 (lower panel), deletion of the chromosomal gene could be achieved, shown by the PCR product at ∼2.6 kb (lanes 3, 4, 5, 10, 11, and 14). Lanes with no PCR product were rescreened by PCR to confirm the genotype and were found to show the wild-type PCR pattern. Lanes M, Hyperladder I (Bioline) molecular size marker.
The tetracysteine and HRDC domains of UvrD2 are dispensable.
Despite our observation and the previous prediction that uvrD2is essential in M. tuberculosis, another report described the isolation of a strain of M. tuberculosisCDC1551 carrying a transposon in uvrD2(12). The insertion point of the transposon was identified as being 1,954 nucleotides into the gene, which lies in the region encoding the C-terminal HRDC domain of the translated protein.
We hypothesized that the conflicting results regarding the essentiality of uvrD2might be due either to inherent strain variation between M. tuberculosis1424 and CDC1551 or, more likely, to the fact that the C-terminal portion of UvrD2 is not required and that the N-terminal region upstream of the transposon would be expressed in the transposon mutant and be sufficient for survival. To investigate this, we decided to use complement switching (20, 32) to identify which protein domains of UvrD2 are essential. This technique exploits the dual integration/excision function of the L5 phage integrase (21, 22), whereby integrated vectors can be excised from the attachment site at a low frequency, regenerating the attBsite and allowing subsequent integration of attP-containing plasmids. To do this, we created alternate complementing constructs in a vector which also inserts into the L5 phage attachment site but which carries a gentamicin rather than a kanamycin resistance marker. By transforming the complemented uvrD2deletion strain and selecting for GmrKanscolonies, it was possible to select for colonies in which the original integrating construct had been replaced by an alternate complement.
Constructs were made to contain the full-length uvrD2gene (pAW26) or truncated versions of the gene lacking just the HRDC domain coding sequence (pAW28, containing the coding sequence for amino acids 1 to 640) or both the tetracysteine and HRDC domain coding sequences (pAW27, containing the coding sequence for amino acids 1 to 600) (Fig. 2). Table 3shows the frequencies of complement switching. Transformation of the empty vector pKP203 into the complemented uvrD2deletion strain gave only 26 colonies with gentamicin resistance, and all of these remained kanamycin resistant, suggesting that they resulted from a random integration of the vector. Transformation with plasmids carrying the wild-type (pAW26) or truncated (pAW27 and pAW28) versions of the uvrD2gene resulted in similar numbers of GmrKanscolonies, suggesting that in each case the complement had been switched. To confirm this, we designed a PCR-based screen which can distinguish between the two complements. A PCR primer designed internal to uvrD2was used in conjunction with PCR primers designed for each vector (pKP186 or pKP203) and confirmed that we had true complement switching, with at least 12 individual colonies checked for each replacement. The replacement of the integrated uvrD2gene with the truncated uvrD21–600gene demonstrates that the essential functions carried out by UvrD2 can be provided by the UvrD domain.
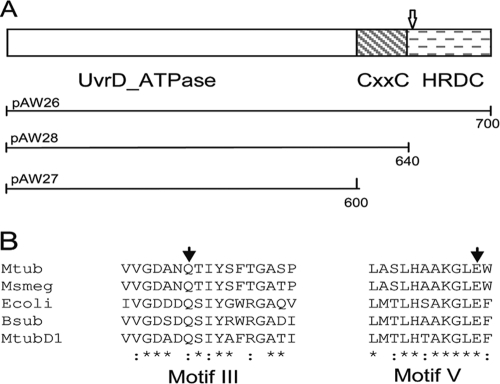
Fig. 2.
Domain structure and sequence alignment of UvrD2. (A) Domain structure of M. tuberculosisUvrD2, showing the N-terminal UvrD ATPase domain and the C-terminal HRDC domain separated by the tetracysteine motif-containing domain. The position of the transposon insertion identified in M. tuberculosisCDC1551 is indicated with an arrow. The extents of the full-length and truncated constructs used in the complement switch experiments are shown below the structure, with the numbers indicating the numbers of amino acid residues. (B) Partial alignment of UvrD2 sequences from M. tuberculosis(Mtub) and M. smegmatis(Msmeg) with those of UvrD homologues from E. coli(Ecoli) and Bacillus subtilis(Bsub) and of UvrD1 from M. tuberculosis(MtubD1). The sequences shown are for motif III and motif V of the ATPase domain, with the conserved residues mutated in this study indicated by filled arrowheads.
Table 3.
Vector switching frequencies of uvrD2allelesa
| Plasmid | Genotype | No. of Gmrtransformantsb | No. of Kanscolonies/no. of transformantsc |
|---|---|---|---|
| pAW26 | uvrD2 | 144 ± 43 | 58/64 |
| pAW27 | uvrD21-600 | 152 ± 10 | 51/64 |
| pAW28 | uvrD21-640 | 163 ± 50 | 55/64 |
| pAW29 | uvrD2(Q266R) | 8 ± 3 | 0/25 |
| pAW30 | uvrD2(E508A) | 90 ± 33 | 47/64 |
| pKP203 | 9 ± 3 | 0/28 |
The complemented ΔuvrD2strain was transformed with 500 ng of the indicated plasmid and with 300 ng pBS-Int. Colonies were selected for gentamicin resistance and then tested for kanamycin sensitivity.
Data shown are means ± standard deviations for gentamicin-resistant transformants obtained from three independent transformations from two separate experiments.
The number of kanamycin-sensitive colonies, indicating true complement replacement, is reported. Twelve kanamycin-sensitive colonies from each successful switch were verified by PCR and sequencing, and all showed that they were true complement replacements.
The helicase activity of UvrD2, but not its ATPase and DNA translocase activities, is dispensable.
Since the NER pathway is dispensable in mycobacteria (5, 30), the fact that UvrD2 is essential suggests that it plays a role outside NER. Because some functions of UvrD homologues do not require helicase or ATPase activity (1, 15), we investigated whether these functions are required by UvrD2.
To test this, we created mutated complementing constructs that should lack these activities by identifying important residues for each activity. The highly conserved glutamine residue in motif III of the UvrD domain is required for ATP hydrolysis in many UvrD homologues (1, 7), while a recent report has shown that changing a glutamate residue in motif V of M. smegmatisUvrD1 can abolish helicase activity while leaving ATP hydrolysis unaffected (29). Therefore, the corresponding Q266R and E508A mutations were made separately in pAW26 to give pAW29 and pAW30, respectively.
Transformation with pAW29 gave no colonies with true replacements, indicating that ATPase activity is necessary for the function of UvrD2 that renders it essential. In contrast, switched complements were obtained with pAW30, although with a lower efficiency than with the full-length or truncated constructs (Table 3). A proportion of the switched colonies were confirmed by PCR screening as described above. The replacement of the complement with the UvrD2 E508A construct shows that the helicase activity of UvrD2 is not required.
These results were confirmed by transforming the constructs bearing the point mutations, along with appropriate wild-type and vector controls, into the single-crossover mutant of the uvrD2targeting construct to create merodiploid strains carrying the mutant or wild-type allele in the attBlocus. Selection for double-crossover transformants resulted only in the wild-type genotype at the chromosomal locus upon complementation with UvrD2 Q266R (data not shown), like the case for the vector control, indicating that ATPase activity is required. In contrast, when strains were complemented with UvrD2 E508A, 44% of the isolated colonies were deleted for uvrD2at its native locus, confirming that helicase activity is not necessary for the essential function of UvrD2.
To confirm that the point mutations tested affected the UvrD2 protein as we predicted, we expressed and purified wild-type UvrD2 and UvrD2 carrying the same mutations as histidine-tagged proteins in E. coli(Fig. 3). Helicase, ATPase, and DNA translocase assays were then performed using the purified proteins.
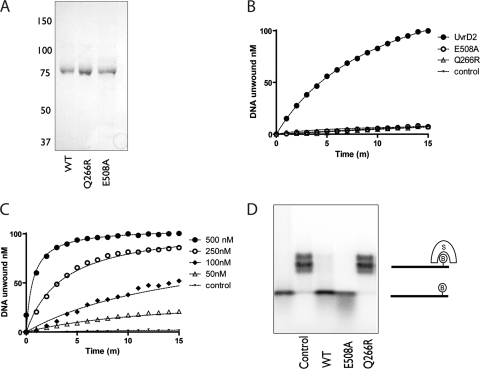
Fig. 3.
Purification and ATPase, helicase, and translocase activities of wild-type UvrD2 and Q266R and E508A mutants of UvrD2. (A) Purified recombinant wild-type M. tuberculosisUvrD2 (WT; lane 1) and the E508A (lane 2) and Q266R (lane 3) mutant derivatives were analyzed by SDS-PAGE. A Coomassie blue-stained gel is shown. The positions of molecular size markers (in kDa) are indicated. (B) DNA unwinding by M. tuberculosisUvrD2. Reaction mixtures contained 100 nM fluorescently labeled DNA substrate, 100 nM UvrD2 or a mutant derivative, 2 mM ATP, and 5 mM MgCl2. Upon unwinding of the substrate, the physical separation of the fluorophore ROX from the quencher BHQ-2 was measured by assaying fluorescence at 612 nm after excitation at 584 nm. Data for a control reaction mixture lacking enzymes are shown. No unwinding of the DNA substrate above background fluorescence was observed for either the Q266R or E508A mutant, with the data overlying the control reaction. (C) Concentration dependence of DNA unwinding by UvrD2 was investigated by varying the concentration of UvrD2 in the reaction mixtures. (D) Displacement of streptavidin (S) from a biotinylated (B) oligonucleotide. A streptavidin (10 nM)-annealed oligonucleotide was incubated with 200 nM UvrD2 or a mutant derivative at 37°C. DNA translocation was initiated by the addition of ATP, and reactions were continued for 15 min. Reaction products were separated in a 15% polyacrylamide gel and visualized by autoradiography.
Helicase activity was measured using a fluorometric assay whereby the unwinding of the synthetic DNA substrate results in the physical separation of a fluorophore from a quencher moiety, resulting in increased fluorescence. Wild-type or mutant UvrD2 proteins were incubated at 37°C with the fluorophore/quencher-labeled DNA substrate to allow DNA bind-ing, followed by the addition of a capture strand (corresponding in sequence to the oligonucleotide with the quencher moiety but lacking the adaptation) to prevent reannealing of separated substrate strands. Reactions were started by the addition of ATP, and DNA unwinding was measured as an increase in fluorescence.
Wild-type M. tuberculosisUvrD2 was competent in unwinding a standard double-stranded DNA (dsDNA) substrate with a 3′ overhang, as has been reported previously for the M. smegmatisenzyme (31), while neither of the mutant proteins was able to do so (Fig. 3). This unwinding was strictly ATP dependent and required magnesium in the buffer, as was seen for UvrD1 (4). DNA unwinding increased with increasing UvrD2 concentrations, and plotting the initial unwinding rate against protein concentration showed a linear increase, again as seen with UvrD1. The initial rate of DNA unwinding (3 nM min−1) was similar to that reported for M. tuberculosisUvrD1 (despite being assayed at a much lower enzyme/substrate ratio) and appears to be consistent with the unwinding observed for M. smegmatisUvrD1 and UvrD2, though initial rates of hydrolysis were not reported (4, 30, 31). Significantly, UvrD2 E508A and UvrD2 Q266R showed no detectable levels of DNA unwinding (Fig. 3B). Increasing concentrations of the mutant forms of UvrD2 were used in helicase assays, but no helicase activity was detected (data not shown).
ATP hydrolysis was measured in a continuous coupled assay in which ATPase activity is linked to NADH oxidation, which can be measured spectrophotometrically. Wild-type UvrD2 showed ATP hydrolysis which was strictly dependent upon the presence of DNA and magnesium. Surprisingly, the steady-state kinetics of ATP hydrolysis showed that UvrD2 had slow ATP hydrolysis (kcat, 0.035 s−1) under standard conditions where it had shown strong helicase activity, despite having an affinity for ATP (Km, 68.0 μM) similar to that of UvrD1 (kcat, 43 s−1; Km, 60.2 μM) (4). ATP hydrolysis was stimulated to similar extents by both ssDNA and dsDNA, and the affinity for ssDNA (KDNA, 0.049 ± 0.018 μM) was similar to that reported for UvrD1 (KDNA, 0.045 ± 0.004 μM) for oligonucleotides of equivalent length (4). We investigated different buffer conditions, DNA substrates, and metal ion specificities to see if the rate of ATP hydrolysis could be improved; however, the conditions reported appear to be optimal. It is not possible to compare these data with M. smegmatisUvrD2, as only endpoint values for ATP hydrolysis were reported (31). UvrD2 E508A had ATPase activity with similar steady-state kinetics (Km, 66.7 μM; kcat, 0.031 s−1) to those of the wild-type protein, with a similar DNA affinity (KDNA, 0.045 ± 0.039 μM), while UvrD2 Q266R had no detectable ATPase activity.
The 3′–5′ translocation of UvrD2 along ssDNA was investigated by its ability to displace streptavidin from a biotinylated oligonucleotide. Wild-type and mutant UvrD2 proteins were incubated with the DNA substrate in the presence of ATP, Mg2+, and biotin to trap the displaced streptavidin. Reactions were quenched, followed by native polyacrylamide gel electrophoresis to separate free oligonucleotides from streptavidin-bound oligonucleotides. UvrD2 and UvrD2 E508A were able to displace streptavidin from the 5′-biotinylated oligonucleotide in an ATP-dependent manner, suggesting that they are able to translocate along the ssDNA substrate, while UvrD2 Q266R showed no such activity (Fig. 3D). In order to exclude the possibility that the Q266R mutation affected protein folding, therefore explaining the lack of activity we observed, we analyzed the recombinant proteins by circular dichroism. UvrD2 and UvrD2 Q266R gave absorption spectra which were indistinguishable (data not shown), showing that the effects of the Q266R mutation on protein function were not caused by protein misfolding.
Thus, the point mutations introduced affected the enzymatic activities of UvrD2 as expected, validating the conclusions reached from the genetic experiments.
DISCUSSION
In this study, we demonstrated conclusively that uvrD2is essential in M. tuberculosisand determined which of its properties are required for its essential function.
Neither the HRDC domain nor the tetracysteine module was needed, providing an explanation for the reported isolation of a transposon mutant of uvrD2, as the transposon is located in the region encoding the HRDC domain (12). Biochemical studies of M. smegmatisUvrD2 (31) have indicated that deletion of the HRDC domain alone does not significantly affect ATPase or helicase activity, whereas the additional removal of the tetracysteine domain eliminates helicase but not ATPase activity. Taking this into account, our observations might suggest that helicase activity is not required for the essential role of UvrD2. However, Sinha et al. (31) found that the addition of Ku, a component of the nonhomologous end-joining DNA repair system, restored helicase function to the truncated UvrD2 protein, and Ku would be present in our genetic experiments.
By introducing point mutations into conserved motifs, we were able to separate these two biochemical properties of UvrD2. One mutation (Q266R) eliminated all detectable activity, while the other (E508A) abolished helicase activity without eliminating the ability of UvrD2 to hydrolyze ATP in a DNA-dependent manner or to translocate along DNA. It remains possible that Ku could enable helicase activity of UvrD2 E508A, but this is unlikely; the ability of Ku to stimulate DNA unwinding by the truncated M. smegmatisUvrD2 protein was investigated on the basis that Ku could stimulate helicase activity in M. smegmatisUvrD1 (30), yet Ku could not stimulate helicase activity in M. smegmatisUvrD1 carrying an E609A mutation, the mutation corresponding to that in M. tuberculosisUvrD2 E508A. The use of these point mutants in complementation experiments clearly demonstrated that helicase activity was not required. In contrast, the ATPase activity of UvrD2 was necessary for the bacterium to survive. The nonessentiality of helicase activity is useful information for directing screening approaches should UvrD2 be pursued as a potential new drug target for M. tuberculosis.
The requirement for ATP hydrolysis appears surprising given the slow kinetics observed for UvrD2. The affinity of UvrD2 for ATP (Km, 68 μM) is similar to those seen for UvrD1 (Km, 60.2 μM) and E. coliUvrD (Km, 53 μM), while the catalytic constants differ greatly (kcat, 0.035 s−1compared to 43 s−1and 95 s−1, respectively). The rate of DNA unwinding shown by UvrD2 is comparable to that seen with UvrD1, suggesting that the purified recombinant protein is fully active. The lack of translocation by UvrD2 Q266R suggests that UvrD2 uses the energy from ATP hydrolysis to drive DNA translocation and that the observed slow ATP hydrolysis by UvrD2 is sufficient to allow DNA translocation. Alternatively, it is possible that the conditions tested did not allow optimal ATP hydrolysis, although the same buffer conditions and substrates allowed DNA unwinding by UvrD2 to occur at rates comparable to those seen with other DNA helicases, and extensive attempts to identify a buffer composition which would allow an increase in the rate of observed ATP hydrolysis were unsuccessful, suggesting that these conditions allow optimal protein activity and that the observed steady-state kinetics are the true values.
The ability of UvrD2 to translocate along ssDNA and displace streptavidin suggests that UvrD2 is capable of translocating along DNA and imparting a force on a molecule blocking its path. The apparent requirement for ATP hydrolysis and/or DNA translocation, but not helicase, activity suggests that the essential function of UvrD2 is most likely to involve some such DNA translocation and, potentially, protein displacement. Mycobacterial UvrD1 can function to inhibit RecA-mediated strand exchange (28); whether UvrD2 also plays a role in regulating recombination remains to be seen. The identification of the crucial role of UvrD2 in M. tuberculosisgrowth and survival will require additional studies.
ACKNOWLEDGMENTS
This work was funded by the European Community(CSI_LTB LSHP-CT-2007-037235) and the UK Medical Research Council(program number U1175 32056).
We thank R. Veyron-Churlet for construction of the pET28-UvrD2 expression clone and Stephen Martin for performing circular dichroism analysis.
Footnotes
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Anand S. P., Zheng H., Bianco P. R., Leuba S. H., Khan S. A. 2007. DNA helicase activity of PcrA is not required for the displacement of RecA protein from DNA or inhibition of RecA-mediated strand exchange. J. Bacteriol. 189:4502–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bruand C., Ehrlich S. D. 2000. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol. Microbiol. 35:204–210 [DOI] [PubMed] [Google Scholar]
- 3. Cole S. T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 4. Curti E., Smerdon S. J., Davis E. O. 2007. Characterization of the helicase activity and substrate specificity of Mycobacterium tuberculosis UvrD. J. Bacteriol. 189:1542–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Darwin K. H., Nathan C. F. 2005. Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect. Immun. 73:4581–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davis E. O., et al. 2002. DNA damage induction of recA in Mycobacterium tuberculosis independently of RecA and LexA. Mol. Microbiol. 46:791–800 [DOI] [PubMed] [Google Scholar]
- 7. Dillingham M. S., Soultanas P., Wigley D. B. 1999. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 27:3310–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dutta N. K., et al. Genetic requirements for the survival of tubercle bacilli in primates. J. Infect. Dis. 201:1743–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flores M. J., Sanchez N., Michel B. 2005. A fork-clearing role for UvrD. Mol. Microbiol. 57:1664–1675 [DOI] [PubMed] [Google Scholar]
- 10. Guthlein C., et al. 2009. Characterization of the mycobacterial NER system reveals novel functions of the uvrD1 helicase. J. Bacteriol. 191:555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guy C. P., et al. 2009. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell 36:654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lamichhane G., et al. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 100:7213–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee M. H., Pascopella L., Jacobs W. R., Jr., Hatfull G. F. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. U. S. A. 88:3111–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lestini R., Michel B. 2008. UvrD and UvrD252 counteract RecQ, RecJ, and RecFOR in a rep mutant of Escherichia coli. J. Bacteriol. 190:5995–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lestini R., Michel B. 2007. UvrD controls the access of recombination proteins to blocked replication forks. EMBO J. 26:3804–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matson S. W., Robertson A. B. 2006. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res. 34:4089–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mendonca V. M., Matson S. W. 1995. Genetic analysis of delta helD and delta uvrD mutations in combination with other genes in the RecF recombination pathway in Escherichia coli: suppression of a ruvB mutation by a uvrD deletion. Genetics 141:443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moolenaar G. F., Moorman C., Goosen N. 2000. Role of the Escherichia coli nucleotide excision repair proteins in DNA replication. J. Bacteriol. 182:5706–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murray H. W., Juangbhanich C. W., Nathan C. F., Cohn Z. A. 1979. Macrophage oxygen-dependent antimicrobial activity. II. The role of oxygen intermediates. J. Exp. Med. 150:950–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pashley C. A., Parish T. 2003. Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 229:211–215 [DOI] [PubMed] [Google Scholar]
- 21. Pena C. E., Kahlenberg J. M., Hatfull G. F. 1999. Protein-DNA complexes in mycobacteriophage L5 integrative recombination. J. Bacteriol. 181:454–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pena C. E., Lee M. H., Pedulla M. L., Hatfull G. F. 1997. Characterization of the mycobacteriophage L5 attachment site, attP. J. Mol. Biol. 266:76–92 [DOI] [PubMed] [Google Scholar]
- 23. Richards J. D., et al. 2008. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J. Biol. Chem. 283:5118–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rickman L., et al. 2005. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 56:1274–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sambrook J., Fritsch E., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Sander P., Meier A., Böttger E. C. 1995. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol. Microbiol. 16:991–1000 [DOI] [PubMed] [Google Scholar]
- 27. Sassetti C. M., Boyd D. H., Rubin E. J. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84 [DOI] [PubMed] [Google Scholar]
- 28. Singh P., et al. Mycobacterium tuberculosis UvrD1 and UvrA proteins suppress DNA strand exchange promoted by cognate and noncognate RecA proteins. Biochemistry 49:4872–4883 [DOI] [PubMed] [Google Scholar]
- 29. Sinha K. M., Glickman M. S., Shuman S. 2009. Mutational analysis of Mycobacterium UvrD1 identifies functional groups required for ATP hydrolysis, DNA unwinding, and chemomechanical coupling. Biochemistry 48:4019–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sinha K. M., Stephanou N. C., Gao F., Glickman M. S., Shuman S. 2007. Mycobacterial UvrD1 is a Ku-dependent DNA helicase that plays a role in multiple DNA repair events, including double-strand break repair. J. Biol. Chem. 282:15114–15125 [DOI] [PubMed] [Google Scholar]
- 31. Sinha K. M., Stephanou N. C., Unciuleac M. C., Glickman M. S., Shuman S. 2008. Domain requirements for DNA unwinding by mycobacterial UvrD2, an essential DNA helicase. Biochemistry 47:9355–9364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Springer B., Sander P., Sedlacek L., Ellrott K., Bottger E. C. 2001. Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int. J. Med. Microbiol. 290:669–675 [DOI] [PubMed] [Google Scholar]
- 33. Stover C. K., et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460 [DOI] [PubMed] [Google Scholar]
- 34. Stuehr D. J., Marletta M. A. 1987. Synthesis of nitrite and nitrate in murine macrophage cell lines. Cancer Res. 47:5590–5594 [PubMed] [Google Scholar]
- 35. Wanner R. M., et al. 2009. The uracil DNA glycosylase UdgB of Mycobacterium smegmatis protects the organism from the mutagenic effects of cytosine and adenine deamination. J. Bacteriol. 191:6312–6319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wanner R. M., Guthlein C., Springer B., Bottger E. C., Ackermann M. 2008. Stabilization of the genome of the mismatch repair deficient Mycobacterium tuberculosis by context-dependent codon choice. BMC Genomics 9:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wards B. J., Collins D. M. 1996. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol. Lett. 145:101–105 [DOI] [PubMed] [Google Scholar]
- 38. WHO 2009. Tuberculosis facts 2009 update. World Health Organization, Geneva, Switzerland [Google Scholar]