Abstract
Summary: The correct translation of mRNA depends critically on the ability to initiate at the right AUG codon. For most mRNAs in eukaryotic cells, this is accomplished by the scanning mechanism, wherein the small (40S) ribosomal subunit attaches to the 5′ end of the mRNA and then inspects the leader base by base for an AUG in a suitable context, using complementarity with the anticodon of methionyl initiator tRNA (Met-tRNAiMet) as the key means of identifying AUG. Over the past decade, a combination of yeast genetics, biochemical analysis in reconstituted systems, and structural biology has enabled great progress in deciphering the mechanism of ribosomal scanning. A robust molecular model now exists, describing the roles of initiation factors, notably eukaryotic initiation factor 1 (eIF1) and eIF1A, in stabilizing an “open” conformation of the 40S subunit with Met-tRNAiMet bound in a low-affinity state conducive to scanning and in triggering rearrangement into a “closed” conformation incompatible with scanning, which features Met-tRNAiMet more tightly bound to the “P” site and base paired with AUG. It has also emerged that multiple DEAD-box RNA helicases participate in producing a single-stranded “landing pad” for the 40S subunit and in removing the secondary structure to enable the mRNA to traverse the 40S mRNA-binding channel in the single-stranded form for base-by-base inspection in the P site.
INTRODUCTION
This review will deal almost exclusively with the process of initiation codon selection in eukaryotes by ribosomal scanning, drawing primarily from studies of mammals and budding yeast, where most of the recent progress has occurred. While compelling evidence for scanning was obtained over 30 years ago, a mechanistic understanding of the process has begun to emerge only recently. I will not cover the various nonscanning modes of initiation that have been described, such as internal initiation, where the small (40S) ribosomal subunit binds directly to the initiation site on the mRNA and dispenses with (at a minimum) the m7G cap and initiation factors that recognize the cap and recruit the ribosome to the 5′ end of the mRNA. Internal initiation is well established for certain internal ribosome entry sites (IRESs) in viral mRNAs, and in some cases their functions have been reconstituted in vitro, and structures have been elucidated at an atomic resolution (58, 82, 98). My goal is to provide readers with a current understanding of the mechanisms that ensure the base-by-base inspection of the 5′ untranslated region (5′UTR) and the stringent recognition of AUG codons in an optimal sequence context during the process of scanning. This should provide the necessary foundation for considering regulatory pathways that target particular steps of the process to modulate gene expression and the functions that must be circumvented or replaced to allow ribosomes to bypass 5′UTR sequences during internal initiation or utilize suboptimal or non-AUG triplets as start codons.
OVERVIEW OF TRANSLATION INITIATION BY THE SCANNING MECHANISM
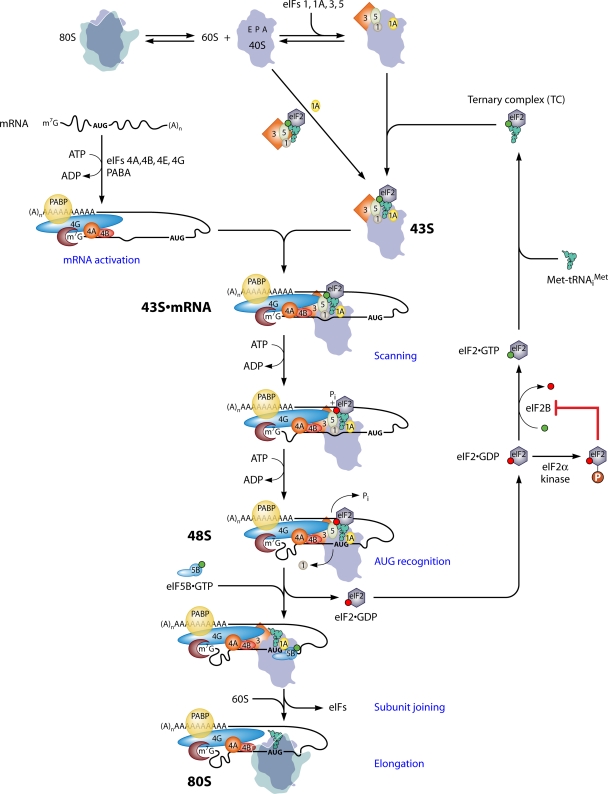
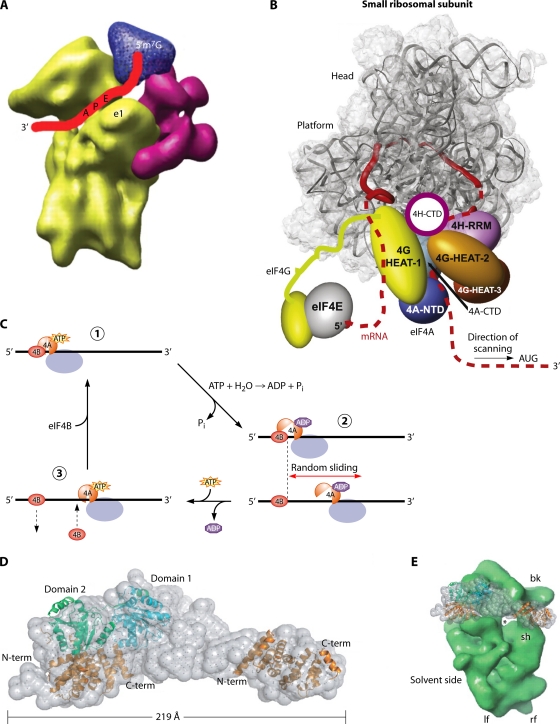
According to the current paradigm, the identification of the initiation codon by the scanning mechanism begins with the assembly of a 43S preinitiation complex (PIC), containing the initiator methionyl tRNA (Met-tRNAiMet or initiator tRNA) in a ternary complex (TC) with the GTP-bound form of eukaryotic initiation factor 2 (eIF2). The assembly of the 43S PIC is stimulated by eIFs 1, 1A, and 5 and the eIF3 complex, which (except for eIF5) are known to bind directly, and cooperatively, to the 40S subunit (Fig. 1). At least in budding yeast, a network of interactions links eIFs 1, 3, and 5 and the TC in a multifactor complex (MFC) (10), and there is evidence that these interactions enhance the formation or stability of the 43S PIC (reviewed in reference 85). The 43S PIC attaches to the mRNA at the capped 5′ end in a manner facilitated by the eIF4F complex, comprised of the cap-binding protein eIF4E, eIF4G, the RNA helicase eIF4A, and the poly(A)-binding protein (PABP). eIF4G is a scaffold protein that harbors binding domains in its N terminus for PABP and eIF4E and in its middle and C-terminal regions for eIF4A and (in mammals) eIF3. The binding domains for eIF4E and PABP in eIF4G, and its RNA-binding activity, enable eIF4G to coordinate independent interactions with mRNA via the cap, the poly(A) tail, and sequences in the mRNA body to assemble a stable, circular eIF4F·mRNA·PABP mRNP, referred to as the “closed-loop” structure. The eIF4G-eIF3 interaction is expected to establish a protein bridge between this activated mRNP and the 43S PIC to stimulate 43S attachment to the mRNA, and the helicase activity of eIF4A, enhanced by its interaction with eIF4G and eIF4B, is thought to generate a single-stranded region in the mRNA for recruiting the 43S PIC (85, 124, 155, 168, 221).
Fig. 1.
Pathway of eukaryotic translation initiation via ribosomal scanning. The process of initiation is depicted as a pathway of reactions (a subset described in blue type), beginning with the dissociation of 80S ribosomes into free 40S and 60S subunits and the assembly of the 43S PIC on the small ribosomal subunit. 80S ribosomes and 40S subunits are depicted as silhouettes of the crystal structures of bacterial 70S and 30S ribosomal species, with approximate locations of the aminoacyl-tRNA (A), peptidyl-tRNA (P), and exit (E) sites labeled in the 40S subunit. eIFs are depicted as shapes labeled by numbers, and GTP or GDP bound to eIF2 in the ternary complex (TC) is depicted as green or red balls, respectively. Two pathways for the recruitment of the TC to the 40S subunit in assembling the 43S PIC are depicted, with one involving the prior incorporation of the TC into the multifactor complex (MFC). mRNA is activated by the binding of eIF4F (eIF4E/eIF4G/eIF4A) to the m7G cap and of PABP to the poly(A) tail of the mRNA, with the attendant production of a single-stranded region at the mRNA 5′ end in a manner facilitated by eIF4B and the ATP-dependent helicase activity of eIF4A. 43S PICs attach to the 5′ end of the mRNA, forming the 43S·mRNA complex, in a manner stabilized by a network of interactions among the mRNA, eIF4G, eIF3, eIF5, eIF4B, and the 40S subunit. The 43S PIC scans the mRNA 5′UTR in a manner facilitated by eIF4A's helicase activity, and the helicases Ded1 and Dhx29 (not shown) also facilitate scanning through stable secondary structures in the 5′UTR. Note that the hydrolysis of the GTP in the TC occurs in the scanning complex, but the release of Pi is blocked until the AUG base pairs with the anticodon of Met-tRNAiMet, with the attendant dissociation of eIF1 from the 40S subunit and the arrest of scanning. eIF2-GDP is released from Met-tRNAiMet, and eIF5B-GTP joins the complex to catalyze the joining of the 60S subunit and the production of an 80S initiation complex competent for elongation. The eIF2-GDP is recycled to the eIF2-GTP by the GEF eIF2B to enable the reassembly of the TC. eIF2B is inhibited by the phosphorylation of eIF2 on Ser-51 of its α subunit by various kinases, activated by different kinds of stress. See the text for more details. (Modified from reference 85 with permission of Cold Spring Harbor Laboratory Press.)
Once bound near the cap, the 43S PIC scans the mRNA leader for an AUG codon in a suitable sequence context, using perfect complementarity with the anticodon (Ac) of initiator tRNA as the means of recognizing an AUG when it enters the peptidyl-tRNA (P) site of the 40S subunit. AUG recognition signals the cessation of scanning and the irreversible hydrolysis of the GTP bound to eIF2 in the TC, dependent on the GTPase-activating protein (GAP) eIF5, to produce a stable 48S PIC. Following the release of eIF2-GDP and other eIFs present in the scanning PIC, the joining of the large (60S) subunit is catalyzed by eIF5B to produce an 80S initiation complex (IC) containing Met-tRNAiMet base paired to AUG in the P site and ready to begin the elongation phase of protein synthesis (85, 168) (Fig. 1).
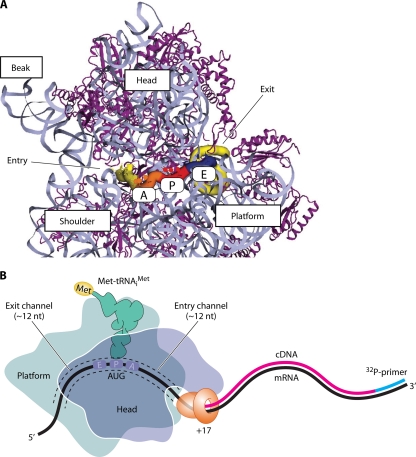
A key aspect of scanning concerns the ability of the 43S PIC to thread along the mRNA to permit the base-by-base inspection of nucleotides in the P site. The path of mRNA through the 70S bacterial ribosome has been defined by X-ray crystallography (230) (Fig. 2A), and it appears to be quite similar for the eukaryotic 43S PIC. This conclusion is based on the strong structural similarity between eukaryotic and prokaryotic ribosomes (20, 178, 197, 208) and analyses of the cross-linking of thiolated bases introduced at different positions in the mRNA to 18S rRNA residues or ribosomal proteins in reconstituted mammalian PICs (172, 173). The mRNA enters from the solvent-exposed “back” side of the small subunit between the head and shoulder domains and occupies a ∼12-nucleotide (nt) entry channel that precedes the A, P, and E decoding sites, which are exposed on the subunit interface surface, and then proceeds through a ∼12-nt exit channel that opens on the back side of the 40S subunit (Fig. 2). As discussed below, it is thought that the ability of mRNA to thread easily through the mRNA-binding channel is dependent on an “open” conformation of the 40S subunit, which is promoted by various initiation factors, particularly eIF1 and eIF1A. In a 48S PIC with AUG in the P site, the leading edge of the ribosome is located ∼17 nt 3′ of the AUG. It is possible to assay the formation of this stable complex by the inhibition of reverse transcription from a DNA primer annealed downstream of the PIC, forming a cDNA product whose length maps the “toeprint” of the ribosome on the mRNA (Fig. 2B) (80, 166).
Fig. 2.
Path of mRNA on the bacterial 70S ribosome. (A) Details of the arrangement of mRNA (yellow/orange/red/blue winding cylinder) relative to the major landmarks of the 30S subunit. The mRNA enters and exits on the solvent face of the 30S subunit (not shown here) and winds around the neck, being exposed on the interface surface of the subunit at the A, P, and E decoding sites. (Modified slightly from reference 48 with permission from Elsevier.) (B) Toeprinting analysis of the bacterial PIC containing Met-tRNAiMet base paired to AUG in the P site. The view is looking down from above the head. Primer extension by reverse transcriptase (RT) from a 32P-labeled DNA primer (blue) annealed to the mRNA (black) downstream of the PIC is inhibited when RT collides with the 30S subunit near the entry channel opening, producing a cDNA (red) ∼17 nt smaller than the distance between the 5′ end of the primer and the AUG. (Adapted from reference 68 with kind permission from Springer Science+Business Media.)
Another critical facet of the scanning process concerns the ability of stable secondary structures to impede the progression of the PIC along the 5′UTR, presumably because RNA hairpins cannot thread through the 40S mRNA entry channel. It is thought that ATP-dependent RNA helicases are required to melt secondary structures and enable the PIC to attach at the 5′ end of the mRNA and then scan every base in the 5′UTR. While eIF4A can perform this function, it now appears that other DEAD-box helicases contribute as well, at least during the scanning stage of the process (1, 22, 85, 157, 168, 174).
FEATURES OF THE SCANNING MECHANISM AND EXCEPTIONS TO THE “FIRST-AUG RULE”
The predominant role of scanning in mammalian translation initiation was established primarily by the seminal studies of Kozak, who thoroughly documented the fact that most eukaryotic mRNAs are monocistronic and also lack additional AUGs upstream of their initiation sites. Combining these findings with observations that the m7G cap stimulates translation and results indicating that ribosomes cannot bind circular mRNAs (109) but can migrate on mRNA after binding at the 5′ end (117), Kozak proposed the scanning hypothesis (108). This model was supported by observations that insertions of secondary structure in the 5′UTR can block translation (110, 164) and by mutational analyses of preproinsulin mRNA showing that 5′-proximal AUG triplets are used preferentially as start sites and that the insertion of a short upstream open reading frame (ORF) (uORF) inhibits initiation at downstream coding sequences (108, 113, 114, 116). Genetic studies by Sherman and colleagues also played an important role in uncovering the scanning process and demonstrating its importance in budding yeast. Their findings established that AUG codons at various locations near the 5′ end of the CYC1 ORF can function effectively as start codons, implying that AUG is the only sequence essential for initiation (192, 193, 201). Those authors also identified a cyc1 null mutation that created a short uORF, showing that a functional start codon must be the 5′-proximal AUG and that ribosomes generally cannot reinitiate after terminating at a uORF in yeast (202).
The scanning hypothesis was modified for mammalian cells after the discovery that sequences immediately surrounding the start codon, particularly at the −3 and +4 positions (relative to the AUG), can have a dramatic effect on initiation frequency, to the point where a 5′-proximal AUG will be bypassed at high frequencies if it resides in an unfavorable context. This exception to the first-AUG rule was called “leaky scanning” (112). A consensus sequence surrounding the start codon was compiled by Kozak for vertebrate mRNAs, 5′-GCCGCC(A/G)CCAUGG-3′ (103), and it was shown that mutations that depart from the consensus at position −3 can reduce initiation on preproinsulin mRNA by more than an order of magnitude in mammalian cells. Relatively lesser effects were observed for substitutions at positions +4, −1, and −2, which were greater in degree when the −3 nucleotide was suboptimal (112). A more recent sequence compilation is in general agreement with these earlier findings but indicated that strong sequence bias is limited to only the first 5 nt upstream of the AUG. The 5′UTRs of mammalian mRNAs are G+C rich throughout ∼100 nt upstream of the AUG, except for the strong preference for an A at position −3, and there is an overrepresentation of C's at positions −1, −2, and −4 and an underrepresentation of U's at positions −1 to −5 (190). Indeed, the presence of all U's surrounding the AUG had been shown earlier to reduce preproinsulin mRNA translation to almost undetectable levels (112).
In budding yeast, in contrast, 5′UTRs are highly biased for A's from positions −1 to −30, with a particularly strong preference for A at position −3 that is comparable to what occurs in mammals (190). Mutations from the consensus at position −3 reduced translation only 2-fold or less for various yeast genes in vivo (17, 37, 229). However, deviations from the consensus do permit the leaky scanning of the first AUG in several native yeast mRNAs (195, 218, 220). Furthermore, deviations from the consensus produce stronger effects on the translation of mitochondrial glycine tRNA synthetase (GRS1), which initiates from a UUG start codon, with a ∼30-fold reduction observed upon substituting the native context, A−3-A−2-A−1-UUG, with the poor context, C−3-G−2-C−1-UUG. Again, substitutions at position −3 had the strongest effects, whereas nt +4 was ineffectual. In fact, this switch in context produced a ∼5-fold change in expression with an AUG start codon at GRS1, even though single-base deviations from the consensus had only small effects (31). Thus, sequence context regulates initiation efficiency in yeast, with an A at position −3 conferring the greatest stimulation, but the dynamic range of context effects is smaller in yeast than in mammals for AUG start codons. The importance of an A at position −3 in both yeast and mammals suggests that this residue makes a conserved interaction with an initiation factor or ribosomal constituent near the ribosomal E site to stabilize the 48S PIC. As discussed below, the α subunit of eIF2 is one candidate for this factor (172).
Not surprisingly, the replacement of the AUG start codon with other triplets confers a dramatic reduction in translational efficiency both in vitro and in cells, although, as noted above, the extent of the reduction depends on the sequence context (31). Most single-base substitutions of AUG to “near-cognate” triplets function at frequencies of ∼1 to 10% of AUG, depending on the gene and the study, except for the AAG and AGG purine substitutions at the second position, which are essentially nonfunctional (39, 99, 162). The yeast genes ALA1 and GRS1 (mentioned above) use UUG or ACG codons upstream of an in-frame AUG to produce longer, mitochondrial forms of their gene products. Interestingly, a genome-wide analysis of 80S ribosome occupancies of yeast mRNA sequences uncovered evidence of the translation of 143 uORFs initiating with near-cognate triplets, which, except for AAG-, AGG-, or ACG-initiated uORFs, appear to be translated at levels comparable to those of AUG-initiated uORFs. Interestingly, it appeared that the levels of translation of these non-AUG uORFs (nAuORFs) generally increased relative to levels of the downstream coding sequences, and could even exceed the latter, under conditions of amino acid starvation (88). However, an analysis of translation rates of lacZ fusions made to two such nAuORFs in GCN4 mRNA confirmed the translation of only one of the two, which occurs in an optimal sequence context, and suggested that its translational efficiency relative to that of GCN4 coding sequences was much lower than expected from the ribosome occupancy data, and it did not increase substantially under starvation conditions (231). It is also unclear why other near-cognate triplets in an optimal context in the leader of GCN4 mRNA, and presumably many other genes, were not detectably translated in vivo (88). Results from a separate study suggested that nucleotides besides the critical −3 position significantly affect initiation from certain near-cognate triplets (30).
The strong bias for A bases in yeast 5′UTRs indicates a low structurogenic potential, and indeed, there are few yeast mRNA leaders genome-wide that are predicted to fold into stable secondary structures (122). Consistent with this, translation in yeast is inhibited more effectively than in mammalian cells by structures of lower stability (2, 17, 35). Increasing the length of the 5′UTR, in one case by ∼1,700 nt, had little effect on translational efficiency in yeast (17, 22, 37), implying that scanning is not rate limiting when the 5′UTR lacks a stable secondary structure. A similar conclusion was made for mammalian extracts, wherein increasing the leader length without introducing structure was found to increase the translational efficiency, and this was attributed to the ability to preload the 5′UTR with multiple scanning PICs (107).
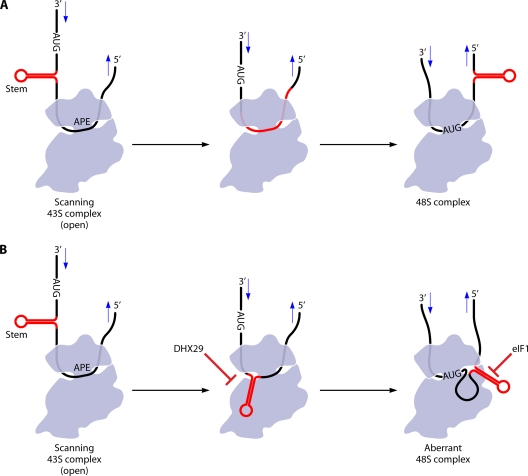
On the other hand, a shortening of the 5′UTR to 21 nt or less reduced the translational efficiency of yeast PGK1 mRNA by 50% (214), and related observations made for reporter mRNAs in mammalian extracts indicated that the recognition of the 5′-proximal AUG codon was inefficient when located too close to the cap and that an AUG further downstream was utilized instead (115). The inefficiency of translation from start codons located 1 to 2 nt from the cap has been exploited in yeast to permit the translation of multiple proteins from the same mRNA initiated from either the cap-proximal AUG (inefficiently) or the next AUG by ribosomes that skip the first AUG (63, 144, 195). For MOD5 mRNA, leaky scanning of the 5′-proximal AUG is dictated by both a short leader length and a suboptimal sequence context (195). A leader length of <15 nt is expected to produce a 48S PIC complex in which the m7G cap will be situated in the mRNA exit channel, and the exit channel will not be fully engaged by nucleotides 5′ of the AUG (Fig. 2B). As discussed below, eIF1 destabilizes 48S PICs with this “abnormality” in vitro (167).
To summarize, mRNAs translated with high efficiencies, both in yeast and in mammals, are expected to contain a 5′UTR of ∼32 nt or more; to be devoid of stable secondary structure, extra AUG codons, and near-cognate triplets in an optimum sequence context; and to contain a favorable context surrounding the functional AUG start codon. The requirement for an optimal context can be mitigated by the presence of a stable secondary structure located 3′ of the start codon, with the strongest stimulation of translation occurring when the base of the stem-loop is 14 nt 3′ of the AUG. As this spacing corresponds to the length of the A site and mRNA entry channel of the ribosome (Fig. 2), the stem-loop is likely stalling the scanning 43S PIC with the AUG positioned in the P site (105).
Although it is generally considered that scanning is unidirectional, there is intriguing evidence that the process can be punctuated by periodic “backwards excursions” of ∼15 nt or less in the 3′-to-5′ direction. It was found that a second AUG located within this interval 3′ to the first AUG in turnip yellow mosaic virus (TYMV) mRNA reduced initiation at the latter, provided that the first AUG occurred in a suboptimal context that allowed for leaky scanning. It was suggested that a fraction of ribosomes that leaky scan the first AUG get a second chance to encounter it during backwards scanning, but this possibility is reduced if they traverse a second AUG with a favorable context during the backwards excursion (136).
Kozak showed that the first-AUG rule can also be violated in mammalian cells when a uORF is located a considerable distance upstream from the coding sequences and provided evidence that ribosomes can resume scanning after translating the uORF and reinitiate downstream if they have sufficient time to reassemble a 43S PIC before reaching the next start codon. The reinitiation efficiency increased with increasing distance between the stop codon of the uORF and the downstream AUG and reached a plateau at a separation of ∼80 nt (106). This principle underlies the translational control of yeast GCN4 mRNA by multiple uORFs, with the added feature that the scanning distance/time required to recover reinitiation competency is increased under amino acid starvation conditions. Because this mechanism represents an intricate example of how the rules of scanning can be exploited to regulate translation and is frequently employed as an in vivo reporter of the rate of TC recruitment, the rate of scanning, or leaky scanning of AUG codons, it is worth mentioning here in some detail.
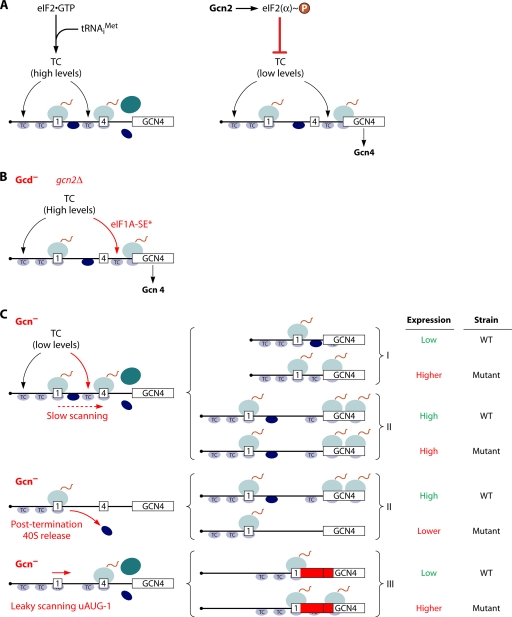
According to the current model (Fig. 3A), ribosomes scanning from the cap translate the first of four uORFs (uORF1), and ∼50% of the posttermination 40S subunits remain attached to the mRNA and resume scanning. Under nonstarvation conditions, essentially all of these scanning 40S subunits rebind the TC; reinitiate at uORF2, uORF3, or uORF4; and then dissociate from the mRNA and fail to translate the GCN4 ORF. The ability of ribosomes to resume scanning following uORF1 translation, and their inability to do so after translating uORFs 2 to 4, is dictated by distinct sequences surrounding the uORF stop codons. In amino-acid-starved cells, the α subunit of eIF2 (eIF2α) is phosphorylated by the kinase Gcn2, converting eIF2 into a competitive inhibitor of its guanine nucleotide exchange factor (GEF), eIF2B, and thus impeding the reassembly of the TC after each round of initiation (Fig. 1). Because of the reduced TC concentration, ∼50% of the 40S subunits scanning from uORF1 now reach uORF4 before rebinding TC and, lacking Met-tRNAiMet, bypass the AUGs at uORFs 2, 3, and 4. Most of these ribosomes rebind the TC before reaching the GCN4 AUG and reinitiate there instead (Fig. 3A). Thus, a reduction of TC levels induces GCN4 translation by allowing a fraction of scanning 40S subunits to bypass the inhibitory uORFs 2 to 4. Key support for this model comes from the finding that progressively increasing the separation between uORF1 to uORF4 progressively reduces GCN4 translation, presumably increasing the fraction of rescanning 40S subunits that rebind the TC before reaching uORF4 (84).
Fig. 3.
Schematic model of GCN4 translational control and mechanisms of Gcd− and Gcn− mutations. (A) Following the translation of uORF1 (boxed 1), posttermination 40S subunits remain attached to the GCN4 mRNA and resume scanning. Under nonstarvation conditions (left), they quickly rebind the TC and reinitiate at uORF4 (boxed 4), and the 80S ribosome dissociates after terminating at uORF4. Under amino acid starvation conditions (right), the concentration of the TC is reduced by eIF2α phosphorylation, such that many 40S ribosomes fail to rebind the TC until scanning past uORF4 and can reinitiate at the GCN4 ORF instead. (B) GCN4 translation is normally constitutively repressed in gcn2Δ cells owing to the inability to phosphorylate eIF2α in response to starvation. However, mutations that reduce the rate of TC loading on 40S subunits, such as substitutions in the eIF1A SE elements (eIF1A-SE*) (see text for details), constitutively derepress GCN4 translation, producing the Gcd− phenotype. (C, left) Defects in different steps of reinitiation pictured here all prevent the induction of GCN4 translation in starved cells despite the reduction in TC levels, conferring the Gcn− phenotype. Mechanisms of Gcn− phenotypes can be discerned by analyzing the expression of the solitary uORF1 constructs depicted on the right. Gcn− mutations that confer slow scanning should increase reinitiation at GCN4 for construct I, in which uORF1 is very close to the GCN4 uORF, by increasing the scanning time available for the reassembly of the PIC, but have little effect on construct II, in which uORF1 is far upstream of GCN4, and the scanning time available for reinitiation is long. Gcn− mutations that confer the release of posttermination 40S subunits or reduce their ability to resume scanning will reduce the expression of constructs I and II. Gcn− mutations that confer a leaky scanning of uORF1 will increase expression from construct III, in which uORF1 is elongated and extensively overlaps the GCN4 ORF, abolishing reinitiation.
It has been possible to identify mutations in yeast that affect reinitiation or scanning by their impairment of GCN4 translation. Because GCN4 encodes a transcriptional activator of amino acid biosynthesis genes (84), such mutations reduce the ability to grow under conditions of amino acid starvation imposed with inhibitors of biosynthetic enzymes (Gcn− phenotype). The first described Gcn− mutations impaired eIF2α phosphorylation, or eliminated the inhibition of eIF2B by eIF2(αP), and the attendant reduction in TC abundance (84). However, there are multiple other defects that can produce Gcn− phenotypes without a reduction in TC abundance (Fig. 3C). These include the leaky scanning of the uORF1 AUG (uAUG1) or the failure to retain 40S subunits after termination at the uORF1 stop codon, which eliminates the pool of reinitiating 40S subunits capable of bypassing uORFs 2 to 4 at low TC levels. Another possibility is a reduced rate of scanning by reinitiating 40S subunits, which provides additional time to rebind the TC by the reinitiating 40S subunits and prevent the bypass of uORFs 2 to 4 at low TC levels. These mechanisms can be distinguished by analyzing the expression of the solitary-uORF1 GCN4-lacZ reporters depicted in Fig. 3C. Mutations have also been obtained with the opposite phenotype, of constitutively derepressing GCN4 translation, in the absence of eIF2 phosphorylation by Gcn2, conferring a Gcd− phenotype. Classical Gcd− mutations reduce the recycling of eIF2 by eIF2B or otherwise impair TC assembly by eIF2; however, this phenotype can also arise from mutations that reduce the rate of TC loading onto 40S subunits at normal TC concentrations, such that a fraction of reinitiating 40S subunits fail to recover the TC until after bypassing uORFs 2 to 4 (Fig. 3B) (84).
Interestingly, a mechanism fundamentally similar to that governing GCN4 translation operates in mammalian cells to regulate the translation of ATF4 (78, 215) and ATF5 (232), which, like GCN4, encode b-ZIP transcription factors that stimulate the transcription of stress-responsive genes. Although there are only two uORFs rather than four, the 5′-proximal uORF is permissive for reinitiation, and the second uORF, which is not permissive for rescanning, must be located an appropriate distance downstream from uORF1. A difference is that the second uORF is considerably longer and overlaps the ATF4/ATF5 start codon, which likely reflects observations that short uORFs are more permissive for reinitiation in mammalian cells than in yeast cells. Having a longer second uORF that overlaps the ATF4/ATF5 AUG codon can be expected to effectively eliminate reinitiation following its translation (92). In fact, the translational control of GCN4 in yeast can be maintained with only uORFs 1 and 4 and with a similarly elongated version of uORF4 (84).
GENETIC IDENTIFICATION OF FACTORS INVOLVED IN ACCURATE AUG SELECTION
The Anticodon of tRNAiMet Is a Critical Determinant of AUG Selection during Scanning
Genetic experiments by Donahue et al. first proved that the Ac of tRNAiMet plays an essential role in start codon recognition in yeast (36), by showing that overproducing mutant tRNAiMet containing a UCC versus a UAC Ac restored the expression of a his4 allele with AGG (but not other triplets) in place of the AUG start codon. The introduction of an extra AGG upstream of the AGG start site abolished his4 expression, indicating that the upstream AGG was recognized preferentially by the mutant tRNAiMet(CCU), a hallmark of linear scanning. Similar results were obtained with mammalian cells (60) and also with a mammalian cell-free system reconstituted with purified ribosomes and initiation factors by using the toeprint assay to locate the 48S PIC (128). Thus, perfect base pairing between the initiator tRNA Ac and the start codon, regardless of the exact sequence of the resulting duplex, is crucial for efficient formation of the 48S PIC.
Kinetic and thermodynamic analyses of TC binding to 40S subunits in vitro suggest that the rate and stability of base pairing between the Ac of tRNAiMet and the start codon are critical determinants of initiation efficiency in yeast. TC binding to the PIC was measured with a reconstituted yeast system containing 40S subunits, eIF1, eIF1A, TC, and a model, short (45-nt) unstructured and uncapped mRNA with a centrally located AUG or non-AUG triplet (In this system, referred to below as the “basic” reconstituted yeast system, 48S PIC assembly likely begins with the random collision of the TC with the 43S·mRNA complex. It was found that all substitutions in the second or third position of AUG, as well as the CUG substitution, dramatically reduced the affinity of the TC for the 43S·mRNA complex, primarily by decreasing the on rate of the reaction by ∼100- to 1,000-fold. Consistent with results cited above, a wild-type (WT) rate of binding was restored for the CUG start codon by using the complementary mutant tRNAiMet(CAG) initiator. A kinetic model was developed, in which the dramatically lower rate of binding observed for the codon-Ac mismatches reflects a strong decrease in the rate of a conformational change that occurs following the initial, diffusion-limited encounter of the TC with the PIC and takes place rapidly only with AUG, UUG, or GUG start codons. While the UUG and GUG mismatches have relatively little effect on the TC on rate, they increase the off rate (as do all mismatches from AUG) and elevate the predicted Kd (dissociation constant) (koff/kon) by ∼10-fold. The range of Kd values for TC binding in these reactions roughly parallels the relative efficiencies of reporter mRNA translation with different near-cognate start codons in yeast cells, suggesting that the stability of the codon-Ac duplex is a key factor determining the probability of start codon selection in vivo, when the TC is sampling triplets as they enter the P site of the scanning PIC. However, other constraints are needed to account for the fact that ACG, AUA, and AUU were utilized only slightly less efficiently than UUG, CUG, and GUG, and for the complete inefficacy of AGG, in reporter translation in cells (99).
eIF1, eIF5, and the Three Subunits of eIF2 All Participate in Stringent AUG Recognition
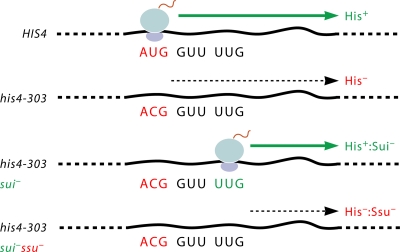
An early breakthrough in the identification of initiation factors involved in AUG recognition also came from Donahue et al. using yeast genetics. Those workers isolated mutations that restored the expression of his4 alleles with a non-AUG start codon by enabling translation to initiate more efficiently at the third, UUG codon of the his4 ORF, reestablishing growth in histidine-free medium (Fig. 4). Such Sui− (suppressor of initiation codon mutant) mutations were obtained for each of the three subunits of eIF2, eIF5, and eIF1, which increased the ratio of initiation at a UUG versus an AUG present at the first codon of HIS4 by roughly an order of magnitude. These findings not only implicated these factors in stringent AUG selection but, as described next, also provided insights into their molecular functions in the process (54, 226).
Fig. 4.
Scheme for identifying Sui− and Ssu− mutations in yeast. See the text for details.
Biochemical analysis revealed that the G31R substitution in eIF5 produced by the Sui− mutation SUI5 increases the ability of eIF5 to stimulate GTP hydrolysis by the TC in a minimal PIC reconstituted with only 40S subunits, TC, AUG, and eIF5, i.e., stimulating its GAP function, consistent with the dominance of its Sui− phenotype in vivo. Analysis of the mutant eIF2 holoproteins containing a Sui− substitution in eIF2β (S264Y, encoded by SUI3-2) and eIF2γ (N135K, encoded by the SUI4 allele of GCD11) showed that the mutations increase the intrinsic (eIF5-independent) GTPase activity of eIF2 and elevate levels of Met-tRNAiMet dissociation from eIF2 in the absence of GTP hydrolysis, respectively (86). These findings suggested that inappropriate selections of UUG start codons could be enhanced by increasing the rate of GTP hydrolysis by the TC, or by enabling the release of Met-tRNAiMet from eIF2-GTP to the P site without GTP hydrolysis, without a perfect AUG-Ac match. The effects of Sui− mutations affecting eIF1 (encoded by SUI1) were not analyzed until much later, but the fact that they are recessive to WT SUI1 (226) suggested that the loss of an eIF1 function enables non-AUG selection, which fits with the current model that the dissociation of eIF1 from the PIC is required for AUG selection.
eIF1 PROMOTES SCANNING, IMPEDES GTP HYDROLYSIS BY THE TC, AND PREVENTS TIGHT BINDING OF Met-tRNAiMet TO THE P SITE AT NON-AUG CODONS
eIF1 Mediates Gated Release of Pi from eIF2-GDP · Pi upon AUG Recognition
Biochemical analysis of eIF1 and eIF5 using the basic yeast reconstituted system described above fundamentally altered our understanding of how AUG recognition regulates GTP hydrolysis by eIF2. Lorsch and colleagues found that fluorescently tagged derivatives of eIF1 and eIF1A exhibit fluorescence resonance energy transfer (FRET) in 43S PICs and that the FRET decays in a two-phase reaction upon the addition of mRNA and the formation of 48S complexes. The rapid phase was assigned to a conformational change that increases the separation between fluorophores, while the slower phase reflects the dissociation of eIF1 from the PIC. Both phases occur more rapidly, or proceed further to completion, with AUG than with non-AUG start codons in the mRNA, demonstrating that AUG recognition triggers the eIF1 dissociation from the PIC (132). When the 43S·mRNA complexes were reconstituted with eIF5, to stimulate GTP hydrolysis by the TC, a proportion of the GTP was hydrolyzed even with non-AUG start codons, but the Pi was released from eIF2-GDP·Pi very slowly for non-AUG complexes versus AUG complexes. Furthermore, the presence of AUG evoked Pi release at a rate essentially identical to that of the eIF1 dissociation, and an eIF1 mutation (G107R) that retards its dissociation from AUG complexes similarly delayed the Pi release (5, 132). These findings indicated that the release of Pi from eIF2-GDP·Pi is a critical step in AUG recognition and that Pi release is triggered by the elimination of eIF1 from the PIC.
The notion that eIF1 is a “gatekeeper” that must be released from the scanning PIC is consonant with genetic findings that the overexpression of eIF1 suppresses the increased UUG initiation in SUI5 (eIF5) or SUI3-2 (eIF2β) Sui− mutants (70, 212). It was envisioned that elevated eIF1 concentrations will shift the equilibrium between 40S-bound and free eIF1 toward the bound state and thereby reduce the rate of Pi release from eIF2-GDP·Pi. This explanation assumes that Pi release does not occur instantaneously upon the eIF1 dissociation, such that the reassociation of eIF1 can restore the original configuration of the scanning complex. Furthermore, biochemical analyses of various Sui− eIF1 mutants revealed reduced eIF1 occupancies in native 40S subunits in vivo and a reduced affinity for 40S subunits in the basic yeast reconstituted system. Importantly, one such mutant (with alanine substitutions at residues 93 to 97) dissociates more rapidly than WT eIF1 from PICs reconstituted with a UUG- or AUG-containing mRNA (32). The overexpression of such eIF1 variants (referred to below as “class I” mutants) diminishes their Sui− phenotypes in vivo, as expected if their inappropriate release from scanning PICs at UUG codons can be prevented at higher concentrations of the mutant proteins. Thus, the accelerated dissociation of eIF1 from the PIC at UUGs represents a straightforward mechanism for the Sui− phenotypes of class I eIF1 mutants.
A biochemical analysis of eIF1 in the mammalian reconstituted system by Pestova and colleagues provided evidence that eIF1 additionally restrains the GAP function of eIF5 at non-AUGs (210), an activity that would reduce the formation of eIF2-GDP·Pi in addition to blocking Pi release in the scanning complex. Indeed, as noted above, eIF2-GTP exists in equilibrium with eIF2-GDP·Pi in yeast PICs reconstituted with eIF5 (5), and the enhanced GAP function and GAP-independent GTPase activity of eIF2 conferred by the SUI5 and SUI3-2 mutations of eIF5 and eIF2β, respectively, suggest that a shift of the equilibrium from eIF2-GTP to eIF2-GDP·Pi can produce inappropriate Pi release at non-AUG codons.
eIF1 Promotes an Open, Scanning-Conducive Conformation of the PIC
The notion that eIF1 acts to block start codon recognition had been proposed independently by Pestova et al. based on the biochemical activities of mammalian eIF1 in fully reconstituted 48S PICs. Using the toeprint assay, those authors found that eIF1 and eIF1A, in addition to TC, eIF3, eIF4A, eIF4B, eIF4F, and ATP, are required to form 48S complexes at the AUG codon of native β-globin mRNA. In the absence of eIFs 1 and 1A, an unstable complex was formed near the 5′ end at the expense of correctly positioned 48S complexes (165), consistent with a defect in scanning from the cap. Remarkably, using an artificial uncapped mRNA with an unstructured 5′UTR, 48S assembly at the AUG could occur in the absence of ATP and the eIF4 factors, in a manner requiring eIF1, eIF1A, eIF3, and the TC, whereas eIF3 and the TC were sufficient when the AUG was very close to the 5′ end. These findings implied that eIF1 and eIF1A promote a conformation of the 40S subunit that is capable of 5′-to-3′ linear scanning, without ATP hydrolysis, provided that no stable secondary structure occurs in the leader (167).
With unstructured mRNA, eIF1 was not required for appreciable 48S assembly at the AUG if eIF4F was provided; however, an increased selection of both near-cognate triplets and AUGs in a suboptimal context or located too close (4 nt) to the cap occurred when eIF1 was omitted (167). Thus, eIF1 was required to restrict start codon selection to AUGs in an optimal context, in accordance with its gatekeeper function deduced from studies of yeast. Indeed, yeast eIF1 can replace mammalian eIF1 in the reconstituted system, and several Sui− yeast eIF1 mutants are defective in promoting scanning on β-globin mRNA and blocking initiation at near-cognate triplets or 5′-proximal AUGs (32). As these scanning assays were conducted without eIF5, the role of eIF1 in discriminating against near-cognate triplets or poor context must be independent of its roles in gated Pi release or restraining the eIF5 GAP function. It was suggested that these functions of eIF1 are derived from its ability to promote an “open” conformation of the 40S subunit, in which the positions of mRNA and initiator tRNA on the ribosome are conducive to scanning but incompatible with start codon selection (167).
Further evidence that eIF1 acts to block the selection of suboptimal start codons came from the mapping of the binding site of mammalian eIF1 on the 40S subunit using directed hydroxyl radical cleavage of 18S rRNA by eIF1 proteins derivatized with Fe(II) at unique surface-exposed cysteines. This analysis predicted binding near the P site on the platform region of the 40S subunit, similar to that determined previously for bacterial initiation factor 3 (IF3) on the 30S subunit (127). This prediction was confirmed with the recent crystal structure of a Tetrahymena 40S·eIF1 complex (178). IF3 in bacteria is known to destabilize the binding of noncognate tRNAs in the P site and to discriminate against 5′-proximal AUGs of leaderless mRNAs. Indeed, IF3 mutations increase non-AUG initiation in the manner observed for eIF1 Sui− mutations in yeast, and the overexpression of IF3 in bacteria inhibits non-AUG initiation, analogous to the effect of eIF1 overexpression in yeast mentioned above (references 128 and 177 and references therein). Thus, it was proposed that eIF1 antagonizes the codon-Ac interaction in the P site in a manner that is overcome efficiently only with a perfect AUG-Ac duplex, an optimal AUG context, and a 5′UTR long enough to interact extensively with the 40S mRNA exit channel. These interactions would promote rearrangement into a stable conformation of initiator tRNA binding that is incompatible with scanning but competent for downstream steps in the pathway, most likely with the attendant release of eIF1 from its 40S-binding site (127).
This proposal was strengthened by the demonstration that eIF1 can bind to bacterial 30S subunits at the same binding site used by IF3 and can replace IF3 in destabilizing bacterial PICs reconstituted with non-AUG codons. Furthermore, IF3 can functionally replace eIF1 in the mammalian reconstituted system, destabilizing 48S PICs formed with AUGs in poor context or near-cognate triplets (128). These findings argue for a common mechanism of AUG selection in bacteria and eukaryotes involving a conformational change in the 40S/30S subunit, induced by eIF1/IF3 binding to the platform, that destabilizes codon-Ac pairing and imposes a requirement for a perfect Ac-AUG duplex.
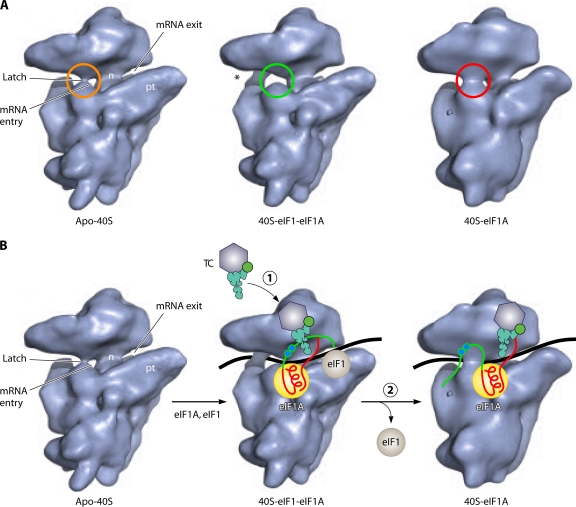
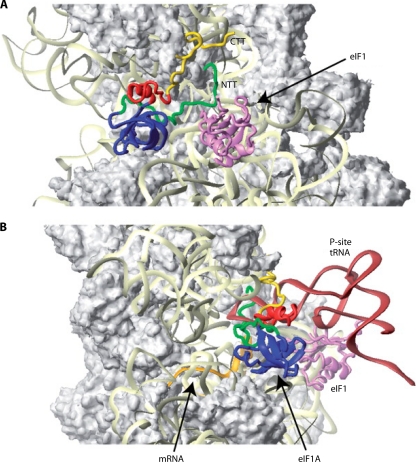
Structural analysis of reconstituted yeast 40S PICs subsequently provided the first direct evidence that eIF1 promotes a rearrangement of the 40S subunit that is conducive to scanning. Cryo-electron microscopy (EM) reconstructions of yeast apo-40S and 40S·eIF1A·eIF1 complexes revealed that eIF1A and eIF1 stabilize an open conformation of the “latch” of the mRNA entry channel. Moreover, the 40S·eIF1A complex, which would mimic the PIC following AUG recognition and eIF1 dissociation, displays a more closed conformation of the latch than that which occurs for apo-40S (Fig. 5A). It was suggested that this open conformation of the latch, stabilized by eIF1A and eIF1, is conducive to scanning, whereas the latch in the closed conformation would be “clamped down” on the mRNA to arrest scanning and enable downstream steps to proceed (159).
Fig. 5.
Cryo-EM model of the yeast eIF1·eIF1A·40S PIC. (A) Cryo-EM reconstructions of free 40S and the indicated complexes with eIF1 or eIF1A, which display moderately closed (apo-40S), open (40S-eIF1-eIF1A), or strongly closed (40S-eIF1A) conformations of the mRNA entry channel latch. (Reprinted from reference 159 with permission from Elsevier.) (B) The images in A have been annotated here with schematics of the mRNA, eIF1A, eIF1, and the TC to summarize the biochemical findings (159) that the rate of TC binding is stimulated by both eIF1 and eIF1A but that the TC is bound more tightly to the PIC in the absence of eIF1, which would prevail with the eIF1 dissociation upon AUG recognition.
Release of eIF1 upon AUG Recognition Evokes Tighter Binding of Met-tRNAiMet to the PIC
As noted above, kinetic and thermodynamic analyses of the basic yeast reconstituted system demonstrated that base pairing with AUG evokes rapid and highly stable TC binding that appears to reflect a dramatic acceleration of a first-order conformational transition in the PIC (99). Further analysis of such reconstituted yeast PICs provided evidence that eIF1 dissociation is a key trigger of this structural rearrangement. Although eIF1 and eIF1A collaborate to increase the rate of TC recruitment, the affinity of the TC for the PIC is actually lower in the presence of eIF1, owing to its greater stimulation of the dissociation rate than the association rate of the TC. Hence, the dissociation of eIF1, elicited by AUG-Ac pairing, should evoke tighter TC binding to the PIC (159). In this view, the TC binds to the 40S subunit in the open conformation (stabilized by eIF1A and eIF1) in a configuration that enables the transient inspection of successive triplets entering the P site during scanning, whereas AUG-Ac pairing and the attendant dissociation of eIF1 enables the TC to bind more tightly to the P site (Fig. 5B).
Consistent with this model, various eIF1 class I Sui− mutants, which dissociate more rapidly than WT eIF1 from the 40S subunit, confer increased rates and affinity of TC binding to the PIC in vitro. In contrast, two eIF1 mutants with basic Arg or Lys substitutions of residue G107 dissociate more slowly from the PIC and exhibit decreased rates and affinity of TC binding (32, 145). Presumably, the latter variants, here dubbed “class II” mutants, function effectively in the first step of TC binding to the open complex but then perform poorly in the second, isomerization step by impeding the transition to the closed complex. In contrast, class I eIF1 mutants accelerate the isomerization reaction by dissociating more rapidly from the 40S subunit. Presumably, the 40S binding defects of the class I mutants, which should slow down the initial binding of the TC to the open conformation, have been suppressed at the saturating concentrations of eIF1 used in these in vitro assays.
In vivo, both class I and class II eIF1 mutants constitutively derepress GCN4 translation, exhibiting the Gcd− phenotype that signifies a defect in TC binding to the 40S subunit (Fig. 3B). The Gcd− phenotype of class I mutants is diminished either by co-overexpressing eIF2 subunits and tRNAiMet (to increase the cellular concentration of the TC) or by overexpressing the mutant eIF1 proteins themselves (32, 145; P. Martin Marcos and A. G. Hinnebusch, unpublished data). These outcomes are expected for such mutations that weaken 40S binding by eIF1, because the reduced 40S occupancy of eIF1 which they engender in vivo (32) will diminish eIF1 function in stimulating TC loading onto the open conformation of the 40S subunit, slowing the first step of TC binding. This defect can be rescued by mass action with increased TC concentrations or by restoring the high-level 40S occupancy of eIF1 by its overexpression. In contrast, the Gcd− phenotype of the class II substitution G107R is not suppressed by the overexpression of the TC or mutant eIF1 (32, 145; Martin Marcos and Hinnebusch, unpublished), consistent with the notion that class II mutations impede eIF1 dissociation and thereby delay the second, isomerization step of TC binding.
As noted above, the Sui− phenotypes of class I eIF1 mutants can be rationalized by their higher-than-WT rates of dissociation from the PIC. Unexpectedly, the class II G107R and G107K (G107R/K) mutants, which dissociate more slowly from the PIC, also confer Sui− phenotypes in vivo. The solution to this paradox seems to be that the G107R/K mutants dissociate more slowly, and retard Pi release, only at AUG codons and, hence, reduce the rate of initiation at AUG without a commensurate reduction at UUG codons. The class I eIF1 mutations, in contrast, increase the UUG-to-AUG initiation ratio primarily by elevating UUG initiation levels. These predictions were supported by the differential effects of class I and class II mutations on the translational efficiencies of luciferase mRNAs with AUG or UUG start codons in mutant extracts. This model can also explain why the Sui− phenotypes of class II eIF1 mutants are not suppressed by their overexpression, which should not correct the inability to dissociate rapidly at AUG (145). The fact that G107R/K mutants discriminate specifically against AUG suggests that eIF1 participates in evaluating the codon-Ac match in the P site and that these substitutions disrupt the recognition or response to a perfect AUG-Ac duplex.
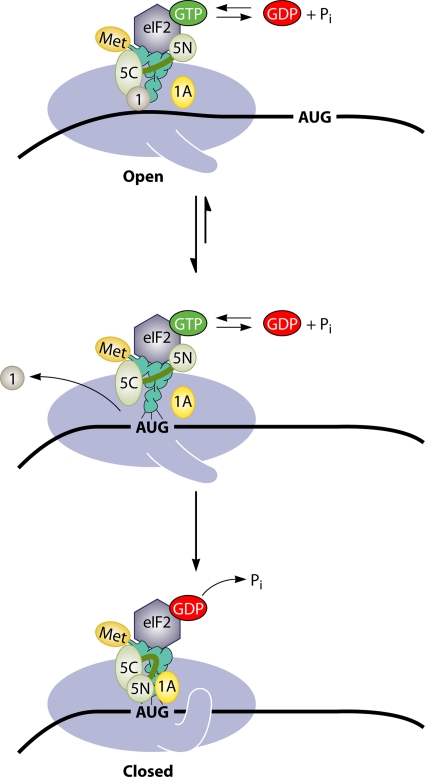
In summary, the analysis of eIF1 suggests that it has multiple functions in the assembly of the PIC, scanning, and AUG recognition. It acts with eIF1A to stabilize an open conformation of the 40S subunit that is conducive to TC recruitment and scanning but incompatible with tight initiator tRNA binding to the P site. eIF1 also restrains the GAP function of eIF5 and impedes Pi release from eIF2-GDP·Pi in the scanning complex. The dissociation of eIF1 upon AUG recognition stabilizes the closed conformation of the 40S subunit, evoking a cessation of scanning, the completion of GTP hydrolysis with Pi release, and a more stable binding of initiator tRNA to the P site (Fig. 6).
Fig. 6.
Schematic model depicting the structural rearrangements of the PIC thought to occur in the transition from the open conformation to the closed conformation upon AUG recognition. (Top) The presence of eIF1 and eIF1A on the 40S subunit stabilizes the open, scanning-conducive conformation of the PIC with an open conformation of the mRNA entry channel latch. TC binding is relatively unstable but in a configuration that allows the base-by-base inspection of the 5′UTR by Met-tRNAiMet in the P site. The GTP in the TC of a fraction of scanning complexes is hydrolyzed to eIF2·GDP·Pi, catalyzed by the N-terminal (GAP) domain of eIF5 (5N), but Pi release is blocked by eIF1. eIF1 maintains its association with the eIF5 CTD (5C), an interaction involved in stabilizing the yeast MFC. (Middle) AUG-Ac pairing evokes eIF1 dissociation from the 40S platform, and this enables isomerization to the closed conformation (Bottom), with the mRNA entry channel latch clamped on the mRNA. The absence of eIF1 in this state also enables tighter binding of TC to the P site and the completion of GTP hydrolysis and dissocation of Pi from eIF2·GDP·Pi. It was proposed that the absence of eIF1 allows the eIF5 NTD to switch partners from the G domain of eIF2γ to a 40S location that might overlap with the eIF1-binding site, enabling the eIF5 NTD interaction with eIF1A in a manner that stabilizes the closed conformation. eIF3 and the eIF4 group of factors were omitted from this diagram for simplicity. (Reproduced from reference 145 with permission from Elsevier.)
Structural Determinants of eIF1 Interactions and Roles in Start Codon Recognition
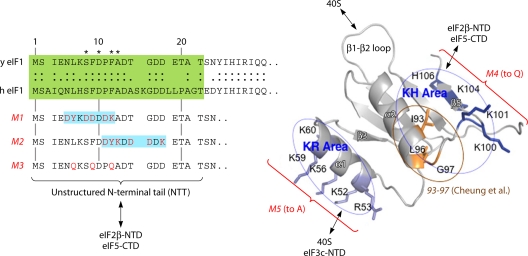
eIF1 interacts directly with both the 40S subunit and three other eIFs in the MFC, and these interactions have been mapped to patches of residues exposed on different surfaces of the protein. eIF1's functions in start codon recognition could involve contributions from some or all of these interactions. The crystal structure of the Tetrahymena 40S·eIF1 complex (178) confirmed the previous prediction that basic residues in helix α1 and the β1-β2 loop of eIF1 interact with residues in helix 44 (h44) and h24 of 18S rRNA in the platform region of the 40S subunit (127). It would be expected that the elimination of these residues would impair eIF1 binding to the 40S subunit and confer a Sui− phenotype in yeast cells. In fact, the Ala substitution of all 5 basic residues of α1 (the −M5 mutation) (Fig. 7) is lethal and impairs 40S binding by eIF1 in vivo but does not confer a dominant Sui− phenotype. The latter finding might be explained by the fact that −M5 impairs eIF1 binding to the c/NIP1 subunit of eIF3 and blocks eIF1 incorporation into the MFC, rendering it unable to compete with WT eIF1 at the stage of PIC assembly (180).
Fig. 7.
Structure and functional domains of yeast eIF1. (Left) The amino acid sequences of the unstructured NTTs of yeast (y) and human (h) eIF1s are aligned and highlighted in green. The substitutions in −M1, −M2, and −M3 mutants of yeast eIF1 are shown below in red, which impair the interaction of eIF1 with the eIF2β NTD and eIF5 CTD in vitro. (Right) Ribbon model of yeast eIF1 indicating the positions of the −M4, −M5, and 93-97 (32) as well as the location of the “KH” and “KR” areas. The effects of the −M5 mutation on the interaction of eIF1 with the eIF3c NTD in vitro or native 40S subunits in vivo and of the −M4 mutation on eIF1 binding to the eIF2β NTD and the eIF5 CTD in vitro (180) are summarized. The interaction of α1 and the β1-β2 loop with the 40S subunit is predicted from structural studies (127, 178). (Reproduced from reference 180 with permission of the publisher.)
The Ala substitution of residues 93 to 97, which alters hydrophobic residues in the C terminus of α2 (Fig. 7), reduces the 40S binding of eIF1 and confers a strong Sui− phenotype (32), but because α2 lies on a solvent-exposed surface in the 40S·eIF1 complex, it likely impairs 40S binding indirectly. The adjacent basic residues in the “KH” (basic-hydrophobic) surface might functionally cooperate with the hydrophobic residues in α2, as their replacement with Gln residues (mutation −M4) (Fig. 7) also reduces 40S binding and, while lethal, confers a dominant Sui− phenotype. As these basic residues mediate eIF1 binding to the N-terminal domain (NTD) of eIF2β (180), perhaps eIF1 binding to eIF2β helps to anchor the TC in a scanning-conducive conformation, and the disruption of the interaction enables a shift to the closed conformation at UUG codons. The KH region basic residues also mediate eIF1 binding to the eIF5 C-terminal domain (CTD), and it was proposed that this interaction is instrumental in coupling eIF1 dissociation from the PIC to AUG recognition (180). However, as 93-97 was shown to accelerate eIF1 dissociation from basic yeast reconstituted 48S PICs lacking eIF5 and eIF3 (32), neither of eIF1's interactions with these other MFC constituents can account completely for the role of helix α2 in triggering eIF1 release upon AUG recognition. Interestingly, the class II eIF1 substitutions G107R/K introduce an additional basic residue into the KH region and (as noted above) impede eIF1 dissociation specifically at AUG codons. It was suggested that by strengthening the association of the eIF1 KH region with the eIF2β NTD or the 40S subunit, the G107K/R substitutions lead to an inappropriate retention of eIF1 in the 48S PIC (145), but this by itself would not account for the AUG specificity of the defect.
The unstructured N-terminal tail (NTT) of eIF1 is dispensable for yeast viability, but its removal or alteration reduces eIF1 binding to the eIF2β NTD and eIF5 CTD in vitro and confers a Gcd− phenotype in vivo. The Gcd− defect of an Ala substitution of residues 9 to 12 is suppressed by the overexpression of the TC, suggesting a reduced rate of TC recruitment, which was confirmed by experiments with the basic yeast reconstituted system (32). The Gcd− phenotype of other NTT mutations (−M1, −M2, and −M3) (Fig. 7) was not suppressed, however, suggesting a defect in AUG recognition at uORFs 2 to 4 by scanning PICs that have already recruited the TC (180). As the latter mutations do not produce a Sui− phenotype, they apparently confer leaky scanning equally at AUG or UUG codons.
Rabl et al. (178) predicted that eIF1 residues in the loop between β1 and β2 are close enough to the mRNA-binding channel to monitor the quality of the codon-Ac duplex. How the recognition of a perfect match signals the dissociation of eIF1 from the 40S platform is an interesting question. Those authors further predicted a clash of eIF1 with the acceptor and D stems of initiator tRNA, which could provide a physical basis for the fact that eIF1 release upon AUG recognition stabilizes initiator tRNA binding to the P site (159). This would also fit with other results, described above and below, indicating that initiator tRNA does not bind deeply in the P site in the open conformation of the PIC.
eIF1 Autoregulates Translation by Discriminating against Poor AUG Context
Consistent with its role in discriminating against non-AUG start codons, bacterial IF3 negatively autoregulates its translation by blocking the utilization of the near-cognate AUU triplet that serves as its start codon (25, 26). It was noticed that all genes encoding eIF1 in eukaryotes contain an AUG in a poor context (140), and Ivanov et al. demonstrated that the overexpression of eIF1 in mammalian cells negatively regulates eIF1 expression in a manner dependent on the poor context of its start codon while having little effect on a mutant eIF1 gene altered to contain the optimum “Kozak” consensus sequence. This provides in vivo evidence that eIF1 destabilizes the PIC at AUG codons in a suboptimal context and that this activity is exploited to maintain eIF1 protein levels within strict limits (91). Presumably, the tight regulation of eIF1 is important because its overexpression would reduce initiation for the small fraction of genes with near-cognate start codons and also the much larger proportion with AUG start codons in a poor context.
EVIDENCE THAT eIF5 PROMOTES eIF1 DISSOCIATION AT START CODONS
The CTD of eIF5 interacts with eIF1, the eIF3c/Nip1 NTD, and the eIF2β NTD to stabilize the yeast MFC, whereas the eIF5 NTD harbors the GAP function (7). The substitution of Arg-15 in eIF5 destroys GAP function in basic reconstituted yeast PICs, and the ability to rescue the growth of a tif5Δ yeast mutant, but does not diminish the eIF5 interaction with recombinant eIF2β or eIF2 holoprotein. Hence, Arg-15 appears to impair the GAP catalytic function rather than substrate binding (5, 44, 160). The eIF2-GDP/eIF5 complex is stabilized by aluminum fluoride (AlF4−), a compound which, combined with GDP, acts as a structural mimic of the transition state of the GTPase reaction (184), and it was suggested that Arg-15 functions as an “Arg finger” that is inserted into the GTP-binding pocket of eIF2γ to stabilize the transition state for GTP hydrolysis (160).
There is both biochemical and genetic evidence that the release of eIF1 upon start codon recognition is stimulated by eIF5. The addition of eIF5 to basic reconstituted yeast PICs increases the normally low rate of eIF1 dissociation from the PIC at the near-cognate triplet AUU but has little effect at AUG, where eIF1 dissociates more rapidly. The extra eIF5 also suppressed the effect of eIF1-G107K, which retards the eIF1 dissociation at AUG, by accelerating the dissociation of this mutant eIF1 at both AUG and AUU codons. The addition of eIF5 increased the rate and extent of TC binding in reactions with eIF1-G107K but not with WT eIF1. These findings fit with the notion that eIF5 enhances the second, isomerization step of TC binding by promoting the eviction of eIF1 from the PIC (145), identifying a role for eIF5 in AUG recognition beyond its GAP function. Consistent with the idea that eIF5 promotes the eIF1 dissociation, the overexpression of eIF5 exacerbates the Sui− phenotypes of eIF1 mutants (145, 212). The latter finding seems to imply that eIF5 has a dynamic interaction with the scanning PIC and that higher concentrations of the factor increase the proportion of scanning PICs in an eIF5-bound state that is more likely to release eIF1 without a perfect AUG-Ac match with initiator tRNA.
Interestingly, there is structural similarity between eIF1 and the eIF5 NTD (41, 71), raising the possibility that eIF5 and eIF1 might compete for binding to the 40S platform. Because eIF5-dependent GTP hydrolysis in the TC occurs in the scanning complex, the GAP domain of eIF5 undoubtedly interacts with the GTP-binding pocket of eIF2γ in the open complex. Nanda et al. speculated that the eIF5 GAP domain might have to dissociate from eIF2γ to allow Pi release from GDP·Pi and that the eIF1 dissociation upon AUG recognition would facilitate this rearrangement by allowing the eIF5 NTD to bind in place of eIF1 on the 40S platform in a manner stabilized by eIF5's interaction with eIF1A (Fig. 6). This would account for the finding that eIF5 can stimulate the eIF1 dissociation and overcome eIF1's destabilizing effect on TC binding (145). An attractive feature of this model is that it provides a mechanism for the coupling of Pi release to eIF1 dissociation, as there is currently no evidence that eIF1 can interact with the eIF2γ G domain and block Pi release directly. It also provides a mechanism for the demonstrated role of an eIF1A-eIF5 interaction in stabilizing the closed 40S conformation (131), by preventing the rebinding of eIF1 to the platform. (More about this eIF1A-eIF5 interaction is presented below.) However, there is no evidence as yet that the eIF5 NTD binds to the 40S subunit, and so eIF5 might promote the eIF1 dissociation by a less direct mechanism.
eIF1A PLAYS A DIRECT ROLE IN SCANNING AND AUG RECOGNITION FROM THE A SITE OF THE 40S SUBUNIT
Unstructured Tails of eIF1A Differentially Modulate Distinct Conformations of the 40S Subunit and Modes of Initiator tRNA Binding
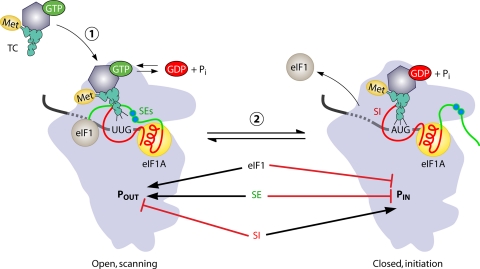
Although eIF1A cooperates with eIF1 to promote scanning in mammalian reconstituted PICs (165) and to stabilize an open conformation of the mRNA entry channel of the yeast 40S subunit (159), no Sui− mutations affecting eIF1A were isolated by Donahue et al. However, the directed mutagenesis of the yeast eIF1A gene (TIF11) revealed that residues in its unstructured C-terminal tail (CTT) are critically required for stringent AUG selection. A Sui− phenotype was observed upon the deletion of the entire CTT plus a portion of the C-terminal helical domain by the ΔC mutation, and this truncation (of residues 108 to 153) also impaired the ability of yeast eIF1A to replace human eIF1A to promote scanning and block initiation at a near-cognate triplet in vitro (69). The scanning-promoting function was localized to two 10-amino-acid (aa) direct repeats in the CTT, dubbed scanning enhancer 1 (SE1) and SE2. The SEs contain pairs of Phe residues as critical constituents (F121 and F123, and F131 and F133) and were shown to function redundantly to block UUG initiation in vivo. While the elimination of all four Phe residues is lethal, a mutant that retains only F131 grows poorly and displays a very high UUG-to-AUG initiation ratio, close to unity (70, 183). It was proposed that the SE elements stabilize the open, scanning conformation of the PIC (Fig. 8) so that their inactivation permits an inappropriate rearrangement to the closed conformation at UUGs.
Fig. 8.
Model depicting the opposite functions of the unstructured tails of eIF1A in modulating the conformational rearrangement of the PIC and distinct modes of TC binding to the open and closed states. The SE elements in the eIF1A CTT (shown in green) stabilize the open conformation of the PIC and the “Pout” mode of TC binding, which is compatible with scanning at non-AUG codons. The SEs also block the full P-site accommodation of initiator tRNA in the “Pin” mode required for start codon selection. eIF1 performs the same functions as the SE elements. The SI elements in the NTT and helical domain of eIF1A (both shown in red) function oppositely and destabilize the open conformation and the Pout mode of TC binding, thus promoting the closed conformation and Pin mode of initiator tRNA binding at AUG codons. Both the dissociation of eIF1 from the 40S subunit and the eviction of the SEs from the P site stabilize the closed conformation and tighter TC binding afforded by the Pin state.
Remarkably, mutations affecting the unstructured NTT of eIF1A have the opposite effect and suppress the elevated levels of UUG initiation conferred by various Sui− mutations, the Ssu− (suppressor of sui) phenotype (Fig. 4), including Sui− SE substitutions of eIF1A itself, the SUI3-2 mutation of eIF2β, and the SUI5 mutation of eIF5 (70, 183). NTT substitutions also increase the leaky scanning of uAUG1 in GCN4 mRNA (Fig. 3C, III) (70), leading to the proposal that they disrupt a scanning inhibitor (SI) function that is required to arrest scanning at AUG or (in Sui− mutants) UUG codons alike. As might be expected, the lethality associated with the inactivation of both SE elements is also suppressed by an Ssu− substitution in the eIF1A SI element (Ala substitution of NTT residues 17 to 21). Thus, the SI element appears to stabilize the closed conformation of the PIC (Fig. 8) such that its inactivation increases the probability of rearrangement to the eIF1-stabilized open conformation and thereby suppresses initiation at UUG codons. Interestingly, eIF1A substitutions that weaken the packing of structured portions of the NTT and CTT against helix α2 have the same genetic properties as substitutions in the SI element in the unstructured portion of the NTT, suggesting that the SI function also depends on the integrity of the helical domain of eIF1A (183).
Several lines of biochemical evidence support the conclusion that SE and SI mutations in eIF1A stabilize the closed and open conformations of the PIC, respectively. First, as noted above, ΔC removes both SEs and impairs scanning in mammalian fully reconstituted PICs (69). Second, SE and SI substitutions have opposite effects on 40S-60S subunit joining in vitro. eIF1 and eIF1A synergistically impede subunit joining, consistent with the idea that the open 40S conformation, promoted by these factors, is incapable of subunit joining (3). The inactivation of both SEs increases the rate of subunit joining, and this effect is suppressed by SI mutations in the NTT (17-21) or the C strand of the helical domain, as expected if SE and SI elements stabilize the open and closed conformations, respectively (183).
A third line of evidence supporting this model is that SE and SI mutations have opposite effects on the affinity of eIF1A for the PIC and on the rotational freedom of the eIF1A C terminus, as determined by measuring the anisotropy of eIF1A C-terminally tagged with fluorescein in basic yeast reconstituted PICs harboring eIF5. The rotational freedom and rate of eIF1A dissociation are both lower in WT PICs reconstituted with AUG than in PICs reconstituted with non-AUG codons, depending on the presence of eIF5, and these differential effects are diminished, or even reversed, by the Sui− mutations SUI5 in eIF5 and ΔC in eIF1A. This finding suggested that eIF1A and eIF5 interact functionally upon AUG recognition in a way that reduces the flexibility of the eIF1A CTT and strengthens eIF1A binding to the PIC. The latter is manifested as an increase in amplitude, or a decrease in the rate constant, of the slower phase of the biphasic eIF1A dissociation kinetics, and the two kinetic phases were postulated to represent the partitioning of PICs between the open and closed conformations (131). In contrast to the Sui− SUI5 and ΔC mutations, which appear to favor the closed conformation, the Ssu− substitutions at residues 17 to 21 and 7 to 11 in the NTT weaken eIF1A binding and increase the mobility of the CTT in both AUG and UUG complexes. Thus, these SI mutations appear to stabilize the open conformation of the PIC regardless of the P-site codon (70). These findings imply that eIF5 functionally interacts with eIF1A to stabilize the closed conformation of the PIC, identifying another non-GAP function for eIF5 in start codon recognition.
A fourth line of evidence that SI mutations stabilize the open conformation is that they decrease the rate of eIF1 dissociation from basic reconstituted PICs. Employing the “loss-of-FRET” assay described above to measure the rapid conformational change that increases separation between eIF1A and eIF1 and the subsequent, slower dissociation of eIF1 (132), it was found that the Ssu− substitution at residues 17 to 21 reduces both phases of the reaction for AUG or UUG complexes, favoring the eIF1-bound, open conformation of the PIC, which is opposite of the effect produced by the Sui− substitution at residues 93 to 97 of eIF1 (32).
A final line of evidence supporting the model is that SE and SI mutations have opposite effects on the rate of TC binding to PICs. Substitutions of the SEs reduce the rate of TC loading, and this defect is diminished by SI mutations in the NTT or C strand, which on their own confer higher-than-WT rates of TC binding in vitro. The same relationships seem to occur in vivo, as the SE mutations derepress GCN4 translation in a manner suppressed by the overexpression of TC components, and this Gcd− phenotype is diminished by SI mutations (183). As the TC binds to the open conformation stabilized by eIFs 1 and 1A (159) (Fig. 5B), it was proposed that SE mutations reduce the rate of the initial, second-order step of TC loading by destabilizing the open conformation of the PIC, whereas SI mutations accelerate TC binding by stabilizing the open conformation (Fig. 8). This model can also explain the ability of SI mutations to suppress the Sui− defects of SE mutations, by stabilizing the open conformation and blocking inappropriate rearrangements to the closed conformation at UUG codons.
Interestingly, in contrast to SI mutations, the overexpression of WT eIF1 suppresses the Sui− but not the Gcd− phenotype of SE mutations, which suggests that the SE elements promote TC binding by a second mechanism besides the stabilization of the open 40S conformation, as the latter should be accomplished effectively by eIF1 overexpression. It was proposed that the direct binding of the SEs to tRNAiMet might specifically stabilize the scanning mode of TC binding that prevails only in the open complex, dubbed the “Pout” configuration, in which initiator tRNA is not fully engaged with the P site. This function of the SEs would be antagonized by the SI elements to enable a rearrangement to the more stable mode of TC binding, with initiator tRNA inserted deeply into the P site, dubbed the “Pin” state, when AUG enters the P site (Fig. 8) (183).
The eIF1A Tails Interact Directly with the P Site
The notion that the eIF1A CTT (where the SE elements reside) acts to stabilize the open conformation and impede initiator tRNA binding deep in the P site was proposed independently by Pestova and colleagues from results of directed hydroxyl radical mapping of eIF1A in reconstituted mammalian 43S complexes. The globular OB domain of eIF1A (18) was mapped to the A site, analogous to the location of its bacterial homolog IF1. Interestingly, the results predicted that the CTT extends out into the P site, threading under the Met-tRNAiMet in a manner that would obstruct the binding of the initiator tRNA anticodon-stem-loop (ASL) in the canonical location in bacterial 70S elongation complexes (Fig. 9), analogous to the Pin state envisioned by Saini et al. (183). Accordingly, the AUG-Ac interaction in the closed complex would likely require the ejection of the eIF1A CTT from the P site (228). In fact, the ejection of the CTT was proposed previously (70), based on its physical displacement from eIF1 in the PIC (132) and its functional interaction with eIF5 (131) upon AUG recognition. Thus, the SEs in the eIF1A CTT might sterically block the “Pin” mode of TC binding in the closed complex in addition to stabilizing the “Pout” state of the open complex. This idea is attractive because SE mutations would then facilitate the open-to-closed transition at UUGs (conferring Sui− phenotypes) in two ways: by destabilizing Pout (183) and also by removing a steric impediment to Pin (228) (Fig. 8).
Fig. 9.
Model of the 48S PIC containing bound eIF1, eIF1A, mRNA, and initiator tRNA. The model is based on directed hydroxyl radical mapping of mammalian eIF1 (127) and eIF1A (228) on 40S subunits and the crystal structure of bacterial 70S elongation complexes containing mRNA and P-site tRNA. The orientation of the tRNA and its location deep in the P site shown here might differ significantly from the eukaryotic scanning 43S or 48S PICs. The image in B is rotated 90° clockwise about the vertical axis from that in A, and the ribosomal protein S13/RpS18e in the head is not shown because it blocks the view of the eIF1A CTT. Ribbon representations of eIF1 are shown in lilac, and the eIF1A domains are shown in blue (OB fold), red (helical domain), green (NTT), and yellow (CTT). mRNA (gold) and tRNA (copper) are shown in the bottom panel only. (Reproduced from reference 228 by permission of Oxford University Press.)
Interestingly, the docking model derived by Pestova et al. for mammalian eIF1A binding to the 40S subunit (228) shows that the helical domain is oriented toward the head and P site and in contact with h31, creating a bridge over the mRNA-binding channel in the A site (Fig. 9B). This is consistent with the closed conformation of the mRNA entry channel latch seen for the cryo-EM model of the yeast 40S·eIF1A complex (Fig. 5A) and with the proposed (SI) function of stabilizing the closed complex ascribed to the helical domain of yeast eIF1A (183). The NTT is predicted to contact the Ac loop of initiator tRNA and mRNA (228), which means that its SI function in stabilizing the Pin configuration of initiator tRNA binding might be exerted directly.
In summary, the analysis of eIF1A suggests that it has multiple functions in the assembly of the PIC, scanning and AUG recognition, executed by its N- and C-terminal tails and helical domain. The SE elements in the CTT act with eIF1 to stabilize an open conformation of the 40S subunit conducive to scanning and a mode of TC binding that blocks the full accommodation of initiator tRNA in the P site. The SI elements in the NTT and helical domain stabilize a closed conformation of the 40S subunit incompatible with scanning, and they promote eIF1 dissociation, eIF1A's interaction with eIF5, and the ejection of the SEs to permit the full accommodation of initiator tRNA in the P site. It can be envisioned that these two states exist in equilibrium in the scanning PIC, enabling the transient sampling of successive codons as they enter the P site, and that the closed conformation is locked in by a perfect AUG-Ac duplex in a favorable sequence context.
The NTT, α2 helical domain, and CTT of eIF1A (18) are all lacking in its bacterial counterpart IF1 (27). The archaeal ortholog (aIF1A) lacks only the C strand and unstructured CTT, and archaea resemble bacteria in using the Shine-Dalgarno (SD) sequence upstream of the start codon to recruit the 30S subunit (49, 129). Thus, SD-facilitated AUG selection appears to be correlated with the absence of SE elements, consistent with their stimulatory role in scanning. Considering that archaea resemble eukaryotes in recruiting initiator tRNA in a TC with aIF2-GTP (163), the NTT and helical domains in aIF1A might play a role in promoting TC binding in the Pin state. Recent in vitro findings suggest that bacterial IF1 acts to stabilize a 30S conformation that is incompatible with subunit joining, which can be overcome by a favorable SD sequence (138). Thus, IF1 carries out a function ascribed to the SEs, of stabilizing a small-subunit conformation incompatible with subunit joining. Accordingly, a region of the OB fold, the main structural element shared between eIF1A and IF1, could augment this aspect of SE function in eukaryotes (183).
INITIATOR tRNA AND 18S rRNA RESIDUES INVOLVED IN AUG RECOGNITION
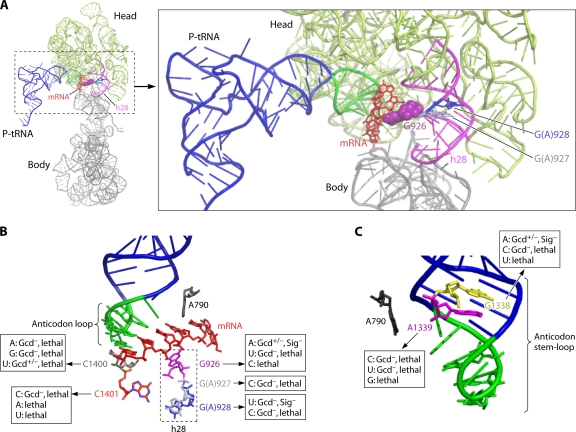
In addition to its Ac, initiator tRNA contains other highly conserved residues not present in elongator tRNAs that could participate in stringent AUG selection (133), notably the three consecutive G·C base pairs in the ASL, G29·C41, G30·C40, and G31·C39. In bacteria, there is evidence that these base pairs, as well as the Ac loop bases, are required for the IF3-mediated stabilization of PICs formed with Met-tRNAiMet versus elongator tRNAs in the P site (79). Interestingly, residues G1338 and A1339 in 16S rRNA are poised to make direct, “A-minor” interactions with the minor grooves of the first and second G·C base pairs of the initiator tRNA ASL (102, 188), and it was proposed that these interactions would be optimal for consecutive G·C base pairs and would stabilize Met-tRNAiMet binding following an IF3-induced conformational change in the rRNA (43). Supporting this hypothesis, all substitutions in G1338 and A1339, except G1338A (which would preserve the A-minor interaction by this base) destabilized Met-tRNAiMet binding to the bacterial 30S subunit in vitro. However, because other tRNAs with G·C base pairs at the first and second positions of the ASL also are relatively more resistant to IF3-mediated ejection from the 30S subunit, it was speculated that an IF3-induced shift of A-minor interactions from G29·C41/G30·C40 to G30·C40/G31·C39 could occur, which would be possible only for initiator tRNA with its three consecutive G·C base pairs (120).
Presumably, the rejection of noninitiator tRNAs is not relevant in eukaryotes, because eIF2 specifically transfers Met-tRNAiMet to the PIC. However, A-minor interactions of G1338/A1339 with the ASL G·C base pairs could still serve to stabilize initiator tRNA binding in the P site. Consistent with this idea, the change of the first and third G·C base pairs in the yeast initiator tRNA ASL to their identities in elongator tRNAeMet, i.e., A29·U41 and U31·U39, eliminates the strong stabilizing effect of AUG on TC binding to basic reconstituted yeast PICs (95). In addition, in the fully reconstituted mammalian system, the alteration of the identities of two or all three of the G·C base pairs led to the formation of 48S PICs that could be destabilized by eIF1 (or bacterial IF3) following eIF5-stimulated GTP hydrolysis in the TC (128). Presumably, eIF1 could reject the mutant tRNAs only after their attachment to eIF2 was severed by GTP hydrolysis. Surprisingly, the A29·U41 and U31·U39 replacements in the ASL do not have a strong effect on yeast cell growth (216), although they were crucial for the ability of elongator tRNAeMet to function during initiation in yeast mutants lacking initiator tRNA (16), and they reduced translation in mammalian cell extracts when introduced into human initiator tRNA (59). Thus, it seems that the G·C identities of the three conserved ASL base pairs enhance initiation but are not essential for the stability of eukaryotic 48S PICs.
Supporting the possibility that the 18S residues corresponding to G1338 and A1339 make A-minor interactions with the ASL G·C base pairs of eukaryotic initiator tRNA, the mutagenesis of episomal ribosomal DNA (rDNA) in a yeast strain lacking the chromosomal rDNA array showed that nearly all substitutions of these residues (G1575 and A1576) are lethal and produce dominant Gcd− phenotypes and leaky scanning of GCN4 uAUG1 in cells coexpressing rRNA from a WT rDNA episome, consistent with defective 48S PIC assembly or AUG recognition in vivo. In contrast, the equivalent of the G1338A substitution, which should preserve A-minor interactions, is viable and confers only a weak Gcd− phenotype (56). Thus, A-minor interactions with the ASL are likely important for initiation in yeast even though there is flexibility in the identities of the base pairs that serve as receptors of the bases G1575 and A1576. This flexibility is consistent with thermodynamic data indicating that the stabilities of A-minor interactions vary relatively little among the four Watson-Crick (W-C) base pairs as receptors (19, 51).
Other conserved residues in eukaryotic initiator tRNA include A54 and A60 in the T loop, and substitutions with the cognate elongator residues (A54U and A60C) were found to suppress the deleterious effect of the U31·U39 replacement of the third ASL G·C base pair upon TC binding in vitro (95). It was suggested that the A54 and A60 replacements could lower the energy barrier to a structural rearrangement of initiator tRNA necessary for its full accommodation in the P site and thereby compensate for the destabilizing effect of the U31·U39 replacement on the 48S PIC (95).
An N6-threonylcarbamoyl modification of A37 (t6A37) of initiator tRNA, immediately adjacent to the Ac triplet, is thought to stabilize the first base pair of the codon-Ac duplex for ANN or UNN codons (4), which includes the decoding of AUG by initiator tRNA. Consistent with this, the inactivation or elimination of yeast proteins that catalyze t6A37 formation, including Sua5 (65, 125) and subunits of the EKC/KEOPS complex, Kae1 and Pcc1 (46, 200), evokes phenotypes indicating an impaired recognition of AUG start codons. These include a general decrease in the initiation rate, a derepression of GCN4 mRNA translation independent of eIF2α phosphorylation (Gcd− phenotype), increased leaky scanning of GCN4 uAUG1, and (for kae1 and pcc1 mutants) synthetic lethality with a mutation in the eIF5 GAP domain (K55E) that also impairs the initiation rate and increases leaky scanning. The fact that the overexpression of the TC did not suppress the leaky-scanning phenotype of the Sua5 depletion is consistent with a failure of initiator tRNA, assembled in the scanning PIC, to stably base pair with GCN4 uAUG1 (125). This mechanism can account for the original identification of sua5 mutations as suppressors of the inhibitory effect of an upstream uORF introduced into CYC1 mRNA (143, 171). Interestingly, kae1 mutations also increase the ratio of GUG to AUG initiation from matched luciferase reporters (64), which can be rationalized by the proposal that the absence of t6A37 does not further impair the recognition of near-cognate triplets with mismatches in the first base pair and thus reduces initiation from AUG but not GUG (nor, presumably, UUG or CUG) codons. This would be akin to the class II Sui− mutations in eIF1 that increase the UUG-to-AUG ratio by selectively decreasing AUG recognition (145).
A random selection for Gcd− mutants of 18S rRNA identified substitutions in helix 28 (h28) that impair TC binding to 40S subunits. Various substitutions that perturb the location or identity of the “bulge” residue in h28 appear to reduce TC binding during reinitiation on GCN4 mRNA and enable the leaky scanning of the GCN4 uORFs during primary initiation events. One such nonlethal mutation, A1152U (corresponding to A928U in 16S rRNA) reduced the rate and stability of TC binding to the mutant 40S subunits in vitro (56). Interestingly, bulge G in h28 contacts the +1 position of the P-site codon (residue A of AUG) in crystal structures of bacterial 70S elongation complexes (102, 188). Directed substitutions in 18S residues corresponding to C1400 and G1401 in 16S rRNA, which also contact the AUG in 70S complexes, confer dominant Gcd− and recessive-lethal phenotypes (Fig. 10). Thus, various P-site residues whose counterparts contact tRNA or mRNA in bacterial elongation complexes likely play an important role in eukaryotic PICs to stabilize initiator tRNA binding to the AUG (56).
Fig. 10.
Substitution of 18S rRNA residues predicted to contact the P-site tRNA or P-site codon in elongating 80S ribosomes confers a phenotype indicating impaired TC binding to the scanning 43S PIC. Shown is the disposition of the bulge G926 of h28 (lilac in panels A and B) and various other 16S rRNA residues that contact the P-site codon or tRNA in a crystal structure of a bacterial 70S complex with mRNA and P-site tRNA. Substitutions of the corresponding residues in yeast 18S rRNA are lethal or confer an Slg− phenotype and in most cases also evoke a dominant Gcd− phenotype (detected in cells also expressing wild-type 18S rRNA), indicating less stable TC binding to the scanning PIC. (Modified from reference 56 with permission of Cold Spring Harbor Laboratory Press.)
A substitution in 18S rRNA (A1193U), corresponding to a residue in the h31 loop of 16S rRNA (A968), located directly below the codon-Ac duplex in bacterial 70S complexes, was shown to increase the leaky scanning of GCN4 uAUG1 and suppress the His+ phenotype of SUI3-2, possibly indicating an Ssu− phenotype. This substitution and another with similar phenotypes in a residue (G875) corresponding to 16S rRNA residue A663 in h22 reduce the binding of multiple eIFs to native 40S subunits and impair the recruitment of Met-tRNAiMet to 40S subunits in cell extracts. It was proposed that both mutations destabilize Met-tRNAiMet binding to AUG in the closed complex either directly (A1193U) or indirectly by reducing the 40S association of eIFs that promote TC binding (G875A) (148).
STRUCTURAL DETERMINANTS OF STRINGENT AUG SELECTION IN SUBUNITS OF eIF2
Sui− Substitutions in eIF2γ Increase UUG Initiation by Perturbing Initiator tRNA Binding to eIF2
As noted above, eIF2 plays a central role in AUG recognition because it anchors Met-tRNAiMet to the 40S subunit, and the hydrolysis of GTP releases Met-tRNAiMet from eIF2-GDP into the P site. As the GTPase activity of eIF2 is very low in the absence of ribosomes, even with eIF5 present (5), AUG recognition is likely dependent on structural features in eIF2 that couple its GTPase activity to interactions with the 40S subunit or eIFs besides eIF5. eIF2 might also contribute to the different modes of Met-tRNAiMet binding to the P site that are thought to characterize the open and closed conformations of the PIC (Fig. 8). Current models for the structure of the TC are based on crystal structures of archaeal orthologs of the eIF2 subunits. This is reasonable because there is considerable sequence conservation between the archaeal and eukaryotic orthologs throughout their lengths, except that eIF2β harbors a sizeable N-terminal extension missing in archaea. Consistently, this segment of eIF2β mediates interactions of eIF2 with eIF5 and the catalytic subunit of eIF2B, both of which lack recognizable archaeal counterparts (12, 45, 121).
Confirming predictions made from the sequence homology of eIF2γ to elongation factor EF-Tu (74, 76), crystal structures of aIF2γ revealed three domains highly similar to the G domain and domains II and III of EF-Tu, including the guanine nucleotide-binding pocket in the G domain (185). In the bacterial EF-Tu/GDPNP/Phe-tRNAPhe complex, the switch 1 (sw1) and switch 2 (sw2) regions contact the γ-phosphate, and in the (hydrolyzed) GDP-bound structure, they adopt a “switch-off” conformation, with domain II disengaged from the G domain. All known structures of apo-, GDP- or GDPNP-bound aIF2γ, in contrast, display a close packing of domain II with the G domain, and the switch regions do not exhibit marked conformational differences (186, 203). Nevertheless, in the structure of an aIF2α/γ-GDPNP heterodimer, the switch regions are apparently closer to the positions that they occupy in the EF-Tu/GDPNP/Phe-tRNAPhe complex than in previous aIF2γ structures, prompting Schmitt et al. to propose a model of Met-tRNAiMet docking to aIF2γ (Fig. 11A). In this model, the “movement” of sw1 evoked by GTP opens a channel between the G domain and domain II that accommodates the methionine and terminal A76 base at the acceptor end of the tRNA, with a conserved Tyr in sw1 (Y51 in Sulfolobus solfataricus) stacking against the methionine (223). This subtle movement of sw1 would presumably enable the positive interactions with methionine that distinguish the GTP- from the GDP-bound states of aIF2/eIF2, deduced from biochemical analyses of eIF2 (96).
Fig. 11.
Interfaces between subunits of archaeal aIF2 and docking tRNA on aIF2γ. (A) Model of Phe-tRNAPhe (in a stick figure) docked onto the S. solfataricus aIF2αγ heterodimer. aIF2γ residues corresponding to those in yeast eIF2γ implicated in tRNA binding by the Sui− substitutions Y142H and G397A are circled. See the text for more details. (Reproduced from reference 223 with slight modifications, with permission from Elsevier.) (B and C) Segments of aIF2α, aIF2γ, and aIF2β at the subunit interfaces of the S. solfataricus heterotrimer of full-length β and γ subunits and domain III of aIF2α. Residues are labeled with a prefix (a, b, or g) indicating their subunit origin (α, β, or γ). (B) Helix α1 in aIF2β is wedged between two helices in the G domain of aIF2γ. The aIF2β residue corresponding to the yeast Sui− substitution Y131 is circled. (C) sw1 in the G domain interacts directly with residues in the ZBD of aIF2β and with domain II of aIF2γ. The ZBD also contacts the ribose ring of the bound GDP (shown in a stick figure). Several ZBD residues near the interface with sw1 are immediately adjacent to residues corresponding to yeast eIF2β residues altered by Sui− substitutions that confer increased GTP hydrolysis by the TC. (Panels B and C are reproduced from reference 224 with slight modifications, with permission of the publisher [copyright 2007 National Academy of Sciences, U.S.A.].)
Consistent with this model, the substitution of the residue in yeast eIF2γ corresponding to aIF2 Y51 by the gcd11-Y142H mutation (Fig. 11A) confers a strong initiation defect and Gcd− phenotype, both suppressible by the overproduction of tRNAiMet (66, 77), and a marked Sui− phenotype (57). Moreover, purified eIF2 containing the gcd11-Y142H subunit shows reduced Met-tRNAiMet binding but normal off rates for GDP and GTP (66). The archaeal docking model also predicts a reduced affinity for Met-tRNAiMet produced by other Sui− Gcd− substitutions (57), including G397A (equivalent of A296 in domain II of aIF2γ [Fig. 11A]), predicted to contact A76 of initiator tRNA, and R510H (equivalent of R406 in domain III [Fig. 11A]), predicted to contact the T stem of initiator tRNA (223). Thus, the Sui− phenotypes of these substitutions could arise from the inappropriate release of initiator tRNA from eIF2-GTP at UUG codons, as described above for the lethal, dominant Sui− allele gcd11-N135K (SUI4) that destabilizes Met-tRNAiMet binding to eIF2-GTP in vitro (86). Indeed, N135K alters a residue in the portion of sw1 that interacts with domain II residues predicted to participate in the binding of A76 of initiator tRNA.
A nonlethal Asp substitution of Asn-135 of yeast eIF2γ likewise confers Sui− and Gcd− phenotypes and destabilizes Met-tRNAiMet binding to eIF2-GTP in vitro. Interestingly, Alone and Dever found that the A208V substitution in sw2 can suppress both the Gcd− and Sui− phenotypes and restore Met-tRNAiMet binding by the eIF2-GTP mutant containing the N135D substitution (6), which supports the model that an elevated rate of Met-tRNAiMet dissociation from eIF2-GTP can produce a Sui− phenotype (86). A different suppressor mutation, A382V in domain II, also restored Met-tRNAiMet binding in vitro, but the N135D A382V double mutant still displayed both Gcd− and Sui− phenotypes in vivo, and the latter phenotype was suppressed by the overexpression of eIF1. To explain these unexpected findings, it was proposed that A382V restores the Met-tRNAiMet-binding pocket, but the initiator tRNA is abnormally anchored to eIF2γ in a manner that perturbs its binding to the P site and evokes eIF1 dissociation at UUG codons (6). A third suppressor of N135D, the A219T substitution in the G domain, destabilizes Met-tRNAiMet binding to eIF2-GTP on its own but surprisingly does not produce a Sui− phenotype. It was proposed that A219T additionally confers an Ssu− defect that compensates for its destabilizing effect on TC binding, and consistent with this, A219T suppresses the Sui− phenotype of the N135D A382V double substitution. It was suggested that A219T causes initiator tRNA to bind in an orientation that restricts the rearrangement to the closed conformation of the PIC (6). These findings are noteworthy in suggesting that the alteration of residues of the G domain or domain II of eIF2γ can alter the fidelity of AUG recognition by affecting the conformation of initiator tRNA binding to the P site.
Substitutions in eIF2β Increase UUG Initiation by Increasing GTP Hydrolysis by the TC
Numerous dominant Sui− substitutions have been isolated in eIF2β, which alter conserved residues in the Zn-binding domain (ZBD) of this protein (29, 55). As noted above, the Sui− substitutions S264Y (SUI3-2) and L254P (SUI3-40) increase eIF5-independent GTP hydrolysis in vitro, and S264Y also increases the dissociation of Met-tRNAiMet from eIF2-GTP in the manner described above for Sui− mutations in eIF2γ. It was proposed that the increased GTP hydrolysis increases the probability that eIF2-GDP dissociates and leaves Met-tRNAiMet base paired with UUG codons, as a way to account for their Sui− phenotypes (86). In the current paradigm (Fig. 6), a shift of the equilibrium toward GDP·Pi in the scanning complex could drive Pi release at UUGs by mass action.
Based on the crystal structure of an aIF2βγ-GDP complex (196), the mechanism of these Sui− mutations seemed mysterious because the ZBD is projecting into the solvent, far from the G domain in aIF2γ. However, a subsequent aIF2αIIIβγ-GDP structure (containing only domain III of aIF2α that anchors it to aIF2γ) revealed a completely different orientation, in which the ZBD interacts with sw1 and the GDP ribose ring in the G domain and the central α/β domain of aIF2β extends into the solvent instead (224) (Fig. 11C). The latter structure is attractive because it places ZBD residues corresponding to those altered by Sui− substitutions in eIF2β in proximity to the GTP-binding pocket (e.g., K118 and A132 [Fig. 11C]), where they could act directly to inhibit GTP hydrolysis. Interestingly, the structure of heterotrimeric aIF2 from a different archaeon showed yet another orientation of the ZBD and α/β domains of aIF2β relative to aIF2γ, although here also the ZBD interacts with the G domain (203).
All of the different heterodimeric and heterotrimeric aIF2 structures have in common that the major contact between β and γ subunits involves the wedging of N-terminal helix α1 of aIF2β between the α4 and α6 helices of the G domain, in a manner involving the stacking of invariant Tyr residues of aIF2β α1 and aIF2γ α4 (Fig. 11B). This helix in aIF2β corresponds to a segment of yeast eIF2β (aa 128 to 159) found to be necessary and sufficient for eIF2γ binding, and Ala substitutions of the conserved Tyr-131 and Ser-132 residues in this domain, corresponding to one of the key Tyr residues mentioned above, impaired binding to eIF2γ in vitro and the interaction of eIF2β with the eIF2γα dimer in vivo (81). Importantly, this SUI3-YS allele conferred Ts− and Sui− phenotypes and was synthetically lethal with the Sui− SUI3-S264Y allele, consistent with the possibility that SUI3-YS elevates levels of GTPase activity. This fits with the idea that the binding of the eIF2β ZBD to eIF2γ mediates the inhibition of GTPase activity, although as the relevant Tyr in aIF2β α1 also interacts with the QNIKE motif in aIF2γ (196), which contacts the guanine base, SUI3-YS might influence GTP hydrolysis more directly.
Both the mammalian eIF2 holocomplex and the β subunit can bind mRNA (reviewed in references 85 and 209), and mRNA binding by yeast eIF2β in vitro was found to require three runs of 7 to 8 Lys residues (K boxes) in the NTD of eIF2β. While the deletion of all three K boxes is lethal, SUI3 alleles retaining any single K box are viable, and interestingly, the removal of K boxes 1 and 2 abolished the Sui− phenotype of the SUI3-S264Y allele (121). Perhaps, the K-box mutations destabilize 48S PICs formed at UUG codons by eliminating the eIF2β-mRNA contact. Alternatively, they might reduce interactions between eIF2 and eIF5, which also depends on the K boxes (12), and thereby suppress the increased GTP hydrolysis displayed by mutant TCs formed with SUI3-S264Y (86).
The functional significance of the extreme conformational flexibility of the ZBD and α/β domains of the β subunit observed for the different aIF2 crystal structures cited above remains unclear. One interesting possibility prompted by the structural similarity between eIF2β and the eIF5 NTD (the GAP domain) is that the flexible eIF2β ZBD might be displaced from the G domain by the analogous ZBD in eIF5 as a means of stimulating GTP hydrolysis. As eIF5 lacks the α1 helix that anchors aIF2β tightly to aIF2γ, eIF5 would be tethered stably to eIF2 by the interaction of its CTD with the eIF2β NTD (12). Considering that an equilibrium between GTP and GDP plus Pi exists in the scanning PIC before AUG recognition (5), the competition for binding the G domain could be highly dynamic and might be shifted in favor of eIF5 by Sui− substitutions in eIF2β. eIF1 also shows structural similarity to the α/β domains in aIF2β and eIF5, leading to the suggestion that eIF1's α/β domain might compete with that in eIF2β for binding to the G domain (41); however, this now seems less likely, considering that the ZBD, and not the α/β domain, of aIF2β contacts the G domain in recent crystal structures of heterotrimeric aIF2 (203, 224). As discussed above, the α/β domains of eIF5 and eIF1 might compete for a binding site on the 40S platform in a way that regulates eIF1 dissociation upon AUG recognition (Fig. 6) (145).
Roles of eIF2α in Initiator tRNA Binding and Recognition of the Start Codon Context
The three-dimensional (3D) structures of archaeal and human eIF2αs (90, 153) indicate the presence of three distinct domains. The C-terminal domain (domain III) interacts directly with domain II of the γ subunit (Fig. 11), near the proposed binding pocket for methionine and A76 of initiator tRNA (223). The N-terminal domain (domain I) of aIF2α contains a β-barrel, with nonspecific RNA-binding activity (225), and interacts with the central, helical domain (domain II) through a hydrophobic core. In the heterotrimeric aIF2 structure, domains I and II of aIF2α make no contacts with other aIF2 subunits, and their orientation relative to domain III is highly variable among published structures (203). In vitro assays suggest that the association with aIF2α domain III increases the affinity of aIF2γ for Met-tRNAiMet by orders of magnitude, whereas the β subunit makes only a small contribution to tRNA binding by aIF2γ (223, 225). The docking model discussed above features no direct contacts between aIF2α and tRNA, and it was envisioned that the α subunit stimulates Met-tRNAiMet binding indirectly by promoting the proper conformation of the tRNA-binding pocket in aIF2γ (223). A very different docking model has been proposed, in which aIF2α also interacts with initiator tRNA and contributes directly to the binding energy (152).
In eukaryotes, only a moderate contribution of eIF2α to Met-tRNAiMet was indicated by the fact that a viable yeast strain could be engineered to lack eIF2α, and in vitro analyses of the eIF2β/γ dimer showed a reduction in the affinity for Met-tRNAiMet of no more than 5-fold (151). Thus, it appeared that eIF2β makes a greater contribution than eIF2α to Met-tRNAiMet binding. However, a recent study of Encephalitozoon cuniculi eIF2 reconstituted from recombinant subunits showed that the α and β subunits both greatly stimulate Met-tRNAiMet binding in dimeric complexes with the γ subunit, but the absence of either one from the heterotrimeric complex only slightly reduces binding. Thus, it seems that eIF2α and eIF2β each make strong, but largely redundant, contributions to Met-tRNAiMet binding by eIF2γ, and it was suggested that each one can stabilize the “on” configuration of the switch regions in eIF2γ (146).
Two Sui− mutations mapping to domain I of yeast eIF2α have been described (P13S and V19F) (38), but their effects on Met-tRNAiMet binding and GTP hydrolysis are unknown. However, there is biochemical evidence that eIF2α plays a direct role in start codon recognition. In reconstituted mammalian PICs, mRNAs with thio-U substitutions at position −3 were cross-linked to eIF2α and Rps5e (172), consistent with the location of an Rps5e segment above the mRNA exit channel (20, 178). Moreover, the replacement of eIF2 with the eIF2βγ heterodimer reduced the efficiency of AUG recognition but also diminished the effect of good context on 48S assembly (172). Thus, eIF2α might mediate a key contribution of the −3 base to the tight binding of Met-tRNAiMet at the AUG codon.
FUNCTIONS OF eIF3 SUBUNITS IN 43S ATTACHMENT, SCANNING, AND START CODON RECOGNITION
The eIF3 complex is a large complex of 13 nonidentical subunits (a to m) in mammals and only 6 subunits (a, b, c, g, h, and j) in budding yeast. A model of mammalian eIF3 binding to the 40S subunit, derived from cryo-EM reconstructions of complexes between eIF3 and the hepatitis C virus IRES and between the IRES and the 40S subunit (198), shows eIF3 binding primarily to the solvent side of the 40S subunit in the vicinity of the exit (E) site (194). Consistent with this finding, in reconstituted mammalian 48S PICs, mRNA replaced with thio-U at position −14 was cross-linked to eIF3a and eIF3d, locating them at the mRNA exit channel (173). The N-terminal domain (NTD) of yeast eIF3a (a/Tif32), which promotes eIF3 binding to native 40S subunits, interacts with Rps0e (211) at the exit channel (20, 178), and the a/Tif32 NTD functionally interacts with sequences upstream of GCN4 uORF1 that apparently must be situated near the exit channel pore to stimulate reinitiation (206). Enzymatic footprinting and hydroxyl radical cleavage by eIF3 in mammalian PICs suggest additional contacts with h16 near the mRNA entry channel (173), and consistent with this, the CTD of yeast a/Tif32 interacts with an rRNA segment encompassing h16 to h18 (211), Rps2e, and Rps3e (33), all situated near the entry channel. Thus, there is increasing evidence that eIF3 subunits are in proximity to the mRNA at both the entry and exit channel openings on the solvent side of the 40S subunit. This would be consistent with a direct role for eIF3 in stabilizing the attachment of the 43S PIC to mRNA.
Supporting this last idea, various eIF3 subunits in mammals (a, d, and g) and yeast (g) bind RNA in vitro (11, 15, 23, 42, 75, 94, 219), and oligonucleotides that bind to the 40S subunit were shown to stabilize the association of mammalian eIF3 (lacking the j subunit) with the 40S subunit in vitro (100). Substitutions in conserved residues in the C-terminal region of yeast eIF3a/Tif32 (the “KERR motif” and “box6”) impair the binding of native mRNAs to 43S PICs in vivo without reducing the 40S binding of the TC or any other 43S components (33). Moreover, yeast eIF3 dramatically stimulates 43S binding to capped mRNAs in vitro, being more critically required than eIF4G (139). Consistent with the prediction that eIF3 extends the mRNA exit channel (173), yeast eIF3 more strongly enhanced 43S binding to a model mRNA with a long leader upstream of AUG (that would protrude from the exit channel) than to one containing a short leader but a long 3′ extension (that would protrude from the entry channel) (139). The a/Tif32 KERR motif and box6 substitutions mentioned above also confer Ssu− phenotypes and reduce the efficiency of scanning through a long 5′UTR in yeast cells. The box6 substitution also impairs scanning through a stem-loop located far downstream from the 5′ end of a reporter mRNA (33). Considering that the a/Tif32 CTD interacts directly with 40S structural elements (h16 and Rps3e) that promote the open conformation of the mRNA channel latch (159), it was suggested that these mutations might impair the opening of the latch, although they could also reduce the function of a helicase at the entry channel that removes the secondary structure ahead of the scanning ribosome.
The C-terminal half of a/Tif32 belongs to a conserved heterotrimeric module within eIF3, containing the eIF3j subunit (j/Hcr1 in yeast) and the N-terminal RNA recognition motif (RRM) domain of eIF3b/Prt1 (b/RRM) (213). j/Hcr1 promotes the 40S binding of eIF3 in mammals (73, 100) and yeast (67, 150, 213), and mammalian eIF3j can bind directly to 40 subunits (73) and mediate 40S binding by the b/RRM in vitro (61). In addition, yeast j/Hcr1 can interact directly with Rps2e (62). The b/RRM probably also promotes the 40S binding of eIF3 via direct interactions with the a/Tif32 CTD (67, 150, 213). Thus, the entire b/RRM-j/Hcr1-a/Tif32 CTD module is likely situated near the mRNA entry channel. The module is stabilized by the interaction of a 35-residue segment of the eIF3j NTD with residues in helix α1 and loop 5 of the b/RRM (61, 62), and substitutions in the b/RRM or j/Hcr1 NTD that disrupt these contacts confer marked leaky scanning of GCN4 uAUG1 (62). However, whereas the deletion of HCR1 confers leaky scanning, it does not produce an Ssu− phenotype (33), suggesting that it reduces equally the recognition of AUG and UUG start codons. The destabilization of the b/RRM-j/Hcr1-a/Tif32 CTD module with the Ssu− KERR and box6 substitutions in the a/Tif32 CTD confers only a small increase in leaky scanning, indicating that alterations of different components of the module can have different consequences on the efficiency and accuracy of AUG recognition.
The CTD of human eIF3j was mapped by site-directed hydroxyl radical probing to the A site and mRNA entry channel on the interface surface of the 40S subunit, and it was found to reduce the affinity of 40S subunits for mRNA in a manner largely overcome by TC binding, without dissociating from the entry channel. It was envisioned that the eIF3j CTD coordinates mRNA binding with the presence of the TC and might also influence 40S-mRNA interactions during scanning (72). The binding of the yeast j/Hcr1 CTD to Rps23 in vitro (62) is consistent with its presence in the mRNA entry channel on the interface side of the yeast 40S subunit (41), and it is possible that the j/Hcr1 CTD toggles between Rps23 contacts on the interface side and Rps2 contacts on the solvent side in performing different roles in PIC assembly or AUG recognition. It should be noted, however, that the elimination of the j/Hcr1 CTD has no effect on yeast growth, except in the presence of the KERR or box6 mutations in the a/Tif32 CTD, and produces only a small increase in leaky scanning. Thus, in yeast the proposed function of eIF3j in regulating mRNA binding near the A site is either noncritical or fully redundant with the KERR and box6 elements of the a/Tif32 CTD. The latter possibility might seem unlikely, considering that the a/Tif32 CTD binds to Rps2 and Rps3, but not Rps23, and thus seems to be restricted to the solvent side of the 40S subunit. The elimination of the j/Hcr1 CTD in the presence of the a/Tif32 KERR/box6 substitutions had nonadditive effects on leaky scanning, which remained modest in the double mutant (33). It remains to be determined what critical aspect(s) of eIF3 function is impacted by this nearly lethal, compound disruption of the eIF3a CTD·eIF3j·eIF3b-RRM module.
The extreme CTD of eIF3b/Prt1 assembles a second autonomous module in yeast eIF3 containing the g/Tif35 and i/Tif34 subunits (13), but g/Tif35 and i/Tif34 were found to be dispensable in vitro for the stimulatory effects of eIF3 on the recruitment of mRNA and the TC and also the translation of a reporter mRNA (169). In vivo, however, the substitution of residues on one surface of the g/Tif35 RRM reduced the efficiency of scanning through a stable stem-loop in the 5′UTR of a reporter mRNA, and the Q258R substitution in i/Tif34 appeared to reduce the rate of scanning by 43S PICs involved in reinitiation downstream of uORF1 on GCN4 mRNA. Thus, the Q258R substitution stimulates reinitiation at GCN4 in a reporter mRNA where uORF1 (as a solitary uORF in the leader) is situated too close to the GCN4 AUG to allow sufficient scanning time for the efficient reassembly of the PIC (Fig. 3C, I). Interestingly, the slow-scanning phenotype of Q258R might be mitigated by eIF1/eIF1A co-overexpression (42). The ability of g/Tif35 to bind specifically to Rps3e and Rps20e suggests that the Prt1 CTD·g/Tif35·i/Tif34 module is situated just above the mRNA entry channel and, hence, could directly impact the efficiency of mRNA entry or its progression into the decoding center.
As noted above, the NTD of eIF3c/Nip1 can interact simultaneously with eIF1 and the “non-GAP” CTD of eIF5 (14, 170) to stabilize the MFC, and the eIF3c-eIF1 interaction is conserved in mammals (71). Thus, the c/Nip1 NTD likely has a role in recruiting eIF1 and eIF5 to the 43S PIC and might also regulate their opposing functions in start codon recognition. Indeed, Ala substitutions of 10 consecutive residues in the yeast c/Nip1 NTD (box12) confer a Sui− phenotype, suppressed by the overexpression of eIF1 and exacerbated by the overexpression of eIF5, and reduce the binding of eIF1 to the c/Nip1 NTD in vitro. It was proposed that the reduced interaction between eIF1 and eIF3c in the scanning 43S PIC would compromise eIF1's ability to inhibit the eIF5 GAP function at UUG codons (212). As noted above (Fig. 7), the c/Nip1 NTD appears to bind the same (α1) surface of eIF1 that contacts the 40S subunit (127, 180), and it is unclear whether eIF1 can bind simultaneously to the 40S subunit and the c/Nip1 NTD. It is possible that eIF1 is recruited by the c/Nip1 NTD and then transferred to the 40S platform. If so, a defect in eIF1 recruitment conferred by the box12 substitution might reduce eIF1 occupancy in the scanning PIC in a manner corrected by eIF1 overexpression (145). The overexpression of eIF1 might saturate both of its binding sites, on the platform and in the c/Nip1 NTD, and this greater eIF1 occupancy could be instrumental to its ability to suppress UUG initiation in a variety of Sui− mutants.
Substitutions in two other strings of 10 aa in the c/Nip1 NTD, box2 (with Ala) and the highly acidic box6 (with Arg; Box6R), appeared to confer Ssu− phenotypes (although the effect on UUG-to-AUG ratios was not determined), and both mutations reduced the binding of the c/Nip1 CTD to eIF5 in vitro, reduced eIF5 occupancy in native PICs, and (for Box6R) conferred a growth defect mitigated by eIF5 overexpression. It was suggested that the reduced GAP function associated with the diminished interaction of eIF5 with the scanning PIC would suppress the increased UUG initiation conferred by Sui− substitutions in eIF1, eIF2β, or eIF5 (212). Again, it is unclear whether the association between the c/Nip1 NTD and the eIF5 CTD, which stabilizes the MFC, is involved only in the initial recruitment of eIF5 or also participates in orienting the eIF5 NTD for its GAP function in the scanning complex.
ROLES OF RNA HELICASES eIF4A, Ded1, AND Dhx29 IN RIBOSOME ATTACHMENT AND SCANNING
eIF4A Helicase Activity Is Activated by eIF4G, eIF4B, and eIF4H
In addition to achieving a conformation of the ribosome conducive to 5′-to-3′ translocation along single-stranded mRNA, efficient scanning on many mRNAs also requires the removal of secondary structures to allow the mRNA to pass through the 40S mRNA entry or exit channels and permit its base-by-base inspection in the P site. The secondary structure must also be removed near the cap to produce a single-stranded “landing pad” for the 43S PIC. In fact, Kozak showed that the insertion of a stem-loop of moderate stability (−30 kcal/mol) 12 nt from the cap effectively blocked 43S attachment to the mRNA, whereas a hairpin of higher stability (−60 kcal/mol) was required to block translation when inserted 72 nt from the cap, where it arrested the progression of scanning 43S PICs (104). Thus, in mammals, the 5′ attachment of the 43S complex appears to be more sensitive than scanning to the secondary structure and thus is more dependent on RNA-unwinding activities. Multiple DEAD-box RNA helicases have been implicated in translation initiation, including eIF4A, Dhx29, and Ded1/Ddx3, and it is not fully understood which ones participate at these two steps, but there is increasing evidence that eIF4A functions in both ribosome attachment and scanning, while Dhx29 and Ded1 act primarily in scanning. DEAD-box helicases do not have processive unwinding activity and can effectively disrupt only relatively short helices by locally melting the duplex strands and then binding to one of the resultant single-stranded regions. This activity requires the ATP-bound form of the enzyme, but the energy of ATP hydrolysis is not required for strand separation per se, which can be catalyzed by Ded1 and eIF4A using certain nonhydrolyzable nucleotides, including ADP-BeFx. Rather, it appears that ATP hydrolysis serves primarily to dissociate the enzyme from the RNA, recycling it for multiple rounds of RNA binding and melting (126, 158).
eIF4A is a typical DEAD-box helicase and thus exhibits RNA-dependent ATPase activity and ATP-dependent duplex-unwinding activity (181). ATP and RNA bind cooperatively to eIF4A (130), consistent with the fact that both RNA and ATP binding by DEAD-box proteins (Dbps) depend on the closed conformation of the N- and C-terminal RecA-like domains, which juxtaposes the conserved determinants of RNA and ATP binding at the domain interface (9, 189). The association of eIF4A with eIF4G in the eIF4F complex not only directs eIF4A to the cap-proximal region of the 5′UTR but also activates its ATPase and helicase activities (181, 187). The interaction of eIF4A with the first of three “HEAT” domains in mammalian eIF4G (HEAT-1), and with the sole HEAT domain in the yeast factor, increases eIF4A's ability to stimulate translation initiation both in vitro and in cells (52, 53, 87, 147, 161). The crystal structure of free eIF4A revealed that the RecA-like domains are widely separated in a fully open conformation (28), whereas an analysis of a complex between eIF4A and the eIF4G HEAT domain in solution led to the proposal that eIF4G functions as a “soft clamp” that stabilizes the active, closed, interdomain conformation of eIF4A by interacting with both the NTD and the CTD of eIF4A via C- and N-terminal α-helices, respectively, in the HEAT domain (154). The crystal structure of the yeast eIF4A/eIF4G-HEAT domain complex revealed that although the RecA domains are held closer together than in free eIF4A, with the conserved DEAD-box motifs facing one another across the cleft, the domains remain disengaged. Moreover, it was predicted that a full domain closure would be incompatible with contacts between the eIF4A NTD and the eIF4G-HEAT CTD observed for the crystal structure. It was proposed that this less extensive, secondary interface between the cIF4A NTD and eIF4G-HEAT CTD, which is not critical for the eIF4A-eIF4G interaction per se, stimulates ATPase activity by preventing the two eIF4A domains from getting trapped in a nonproductive open conformation following ATP hydrolysis (187).
More recently, evidence was presented that the “half-open” conformation observed for the eIF4A/eIF4G-HEAT crystal structure occurs in solution and promotes ATP hydrolysis, dependent on eIF4G residues at the secondary interface. It was suggested that this conformation comes into play following ATP hydrolysis to facilitate the release of Pi from ADP·Pi, which should trigger the dissociation of the RNA and also promote the subsequent ADP-ATP exchange necessary for the next reaction cycle (83). This proposal bears some similarity to the roles advanced for Gle1, which structurally resembles the eIF4G HEAT domain, and Nup159 in the reaction cycle of the DEAD-box helicase Dbp5 in stimulating mRNA export from the nucleus (141, 152a).
The ATPase and helicase activities of eIF4A are also stimulated by eIF4B and eIF4H (181, 182), which share sequence similarity across the length of eIF4H. Both eIF4H and eIF4B bind to eIF4A with low affinity, and this interaction is stabilized by ATP and single-stranded RNA (134, 149, 182). As these two ligands stabilize the closed conformation of eIF4A, and considering other evidence that eIF4H can interact with both domains of eIF4A (134, 182), it is possible that eIF4B and eIF4H stimulate eIF4A helicase activity by enhancing domain closure in the manner described above for eIF4G. However, eIF4B also has single-stranded-RNA-binding activity, and there is evidence that it stabilizes eIF4A binding to RNA by making transient interactions with nucleotides flanking a 9- to 10-nt “core” eIF4A-binding site in an eIF4B-eIF4A-RNA complex (182). This activity could enable eIF4B to help recruit eIF4A to single-stranded stretches in a structured mRNA substrate, and it could also stabilize the single-stranded RNA products of the helicase reaction and prevent reannealing. Assuming a fixed orientation of eIF4B relative to that of eIF4A in the ternary complex with RNA, and a fixed 5′-to-3′ orientation of mRNA binding to eIF4A, the latter activity could provide 5′-to-3′ directionality to the duplex-unwinding reaction (134).
eIF4F Stimulates 43S PIC Attachment to mRNA
The presence of eIF4A in the cap-binding complex eIF4F makes it the prime candidate for the helicase that generates a single-stranded binding site for the 43S PIC at the 5′ end of mRNA. The tight binding of eIF4F to the capped 5′ end is dependent not only on the eIF4E-cap interaction and eIF4E binding to eIF4G (168) but also on the RNA-binding activity of eIF4G (21, 155, 221) and the direct interaction of eIF4G with PABP bound to the poly(A) tail (155, 205, 207). Although the eIF4E-cap interaction adds little to the binding affinity of purified eIF4F for naked mRNA (97), it differs from the eIF4G-mRNA interaction in not having to compete with other general RNA-binding proteins and, thus, can presumably direct eIF4F binding to the capped 5′ end of mRNA in vivo. The ability of eIF4G to also interact with the PABP-bound poly(A) tail provides another means of outcompeting general RNA-binding proteins (205). Thus, eIF4G's ability to simultaneously interact with eIF4E·cap, PABP·poly(A) tail, and the body of mRNA promotes the assembly of a highly stable eIF4F complex at the mRNA 5′ end. Although the eIF4F-PABP interaction enables the joining of the cap and the poly(A) tail in a circle, it remains unclear whether mRNA circularization per se is critical for efficient initiation or, instead, if the eIF4G-PABP interaction simply represents one of several interactions that stabilize eIF4G binding to mRNA, which is likely the critical event in promoting 43S attachment. The latter view is supported by findings in yeast that the PABP-eIF4G interaction is dispensable for WT cell growth and becomes important for viability only when the eIF4E-cap interaction and an RNA-binding domain in the eIF4G NTD are simultaneously ablated (155, 207).
Evidence that the eIF4A helicase activity promotes ribosome attachment to mRNAs with cap-proximal secondary structures is provided by the finding that dominant negative, catalytically inactive eIF4A proteins impair translation in extracts to a greater extent for reporter mRNAs with cap-proximal secondary structures than for constructs lacking such structures. However, the translation of the unstructured reporter was also significantly reduced in a manner attributed to an impaired eIF4F binding to the capped mRNA and a reduction in 43S-mRNA attachment (204). Similarly, the translation of reporter mRNAs with 5′UTRs of only 45 or 8 nt was impaired in eIF4A-dependent yeast extracts when supplemented with catalytically defective eIF4A mutants, indicating that even mRNAs with very short or unstructured leaders are dependent on eIF4A helicase activity. It was speculated that eIF4A is needed to dissociate RNA-RNA interactions besides obvious stem-loops in the 5′UTR to enable ribosome attachment (24).
The eIF4A cofactor eIF4B is dispensable in yeast, but its elimination greatly reduces cell growth, especially at low temperatures. In cells lacking Tif3/eIF4B, the translational efficiency of reporter mRNA was relatively more diminished at lower temperatures and when it harbored a stem-loop (−14.1 kcal/mol) inserted 22 nt from the cap, although the unstructured reporter was also significantly impaired (8). Given its cap-proximal location, this stem-loop likely impeded 43S attachment. The depletion of eIF4B in mammalian cells also appears to selectively inhibit the translation initiation of mRNAs with more structured 5′UTRs, but whether 43S attachment, scanning, or both were impaired for these mRNAs is unclear (191). Hence, evidence is lacking that Tif3/eIF4B is critical for scanning in vivo. In fact, experiments with yeast suggest that Tif3 is dispensable for highly processive scanning through an extremely long 5′UTR (22).
As noted above, Pestova et al. showed that the assembly of 48S PICs on uncapped, synthetic mRNA with an unstructured 5′UTR required only eIFs 1, 1A, and 3 and the TC (167), whereas 48S assembly on native β-globin mRNA also required eIF4F, eIF4A, eIF4B, and ATP, and that the omission of eIFs 1 and 1A led to an aberrant PIC arrested in the globin mRNA 5′UTR (165). These findings implied that the 43S attachment to the structured 5′ end of β-globin mRNA requires eIF4F, eIF4B, and ATP hydrolysis by eIF4A and that subsequent scanning to the AUG codon additionally requires eIFs 1 and 1A. Dmitriev et al. found that eIF4B was crucial for 48S assembly on a derivative of β-globin containing a G-C-rich sequence that sequesters the 5′ end in the secondary structure, further implicating eIF4B in 43S attachment to a structured 5′UTR (50). Similarly, Lorsch et al. showed that eIF4F and eIF4B/Tif3 are both required in a yeast fully reconstituted system for the rapid formation of stable 48S PICs on native capped mRNAs (RPL41A and DAD4) but not on a capped, unstructured, synthetic mRNA, which required only eIFs 1, 1A, and 3 and the TC. As these native mRNAs have short (22- to 23-nt) 5′UTRs, it seems likely that 43S attachment was being stimulated by eIF4F and eIF4B/Tif3. Interestingly, 48S PICs could be formed on uncapped RPL41A and DAD4 mRNAs without eIF4F and eIF4B, dependent on eIF3, but the complexes appeared to be relatively unstable. This and other observations led to the proposal that the triphosphate portion of the cap inhibits 48S PIC assembly by a nonproductive “eIF3-only” pathway and imposes a requirement for the eIF4 factors to form productive 48S PICs (139).
In addition to recruiting eIF4A to the mRNA 5′ end and stimulating its ability to generate a single-stranded binding site for the ribosome, it is thought that eIF4G also actively promotes 43S attachment as a molecular bridge by interacting simultaneously with mRNA and factors bound to the PIC, including eIF3 in mammals and eIF5 and eIF1 in yeast. Mammalian eIF3 interacts with an internal segment of eIF4G located C terminal to the HEAT-1 domain (87, 101, 119, 142). It was shown that the mammalian eIF3e subunit can bind eIF4G directly and that excess free eIF3e competes with the eIF3 holocomplex for eIF4G binding and also impairs translation initiation and the eIF4G association with 40S PICs in cell extracts, all consistent with the idea that the eIF3e-eIF4G interaction promotes 43S PIC attachment to mRNA (124). It was proposed that mammalian eIF4B can also stimulate ribosome attachment to the mRNA by binding simultaneously to mRNA through its C-terminal, Arg-rich, RNA-binding domain and to 18S rRNA through its N-terminal RRM (137).
Neither eIF3e nor the eIF3-binding segment of eIF4G is conserved in yeast (134), and yeast eIF3 and eIF4G do not interact directly (139). However, yeast eIF4G and eIF5 interact tightly (139), and the eIF5 CTD can bridge the interaction between eIF4G2 and the eIF3c/Nip1 NTD (eIF5's partner in eIF3) and stimulate the eIF4G-eIF3 association in cell extracts. A mutation in the eIF5 CTD (tif5-7Ala) that disrupts its interactions with eIF4G2 and the c/Nip1 NTD impairs the binding of native mRNA (MFA2) to 40S subunits in cell extracts in a manner rescued by WT eIF5 (14). Thus, the eIF4G2-eIF5 association might substitute for the eIF4G-eIF3 interaction to promote 43S PIC-mRNA attachment in yeast. This stimulatory function of eIF5 has not been observed thus far in the yeast fully reconstituted system (139). Moreover, the tif5-7Ala mutation (14), or even the depletion of eIF5 in a yeast degron mutant (94), leads to the accumulation (not the depletion) of native 48S complexes. The latter findings suggest that the eIF5 functions in start codon recognition and GTP hydrolysis by the TC, prerequisites for 60S subunit joining to the 48S PIC, are more rate limiting in vivo than its participation in 43S attachment to mRNA. As yeast eIF4B/Tif3 can bind directly to eIF3 (139), it might also form a bridge between the eIF4F·mRNP and the 43S PIC and function redundantly with the eIF5-eIF4G interaction.
Surprisingly, the depletion of eIF4G1 in a yeast degron mutant lacking eIF4G2 did not reduce the binding of RPL41A mRNA to native 43S PICs in yeast cells, although the depletion of eIF2 or eIF3 did have this effect (94). This finding is mirrored by results from the yeast fully reconstituted system, where the omission of eIF4G from reaction mixtures containing eIF3, eIF4A, and eIF4B (in addition to eIFs 1 and 1A and the TC) reduced the rate by 20-fold but did not alter the endpoint of 48S PIC assembly, at least for RPL41A mRNA, whereas no complexes were produced even after prolonged incubation without eIF3 (139). In addition, unlike the depletion of eIF3, the depletion of eIF4G from yeast cells does not abolish protein synthesis but reduces the rate to only ∼25% of the WT rate. Microarray analyses of polysomal mRNAs did not identify any mRNAs whose translation is dramatically impaired upon the depletion of eIF4G, although a widespread narrowing of translational efficiencies occurred, with many of the most efficiently translated mRNAs declining and many of the least efficient mRNAs increasing in efficiency. (The latter increase was explained as the result of reduced competition with the most efficiently translated mRNAs for limiting 43S PICs.) Thus, it appears that eIF4G is rate enhancing rather than essential for the translation of most mRNAs in yeast but is crucial for maintaining the proper spectrum of translational efficiencies across the genome (156). Similar conclusions were reached earlier for the depletion of eIF4G in mammalian cells (179). Unexpectedly, the mRNAs whose efficiencies were reduced the most upon eIF4G depletion in yeast had relatively short 5′UTRs devoid of stable secondary structures, which is typical of most yeast mRNAs, whereas those translated relatively better in the absence of eIF4G had longer-than-average 5′UTRs. It was suggested that this outcome is consistent with the possibility that eIF4G is most critically required for 43S attachment rather than scanning through long, unstructured 5′UTRs, which might depend on Ded1 instead (156).
The observations that eIF4G appears to be stimulatory rather than essential for the translation of most mRNAs in cells (40, 156, 179) are consonant with the findings mentioned above that reconstituted mammalian 43S PICs lacking eIF4F can attach to the 5′ end and scan to the start codon of uncapped mRNAs harboring unstructured 5′UTRs. Indeed, 5′-end attachment was achieved with only eIF3 and the TC for such mRNAs containing 5′-proximal AUGs, and scanning to AUGs further downstream additionally required only eIFs 1 and 1A (167). The mechanism of eIF4F-independent 5′-end attachment by the 43S PIC remains uncharacterized. In addition, it is unclear whether scanning occurs with 5′-to-3′ directionality in the absence of eIF4 factors or, rather, is more akin to random bidirectional diffusion that requires only the open conformation of the 40S subunit imposed by eIFs 1 and 1A. In any event, it appears that initiation at the correct AUG can occur at appreciable levels, even if at considerably lower rates, in the absence of eIF4G for the bulk of mRNAs in living cells.
eIF4F Promotes Scanning through mRNA Secondary Structures
As noted above, 48S PIC assembly on a synthetic unstructured mRNA in the mammalian reconstituted system does not require eIF4F, eIF4A, or eIF4B (167). However, the eIF4 factors and ATP were required for 48S assembly when a stem-loop of moderate stability (−13.1 kcal/mol) was inserted 43 nt from the 5′ end of the unstructured mRNA, providing direct evidence that eIF4F and ATP hydrolysis by eIF4A stimulate scanning through the secondary structure, in addition to promoting 43S attachment to the mRNA. Consistent with a role for eIF4G in scanning, it was shown that a segment of mammalian eIF4G located N terminal to HEAT-1, which contains RNA-binding activity, is required for scanning to the AUG codon after 40S binding to internal ribosome entry sites in certain viral mRNAs (176). In addition, it was demonstrated that reinitiation following the translation of a short uORF in mammalian cell extracts depends on the involvement of eIF4G in the prior translation of the uORF, suggesting that eIF4G remains associated with the ribosome during the translation of the uORF and stimulates reinitiation downstream (175). It is unclear, however, whether eIF4G was required to stabilize the interaction of the posttermination 40S subunits with the mRNA (analogous to 43S attachment at the cap) or for their scanning proficiency. In a related finding, substitutions in yeast eIF4G2 that weaken its binding to eIF4E or eIF4A appear to reduce the rate of scanning by 43S PICs engaged in reinitiation on GCN4 mRNA and, thus, located far downstream from the 5′ cap (217). eIF4G might promote scanning as a component of eIF4F that is still bound to the cap structure, as depicted in Fig. 1 and discussed below.
Models of the Scanning PIC
The integration of separate cryo-EM models of mammalian eIF3-40S and eIF3-eIF4G complexes led to the prediction that eIF4G interacts with the PIC near the mRNA exit channel of the 40S subunit, which would position eIF4F upstream (5′) of the scanning PIC (Fig. 12A). It was proposed that eIF4F would prevent the 3′-to-5′ backsliding of the PIC rather than the unwinding of the secondary structure in advance of the ribosome (194). This idea was elaborated by Spirin (199), who proposed that eIF4A and eIF4B undergo cycles of binding to and dissociation from the mRNA sequences located immediately upstream of the PIC to impose 5′-to-3′ directionality on scanning. The eIF4G would travel with the scanning PIC, owing to the eIF3-eIF4G interaction, and deliver eIF4B-eIF4A·ATP complexes to the single-stranded mRNA as it emerges from the exit channel. ATP hydrolysis would release eIF4A from the mRNA, but the eIF4B would remain bound and block backwards movement by the PIC. When the ribosome moves again in the 3′ direction, the cycle would repeat itself. This would constitute a “Brownian ratchet” mechanism, with eIF4B acting as the pawl and the energy of ATP hydrolysis being expended by eIF4A upstream of the PIC (Fig. 12C).
Fig. 12.
Models of the scanning PIC. (A) Cryo-EM model of mammalian eIF3 (magenta) and eIF4G (blue) binding to the 40S subunit (yellow), annotated to depict the path of the mRNA (red ribbon) in the decoding center and exit channel (Reproduced from reference 194 with permission from AAAS.) (B) Model for the scanning PIC in which the eIF4G N-terminal region remains bound to the cap-eIF4E assembly and the HEAT-1 domain of eIF4G interacts with mRNA sequences immediately 5′ of the 40S exit channel, causing the mRNA to form a loop between the cap and HEAT-1 that grows in size as scanning proceeds. eIF4A, bound to HEAT-1, interacts with mRNA sequences located 3′ of the PIC to destabilize the secondary structure before it reaches the 40S subunit, and eIF4H prevents the reannealing of the unwound, single-stranded nucleotides before they enter the entry channel pore. (Reproduced from reference 134 with permission from Elsevier.) (C) A Brownian ratchet mechanism of scanning in which eIF4A undergoes cycles of binding and dissociation from the mRNA, driven by ATP binding and hydrolysis, near the mRNA exit channel (where it is positioned by eIF4G). eIF4B interacts with the eIF4A·ATP·mRNA complex but dissociates from the mRNA more slowly than does eIF4A·ADP, allowing it to act as a pawl to prevent 3′-to-5′ backsliding. Thus, the random sliding of the ribosome can occur only in the 3′ direction and is fixed at the new location by the next cycle of eIF4A·ATP·mRNA·eIF4B complex assembly. (Reproduced from reference 199 with permission of the publisher [copyright 2009 American Chemical Society].) (D, left) Docking of the crystal structure of yeast eIF4A to eIF4G and the HEAT-1 domain of human eIF4G1 into the SAXS envelope of the complex between eIF4A and the eIF4G fragment containing HEAT domains 1 to 3. (Right) Comparison of the sizes of the 40S subunit and the eIF4A-eIF4G complex. (Reproduced from reference 149 by permission of Oxford University Press.)
A different model proposed by Marintchev et al. (134) envisions that eIF4G can span the mRNA exit and entry channels on the 40S subunit and position eIF4A at the entry channel for the unwinding of the structure ahead of the ribosome while also interacting with mRNA as it emerges from the exit channel via the HEAT-1 domain (Fig. 12B). eIF4B (or eIF4H) would be located behind eIF4A but in front of the ribosome to prevent the reannealing of the unwound mRNA until it feeds into the entry channel, imposing 5′-to-3′ directionality. In this model, the energy of ATP hydrolysis is expended downstream of the PIC. It was envisioned that eIF4F could remain engaged with the cap during scanning, and the already-scanned nucleotides would form a loop between the cap-eIF4F assembly and the ribosome that expands as scanning proceeds (Fig. 12B) (134). This model fits with the conclusions reached from the reconstituted mammalian system, that eIF4F, eIF4A, and eIF4B are required for both ribosome attachment (to β-globin mRNA) and scanning through stem-loops at cap-distal locations (165, 167). The location of the HEAT-1 domain of eIF4G in the model shown in Fig. 12B differs significantly from that predicted by the EM analysis shown in Fig. 12A in residing below, rather than above, the 40S platform, which is more consistent with sites of hydroxyl radical cleavage directed from eIF4G HEAT-1 in reconstituted mammalian 43S PICs. These cleavage sites occurred primarily in helix 2 of expansion segment 6 (ES-6) in 18S rRNA, near the “left foot” of the 40S subunit (227).
Small-angle X-ray scattering (SAXS) analysis of the complex between eIF4A and a segment of mammalian eIF4G containing HEAT-1 and HEAT-2 revealed an elongated, rigid structure of ∼220 Å in length, easily capable of spanning the ∼50 Å separating the entry and exit channel openings on the 40S subunit (Fig. 12D). However, at odds with one aspect of the model described by Marintchev et al. (134), it was found that eIF4G and eIF4B bind to eIF4A in a mutually exclusive manner but that a prior association with eIF4G is required for eIF4A to form a stable mRNP with eIF4B. This led to the proposal that eIF4A is recruited to eIF4F, isomerizes to its closed conformation upon the interaction with eIF4G, and then transfers from eIF4G to an eIF4B molecule bound to the mRNA downstream of the PIC, with the activation of eIF4A helicase activity and the attendant disruption of RNA duplexes at the mRNA entry channel pore (149).
The latter model and that depicted in Fig. 12B, which envision eIF4A and eIF4B working at the mRNA entry channel ahead of the scanning PIC, have to take into account the fact that in toeprint analyses of reconstituted 48S PICs, reverse transcriptase (RT) proceeds all the way to the leading edge of the 40S subunit (139, 165). Similarly, analyses of mRNA fragments protected from RNase digestion in 48S complexes, carried out in the 1970s, did not detect any factors bound to the mRNA downstream of the 40S subunit that can block RNase accessibility, whereas additional residues 5′ of the ribosome were protected from digestion (111, 118, 123). The latter protection can be explained by the stable association of eIF4G with the mRNA immediately upstream of the exit channel, whereas the lack of protection on the downstream side of the PIC seems to indicate that the proposed association of eIF4G, eIF4B, and eIF4A (or other helicases) with mRNA at the entry channel would be very dynamic. Finally, considering that eIF4B/Tif3 is nonessential in yeast (8) and that the severe depletion of eIF4G reduces the rate considerably but does not abolish translation initiation in yeast or mammalian cells (156, 179), these models presumably depict the most efficient, but not the sole, PIC configuration capable of 5′-end attachment and scanning in vivo.
Additional DEAD-Box Helicases, Dhx29 and Ded1, Promote Ribosomal Scanning through Strong Secondary Structures
While there is strong evidence that eIF4F, eIF4A, and eIF4B can stimulate scanning through stem-loop structures, it is clear that other helicases not associated with the cap also have this capacity. Ded1 is a DEAD-box helicase in budding yeast (89, 222) that is essential for cell viability and initiation on the majority of yeast mRNAs in vivo and is also required for translation initiation in cell extracts (34). Furthermore, Ded1 appears to functionally overlap with eIF4F in vivo, as its overexpression suppresses the growth defect of an eIF4E mutation (cdc33-1), and a ded1 mutation exacerbates the growth defects of mutations in eIF4E, eIF4A, eIF4B, or eIF4G (47). Interestingly, the overexpression of Dbp1, 72% identical to Ded1, suppresses the lethality of a ded1Δ mutation (93), raising the possibility of multiple RNA helicases functioning in translation initiation in yeast. Consistent with this, a Ts− ded1 mutation and the deletion of DBP1 each had stronger effects than a Ts− mutation affecting eIF4A or the deletion of eIF4B/Tif3 on the translation of a reporter mRNA harboring an extended 5′UTR. This led to the proposal that Ded1 and Dbp1 are more important than eIF4A/eIF4B for processive scanning in yeast. Indeed, Ded1 was found to be more potent than the eIF4A/eIF4B combination in the unwinding of RNA duplexes in vitro (135), although the stimulatory effect of eIF4G on eIF4A activity was absent in that study. In addition, the ded1 mutation impairs the efficiency of scanning through a stem-loop inserted far downstream from the 5′ cap of a reporter mRNA (33). Thus, it appears that Ded1 is at least as important as eIF4A in promoting ribosomal scanning through long or structured 5′UTRs in yeast cells.
In the reconstituted mammalian system, it was found that that eIF4F, eIF4A, and eIF4B functioned poorly to stimulate scanning through highly stable stem-loops, of ΔG ≤−19 kcal/mol, inserted into a synthetic unstructured 5′UTR. This deficiency led Pestova and colleagues to purify another DEAD-box helicase, Dhx29, by its ability to function synergistically with the eIF4 proteins and stimulate 48S assembly in the presence of such strong stems (174). Purified yeast Ded1 could also function in this manner but less efficiently than Dhx29 for the most stable stems, whereas the human Ded1 homolog, Ddx3, was inactive in this assay (1). Interestingly, neither Dhx29 nor Ded1 could replace the eIF4 factors and support 48S PIC assembly on β-globin mRNA, which was attributed to their inability (in contrast to eIF4F) to stimulate 43S PIC attachment to an mRNA with a cap-proximal structure. Using synthetic mRNAs with unstructured cap-proximal sequences, for which 43S attachment can occur without eIF4 factors, and containing highly stable, cap-distal stems, Dxh29 and Ded1 each could support 48S assembly in the absence of eIF4 factors. Thus, it appeared that Dhx29 and Ded1 specifically stimulate scanning through secondary structures, whereas the eIF4 factors enhance both 43S attachment and scanning but are less effective than Dhx29 and Ded1 in promoting scanning through highly stable stems (1).
Interestingly, in the absence of Dhx29/Ded1, stem-loops that are too stable to be unwound by eIF4 factors alone were found to bypass the mRNA entry channel and become lodged in the exit channel of the scanning PIC. It appeared that mRNA continues to feed into the ribosome and loops out from the decoding center until an AUG enters the P site. This can lead to the formation of a stable 48S PIC but only in the absence of eIF1. It was proposed that a defective interaction with the exit channel upstream of the AUG, provoked by the stem-loop, was recognized as being aberrant and was destabilized by eIF1, even after the stem-loop had cleared the ribosome. This was likened to the role of eIF1 in destabilizing PICs with poor context or a very short 5′UTR (1). The bypass of the entry channel by a stable stem could be suppressed by the addition of Dhx29 or (to a lesser extent) Ded1, which presumably unwound the stem and fed it into the 40S entry channel in a single-stranded form. An interesting implication of these findings is that if a stem-loop is not removed by Dhx29 and bypasses the entry channel, eIF1 will be responsible for the subsequent inhibition of initiation exerted by this structure (Fig. 13). Hence, the downregulation of eIF1 could mitigate the inhibitory effects of strong stems in the 5′UTRs of mRNAs. A stem-loop located downstream of an AUG can also bypass the entry channel and occupy the A site in a 48S PIC arrested at the AUG codon, shifting the location of the toeprint to a position 11 to 12 nt from the 3′ base of the stem, i.e., the length of the entry channel. Dhx29 prevents this aberrant structure from forming as well and, interestingly, can destabilize it even when added to the reaction mixture after its formation (1).
Fig. 13.
Model depicting the bypass of a highly stable stem-loop by the scanning PIC in the presence of eIF4F but in the absence of helicase Dhx29. (A) A stem of moderate stability is unwound by eIF4F, eIF4A, and eIF4B and fed into the entry channel in a single-stranded form, producing a functional 48S PIC at the AUG. (B) A highly stable stem is not unwound effectively by the eIF4 factors alone, as this requires Dhx29 helicase function, bypasses the entry channel, and gets stuck in the exit channel. The mRNA spools into the decoding center, and a 48S PIC can be formed when AUG enters the P site provided that eIF1 is absent, as eIF1 destabilizes such aberrant complexes. (Reproduced from reference 1 by permission from Macmillan Publishers Ltd., copyright 2011.)
Remarkably, Dhx29 is found stably associated with 40S subunits and protects 18S rRNA residues at the top of h16 from chemical modification (174). This suggests that Dhx29 binds near the opening to the mRNA entry channel, consistent with a direct role in the unwinding of helices in advance of the scanning PIC. As noted above, Dhx29 can dissociate aberrant 48S PICs containing a strong stem-loop in the A site. Dhx29 can also destabilize aberrant complexes in which RT penetrates into the PIC 8 nt farther than normal, which has been attributed to an improper interaction of nucleotides 3′ of the AUG in the entry channel. Because Dhx29 can destabilize these preformed aberrant complexes, it was proposed that it can structurally remodel the PIC, rather than simply removing the mRNA secondary structure. Because Dhx29 is not a processive helicase, and its ATPase activity was stimulated more by the PIC than by RNA, it was speculated that Dhx29 destabilizes a stem-loop by stimulating the opening and closing of the mRNA entry channel, enabling single-stranded bases melted from the base of the stem to be captured in the entry channel. Because yeast Ded1 does not bind stably to the 40S subunit and could not function like Dhx29 in preventing strong stems located downstream of an AUG from bypassing the entry channel, it was suggested that Ded1 functions by a different, unknown mechanism (1).
The silencing of Dhx29 in cultured cells reduced the overall translation by ∼50% and produced an obvious reduction in the bulk polysome size, consistent with a decreased rate of initiation (157). Analyses of the polysome distributions of four different native mRNAs indicated that the initiation rate was impaired only for the two mRNAs with highly structured 5′UTRs. However, the substantial reduction in total protein synthesis evoked by Dhx29 seems to indicate that not merely a small fraction of mRNAs burdened with highly stable 5′UTR structures are dependent on this helicase for optimal translation initiation in cells.
PERSPECTIVES
The last decade has seen great progress in deciphering the mechanism of ribosomal scanning, with key contributions from the mammalian and yeast reconstituted systems, yeast genetics, and structural biology. Of course, many important questions remain to be answered. The model that eIF1 and eIF1A promote an open 40S conformation conducive to scanning but incompatible with subunit joining, and the importance of eIF1 and the eIF1A SE elements in achieving this conformation, appears robust, but how these factors accomplish this feat, and open the latch on the mRNA entry channel, is unknown. Biochemical studies should be pursued to test the prediction that the eIF1A CTT interacts directly with initiator tRNA or eIF1 to stabilize the scanning mode of TC binding, is ejected from the P site, and interacts with eIF5 upon AUG recognition. Exploiting the known Sui− substitutions in the SEs should facilitate the analysis of conformational rearrangements of the CTT and initiator tRNA in the P site between the open and closed conformations. It should also be examined whether the NTT and the helical domain of eIF1A interact with the entry channel latch, P site, or initiator tRNA in a manner affected by Ssu− substitutions in these SI elements, to elucidate their roles in the transition from open to closed 40S conformations. The identification of new Sui− and Ssu− substitutions in eIF1 and the characterization of them biochemically in the reconstituted system should help identify the eIF1 domains involved in stabilizing the scanning 40S conformation, impeding Pi release, destabilizing initiator tRNA binding, and detecting the AUG-Ac duplex, in addition to controlling its own dissociation from the 40S subunit upon AUG recognition. It is critical to determine whether specific residues in initiator tRNA have important functions in start codon recognition in vivo and if they interact with rRNA residues to stabilize initiator tRNA binding. It is intriguing to consider that the formation of the AUG-Ac duplex might stabilize a conformational change in the initiator tRNA, or evoke a conformational change in the rRNA, that helps to evict eIF1 from the platform, and mutational analysis of the 18S rRNA in yeast should aid in exploring this possibility. It would also be fascinating if a conformational change in the initiator tRNA or rRNA was transmitted to eIF2 to stimulate Pi release from the GDP·Pi bound to the G domain of eIF2γ.
If the eIF1A interaction with the eIF5 NTD contributes to ejecting eIF1 and allowing Pi release, as envisioned in Fig. 6, it should be possible to identify Ssu− and Sui− substitutions in eIF1A and eIF5 that impair and facilitate, respectively, interactions with one another or interactions of the eIF5 NTD with the 40S subunit. In parallel, biochemical experiments should proceed to map the putative interactions of the eIF5 NTD with the Pi-binding pocket in eIF2γ, eIF1A, and the 40S subunit. It is necessary to understand exactly how eIF2β blocks the GTPase activity of eIF2 in a manner that can be overcome by the eIF5 GAP domain and to determine if eIF5 also impedes Pi release in the scanning complex. Whether the eIF2 subunits in the TC interact directly with 18S rRNA residues or ribosomal proteins and if these interactions differ between the open and closed 40S conformations are other questions of obvious importance. Biochemical studies here would benefit from having additional (or even the first) Sui− or Ssu− substitutions in eIF2 subunits or different ribosomal components present in the decoding center. It also seems possible that eIF2 subunits directly influence the distinct modes of initiator tRNA binding predicted for the open and closed 40S conformations (Fig. 8). Whether eIF2 interacts directly with the mRNA, particularly at the −3 and +4 nucleotides, to regulate start codon recognition is important to address, as is the involvement of ribosomal components near the 40S exit site/channel in recognizing the optimum AUG context. Clearly, all of the outstanding issues regarding the mechanism of ribosomal scanning and AUG recognition would be illuminated dramatically by new high-resolution structures of PICs containing different combinations of factors or representing different stages of the process. Single-molecule studies with fluorescently tagged factors should also be very useful in detecting, and kinetically characterizing, rearrangements that occur in the PIC during scanning and upon AUG recognition.
It is now clear that eIF3 has a critical role in 43S-mRNA attachment, independent of bridging eIF4F and the 40S subunit, and there are genetic data implicating eIF3 in modulating the rate of scanning and the efficiency or accuracy of AUG recognition. There is much to be learned about eIF3's contacts with the 40S subunit and its role(s) in recruiting mRNA, regulating 40S conformational transitions, and opening or closing the mRNA channel openings on the solvent side of the 40S subunit. It is also important to determine whether eIF3's connections to eIF1, the eIF5 CTD, and the eIF2β NTD, which stabilize the MFC, remain intact in the 43S·mRNA complex and regulate the functions of these factors during scanning. The possible roles of ribosomal proteins and rRNA elements at the mRNA entry and exit pores in scanning must also be defined, and the GCN4 reporters that reveal reduced rates of scanning could be useful here. It is certain that other RNA helicases besides eIF4A function in translation initiation, although Dhx29 and Ded1 might be specialized to promote processive scanning through long or structured 5′UTRs, whereas the creation of a single-stranded binding site for the 43S subunit near the cap could depend primarily on eIF4F. The relative importance of the different helicases for 43S attachment versus scanning needs to be defined in vivo, and a genome-wide analysis of their requirements for the activation and translation of native mRNAs would be very enlightening. There is still much to learn about how the functions of the helicases are coordinated with the scanning PIC, including how Ded1 is recruited and how Dhx29 unwinds the secondary structure. It is also important to determine whether eIF4B and eIF4H have roles in 43S-mRNA attachment and scanning that are independent of their functions as eIF4A cofactors.
ACKNOWLEDGMENTS
I am grateful to past and present members of my laboratory, to my collaborators Tom Dever and Jon Lorsch, and to other colleagues in the field, particularly Tatyana Pestova, Nahum Sonenberg, and Richard Jackson, for the many insights and ideas that have emerged in discussions. In addition, the referees' comments were extremely valuable and led to significant improvements of this review.
REFERENCES
- 1. Abaeva I. S., Marintchev A., Pisareva V. P., Hellen C. U., Pestova T. V. 2011. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 30:115–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. 1991. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol. 11:486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Acker M. G., Shin B. S., Dever T. E., Lorsch J. R. 2006. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem. 281:8469–8475 [DOI] [PubMed] [Google Scholar]
- 4. Agris P. F. 2008. Bringing order to translation: the contributions of tRNA anticodon-domain modifications. EMBO Rep. 9:629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Algire M. A., Maag D., Lorsch J. R. 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20:251–262 [DOI] [PubMed] [Google Scholar]
- 6. Alone P. V., Cao C., Dever T. E. 2008. Translation initiation factor 2gamma mutant alters start codon selection independent of Met-tRNA binding. Mol. Cell. Biol. 28:6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alone P. V., Dever T. E. 2006. Direct binding of translation initiation factor eIF2gamma-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2B epsilon. J. Biol. Chem. 281:12636–12644 [DOI] [PubMed] [Google Scholar]
- 8. Altmann M., et al. 1993. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 12:3997–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andersen C. B., et al. 2006. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313:1968–1972 [DOI] [PubMed] [Google Scholar]
- 10. Asano K., Clayton J., Shalev A., Hinnebusch A. G. 2000. A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 14:2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Asano K., Kinzy T. G., Merrick W. C., Hershey J. W. B. 1997. Conservation and diversity of eukaryotic translation initiation factor eIF3. J. Biol. Chem. 272:1101–1109 [DOI] [PubMed] [Google Scholar]
- 12. Asano K., Krishnamoorthy T., Phan L., Pavitt G. D., Hinnebusch A. G. 1999. Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 18:1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Asano K., Phan L., Anderson J., Hinnebusch A. G. 1998. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:18573–18585 [DOI] [PubMed] [Google Scholar]
- 14. Asano K., et al. 2001. Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 20:2326–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Asano K., et al. 1997. Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits: possible roles in RNA binding and macromolecular assembly. J. Biol. Chem. 272:27042–27052 [DOI] [PubMed] [Google Scholar]
- 16. Astrom S. U., von Pawel-Rammingen U., Bystrom A. S. 1993. The yeast initiator tRNAMet can act as an elongator tRNAMet in vivo. J. Mol. Biol. 233:43–58 [DOI] [PubMed] [Google Scholar]
- 17. Baim S. B., Sherman F. 1988. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 8:1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Battiste J. B., Pestova T. V., Hellen C. U. T., Wagner G. 2000. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell 5:109–119 [DOI] [PubMed] [Google Scholar]
- 19. Battle D. J., Doudna J. A. 2002. Specificity of RNA-RNA helix recognition. Proc. Natl. Acad. Sci. U. S. A. 99:11676–11681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ben-Shem A., Jenner L., Yusupova G., Yusupov M. 2010. Crystal structure of the eukaryotic ribosome. Science 330:1203–1209 [DOI] [PubMed] [Google Scholar]
- 21. Berset C., Zurbriggen A., Djafarzadeh S., Altmann M., Trachsel H. 2003. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA 9:871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E. 2004. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol. 51:987–1001 [DOI] [PubMed] [Google Scholar]
- 23. Block K. L., Vornlocher H. P., Hershey J. W. B. 1998. Characterization of cDNAs encoding the p44 and p35 subunits of human translation initiation factor eIF3. J. Biol. Chem. 273:31901–31908 [DOI] [PubMed] [Google Scholar]
- 24. Blum S., et al. 1992. ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 89:7664–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Butler J. S., Springer M., Dondon J., Graffe M., Grunberg-Manago M. 1986. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J. Mol. Biol. 192:767–780 [DOI] [PubMed] [Google Scholar]
- 26. Butler J. S., Springer M., Grunberg-Manago M. 1987. AUU-to-AUG mutation in the initiator codon of the translation initiation factor IF3 abolishes translational autocontrol of its own gene (infC) in vivo. Proc. Natl. Acad. Sci. U. S. A. 84:4022–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carter A. P., et al. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291:498–501 [DOI] [PubMed] [Google Scholar]
- 28. Caruthers J. M., Johnson E. R., McKay D. B. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. U. S. A. 97:13080–13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Castilho-Valavicius B., Thompson G. M., Donahue T. F. 1992. Mutation analysis of the Cys-X2-Cys-X19-Cys-X2-Cys motif in the α subunit of eukaryotic translation factor 2. Gene Expr. 2:297–309 [PMC free article] [PubMed] [Google Scholar]
- 30. Chang C. P., Chen S. J., Lin C. H., Wang T. L., Wang C. C. 2010. A single sequence context cannot satisfy all non-AUG initiator codons in yeast. BMC Microbiol. 10:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen S. J., Lin G., Chang K. J., Yeh L. S., Wang C. C. 2008. Translational efficiency of a non-AUG initiation codon is significantly affected by its sequence context in yeast. J. Biol. Chem. 283:3173–3180 [DOI] [PubMed] [Google Scholar]
- 32. Cheung Y. N., et al. 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 21:1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chiu W. L., et al. 2010. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell. Biol. 30:4415–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chuang R. Y., Weaver P. L., Liu Z., Chang T. H. 1997. Requirement of the DEAD-box protein ded1p for mRNA translation. Science 275:1468–1471 [DOI] [PubMed] [Google Scholar]
- 35. Cigan A. M., Donahue T. F. 1987. Sequence and structural features associated with translational initiator regions in yeast—a review. Gene 59:1–13 [DOI] [PubMed] [Google Scholar]
- 36. Cigan A. M., Feng L., Donahue T. F. 1988. tRNAiMet functions in directing the scanning ribosome to the start site of translation. Science 242:93–97 [DOI] [PubMed] [Google Scholar]
- 37. Cigan A. M., Pabich E. K., Donahue T. F. 1988. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2964–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cigan A. M., Pabich E. K., Feng L., Donahue T. F. 1989. Yeast translation initiation suppressor sui2 encodes the α subunit of eukaryotic initiation factor 2 and shares identity with the human α subunit. Proc. Natl. Acad. Sci. U. S. A. 86:2784–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clements J. M., Laz T. M., Sherman F. 1988. Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4533–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coldwell M. J., Morley S. J. 2006. Specific isoforms of translation initiation factor 4GI show differences in translational activity. Mol. Cell. Biol. 26:8448–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Conte M. R., et al. 2006. Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry 45:4550–4558 [DOI] [PubMed] [Google Scholar]
- 42. Cuchalova L., et al. 2010. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol. 30:4671–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dallas A., Noller H. F. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8:855–864 [DOI] [PubMed] [Google Scholar]
- 44. Das S., Ghosh R., Maitra U. 2001. Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J. Biol. Chem. 276:6720–6726 [DOI] [PubMed] [Google Scholar]
- 45. Das S., Maitra U. 2000. Mutational analysis of mammalian translation initiation factor 5 (eIF5): role of interaction between the beta subunit of eIF2 and eIF5 in eIF5 function in vitro and in vivo. Mol. Cell. Biol. 20:3942–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Daugeron M. C., et al. 2011. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkr178. [DOI] [PMC free article] [PubMed]
- 47. de la Cruz J., Iost I., Kressler D., Linder P. 1997. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 94:5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Demeshkina N., Jenner L., Yusupova G., Yusupov M. 2010. Interactions of the ribosome with mRNA and tRNA. Curr. Opin. Struct. Biol. 20:325–332 [DOI] [PubMed] [Google Scholar]
- 49. Dennis P. P. 1997. Ancient ciphers: translation in archaea. Cell 89:1007–1010 [DOI] [PubMed] [Google Scholar]
- 50. Dmitriev S. E., Terenin I. M., Dunaevsky Y. E., Merrick W. C., Shatsky I. N. 2003. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol. Cell. Biol. 23:8925–8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Doherty E. A., Batey R. T., Masquida B., Doudna J. A. 2001. A universal mode of helix packing in RNA. Nat. Struct. Biol. 8:339–343 [DOI] [PubMed] [Google Scholar]
- 52. Dominguez D., Altmann M., Benz J., Baumann U., Trachsel H. 1999. Interaction of translation initiation factor eIF4G with eIF4A in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274:26720–26726 [DOI] [PubMed] [Google Scholar]
- 53. Dominguez D., Kislig E., Altmann M., Trachsel H. 2001. Structural and functional similarities between the central eukaryotic initiation factor (eIF)4A-binding domain of mammalian eIF4G and the eIF4A-binding domain of yeast eIF4G. Biochem. J. 355:223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Donahue T. 2000. Genetic approaches to translation initiation in Saccharomyces cerevisiae, p. 487–502. In Sonenberg N., Hershey J. W. B., Mathews M. B. (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 55. Donahue T. F., Cigan A. M., Pabich E. K., Castilho-Valavicius B. 1988. Mutations at a Zn(II) finger motif in the yeast elF-2β gene alter ribosomal start-site selection during the scanning process. Cell 54:621–632 [DOI] [PubMed] [Google Scholar]
- 56. Dong J., et al. 2008. Genetic identification of yeast 18S rRNA residues required for efficient recruitment of initiator tRNA(Met) and AUG selection. Genes Dev. 22:2242–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dorris D. R., Erickson F. L., Hannig E. M. 1995. Mutations in GCD11, the structural gene for eIF-2γ in yeast, alter translational regulation of GCN4 and the selection of the start site for protein synthesis. EMBO J. 14:2239–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Doudna J. A., Sarnow P. 2007. Translation initiation by viral internal ribosome entry sites, p. 129–154. In Mathews M., Sonenberg N., Hershey J. W. B. (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 59. Drabkin H. J., Helk B., RajBhandary U. L. 1993. The role of nucleotides conserved in eukaryotic initiator methionine tRNAs in initiation of protein synthesis. J. Biol. Chem. 268:25221–25228 [PubMed] [Google Scholar]
- 60. Drabkin H. J., RajBhandary U. L. 1998. Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol. Cell. Biol. 18:5140–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Elantak L., Tzakos A. G., Locker N., Lukavsky P. J. 2007. Structure of eIF3b-RNA and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40S ribosomal subunit. J. Biol. Chem. 282:8165–8174 [DOI] [PubMed] [Google Scholar]
- 62. Elantak L., et al. 2010. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J. Mol. Biol. 396:1097–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ellis S. R., Hopper A. K., Martin N. C. 1987. Amino-terminal extension generated from an upstream AUG codon is not required for mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 84:5172–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. El Yacoubi B., et al. 2011. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 30:882–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. El Yacoubi B., et al. 2009. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 37:2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Erickson F. L., Hannig E. M. 1996. Ligand interactions with eukaryotic translation initiation factor 2: role of the γ-subunit. EMBO J. 15:6311–6320 [PMC free article] [PubMed] [Google Scholar]
- 67. Evans D. R. H., et al. 1995. Mutational analysis of the Prt1 protein subunit of yeast translation initiation factor 3. Mol. Cell. Biol. 15:4525–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fechter P., et al. 2009. Ribosomal initiation complexes probed by toeprinting and effect of trans-acting translational regulators in bacteria. Methods Mol. Biol. 540:247–263 [DOI] [PubMed] [Google Scholar]
- 69. Fekete C. A., et al. 2005. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J. 24:3588–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fekete C. A., et al. 2007. N-and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO 26:1602–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fletcher C. M., Pestova T. V., Hellen C. U. T., Wagner G. 1999. Structure and interactions of the translation initiation factor eIF1. EMBO J. 18:2631–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fraser C. S., Berry K. E., Hershey J. W., Doudna J. A. 2007. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol. Cell 26:811–819 [DOI] [PubMed] [Google Scholar]
- 73. Fraser C. S., et al. 2004. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J. Biol. Chem. 279:8946–8956 [DOI] [PubMed] [Google Scholar]
- 74. Gaspar N. J., et al. 1994. Translation initiation factor eIF-2. Cloning and expression of the human cDNA encoding the γ-subunit. J. Biol. Chem. 269:3415–3422 [PubMed] [Google Scholar]
- 75. Hanachi P., Hershey J. W. B., Vornlocher H. P. 1999. Characterization of the p33 subunit of eukaryotic translation initiation factor-3 from Saccharomyces cerevisiae. J. Biol. Chem. 274:8546–8553 [DOI] [PubMed] [Google Scholar]
- 76. Hannig E. M., Cigan A. M., Freeman B. A., Kinzy T. G. 1992. GCD11, a negative regulator of GCN4 expression, encodes the γ subunit of eIF-2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Harashima S., Hinnebusch A. G. 1986. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 6:3990–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Harding H. P., et al. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099–1108 [DOI] [PubMed] [Google Scholar]
- 79. Hartz D., Binkley J., Hollingsworth T., Gold L. 1990. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev. 4:1790–1800 [DOI] [PubMed] [Google Scholar]
- 80. Hartz D., McPheeters D. S., Traut R., Gold L. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164:419–425 [DOI] [PubMed] [Google Scholar]
- 81. Hashimoto N. N., Carnevalli L. S., Castilho B. A. 2002. Translation initiation at non-AUG codons mediated by weakened association of eukaryotic initiation factor (eIF) 2 subunits. Biochem. J. 367(Pt. 2):359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hellen C. U. 2009. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta 1789:558–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hilbert M., Kebbel F., Gubaev A., Klostermeier D. 2011. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 39:2260–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hinnebusch A. G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59:407–450 [DOI] [PubMed] [Google Scholar]
- 85. Hinnebusch A. G., Dever T. E., Asano K. 2007. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae, p. 225–268. In Mathews M., Sonenberg N., Hershey J. W. B. (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 86. Huang H., Yoon H., Hannig E. M., Donahue T. F. 1997. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 11:2396–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Imataka H., Sonenberg N. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17:6940–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Iost I., Dreyfus M., Linder P. 1999. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274:17677–17683 [DOI] [PubMed] [Google Scholar]
- 90. Ito T., Marintchev A., Wagner G. 2004. Solution structure of human initiation factor eIF2alpha reveals homology to the elongation factor eEF1B. Structure 12:1693–1704 [DOI] [PubMed] [Google Scholar]
- 91. Ivanov I. P., Loughran G., Sachs M. S., Atkins J. F. 2010. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc. Natl. Acad. Sci. U. S. A. 107:18056–18060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jackson R. J., Kaminski A., Poyry T. A. A. 2007. Coupled termination-reinitiation events in mRNA translation, p. 197–224. In Mathews M., Sonenberg N., Hershey J. W. B. (ed.), Translational control in biology and medicine Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 93. Jamieson D. J., Beggs J. D. 1991. A suppressor of yeast spp81/ded1 mutations encodes a very similar putative ATP-dependent RNA helicase. Mol. Microbiol. 5:805–812 [DOI] [PubMed] [Google Scholar]
- 94. Jivotovskaya A. V., Valasek L., Hinnebusch A. G., Nielsen K. H. 2006. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 26:1355–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kapp L. D., Kolitz S. E., Lorsch J. R. 2006. Yeast initiator tRNA identity elements cooperate to influence multiple steps of translation initiation. RNA 12:751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kapp L. D., Lorsch J. R. 2004. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 335:923–936 [DOI] [PubMed] [Google Scholar]
- 97. Kaye N. M., Emmett K. J., Merrick W. C., Jankowsky E. 2009. Intrinsic RNA binding by the eukaryotic initiation factor 4F depends on a minimal RNA length but not on the m7G cap. J. Biol. Chem. 284:17742–17750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kieft J. S. 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 33:274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kolitz S. E., Takacs J. E., Lorsch J. R. 2009. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA 15:138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kolupaeva V. G., Unbehaun A., Lomakin I. B., Hellen C. U., Pestova T. V. 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11:470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Korneeva N. L., Lamphear B. J., Hennigan F. L., Rhoads R. E. 2000. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem. 275:41369–41376 [DOI] [PubMed] [Google Scholar]
- 102. Korostelev A., Trakhanov S., Laurberg M., Noller H. F. 2006. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126:1065–1077 [DOI] [PubMed] [Google Scholar]
- 103. Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125–8149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kozak M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 9:5134–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kozak M. 1990. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U. S. A. 87:8301–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kozak M. 1987. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 7:3438–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kozak M. 1991. Effects of long 5′ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr. 1:117–125 [PMC free article] [PubMed] [Google Scholar]
- 108. Kozak M. 1978. How do eucaryotic ribosomes select initiation regions in mRNA? Cell 15:1109–1123 [DOI] [PubMed] [Google Scholar]
- 109. Kozak M. 1979. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature 280:82–85 [DOI] [PubMed] [Google Scholar]
- 110. Kozak M. 1986. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U. S. A. 83:2850–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kozak M. 1977. Nucleotide sequences of 5′-terminal ribosome-protected initiation regions from two reovirus messages. Nature 269:391–394 [DOI] [PubMed] [Google Scholar]
- 112. Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283–292 [DOI] [PubMed] [Google Scholar]
- 113. Kozak M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kozak M. 1984. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 12:3873–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kozak M. 1991. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1:111–115 [PMC free article] [PubMed] [Google Scholar]
- 116. Kozak M. 1983. Translation of insulin-related polypeptides from messenger RNAs with tandemly reiterated copies of the ribosome binding site. Cell 34:971–978 [DOI] [PubMed] [Google Scholar]
- 117. Kozak M., Shatkin A. J. 1978. Migration of 40S ribosomal subunit on mRNA in the presence of edeine. J. Biol. Chem. 253:6568–6577 [PubMed] [Google Scholar]
- 118. Kozak M., Shatkin A. J. 1977. Sequences and properties of two ribosome binding sites from the small size class of reovirus mRNA. J. Biol. Chem. 252:6895–6908 [PubMed] [Google Scholar]
- 119. Lamphear B. J., Kirchweger R., Skern T., Rhoads R. E. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J. Biol. Chem. 270:21975–21983 [DOI] [PubMed] [Google Scholar]
- 120. Lancaster L., Noller H. F. 2005. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell 20:623–632 [DOI] [PubMed] [Google Scholar]
- 121. Laurino J. P., Thompson G. M., Pacheco E., Castilho B. A. 1999. The β subunit of eukaryotic translation initiation factor 2 binds mRNA through the lysine repeats and a region comprising the C2-C2 motif. Mol. Cell. Biol. 19:173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lawless C., et al. 2009. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics 10:7–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lazarowitz S. G., Robertson H. D. 1977. Initiator regions from the small size class of reovirus mRNA protected by rabbit reticulocyte ribosomes. J. Biol. Chem. 252:7842–7849 [PubMed] [Google Scholar]
- 124. LeFebvre A. K., et al. 2006. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 281:22917–22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Lin C. A., Ellis S. R., True H. L. 2009. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell. Biol. 30:354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Liu F., Putnam A., Jankowsky E. 2008. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. U. S. A. 105:20209–20214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lomakin I. B., Kolupaeva V. G., Marintchev A., Wagner G., Pestova T. V. 2003. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 17:2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lomakin I. B., Shirokikh N. E., Yusupov M. M., Hellen C. U., Pestova T. V. 2006. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 25:196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Londei P. 2005. Evolution of translational initiation: new insights from the archaea. FEMS Microbiol. Rev. 29:185–200 [DOI] [PubMed] [Google Scholar]
- 130. Lorsch J. R., Herschlag D. 1998. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry 37:2180–2193 [DOI] [PubMed] [Google Scholar]
- 131. Maag D., Algire M. A., Lorsch J. R. 2006. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 356:724–737 [DOI] [PubMed] [Google Scholar]
- 132. Maag D., Fekete C. A., Gryczynski Z., Lorsch J. R. 2005. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell 17:265–275 [DOI] [PubMed] [Google Scholar]
- 133. Marck C., Grosjean H. 2002. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8:1189–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Marintchev A., et al. 2009. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136:447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Marsden S., Nardelli M., Linder P., McCarthy J. E. 2006. Unwinding single RNA molecules using helicases involved in eukaryotic translation initiation. J. Mol. Biol. 361:327–335 [DOI] [PubMed] [Google Scholar]
- 136. Matsuda D., Dreher T. W. 2006. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA 12:1338–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Methot N., Pickett G., Keene J. D., Sonenberg N. 1996. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA remotif. RNA 2:38–50 [PMC free article] [PubMed] [Google Scholar]
- 138. Milon P., Konevega A. L., Gualerzi C. O., Rodnina M. V. 2008. Kinetic checkpoint at a late step in translation initiation. Mol. Cell 30:712–720 [DOI] [PubMed] [Google Scholar]
- 139. Mitchell S. F., et al. 2010. the 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and block an alternative pathway. Mol. Cell 39:950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Miyasaka H., Endo S., Shimizu H. 2010. Eukaryotic translation initiation factor 1 (eIF1), the inspector of good AUG context for translation initiation, has an extremely bad AUG context. J. Biosci. Bioeng. 109:635–637 [DOI] [PubMed] [Google Scholar]
- 141. Montpetit B., et al. 2011. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472:238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Morino S., Imataka H., Svitkin Y. V., Pestova T. V., Sonenberg N. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20:468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Na J. G., Pinto I., Hampsey M. 1992. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131:791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Najarian D., Dihanich M. E., Martin N. C., Hopper A. K. 1987. DNA sequence and transcript mapping of MOD5: features of the 5′ region which suggest two translational starts. Mol. Cell. Biol. 7:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Nanda J. S., et al. 2009. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 394:268–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Naveau M., Lazennec-Schurdevin C., Panvert M., Mechulam Y., Schmitt E. 2010. tRNA binding properties of eukaryotic translation initiation factor 2 from Encephalitozoon cuniculi. Biochemistry 49:8680–8688 [DOI] [PubMed] [Google Scholar]
- 147. Neff C. L., Sachs A. B. 1999. Eukaryotic translation initiation factors eIF4G and eIF4A from Saccharomyces cerevisiae physically and functionally interact. Mol. Cell. Biol. 19:5557–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Nemoto N., et al. 2010. Yeast 18 S rRNA is directly involved in the ribosomal response to stringent AUG selection during translation initiation. J. Biol. Chem. 285:32200–32212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Nielsen K. H., et al. 2011. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res. 39:2678–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Nielsen K. H., Valasek L., Sykes C., Jivotovskaya A., Hinnebusch A. G. 2006. Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast. Mol. Cell. Biol. 26:2984–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Nika J., Rippel S., Hannig E. M. 2001. Biochemical analysis of the eIF2beta gamma complex reveals a structural function for eIF2alpha in catalyzed nucleotide exchange. J. Biol. Chem. 276:1051–1056 [DOI] [PubMed] [Google Scholar]
- 152. Nikonov O., et al. 2007. New insights into the interactions of the translation initiation factor 2 from archaea with guanine nucleotides and initiator tRNA. J. Mol. Biol. 373:328–336 [DOI] [PubMed] [Google Scholar]
- 152a. Noble K. N., et al. 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 25:1065–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Nonato M. C., Widom J., Clardy J. 2002. Crystal structure of the N-terminal segment of human eukaryotic translation initiation factor 2alpha. J. Biol. Chem. 277:17057–17061 [DOI] [PubMed] [Google Scholar]
- 154. Oberer M., Marintchev A., Wagner G. 2005. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 19:2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Park E., et al. 2011. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1 PABP mRNPs in vivo. EMBO J. 30:302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Park E. H., Zhang F., Warringer J., Sunnerhagen P., Hinnebusch A. G. 2011. Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics 12:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Parsyan A., et al. 2009. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 106:22217–22222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Parsyan A., et al. 2011. mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 12:235–245 [DOI] [PubMed] [Google Scholar]
- 159. Passmore L. A., et al. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell 26:41–50 [DOI] [PubMed] [Google Scholar]
- 160. Paulin F. E., Campbell L. E., O'Brien K., Loughlin J., Proud C. G. 2001. Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase-activator protein. Curr. Biol. 11:55–59 [DOI] [PubMed] [Google Scholar]
- 161. Pause A., Methot N., Svitkin Y., Merrick W. C., Sonenberg N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Peabody D. S. 1989. Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 264:5031–5035 [PubMed] [Google Scholar]
- 163. Pedulla N., et al. 2005. The archaeal eIF2 homologue: functional properties of an ancient translation initiation factor. Nucleic Acids Res. 33:1804–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Pelletier J., Sonenberg N. 1985. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40:515–526 [DOI] [PubMed] [Google Scholar]
- 165. Pestova T. V., Borukhov S. I., Hellen C. U. T. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394:854–859 [DOI] [PubMed] [Google Scholar]
- 166. Pestova T. V., Hellen C. U. T., Shatsky I. V. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Pestova T. V., Kolupaeva V. G. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16:2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Pestova T. V., Lorsch J. R., Hellen C. U. T. 2007. The mechanism of translation initiation in eukaryotes, p. 87–128. In Mathews M., Sonenberg N., Hershey J. W. B. (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 169. Phan L., Schoenfeld L. W., Valášek L., Nielsen K. H., Hinnebusch A. G. 2001. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNAiMet. EMBO J. 20:2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Phan L., et al. 1998. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol. 18:4935–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Pinto I., Na J. G., Sherman F., Hampsey M. 1992. cis- and trans-acting suppressors of a translation initiation defect at the cyc1 locus of Saccharomyces cerevisiae. Genetics 132:97–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Pisarev A. V., et al. 2006. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 20:624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Pisarev A. V., Kolupaeva V. G., Yusupov M. M., Hellen C. U., Pestova T. V. 2008. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 27:1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Pisareva V. P., Pisarev A. V., Komar A. A., Hellen C. U., Pestova T. V. 2008. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135:1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Poyry T. A., Kaminski A., Jackson R. J. 2004. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 18:62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Prevot D., et al. 2003. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 22:1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Qin D., Fredrick K. 2009. Control of translation initiation involves a factor-induced rearrangement of helix 44 of 16S rRNA. Mol. Microbiol. 71:1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Rabl J., Leibundgut M., Ataide S. F., Haag A., Ban N. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331:730–736 [DOI] [PubMed] [Google Scholar]
- 179. Ramirez-Valle F., Braunstein S., Zavadil J., Formenti S. C., Schneider R. J. 2008. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 181:293–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Reibarkh M., et al. 2008. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J. Biol. Chem. 283:1094–1103 [DOI] [PubMed] [Google Scholar]
- 181. Rogers G. W., Jr., Komar A. A., Merrick W. C. 2002. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 72:307–331 [DOI] [PubMed] [Google Scholar]
- 182. Rozovsky N., Butterworth A. C., Moore M. J. 2008. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA 14:2136–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Saini A. K., Nanda J. S., Lorsch J. R., Hinnebusch A. G. 2010. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i) (Met) binding to the ribosome. Genes Dev. 24:97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Scheffzek K., Ahmadian M. R., Wittinghofer A. 1998. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci. 23:257–262 [DOI] [PubMed] [Google Scholar]
- 185. Schmitt E., Blanquet S., Mechulam Y. 2002. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 21:1821–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Schmitt E., Naveau M., Mechulam Y. 2010. Eukaryotic and archaeal translation initiation factor 2: a heterotrimeric tRNA carrier. FEBS Lett. 584:405–412 [DOI] [PubMed] [Google Scholar]
- 187. Schutz P., et al. 2008. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 105:9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Selmer M., et al. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313:1935–1942 [DOI] [PubMed] [Google Scholar]
- 189. Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S. 2006. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125:287–300 [DOI] [PubMed] [Google Scholar]
- 190. Shabalina S. A., Ogurtsov A. Y., Rogozin I. B., Koonin E. V., Lipman D. J. 2004. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Res. 32:1774–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Shahbazian D., et al. 2010. Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol. Cell. Biol. 30:1478–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Sherman F., Stewart J. W. 1982. Mutations altering initiation of translation of yeast iso-1-cytochrome c; contrasts between the eukaryotic and prokaryotic initiation process, p. 301–334. In Strathern J. N., Jones E. W., Broach J. R. (ed.), The molecular biology of the yeast Saccharomyces metabolism and gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 193. Sherman F., Stewart J. W., Schweingruber A. M. 1980. Mutants of yeast initiating translation of iso-1-cytochrome c within a region spanning 37 nucleotides. Cell 20:215–222 [DOI] [PubMed] [Google Scholar]
- 194. Siridechadilok B., Fraser C. S., Hall R. J., Doudna J. A., Nogales E. 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310:1513–1515 [DOI] [PubMed] [Google Scholar]
- 195. Slusher L. B., Gillman E. C., Martin N. C., Hopper A. K. 1991. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc. Natl. Acad. Sci. U. S. A. 88:9789–9793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Sokabe M., Yao M., Sakai N., Toya S., Tanaka I. 2006. Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region. Proc. Natl. Acad. Sci. U. S. A. 103:13016–13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Spahn C. M., et al. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA ribosome and subunit-subunit interactions. Cell 107:373–386 [DOI] [PubMed] [Google Scholar]
- 198. Spahn C. M., et al. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959–1962 [DOI] [PubMed] [Google Scholar]
- 199. Spirin A. S. 2009. How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian ratchet model. Biochemistry 48:10688–1069219835415 [Google Scholar]
- 200. Srinivasan M., et al. 2011. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 30:873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Stewart J. W., Sherman F., Shipman N. A., Jackson M. 1971. Identification and mutational relocation of the AUG codon initiating translation of iso-1-cytochrome c in yeast. J. Biol. Chem. 246:7429–7445 [PubMed] [Google Scholar]
- 202. Stiles J. I., et al. 1981. DNA sequence of a mutation in the leader region of the yeast iso-1-cytochrome c mRNA. Cell 25:277–284 [DOI] [PubMed] [Google Scholar]
- 203. Stolboushkina E., et al. 2008. Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the alpha- and beta-subunits. J. Mol. Biol. 382:680–691 [DOI] [PubMed] [Google Scholar]
- 204. Svitkin Y., et al. 2001. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7:382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Svitkin Y. V., et al. 2009. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J. 28:58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Szamecz B., et al. 2008. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 22:2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Tarun S. Z., Wells S. E., Deardorff J. A., Sachs A. B. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. U. S. A. 94:9046–9051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Taylor D. J., et al. 2009. Comprehensive molecular structure of the eukaryotic ribosome. Structure 17:1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Trachsel H. 1996. Binding of initiator methionyl-tRNA to ribosomes, p. 113–138. In Hershey J. W. B., Mathews M.B., Sonenberg N. (ed.), Translational control. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 210. Unbehaun A., Borukhov S. I., Hellen C. U., Pestova T. V. 2004. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 18:3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Valášek L., et al. 2003. The yeast eIF3 subunits TIF32/a and NIP1/c and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 17:786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Valasek L., Nielsen K. H., Zhang F., Fekete C. A., Hinnebusch A. G. 2004. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 24:9437–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Valášek L., Phan L., Schoenfeld L. W., Valásková V., Hinnebusch A. G. 2001. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 20:891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. van den Heuvel J. J., Bergkamp R. J. M., Planta R. J., Raue H. A. 1989. Effect of deletions in the 5′-noncoding region on the translational efficiency of phosphoglycerate kinase mRNA in yeast. Gene 79:83–95 [DOI] [PubMed] [Google Scholar]
- 215. Vattem K. M., Wek R. C. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 101:11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. von Pawel-Rammingen U., Åström S., Byström A. S. 1992. Mutational analysis of conserved positions potentially important for initiator tRNA function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:1432–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Watanabe R., et al. 2010. The eukaryotic initiation factor (eIF) 4G HEAT domain promotes translation re-initiation in yeast both dependent on and independent of eIF4A mRNA helicase. J. Biol. Chem. 285:21922–21933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Werner M., Feller A., Messenguy F., Pierard A. 1987. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell 49:805–813 [DOI] [PubMed] [Google Scholar]
- 219. Westermann P., Nygard O. 1984. Cross-linking of mRNA to initiation factor eIF-3, 24 kDa cap binding protein and ribosomal proteins S1, S3/3a, S6 and S11 within the 48S pre-initiation complex. Nucleic Acids Res. 12:8887–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Wolfe C. L., Lou Y. C., Hopper A. K., Martin N. C. 1994. Interplay of heterogeneous transcriptional start sites and translational selection of AUGs dictate the production of mitochondrial and cytosolic/nuclear tRNA nucleotidyltransferase from the same gene in yeast. J. Biol. Chem. 269:13361–13366 [PubMed] [Google Scholar]
- 221. Yanagiya A., et al. 2009. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol. Cell. Biol. 29:1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Yang Q., Jankowsky E. 2005. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry 44:13591–13601 [DOI] [PubMed] [Google Scholar]
- 223. Yatime L., Mechulam Y., Blanquet S., Schmitt E. 2006. Structural switch of the gamma subunit in an archaeal aIF2 alpha gamma heterodimer. Structure 14:119–128 [DOI] [PubMed] [Google Scholar]
- 224. Yatime L., Mechulam Y., Blanquet S., Schmitt E. 2007. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc. Natl. Acad. Sci. U. S. A. 104:18445–18450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Yatime L., Schmitt E., Blanquet S., Mechulam Y. 2004. Functional molecular mapping of archaeal translation initiation factor 2. J. Biol. Chem. 279:15984–15993 [DOI] [PubMed] [Google Scholar]
- 226. Yoon H. J., Donahue T. F. 1992. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol. Cell. Biol. 12:248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Yu Y., Abaeva I. S., Marintchev A., Pestova T. V., Hellen C. U. 2011. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 39:4851–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Yu Y., et al. 2009. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 37:5167–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Yun D. F., Laz T. M., Clements J. M., Sherman F. 1996. mRNA sequences influencing translation and the selection of AUG initiator codons in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 19:1225–1239 [DOI] [PubMed] [Google Scholar]
- 230. Yusupova G. Z., Yusupov M. M., Cate J. H., Noller H. F. 2001. The path of mRNA through the ribosome. Cell 106:233–241 [DOI] [PubMed] [Google Scholar]
- 231. Zhang F., Hinnebusch A. G. 2011. An upstream ORF with non-AUG start codon is translated in vivo but dispensable for translational control of GCN4 mRNA. Nucleic Acids Res. 39:68–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Zhou D., et al. 2008. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 283:7064–7073 [DOI] [PubMed] [Google Scholar]