Abstract
Summary: The interactions and processes which structure prokaryotic cytoplasm (water, ions, metabolites, and biomacromolecules) and ensure the fidelity of the cell cycle are reviewed from a physicochemical perspective. Recent spectroscopic and biological evidence shows that water has no active structuring role in the cytoplasm, an unnecessary notion still entertained in the literature; water acts only as a normal solvent and biochemical reactant. Subcellular structuring arises from localizations and interactions of biomacromolecules and from the growth and modifications of their surfaces by catalytic reactions. Biomacromolecular crowding is a fundamental physicochemical characteristic of cells in vivo. Though some biochemical and physiological effects of crowding (excluded volume effect) have been documented, crowding assays with polyglycols, dextrans, etc., do not properly mimic the compositional variety of biomacromolecules in vivo. In vitro crowding assays are now being designed with proteins, which better reflect biomacromolecular environments in vivo, allowing for hydrophobic bonding and screened electrostatic interactions. I elaborate further the concept of complex vectorial biochemistry, where crowded biomacromolecules structure the cytosol into electrolyte pathways and nanopools that electrochemically “wire” the cell. Noncovalent attractions between biomacromolecules transiently supercrowd biomacromolecules into vectorial, semiconducting multiplexes with a high (35 to 95%)-volume fraction of biomacromolecules; consequently, reservoirs of less crowded cytosol appear in order to maintain the experimental average crowding of ∼25% volume fraction. This nonuniform crowding model allows for fast diffusion of biomacromolecules in the uncrowded cytosolic reservoirs, while the supercrowded vectorial multiplexes conserve the remarkable repeatability of the cell cycle by preventing confusing cross talk of concurrent biochemical reactions.
Are the smallest particles of living matter which still exhibit all its functions of the order of magnitude of molecules and atoms, or are they of different order? The first step toward an answer to this question was accomplished by Moritz Nussbaum, who found that if an Infusorian be divided into two pieces, one with and one without a nucleus, only the [former] will continue to live and perform all the functions of self-preservation and development which are characteristic of living organisms. This shows that not only more than two definite substances, but two different structural elements, are needed for life.
—Jacques Loeb, 1906 (122)
INTRODUCTION
Even though phylogenetics (155, 185, 248–254, 268) has resolved prokaryotes in the domains Bacteria and Archaea as the most deeply rooted group of organisms in the evolutionary tree, we have a surprisingly rudimentary understanding of physicochemical processes essential for these organisms' cellular life. The simplest model of a prokaryotic cell has been described as a semipermeable “bag” that encloses catalytic reaction systems of diffusing metabolites and biomacromolecules (13, 139, 140, 146, 189, 234, 243). Though the bag itself has been known to be a highly structured entity—the “fluid mosaic” of integral membrane proteins energized by the membrane potential (146, 199) and supported by the cell wall and extracytoplasmic layers (196)—the contents of the bag showed only the chemically distinct nucleoid (177, 228) and the ribosomes (198) in an unstructured cytoplasm when electron microscopy became established (35, 188). Because cells in vivo are crowded with biomacromolecules, typically about a 20 to 30% volume fraction (77, 78, 265), the bag model has been challenged by concepts of higher-order structural elements derived from biochemical and physiological functions. For example, “metabolons” and “quinary” protein structures have been suggested to channel metabolites directly from one catalytic site to another (135, 179, 207, 208, 242), and a number of “hyperstructures” (154) have been enumerated to exemplify the concept of modular cytology (91). Such structural and functional elements are intuitively appealing because they could represent higher-order building blocks that make up a cell. However, they raise new questions, for example, which physicochemical mechanisms assemble and localize such larger modules, and how do they interact and communicate with each other during the cell cycle?
In the last 15 years or so, microbiologists have demonstrated experimentally that bacterial proteins and nucleic acids become localized temporarily at specific cytoplasmic “addresses” (17, 106, 125, 191, 192), where they enable biochemical and physiological functions such as transcription, translation, plasmid and chromosome replication and segregation, and septum formation during cell division and sporulation (6, 31–34, 36, 53–55, 60, 71–73, 80, 120, 121, 127, 130, 134, 137, 138, 150, 152, 153, 183, 195, 220–223, 225, 230, 235, 256). In particular, the recent discoveries of bacterial cytoskeleton genes and their protein products in the cells of Bacillus subtilis, Escherichia coli, Caulobacter crescentus, and Prosthecobacter dejongeii have changed our view of the bacterial cell from a bag of enzymes to a heterogeneous but well-organized assembly of dynamic biomacromolecules (17, 103, 106, 112, 126, 187). The newly discovered cytoskeleton proteins were found to assist in the remodeling of the cell envelope and in the replication and segregation of the nucleoid, and they establish the polarity of the cell during the cell cycle; some of them tend to localize underneath the plasma membrane, sometimes in a dynamic helical fashion (33, 106). They are functionally similar to the well-known cytoskeleton proteins within larger eukaryotic cells (153, 173, 212, 224, 227), though prokaryotic cytoskeletal proteins need not shape and localize organelles—one of the important functions of the eukaryotic cytoskeleton.
It now appears that all cells, and possibly all organelles, have structuring proteins of various kinds, often peripherally associated with or penetrating into the inner (cytoplasmic) sheet of the phospholipid bilayer and contributing to the overall mechanical strength and shape of a cell or organelle. Some examples of such eukaryotic skeletal and scaffolding proteins are actins and myosins in the cytoplasm of contractile cells (99, 173, 212), actins and lamins in the eukaryotic nucleus (16, 75, 214), articulins of the single-celled eukaryote Euglena gracilis (132), reticulons of peripheral endoplasmic reticulum (236, 237), various spectrins underneath the plasma membrane in many kinds of cells (particularly erythrocytes and neurons) (9, 12), golgins of the Golgi apparatus (197), and a new filament-forming protein that localizes in the intermembrane space of mitochondria and is related to bacterial penicillin-binding proteins and hence to the biogenesis of the bacterial cell wall (167). In addition, homologs of the tubulin-like bacterial protein FtsZ play a role in chloroplast division (260). As a common feature, the prokaryotic and eukaryotic cytoskeleton proteins utilize ionic ATP and/or GTP, which points to electrostatic interactions participating in active (energy-requiring) mechanisms to effect structuring functions (28, 66); for example, ATP-binding sites are required for proper cell division (5). Thus, at the subcellular and suborganelle levels (∼50 to 500 nm) of crowded and interacting biomacromolecules, the distinction between prokaryotic and eukaryotic cells is beginning to be erased (60, 120, 125, 153).
The discovery of cytoskeleton proteins within bacterial cells underscores the view that a prokaryotic cell is a self-structuring dynamic device whose “engineering design” has yet to be uncovered (1). The basic question then is this: how do biomacromolecules know where and when to go as the cell grows, changes its shape, and eventually divides? Or, in physicochemical terms, what are the structuring mechanisms that guide water molecules, ions, small metabolites, and larger (slower) biomacromolecules and their assemblies (biomacromolecular machines) as they are being synthesized and localized within the growing volume of the cell? Clearly, a complex spatial and temporal hierarchy of biochemical reactions and noncovalent interactions operates in conjunction with physicochemical gradients across the cell envelope. For prokaryotic cells, the spatial and temporal hierarchy spans over 9 orders of magnitude of time (picoseconds to seconds) and over 3 orders of magnitude of size (nanometers to micrometers). These timescales and size ranges are related to the motions of molecules, ions, biomacromolecules, and their large biochemical complexes and structures, e.g., from the picosecond persistence of hydrogen bonds to the microsecond and millisecond timescales of protein conformational changes, enzymatic reactions, and protein diffusion and to the morphological and motile motions of the whole cell, which occur on timescales of seconds and minutes.
Partly from a historical perspective, I first review the roles of water, ions, and biomacromolecular crowding in subcellular structuring, with an emphasis on classical noncovalent inter- actions of physical chemistry, such as hydrogen bonding, screened electrostatic interactions, hydrophobic bonding, and excluded volume. On account of the attractive noncovalent interactions, I then theorize that biomacromolecules become transiently supercrowded (∼35 to 95% volume fraction) into vectorial, electrolytically semiconducting multiplexes that coexist with reservoirs of less crowded cytosol. Some experimental observations in support of this model are discussed, with implications for further work.
WATER IN SUBCELLULAR STRUCTURING
A Quick Look Back
Physicochemical properties of water have long been recognized as essential for the origin of living systems (215) and for the evolution of cellular organisms, as Henderson argued already in 1913 (10, 38, 39, 64, 92). While Henderson realized the importance of specific properties of pure water, such as its high heat capacity and density, it is less appreciated that he equally stressed the importance of electrolyte buffers (hence the Henderson-Hasselbalch buffer equation) and of the high content of salts in seawater. Historically, Henderson's life-supporting properties of seawater (a multi-ionic electrolyte) are related to the triggering of parthenogenesis of sea urchin eggs by treatments with a specific mixture of sodium and potassium salts (122, 123, 162); this discovery foreshadowed the existence of ion-selective pumps in cellular membranes (79).
These early advances in the recognition of the role of water and ions in life phenomena were made with a still imperfect understanding of the differences between weak electrolytes (described by dissociation constants) and strong electrolytes, to which dissociation constant formalism does not apply (233); the highly nonideal behavior of strong electrolytes was understood in the early 1920s, within the framework of the interionic attraction theory based on the Poisson-Boltzmann equation (43, 178; L. Pauling, personal communication). At that time, some colloids, particularly natural rubber latex, cellulose, and proteins, also became understood as very large, covalently bonded single molecules that gave rise to various aqueous (and nonaqueous) gels and to polymer chemistry as a distinct chemical discipline (209, 216, 219). Until then, such soluble polymers were regarded as associated colloids made up of low-molecular-weight molecules such as surfactants.
While physical chemistry already recognized the equal roles of water and ions in cell biology at the beginning of the 20th century (92, 122, 123), recent emphasis on “cellular” water has led to some esoteric experimentation and improbable interpretations of data. Thus, water is imagined to be biophilic in an active cytoplasmic matrix that “controls” biomacromolecules (10, 38, 194); such intracellular water is thought to bring about unusual effects, such as the appearance of “ice-like,” denser water molecules that structure the cytoplasm (39, 194); the memory of such structured water that enables transmission of biological information (10, 128); the misunderstood behavior of aqueous gels, with their “do-it-yourself” ion-selecting mechanisms (168); the strangeness of liquid water that separates into lower- and higher-density fractions (244, 245); and the subtle temperature sensitivity exhibited by structured vicinal water (50). Such improbable effects and interpretations still resonate within current literature (10, 38), though it is now widely held that water is simply a (unique) solvent and a (crucial) biochemical reactant; indeed, the cytoplasm is currently viewed as a crowded but watery environment (234).
Since the aqueous phase inside the cell is a rather concentrated aqueous electrolyte solution and the average crowding of biomacromolecules is severe (20 to 50% volume fraction), the physicochemical interactions of solutes and confining biomacromolecular surfaces with water molecules become dominant and therefore also chemically specific (206). While intracellular water is obviously modified compared to pure water, its modification reflects its noncovalent interactions with other chemistries, e.g., some water molecules may be modified by hydrogen bonding to polar uncharged surfaces (e.g., hydroxy-, amino-, or amido- surfaces), other water molecules are modified by charge-dipole interactions with charged surfaces (e.g., phosphates of nucleic acids or acidic or basic amino acid residues), and still others are modified by interactions with hydrophobic surfaces (methyl, methylene, and aromatic surfaces), all of which are present inside the crowded cell; additionally, water acts as a solvent for free ions and other small metabolite molecules. Hence, taken as a whole, modified intracellular water reflects some average of the interactions of water molecules with many different molecular and ionic moieties and with itself; water “structure” and its “breaking and making” are thus not well defined (nor supported by the thermodynamics of model two-component solutions in vitro [11]). Hence, the notion of modified intracellular water needs to be constrained by timescales of its molecular motions.
The Picosecond Hydration of Biomacromolecules
In the last 20 years, there has been significant progress in various spectroscopies (infrared, nuclear magnetic resonance [NMR], and neutron-scattering methods) and in computer simulations, and this has allowed more extensive and precise determinations of the timescales of biomacromolecular hydration (8, 19, 30, 56, 86, 100, 101, 119, 156–159, 163, 232). Essentially, all biomacromolecular hydration interactions take place on timescales of picoseconds, and only a negligible amount of water is buried and immobilized within the interiors of proteins; in other words, cellular water is not ice-like, even though the rotational and translational motions of water molecules are strongly correlated. It is the tetrahedral structure of water molecules which brings about the three-dimensional cohesiveness of the hydrogen-bonded network. The consensus that has now emerged shows that about 80 to 90% of water dynamics inside the cell is the same as in pure water or in dilute solutions, with about 10 to 20% water slowed down by a factor of (only) 2 to 10; this slightly slower hydrogen bonding likely arises from restricted molecular motions of hydrogen-bonding groups attached to polymer backbones. Another important recent finding shows that the dynamics of hydrogen-bonded water in very concentrated salt solutions (where all water molecules interact strongly with anions and cations) is slowed down by a factor of only 3 compared to that of pure water (159). Furthermore, the switching of energetically small native protein substates also occurs in the picosecond time range (97). In other words, protein surfaces explore many energy substates as fast as they renew their hydrogen bonding with water molecules—and perhaps this is one aspect that contributes to the fitness of water for both biochemical reactions and hydration stabilization. Even when water is confined within nanopools surrounded by a hydrophobic phase, its rotational motion is also slowed down by a factor of only ∼2 and is independent of whether the confining surfaces are charged or electrically neutral (62).
The very fast picosecond dynamics of water fits well with the already well-established and surprisingly fast diffusion of metabolites and biomacromolecules in cells and organelles (and in corresponding systems in vitro), as recently reviewed (48, 140), and is corroborated by similar results for diffusion in vitro in nanopores of complex polymeric coacervates, which is also slowed down by a factor of only about 10 compared to that of pure water (40, 108, 109).
The uniqueness of water as a solvent lies chiefly in its small size, with distinctive electron cloud distributions that can be modeled to various degrees of sophistication (47). These properties largely account for its strong hydrogen-bonding capacity (leading to “hydrophobic bonding” of solutes and surfaces that do not have hydrogen-bonding functionality), high degree of polarizability (large dielectric constant to stabilize charged functionalities), and unusually large degree of self-ionization (ability to act as both a base and an acid). Thus, water's only “active” role is that of a participant in biochemical reactions, because biomacromolecular machines that catalyze such reactions are synthesized, assembled, and localized on much longer timescales than the picosecond persistence of hydrogen bonds. Consequently, subcellular structuring of cells arises from (much slower) translational molecular motions and noncovalent interactions of large biomacromolecules, not from ice-like or bound water. The hydration of biomacromolecular surfaces is so fast that it can be considered equilibrated (decoupled) with respect to cellular physiological processes, such as localizations of proteins and nucleic acids within the cell.
Interestingly, transport properties, e.g., diffusibility of small molecules in water, appear unremarkable when compared to such properties measured in other solvents (94). Such dynamic properties of water and aqueous solutions, including viscosity, come into play during morphogenesis and cellular osmotic flows (147, 243, 267) and are as important to the workings of the cell as its often-invoked equilibrium thermodynamic properties. This is particularly related to the hydrophobic effect, which remains elusive to characterize clearly (47, 95, 201, 217, 218) and is discussed below in relation to hyperthermophiles.
Prokaryotes in “hot water.”
While the spectroscopic evidence strongly argues against descriptions of water as an active biophilic solvent, there is also compelling, though less direct, biological evidence: the persistence and evolution (and possible origin) of prokaryotic life in extreme environments (29, 81, 133, 210, 215). In particular, the environment of very hot water at high pressures, high salinities, and both extremes of the pH range—where water has significantly different physicochemical properties from those of water under ambient conditions of around 20°C and 105 Pa (one atmosphere pressure)—argues against any subtle roles for water in cellular biology. For instance, it has been known for a long time that the thermodynamic origin of the hydrophobic effect changes from entropic (melting of picosecond-persisting “icebergs,” if one is given to such structural interpretation) to enthalpic (strengthening of water hydrogen bonds) as one goes from cool to hot water in the 0 to 100°C range (201). Though this is undoubtedly a valid observation consistent with the temperature trends of thermodynamic entropies and enthalpies of the hydrophobic effect, its relevance to our understanding of living systems is somewhat tenuous: lipid membranes and associated biomacromolecules do exist and evolve in both very cold and very hot water. Indeed, new extremophiles continue to be discovered on an unprecedented geological scale, in environments ranging from very cold briny lakes in Antarctica to very hot and deep seafloor vents (61, 133, 142, 262).
From a physicochemical point of view, life most likely arose in hot salty water (133, 205) and then spread out into a wide range of temperatures and pressures following Earth's cycles of geophysical cooling and heating over billions of years (215). In this context, in addition to the unique thermodynamic properties of water that give rise to the hydrophobic effect, its dielectric properties are also noteworthy: the dielectric constant of water decreases at about the same rate that the temperature increases (on the Kelvin scale). This unique dependence makes the Debye-Hückel-type screened electrostatic forces essentially independent of temperature over a very wide range of temperatures (0 to 100°C); hence, hydrated, charge-stabilized biomacromolecules (nucleic acids and proteins) could evolve their catalytic properties (tertiary and quaternary structures) in aqueous environments, independent of geophysical temperature cycles, particularly diurnal cycles. This circumstance has made the “blind watchmaker's” task (42), i.e., molecular evolution's task, of having to deal with cold nights and hot days considerably easier; enzymes could then evolve fast at higher temperatures with a decreased tendency to randomly agglomerate (211).
Cytosol: the Electrolytic Matrix of Life
The role of the matrix of life can be ascribed more precisely to the cellular multicomponent aqueous solution, the cytosol, rather than to water as such. The cytosol is a higher-level low-molecular-weight (low-viscosity) cytoplasmic medium—in essence a relatively concentrated (>0.1 M) aqueous solution of ions, both geochemical inorganic ions (e.g., sodium, potassium, phosphate, and bicarbonate) and genome-derived ions (ATP and other phosphates and carboxylates of biochemical pathways). The main electrolyte in prokaryotes is often potassium glutamate, though the overall ionic composition depends on the changes of composition of the nutrient solution during the cell cycle and under extracellular stresses (20, 52). The cytosol may contain a fraction of biomacromolecules, which can be defined only operationally during the break-up and separation of biomacromolecules from a population of cells (265). (The term “cytosol” has a somewhat variable meaning, e.g., in eukaryotic cells, it is the medium outside the organelles and is also crowded with biomacromolecules; the cytosol is best defined operationally as the liquid part of the cell, with a low content of biomacromolecules.)
Simplified compositions of cytosol of prescribed ionic strength and buffering capacity are stipulated in the extensive compilations of experimental protocols of biochemistry, molecular biology, and genetic engineering (7); these electrolyte compositions have been found empirically to be necessary for the biochemical activity of biomacromolecules in vitro (e.g., folding of proteins and enzymatic reactions) and for the precise self-construction of their functional quaternary complexes (biomacromolecular machines), including the self-construction of ribosomes or even viruses. (Self-construction is a process that requires energy [and catalysts] to surmount thermodynamic and kinetic barriers [90]; self-assembly is a thermodynamically spontaneous and kinetically unhindered process.)
THE CELL IN VIVO
In considering the functioning of a prokaryotic cell in vivo, the phenomenon of biomacromolecular crowding is critical from the points of view of both reactive biochemistry and spatial structuring (biomacromolecular localization) inside the cell (56–58, 77, 78, 104, 144, 145, 205, 206, 263, 264). Below I review some aspects of biomacromolecular crowding, which remains a useful biochemical designation that implies a very large number of interacting biopolymers (and biomolecules) at individually low concentrations. Under such multicomponent conditions, classical physicochemical concentration terms, such as molarity or mole fraction, begin to lose their usefulness (173), hence the term “crowding.”
Biomacromolecular Crowding
Originally, biomacromolecular crowding (the excluded volume effect from a physicochemical standpoint) was used to account for the nonideality (e.g., osmotic pressure) of concentrated (crowded) protein solutions such as hemoglobin (144, 181); such crowdedness is typical of red blood cells and of cells in vivo compared to the case in classical biochemical assays, which are conducted in much more dilute biomacromolecular solutions. The crowding effect was also used as an expedient in designing enzymatic assays to demonstrate the initiation of bacterial DNA replication in vitro; it proved crucial to crowd the assay medium with synthetic hydrophilic polymers (67). Later, the assay medium could be made to work by crowding with (biologically more relevant) proteins (114). Thus, biomacromolecular crowding has become a recognized variable for in vitro biochemical assays to investigate how large concentrations of biomacromolecules might affect their catalytic activities in vivo (176). Typically, such assay protocols have used high concentrations of various (uncharged) hydrophilic polymers (5 to 35% volume fraction), such as high-molecular-mass polyethylene glycols (PEGs), dextrans and other polymeric derivatives of sugars, polyvinylpyrrolidone (PVP), and similar water-soluble polymers; these synthetic polymers have become known as crowders that simulate high-volume fractions of biomacromolecules in cells in vivo (58).
While crowding is undoubtedly present as an analytical fact in cells in vivo (265), its effects on the structuring of cells and on the catalytic reactivity of biomacromolecules (folding of proteins and their assembly into “machines” and “factories,” often with nucleic acids) have proved harder to generalize (69) and have remained a topic of investigations. For example, in the case of intrinsically disordered proteins (51, 226), there are reports of some proteins gaining structure extensively, while others appear quite resistant to folding under the influence of crowders; other proteins may gain structure only on a modest scale (44, 63, 136, 148, 151, 172, 184). Such results are not necessarily conflicting; rather, they reflect the difficulty of extracting the theoretical nonspecific excluded volume effect from other interactions of biomacromolecules (104, 141, 186, 240).
As well appreciated in many textbooks (2), there is an immense variety of proteins that can be synthesized from 20 amino acids: some water soluble (random coil proteins), some water insoluble (crystallized or amorphous proteins), and many in between (water-swellable and water-dispersible proteins), folding or folded into their functional conformations in vitro or in vivo. When proteins are crowded, they interact mutually by repulsive excluded volume interactions, by attractive hydrophobic interactions, and by repulsive and attractive electrostatic interactions; the latter are modulated by pH (buffers), by the ionic strength of the aqueous medium, and often by specific ions, such as potassium or magnesium. The main advantage of the hydrophilic crowders is their extensive bonding with water molecules, which diminishes their specific interactions with biomacromolecules; hence, such crowders have theoretically represented the best option for studying the nonspecific excluded volume effect, for example, in relation to bringing about more compact protein conformations or protein misfolding (21, 165, 172, 180). However, it was recognized recently that the effects of crowders can in fact be specific and ought to be treated carefully on a case-by-case basis (104, 141, 240). Also, since the crowders are highly hydrated and hence “soft,” the model of hard spheres (excluded volume) for computer simulations is questionable (56).
From a microbiological structuring point of view, cells in vivo do not generally have large concentrations (over 10% volume fraction) of such high-molecular-weight, highly hydrophilic crowders; because these crowders have typically been used as single compounds, they do not reflect the multicomponent spatiotemporal structuring of the cell in vivo, as currently envisaged graphically (78, 205); indeed, it has been suggested that in vitro crowding studies with current hydrophilic crowders may have only peripheral relevance to the in vivo situation (56). The use of more realistic crowding agents that would be more representative of cytoplasmic chemistry (proteins and nucleic acids) often leads to uncontrolled biomacromolecular agglomerations under conditions with a large amount of crowding. Similar agglomerates, described as inclusion bodies, are sometimes encountered in biotechnological exploitations of recombinant expression of proteins (200). Similarly, genetically expressed green fluorescent protein (GFP) fusions may be sensitive to experimental conditions and may agglomerate into larger entities (139, 140), possibly via weak hydrophobic attractions inherently arising from the structure of GFP (164).
Hyperosmotic stress-induced crowding.
Another phenomenon where the effects of biomacromolecular crowding become apparent is the response of a prokaryotic cell population to hyperosmotic external stresses, brought about by highly concentrated solutions of membrane-impermeant solutes such as sodium chloride or sucrose (41, 149, 169, 170, 182, 229, 231, 257–259). As water molecules pass osmotically through the membrane into the external solution, the biomacromolecular crowding and concentrations of low-molecular-weight ions and metabolites increase; in order to better balance the osmotic pressure across the cell envelope and thereby to prevent or delay shrinkage (and eventually plasmolysis and cell death), the cell in vivo quickly activates membrane transporters to import compatible solutes if they are available externally (such as potassium salts, glycine-betaine, proline, trehalose, and others). As a secondary line of defense, which has an inherent time delay, the cell can activate genetic circuits to synthesize enzymes that then catalyze the synthesis of compatible solutes de novo (257).
Under hyperosmotic conditions, biomacromolecular crowding becomes more severe, increasing from an ∼25% volume fraction to an ∼50% volume fraction, but its effects on the subcellular structuring are complex, because all noncovalent interactions in the cell are affected, rather than just a few. For instance, the binding of the lac repressor to DNA was found to be very strongly dependent on the ionic content in the (uncrowded) in vitro assay (45), suggesting a severe disruption of biochemical reactions in vivo under hyperosmotic conditions when the cytoplasmic ionic content increases. Surprisingly, E. coli in vivo could adjust to a new hyperosmotic external medium and could even grow and multiply, albeit at a lower rate. Clearly, biomacromolecular crowding in vivo somehow compensates for the ionic dependence of DNA-protein interactions in vitro, though how this happens from a physicochemical standpoint is not clear (37, 174). Recent experiments showed that the growth rate and survival also depend on how fast the external osmolarity is raised; at low rates, cells can adjust and survive, but at high rates, cells are more likely to die (113).
From a structuring or engineering point of view, the cell has a better chance of survival if the spatial relations of key biomacromolecular structures within the cell remain more or less the same and have time to adjust than when the cytoplasmic structuring is disrupted suddenly by a large stress. In still more stressful anhydrobiotic experiments, the preservation of subcellular integrity is necessary for survival upon rewetting (4, 18, 171). Ultimately, this is a broader issue of the response of metabolic and genetic circuits to extracellular conditions when such circuits become temporally and spatially stressed; such circuits then readjust by new conformations and localizations of proteins and nucleic acids and by the remodeling of the cell envelope. In this situation, electrostatic interactions (dependent on ionic strength, pH, and specific ions) are known to play important, though not well-understood roles, for example, in protein-nucleic acid interactions related to gene expression (49, 84, 85, 93, 96, 238). How are such metabolic and genetic circuits realized in a cell in vivo? How do they restructure, dissipate, and reappear in response to extracellular inputs?
Complex Vectorial Biochemistry
One way to think about crowded biomacromolecules in vivo is to treat them as reacting and colloidally interacting particles within the cytosol rather than as dissolved molecular species in a homogeneous solution that obey the classical relationships of chemical thermodynamics and kinetics. While association (binding) constants provide useful descriptions of dilute, in vitro biochemistry, such as the mechanisms and rates of protein or RNA catalyses, the experimental kinetic and equilibrium constants are not actually constant; typically, they depend on pH (buffer) and ionic strength (83) and often depend on the chemical specificity of ions and, in general, on the nonideality of such solutions (104, 144). Hence, such formalisms do not easily allow for the severe crowding of many noncovalently interacting and chemically reacting biomacromolecules in vivo, some of which are present at low copy numbers. Taking such a colloidal view of in vivo crowding suggests that biomacromolecules and their complexes divide the cytosol into a topologically complementary system of liquid nanopools and pathways (see the two-dimensional cartoon in Fig. 1, discussed below). This crowded state of living matter is further complicated by being away from physicochemical equilibrium, both internally and against the extracellular milieu.
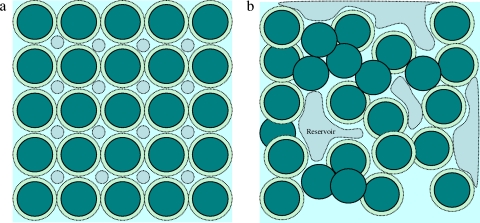
Fig. 1.
(a) General origin of complementarity of electrolyte pathways to crowded charged particles at a 25% volume fraction (over 50% with Debye lengths added). There is no bulk concentration of solutes dissolved in water. The small circles emphasize that ionic distributions are nonuniform everywhere throughout the volume of crowded spheres. (b) Hydrophobic and screened electrostatic attractions bring about complex structures with a very-high-volume fraction of particles (multiplexes), with semiconducting pathways and nanopools and larger reservoirs with quasi-bulk concentrations of ions and low-molecular-weight dissolved compounds.
Thus, a great variety of biomacromolecules must maintain their functional conformations and colloidal stability under crowded conditions in vivo—they must not haphazardly agglomerate, or dissolve, i.e., denature, and lose their catalytic activity. How do biomacromolecules accomplish this task of maintaining their functional stability in vivo? It is theorized, from a classical physical chemistry standpoint, that biomacromolecular surfaces are stabilized by hydrogen bonding with water molecules, by screened electrostatic repulsions, and by hard excluded volume repulsions, all of which must act over commensurate ranges of distances to achieve their mutually complementary stabilizing effects (205, 206). The term “complex vectorial biochemistry” was coined to describe such stabilization of crowded and reacting macromolecules and suggested that the onset of complex vectorial biochemistry was a distinct (emergent) transition between the physicochemical and biochemical evolution of the early Earth during the Hadean eon (205). Below, I briefly recapitulate the ideas giving rise to complex vectorial chemistry of charged crowded surfaces.
In Fig. 1a, the two-dimensional cartoon shows the crowding of an ∼25% volume fraction of (uniformly) charged 5-nm spheres (proteins) stabilized by an aqueous Debye length of 0.7 nm (corresponding to the typical ionic strength of the cytoplasm); the spheres essentially touch when they are extended by the Debye length, which indicates their strong interactions mediated by the ionic strength of the liquid. If the Debye length were included in the size of the spheres, the crowding would represent a very large, 52.4% volume fraction (simple cubic packing of one sphere per cube). The key point is that the nanoparticles and the aqueous phase become topologically and physicochemically complementary, forming a single interfacial system, with not enough space anywhere for low-molecular-weight ions and metabolites to assume bulk concentrations. In Fig. 1b, some of the spheres stick together via attractive hydrophobic bonding and screened electrostatic attractions, thereby creating a more realistic but complex system of crowded biomacromolecules. In this case, larger reservoirs can appear with quasi-bulk concentrations of ions and metabolites. This structural complexity is further increased by the reactive character of biomacromolecular surfaces.
The surfaces of biomacromolecules are not really mechanically hard—their hydrated surfaces execute very fast motions on timescales of femtoseconds to nanoseconds, which enable catalytic biochemical reactions, as discussed earlier. Thus, in addition to noncovalent attractive and repulsive forces, the subcellular structuring is also determined by three reactive mechanisms: (i) biopolymerizations and biopolymer degradations that extend or reduce existing biomacromolecular surfaces (173); (ii) reactive modifications of existing surfaces for greater hydrophilicity (e.g., phosphorylation) or for greater hydrophobicity (e.g., methylation), which change attractive and repulsive interactions between biomacromolecules (biochemical signaling); and (iii) catalytic syntheses of low-molecular-weight metabolic intermediates, such as phosphate esters, amino acids, lipids, nucleotides, etc. All three reactive mechanisms require the supply of low-molecular-weight reactants, which is handled through the electrolyte nanopools and pathways shaped by the surfaces of crowded biomacromolecules. In this way, also, confusing cross talk between many concurrent biochemical reactions is prevented or at least reduced during the cell cycle, as biochemical reactions become sequestered vectorially in three dimensions. This view is not unlike the concept of metabolons, which arose from the “unnaturally large” size of many biomacromolecules compared to their actual catalytic sites (207, 208); however, the transient vectorial nature of electrolyte pathways and nanopools is simply the consequence of high crowding and attractive noncovalent interactions.
Zooming Out from Nanometers to Micrometers
In the next sections, I “zoom out” from molecular interactions on the nanometer/microsecond scale to a dynamically structured, prokaryotic cell on the micrometer/second scale. First, I briefly review the semiconducting character of two charged (bio)surfaces, and then I consider the effects of noncovalent physicochemical interactions (repulsive and attractive) on larger-scale cellular structuring.
Biomacromolecular semiconductors.
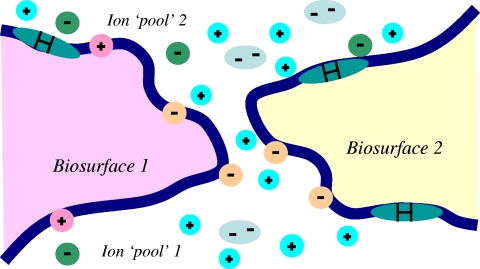
In Fig. 2, the two-dimensional cartoon shows an electrolytic pathway between two charged (e.g., phosphorylated) biosurfaces that connect two electrolyte nanopools (170, 204, 206). On a scale of nanoseconds to microseconds, the overall shape of the surfaces is averaged to a more or less constant (functionally folded) shape that is stabilized (equilibrated) by hydrogen bonding with water molecules, as represented by the thick line. Such a surface generally also contains attractive hydrophobic patches (CH3, CH2, CH, and aromatic rings), designated “H,” with no ability to become stabilized by hydrogen bonding, as well as sites with electrostatic positive and negative charges; the latter cause the redistribution of cations and anions around them—the Debye-Hückel screened electrostatic forces. The ions are drawn with distinct boundaries, though on this timescale of about microseconds and longer, they are smoothed out into volume charge densities in an aqueous dielectric (in water).
Fig. 2.
Cartoon of an electrolyte pathway that is permeable to cations only, with an active electrochemical gradient between the ionic pools. Surface potential reaches over −25.7 mV in the pathway at 25°C.
If the charged surfaces are approximated by two charged planes, it can be calculated that the electrostatic repulsions in water can be very large and, under certain conditions, nearly independent of ionic strength (202–204). Furthermore, depending on the calculated (negative) potential distributions between the surfaces, the planar electrolyte gap can become conducting to all ionic species (cations and monovalent and divalent anions) or only to cations and monovalent anions or to cations (170, 206). Thus, on a scale of microseconds and longer, the cytosol can be regarded as a buffered, electrically conducting fluid that carries ions (both inorganic and genome-derived) throughout the cell.
This electrolytic view of the cytosol agrees with the notion that a cell has some characteristics of computer hardware and software operation (25, 27, 111). Although Brownian diffusion is absolutely necessary for cellular metabolism and signaling and appears generally fast enough in vivo, though slower than the case in vitro (13, 48, 59, 116, 140, 234), particularly under osmotic stress (113, 139, 229), it is not sufficient for computer-like operation of the cell. Electrostatic potentials arising from transport of ions, modulated by covalently bound charges at biomacromolecular surfaces, must play a role (105), albeit one that is not well understood as yet, just as electrical potentials perform electronic computations in silicone-based semiconductors of integrated circuits. The predominance of charged molecules and polymers within cells, including the role of anionic lipids within plasma membranes, underscores their electrochemical functioning (28, 80, 170, 206, 231, 259, 261).
Supercrowded systems of catalytic semiconductors.
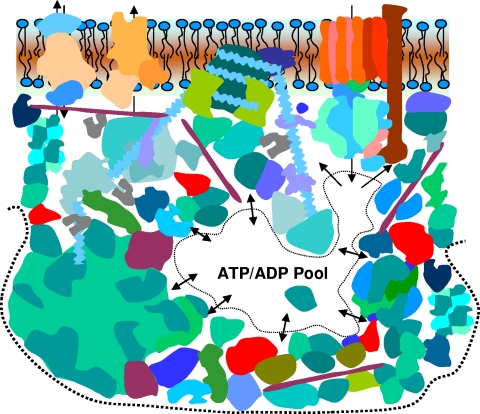
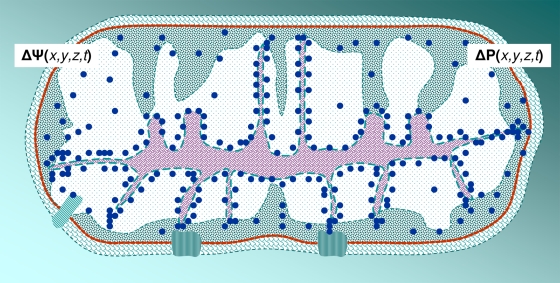
Zooming out from two interacting biomacromolecular surfaces to a somewhat larger scale, Fig. 3 depicts an arbitrary, though more realistic, system of electrolytic semiconducting pathways and electrolyte nanopools; they are shown adjacent to the lipid bilayer with integral membrane proteins (the cell wall and extracellular layers are not shown), where the lowering of the dielectric constant (increase of electrostatic forces) complicates the relationship between hydrophobic and electrostatic forces (117). This cartoon attempts to show some important biomachinery (78), for example, ATP synthase in the membrane in the upper right corner, a ribosome in the lower bottom corner, and long fibrous cytoskeleton proteins as well as a length of DNA wound up and anchored by a protein in the cytoplasmic side of the membrane.
Fig. 3.
System of supercrowded spatiotemporal semiconductors “spot welded” by attractive noncovalent forces (screened electrostatic and hydrophobic forces), giving a multiplex of electrolyte nanopools and electrolyte pathways fed from larger electrolyte reservoirs, such as the ATP/ADP reservoir.
The biomacromolecules are drawn under supercrowded conditions, with higher than the average 20 to 30% volume fraction that is regarded as representative of living systems (48, 140, 265); reservoirs with lower than the average 20 to 30% volume fraction of biomacromolecules then appear within the cytoplasm, for example, the ATP/ADP reservoir, into which ATP synthase gives off ATP ions, as shown in Fig. 3. Such a supercrowded assembly of biomacromolecules can be called a multiplex, as I explain and discuss below.
(i) Physicochemical rationalizations.
The foremost physicochemical reason for the supercrowding of biomacromolecules is the chemical inhomogeneity of their surfaces, resulting not only in stabilizing repulsions but also in attractive noncovalent association of biomacromolecules. From a physicochemical standpoint, biomacromolecular surfaces are encoded by the genome to have four distinct physicochemical patches: (i) uncharged hydrophilic, (ii) negatively charged, (iii) positively charged, and (iv) hydrophobic. These surface chemistries and their interactions with water molecules and ions determine both the functional folded structure of individual biomacromolecules and the attractions and repulsions between themselves.
What will be the outcome of balancing such attractive and repulsive noncovalent forces on the structure of crowding biomacromolecules in vivo? First, when and where repulsive (stabilizing) forces “win,” haphazard biomacromolecular agglomerations will be prevented. Such forces are the excluded volume effect (hydration of biosurfaces) and screened electrostatic repulsions that ensure that the electrolyte pathways remain open and electrolytically conducting. Second, when and where noncovalent attractive forces win, there will be noncovalent formation of (weak) cross-links. Such forces are screened electrostatic attractions between positive and negative patches and hydrophobic bonding between two hydrophobic patches on different surfaces. The attractive noncovalent forces bring about biomacromolecular cluster formation (phase separations), as demonstrated in vitro with protein and colloidal systems (22, 40, 57, 68, 82, 109, 110, 124, 166, 213, 239); another example is the phenomenon of weak association of proteins via their hydrophobic patches, for example, the dimerization of GFP (68, 164). Also, as discussed earlier in relation to biomacromolecular crowding, recent experiments on stabilization (folding) of proteins under crowded conditions (104, 141, 186, 240) show attractive interactions in addition to the repulsive effect of excluded volume. Attractive clustering interactions have also been deduced from the results of genomewide proteomic screens; it has been suggested that proteins may in fact assemble into larger physical entities that fall apart and reform during the cell cycle in a stepwise fashion (115); thus, proteins can be reused during the cell cycle to participate in the formation of different multiplexes (they are multifunctional, being promiscuous and “moonlighting” [102]).
Such a weakly “cross-linked,” large-scale association of supercrowded biomacromolecules represents a multiplex. This term accounts for many different biomacromolecules involved and for the multiplexing pathways between biomacromolecular surfaces, i.e., signal transmission channels through which different ionic metabolites (different signals) can travel sequentially and concurrently (Fig. 3 and 4). The rate of formation of such multiplexes could be quite high, given the remarkable accelerating effect of screened electrostatic attractions on the rate of association of proteins (190).
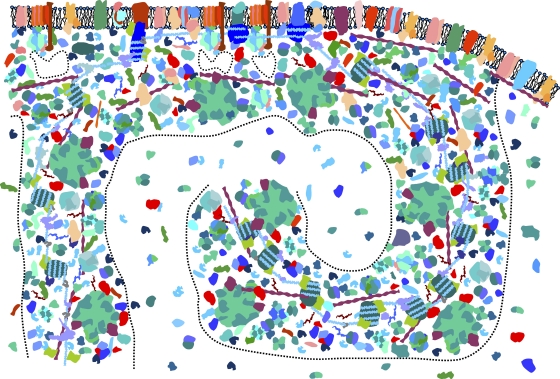
Fig. 4.
Model of unequal biomacromolecular crowding in vivo at a larger scale. A supercrowded multiplex with electrolyte nanopools and semiconducting pathways and uncrowded reservoirs of cytosolic electrolyte (quasi-bulk concentrations) with a low content of biomacromolecules is shown.
Figure 3 shows crowded biomacromolecules forming a multiplex system of interconnected electrolytic pathways and nanopools and an ATP/ADP reservoir nearly empty of biomacromolecules. How could such a network work physicochemically? As an example, the following scenario demonstrates an efficient distribution of ATP related to the powering of the cell: ATP, being a divalent anion (with 2 of the 4 charges complexed with magnesium), flows out of its reservoir only via dephosphorylated (zero and low negative potential) pathways but can return as ADP (monovalent ion) through both phosphorylated and dephosphorylated pathways. In other words, phosphorylation of an electrolytic pathway creates a more negative potential within the pathway that blocks ATP (or any divalent anion) from going where it might go wastefully just by random thermal diffusion. Such a multiplex in effect creates localized vectorial reaction loops (transient networks) that can efficiently deliver ATP (and other required metabolites) to various catalytic sites. This network has the character of a packet-switching information transmission, since ATP ions are not delivered sequentially through just one (predetermined) pathway (analogous to a telephone line) but in a parallel manner by many available pathways (analogous to packet-switching routing of Internet information packets). The advantage of packet-switching networks is that when a few pathways become inoperable (poisoned), the network does not crash; in this way, the dynamic stability of processes which keep the genome alive and evolving becomes quite robust. The semiconducting pathways can be controlled (open or closed) by biochemical signaling cascades that restructure or even redisperse the multiplex by phosphorylations, methylations, acetylations, etc., in order to effectively respond to extracellular conditions (availability of nutrients, water activity, temperature variations, etc.). Inhomogeneous distribution of ATP has been measured in the cytoplasm of eukaryotic oocytes (143).
Though the above description of noncovalent forces focuses on specific attractions arising from coulombic and hydrophobic interactions, the strength of these attractions could be increased by the nonspecific entropic force (depletion force) arising from an unequal distribution of sizes and shapes of crowded biomacromolecules (129). However, such nonspecific colloidal forces, including van der Waals forces, cannot be related directly to the chemical inhomogeneities of biomacromolecular surfaces (which in turn are determined by the genome) and thus cannot readily illuminate the detailed functioning of the cell. The nonspecific depletion forces thus provide a general attraction that could come more strongly into play in slowly metabolizing (stationary) cells and in their transitions toward more anhydrobiotic, nonmetabolizing states, such as bacterial spores, when crowded biomacromolecules have to be “frozen” in revivable configurations (18, 65); also, in osmotically balanced bacterial L forms, such attractions could aid the structural stability of the cell (3, 46, 118).
(ii) Reservoirs of cytosol.
Though chemical inhomogeneities of biomacromolecular surfaces provide an a priori physicochemical rationale for the model of supercrowded multiplexes, there is also experimental evidence that points to a similar concept: the relatively fast Brownian diffusion of various biomacromolecules and their GFP fusions within the cytoplasm of different cells and organelles (48, 59, 113, 116, 139, 140, 234). To rationalize such results, it has been suggested that mitochondria (highly crowded organelles) tend to supercrowd biomacromolecules mostly to the cytoplasmic side of the membrane, leaving the interior relatively empty for biomacromolecular diffusion (160). The newly discovered prokaryotic cytoskeleton proteins, as well as some eukaryotic scaffolding proteins (see Introduction), are reported to also associate with the cytoplasmic side of the membrane. This view is adopted as the key feature of the structuring of cells and organelles, where supercrowding of biomacromolecules toward the cytoplasmic side of the membrane creates reservoirs of cytosol with low biomacromolecular content, in effect creating space for the deployment of the genome (where appropriate). Such a model is somewhat different from current descriptions of crowded cells, with undifferentiated biomacromolecular crowding of a 20 to 30% volume fraction (77, 78).
In a broader sense, the suggested model of the semiconducting multiplexes and larger reservoirs of cytosol reconciles two somewhat contradictory experimental observations: (i) the generally fast, chaotic Brownian diffusion of biomacromolecules in vivo and (ii) the fidelity of the cell cycle through localization and structuring of biomacromolecules. In this model, the transiently structured, vectorially crowded multiplexes sequester concurrent biochemical reactions during the cell cycle (prevent and control their cross talk), while the uncrowded cytosol allows for fast diffusion (ensuring fast metabolic growth). In such a model, signaling reactions such as (de)phosphorylations and (de)methylations control the dynamics of multiplexes, i.e., their network formation, restructuring, and dispersal, thereby linking biochemical signaling with metabolism.
The semiconducting prokaryotic scaffold.
Zooming out now to visualize the cell in toto (Fig. 5), the well-known structural elements of prokaryotic cells—the nucleoid, ribosomes, and the cell envelope—come into focus. How are these complex cytological structures organized and related to the multiplexes at this coarse microscopic level? Though at this point the multiplexes are only hypothesized physicochemical entities, they may represent biochemical or physiological constructs or hyperstructures (154) at appropriate times during the cell cycle, such as the divisome or stressosome, derived from physiological behavior; other “-omes” could be found by analyses of genomewide interactomes of system biology (76, 87, 131, 175). For instance, the divisome involves a large number of proteins situated in the cytoplasm, in the membrane, and extracellularly, possibly forming a large multiplex that spans the cell envelope. Signaling proteins involved in bacterial chemotaxis phenomena (23, 26) could also be associated physically into a multiplex spanning the cell envelope.
Fig. 5.
Physicochemical model of a cell, characterized by electrostatic and pressure differences across the cell envelope and between the cytosolic reservoirs and the nucleoid (not shown). The membrane is red, with attached supercrowded multiplexes and nucleoid excrescences on the inside and the cell wall and extracellular layers on the outside. Ribosomes (blue circles) are distributed mostly along the periphery of the nucleoid, which is connected spatiotemporally to the cell envelope. Signaling receptors are shown to span the cell envelope (bottom and left); they convert extracellular physicochemical signals into cytoplasmic signals that are transmitted vectorially to specific cytoplasmic “addresses” during the cell cycle.
This rather complicated situation of too many different biomacromolecules associating precisely into multiplexes of different sizes, persistence, and localizations during the cell cycle can be simplified by assuming that there are only two (very large and time-evolving) multiplexes, which in effect define a binary (see Loeb's observation at the beginning of this report) prokaryotic scaffold. It consists of (i) the genetic part, the structured, replicating, and transcribing nucleoid that makes DNA loops or excrescences that form multiplexes (communicate electrolytically and vectorially) with the cell envelope; and (ii) the somatic part, the cell envelope multiplexed at the cytoplasmic side with a sizeable fraction of cytoplasmic proteins, leaving behind reservoirs of uncrowded cytosol.
Such a cell model (Fig. 5) is based on recent progress in the understanding of subcellular structuring of prokaryotes, where nucleoid/ribosome/cell envelope connections synchronize gene expression, chromosome orientation, replication, and segregation with cell septation and division during the cell cycle (14, 15, 24, 28, 34, 70, 73, 80, 98, 120, 121, 193, 225, 235, 255). The nucleoid excrescences and their anchoring at the cytoplasmic side of the cell envelope are qualitatively consistent with the subdiffusive motion (non-Brownian) of chromosomal loci (241). This model also fits with recent diffusion data on proteins of various sizes, where large proteins and their complexes are significantly slowed, or even rendered immobile, by the mesh-like excrescences of the nucleoid (or other large structures), particularly under osmotic stress, when the cytosolic reservoirs become smaller or disappear (113, 116, 139, 140, 229). The ribosome assembly and positioning is an active process that requires energy (28); in Fig. 5, the ribosomes (107, 246, 247) are positioned for transertion of membrane-bound proteins (255), for insertion of proteins needed for its replication, compaction, and segregation into the nucleoid, and for ejection of soluble (globular and unstructured) proteins into the cytosol. When assembled, ribosomes can be repositioned further by active transcription (134).
The topology and supercrowdedness of the cellular scaffold are variable during the cell cycle, responding to the physicochemical variables of the nutrient solution outside (composition, temperature, and pressure). One feature of the model is a dynamic system of reservoirs of uncrowded cytosol of variable continuity and variable but quasi-bulk compositions, giving rise to electrochemical (Donnan) and pressure (osmotic) differences within the cell and against the nutrient solution; such gradients are likely to play a role in cellular morphogenesis and motion at hundreds of nanometers and larger scales (88–90). This aspect of the model is akin to the recently proposed poroelastic model of the cytoplasm of eukaryotic cells (147).
CONCLUSIONS AND OUTLOOK
The physicochemical interactions and processes that spatially and temporally structure prokaryotic cytoplasm in vivo (water, ions, low-molecular-weight metabolites, and biomacromolecules) must ultimately ensure the fidelity and successful functioning of the cell cycle. These physicochemical processes are complicated and are not well understood as yet (88–90), though progress is being made in some areas. In particular, the notion of intracellular water as an active structuring cytoplasmic medium with subtle effects is unnecessary. There is enough evidence to conclude that water is simply a solvent that enables molecular evolution over wide ranges of temperatures; the extremely fast picosecond hydration of biomacromolecular surfaces and the temperature independence of screened electrostatic interactions explain to a large degree why this is so.
The hallmark of a living system is the high degree of crowding of a large number of different biomacromolecules confined by a semipermeable cell envelope. New research shows that dextrans, polyglycols, and similar crowding polymers, which traditionally have been employed in crowding assays as inert compounds, can in fact interact specifically with other biomacromolecules; they are thus less suitable as model crowding polymers than previously thought. Moreover, no similar crowders are normally present in prokaryotes in vivo. Currently, in vitro crowding assays are being designed with proteins, which better reflect biomacromolecular environments in vivo; this new research could also illuminate multicomponent biomacromolecular clustering arising from hydrophobic and coulombic attractions.
In considering biomacromolecules as colloidal particles, their high level of crowding in vivo leads straightforwardly to the hypothesis of complex vectorial (bio)chemistry. Simple analysis shows that high biomacromolecular crowding divides the aqueous phase (cytosol) into a vectorial, interfacial system that is topologically and physicochemically complementary to the chemistry of biomacromolecular surfaces; in this system, low-molecular-weight metabolites have insufficient free volume to attain bulk compositions independent of position. Since biomacromolecules have genome-encoded chemical inhomogeneities in their surfaces, they can associate by many weak bonds into supercrowded clusters or multiplexes (35 to 95% volume fraction). Within the multiplexes, the electrolyte nanopools and pathways can theoretically transport ions in a semiconducting, vectorial manner. Consequently, the cytoplasm is not uniformly crowded at a 20 to 30% volume fraction, but the supercrowded multiplexes create relatively uncrowded reservoirs of cytosol (0 to 20% volume fraction) through which biomacromolecules can diffuse more or less unhindered.
The above considerations and recent microbiological advances in biomacromolecular localization in vivo allow the construction of a generic model of a prokaryotic cell, consisting of a supercrowded (multiplexed) prokaryotic scaffold where the nucleoid is connected to the cell envelope via its excrescences. The ribosomes are located on the cytoplasmic periphery of the scaffold and the cytosol. Most proteins are multiplexed into clusters associated with the cell envelope and the nucleoid; they cross the cytosolic reservoirs by a diffusion-to-capture mechanism. This model ensures the fidelity of the cell cycle by sequestering biochemical reactions in multiplexes, thereby avoiding confusing reactions in unstructured prokaryotic cytoplasm; a great deal of genome-based structural information is thus inherited in the cell cycle. This information is lost when individual biomacromolecules are isolated in vitro, and it is not recovered in current in vitro crowding assays. New experiments and techniques are needed to distinguish between the current cytoplasmic model of average crowding and models of unequal crowding (supercrowded biomacromolecular clusters and uncrowded reservoirs of cytosol). Possibly, one could attempt to isolate multiplexes and nucleoids through a controlled break-up of bacterial L forms in a properly formulated medium, analogous to the isolation of nucleoids (266).
There is now probably enough biochemical, genetic, and physiological information to attempt to construct a coarse spatiotemporal model of a simplified (ideal?) cell in toto (74, 87, 107, 110, 120, 140, 246, 247, 259) as it progresses through the cell cycle—perhaps starting at a 1-s resolution for one division per hour and then increasing the temporal resolution toward milliseconds and microseconds while zooming in on important phenomena and regions of the cell. Such a model could summarize current knowledge of the flow of physicochemical information from the nutrient solution and its transformation through the cell envelope to activate and maintain gene expression and genome replication, leading to cell volume growth and eventual cell fission. Such a model of a simplified prokaryotic cell cycle could also uncover the “machine language” of electrochemical and osmotic potentials (that enables the DNA/RNA/protein language of molecular biology), as well as the phenomena of cell morphogenesis and motility. Such an endeavor would require a collaborative effort of different disciplines (1).
ADDENDUM IN PROOF
The literature related to the structuring of cells during their growth and division is scattered in many general and specialized journals about biology, chemistry, and physics; hence, some references were likely missed in this review, for which I apologize. For example, there is a great diversity of protein-protein interactions, from very strong to weak and ultraweak (I. M. A. Nooren and J. M. Thornton, EMBO J. 22:3486–3492, 2003). The weaker interactions have been described as biomacromolecular clustering, phase separation, compartmentation, and gel formation, or in the present case as “multiplex” formation. Currently, new methods for their structural and transient characterizations are being developed, e.g., NMR and analytical ultracentrifuge (J. Vaynberg and J. Qin, Trends Biotechnol. 24:22–27, 2006; A. J. Rowe, Methods 54:157–166, 2011).
ACKNOWLEDGMENTS
I thank Gary Pielak for very useful comments on the manuscript and for allowing me to view his latest manuscripts on biomacromolecular crowding before publication. I also thank George Fox, Michael Russell, and Dennis Bray for comments and for suggesting additional references. I am greatly indebted to Janet Wood and Bert Poolman for introducing me to the physicochemical issues of cellular osmoregulation in bacteria and for stimulating discussions. The manuscript was substantially improved by adopting comments and suggestions of anonymous reviewers; some are used verbatim.
The management of Mallard Creek Polymers, Inc., is thanked for providing support.
REFERENCES
- 1. Alberts B. 1998. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92:291–293 [DOI] [PubMed] [Google Scholar]
- 2. Alberts B., et al. 2002. Molecular biology of the cell. Garland Science, New York, NY [Google Scholar]
- 3. Allan E. J., Hoischen C., Gumpert J. 2009. Bacterial L-forms. Adv. Appl. Microbiol. 68:1–39 [DOI] [PubMed] [Google Scholar]
- 4. Alpert P. 2006. Constraints of tolerance: why are desiccation-tolerant organisms so small or rare? J. Exp. Biol. 209:1575–1584 [DOI] [PubMed] [Google Scholar]
- 5. Arends S. J. R., Kustusch R. J., Weiss D. S. 2009. ATP-binding site lesions in FtsE impair cell division. J. Bacteriol. 191:3772–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Assumes N., Kuhn J. R., Jacobs-Wagner C. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705–713 [DOI] [PubMed] [Google Scholar]
- 7. Ausubel F. M., et al. (ed.). 2002. Short protocols in molecular biology. John Wiley & Sons, New York, NY [Google Scholar]
- 8. Bagchi B. 2005. Water dynamics in the hydration layer around proteins and micelles. Chem. Rev. 105:3197–3219 [DOI] [PubMed] [Google Scholar]
- 9. Baines A. J. 2009. Evolution of spectrin function in cytoskeletal and membrane networks. Biochem. Soc. Trans. 37:796–803 [DOI] [PubMed] [Google Scholar]
- 10. Ball P. 2008. Water as an active constituent in cell biology. Chem. Rev. 108:74–108 [DOI] [PubMed] [Google Scholar]
- 11. Batchelor J. D., Olteanu A., Tripathy A., Pielak G. J. 2004. Impact of protein denaturants and stabilizers on water structure. J. Am. Chem. Soc. 126:1958–1961 [DOI] [PubMed] [Google Scholar]
- 12. Bennett V., Healy J. 2009. Membrane domains based on ankyrin and spectrin associated with cell-cell interactions. Cold Spring Harb. Perspect. Biol. 1:a003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berg H. C. 1993. Random walks in biology. Princeton University Press, Princeton, NJ [Google Scholar]
- 14. Berger M., et al. 2010. Coordination of genomic structure and transcription by the main bacterial nucleoid-associated protein HU. EMBO Rep. 11:59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berlatzky I. A., Rouvinski A., Ben-Yehuda S. 2008. Spatial organization of a replicating bacterial chromosome. Proc. Natl. Acad. Sci. U. S. A. 105:14136–14140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bettinger B. T., Gilbert D. M., Amberg D. C. 2004. Acting up in the nucleus. Nat. Rev. Mol. Cell. Biol. 5:410–415 [DOI] [PubMed] [Google Scholar]
- 17. Bi E. F., Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164 [DOI] [PubMed] [Google Scholar]
- 18. Billi D. 2009. Subcellular integrities in Chroococcidiopsis sp. CCMEE 029 survivors after prolonged desiccation revealed by molecular probes and genome stability assays. Extremophiles 13:49–57 [DOI] [PubMed] [Google Scholar]
- 19. Boehr D. D., Dyson H. J., Wright P. E. 2006. An NMR perspective on enzyme dynamics. Chem. Rev. 106:3055–3079 [DOI] [PubMed] [Google Scholar]
- 20. Booth I. R., Pourkomailian B., McLaggan D., Koo S.-P. 1994. Mechanisms controlling compatible solute accumulation: a consideration of the genetics and physiology of bacterial osmoregulation. J. Food Eng. 22:381–397 [Google Scholar]
- 21. Boquist M., Gröbner G. 2007. Misfolding of amyloidogenic proteins at membrane surfaces: the impact of macromolecular crowding. J. Am. Chem. Soc. 129:14848–14849 [DOI] [PubMed] [Google Scholar]
- 22. Borrega R., Tribet C., Audebert R. 1999. Reversible gelation in hydrophobic polyelectrolyte/protein mixtures: example of cross-links between soft and hard colloids. Macromolecules 32:7798–7806 [Google Scholar]
- 23. Bourret R. B., Stock A. M. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625–9628 [DOI] [PubMed] [Google Scholar]
- 24. Brandt F., et al. 2009. The native 3D organization of bacterial polysomes. Cell 136:261–271 [DOI] [PubMed] [Google Scholar]
- 25. Bray D. 1995. Protein molecules as computational elements in living cells. Nature 376:307–312 [DOI] [PubMed] [Google Scholar]
- 26. Bray D. 1998. Signaling complexes: biophysical constraints on intracellular communication. Annu. Rev. Biophys. Biomol. Struct. 27:59–75 [DOI] [PubMed] [Google Scholar]
- 27. Bray D. 2009. Wetware: a computer in every living cell. Yale University Press, New Haven, CT [Google Scholar]
- 28. Britton R. A. 2009. Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 63:155–176 [DOI] [PubMed] [Google Scholar]
- 29. Brock T. D., Brock K. M., Belly R. T., Weiss R. L. 1972. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 84:54–68 [DOI] [PubMed] [Google Scholar]
- 30. Bryant R. G. 2010. Dynamics of water in and around proteins characterized by 1H-spin-lattice relaxometry. C. R. Phys. 11:128–135 [Google Scholar]
- 31. Cabeen M. T., Jacobs-Wagner C. 2007. Skin and bones: the bacterial cytoskeleton, cell wall and cell morphogenesis. J. Cell Biol. 179:381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cabeen M. T., et al. 2009. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 28:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cabeen M. T., Jacobs-Wagner C. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3:601–610 [DOI] [PubMed] [Google Scholar]
- 34. Cabrera J. E., Cagliero C., Quan S., Squires C. L., Jin D. J. 2009. Active transcription of rRNA operons condenses the nucleoid in Escherichia coli: examining the effect of transcription on nucleoid structure in the absence of transertion. J. Bacteriol. 191:4180–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Campbell I. D. 2008. The Croonian lecture 2006: structure of the living cell. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363:2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carballido-López R. 2006. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 70:888–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cayley S., Lewis B. A., Guttman H. J., Record M. T., Jr 1991. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J. Mol. Biol. 222:281–300 [DOI] [PubMed] [Google Scholar]
- 38. Chaplin M. 2006. Do we underestimate the importance of water in cell biology? Nat. Rev. Mol. Cell. Biol. 7:861–866 [DOI] [PubMed] [Google Scholar]
- 39. Clegg J. S. 1984. Intracellular water and the cytomatrix: some methods of study and current views. J. Cell Biol. 99:167s–171s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cooper C. L., Dubin P. L., Kayitmazer A. B., Turksen S. 2005. Polyelectrolyte-protein complexes. Curr. Opin. Colloid Interface Sci. 10:52–78 [Google Scholar]
- 41. Csonka L. N. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Mol. Biol. Rev. 53:121–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dawkins R. 1996. The blind watchmaker: why the evidence of evolution reveals a universe without design. W. W. Norton & Co., New York, NY [Google Scholar]
- 43. Debye P., Hückel E. 1923. Zur Theorie der Elektrolyte. Phys. Z. 24:185–206 [Google Scholar]
- 44. Dedmon M. M., Patel C. N., Gregory B., Young G. B., Pielak G. J. 2002. FlgM gains structure in living cells. Proc. Natl. Acad. Sci. U. S. A. 99:12681–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. deHaseth P. L., Lohman T. M., Record M. T., Jr 1977. Nonspecific interaction of lac repressor with DNA: an association reaction driven by counterion release. Biochemistry 16:4783–4790 [DOI] [PubMed] [Google Scholar]
- 46. Diennes L., Sharp J. T. 1956. The role of high electrolyte concentration in the production and growth of L forms of bacteria. J. Bacteriol. 71:208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dill K. A., Truskett T. M., Vlachy V., Hribar-Lee B. 2005. Modelling water, hydrophobic effect and ion solvation. Annu. Rev. Biophys. Biomol. Struct. 34:173–199 [DOI] [PubMed] [Google Scholar]
- 48. Dix J. A., Verkman A. S. 2008. Crowding effects on diffusion in solutions and cells. Annu. Rev. Biophys. 37:247–263 [DOI] [PubMed] [Google Scholar]
- 49. Draper D. E. 2004. A guide to ions and RNA structure. RNA 10:335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drost-Hansen W. 2001. Temperature effects on cell functioning—a critical role for vicinal water. Cell. Mol. Biol. (Noisy-le-Grand) 47:865–883 [PubMed] [Google Scholar]
- 51. Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradović Z. 2002. Intrinsic disorder and protein function. Biochemistry 41:6573–6582 [DOI] [PubMed] [Google Scholar]
- 52. Dworkin J., Losick R. 2001. Linking nutritional status to gene activation and development. Genes Dev. 15:1051–1054 [DOI] [PubMed] [Google Scholar]
- 53. Dworkin J. 2009. Cellular polarity in prokaryotic organisms. Cold Spring Harb. Perspect. Biol. 1:a003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dye N. A., Shapiro L. 2007. The push and pull of the bacterial cytoskeleton. Trends Cell Biol. 17:239–245 [DOI] [PubMed] [Google Scholar]
- 55. Ebersbach G., Jacobs-Wagner C. 2007. Exploration into the spatial and temporal mechanisms of bacterial polarity. Trends Microbiol. 15:101–108 [DOI] [PubMed] [Google Scholar]
- 56. Elcock A. H. 2010. Models of macromolecular crowding effects and the need for quantitative comparisons with experiment. Curr. Opin. Struct. Biol. 20:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ellis R. J., Minton A. P. 2006. Protein aggregation in crowded environments. Biol. Chem. 387:485–497 [DOI] [PubMed] [Google Scholar]
- 58. Ellis R. J. 2001. Macromolecular crowding—obvious but underappreciated. Trends Biochem. Sci. 26:597–604 [DOI] [PubMed] [Google Scholar]
- 59. Ellowitz M. B., Surette M. G., Wolf P.-E., Stock J. B., Leibler S. 1999. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 181:197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Errington J. 2003. Dynamic proteins and a cytoskeleton in bacteria. Nat. Cell Biol. 5:175–178 [DOI] [PubMed] [Google Scholar]
- 61. Fang J., Zhang L., Bazylinski D. A. 2010. Deep-sea piezosphere and piezophiles: geomicrobiology and biogeochemistry. Trends Microbiol. 18:413–422 [DOI] [PubMed] [Google Scholar]
- 62. Fenn E. E., Wong D. B., Fayer M. D. 2009. Water dynamics at neutral and ionic interfaces. Proc. Natl. Acad. Sci. U. S. A. 106:15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Flaugh S. L., Lumb K. J. 2001. Effects of macromolecular crowding on the intrinsically disordered proteins c-Fos and p27. Biomacromolecules 2:538–540 [DOI] [PubMed] [Google Scholar]
- 64. Franks F. 1983. Water. The Royal Society of Chemistry, London, United Kingdom [Google Scholar]
- 65. Frenkiel-Krispin D., et al. 2004. Nucleoid restructuring in stationary-state bacteria. Mol. Microbiol. 51:395–405 [DOI] [PubMed] [Google Scholar]
- 66. Fujiwara I., Vavylonis D., Pollard T. D. 2007. Polymerization kinetics of ADP- and ADPi-actin determined by fluorescence microscopy. Proc. Natl. Acad. Sci. U. S. A. 104:8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fuller R. S., Kaguni J. M., Kornberg A. A. 1981. Enzymatic replication of the origin of Escherichia coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 78:7370–7374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gao J. Y., Dubin P. L. 1999. Binding of proteins to copolymers of varying hydrophobicity. Biopolymers 49:185–193 [DOI] [PubMed] [Google Scholar]
- 69. Garner M. M., Burg M. B. 1994. Macromolecular crowding and confinement in cells exposed to hypertonicity. Am. J. Physiol. 266:C877–C892 [DOI] [PubMed] [Google Scholar]
- 70. Garner E. C., Campbell C. S., Weibel D. B., Mullins R. D. 2007. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315:1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gerdes K., Moeller-Jensen J., Ebersbach G., Kruse T., Nordström K. 2004. Bacterial mitotic machineries. Cell 116:359–366 [DOI] [PubMed] [Google Scholar]
- 72. Gitai Z. 2005. The new bacterial cell biology: moving parts and subcellular architecture. Cell 120:577–586 [DOI] [PubMed] [Google Scholar]
- 73. Gitai Z., Thanbichler M., Shapiro L. 2005. The choreographed dynamics of bacterial chromosomes. Trends Microbiol. 13:221–228 [DOI] [PubMed] [Google Scholar]
- 74. Glass J. I., et al. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U. S. A. 103:425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Goldman R. D., Gruenbaum Y., Moir R. D., Shumaker D. K., Spann T. P. 2002. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16:533–547 [DOI] [PubMed] [Google Scholar]
- 76. Gonzalez M. D., Beckwith J. 2009. Divisome under construction: distinct domains of the small membrane protein FtsB are necessary for interaction with multiple cell division proteins. J. Bacteriol. 191:2815–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Goodsell D. S. 1991. Inside a living cell. Trends Biochem. Sci. 16:203–206 [DOI] [PubMed] [Google Scholar]
- 78. Goodsell D. S. 2010. The machinery of life. Springer Science + Business Media, New York, NY [Google Scholar]
- 79. Gouaux E., MacKinnon R. 2005. Principles of selective ion transport in channels and pumps. Science 310:1461–1465 [DOI] [PubMed] [Google Scholar]
- 80. Gralla J. D. 2005. Escherichia coli rRNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol. Microbiol. 55:973–977 [DOI] [PubMed] [Google Scholar]
- 81. Gribaldo S., Brochier-Armanet C. 2006. The origin and evolution of Archaea: a state of the art. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gummel J., Boué F., Clemens D., Cousin F. 2008. Finite size and inner structure controlled by electrostatic screening in globular complexes of proteins and polyelectrolyte. Soft Matter 4:1653–1664 [DOI] [PubMed] [Google Scholar]
- 83. Gutfreund H. 1995. Kinetics for the life sciences: receptors, transmitters and catalysts. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 84. Halford S. E., Marko J. F. 2004. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32:3040–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Halford S. E. 2009. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 37:343–348 [DOI] [PubMed] [Google Scholar]
- 86. Halle B. 2004. Protein hydration dynamics in solution: a critical survey. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:1207–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Han M. J., Lee S. Y. 2006. The Escherichia coli proteome: past, present, and future prospects. Mol. Biol. Rev. 70:362–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Harold F. M. 2007. Bacterial morphogenesis: learning how cells make cells. Curr. Opin. Microbiol. 10:591–595 [DOI] [PubMed] [Google Scholar]
- 89. Harold F. M. 2001. The way of the cell: molecules, organisms, and the order of life. Oxford University Press, New York, NY [Google Scholar]
- 90. Harold F. M. 2005. Molecules into cells: specifying spatial architecture. Microbiol. Mol. Biol. Rev. 69:544–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hartwell L. H., Hopfield J. J., Leibler S., Murray A. W. 1999. From molecular to modular cell biology. Nature 402:C47–C52 [DOI] [PubMed] [Google Scholar]
- 92. Henderson L. J. 1913. The fitness of the environment. MacMillan, New York, NY [Google Scholar]
- 93. Higgins C. F., Cairney J., Stirling D. A., Sutherland L., Booth I. R. 1987. Osmotic regulation of gene expression: ionic strength as an intracellular signal? Trends Biochem. Sci. 12:339–344 [Google Scholar]
- 94. Hildebrand J. H. 1969. Relative diffusivities of methane in water and carbon tetrachloride. Proc. Natl. Acad. Sci. U. S. A. 64:1329–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hildebrand J. H. 1979. Is there a “hydrophobic effect”? Proc. Natl. Acad. Sci. U. S. A. 76:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hud N. V. 2008. Nucleic acid metal ion interactions. Royal Society of Chemistry, London, United Kingdom [Google Scholar]
- 97. Ishikawa H., Kwak K., Chung J. K., Kim S., Fayer M. D. 2008. Direct observation of fast protein conformational switching. Proc. Natl. Acad. Sci. U. S. A. 105:8619–8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jacob F., Brenner S., Cusin F. 1963. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 23:329–348 [Google Scholar]
- 99. Janmey P. A., Peetermans J., Zaner K. S., Stossel T. P., Tanaka T. 1986. Structure and mobility of actin filaments as measured by quasielastic light scattering, viscometry and electron microscopy. J. Biol. Chem. 261:8357–8362 [PubMed] [Google Scholar]
- 100. Jasnin M., Molin M., Haertlein M., Zaccai G., Tehei M. 2008. Down to atomic-scale intracellular water dynamics. EMBO Rep. 9:543–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jasnin M. 2009. Atomic-scale dynamics inside living cells explored by neutron. J. R. Soc. Interface 6:S611–S617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jeffrey C. J. 1999. Moonlighting proteins. Trends Biochem. Sci. 24:8–11 [DOI] [PubMed] [Google Scholar]
- 103. Jenkins C., et al. 2002. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc. Natl. Acad. Sci. U. S. A. 99:17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jiao M., Li H.-T., Chen J., Minton A. P., Liang Y. 2010. Attractive protein-polymer interactions markedly alter the effect of macromolecular crowding on protein association equilibria. Biophys. J. 99:914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jo S., Vargyas M., Vasko-Szedlar J., Roux B., Im W. 2008. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 36:W270–W275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jones L. J., Carballido-López R., Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922 [DOI] [PubMed] [Google Scholar]
- 107. Katcznaowska M., Rydén-Aulin M. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71:477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kausik R., et al. 2009. Local water dynamics in coacervated polyelectrolytes monitored through dynamic nuclear polarization-enhanced 1H NMR. Macromolecules 42:7404–7412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kayitmazer A. B., et al. 2007. Mesophase separation and probe dynamics in protein-polyelectrolyte coacervates. Soft Matter 3:1064–1076 [DOI] [PubMed] [Google Scholar]
- 110. Keighron J. D., Keating C. D. 2010. Towards a minimal cytoplasm, p. 3–30 In Luisi P. L., Stano P. (ed.), The minimal cell: the biophysics of cell compartment and the origin of cell functionality. Springer Science + Business Media, New York, NY [Google Scholar]
- 111. Kell D. B. 2006. Metabolomics, modelling and machine learning in systems biology—towards an understanding of the languages of cells. Drug Discov. Today 11:1085–1092 [DOI] [PubMed] [Google Scholar]
- 112. Kim S. Y., Gitai Z., Kinkhabwala A., Shapiro L., Moerner W. E. 2006. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 103:10929–10934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Konopka M. C., et al. 2009. Cytoplasmic protein mobility in osmotically stressed Escherichia coli. J. Bacteriol. 191:231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kornberg A. 2000. Ten commandments: lessons from the enzymology of DNA replication. J. Bacteriol. 182:3613–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kühner S., et al. 2009. Proteome organization in a genome-reduced bacterium. Science 326:1235–1240 [DOI] [PubMed] [Google Scholar]
- 116. Kumar M., Mommer M. S., Sourjik V. 2010. Mobility of cytoplasmic, membrane, and DNA-binding proteins in Escherichia coli. Biophys. J. 98:552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ladokhin A. S., White S. H. 2001. Protein chemistry at membrane interfaces: non-additivity of electrostatic and hydrophobic interactions. J. Mol. Biol. 309:543–552 [DOI] [PubMed] [Google Scholar]
- 118. Leaver M., Domínguez-Cuevas P., Coxhead J. M., Daniel R. A., Errington J. 2009. Life without a wall or division machine in Bacillus subtilis. Nature 457:849–853 [DOI] [PubMed] [Google Scholar]
- 119. Levitt M., Park B. H. 1993. Water: now you see it, now you don't. Structure 1:223–226 [DOI] [PubMed] [Google Scholar]
- 120. Lewis P. J. 2008. Subcellular organization in bacteria, p. 1–42 In Sharoud W. E. (ed.), Bacterial physiology: a molecular approach. Springer-Verlag, Berlin, Germany [Google Scholar]
- 121. Llopis P. M., et al. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Loeb J. 1906. The dynamics of living matter, p. 16 and 29 The Columbia University Press, New York, NY [Google Scholar]
- 123. Loeb J. 1899. On the nature of the process of fertilization and the artificial production of normal larvae (plutei) from the unfertilized eggs of the sea urchin. Am. J. Physiol. 3:135–138 [Google Scholar]
- 124. Long M. S., Jones C. D., Helfrich M. R., Mangeney-Slavin L. K., Keating C. D. 2005. Dynamic microcompartmentation in synthetic cells. Proc. Natl. Acad. Sci. U. S. A. 102:5920–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Losick R., Shapiro L. 1999. Changing views on the nature of the bacterial cell: from biochemistry to cytology. J. Bacteriol. 181:4143–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lutkenhaus J. 2003. Another cytoskeleton in the closet. Cell 115:648–650 [DOI] [PubMed] [Google Scholar]
- 127. Lybarger S. R., Maddock J. R. 2001. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 183:3261–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Maddox J., Randi J., Stewart W. W. 1988. High dilution experiments a delusion. Nature 334:287–290 [DOI] [PubMed] [Google Scholar]
- 129. Marenduzzo D., Finan K., Cook P. R. 2006. The depletion attraction: an underappreciated force driving cellular organization. J. Cell Biol. 175:681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Margolin W. 2009. Sculpting the bacterial cell. Curr. Biol. 19:R812–R822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Marles-Wright J., Lewis J. L. 2008. The Bacillus subtilis stressosome. Commun. Integr. Biol. 1:182–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Marrs J. A., Bouck G. B. 1992. The two major membrane skeletal proteins (articulins) of Euglena gracilis define a new class of cytoskeletal proteins. J. Cell Biol. 118:1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Martin W., Baross J., Kelley D., Russell M. J. 2008. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 6:805–814 [DOI] [PubMed] [Google Scholar]
- 134. Mascarenhas J., Weber M. H. W., Graumann P. L. 2001. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2:685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Mathews C. K. 1993. The cell—a bag of enzymes or a network of channels? J. Bacteriol. 175:6377–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. McNulty B. C., Young G. B., Pielak G. J. 2006. Macromolecular crowding in the Escherichia coli periplasm maintains alpha-synuclein disorder. J. Mol. Biol. 355:893–897 [DOI] [PubMed] [Google Scholar]
- 137. Michie K. A., Löwe J. 2006. Dynamic filaments of the bacterial cytoskeleton. Annu. Rev. Biochem. 75:467–492 [DOI] [PubMed] [Google Scholar]
- 138. Mignot T., Shaevitz J. W. 2008. Active and passive mechanisms of intracellular transport and localization in bacteria. Curr. Opin. Microbiol. 11:580–585 [DOI] [PubMed] [Google Scholar]
- 139. Mika J. T., van der Bogaart G., Veenhof L., Krasnikov V., Poolman B. 2010. Molecular sieving properties of the cytoplasm of Escherichia coli and consequences of osmotic stress. Mol. Microbiol. 77:200–207 [DOI] [PubMed] [Google Scholar]
- 140. Mika J. T., Poolman B. 2011. Macromolecule diffusion and confinement in prokaryotic cells. Curr. Opin. Biotechnol. 22:117–126 [DOI] [PubMed] [Google Scholar]
- 141. Miklos A. C., Li C., Sharaf N. G., Pielak G. J. 2010. Volume exclusion and soft interaction effects on protein stability under crowded conditions. Biochemistry 49:6984–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Mikucki J. A., et al. 2009. A contemporary microbially maintained subglacial ferrous “ocean.” Science 324:397–400 [DOI] [PubMed] [Google Scholar]
- 143. Miller D. S., Horowitz S. B. 1986. Intracellular compartmentalization of ATP. J. Biol. Chem. 261:13911–13915 [PubMed] [Google Scholar]
- 144. Minton A. P. 1983. The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences. Mol. Cell. Biochem. 55:119–140 [DOI] [PubMed] [Google Scholar]
- 145. Minton A. P. 2001. The influence of macromolecular crowding and confinement on biochemical reactions in physiological media. J. Biol. Chem. 276:10577–10580 [DOI] [PubMed] [Google Scholar]
- 146. Mitchell P. 1979. Compartmentation and communication in living systems. Ligand conduction: a general catalytic principle in chemical, osmotic, and chemiosmotic reaction systems. Eur. J. Biochem. 95:1–20 [DOI] [PubMed] [Google Scholar]
- 147. Mitchison T. J., Charras G. T., Mahadevan L. 2008. Implications of a poroelastic cytoplasm for the dynamics of animal cell shape. Semin. Cell Dev. Biol. 19:215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Molloy R. G., et al. 2010. Aquifex aeolicus FlgM protein exhibits a temperature-dependent disordered nature. Biochim. Biophys. Acta 1804:1457–1466 [DOI] [PubMed] [Google Scholar]
- 149. Morbach S., Krämer R. 2002. Body shaping under water stress: osmosensing and osmoregulation of solute transport in bacteria. Chembiochem 3:384–397 [DOI] [PubMed] [Google Scholar]
- 150. Morris D. M., Jensen G. J. 2008. Toward a biomechanical understanding of whole bacterial cells. Annu. Rev. Biochem. 77:583–613 [DOI] [PubMed] [Google Scholar]
- 151. Mouillon J.-M., Eriksson S. K., Harryson P. 2008. Mimicking the plant cell interior under water stress by macromolecular crowding: disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol. 148:1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Nanninga N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Nanninga N. 2001. Cytokinesis in prokaryotes and eukaryotes: common principles and different solutions. Microbiol. Mol. Biol. Rev. 65:319–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Norris V., et al. 2007. Functional taxonomy of bacterial hyperstructures. Microbiol. Mol. Biol. Rev. 71:230–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Pace N. R. 2009. Mapping the tree of life: progress and prospects. Microbiol. Mol. Biol. Rev. 73:565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Paciaroni A., et al. 2009. Coupled relaxations at the protein-water interface in the picosecond time scale. J. R. Soc. Interface 6:S635–S640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Pal S. K., Zewail A. H. 2004. Dynamics of water in biological recognition. Chem. Rev. 104:2099–2123 [DOI] [PubMed] [Google Scholar]
- 158. Pal S. K., Peon J., Bagchi B., Zewail A. H. 2002. Biological water: femtosecond dynamics of macromolecular hydration. J. Phys. Chem. B 106:12376–12395 [Google Scholar]
- 159. Park S., Moilanen D. E., Fayer M. D. 2008. Water dynamics—the effects of ions and nanoconfinement. J. Phys. Chem. B 112:5279–5290 [DOI] [PubMed] [Google Scholar]
- 160. Partikian A., Ölveczky B., Swaminathan R., Li Y. X., Verkman A. S. 1998. Rapid diffusion of green fluorescent protein in the mitochondrial matrix. J. Cell Biol. 140:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Reference deleted.
- 162. Pauly P. J. 1987. Controlling life: Jacques Loeb and the engineering ideal in biology. Oxford University Press, New York, NY [Google Scholar]
- 163. Persson E., Halle B. 2008. Cell water dynamics on multiple time scales. Proc. Natl. Acad. Sci. U. S. A. 105:6266–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Phillips J. N., Jr. 1997. Structure and dynamics of green fluorescent protein. Curr. Opin. Struct. Biol. 7:821–827 [DOI] [PubMed] [Google Scholar]
- 165. Pielak G. J., Miklos A. C. 2010. Crowding and function reunite. Proc. Natl. Acad. Sci. U. S. A. 107:17457–17458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Pielak G. J. 2005. A model of intracellular organization. Proc. Natl. Acad. Sci. U. S. A. 102:5901–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Polianskyte Z., et al. 2009. LACTB is a filament-forming protein localized in mitochondria. Proc. Natl. Acad. Sci. U. S. A. 106:18960–18965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Pollack G. H. 2001. Cells, gels and the engines of life. A new, unifying approach to cell function. Ebner & Sons, Seattle, WA [Google Scholar]
- 169. Poolman B., Glaasker E. 1998. Regulation of compatible solutes accumulation in bacteria. Mol. Microbiol. 29:397–407 [DOI] [PubMed] [Google Scholar]
- 170. Poolman B., Spitzer J. J., Wood J. M. 2004. Bacterial osmosensing: roles of membrane structure and electrostatics in lipid-protein and protein-protein interactions. Biophys. Biochim. Acta 1666:88–104 [DOI] [PubMed] [Google Scholar]
- 171. Potts M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Mol. Biol. Rev. 58:755–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Qu Y., Bolen D. W. 2002. Efficacy of macromolecular crowding in forcing proteins to fold. Biophys. Chem. 101–102:155–165 [DOI] [PubMed] [Google Scholar]
- 173. Rafelski S. M., Theriot J. A. 2004. Crawling toward a unified model of cell motility: spatial and temporal regulation of actin dynamics. Annu. Rev. Biochem. 73:209–239 [DOI] [PubMed] [Google Scholar]
- 174. Record M. T., Courtenay E. S., Cayley S., Guttman J. H. 1998. Biophysical compensation mechanisms buffering E. coli protein-nucleic acid interactions against changing environments. Trends Biochem. Sci. 23:190–194 [DOI] [PubMed] [Google Scholar]
- 175. Reeves A., Martinez L., Haldenwang W. 2010. Expression of, and in vivo stressosome formation of, single members of the RsbR protein family in Bacillus subtilis. Microbiology 156:990–998 [DOI] [PubMed] [Google Scholar]
- 176. Rivas G., Ferrone F., Herzfeld J. J. 2004. Life in a crowded world. EMBO Rep. 5:23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Robinow C., Kellenberger E. 1994. The bacterial nucleoid revisited. Microbiol. Mol. Biol. Rev. 58:211–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Robinson R. A., Stokes R. H. 1959. Electrolyte solutions. Butterworths, London, United Kingdom [Google Scholar]
- 179. Rohwer J. M., Potsma P. W., Kholodenko B. N., Westerhoff H. V. 1998. Implications of macromolecular crowding for signal transduction and metabolic channeling. Proc. Natl. Acad. Sci. U. S. A. 95:10547–10552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Roque A., Ponte I., Suau P. P. 2007. Macromolecular crowding induces a molten globule state in the C-terminal domain of histone H1. Biophys. J. 93:2170–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ross P. D., Minton A. P. 1977. Analysis of nonideal behavior in concentrated hemoglobin solutions. J. Mol. Biol. 112:437–452 [DOI] [PubMed] [Google Scholar]
- 182. Rübenhagen R., Rönsch H., Jung H., Krämer R., Morbach S. 2000. Osmosensor and osmoregulator properties of the betaine carrier BetP from Corynebacterium glutamicum in proteoliposomes. J. Biol. Chem. 275:735–741 [DOI] [PubMed] [Google Scholar]
- 183. Rudner D. Z., Losick R. 2009. Protein subcellular localization in bacteria. Cold Spring Harb. Perspect. Biol. 2:a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Samiotakis A., Wittung-Stafshede P., Cheung M. S. 2009. Folding, stability and shape of proteins in crowded environments: experimental and computational approaches. Int. J. Mol. Sci. 10:572–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Sapp J. 2005. The prokaryote-eukaryote dichotomy: meanings and mythology. Microbiol. Mol. Biol. Rev. 69:292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Schlesinger A. P., Wang Y., Tadeo X., Millet O., Pielak G. J. 2011. Macromolecular crowding fails to fold a globular protein in cells. J. Am. Chem. Soc. 133:8082–8085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Schlieper D., Oliva M. A., Andreu J. M., Löwe J. 2005. Structure of bacterial tubulin BtubA/B: evidence for horizontal gene transfer. Proc. Natl. Acad. Sci. U. S. A. 102:9170–9175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Schliwa M. 2002. The evolving complexity of cytoplasmic structure. Nat. Rev. Mol. Cell. Biol. 3:2–6 [DOI] [PubMed] [Google Scholar]
- 189. Schnell S., Turner T. E. 2004. Reaction kinetics in intracellular environments with macromolecular crowding: simulations and rate laws. Prog. Biophys. Mol. Biol. 85:235–260 [DOI] [PubMed] [Google Scholar]
- 190. Schreiber G., Fersht A. R. 1996. Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 3:427–431 [DOI] [PubMed] [Google Scholar]
- 191. Shapiro L., Losick R. 1997. Protein localization and cell fate in bacteria. Science 276:712–718 [DOI] [PubMed] [Google Scholar]
- 192. Shapiro L., Losick R. R. 2000. Dynamic spatial regulation in the bacterial cell. Cell 100:89–98 [DOI] [PubMed] [Google Scholar]
- 193. Shapiro L., McAdams H., Losick R. 2009. Why and how bacteria localize proteins. Science 326:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Shepherd V. A. 2006. The cytomatrix as a cooperative system of macromolecular and water networks. Curr. Top. Dev. Biol. 75:173–223 [DOI] [PubMed] [Google Scholar]
- 195. Shih Y.-L., Rothfield L. 2006. The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 70:729–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Shilhavy T., Kahne J. D., Walker S. 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Short B., Haas A., Barr F. A. 2005. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim. Biophys. Acta 1744:383–395 [DOI] [PubMed] [Google Scholar]
- 198. Siekevitz P., Zamecnik P. C. 1981. Ribosomes and protein synthesis. J. Cell Biol. 91:53s–65s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Singer S. J., Nicholson G. L. 1972. The fluid mosaic model of the structure of cell membranes. Science 175:720–731 [DOI] [PubMed] [Google Scholar]
- 200. Sörensen H. P., Mortensen K. K. 2005. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Factories 4:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Southall N. T., Dill K. A., Haymet A. D. J. 2002. A view of the hydrophobic effect. J. Phys. Chem. B 106:521–533 [Google Scholar]
- 202. Spitzer J. J. 1984. A re-interpretation of hydration forces. Nature 310:396–397 [Google Scholar]
- 203. Spitzer J. J. 1992. Theory of dissociative electrical double layers: the limit of close separations and “hydration” forces. Langmuir 8:1659–1662 [Google Scholar]
- 204. Spitzer J. J. 2003. Maxwellian double layer forces: from infinity to contact. Langmuir 19:7099–7111 [Google Scholar]
- 205. Spitzer J. J., Poolman B. 2009. The role of biomacromolecular crowding, ionic strength and physicochemical gradients in the complexities of life's emergence. Microbiol. Mol. Biol. Rev. 73:371–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Spitzer J. J., Poolman B. 2005. Electrochemical structure of the crowded cytoplasm. Trends Biochem. Sci. 30:536–541 [DOI] [PubMed] [Google Scholar]
- 207. Srere P. A. 1984. Why are enzymes so big? Trends Biochem. Sci. 9:387–390 [Google Scholar]
- 208. Srere P. A. 1985. The metabolon? Trends Biochem. Sci. 10:109–110 [Google Scholar]
- 209. Staundinger H. 1920. Über polymerisation. Ber. Dtsch. Chem. Ges. 53:1073–1085 [Google Scholar]
- 210. Stetter K. O. 1982. Ultrathin mycelia-forming organisms from submarine volcanic areas having an optimum growth temperature of 105°C. Nature 300:258–260 [Google Scholar]
- 211. Stockbridge R. B., Lewis C. A., Jr., Yuan Y., Wolfenden R. 2010. Impact of temperature on the time required for the establishment of primordial biochemistry, and for the evolution of enzymes. Proc. Natl. Acad. Sci. U. S. A. 107:22102–22105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Stossel T. P. 1984. Contribution of actin to the structure of the cytoplasmic matrix. J. Cell Biol. 99:15s–21s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Stradner A., et al. 2004. Equilibrium cluster formation in concentrated protein solutions and colloids. Nature 432:492–495 [DOI] [PubMed] [Google Scholar]
- 214. Stuurman N., Heins S., Aebi U. 1998. Nuclear lamins: their structure, assembly, and interactions. J. Struct. Biol. 122:42–66 [DOI] [PubMed] [Google Scholar]
- 215. Sullivan W. T., III, Baross J. A. (ed.). 2007. Planets and life: the emerging science of astrobiology. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 216. Svedberg T., Fåhraeus R. 1926. A new method for the determination of the molecular weight of the proteins. J. Am. Chem. Soc. 48:430–438 [Google Scholar]
- 217. Tanford C. 1978. The hydrophobic effect and the organization of living matter. Science 200:1012–1018 [DOI] [PubMed] [Google Scholar]
- 218. Tanford C. 1980. The hydrophobic effect: formation of micelles and biological membranes. John Wiley & Sons, New York, NY [Google Scholar]
- 219. Tanford C., Reynolds J. 2001. Nature's robots. Oxford University Press, Oxford, England [Google Scholar]
- 220. Thanbichler M., Shapiro L. 2008. Getting organized—how bacterial cells move proteins and DNA. Nat. Rev. Microbiol. 6:28–40 [DOI] [PubMed] [Google Scholar]
- 221. Thanbichler M., Wang S. C., Shapiro L. 2005. The bacterial nucleoid: a highly organized and dynamic structure. J. Cell. Biochem. 96:506–521 [DOI] [PubMed] [Google Scholar]
- 222. Thanbichler M., Viollier P. H., Shapiro L. 2005. The structure and function of bacterial chromosome. Curr. Opin. Genet. Dev. 15:153–162 [DOI] [PubMed] [Google Scholar]
- 223. Thanbichler M. 2009. Synchronization of chromosome dynamics and cell division in bacteria. Cold Spring Harb. Perspect. Biol. doi: 10.1101/cshperpect.a000331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Theriot J. A. 2000. The polymerization motor. Traffic 1:19–28 [DOI] [PubMed] [Google Scholar]
- 225. Toro E., Shapiro L. 2010. Bacterial chromosome organization and segregation. Cold Spring Harb. Perspect. Biol. 2:a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Uversky V. N. 2009. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter-ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 28:305–325 [DOI] [PubMed] [Google Scholar]
- 227. Vale R. D. 2003. The molecular motor toolbox for intracellular transport. Cell 112:467–480 [DOI] [PubMed] [Google Scholar]
- 228. Valkenburg J. A. C., Woldringh C. L. 1984. Phase separation between nucleoid and cytoplasm in Escherichia coli as defined by immersive refractometry. J. Bacteriol. 160:1151–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. van den Bogaart G., Heermans N., Krasnikov V., Poolman B. 2007. Protein mobility and diffusive barriers in Escherichia coli: consequences of osmotic stress. Mol. Microbiol. 64:858–871 [DOI] [PubMed] [Google Scholar]
- 230. van den Ent F., Amos L. A., Löwe J. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39–44 [DOI] [PubMed] [Google Scholar]
- 231. van der Heide T., Stuart M. C. A., Poolman B. 2001. On the osmotic signal and osmosensing mechanism of an ABC transport system for glycine betaine. EMBO J. 20:7022–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Van-Quynh A., Willson S., Bryant R. G. 2003. Protein reorientation and bound water molecules measured by 1H magnetic spin-lattice relaxation. Biophys. J. 84:558–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. van't Hoff J. H. 1888. The function of osmotic pressure in the analogy between solutions and gases. Phil. Mag. 26:81–125 [Google Scholar]
- 234. Verkman A. S. 2002. Solute and macromolecular diffusion in cellular aqueous compartments. Trends Biochem. Sci. 27:27–33 [DOI] [PubMed] [Google Scholar]
- 235. Viollier P. H., Shapiro L. 2004. Spatial complexity of mechanisms controlling a bacterial cell cycle. Curr. Opin. Microbiol. 7:572–578 [DOI] [PubMed] [Google Scholar]
- 236. Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. Y. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573–586 [DOI] [PubMed] [Google Scholar]
- 237. Voeltz G. K., Prinz W. A. 2007. Sheets, ribbons and tubules—how organelles get their shape. Nat. Mol. Cell. Biol. 8:258–264 [DOI] [PubMed] [Google Scholar]
- 238. von Hippel P. H. 2007. From “simple” DNA-protein interactions to the macromolecular machines of gene expression. Annu. Rev. Biophys. Biomol. Struct. 36:79–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Walter H., Brooks D. E. 1995. Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. 361:135–139 [DOI] [PubMed] [Google Scholar]
- 240. Wang Y., Li C., Pielak G. J. 2010. Effects of proteins on protein diffusion. J. Am. Chem. Soc. 132:9392–9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Weber S. C., Spakowitz A. J., Theriot J. A. 2010. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys. Rev. Lett. 204:238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Welch G. R., Easterby J. S. 1994. Metabolic channeling versus free diffusion: transition-time analysis. Trends Biochem. Sci. 19:193–197 [DOI] [PubMed] [Google Scholar]
- 243. Westerhoff H. V., Welch G. R. 1992. Enzyme organization and the direction of metabolic flow: physicochemical considerations. Curr. Top. Cell. Regul. 33:361–390 [DOI] [PubMed] [Google Scholar]
- 244. Wiggins P. 1990. Role of water in some biological processes. Microbiol. Mol. Biol. Rev. 54:432–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Wiggins P. 2001. High and low density intracellular water. Cell. Mol. Biol. (Noisy-le-Grand) 47:735–744 [PubMed] [Google Scholar]
- 246. Williamson J. R. 2008. Biophysical studies of bacterial ribosome assembly. Curr. Opin. Struct. Biol. 18:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Wilson D. N., Nierhaus K. H. 2007. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 42:187–219 [DOI] [PubMed] [Google Scholar]
- 248. Woese C. R. 1998. The universal ancestor. Proc. Natl. Acad. Sci. U. S. A. 95:6854–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Woese C. R. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. U. S. A. 97:8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Woese C. R. 2002. On the evolution of cells. Proc. Natl. Acad. Sci. U. S. A. 99:8742–8747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Woese C. R. 2004. A new biology for a new century. Microbiol. Mol. Biol. Rev. 68:173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Woese C. R. 2009. How the microbial world saved evolution from the Scylla of molecular biology and the Charybdis of the modern synthesis. Microbiol. Mol. Biol. Rev. 73:14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Woese C. R., Fox G. E. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 74:5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Woese C. R., Kandler O., Wheelis M. L. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U. S. A. 87:4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Woldringh C. L. 2002. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol. Microbiol. 45:17–29 [DOI] [PubMed] [Google Scholar]
- 256. Woldringh C. L., Nanninga N. 2006. Structural and physical aspects of bacterial chromosome segregation. J. Struct. Biol. 156:273–283 [DOI] [PubMed] [Google Scholar]
- 257. Wood J. M. 1999. Osmosensing by bacteria: signals and membrane based sensors. Microbiol. Mol. Biol. Rev. 63:230–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Wood J. 2007. Bacterial osmosensing transporters. Methods Enzymol. 428:77–107 [DOI] [PubMed] [Google Scholar]
- 259. Wood J. 2010. Osmotic stress, p. 133–156 In Storz G., Hengge R. (ed.), Bacterial stress responses, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 260. Yang Y., Glynn J. M., Olson B. J. S. C., Schmitz A. J., Osteryoung K. W. 2008. Plastid division: across time and space. Curr. Opin. Plant Biol. 11:577–584 [DOI] [PubMed] [Google Scholar]
- 261. Yeung T., et al. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science 319:210–213 [DOI] [PubMed] [Google Scholar]
- 262. Zeng X., et al. 2009. Pyrococcus CH1, an obligate piezophilic hyperthermophile: extending the upper pressure-temperature limits for life. ISME J. 3:873–876 [DOI] [PubMed] [Google Scholar]
- 263. Zhou H.-X., Rivas G., Minton A. P. 2008. Macromolecular crowding and confinement: biochemical, biophysical and potential physiological consequences. Annu. Rev. Biophys. 37:375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Zimmerman S. B., Minton A. P. 1993. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 22:27–65 [DOI] [PubMed] [Google Scholar]
- 265. Zimmerman S. B., Trach O. 1991. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 222:599–620 [DOI] [PubMed] [Google Scholar]
- 266. Zimmerman S. B., Murphy L. D. 2001. Release of compact nucleoids with characteristic shapes from Escherichia coli. J. Bacteriol. 183:5041–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Zonia L., Munnik T. 2007. Life under pressure: hydrostatic pressure in cell growth and function. Trends Plant Sci. 12:90–97 [DOI] [PubMed] [Google Scholar]
- 268. Zuckerkandl E., Pauling L. 1965. Molecules as documents of evolutionary history. J. Theor. Biol. 8:357–366 [DOI] [PubMed] [Google Scholar]