Abstract
Summary: The discovery of a new class of cytosolic receptors recognizing viral RNA, called the RIG-like receptors (RLRs), has revolutionized our understanding of the interplay between viruses and host cells. A tremendous amount of work has been accumulating to decipher the RNA moieties required for an RLR agonist, the signal transduction pathway leading to activation of the innate immunity orchestrated by type I interferon (IFN), the cellular and viral regulators of this pathway, and the viral inhibitors of the innate immune response. Previous reviews have focused on the RLR signaling pathway and on the negative regulation of the interferon response by viral proteins. The focus of this review is to put this knowledge in the context of the virus replication cycle within a cell. Likewise, there has been an expansion of knowledge about the role of innate immunity in the pathophysiology of viral infection. As a consequence, some discrepancies have arisen between the current models of cell-intrinsic innate immunity and current knowledge of virus biology. This holds particularly true for the nonsegmented negative-strand viruses (Mononegavirales), which paradoxically have been largely used to build presently available models. The aim of this review is to bridge the gap between the virology and innate immunity to favor the rational building of a relevant model(s) describing the interplay between Mononegavirales and the innate immune system.
INTRODUCTION
Being devoid of any energy source and metabolism, viruses must rely on a host cell for expression and replication. Because virus infection often leads to cell and/or tissue injury, debilitation, and/or death, the animal kingdom has developed through evolution specific mechanisms to either prevent, limit, control, or cure viral infections. The first line of defense is innate immunity, which produces immediate, but nonspecific, immune responses. Because it requires some time to be active after first encountering a pathogen, adaptive immunity represents a second line of defense. Assumed by the lymphoid system, it is characterized by extraordinary specificity and memory. There is a strong cross talk between innate and adaptive immunity that makes them not only complementary but also synergistic. Innate immunity comprises several mechanisms, such as the complement system and the “cellular” and the “cell-intrinsic” (i.e., intracellular) innate immunity. The complement system is an automatic and autonomous enzymatic cascade that is quickly activated upon contact. The main actors of cellular innate immunity, as far as viruses are concerned, are natural killer (NK) cells, which are able to recognize virus-infected cells exhibiting anomalous expression of major histocompatibility complex (MHC) molecules. The cell-intrinsic innate immunity consists of the activation of specific genes resulting in an antiviral state of both infected and neighbor cells via autocrine and paracrine signaling, respectively. More than half a century ago, interferon (IFN), later classified as type I IFN, was discovered as the cytokine induced by viral infection and able to activate an antiviral state (114, 115). It then took almost 50 years before C. Janeway et al. proposed that the innate immune system recognizes pathogens by expressing dedicated receptors able to bind to microbe-associated molecular patterns (MAMPs), which are known as pattern recognition receptors (PRRs) (118). Three subsets of PRRs, the Toll-like receptors (TLRs), the Rig-I like receptors (RLRs), and the Nod-like receptors (NLRs), can be engaged in recognition of viruses. TLRs were first demonstrated as innate immunity receptors in insects (242). They are located at the cell surface and/or in endosomes, and their expression is heterogeneous and mostly restricted to a few cell subsets. The cytosolic NLRs can recognize some virus families, particularly DNA viruses (122). The RLRs are cytoplasmic and expressed by all nucleated cells. Thus, they play a critical role in the early recognition of viruses in any tissue. For Chordata, double-stranded RNA (dsRNA) is the MAMP commonly recognized by the RLRs, hence their key role in recognizing infections by RNA viruses.
Among viruses possessing RNA genomes, the order of negative-single-strand viruses (Mononegavirales) encompasses many human and animal pathogens causing severe disease, such as measles virus (MeV), rinderpest virus (RPV), vesicular stomatitis virus (VSV), Ebola virus (EBOV), rabies virus (RABV), Nipah virus, and respiratory syncytial virus (RSV), to name a few. Except for in the Bornaviridae family, the virus cycle is entirely cytoplasmic. They all share a transcription and replication process that is unique in the living world. The viral polymerase uses a helicoidal nucleoprotein complex or nucleocapsid as a template instead of naked RNA. Despite their limited genome size and close relationship, these viruses have developed extraordinarily diverse strategies to escape detection by RLRs, to impair interferon signaling, and/or to counteract cellular antiviral effectors.
This review aims at integrating the most recent knowledge about the RLR-dependent activation of IFN, the replication of the Mononegavirales, and how they interact with each other. Their relevance in the physiopathology of diseases induced by prototypic viruses will then be discussed.
PRRs AND IFN RESPONSE
Viral nucleic acids, which may be DNA, single-stranded RNA (ssRNA), or double-stranded RNA (dsRNA), are the main common source of MAMPs that animal cells can detect. The nucleic acids are recognized in the endosomal compartment by TLR3, -7, -8, and -9 and possibly at the cell surface by TLR3, with the following specificities: DNA⇔TLR9, ssRNA⇔TLR7/8, and dsRNA⇔TLR3 (125). The intracellular RLRs selectively detect RNA-bearing moieties that are normally absent from the cytoplasm (see below). This nucleic acid recognition induces the activation of the genes encoding IFN-β and IFN-α1 (human; α4 in mouse), the only IFN genes that are directly activated by the RLRs through the downstream building of an enhanceosome made of AP1 (ATF-2/c-Jun), two interferon regulatory factor 3 (IRF3)/IRF7 heterodimers, and NF-κB (p50/p65 RelA) (211). The concomitant activation of IRF3 and NF-κB also results in the activation of a subset of cytokines with inflammatory properties. The transcriptional activation of all other IFN-α subtypes requires IRF7 homodimers, which are available only in cells expressing high levels of IRF7 (87). This constitutively occurs in plasmacytoid dendritic cells (pDCs) and endows them with the unique ability to secrete massive amounts of IFN-α after stimulation of their TLRs (37). The IRF7 gene is also a member of the interferon-stimulated genes (ISGs) that are activated downstream of the type I interferon receptor (IFNAR) in cells exposed to IFN-α/β (87). Thus, IRF7 mediates the IFN-α/β secretion amplification loop that occurs upon infection of cells primed with IFN. The binding of IFN to IFNAR activates a signaling cascade that leads to the phosphorylation and heterodimerization of STAT-1 and STAT2 transcription factors and their association with IRF9 to constitute the ISGF3-enhancing complex, which is translocated into the nucleus to mediate the activation of several hundred ISGs (87). ISGs comprise PRRs and modulators of the signaling pathway, as well as transcription factors responsible for the amplification loop and antiviral effectors (245). Of note, some of the ISGs can also be directly activated downstream of the RLRs because of the large combinatorial diversity of the transcription factors of the innate immune response and the heterogeneity of the various promoters (90). In addition, upon RLR-mediated activation of TRAF2 and TRAF6, part of IRF3 independently associates with Bax, and the complex is translocated to the mitochondria to induce cell apoptosis (46). This IRF3/Bax apoptosis pathway is part of the antiviral response, as shown by enhanced Sendai virus (SeV) and VSV replication in Bax-deficient cells (47). An antiviral state can be also induced by long dsRNA in the absence of RLR signaling and IFN activation. This points to a yet-unknown mechanism of intracellular detection of viral RNA, although the possible role of the RNA-dependent P1/eIF-2α protein kinase (PKR) has not been investigated (62).
STRUCTURE OF RLRs
Three members constitute the RLR family, i.e., RIG-I (retinoic acid-inducible gene I or DDX58), MDA5 (melanoma differentiation-associated gene 5), and LGP2 (laboratory of genetics and physiology) (305, 310). They are RNA helicases belonging to the helicase 2 superfamily, which is characterized by seven conserved sequences (I, Ia, and II to VI) (10). Upon recognition of their RNA agonist(s), RIG-I and MDA5 switch on the innate immunity program, whereas LGP2 acts as a regulator (139, 140, 247, 289, 305, 310). RIG-I and MDA5 have two caspase activation and recruitment domains (CARDs) at their N termini, and the three RLRs have homologous C-terminal domain (CTDs) with similar fold and RNA binding properties (158, 165, 194, 273, 296). Although only the CTD of RIG-I accommodates a 5′-triphosphate end (5′ppp), all RLRs preferentially bind to dsRNA with blunt ends, as shown biochemically and structurally. Four conserved cysteine residues and positively charged residues account for the RNA binding of the CTDs, mainly via electrostatic bonds. The helicase domain is highly conserved between the RLRs, with 41%, 35%, and 31% homology between MDA5 and LGP2, RIG-I and MDA5, and RIG-I and LGP2, respectively (313). This domain binds RNA (56, 83), and several point mutations in the conserved helicase motifs abolish RNA binding (10, 221). The binding of dsRNA activates an ATPase activity (56, 179), which is dispensable for RNA binding (221) but required for RIG-I conformational change (246) and signal transduction (312). This domain seems to have a translocase rather than a helicase activity. This allows an ATP-dependent dynamic and processive interaction along the dsRNA without separation of the two RNA strands (195). The ATPase is negatively regulated by the CARDs and alleviated upon RNA binding to the CTD of RIG-I (83, 246) and/or to the helicase domain (177).
RNA PATTERNS RECOGNIZED BY RLRs AND THEIR EXCLUSION FROM THE CYTOSOL
RNA Patterns of RLR Agonists
Over half a century ago, dsRNA isolated from reovirus particles was identified as a potent inducer of IFN in vivo (287). In vitro, the intracellular delivery of dsRNA is much more efficient, and the involvement of a specific intracellular receptor was thus postulated (53). Indeed, dsRNAs are recognized by the three RLRs. However, there are subtle differences in their preferred dsRNA structures. The optimal agonist for RIG-I is a blunt-ended dsRNA at least 24 nucleotides (nt) long with a 5′-triphosphate end (249, 251), with a preference for a small bulge due to one mismatch located a few nucleotides downstream (177). This is supported by the crystallographic structure of the RIG-I CTD in complex with 5′ppp-dsRNA (165, 296). The affinity of a 14-bp-long 5′ppp-dsRNA is in the range of 200 pM. This high affinity explains the ability of a cell with low basal RIG-I expression to respond to a virus infection (312). Whereas the dsRNA moiety seems to be an absolute requirement, long dsRNA devoid of 5′ppp can also bind RIG-I. This gives rise to signaling (124, 147) according to a RIG-I activation model that is distinct from that induced by blunt-ended 5′ppp-dsRNA, but the details of this signaling remain to be defined (177). Likewise some variation in the RNA structure, such as a 5′ and/or 3′ overhang over 1 to 2 nucleotides and small mismatches in the dsRNA, may be more or less tolerated (178, 249, 251). However, short dsRNA with an overhanging 5′ppp nucleotide acts in vivo as a RIG-I decoy for IFN activation, although activating the ATPase activity of RIG-I in vitro (177). The LGP2 CTD and MDA5 CTD lack the residues responsible for 5′ppp binding. Their ligands are dsRNAs, with a preference for blunt ends in the case of LGP2 (159, 217, 246, 311). In cells, MDA5 prefers long dsRNA (>1 kb) with complex structures (216). MDA5 is also selectively activated by mRNA with a cap devoid of ribose 2′ O methylation, which appears to be a major MDA5 agonist motif (319).
Exclusion of RLR Agonist Patterns from Cytosolic RNA
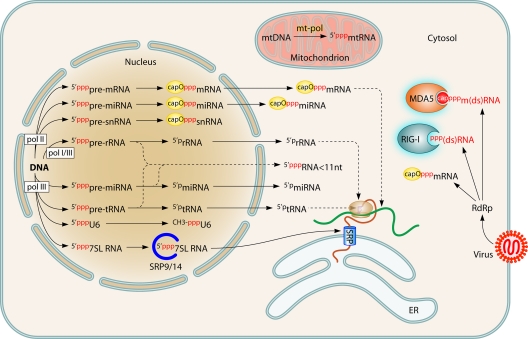
Nascent RNA made by a cellular or viral polymerase is necessarily 5′ppp ended because of the 5′-to-3′ nucleotide polymerization, with the notable exception of the polymerase of Picornaviridae, which uses a protein (VPg) primer. RNAs that are long enough have secondary structures including dsRNA stretches, and thus any cellular self-RNA can potentially be sensed by the RLRs. Why does this not occur? During evolution, eukaryotic cells have wrapped all their transcription machineries within the nucleus and the mitochondria, thus avoiding cytosol influx with long 5′ppp-RNA (70, 241) (Fig. 1). While mitochondrial RNA cannot escape this organelle, there is a continuous flow of nucleus-derived RNA in the cytosol. However, almost any primary nuclear transcripts from polymerase I, II, or III have their 5′ ends immediately processed early in RNA elongation, either by early cleavage of the first few nucleotides or by cotranscriptional modifications such as cap addition and 2′ O-ribose methylation of the first transcribed nucleotide by the methyltransferase hMTr1 (also known as FTSJD2 and ISG95) (15). As a consequence, any cytosolic export of 5′ppp-RNA of a size exceeding a few nucleotides is avoided. The only known exception is the 7SL RNA, a structural element of the signal recognition particle (SRP) or translocon. This RNA likely avoids recognition of its 5′ppp end by RIG-I because of its early association with translocon protein subunits upon transit through the nucleoli (300). Likewise, cellular transcripts having O-methylated caps escape recognition by MDA5 (319).
Fig. 1.
Exclusion of self-RNA agonists of RLRs from the cytosol. The cytosol is kept free from any 5′ppp-ended RNA, with the exception of the 7SL RNA, the 5′ end of which is shielded by SRP components before exit from the nucleolus. All other nuclear transcripts either are capped with ribose 2′ O methylation and guanosine N-7 methylation or are cleaved shortly after the beginning of RNA synthesis. Mitochondrial transcripts do not exit from this organelle. The polymerase (RdRp) from cytosolic members of Mononegavirales produces capped mRNAs with ribose 2′ O methylation and guanosine N-7 methylation, as well as 5′ppp transcripts that are recognized by and activate RIG-I. These viruses also produce 5′ppp genomes and 5′ppp antigenomes. However, their 5′ppp termini are embedded in N protein subunits multimerized into the helicoidal nucleocapsid (not shown). (Adapted from reference 85 with kind permission from Springer Science + Business Media.)
Furthermore, the basal levels of the RLRs are rather low. This reduces the probability of inadvertently becoming activated. Thus, the RLRs do not directly recognize an incoming pathogen but rather recognize the RNA it is synthesizing in a cell compartment where such activity is normally absent.
MODEL OF RLR ACTIVATION
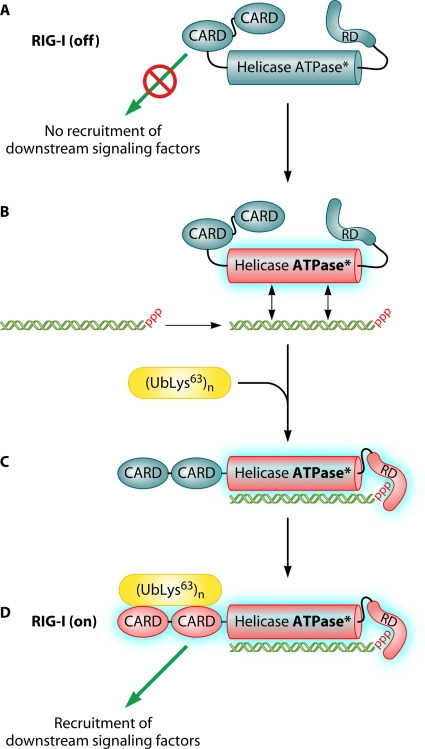
Thanks to large amounts of data, a recently revised model of RIG-I activation can be drawn (Fig. 2) (177). In the absence of an RNA agonist, the molecule is inactive, possibly because of intramolecular interactions (Fig. 2). dsRNA would bind to the helicase domain (83), allowing activation of RIG-I ATPase activity (83, 177, 195) that fuels RIG-I translocation along the nucleic acid (195). If the dsRNA is short and bears a 5′ppp end, the binding is strongly stabilized by the electrostatic interaction of the triphosphate with the CTD (56, 165, 177, 296). RNA binding appears to induce dimerization of RIG-I, which would enable RIG-I function (56, 159, 230, 246, 251, 278, 311). However, experimental evidence for RIG-I dimerization is not conclusive, because the dimeric status of the dsRNA made of two complementary strands with 5′ppp ends and/or RIG-I engagement into a high-molecular-weight complex with the membrane-anchored IPS-1 (interferon promoter-stimulating 1) has not been properly addressed. This changes the conformation of RIG-I (246) and exposes the N-terminal CARDs for binding to an unanchored short polyubiquitin Lys63 chain(s) (316). The source of the polyubiquitin chains seems to be the E3 ubiquitin ligase TRIM25 (tripartite motif containing 25). TRIM25 also associates with the first CARD (80, 81). Alternatively, TRIM25 first associates with the CARD to produce the polyubiquitin chain, which then binds to the CARD tandem and possibly displaces TRIM25. The CARD-polyubiquitin chain complex then recruits the CARDs of the IPS-1 (316), possibly because the polyubiquitin chain bridges the CARDs of RIG-I and IPS-1. The activated IPS-1 transduces the signal that induces the interferon response (127, 186, 260, 308). The activation of MDA5 is less well known, but available data support a similar scheme. Since 2′ O methylation is a motif allowing the discrimination of self and nonself transcripts recognized by MDA5 (319), it is tempting to predict the recognition of an RNA cap devoid of O-methyl substitutions by the CTD of MDA5 by analogy with RIG-I recognition of 5′ppp-RNA.
Fig. 2.
Model of RIG-I activation. (A) In its resting state, the RIG-I RD prevents CARD-mediated recruitment of downstream signaling factors. (B) Upon encountering a 5′ppp-dsRNA, the RIG-I helicase domain binds to the dsRNA and activates its ATP-dependent translocase activity. (C) This allows the RD to bind to the dsRNA blunt and 5′ppp ends. (D) 5′ppp recognition by the RD allows the CARDs to recruit downstream signaling factors.
PREFERENTIAL VIRUS RECOGNITION BY RLRs
The use of cells and/or transgenic mice with selective defects in RIG-I or MDA5 has revealed that each RLR can be preferentially involved in the recognition and control of distinct virus families. RIG-I acts as a sensor of most Mononegavirales, including measles virus (221), rabies virus, Ebola virus, Nipah virus, and respiratory syncytial virus (164), and Sendai virus and vesicular stomatitis virus (305), but not members of the Bornaviridae family, which uniquely transcribe and replicate in the nucleus (94). RIG-I also senses segmented negative-strand RNA viruses (Orthomyxoviridae) (164), some positive-strand RNA viruses (Flaviviridae, including hepatitis C virus and dengue virus) (164), and dsRNA viruses (reovirus) (164) (54). MDA5 senses Picornaviridae, dengue virus, and reovirus (164). However, when a preference is observed, it is unlikely to be exclusive. For example, a minor contribution of MDA5 can be found for measles virus and Sendai virus detection (112, 314).
RLR AGONISTS FROM MONONEGAVIRALES
So far no viral RNA recognized by a given RLR in infected cells has been formally identified with certainty. To hypothesize which viral RNA species can be the best candidates as RLR agonists in “real life,” one should consider which RNA species and structures are made during the virus infection cycle.
Virus Replication Cycle
Mononegavirales are enveloped viruses that share many common features. Their genome organization is well conserved throughout this virus order, with genes encoding a nucleoprotein (N), a phosphoprotein (P), a matrix protein (M), an attachment protein differently named H (hemagglutinin), HN (hemagglutinin-neuraminidase), or G (glycoprotein) for different viruses, a fusion protein (F) (lacking in the Rhabdoviridae and Filoviridae families), and a large L RNA-dependent RNA polymerase (RdRp) (Fig. 3). Virus families differ from each other by the presence or absence of additional genes or by multiple strategies for coding from the P gene, including an alternative reading frame (C and Y proteins) or RNA editing (V, D, and W proteins) (144).
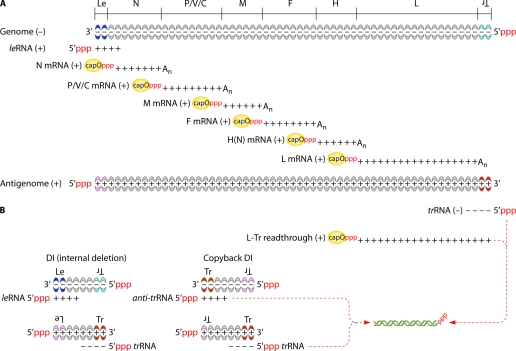
Fig. 3.
Viral RNA species from measles virus as a prototype for Mononegavirales. (A) RNA products involved in virus replication. The viral polymerase enters at the 3′ end of the encapsidated genome to transcribe successively the 6 genes. In the case of the antigenome, only the trRNA is transcribed. All of the transcripts except the leRNA and trRNA are capped, ribose O methylated, and polyadenylated. The transcriptase pauses at every intergenic junction to polyadenylate and terminate the upstream transcript and then resumes transcription of the downstream transcript, except at the end of L gene, where transcription stops at the L polyadenylation site. Replication differs from transcription by continuous RNA synthesis (i.e., intergenic junctions are ignored), lack of capping and polyadenylation, and concomitant encapsidation of the newly synthesized RNA into a regular polymer of N protein. (B) Potential source of 5′ppp-dsRNA due to transcriptase and/or replicase errors. Note that any free 5′ppp-ssRNA can have secondary structures resulting in 5′ppp-dsRNA moieties.
The virus cycle starts by the binding of viral envelope glycoproteins to a plasma membrane receptor. The G or H(N)F complex mediates the fusion of the viral envelope with the plasma membrane (entry at neutral pH) or, after endocytosis or as more recently recognized by macropinocytosis (199, 214), with endosomal membranes (entry at acidic pH). Schematically, upon cytosolic delivery of the ribonucleoprotein (RNP) and likely dissociation from the M lattice, the incoming P+L polymerase complex starts to transcribe all genes from the genomic RNA of negative polarity (144) without a measurable time lag (219) (Fig. 3). The transcript from every gene is capped and polyadenylated by the polymerase. When arriving at an intergenic junction, the transcriptase stops at the gene end and mostly resumes RNA synthesis at the next downstream gene start. New P+L transcriptases translated from the primary transcripts accumulate and enhance the transcription rate until enough N protein chaperoned by the P protein accumulates to form an N·P complex. This complex is the substrate of the replicase complex made of N, P, and L proteins. The replication consists of uninterrupted synthesis of complementary antigenomic RNA strands of positive polarity which are concomitantly encapsidated by N subunits that oligomerize around the RNA to constitute the helicoidal nucleocapsid (NC). The new antigenomic NC serves as a template for the synthesis of the genomic RNA, which is also concomitantly encapsidated. Particle assembly then proceeds with the M-mediated wrapping of NC into an envelope derived from the plasma membrane enriched in viral glycoproteins (85, 144). The encapsidation of RNA into NC is necessary and sufficient for incorporation into a viral particle. The virions contained both genomes and antigenomes but no viral transcript (219).
Besides the viral mRNA, two small transcripts can be synthesized, the positive-stranded leader (leRNA) and negative-stranded trailer (trRNA) (150, 151, 231). They are transcribed from the genome 3′ end, which contains the transcription and genomic promoters, and from the antigenome 3′ end, encompassing the antigenomic promoter, respectively (54, 105). The trRNA is the unique antigenome transcript since the antigenome lacks a transcription promoter allowing the expression of downstream sequences (116, 147). In the absence of protein synthesis, which prevents replication, the amount of leRNA increases (27). leRNA and trRNA are neither capped nor polyadenylated (54). They both contain the encapsidation signal that drives N binding and polymerization around the genome and antigenome (25, 27).
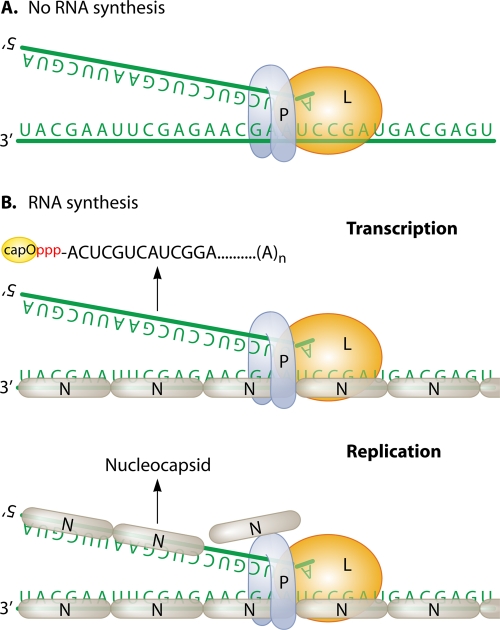
Viral polymerase attachment to and progression along the genome and antigenome strictly require the RNA template to be encapsidated (Fig. 4). The viral polymerase does not bind directly to the RNA template but relies on being in a complex with the phosphoprotein (P). P protein mediates the recognition of the NC by dynamically binding to nucleoprotein subunits in a predicted staircase way (144, 163, 240, 302). Neither viral genome RNA nor antigenome RNA is infectious unless N, P, and L are provided in trans (55). The three-dimensional (3D) structures of the nucleocapsids from rabies virus (RABV) (7), vesicular stomatitis virus (VSV) (92), and respiratory syncytial virus (RSV) (279) show how the RNA is embedded between two domains of the N protein. Every N subunit covers an exact number of nucleotides. For example, N protein from paramyxovirus covers 6 nucleotides, and the genome length has to follow the “rule of six”; i.e., the total number of nucleotides should strictly be a multiple of 6 (36, 95, 137, 138, 213, 265). Consequently, the genome and antigenome are unlikely to be found “naked” in infected cells and thus cannot anneal to each other or to viral transcripts. RNA is largely inaccessible within the NC as shown biochemically by resistance to nuclease degradation (27) (unless RNA is dissociated by extra energy [91]) and as shown functionally in infected cells by resistance to silencing by interfering RNA while viral transcripts are readily targeted (19, 190, 237).
Fig. 4.
Uniqueness of RNA synthesis by Mononegavirales. (A) Viral polymerase L and its cofactor P is unable to synthesize RNA on naked genomic RNA. (B) The genome (and antigenome) is entirely covered by a continuous noncovalent polymer of N protein subunits. P anchors L polymerase onto the nucleocapsid template and mediates its traveling along the nucleocapsid while transcribing (upper chart) or replicating (lower chart). See the legend to Fig. 3 for details of the RNAs that are produced.
The RdRp machinery is not perfect. It also produces other RNA species that appear to be useless (Fig. 3). Read-through transcripts occur at a frequency of a few percent, resulting in bi- or tricistronic RNAs and leader-N and L-antitrailer RNAs (43). An abortive 5′ genome and antigenome gives rises to short nucleocapsids unable to be duplicated (219). Finally, with a high multiplicity of infection, the RdRp can jump to another template during the replication step, resulting in nucleocapsids with large internal deletions. These are the defective interfering (DI) particles, which compete with genome replication by taking less time to replicate than the full-length genome. Copy-back DI particles are often observed with the strong antigenomic promoter on the 3′ ends of both strands (191), resulting in more potent interfering efficiency.
Viral RNA Agonists for RLRs
Source of RIG-I and MDA5 selective agonists.
What could be the viral source of 5′ppp-ended RNA? At this stage, it should be emphasized that the transfection of RNAs extracted from virions or infected cells brings only limited and potentially misleading information about the real viral RNA acting as an RLR agonist in vivo. Indeed, when extracted, denatured, and purified, both positive and negative RNA strands are present and easily anneal to each other into dsRNA that may never exist in an infected cell. Unfortunately, many major papers published in the field have relied mainly, if not solely, on such experiments, leading to the overstatement that viral genomes are the RLR agonists.
(i) Encapsidation of genome and antigenome prevents RNA annealing and exposure of 5′ppp ends.
The genome replication of Mononegavirales is primer independent, and the initiation of RNA synthesis starts with a single nucleoside triphosphate; for example, it is always ATP for Paramyxoviridae (204). As a result, their genomes and antigenomes are 5′ppp ended (149) and potently strong RIG-I agonists. Indeed, purified protein-free RNAs from viruses, i.e., a mixture of genome and antigenome RNAs that likely anneal to each other, are good inducers of RIG-I-mediated IFN response (94, 106). However, they are never found as free RNA but are found exclusively as part of the nucleocapsid (NC) in CsCl gradients (290). Although trace amounts of protein-free genome/antigenome can be found in a CsCl pellet, their actual presence in infected cells remains doubtful. Indeed, during gradient purification, a small fraction of NCs can likely be denatured by the high salt concentration. Shielded within the helicoidal homopolymeric nucleoprotein, the intracellular genomes and antigenomes are hardly available for recognition by the RLR, as shown by the inability of NC from UV-inactivated virus to be intracellularly delivered to activate the IFN-β gene (221). However, SeV genomic RNA has been proposed as a RIG-I ligand in infected cells, since 211-nt-long primer extension products encompassing the end of the L gene and trRNA are enriched in RIG-I immunoprecipitates (233). Likewise, deep sequencing of RNA bound to endogenous RIG-I, supported by PCR identification, suggests that the viral agonists are DI genomes and antigenomes (14). However, in both cases, the interpretation is obscured by the lack of further characterization of this RNA (free or NC associated?) and poor analysis of short transcripts.
(ii) Viral coding transcripts are capped and 2′ O methylated.
Viral transcription also starts with a single 5′ppp nucleotide as a primer, but mRNAs are capped shortly after their initiation, since a 31-nt-long transcript is fully capped in vitro (Fig. 3) (280). Upon inhibition of capping, the elongation of the 5′ppp transcript aborts after a 100- to 400-nt elongation (157, 162). Both cap addition and guanyl-7-N methylation are signals that tightly control the polyadenylation of the transcript (and hence its stability) and the recognition of intergenic junctions for transcription of the downstream genes (156). The viral capping process also includes ribose 2′ O methylation that precedes the guanyl-7 methylation (226). Whether any capping failure occurs during a natural infection is unknown. Such failure would be predicted to enhance IFN activation and virus attenuation, as occurred for a recombinant coronavirus devoid of ribose 2′-O-methylase activity in vitro and in vivo (319). Since full-length ∼1.7-kb MeV N mRNA linearly accumulates over 8 h due to a constant number of incoming transcriptases (219, 220), such a failure should remain at least below 10% (i.e., within the detection limits of the measurement). When expressed individually from a plasmid, none of the viral coding transcripts induces an IFN-β response (221). However, a short region of L mRNA from another paramyxovirus, parainfluenza virus 5 (PIV5), seems to be able to activate the IFN-β gene via an RNase L and MDA5 pathway (169).
(iii) Small noncoding viral transcripts remain 5′ppp.
leRNA and trRNA transcripts remain 5′ppp (54). In SeV- or VSV-infected cells, only limited amounts of free leRNA and/or trRNA species (31, 55, and 65 nt long) can be detected, likely because of lack of stability (151, 290). Late in the infection, ∼75% of them are found to be associated with N protein, with a buoyancy similar to that of NC on CsCl gradients (26). More abundant free short leRNA N read-through transcripts (<300 nt long) can be also detected (290). These are distinct from the encapsidated complete (∼2 kb long) leRNA N read-through RNAs corresponding to abortive replication products (40, 219, 290). As their abundance remains largely unchanged when replication is blocked, they likely represent uncapped transcripts, the elongation of which has prematurely stopped (290). The elongation checkpoint for RSV polymerase is at ∼50 nucleotides (162). The aborted transcription could be due to the presence of leRNA sequence and/or the lack of capping. Indeed, the priming of the transcriptase activity requires the recognition of the unique bipartite transcriptase promoter located within leRNA and N 5′ untranslated regions (5′UTRs) of the genome (136, 302).
(iv) IFN-β gene activation is related to virus transcription.
When considering many functional investigations performed with virus-infected cells, one can argue for the recognition of a viral transcript by RIG-I during a paramyxovirus infection. In MeV, the kinetics of IFN-β gene activation parallels that of viral transcription and not replication. Furthermore, a replication-disabled MeV that still transcribes activates the IFN-β response in a dose-dependent manner (221), in agreement with the pioneering observations made with VSV (176). A parainfluenza virus 5 (PIV5) expressing a P protein unable to be phosphorylated by polo-like kinase 1 (PLK1) displays both enhanced transcription and stronger activation of the IFN-β and cytokine genes (271, 282). Likewise, two mutations in the genomic promoter result in both enhanced viral transcription and interferon activation (172). Conversely, an SeV variant producing smaller amounts of free leader and leader-N read-through transcripts is a poorer activator of the IFN response (269, 290). Accordingly, the replacement of the genomic promoter with the stronger antigenomic promoter upregulates both viral transcription and the IFN response (116, 147). Finally, the transfection of short viral transcripts made in vitro from permeabilized Mononegavirales virions and likely corresponding to leRNAs activates the IFN-β gene (20, 221), as does leRNA transcribed from DNA in the cytosol but not in the nucleus (221).
(v) Cellular proteins bind to short noncoding viral 5′ppp transcripts.
trRNA and leRNA are able to recruit two cellular proteins, TIAR (T cell-activated intracellular antigen related 1) and La autoantigen, respectively. TIAR bound to SeV trRNA modulates the induction of apoptosis (116, 303). leRNA from numerous Mononegavirales, including VSV, PIV3, rabies virus, RSV, and rinderpest virus (RPV), have been shown to bind to La protein in vitro and in infected cells (20, 59, 142, 143, 225, 306). Infection with RSV or RPV, leads to the translocation of La protein from the nucleus into the cytoplasm (20, 225). Efficient transcription of RSV requires La expression in an IFN-independent manner (20). La protein can also compete with RIG-I for binding to RSV leRNA. The silencing of La expression favors leRNA binding to RIG-I and enhances the IFN-β response. The inhibition of La expression lowered the production of SeV, an IFN-sensitive virus, because of the additive effects of the IFN-independent decrease of viral transcription and the enhanced IFN response mediated by RIG-I (20) (Fig. 5). La autoantigen belongs to the RNA recognition motif (RRM) family of ribonucleoproteins and regulates the metabolism of cellular and viral RNAs (29). Its La domain mediates the binding to a 3′OH-UUU motif of RNA polymerase III (Pol III) transcripts and prevents their digestion by exonucleases (281). The adjacent RRM1 secondary RNA binding domain (108) binds to mRNA as part of an RNA regulon to coordinate ribosome biogenesis (128). Mononegavirales leRNAs lack the RNA 3′OH-UUU motif responsible for the high-affinity binding to La protein (108, 281). Instead, leRNA binds to the RRM1 La domain (20) with a high affinity (80 nM range) (225). The 5′ppp end enhances the RNA-La interaction (71). leRNA and La protein complexes are stable in CsCl gradients (20, 225) and have a buoyant density similar to that of viral NC (20). At the beginning of RSV infection, the leRNA associates with La, and later it becomes predominantly associated with N protein (20). At early times postinfection, primary transcription results in the synthesis of leRNA, which is concurrently bound by La and RIG-I, with an advantage for La. Moreover, La could also unwind any dsRNA feature (109) within the viral RNA, preventing its recognition by RIG-I. At the later step of concomitant replication and transcription, preferential encapsidation of leRNA would occur. PKR may be an alternative competitor for RNA agonist recognition by RIG-I, since it binds to and is activated by dsRNA with a 5′ppp end (198).
Fig. 5.
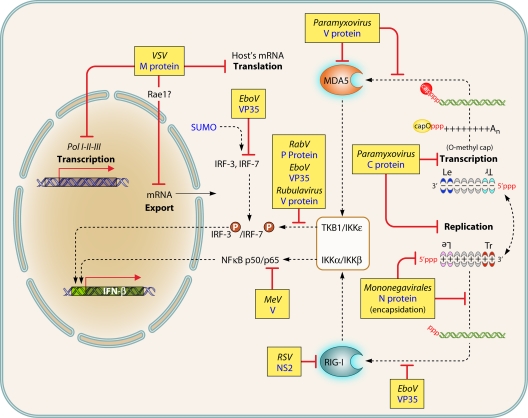
Viral evasion strategies to escape detection. T-ended red lines indicate either blockade or competition for binding, with dashed lines for reported but not formally proved mechanism of action. Black dashed arrows indicate pathways and/or shuttling. P in red circles indicates phosphorylation. See the text for further details and references.
Source of dsRNA. (i) Mononegavirales avoid producing dsRNA.
RIG-I dsRNA agonists can be a single-strand RNA (ssRNA) with intramolecular dsRNA secondary structures or two annealed complementary ssRNAs. It should be stressed that in cells infected with negative-strand RNA viruses, dsRNAs >40 bp long are not detected by a specific antibody, whereas such dsRNAs are easily found after infection with positive-strand RNA viruses (299). Coinfection with two recombinant SeVs expressing green fluorescent protein (GFP) sense mRNA and its complementary antisense mRNA, respectively, strongly activates the IFN-β gene through a RIG-I-dependent pathway (100, 270). This suggests that long complementary GFP dsRNAs without 5′ppp ends may also be activators of RIG-I. Alternatively, the presence of one transcript may have prevented the capping of its nascent complementary strand to form dsRNAs with 5′ppp ends.
(ii) Production of DI particles leads to IFN activation.
VSV stocks rich in defective interfering (DI) particles are potent inducers of IFN secretion (174). The genomes of most of these DI particles have large internal deletions and contain the stronger antigenomic promoter on both negative and positive strands (copy-back DI particles). Physically purified ∼250-nt-long copy-back DI particles free from infectious VSV are strong IFN inducers, with one DI particle per cell inducing a quantum yield of IFN (174). Likewise, while a carefully cloned SeV is a poor inducer of the IFN response, SeV stocks contaminated with DI particles or copy-back DI particles are good stimulators. Notably, the IFN-stimulatory activity is stronger for copy-back DI particles (268) (Fig. 3). In addition, the ability of DI particles to activate the IFN-β gene is inhibited by UV irradiation of the viral stock in a dose-responsive manner (268), as observed for the UV sensitivity of MeV N transcription (221). Moreover, the replication of full-length SeV and MeV displays higher sensitivity to UV (note that SeV DI RNA and MeV N mRNA as well as their corresponding genomes are of comparable ∼1.4- to 1.6-kb and ∼16-kb sizes, respectively). Although this was interpreted as reflecting the levels of DI replication, the free or encapsidated status of the DI RNA produced from UV-irradiated virus stock was not reported.
What are the RNA species that are synthesized from DI genomes and antigenomes? Fully encapsidated DI genomes and antigenomes are replicated, but their 5′ppp ends are shielded (170) and are unlikely to be available for recognition by RIG-I (Fig. 3). Genomic and antigenomic DI RNAs may fail to be encapsidated, since ∼5% of DI (anti)genomes sediment as free RNAs through CsCl gradients. Upon self-hybridization into panhandle RNA structures, because the 5′ and 3′ ends of copy-back DI particles are complementary, or upon genome-antigenome hybridization, they could make 5′ppp and blunt-ended dsRNA (268). leRNA and trRNA (or complementary trRNA [ctrRNA] for copy-back DI particles) can be produced during transcription. trRNA/ctrRNA hybrids would represent perfect RIG-I agonists, forming 5′ppp-ended ∼50-nt-long dsRNA (Fig. 3). In favor of the latter, the susceptibility of the replication of VSV copy-back ∼250-nt-long DI particles (determined by their interfering capacities) to UV inactivation is much higher than that of their ability to activate the IFN response. Moreover, internal-deletion DI particles with virus-like leader and trailer promoters failed to induce the IFN response (174). In addition, dsRNA forms over 40 to 50 bp long have not been detected intracellularly by the J2 antibody, indicating their low abundance or absence (276). Importantly, leRNAs and trRNAs of Mononegavirales are not complementary to each other, and (anti)genomes cannot join their 5′ and 3′ ends into a panhandle structure as is too often asserted in the literature.
(iii) Alternative sources of viral dsRNA.
Whether leRNA and/or trRNA, or short leRNA-N read-through transcripts, fold into secondary structures with long enough dsRNA regions at suitable distances from the 5′ppp end to act as RIG-I agonists remains to be determined. Alternative candidates are (i) read-through L-trRNA mRNA (43, 93, 100, 181, 304) hybridized to 5′ppp-trRNA (Fig. 3) and (ii) self-complementary NC-free uncapped RNA resulting from copy choice mechanisms involving a jump of the polymerase to an adjacent antigenome end, as occurs for the generation of copy-back DI particles (232).
In Drosophila, infection by VSV is controlled by the RNA interference (RNAi) system. VSV-derived small RNAs cover the whole genome and equally map the genome and antigenome (192). This suggests that they result from the processing of complete genome/antigenome dsRNA, (or genome hybridized with mRNA from every gene) by the RNase III Dicer-2. Because direct evidence of such viral dsRNA in infected cells is critically lacking and difficult to reconcile with the replication process of Mononegavirales, one can imagine the synthesis of negative-strand RNA complementary to the viral mRNAs by the RNA-dependent RNA polymerase activity of the elongator protein 1 (Elp1, also called IκB kinase-associated protein [IKAP]) recently identified in Drosophila (161). Interestingly, this cellular RdRp is highly conserved throughout the eukaryotic kingdom, including human. Elp1/IKAP exerts its RdRp activity using ssRNA as a template in both primer-independent and primer-dependent reactions. Moreover, it is devoid of back-primed RNA synthesis, thus precluding multiple rounds of autocopying and amplification of the dsRNA. Capped and polyadenylated ssRNA is a suitable template for generating a cRNA that is full length (161). Since Elp1/IKAP is localized in the cytoplasm (119), where Mononegavirales replicates, it could lead to the production of blunt-ended dsRNAs made of the 5′ppp strand annealed to the viral cap mRNA used as a template, i.e., a potential agonist for RIG-I.
AVOIDING PRODUCTION OF RNA AGONISTS FOR RLRs
RNA viruses, which rely mostly on the cytosol for transcription, replication, and/or assembly, have selected several strategies to avoid the production of 5′ppp-ended dsRNA and transcripts with caps devoid of O methylation (Fig. 5).
The 5′ ends of viral transcripts are fully capped either by an autonomous capping process (in Mononegavirales) or by snatching caps from cellular mRNAs (e.g., in Orthomyxoviridae). The L protein is a multimodular enzyme containing RNA polymerase activity, polyribonucleotidyltransferase activity that transfers 5′p-RNA onto a GDP acceptor through a covalent L-pRNA intermediate (205, 206), and methylase activity. The last is responsible for the guanyl-N-7- and ribose 2′ O methylations. From alignment of L proteins from a member of each family using the Muscle algorithm, key residues for each activity appear to be fully conserved throughout the whole Mononegavirales order. Interestingly, L polymerase from Bornaviridae is an exception, as it is predicted to lack guanyltransferase and methylase activities. However, since transcription of this family occurs in the nucleus, the endogenous nuclear enzymes can be recruited to ensure proper capping and 2′ O methylation of the viral transcripts before their export into the cytosol. Thus, all viruses protect themselves from MDA5 recognition by having their mRNA be O ribose methylated.
To avoid exposure of a 5′ppp motif at the (anti)genome end, RNA viruses can trim it by encoding a phosphatase or a nuclease (e.g., in Bunyaviridae and Bornaviridae, respectively) (94, 252). Alternatively, the 5′ end may be covalently linked to a viral protein (in Picornaviridae) or finally shielded within a NC (in Mononegavirales). Indeed, by associating with nascent 5′ppp-ended genome and antigenome RNAs and covering every single nucleotide, the N protein acts as a major inhibitor of RIG-I recognition and IFN-β activation. It shields the 5′ppp end away from the regulatory domain of RIG-I, and it prevents the annealing of genome-antigenome and/or mRNAs-genome complexes into dsRNA.
Unless contaminated with DI particles, PIV5 lacking the V antagonist of the interferon response does not activate the IFN response in most cells it infects, as if the polymerase avoids producing any agonist of the RLRs (131). Thus, the IFN response is most likely due to the production of RLR agonists wrongly made during viral transcription and/or replication.
That the virus avoids production of dsRNA has been experimentally challenged by using a recombinant ambisense SeV. A transcription unit encoding chloramphenicol acetyltransferase (CAT) has been added at the 3′end of the antigenome, i.e., in the reverse orientation, by duplication of the genomic replication and transcription promoter. In IFN-competent cells, this virus grows poorly and quickly loses the expression of the antisense transcript. The likely explanation is that the plus-sense L-CAT read-through transcript and the minus-sense CAT-L read-through transcript annealed into dsRNA. This dsRNA strongly activates the antiviral response, which leads to selection of SeV with a disabled antisense transcription promoter (147). Such ambisense SeV and RABV can be easily rescued and grow well only in IFN-disabled cells (76, 147), indicating that there is strong selective pressure for avoiding the ambisense coding strategy by Mononegavirales due to RLR-dependent innate immunity.
SIGNAL TRANSDUCTION BY RLRs AND REGULATION
Signal Transduction by RLRs
Upon binding to their RNA agonists, the RLRs associate with IPS-1, also known as MAVS (mitochondrial antiviral signaling), VISA (virus-induced signaling adaptor), and CARDIF (CARD adaptor inducing IFN-β). IPS-1 acts as the central node between sensing abnormal RNA and inducing the cellular innate immune response. It is recruited by homotypic interaction of its CARD with RLR counterparts and is anchored at the outer layer of mitochondria and peroxisomes. The CARD-CARD interaction induces IPS-1 dimerization and recruitment of several factors, including members of the tumor necrosis factor receptor-associated factor (TRAF) family. TRAFs in turn recruit inhibitors of NF-κB kinases (IKK) and TANK binding kinase 1 (TBK1), which will phosphorylate IκB and the interferon-regulating factors IRF3 and IRF7, respectively. NF-κB, liberated from IκB, and phosphorylated IRF3/IRF7 associate with AP-1 to constitute the enhanceosome that activates the IFN-β gene. Our knowledge of cellular components of the transduction machinery and the complexity of their intertwined functions is ever growing, and these are the subject of regular updated reviews (207, 277, 305, 311).
Regulation of RLR-Mediated Signal Transduction
The interferon system is absolutely required for the control of almost any viral infection, as shown by the exquisite sensitivity of mice with debilitated IFNAR genes to many viruses (193), including those (e.g., measles virus) normally restricted to another species (259). The IFN system should be easily and quickly switched on to disseminate an antiviral state so as to restrain the virus expansion. Because of its inflammatory contribution and adverse effects (285), the IFN response should also be dampened and extinguished at the end of the viral infection, hence its tight regulation by numerous cellular factors acting at almost every step of the response.
Three mechanisms are proposed to keep RIG-I inactive in healthy cells from humans and primates: phosphorylation on serine 8 of the CARD1 domain (203), phosphorylation on serines 854 and 855 and threonine 770 by casein kinase II of the RD domain (272), and binding to the ARF-like 16 protein (ARL16), which prevents the association with RNA (309).
A positive regulation occurs very early, with the quickest activation of a subset of IPS-1 located on the peroxisomes leading to immediate expression of antiviral inflammatory genes. It acts as a potent amplifier of the slightly delayed mitochondrial IPS-1-mediated signaling (64). The RLRs are induced by the IFN/IFNAR signaling to increase cellular sensitivity to the virus invasion. A spliced form, RIG-I SV, coding for RIG-I with a truncated first CARD, appears later and acts as a negative feedback inhibitor (80). The third RLR member, LGP2, binds to dsRNA but lacks the CARDs. It regulates the responses mediated by RIG-I and MDA5 either positively or negatively, although its role requires further clarification (247, 289). ZAPS, a short isoform of the zinc finger CCCH-type antiviral protein 1 (also called PARP-13), is an ISG that potentiates RIG-I activation by 5′ppp-RNA according to a positive feedback mechanism (101).
A deubiquitinase, cylindromatosis (CYLD), and E3 ubiquitin ligase ring finger 125 (RNF125) act as negative regulators of RIG-I and MDA5 by cleaving key ubiquitins involved in the recruitment of the downstream signaling cascade and by tagging the molecules for proteasome-mediated degradation (see reference 311) for a review). MDA5 is kept inactive by the interaction of its CARDs with the dihydroxyacetone kinase (DAK) (311). RIG-I is negatively regulated by its feedback conjugation with ISG15 (133, 317) and the linear ubiquitin complex, which both prevents association of RIG-I with TRIM25 and targets the latter for proteasomal degradation (113). The association of the Atg5/Atg12 heterodimer, a component of the autophagy system, with the CARDs of RIG-I, MDA5, and IPS1 inactivates the complex (311).
At least half a dozen factors tightly regulate IPS-1 activity at the surface of the mitochondria (see references 207, 277, 305, and 311) for reviews), including proteins that control mitochondrial fusion (41, 208). The TRAF family is also controlled by deubiquitination and ubiquitination through interaction with deubiquitinating enzyme A (DUBA) and A20 (see references 207, 277, 305, and 311) for reviews). Other mechanisms regulate downstream signaling molecules. For example, the ubiquitin E3 ligase RAUL represses the expression of IRF3 and IRF7 (315), and the recruitment of coactivators to IRF3 is antagonized by the transcription factor MafB (132). Even the stability of the IFN-β mRNA is under control, since PKR prevents the deadenylation of its poly(A) tail. This can explain conflicting reports about the importance of this enzyme in the activation of the IFN-β gene, although upon infection by a paramyxovirus, the latter occurs downstream from RIG-I and independently from PKR (256).
VIRAL SUPRESSORS OF INNATE ANTIVIRAL SIGNALING
An early strong innate immune response of a host cell would result in the abortion of a virus infection. Likewise, the quick spreading of an antiviral state induced by the IFN cytokine represents a powerful barrier for the dissemination of the virus throughout the organism. Successful viruses have been evolutionarily selected to express countermeasures efficient enough for escape from the antiviral cellular defenses. The range of virus countermeasures is almost as wide as the innate immune system is complex, from shielding the MAMPs from recognition by the PRRs (Fig. 5) to abolishing IFNAR-dependent signaling (Fig. 6). Despite some common features, counteracting activities are highly variable among the Mononegavirales; there are multiple mechanisms for a given virus, and they are exerted by different viral proteins that are themselves multifunctional. Each viral countermeasure is usually dispensable for virus growth, and even more, their absence can facilitate virus replication, particularly in cells exhibiting some innate immunity defect. Strikingly, every countermeasure acts as a critical virulence factor in vivo. The intracellular innate immune response pathway will be used as a guideline for their overview.
Fig. 6.
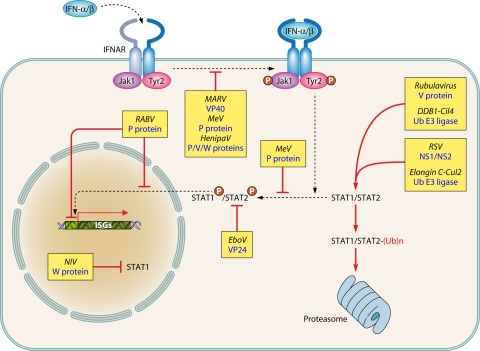
Viral evasion strategies to prevent activation of an antiviral state mediated by type I IFN binding to IFNAR. T-ended red arrows indicate either blockade or competition for binding. Black dashed arrows indicate pathways and/or shuttling. P in red circles indicates phosphorylation. Ub, ubiquitin. See the text for further details and references.
Blockade of the IFN Induction Pathway
Mononegavirales can either mask the RNA structures that are recognized by the RLRs and/or minimize their production (Fig. 5). As discussed above, the encapsidation of the genome and antigenome prevents dsRNA formation and shields the 3′ppp ends. Likewise, adding a cap to their mRNA obviates 3′ppp end exposure. The transcription of paramyxovirus is finely tuned by the intrinsic strength of the transcription promoter, thus limiting the activation of IFN (172, 269, 290). The transcription is also negatively regulated by paramyxovirus C proteins and RSV NS1 protein, which limit the production of transcript agonists of RIG-I (8, 13, 34, 57, 104, 160, 171, 238). In the absence of C protein, dsRNA accumulates in cells infected by PIV1 and SeV (28, 276), with a concomitant activation of the PKR and phosphorylation of eIF2α. This results in decreased protein synthesis, as also observed with C-deleted MeV (184, 196, 283), and likely leads to an imbalance in the N protein/(anti)genome ratio possibly favoring naked complementary viral RNAs (28). The deamination of (5′ppp)dsRNA by the cellular adenosine deaminase ADAR1 under the control of C protein may also contribute to lower both PKR and RIG-I activation (284). The polymerase subunit of Filoviridae, called VP35, has a C-terminal domain that binds dsRNA (38). The crystal structure of the C-terminal domain consists of a unique fold that differs from other viral dsRNA binding proteins, with a cluster of basic residues contacting the dsRNA (155). VP35 forms an asymmetric dimer on dsRNA binding, with two monomers binding to the backbone and capping the terminus, respectively. The model predicts the cooperative recognition of dsRNA, with the binding of the first VP35 monomer assisting that of the next one (134). Mutations that abolish RNA binding activity reduce the ability of VP35 to inhibit IFN production (38) and dramatically reduce the virulence of recombinant EBOVs in mice and guinea pigs (96, 224). Interestingly, although the optimal anti-IFN function of VP35 requires oligomerization mediated by the N-terminal domain (234), this activity is independent from its function in RNA synthesis (38, 96–98).
The RNA sensors can be directly targeted by viral proteins (Fig. 5). RSV NS2 protein binds to the N-terminal CARD of RIG-I and prevents the recruitment of IPS-1 by competition (160). The C-terminal domains of V proteins from all paramyxoviruses tested so far (except possibly RPV, which has been tested only on human MDA5 [30]) bind MDA5 and prevent MDA5-mediated activation of the IFN-β gene according to a likely common mechanism (50). This domain of V is an evolutionarily conserved 49- to 68-amino-acid region that coordinates two zinc atoms and mediates the binding to the helicase domain of MDA5 through the conserved RRE[W,V,I]S[L,I](x)6V peptide motif (248). Consequently, the binding of V protein competes with binding of dsRNA, and the oligomerization and ATPase activity of MDA5 are prevented (51, 212). The high efficiency of MDA5 inhibition by V protein explains, at least in part, the apparent low contribution of MDA5 in mediating the activation of the IFN-β gene by paramyxoviruses. The effects of MDA5 can be observed only when RIG-I is silenced (86, 112). However, a recombinant hPIV2 encoding a V mutant unable to bind to MDA5 is attenuated in vivo (248). V protein binds also to LGP2. VP35 interacts directly with the kinase domains of TBK1 and IKKε and acts as a competitive substrate for phosphorylation. In addition, VP35 promotes the conjugation of IRF7 with SUMO, a small ubiquitin-like protein that represses the transcriptional activity of IRF7. VP35 acts as a scaffold between IRF7 and the SUMO E2 and E3 enzymes Ubc9 and PIAS1, respectively (45). Measles virus V protein binds to RelA (NF-κB p65 subunit) and prevents its nuclear translocation. Thus, V suppresses NF-κB activity that is required for many cytokine responses. Binding sites map to the C-terminal domain of V and the RHD N-terminal domain of RelA, which are responsible for dimerization, nuclear import, and DNA binding (255) that are predicted to inhibit its regulatory activities (212).
Rubulavirus V protein, RABV P protein, and filovirus VP35 prevent the phosphorylation of IRF3 by TBK1 and IKKε (31, 166, 223, 239) (Fig. 5).
C proteins from RPV (30), MeV (262), and SeV (82, 89, 270) inhibit the activation of the IFN-β gene mediated by RIG-I stimulated by an artificial RNA agonist (30, 262, 270) or by viral infection. This effect is moderate and variable according to the virus strain (30, 77) and is also observed in cells devoid of the IFN/IFNAR amplification loop (30). The RPV C protein acts downstream of the phosphorylation, dimerization, and nuclear import of IRF3, i.e., possibly at the level of the IFN enhanceosome (Fig. 5). This would require C protein shuttling into the nucleus. Accordingly, the closely related MeV C protein is endowed with such shuttling properties (201), but the underlying mechanism is unknown.
The VSV matrix (M) protein induces a global inhibition of host gene expression (73) (Fig. 5). This activity is genetically separable from its role in virus structure and assembly as shown by M mutants that are selectively defective (5, 22, 75, 78). When expressed alone by transfection, M protein inhibits transcription by all three host RNA polymerases (3) and the nuclear-cytoplasmic transport of host RNA (102, 294). This blocks the membrane-associated Akt/mTor signaling, as early as 2 h postinfection, which corresponds to the onset of detectable M protein and requires virus transcription (66). Translation of host mRNA is also impaired in VSV-infected cells, due in part to the activity of M protein (5, 21). VSV M protein lacks enzymatic activity, and its inhibitory activity is likely mediated by binding to host factors and interfering with their normal function. One factor is Rae1 (72), which is implicated in mRNA transport and regulates the mitotic spindle assembly and mitotic checkpoint regulation (9, 24, 307). M protein and Rae1 are found associated in complexes made of several proteins, including Nup98, hnRNP-U, and E1B-AP5, that are involved in mRNA transport and other cellular functions (44, 72, 294). Binding of M protein to the Rae1-Nup98 complex is thought to be responsible for the inhibition of nuclear-cytoplasmic RNA transport, although Rae1 is not essential for mRNA transport (9, 24, 307). The broad distribution of Rae1 throughout the cytoplasm and the nucleus suggests its involvement in several steps in host gene expression.
Because of the global inhibition of host gene expression by VSV M protein, cells infected with wild-type (wt) VSV produce very little IFN or other cytokines. Likewise, treatment of VSV-infected cells with IFN has relatively little effect on virus yield. However, once the antiviral state becomes established by pretreating cells with IFN, primary transcription of VSV is inhibited (173, 175) and the virus does not have a mechanism to suppress the response to IFN. Hence, the very peculiar phenotype of VSV, which is probably the most efficient virus in prevention of both the activation of the IFN-β gene and the establishment of an antiviral state and the least capable of invading cells that have activated antiviral defenses.
The inhibition of host gene expression by M protein is not essential for virus replication in a single-cycle infection, and many M protein mutants of VSV that are defective in their ability to inhibit host gene expression replicate as well as or better than wt viruses in most cell types in vitro. However, they are unable to perform multiple cycles of infection of cells that are competent to produce and respond to IFNs (2, 5, 267). Accordingly, such M protein mutant viruses are dramatically attenuated in intact animal hosts (2, 267). Following intranasal inoculation, M protein mutant VSV replicates in the nasal epithelium. However, in contrast to the wt VSV, this mutant is rapidly cleared and does not invade the central nervous system (CNS) (4), because of the local production of type I IFNs that it induces (286).
Suppression of the Response to IFN
The IFN-α/β receptor, IFNAR, is made of two subunits, the cytosolic tails of which recruit the Jak1 and Tyk2 tyrosine kinases. Upon IFN binding, these kinases are activated and become autophosphorylated. They then phosphorylate STAT1 and STAT2, which heterodimerize and associate with IRF9 to form the ISGF3 transcription complex. ISGF3 is translocated into the nucleus to bind to the interferon-sensitive response element (ISRE) of the ISGs (229). This pathway is specifically targeted by many Mononegavirales according to strategies with features that can be common or subtly distinct (Fig. 6).
The autophosphorylation of Jak1 and Tyk2 and the phosphorylation of STAT1 and STAT2 are inhibited by the matrix VP40 of Marburg virus (MARV) by an unknown mechanism (288). Measles virus V protein binds to Jak1 and prevents STAT1 phosphorylation (35). Measles virus and other morbillivirus V and P proteins bind to STAT1 via a conserved tyrosine motif present on their shared N-terminal domain (61), away from the Jak1 binding site. The unique C-terminal domain of V binds to STAT2 via two conserved tryptophan residues (35) that are located away from the MDA5 binding site (227, 248). Together with the ability of V protein to form macromolecular complexes including STAT1, STAT2, and IRF9 (210), this suggests that V protein acts as a molecular scaffold, which prevents both STAT1/2 phosphorylation (275) and nuclear translocation of the ISGF3 complex. A single mutation that abolishes STAT1 binding results in a virus exhibiting an attenuated phenotype in vivo (60). Henipavirus P, V, and W proteins similarly sequester STATs away from activation. In addition, the W protein possesses a potent nuclear localization signal that interacts with karyopherin α3 and α5, pointing to a possible regulatory function at the level of nuclear-cytoplasmic transport (see reference 228) for a review).
STATs can be targeted for proteasome degradation (Fig. 6). V proteins from the Rubulavirus genus, such as mumps virus and PIV5, bind STAT1/STAT2 complexes and recruit other cellular components to form an active ubiquitin E3 ligase. Through independent interacting sites, V binds to DDB1 to recruit the cullin family member Cul4A. The C-terminal domain of V protein mediates homo- and hetero-oligomerization of a spherical molecular complex large enough to be seen in the electron microscope. V protein acts as an enzymatic scaffold that hijacks cellular E3 ligase components. It uses STAT2 as a substrate adaptor for the ubiquitination of STAT1 by DDB1/Cul4A, leading to its proteosomal degradation (see reference 228 for a review). The pneumovirus genome includes two separate genes, NS1 and NS2. Via a consensus binding site, NS1 assembles with another E3 ligase complex made of elongin C and cullin 2. The NS1/NS2-elongin/cullin E3 ligase complex is thought to recruit STAT2 for ubiquitination and proteasomal degradation (68). The cumulative inhibitory effect on RIG-I (160) and IFNAR signaling (68) likely explains the particular resistance of RSV to the IFN system (86, 88).
Further down the signaling pathway, the sequestration can occur before or after the phosphorylation of STATs (Fig. 6). NiV W protein binds to inactive STAT1 and sequesters it into the nucleus (52). The binding of RABV P protein to phosphorylated STAT1 and STAT2 prevents their translocation to the nucleus by anchoring the phospho-STAT1 complex onto microtubules (32, 188, 189, 291, 292). This effect requires P export from the nucleus and is critical for RABV pathogenicity in vivo (117). The STAT1/STAT2 complexes that may escape sequestration in the cytoplasm are prevented from binding to their DNA targets by their association with nuclear truncated P protein isoforms (292). An alternative strategy is used by the EBOV VP24 membrane-associated protein by binding karyopherins α1, α5, and α6. These are intracellular receptors that recognize the nuclear localization signal on phospho-STAT1. Thus, VP24 likely acts as a competitive inhibitor that prevents the nuclear import of STAT1/STAT2 complexes (182, 235, 236).
CONTROL OF VIRUS INFECTION BY IFN-INDUCED ANTIVIRAL EFFECTORS
The antiviral activity mediated by the type I IFN response is complex, with both induction of a cell-intrinsic antiviral cellular program and stimulation of the adaptive immunity (197). The IFN-induced antiviral state against a given virus is mediated by the cooperative action of multiple antiviral molecules (245). So far, five major innate antiviral mechanisms have been unraveled: (i) RNA modification; (ii) translation impairment, which is likely of major importance owing to the prominent impact of translation on the expression of cellular protein (258); (iii) protein tagging; (iv) inhibition of nucleocapsid assembly; (iv) prevention of virus release; and (v) blockade of virus entry. It is beyond the scope of this review to describe in detail every antiviral effector. Indeed, every type of virus is susceptible to a particular set of antiviral effectors (254). Instead, VSV, a virus highly sensitive to IFN, will be used as a model to illustrate the complexity and redundancies of effectors.
The adenosine deaminase acting on RNA (ADAR1) is an RNA-editing enzyme that catalyzes the conversion of adenosine to inosine. Inosine is recognized as a guanosine by translation and polymerase machineries. Hence, mutations can accumulate on viral and cellular RNAs, including noncoding RNAs (202), with multiple effects, as shown by the essential role of ADAR1 to maintain fetal and adult hematopoiesis (99). The IFN-inducible ADAR1 p150 isoform is inactive against VSV, while being a restriction factor for paramyxoviruses (298). In the latter case, one can speculate about the possible involvement of ADAR1 in the A-to-G hyperediting of the MeV M gene observed in brain tissue from cases of measles inclusion body encephalitis and subacute sclerosis panencephalitis (42). Likewise, VSV is poorly sensitive to the RNA degradation mediated by the 2,5-oligoadenylate synthetase/RNase L pathway (63, 130, 264). PKR, which inactivates the translation elongation factor eIF2 by phosphorylation, is required to control VSV infection in vivo (266) but not in vitro (130). The ubiquitin-like ISG15 is dispensable (209). Mx GTPases postulated to act on viral nucleocapsids are partially required for controlling VSV (145, 257) and rabies virus but not paramyxoviruses (152–154). Interestingly, this effect is species dependent, with chicken Mx being inactive against VSV and SeV (16). Tetherin inhibits VSV release from infected cells as it does for many viruses, including Filoviridae (301). The interferon-induced transmembrane protein 3 (IITM3) blocks VSV entry at postendocytosis step (301). Interestingly, the two latter antiviral effectors act also as restriction factors due to a basal expression of already-active molecules (301). Additional antiviral effectors active on VSV include IFIT3 (IFN-induced protein with tetratricopeptide repeats 3) (250) and ISG20 (69) via translation inhibition (58) and an unknown mechanism, respectively.
INFECTION DRIVEN BY INTERPLAY BETWEEN VIRUS AND INNATE IMMUNITY
RLRs Are the Primary Major Players in Recognizing Mononegavirales
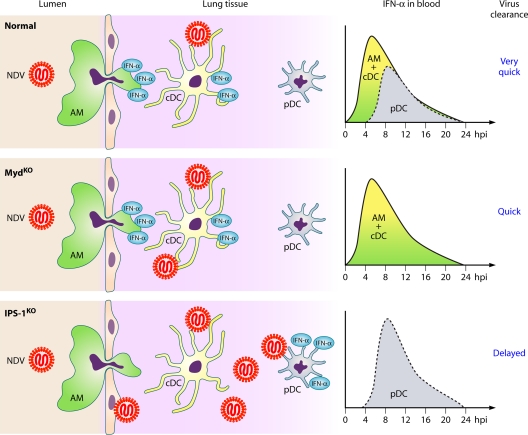
Mice that lack the signaling nodes, i.e., IPS-1 for RLRs and Myd88 for TLR, have been used to decipher the respective role of these two types of PRRs in recognizing negative-strand RNA viruses. In vitro, pDCs are the only cells where the TLR/Myd88/IRF7 pathway is preferentially used, whereas in all other cell types, including myeloid DCs and macrophages, the RLR/IPS-1/IRF3 pathway is exclusively used (17, 123, 126, 141, 318). Thanks to the seminal work of Kumagai et al., the kinetics of IFN activation in various cell types during virus infection of a transgenic mouse expressing a fluorescent protein under control of the promoter of IFN-α6 has been unraveled (Fig. 7) (141). After pulmonary infection of these mice with Newcastle disease virus (NDV), alveolar macrophages and some conventional DCs (cDCs) present in the lung and in the draining lymph node, but not pDCs, are IFN producers via the IPS-1 pathway. After an initial local replication, the NDV infection is rapidly cleared. In MyD88−/− mice, the circulating IFN production and viral clearance are modestly reduced, indicating that some TLR-dependent activation also occurs upon NDV infection. If alveolar macrophages are locally destroyed by chemical instillation or if the IPS-1 gene is knocked down, pDCs become the major IFN producers via a Myd88 pathway. However, their activation and IFN production are delayed by a few hours, while NDV replication is strongly enhanced. NDV clearance is strongly delayed particularly in IPS-1−/− mice. When mice are infected intranasally with the mouse pathogen SeV, alveolar macrophages and cDCs fail to produce IFN, but pDCs are strongly activated to produce IFN and mice die of SeV infection. In marked contrast, infection with a C-deficient SeV resembles the NDV infection, with exclusive IFN production by alveolar macrophages and cDCs. Correspondingly, the C-deficient SeV is successfully cleared. Thus, virulence is determined by the ability of the virus to primarily subvert the RLR/IPS-1 pathway in alveolar macrophages and cDCs. When the first line of IFN response is inefficient in controlling the infection, pDCs act as a second line of defense by producing large amounts of IFN. However, the route of infection is a critical issue. Indeed, after nonnatural intravenous and intramuscular injections, the Myd88-dependent IFN production by pDCs becomes more critical in controlling the virus burden (141, 146, 318). The relative importance of the RLR/IPS-1 and TLR/Myd88 pathways can differ according to the virus species. For example, infection by RSV activates both pathways, with a prominent role of TLR/Myd88 in virus clearance, possibly because of the ability of RSV to sustain growth even in the presence of some IFN (17).
Fig. 7.
Primary role of the RLR/IPS-1 pathway in the control of lung infection by paramyxoviruses. (Top) Intranasal infection of normal mice with NDV results in IFN-α activation by the alveolar macrophages (AM) and conventional dendritic cells (cDC), leading to a limited virus burden locally with rapid virus clearance; consequently, local plasmacytoid DCs (pDC) are poorly activated to produce IFN-α. (Middle) In infected Myd88−/− mice, the deficient TLR/Myd88 pathway prevents pDC from producing IFN-α. The virus burden, IFN-α production, and virus clearance are subnormal. (Bottom) In infected IPS-1−/− mice, the deficient RLR/IPS-1 pathway prevents both AM and cDCs from producing IFN-α. The virus burden increases, and the IFN-α production and virus clearance are delayed. (Based in part on data from reference 141.)
The TLR pathway can be the target of viral countermeasures. V proteins from paramyxoviruses bind to IRF7 via a tryptophan-rich region (135), leading to a block of TLR7/TLR9 signaling in a reconstitution system. V from MeV also interacts with the upstream kinase IKKα (215). Whether such a viral countermeasure is operative in pDCs and in vivo remains an open question. Indeed, ex vivo, the infection of human pDCs by MeV is associated with massive expression of IFN, but viral gene expression is hardly detectable, thus raising the question of whether there is any intracellular expression of the nonstructural V protein (65). Likewise, in vivo, when mouse alveolar macrophages are destroyed, SeV induces a strong TLR/Myd88-dependent response in pDCs despite expressing both functional C and V proteins (141).
Besides its role as a secondary IFN-mediated line of defense, the TLR/Myd88 pathway is critically involved in the induction of inflammation, activation of Th1, and stimulation of antibody responses (126, 146, 318). Thus, by virtue of the multiple roles of type I IFN in the adaptive response, the interplay between RLRs and TLRs ensures the robustness of the immune response (126).
Upon recognition of RSV, optimal production of type I IFN also involves Nod2, a component of the inflammasome (244). However, a direct recognition of viral RNA by Nod2 remains to be demonstrated (122), and the lack of basal expression of Nod2 precludes any primary role in detecting viral transcription (244). Thus, Nod2 is more likely a positive regulator of the RLR pathway. Future studies will decipher the cross talk between the different PRR pathways to unravel their respective contributions in recognizing every virus family.
Strangely, in studies aimed at deciphering the respective roles of RLRs and TLRs, the amounts of available viral particles at various times postinfection have been largely ignored. In natural infections, the amounts of infectious virus reaching the airway mucosae (the main route of entry for most Mononegavirales) are likely to be very low, on the order of only a few infectious units (range of ca. 1 to 100 infectious units). Such a low virus load is unlikely to be able to reach enough pDCs deeply embedded in the mucosae to give rise to a potent IFN response. Instead, alveolar macrophages and local cDCs (and possibly airway epithelial cells) are likely to become infected, as demonstrated experimentally for NDV, VSV, and SeV (141) and MeV (148). If the virus does not counteract the RLR-mediated IFN production by alveolar macrophages, the infection is largely abortive. In this case, the virus progeny is too limited to reach and activate a large number of pDCs. If the virus efficiently counteracts the RLR-mediated IFN production and/or the IFN-induced antiviral state within the alveolar macrophages, there is a large production of virus particles that become available to (i) infect many other permissive cells and (ii) reach numerous pDCs to activate their program of TLR-mediated IFN production. Thus, the outcome of a virus infection strongly and successively depends on its ability to prevent RLR-mediated IFN response, to possibly counteract the TLR-mediated response of the pDCs, and to escape the late adaptive immune response. Because Mononegavirales seem to be much more equipped with countermeasures against the RLR pathway than with those against the TLR pathway, their recognition by RLRs is likely of critical importance in determining the outcome of the infection.
Successful Infection despite IFN Response
Most, if not all, Mononegavirales are paradoxically inducers of an IFN response that can prevent their dissemination to neighboring cells. Multiple strategies and/or opportunities allow virus spreading that accounts for virus-induced diseases. Several examples can illustrate the complexity of the interplay between the virus and innate immunity.
A highly virulent virus can elicit a strong innate immune response in the host. Virulent NDV, but not an attenuated vaccine strain, induces early and strong IFN and inflammatory responses in chickens, both in vitro and in vivo (243). Possible explanations for this discrepancy are different kinetics of virus infection and IFN production by bystander noninfected cells and/or the use of IFN and cytokines to increase the number of cells that are the major target for NDV replication.
Because of their transcription activity in the cytosol, paramyxoviruses appear to intrinsically activate the intracellular innate immunity upon entry. However, the transcription promoter of wild-type PIV5 (formerly simian virus 5 [SV5]) has evolved to synthesize low levels of RNA at early times postinfection, thus limiting the IFN response (172). Furthermore, analysis of viruses isolated from individual plaques generated by infection at low multiplicity surprisingly revealed that an IFN response was induced by viruses from a minority of plaques (49), as if there is a virus polymorphism at the population level, with a minority of IFN inducers and a majority of noninducers of IFN. The virus can also propagate into adjacent cells in the antiviral state. The replication cycle of PIV5 is severely slowed in IFN-primed cells. However, some viral transcription and replication can occur, resulting in the progressive degradation of STAT1. Consequently, the IFN stimulation progressively vanishes and permits the delayed onset of efficient PIV5 replication. To escape neutralization by the antiviral state, the incoming nucleocapsid initially forms a small cytoplasmic body that could shield the viral transcription machinery from cellular antiviral activity (39). Kinetics analysis and mathematical modeling (261) and maintenance of IFN signaling during henipavirus infection of human cells (293) further argue for a subtle and dynamic equilibrium between activation of intracellular innate immunity and virus replication.
Produce Virus and/or Type I IFN?
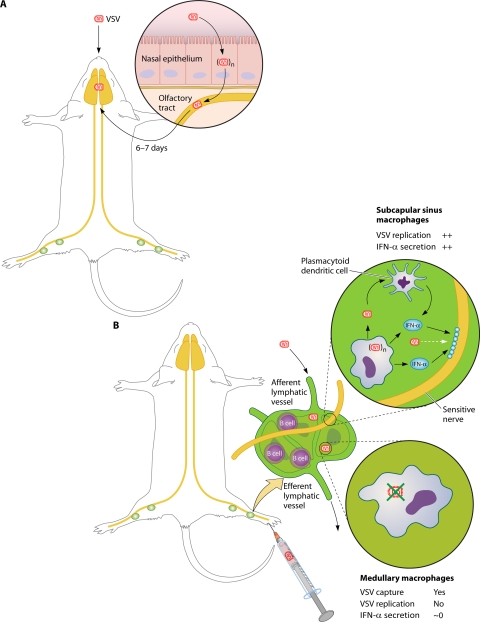
As discussed above, every member of the Mononegavirales is able to dampen the production of cytokines and IFNs and to prevent the onset of an antiviral state. Yet, in most cases, high levels of IFNs and other cytokines are produced systemically and/or locally at the site of infection. Indeed, the inflammatory response very often contributes to the virus-induced pathology, while direct cytopathic effects may not have major adverse effects. What then are the sources of these cytokines? VSV infection of mice provides two complementary answers. Before describing them, it is remarkable that in vitro, almost no animal cell resists VSV infection, likely because of the use of a ubiquitous but undefined cellular receptor(s) that requires the expression of the endoplasmic reticulum chaperone gp96 (23). In contrast, in vivo, VSV is restricted to few tissue-specific cells, and invasion of nonnatural hosts is not so frequent.
After intranasal inoculation (Fig. 8 A), VSV spreads from the nasal epithelium through the olfactory tract to the CNS over about 6 to 7 days. Nasal epithelial cells and neurons are the main producers of virus during this process (110, 218). Some virus can occasionally be found in the lungs, draining lymph nodes, and spleen. In contrast, type I IFN is mostly absent from the CNS but can be detected in the serum, spleen, and lungs (286). A subset of plasmacytoid dendritic cells in the spleen is likely the main source of this IFN (11). These cells are relatively resistant to the inhibition of host gene expression by VSV M protein, as are also the TLR7+ myeloid dendritic cells (6, 295). Consequently, they can respond to VSV infection by producing type I IFN and other cytokines, although they produce only a limited amount of infectious virus (6). However, the systemic IFN is generally unable to prevent CNS invasion by VSV, likely because of its inefficient penetration into this tissue. Indeed, the intranasal inoculation of M protein mutant VSV induces a local production of IFN and other cytokines that prevents neuroinvasion (286). The extent of morbidity and mortality resulting from intranasal infection with virulent VSV is somehow variable (12, 67, 286), due to possible local control of the VSV infection. Indeed, astrocytes and microglial cells also express TLR7 (33). They are partially resistant to the inhibitory effect of VSV M protein, are poor producers of infectious virus, and can produce IFN and cytokines in response to VSV infection (18, 48, 79).
Fig. 8.
Influence of route of inoculation on the ability of VSV to invade mouse brain. (A) Neuroinvasion from nasal infection with VSV; (B) mechanism preventing neuroinvasion after peripheral inoculation with VSV. See the text for details and references.
Peripheral inoculation of low doses of VSV does not lead to neuroinvasion unless the mouse is immunocompromised, e.g., due to the lack of IFNAR (Fig. 8B). The only cells that replicate the virus are the subcapsular sinus macrophages of the draining lymph node, with macrophages of the medulla being nonpermissive (111). Subcapsular sinus macrophages both are permissive for VSV infection and respond by producing high levels of type I IFN. They also recruit IFN-producing plasmacytoid dendritic cells. This local strong IFN response prevents VSV neuroinvasion via the infection of the intranodal nerves. The high efficiency of this cellular barrier is illustrated by the successful VSV neuroinvasion upon local destruction of these macrophages that permits the infection of the intranodal nerves. This demonstrates how the outcome of a viral infection can depend on a cell type that is highly permissive to virus replication and consequently produces high levels of type I IFN.
A Stealth Pathway for Successful Neuroinvasion
Peripheral inoculation of RABV results in systemic and local type I IFN production in the CNS (120, 121, 185). Dendritic cells are poor producers of infectious virus but respond to RABV infection by producing large amounts of IFN, mostly via a RIG-I-mediated activation pathway. Thus, the activation pathway of innate immunity in dendritic cells is resistant to the inhibition by RABV P protein (74). Importantly, field strains of RABV are more virulent than vaccine strains, and this correlates with a reduced level of viral gene expression and a correspondingly poor antiviral response (187, 297). Thus, the pathogenicity requires that RABV associates a potent suppressive activity against the IFN response of permissive cells with a reduced viral gene expression in the cells of the innate immune system to limit the IFN response. In other words, RABV “[uses] stealth to reach the brain” (253).
Cell Diversity and Genetic Variability of Virus and Host
There are likely additional cells of the innate immune system that are capable of responding to virus infection without being directly infected or by being abortively infected and thus are not subject to viral inhibitory mechanisms. For example, natural killer cells and other lymphoid cells are largely nonpermissive for infection by some viruses yet have mechanisms to recognize and respond to virus-infected cells. Another source of such cells would be those normally permissive to virus infection but rendered nonpermissive by their response to type I IFN. For example, most conventional dendritic cells are susceptible to VSV infection but are unable to respond due to the inhibitory effects of M protein (1, 6, 168, 295). However, dendritic cells that have been exposed to type I IFN could be abortively infected and in turn become an additional source of IFN and cytokines (168). Cells of the innate immune system that are capable of responding to virus-induced tissue damage and other “danger signals” in addition to viral products also likely contribute to the development of the antiviral defenses (183). Because of the tissue-specific distribution of these cell types and the heterogeneity of their response (183), their role in tissue-specific responses to viral tropism and pathogenesis deserves future investigation.
Last but not least, the outcome of the interplay between the innate immunity and a virus results from the (non)complementarity of the genetic polymorphism of the innate immunity machinery of the host and that of the virus. For example, human primary dendritic cells and epithelial cells display a large variety of levels of their IFN responses depending on the infecting strain of measles virus (65, 129, 274), and a mutation in V protein that is deleterious for counteracting MDA5 can contribute to the attenuation phenotype of the vaccine strain. Likewise, for a given virus, dendritic and/or peripheral blood cells from different donors also produce a wide range of IFN levels (65, 200). Accordingly, allelic polymorphism of genes belonging to the innate immune system is increasingly observed, as in the case of RIG-I (107, 222, 263). The high frequency of allelic variants of RLRs with variable activity in the human population could reflect their particular adaptation to respond to some viruses and/or their involvement in autoimmune disease associated with viral infection (263).
CONCLUSION
The outcome of a virus infection, from abortive (asymptomatic) to disastrous (illness and death), relies primarily on a complex interplay between virus replication and its control by the cellular innate antiviral response. The innate immune response plays a crucial role both as an antiviral shield and as a contributor to the pathogenicity. Indeed, the adaptive response alone cannot control Mononegavirales infections in the absence of an intact IFN system. This system involves a growing number of factors engaged in an amazingly complex molecular choreography finely tuned to recognize subtle indicators of virus infection. RNA viruses have coevolved by developing multiple and redundant strategies designed to avoid the exposure of abnormal RNA patterns and to block the activation of innate immunity by interfering with the pathways controlling the induction of, and the response to, type I IFN. The heterogeneity of innate antiviral responses of tissue-specific cell subsets (167) and the variability of their permissiveness to viruses add another level of complexity in vivo. Hence, there are multiple ways of controlling virus infection and multiple strategies for escape from the viruses according to the route of entry into the body.
What can be expected from future investigations? Most of the factors involved in the molecular choreography of innate immunity and virus replication should soon be known, but knowledge of the sophisticated dynamics that link them will require much more intensive investigation. Caution will have to be taken to consider any data in the precise context of the virus-host combination to ensure the correctness of the proposed model. This will require full knowledge of (i) the tissue-specific cell subtypes that are virus targets or direct innate immunity players, (ii) the genetic background of the host, possibly at the tissue-specific cell subtype level, and (iii) the multiallelism of the virus genome and its evolution during tissue propagation, with selection of either the genomic variants with highest fitness or a viral subpopulation as a whole according to the quasispecies theory (103, 180). Finally, the extension of such searches in the context of species restriction of virus will uncover how a given species has so far escaped sensitivity to infection by a given virus and, conversely, how a nonnatural host can be exquisitely sensitive to a virus naturally hosted by another species. This holds particularly true for viruses that use universal cellular receptors and are able to grow in vitro in cells from a large set of animal species, such as, for example, VSV and SeV in the former case, and Ebola virus and Hendra/Nipah viruses in the latter case (84).
ACKNOWLEDGMENTS
This review would not have been possible without the support of the CNRS (D.G.), grant ANR-08-PCV-0020-03 AFF-IDP (D.G.), and NIH grants AI032983 and AI084886 (D.S.L.), allowing experimental work that has driven our overall analysis.
We thank all of our past and present collaborators for numerous clever contributions cited in this review, many colleagues (in particular D. Kolakofsky and S. Cusack) for very fruitful discussions, and B. Grigorov, P. Lawrence, C. Mathieu, and anonymous reviewers for their critical inputs to the manuscript. We acknowledge the artwork of P. Lane. Last but not least, we apologize for being unable to refer to numerous works from colleagues because of space limitation.
REFERENCES
- 1. Ahmed M., Brzoza K. L., Hiltbold E. M. 2006. Matrix protein mutant of vesicular stomatitis virus stimulates maturation of myeloid dendritic cells. J. Virol. 80:2194–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed M., Cramer S. D., Lyles D. S. 2004. Sensitivity of prostate tumors to wild type and M protein mutant vesicular stomatitis viruses. Virology 330:34–49 [DOI] [PubMed] [Google Scholar]
- 3. Ahmed M., Lyles D. S. 1998. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J. Virol. 72:8413–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahmed M., Marino T. R., Puckett S., Kock N. D., Lyles D. S. 2008. Immune response in the absence of neurovirulence in mice infected with M protein mutant vesicular stomatitis virus. J. Virol. 82:9273–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahmed M., et al. 2003. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J. Virol. 77:4646–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ahmed M., et al. 2009. Vesicular stomatitis virus M protein mutant stimulates maturation of Toll-like receptor 7 (TLR7)-positive dendritic cells through TLR-dependent and -independent mechanisms. J. Virol. 83:2962–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Albertini A. A., et al. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360–363 [DOI] [PubMed] [Google Scholar]
- 8. Atreya P. L., Peeples M. E., Collins P. L. 1998. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J. Virol. 72:1452–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Babu J. R., et al. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160:341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bamming D., Horvath C. M. 2009. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 284:9700–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barchet W., et al. 2002. Virus-induced interferon alpha production by a dendritic cell subset in the absence of feedback signaling in vivo. J. Exp. Med. 195:507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barna M., Komatsu T., Bi Z., Reiss C. S. 1996. Sex differences in susceptibility to viral infection of the central nervous system. J. Neuroimmunol. 67:31–39 [DOI] [PubMed] [Google Scholar]
- 13. Baron M. D., Barrett T. 2000. Rinderpest viruses lacking the C and V proteins show specific defects in growth and transcription of viral RNAs. J. Virol. 74:2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baum A., Sachidanandam R., Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Belanger F., Stepinski J., Darzynkiewicz E., Pelletier J. 2010. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem. 285:33037–33044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bernasconi D., Schultz U., Staeheli P. 1995. The interferon-induced Mx protein of chickens lacks antiviral activity. J. Interferon Cytokine Res. 15:47–53 [DOI] [PubMed] [Google Scholar]
- 17. Bhoj V. G., et al. 2008. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc. Natl. Acad. Sci. U. S. A. 105:14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bi Z., Barna M., Komatsu T., Reiss C. S. 1995. Vesicular stomatitis virus infection of the central nervous system activates both innate and acquired immunity. J. Virol. 69:6466–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bitko V., Barik S. 2001. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bitko V., Musiyenko A., Bayfield M. A., Maraia R. J., Barik S. 2008. Cellular La protein shields nonsegmented negative-strand RNA viral leader RNA from RIG-I and enhances virus growth by diverse mechanisms. J. Virol. 82:7977–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Black B. L., Brewer G., Lyles D. S. 1994. Effect of vesicular stomatitis virus matrix protein on host-directed translation in vivo. J. Virol. 68:555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Black B. L., Rhodes R. B., McKenzie M., Lyles D. S. 1993. The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly. J. Virol. 67:4814–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bloor S., Maelfait J., Krumbach R., Beyaert R., Randow F. 2010. Endoplasmic reticulum chaperone gp96 is essential for infection with vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 107:6970–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blower M. D., Nachury M., Heald R., Weis K. 2005. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121:223–234 [DOI] [PubMed] [Google Scholar]
- 25. Blumberg B. M., Giorgi C., Kolakofsky D. 1983. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell 32:559–567 [DOI] [PubMed] [Google Scholar]
- 26. Blumberg B. M., Kolakofsky D. 1981. Intracellular vesicular stomatitis virus leader RNAs are found in nucleocapsid structures. J. Virol. 40:568–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blumberg B. M., Leppert M., Kolakofsky D. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23:837–845 [DOI] [PubMed] [Google Scholar]
- 28. Boonyaratanakornkit J., et al. 2011. The C proteins of human parainfluenza virus type 1 limit double-stranded RNA accumulation that would otherwise trigger activation of MDA5 and protein kinase R. J. Virol. 85:1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bousquet-Antonelli C., Deragon J. M. 2009. A comprehensive analysis of the La-motif protein superfamily. RNA 15:750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boxer E. L., Nanda S. K., Baron M. D. 2009. The rinderpest virus non-structural C protein blocks the induction of type 1 interferon. Virology 385:134–142 [DOI] [PubMed] [Google Scholar]
- 31. Brzozka K., Finke S., Conzelmann K. K. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79:7673–7681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brzozka K., Finke S., Conzelmann K. K. 2006. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J. Virol. 80:2675–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Butchi N. B., Du M., Peterson K. E. 2010. Interactions between TLR7 and TLR9 agonists and receptors regulate innate immune responses by astrocytes and microglia. Glia 58:650–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cadd T., et al. 1996. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J. Virol. 70:5067–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caignard G., et al. 2007. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/beta signaling. Virology 368:351–362 [DOI] [PubMed] [Google Scholar]
- 36. Calain P., Roux L. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 67:4822–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cao W., Liu Y. J. 2007. Innate immune functions of plasmacytoid dendritic cells. Curr. Opin. Immunol. 19:24–30 [DOI] [PubMed] [Google Scholar]
- 38. Cardenas W. B., et al. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80:5168–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carlos T. S., Young D. F., Schneider M., Simas J. P., Randall R. E. 2009. Parainfluenza virus 5 genomes are located in viral cytoplasmic bodies whilst the virus dismantles the interferon-induced antiviral state of cells. J. Gen. Virol. 90:2147–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Castaneda S. J., Wong T. C. 1990. Leader sequence distinguishes between translatable and encapsidated measles virus RNAs. J. Virol. 64:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Castanier C., Garcin D., Vazquez A., Arnoult D. 2010. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 11:133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cattaneo R., Billeter M. A. 1992. Mutations and A/I hypermutations in measles virus persistent infections. Curr. Top. Microbiol. Immunol. 176:63–74 [DOI] [PubMed] [Google Scholar]
- 43. Cattaneo R., et al. 1987. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6:681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chakraborty P., et al. 2009. Vesicular stomatitis virus inhibits mitotic progression and triggers cell death. EMBO Rep. 10:1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang T. H., et al. 2009. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 5:e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chattopadhyay S., et al. 2010. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 29:1762–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chattopadhyay S., Yamashita M., Zhang Y., Sen G. C. 2011. The IRF-3/Bax-mediated apoptotic pathway, activated by viral cytoplasmic RNA and DNA, inhibits virus replication. J. Virol. 85:3708–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chauhan V. S., et al. 2010. Vesicular stomatitis virus infects resident cells of the central nervous system and induces replication-dependent inflammatory responses. Virology 400:187–196 [DOI] [PubMed] [Google Scholar]
- 49. Chen S., et al. 2010. Heterocellular induction of interferon by negative-sense RNA viruses. Virology 407:247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Childs K., et al. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200 [DOI] [PubMed] [Google Scholar]
- 51. Childs K. S., Andrejeva J., Randall R. E., Goodbourn S. 2009. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. J. Virol. 83:1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ciancanelli M. J., Volchkova V. A., Shaw M. L., Volchkov V. E., Basler C. F. 2009. Nipah virus sequesters inactive STAT1 in the nucleus via a P gene-encoded mechanism. J. Virol. 83:7828–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Colby C., Chamberlin M. J. 1969. The specificity of interferon induction in chick embryo cells by helical RNA. Proc. Natl. Acad. Sci. U. S. A. 63:160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Colonno R. J., Banerjee A. K. 1976. A unique RNA species involved in initiation of vesicular stomatitis virus RNA transcription in vitro. Cell 8:197–204 [DOI] [PubMed] [Google Scholar]
- 55. Conzelmann K. K. 2005. Transcriptional activation of alpha/beta interferon genes: interference by nonsegmented negative-strand RNA viruses. J. Virol. 79:5241–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cui S., et al. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169–179 [DOI] [PubMed] [Google Scholar]
- 57. Curran J., Marq J. B., Kolakofsky D. 1992. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189:647–656 [DOI] [PubMed] [Google Scholar]
- 58. Daffis S., et al. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. De B. P., Gupta S., Zhao H., Drazba J. A., Banerjee A. K. 1996. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and LA protein with cis-acting RNAs of human parainfluenza virus type 3. J. Biol. Chem. 271:24728–24735 [DOI] [PubMed] [Google Scholar]
- 60. Devaux P., et al. 2011. A recombinant measles virus unable to antagonize STAT1 function cannot control inflammation and is attenuated in rhesus monkeys. J. Virol. 85:348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Devaux P., von Messling V., Songsungthong W., Springfeld C., Cattaneo R. 2007. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology 360:72–83 [DOI] [PubMed] [Google Scholar]
- 62. DeWitte-Orr S. J., et al. 2009. Long double-stranded RNA induces an antiviral response independent of IFN regulatory factor 3, IFN-beta promoter stimulator 1, and IFN. J. Immunol. 183:6545–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Diaz-Guerra M., Rivas C., Esteban M. 1997. Inducible expression of the 2-5A synthetase/RNase L system results in inhibition of vaccinia virus replication. Virology 227:220–228 [DOI] [PubMed] [Google Scholar]
- 64. Dixit E., et al. 2010. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141:668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Duhen T., et al. 2010. Cellular receptors, differentiation and endocytosis requirements are key factors for type I IFN response by human epithelial, conventional and plasmacytoid dendritic infected cells by measles virus. Virus Res. 152:115–125 [DOI] [PubMed] [Google Scholar]
- 66. Dunn E. F., Connor J. H. 2011. Dominant inhibition of Akt/PKB signaling by the matrix protein of a negative-strand RNA virus. J. Virol. 85:422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Durbin R. K., Mertz S. E., Koromilas A. E., Durbin J. E. 2002. PKR protection against intranasal vesicular stomatitis virus infection is mouse strain dependent. Viral Immunol. 15:41–51 [DOI] [PubMed] [Google Scholar]
- 68. Elliott J., et al. 2007. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J. Virol. 81:3428–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Espert L., et al. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 278:16151–16158 [DOI] [PubMed] [Google Scholar]
- 70. Falkenberg M., Larsson N. G., Gustafsson C. M. 2007. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 76:679–699 [DOI] [PubMed] [Google Scholar]
- 71. Fan H., Goodier J. L., Chamberlain J. R., Engelke D. R., Maraia R. J. 1998. 5′ processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol. Cell. Biol. 18:3201–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Faria P. A., et al. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17:93–102 [DOI] [PubMed] [Google Scholar]
- 73. Faul E. J., Lyles D. S., Schnell M. J. 2009. Interferon response and viral evasion by members of the family Rhabdoviridae. Viruses 1:832–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Faul E. J., et al. 2010. Rabies virus infection induces type I interferon production in an IPS-1 dependent manner while dendritic cell activation relies on IFNAR signaling. PLoS Pathog. 6:e1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ferran M. C., Lucas-Lenard J. M. 1997. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J. Virol. 71:371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Finke S., Conzelmann K. K. 1997. Ambisense gene expression from recombinant rabies virus: random packaging of positive- and negative-strand ribonucleoprotein complexes into rabies virions. J. Virol. 71:7281–7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fontana J. M., Bankamp B., Bellini W. J., Rota P. A. 2008. Regulation of interferon signaling by the C and V proteins from attenuated and wild-type strains of measles virus. Virology 374:71–81 [DOI] [PubMed] [Google Scholar]
- 78. Francoeur A. M., Poliquin L., Stanners C. P. 1987. The isolation of interferon-inducing mutants of vesicular stomatitis virus with altered viral P function for the inhibition of total protein synthesis. Virology 160:236–245 [DOI] [PubMed] [Google Scholar]
- 79. Furr S. R., Chauhan V. S., Sterka D., Jr., Grdzelishvili V., Marriott I. 2008. Characterization of retinoic acid-inducible gene-I expression in primary murine glia following exposure to vesicular stomatitis virus. J. Neurovirol. 14:503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gack M. U., et al. 2008. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. U. S. A. 105:16743–16748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gack M. U., et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920 [DOI] [PubMed] [Google Scholar]
- 82. Garcin D., Latorre P., Kolakofsky D. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gee P., et al. 2008. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J. Biol. Chem. 283:9488–9496 [DOI] [PubMed] [Google Scholar]
- 84. Gerlier D. Emerging zoonotic viruses: new lessons on receptor and entry mechanisms. Curr. Opin. Virol. 1:27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gerlier D., Valentin H. 2009. Measles virus interaction with host cells and impact on innate immunity. Curr. Top. Microbiol. Immunol. 329:163–191 [DOI] [PubMed] [Google Scholar]
- 86. Gitlin L., et al. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6:e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Goodbourn S., Didcock L., Randall R. E. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341–2364 [DOI] [PubMed] [Google Scholar]
- 88. Goodbourn S., Randall R. E. 2009. The regulation of type I interferon production by paramyxoviruses. J. Interferon Cytokine Res. 29:539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gotoh B., et al. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205–210 [DOI] [PubMed] [Google Scholar]
- 90. Grandvaux N., Servant M. J., tenOever B., et al. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Green T. J., et al. 2011. Access to RNA encapsidated in the nucleocapsid of vesicular stomatitis virus. J. Virol. 85:2714–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Green T. J., Zhang X., Wertz G. W., Luo M. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313:357–360 [DOI] [PubMed] [Google Scholar]
- 93. Gupta K. C., Kingsbury D. W. 1985. Polytranscripts of Sendai virus do not contain intervening polyadenylate sequences. Virology 141:102–109 [DOI] [PubMed] [Google Scholar]
- 94. Habjan M., et al. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3:e2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Halpin K., Bankamp B., Harcourt B. H., Bellini W. J., Rota P. A. 2004. Nipah virus conforms to the rule of six in a minigenome replication assay. J. Gen. Virol. 85:701–707 [DOI] [PubMed] [Google Scholar]
- 96. Hartman A. L., et al. 2008. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of Ebola virus. J. Virol. 82:2699–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hartman A. L., Dover J. E., Towner J. S., Nichol S. T. 2006. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J. Virol. 80:6430–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hartman A. L., Ling L., Nichol S. T., Hibberd M. L. 2008. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J. Virol. 82:5348–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hartner J. C., Walkley C. R., Lu J., Orkin S. H. 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 10:109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hausmann S., Marq J. B., Tapparel C., Kolakofsky D., Garcin D. 2008. RIG-I and dsRNA-induced IFNbeta activation. PLoS One 3:e3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hayakawa S., et al. 2011. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat. Immunol. 12:37–44 [DOI] [PubMed] [Google Scholar]
- 102. Her L. S., Lund E., Dahlberg J. E. 1997. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science 276:1845–1848 [DOI] [PubMed] [Google Scholar]
- 103. Holmes E. C. 2010. The RNA virus quasispecies: fact or fiction? J. Mol. Biol. 400:271–273 [DOI] [PubMed] [Google Scholar]
- 104. Horikami S. M., Hector R. E., Smallwood S., Moyer S. A. 1997. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology 235:261–270 [DOI] [PubMed] [Google Scholar]
- 105. Horikami S. M., Moyer S. A. 1991. Synthesis of leader RNA and editing of the P mRNA during transcription by purified measles virus. J. Virol. 65:5342–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hornung V., et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997 [DOI] [PubMed] [Google Scholar]
- 107. Hu J., et al. 2010. A common polymorphism in the caspase recruitment domain of RIG-I modifies the innate immune response of human dendritic cells. J. Immunol. 185:424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Huang Y., Bayfield M. A., Intine R. V., Maraia R. J. 2006. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat. Struct. Mol. Biol. 13:611–618 [DOI] [PubMed] [Google Scholar]
- 109. Huhn P., Pruijn G. J., van Venrooij W. J., Bachmann M. 1997. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res. 25:410–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Huneycutt B. S., et al. 1994. Distribution of vesicular stomatitis virus proteins in the brains of BALB/c mice following intranasal inoculation: an immunohistochemical analysis. Brain Res. 635:81–95 [DOI] [PubMed] [Google Scholar]
- 111. Iannacone M., et al. 2010. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature 465:1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ikegame S., et al. 2010. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J. Virol. 84:372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Inn K. S., et al. 2011. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell 41:354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Isaacs A., Lindenmann J. 1957. Virus interference. I. The interferon. Proc. R Soc. Lond. B Biol. Sci. 147:258–267 [PubMed] [Google Scholar]
- 115. Isaacs A., Lindenmann J., Valentine R. C. 1957. Virus interference. II. Some properties of interferon. Proc. R Soc. Lond. B Biol. Sci. 147:268–273 [PubMed] [Google Scholar]
- 116. Iseni F., et al. 2002. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 21:5141–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ito N., et al. 2010. Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J. Virol. 84:6699–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Janeway C. A., Jr, Goodnow C. C., Medzhitov R. 1996. Danger—pathogen on the premises! Immunological tolerance. Curr. Biol. 6:519–522 [DOI] [PubMed] [Google Scholar]
- 119. Johansen L. D., et al. 2008. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J. Cell Sci. 121:854–864 [DOI] [PubMed] [Google Scholar]
- 120. Johnson N., et al. 2008. Inflammatory responses in the nervous system of mice infected with a street isolate of rabies virus. Dev. Biol. (Basel) 131:65–72 [PubMed] [Google Scholar]
- 121. Johnson N., et al. 2006. Lyssavirus infection activates interferon gene expression in the brain. J. Gen. Virol. 87:2663–2667 [DOI] [PubMed] [Google Scholar]
- 122. Kanneganti T. D. 2010. Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 10:688–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kato H., et al. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19–28 [DOI] [PubMed] [Google Scholar]
- 124. Kato H., et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kawai T., Akira S. 2007. TLR signaling. Semin. Immunol. 19:24–32 [DOI] [PubMed] [Google Scholar]
- 126. Kawai T., Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650 [DOI] [PubMed] [Google Scholar]
- 127. Kawai T., et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 128. Keene J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8:533–543 [DOI] [PubMed] [Google Scholar]
- 129. Kessler J. R., Kremer J. R., Muller C. P. 2011. Interplay of measles virus with early induced cytokines reveals different wild type phenotypes. Virus Res. 155:195–202 [DOI] [PubMed] [Google Scholar]
- 130. Khabar K. S., et al. 2000. Effect of deficiency of the double-stranded RNA-dependent protein kinase, PKR, on antiviral resistance in the presence or absence of RNase L: HSV-1 replication is particularly sensitive to deficiency of the major IFN-mediated enzymes. J. Interferon Cytokine Res. 20:653–659 [DOI] [PubMed] [Google Scholar]
- 131. Killip M. J., et al. 2011. Failure to activate the IFN-beta promoter by a paramyxovirus lacking an interferon antagonist. Virology 415:39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kim H., Seed B. 2010. The transcription factor MafB antagonizes antiviral responses by blocking recruitment of coactivators to the transcription factor IRF3. Nat. Immunol. 11:743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kim M. J., Hwang S. Y., Imaizumi T., Yoo J. Y. 2008. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 82:1474–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kimberlin C. R., et al. 2010. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. U. S. A. 107:314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kitagawa Y., et al. 2011. A tryptophan-rich motif in the human parainfluenza virus type 2 V protein is critical for the blockade of Toll-like receptor 7 (TLR7)- and TLR9-dependent signaling. J. Virol. 85:4606–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kolakofsky D., Le Mercier P., Iseni F., Garcin D. 2004. Viral RNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318:463–473 [DOI] [PubMed] [Google Scholar]
- 137. Kolakofsky D., et al. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J. Virol. 72:891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kolakofsky D., Roux L., Garcin D., Ruigrok R. W. 2005. Paramyxovirus mRNA editing, the “rule of six” and error catastrophe: a hypothesis. J. Gen. Virol. 86:1869–1877 [DOI] [PubMed] [Google Scholar]
- 139. Komuro A., Bamming D., Horvath C. M. 2008. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine 43:350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Komuro A., Horvath C. M. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 80:12332–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kumagai Y., et al. 2007. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27:240–252 [DOI] [PubMed] [Google Scholar]
- 142. Kurilla M. G., Cabradilla C. D., Holloway B. P., Keene J. D. 1984. Nucleotide sequence and host La protein interactions of rabies virus leader RNA. J. Virol. 50:773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kurilla M. G., Keene J. D. 1983. The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell 34:837–845 [DOI] [PubMed] [Google Scholar]
- 144. Lamb R. A., Parks G. D. 2007. Paramyxoviridae: the viruses and their replication, p. 1449–1496 In Fields B. N., Howley P. M. (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 145. Landis H., et al. 1998. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J. Virol. 72:1516–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Lang K. S., et al. 2007. MyD88 protects from lethal encephalitis during infection with vesicular stomatitis virus. Eur. J. Immunol. 37:2434–2440 [DOI] [PubMed] [Google Scholar]
- 147. Le Mercier P., Garcin D., Hausmann S., Kolakofsky D. 2002. Ambisense Sendai viruses are inherently unstable but are useful to study viral RNA synthesis. J. Virol. 76:5492–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Lemon K., et al. 2011. Early target cells of measles virus after aerosol infection of non-human primates. PLoS Pathog. 7:e1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Leppert M., Kolakofsky D. 1978. 5′ terminus of defective and nondefective Sendai viral genomes is ppp Ap. J. Virol. 25:427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Leppert M., Kolakofsky D. 1980. Effect of defective interfering particles on plus- and minus-strand leader RNAs in vesicular stomatitis virus-infected cells. J. Virol. 35:704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Leppert M., Rittenhouse L., Perrault J., Summers D. F., Kolakofsky D. 1979. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell 18:735–747 [DOI] [PubMed] [Google Scholar]
- 152. Leroy M., Baise E., Pire G., Gerardin J., Desmecht D. 2005. Resistance of paramyxoviridae to type I interferon-induced Bos taurus Mx1 dynamin. J. Interferon Cytokine Res. 25:192–201 [DOI] [PubMed] [Google Scholar]
- 153. Leroy M., Pire G., Baise E., Desmecht D. 2006. Expression of the interferon-alpha/beta-inducible bovine Mx1 dynamin interferes with replication of rabies virus. Neurobiol. Dis. 21:515–521 [DOI] [PubMed] [Google Scholar]
- 154. Leroy M. P., Baise E. A., Pire G. A., Desmecht D. J. 2007. Contribution of MX dynamin, oligoadenylate synthetase, and protein kinase R to anti-paramyxovirus activity of type 1 interferons in vitro. Am. J. Vet. Res. 68:988–994 [DOI] [PubMed] [Google Scholar]
- 155. Leung D. W., et al. 2009. Structure of the Ebola VP35 interferon inhibitory domain. Proc. Natl. Acad. Sci. U. S. A. 106:411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Li J., Rahmeh A., Brusic V., Whelan S. P. 2009. Opposing effects of inhibiting cap addition and cap methylation on polyadenylation during vesicular stomatitis virus mRNA synthesis. J. Virol. 83:1930–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Li J., Wang J. T., Whelan S. P. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 103:8493–8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Li X., et al. 2009. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch. Biochem. Biophys. 488:23–33 [DOI] [PubMed] [Google Scholar]
- 159. Li X., et al. 2009. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 284:13881–13891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Ling Z., Tran K. C., Teng M. N. 2009. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J. Virol. 83:3734–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Lipardi C., Paterson B. M. 2009. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc. Natl. Acad. Sci. U. S. A. 106:15645–15650 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 162. Liuzzi M., et al. 2005. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J. Virol. 79:13105–13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Longhi S. 2009. Nucleocapsid structure and function. Curr. Top. Microbiol. Immunol. 329:103–128 [DOI] [PubMed] [Google Scholar]
- 164. Loo Y. M., et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Lu C., et al. 2010. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure 18:1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Lu L. L., Puri M., Horvath C. M., Sen G. C. 2008. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappaB kinase epsilon (IKKe)/TBK1. J. Biol. Chem. 283:14269–14276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Luber C. A., et al. 2010. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32:279–289 [DOI] [PubMed] [Google Scholar]
- 168. Ludewig B., et al. 2000. Induction of optimal anti-viral neutralizing B cell responses by dendritic cells requires transport and release of virus particles in secondary lymphoid organs. Eur. J. Immunol. 30:185–196 [DOI] [PubMed] [Google Scholar]
- 169. Luthra P., Sun D., Silverman R. H., He B. 2011. Activation of IFN-β expression by a viral mRNA through RNase L and MDA5. Proc. Natl. Acad. Sci. U. S. A. 108:2118–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Lynch S., Kolakofsky D. 1978. Ends of the RNA within Sendai virus defective interfering nucleocapsids are not free. J. Virol. 28:584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Malur A. G., Hoffman M. A., Banerjee A. K. 2004. The human parainfluenza virus type 3 (HPIV 3) C protein inhibits viral transcription. Virus Res. 99:199–204 [DOI] [PubMed] [Google Scholar]
- 172. Manuse M. J., Parks G. D. 2009. Role for the paramyxovirus genomic promoter in limiting host cell antiviral responses and cell killing. J. Virol. 83:9057–9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Marcus P. I., Engelhardt D. L., Hunt J. M., Sekellick M. J. 1971. Interferon action: inhibition of vesicular stomatitis virus RNA synthesis induced by virion-bound polymerase. Science 174:593–598 [DOI] [PubMed] [Google Scholar]
- 174. Marcus P. I., Sekellick M. J. 1977. Defective interfering particles with covalently linked [+/−]RNA induce interferon. Nature 266:815–819 [DOI] [PubMed] [Google Scholar]
- 175. Marcus P. I., Sekellick M. J. 1978. Interferon action. III. The rate of primary transcription of vesicular stomatitis virus is inhibited by interferon action. J. Gen. Virol. 38:391–408 [DOI] [PubMed] [Google Scholar]
- 176. Marcus P. I., Sekellick M. J. 1980. Interferon induction by viruses. III. Vesicular stomatitis virus: interferon-inducing particle activity requires partial transcription of gene N. J. Gen. Virol. 47:89–96 [DOI] [PubMed] [Google Scholar]
- 177. Marq J. B., Hausmann S., Veillard N., Kolakofsky D., Garcin D. 2011. Short double-stranded RNAs with an overhanging 5′ ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. J. Biol. Chem. 286:6108–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Marq J. B., Kolakofsky D., Garcin D. 2010. Unpaired 5′ ppp-nucleotides, as found in arenavirus double-stranded RNA panhandles, are not recognized by RIG-I. J. Biol. Chem. 285:18208–18216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Marques J. T., et al. 2006. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 24:559–565 [DOI] [PubMed] [Google Scholar]
- 180. Mas A., Lopez-Galindez C., Cacho I., Gomez J., Martinez M. A. 2010. Unfinished stories on viral quasispecies and Darwinian views of evolution. J. Mol. Biol. 397:865–877 [DOI] [PubMed] [Google Scholar]
- 181. Masters P. S., Samuel C. E. 1984. Detection of in vivo synthesis of polycistronic mRNAs of vesicular stomatitis virus. Virology 134:277–286 [DOI] [PubMed] [Google Scholar]
- 182. Mateo M., Reid S. P., Leung L. W., Basler C. F., Volchkov V. E. 2010. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J. Virol. 84:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Matzinger P. 2002. The danger model: a renewed sense of self. Science 296:301–305 [DOI] [PubMed] [Google Scholar]
- 184. McAllister C. S., et al. 2010. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J. Virol. 84:380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Mendonca R. Z., Pereira C. A. 1994. Relationship of interferon synthesis and the resistance of mice infected with street rabies virus. Braz. J. Med. Biol. Res. 27:691–695 [PubMed] [Google Scholar]
- 186. Meylan E., et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 187. Morimoto K., Foley H. D., McGettigan J. P., Schnell M. J., Dietzschold B. 2000. Reinvestigation of the role of the rabies virus glycoprotein in viral pathogenesis using a reverse genetics approach. J. Neurovirol. 6:373–381 [DOI] [PubMed] [Google Scholar]
- 188. Moseley G. W., Filmer R. P., DeJesus M. A., Jans D. A. 2007. Nucleocytoplasmic distribution of rabies virus P-protein is regulated by phosphorylation adjacent to C-terminal nuclear import and export signals. Biochemistry 46:12053–12061 [DOI] [PubMed] [Google Scholar]
- 189. Moseley G. W., et al. 2009. Dual modes of rabies P-protein association with microtubules: a novel strategy to suppress the antiviral response. J. Cell Sci. 122:3652–3662 [DOI] [PubMed] [Google Scholar]
- 190. Mottet-Osman G., et al. 2007. Suppression of the Sendai virus M protein through a novel short interfering RNA approach inhibits viral particle production but does not affect viral RNA synthesis. J. Virol. 81:2861–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Mottet G., Curran J., Roux L. 1990. Intracellular stability of nonreplicating paramyxovirus nucleocapsids. Virology 176:1–7 [DOI] [PubMed] [Google Scholar]
- 192. Mueller S., et al. 2010. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 107:19390–19395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Muller U., et al. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921 [DOI] [PubMed] [Google Scholar]
- 194. Murali A., et al. 2008. Structure and function of LGP2, a DEX(D/H) helicase that regulates the innate immunity response. J. Biol. Chem. 283:15825–15833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Myong S., et al. 2009. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323:1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Nakatsu Y., Takeda M., Ohno S., Koga R., Yanagi Y. 2006. Translational inhibition and increased interferon induction in cells infected with C protein-deficient measles virus. J. Virol. 80:11861–11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Nakayama Y., et al. 2010. Role of PKR and type I IFNs in viral control during primary and secondary infection. PLoS Pathog. 6:e1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Nallagatla S. R., et al. 2007. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 318:1455–1458 [DOI] [PubMed] [Google Scholar]
- 199. Nanbo A., et al. 2010. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 6:e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Naniche D., et al. 2000. Evasion of host defenses by measles virus: wild-type measles virus infection interferes with induction of alpha/beta interferon production. J. Virol. 74:7478–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Nishie T., Nagata K., Takeuchi K. 2007. The C protein of wild-type measles virus has the ability to shuttle between the nucleus and the cytoplasm. Microbes Infect. 9:344–354 [DOI] [PubMed] [Google Scholar]
- 202. Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79:321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Nistal-Villan E., et al. 2010. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J. Biol. Chem. 285:20252–20261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Noton S. L., Cowton V. M., Zack C. R., McGivern D. R., Fearns R. 2010. Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion. Proc. Natl. Acad. Sci. U. S. A. 107:10226–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Ogino T., Banerjee A. K. 2010. The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity. J. Gen. Virol. 91:1311–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Ogino T., Yadav S. P., Banerjee A. K. 2010. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 107:3463–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. O'Neill L. A., Bowie A. G. 2010. Sensing and signaling in antiviral innate immunity. Curr. Biol. 20:R328–333 [DOI] [PubMed] [Google Scholar]
- 208. Onoguchi K., et al. 2010. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog. 6:e1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Osiak A., Utermohlen O., Niendorf S., Horak I., Knobeloch K. P. 2005. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell. Biol. 25:6338–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Palosaari H., Parisien J. P., Rodriguez J. J., Ulane C. M., Horvath C. M. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Panne D. 2008. The enhanceosome. Curr. Opin. Struct. Biol. 18:236–242 [DOI] [PubMed] [Google Scholar]
- 212. Parisien J. P., et al. 2009. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83:7252–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Peeters B. P., Gruijthuijsen Y. K., de Leeuw O. S., Gielkens A. L. 2000. Genome replication of Newcastle disease virus: involvement of the rule-of-six. Arch. Virol. 145:1829–1845 [DOI] [PubMed] [Google Scholar]
- 214. Pernet O., Pohl C., Ainouze M., Kweder H., Buckland R. 2009. Nipah virus entry can occur by macropinocytosis. Virology 395:298–311 [DOI] [PubMed] [Google Scholar]
- 215. Pfaller C. K., Conzelmann K. K. 2008. Measles virus V protein is a decoy substrate for IkappaB kinase alpha and prevents Toll-like receptor 7/9-mediated interferon induction. J. Virol. 82:12365–12373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Pichlmair A., et al. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Pippig D. A., et al. 2009. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 37:2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Plakhov I. V., Arlund E. E., Aoki C., Reiss C. S. 1995. The earliest events in vesicular stomatitis virus infection of the murine olfactory neuroepithelium and entry of the central nervous system. Virology 209:257–262 [DOI] [PubMed] [Google Scholar]
- 219. Plumet S., Duprex W. P., Gerlier D. 2005. Dynamics of viral RNA synthesis during measles virus infection. J. Virol. 79:6900–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Plumet S., Gerlier D. 2005. Optimized SYBR green real-time PCR assay to quantify the absolute copy number of measles virus RNAs using gene specific primers. J. Virol. Methods 128:79–87 [DOI] [PubMed] [Google Scholar]
- 221. Plumet S., et al. 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS One 2:e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Pothlichet J., et al. 2009. Study of human RIG-I polymorphisms identifies two variants with an opposite impact on the antiviral immune response. PLoS One 4:e7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Prins K. C., Cardenas W. B., Basler C. F. 2009. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J. Virol. 83:3069–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Prins K. C., et al. 2010. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J. Virol. 84:3004–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Raha T., Pudi R., Das S., Shaila M. S. 2004. Leader RNA of Rinderpest virus binds specifically with cellular La protein: a possible role in virus replication. Virus Res. 104:101–109 [DOI] [PubMed] [Google Scholar]
- 226. Rahmeh A. A., Li J., Kranzusch P. J., Whelan S. P. 2009. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 83:11043–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Ramachandran A., Horvath C. M. 2010. Dissociation of paramyxovirus interferon evasion activities: universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J. Virol. 84:11152–11163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Ramachandran A., Horvath C. M. 2009. Paramyxovirus disruption of interferon signal transduction: STATus report. J. Interferon Cytokine Res. 29:531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Randall R. E., Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 230. Ranjith-Kumar C. T., et al. 2009. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J. Biol. Chem. 284:1155–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Rao D. D., Huang A. S. 1979. Synthesis of a small RNA in cells coinfected by standard and defective interfering particles of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 76:3742–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Re G. G., Morgan E. M., Kingsbury D. W. 1985. Nucleotide sequences responsible for generation of internally deleted Sendai virus defective interfering genomes. Virology 146:27–37 [DOI] [PubMed] [Google Scholar]
- 233. Rehwinkel J., et al. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408 [DOI] [PubMed] [Google Scholar]
- 234. Reid S. P., Cardenas W. B., Basler C. F. 2005. Homo-oligomerization facilitates the interferon-antagonist activity of the Ebolavirus VP35 protein. Virology 341:179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Reid S. P., et al. 2006. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Reid S. P., Valmas C., Martinez O., Sanchez F. M., Basler C. F. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 81:13469–13477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Reuter T., Weissbrich B., Schneider-Schaulies S., Schneider-Schaulies J. 2006. RNA interference with measles virus N, P, and L mRNAs efficiently prevents and with matrix protein mRNA enhances viral transcription. J. Virol. 80:5951–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Reutter G. L., Cortese-Grogan C., Wilson J., Moyer S. A. 2001. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology 285:100–109 [DOI] [PubMed] [Google Scholar]
- 239. Rieder M., et al. 2011. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J. Virol. 85:842–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Rima B. K., Duprex W. P. 2009. The measles virus replication cycle. Curr. Top. Microbiol. Immunol. 329:77–102 [DOI] [PubMed] [Google Scholar]
- 241. Rodriguez M. S., Dargemont C., Stutz F. 2004. Nuclear export of RNA. Biol. Cell 96:639–655 [DOI] [PubMed] [Google Scholar]
- 242. Rosetto M., Engstrom Y., Baldari C. T., Telford J. L., Hultmark D. 1995. Signals from the IL-1 receptor homolog, Toll, can activate an immune response in a Drosophila hemocyte cell line. Biochem. Biophys. Res. Commun. 209:111–116 [DOI] [PubMed] [Google Scholar]
- 243. Rue C. A., et al. 2011. Virulent Newcastle disease virus elicits a strong innate immune response in chickens. J. Gen. Virol. 92:931–939 [DOI] [PubMed] [Google Scholar]
- 244. Sabbah A., et al. 2009. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 10:1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Sadler A. J., Williams B. R. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Saito T., et al. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104:582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Satoh T., et al. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. 107:1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Schaap-Nutt A., et al. 2011. Identification of human parainfluenza virus type 2 (HPIV-2) V protein amino acid residues that reduce binding of V to MDA5 and attenuate HPIV-2 replication in nonhuman primates. J. Virol. 85:4007–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Schlee M., et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Schmeisser H., et al. 2010. Identification of alpha interferon-induced genes associated with antiviral activity in Daudi cells and characterization of IFIT3 as a novel antiviral gene. J. Virol. 84:10671–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Schmidt A., et al. 2009. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U. S. A. 106:12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Schneider U., Schwemmle M., Staeheli P. 2005. Genome trimming: a unique strategy for replication control employed by Borna disease virus. Proc. Natl. Acad. Sci. U. S. A. 102:3441–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Schnell M. J., McGettigan J. P., Wirblich C., Papaneri A. 2010. The cell biology of rabies virus: using stealth to reach the brain. Nat. Rev. Microbiol. 8:51–61 [DOI] [PubMed] [Google Scholar]
- 254. Schoggins J. W., et al. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Schuhmann K. M., Pfaller C. K., Conzelmann K. K. 2011. The measles virus V protein binds to p65 (RelA) to suppress NF-kappaB activity. J. Virol. 85:3162–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Schulz O., et al. 2010. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe 7:354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Schuster A., et al. 1996. Expression of the human MxA protein is associated with hyperphosphorylation of VSV P protein in human neural cells. Virology 220:241–245 [DOI] [PubMed] [Google Scholar]
- 258. Schwanhausser B., et al. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342 [DOI] [PubMed] [Google Scholar]
- 259. Sellin C. I., Horvat B. 2009. Current animal models: transgenic animal models for the study of measles pathogenesis. Curr. Top. Microbiol. Immunol. 330:111–127 [DOI] [PubMed] [Google Scholar]
- 260. Seth R. B., Sun L., Ea C. K., Chen Z. J. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 261. Seto J., et al. 2010. Novel Nipah virus immune-antagonism strategy revealed by experimental and computational study. J. Virol. 84:10965–10973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Shaffer J. A., Bellini W. J., Rota P. A. 2003. The C protein of measles virus inhibits the type I interferon response. Virology 315:389–397 [DOI] [PubMed] [Google Scholar]
- 263. Shigemoto T., et al. 2009. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J. Biol. Chem. 284:13348–13354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Silverman R. H. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81:12720–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Skiadopoulos M. H., et al. 2003. The genome length of human parainfluenza virus type 2 follows the rule of six, and recombinant viruses recovered from non-polyhexameric-length antigenomic cDNAs contain a biased distribution of correcting mutations. J. Virol. 77:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Stojdl D. F., et al. 2000. The murine double-stranded RNA-dependent protein kinase PKR is required for resistance to vesicular stomatitis virus. J. Virol. 74:9580–9585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Stojdl D. F., et al. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275 [DOI] [PubMed] [Google Scholar]
- 268. Strahle L., Garcin D., Kolakofsky D. 2006. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351:101–111 [DOI] [PubMed] [Google Scholar]
- 269. Strahle L., Garcin D., Le Mercier P., Schlaak J. F., Kolakofsky D. 2003. Sendai virus targets inflammatory responses, as well as the interferon-induced antiviral state, in a multifaceted manner. J. Virol. 77:7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Strahle L., et al. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J. Virol. 81:12227–12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Sun D., Luthra P., Li Z., He B. 2009. PLK1 down-regulates parainfluenza virus 5 gene expression. PLoS Pathog. 5:e1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Sun Z., Ren H., Liu Y., Teeling J. L., Gu J. 2011. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J. Virol. 85:1036–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Takahasi K., et al. 2009. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J. Biol. Chem. 284:17465–17474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Takaki H., et al. 2011. Strain-to-strain difference of V protein of measles virus affects MDA5-mediated IFN-beta-inducing potential. Mol. Immunol. 48:497–504 [DOI] [PubMed] [Google Scholar]
- 275. Takeuchi K., Kadota S. I., Takeda M., Miyajima N., Nagata K. 2003. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS Lett. 545:177–182 [DOI] [PubMed] [Google Scholar]
- 276. Takeuchi K., Komatsu T., Kitagawa Y., Sada K., Gotoh B. 2008. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J. Virol. 82:10102–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Takeuchi O., Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820 [DOI] [PubMed] [Google Scholar]
- 278. Takeuchi O., Akira S. 2007. Recognition of viruses by innate immunity. Immunol. Rev. 220:214–224 [DOI] [PubMed] [Google Scholar]
- 279. Tawar R. G., et al. 2009. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326:1279–1283 [DOI] [PubMed] [Google Scholar]
- 280. Tekes G., Rahmeh A. A., Whelan S. P. J. 2011. A freeze frame view of vesicular stomatitis virus transcription defines a minimal length of RNA for 5′ processing. PLoS Pathog. 7:e1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Teplova M., et al. 2006. Structural basis for recognition and sequestration of UUU(OH) 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell 21:75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Timani K. A., et al. 2008. A single amino acid residue change in the P protein of parainfluenza virus 5 elevates viral gene expression. J. Virol. 82:9123–9133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Toth A. M., Devaux P., Cattaneo R., Samuel C. E. 2009. Protein kinase PKR mediates the apoptosis induction and growth restriction phenotypes of C protein-deficient measles virus. J. Virol. 83:961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Toth A. M., Li Z., Cattaneo R., Samuel C. E. 2009. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J. Biol. Chem. 284:29350–29356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Trinchieri G. 2010. Type I interferon: friend or foe? J. Exp. Med. 207:2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Trottier M. D., Lyles D. S., Reiss C. S. 2007. Peripheral, but not central nervous system, type I interferon expression in mice in response to intranasal vesicular stomatitis virus infection. J. Neurovirol. 13:433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Tytell A. A., Lampson G. P., Field A. K., Hilleman M. R. 1967. Inducers of interferon and host resistance. 3. Double-stranded RNA from reovirus type 3 virions (reo 3-RNA). Proc. Natl. Acad. Sci. U. S. A. 58:1719–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Valmas C., et al. 2010. Marburg virus evades interferon responses by a mechanism distinct from Ebola virus. PLoS Pathog. 6:e1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Venkataraman T., et al. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178:6444–6455 [DOI] [PubMed] [Google Scholar]
- 290. Vidal S., Kolakofsky D. 1989. Modified model for the switch from Sendai virus transcription to replication. J. Virol. 63:1951–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Vidy A., Chelbi-Alix M., Blondel D. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J. Virol. 79:14411–14420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Vidy A., El Bougrini J., Chelbi-Alix M. K., Blondel D. 2007. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J. Virol. 81:4255–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Virtue E. R., Marsh G. A., Wang L. F. 2011. Interferon signaling remains functional during henipavirus infection of human cell lines. J. Virol. 85:4031–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. von Kobbe C., et al. 2000. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell 6:1243–1252 [DOI] [PubMed] [Google Scholar]
- 295. Waibler Z., Detje C. N., Bell J. C., Kalinke U. 2007. Matrix protein mediated shutdown of host cell metabolism limits vesicular stomatitis virus-induced interferon-alpha responses to plasmacytoid dendritic cells. Immunobiology 212:887–894 [DOI] [PubMed] [Google Scholar]
- 296. Wang Y., et al. 2010. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 17:781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Wang Z. W., et al. 2005. Attenuated rabies virus activates, while pathogenic rabies virus evades, the host innate immune responses in the central nervous system. J. Virol. 79:12554–12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Ward S. V., et al. 2011. RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc. Natl. Acad. Sci. U. S. A. 108:331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Weber F., Wagner V., Rasmussen S. B., Hartmann R., Paludan S. R. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Weichenrieder O., Wild K., Strub K., Cusack S. 2000. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408:167–173 [DOI] [PubMed] [Google Scholar]
- 301. Weidner J. M., et al. 2010. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84:12646–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Whelan S. P., Barr J. N., Wertz G. W. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:61–119 [DOI] [PubMed] [Google Scholar]
- 303. Wiegand M., Bossow S., Neubert W. J. 2005. Sendai virus trailer RNA simultaneously blocks two apoptosis-inducing mechanisms in a cell type-dependent manner. J. Gen. Virol. 86:2305–2314 [DOI] [PubMed] [Google Scholar]
- 304. Wilde A., McQuain C., Morrison T. 1986. Identification of the sequence content of four polycistronic transcripts synthesized in Newcastle disease virus infected cells. Virus Res. 5:77–95 [DOI] [PubMed] [Google Scholar]
- 305. Wilkins C., Gale M., Jr 2010. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Wilusz J., Kurilla M. G., Keene J. D. 1983. A host protein (La) binds to a unique species of minus-sense leader RNA during replication of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 80:5827–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Wong R. W., Blobel G., Coutavas E. 2006. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. U. S. A. 103:19783–19787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Xu L. G., et al. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740 [DOI] [PubMed] [Google Scholar]
- 309. Yang Y. K., et al. 2011. ARF-like protein 16 (ARL16) inhibits RIG-I by binding with its C-terminal domain in a GTP-dependent manner. J. Biol. Chem. 286:10568–10580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Yoneyama M., Fujita T. 2010. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 20:4–22 [DOI] [PubMed] [Google Scholar]
- 311. Yoneyama M., Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227:54–65 [DOI] [PubMed] [Google Scholar]
- 312. Yoneyama M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 313. Yoneyama M., Onomoto K., Fujita T. 2008. Cytoplasmic recognition of RNA. Adv. Drug Deliv. Rev. 60:841–846 [DOI] [PubMed] [Google Scholar]
- 314. Yount J. S., Gitlin L., Moran T. M., Lopez C. B. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J. Immunol. 180:4910–4918 [DOI] [PubMed] [Google Scholar]
- 315. Yu Y., Hayward G. S. 2010. The ubiquitin E3 ligase RAUL negatively regulates type I interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33:863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Zeng W., et al. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141:315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Zhao C., Denison C., Huibregtse J. M., Gygi S., Krug R. M. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 102:10200–10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Zhou S., et al. 2007. Role of MyD88 in route-dependent susceptibility to vesicular stomatitis virus infection. J. Immunol. 178:5173–5181 [DOI] [PubMed] [Google Scholar]
- 319. Zust R., et al. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]