Abstract
A new class of inflammatory CD4+ T cells that produce interleukin-17 (IL-17) (termed Th17) has been identified, which plays a critical role in numerous inflammatory conditions and autoimmune diseases. The active form of vitamin D, 1,25-dihydroxyvitamin D3 [1,25(OH)2D3], has a direct repressive effect on the expression of IL-17A in both human and mouse T cells. In vivo treatment of mice with ongoing experimental autoimmune encephalomyelitis (EAE; a mouse model of multiple sclerosis) diminishes paralysis and progression of the disease and reduces IL-17A-secreting CD4+ T cells in the periphery and central nervous system (CNS). The mechanism of 1,25(OH)2D3 repression of IL-17A expression was found to be transcriptional repression, mediated by the vitamin D receptor (VDR). Transcription assays, gel shifting, and chromatin immunoprecipitation (ChIP) assays indicate that the negative effect of 1,25(OH)2D3 on IL-17A involves blocking of nuclear factor for activated T cells (NFAT), recruitment of histone deacetylase (HDAC), sequestration of Runt-related transcription factor 1 (Runx1) by 1,25(OH)2D3/VDR, and a direct effect of 1,25(OH)2D3 on induction of Foxp3. Our results describe novel mechanisms and new concepts with regard to vitamin D and the immune system and suggest therapeutic targets for the control of autoimmune diseases.
INTRODUCTION
Interleukin-17A (IL-17A)-producing T cells are a subset of CD4+ T cell lineage, termed Th17, distinct from Th1, Th2, and T regulatory (Treg) subsets (52). IL-17 is involved in the pathogenesis of autoimmune inflammation and has been implicated in numerous autoimmune diseases, including systemic lupus erythematosus, rheumatoid arthritis, and multiple sclerosis (MS) (10, 21, 26, 41). IL-17 mRNA and protein levels in patients with MS have been shown to be increased in mononuclear cells isolated from blood, in cerebrospinal fluid, and in brain lesions (39, 41). IL-17 is also increased in lymphocytes derived from mice with experimental autoimmune encephalomyelitis (EAE; mouse model for multiple sclerosis) (33). In IL-17A knockout (KO) mice, EAE is markedly suppressed, indicating that IL-17 contributes to the development of EAE (33). Although it has been reported that the transcription factors nuclear factor for activated T cells (NFAT), retinoid orphan nuclear receptor γt (RORγt), and Runt-related transcription factor 1 (Runx1) are important for the T cell receptor (TCR)-mediated transcriptional regulation of IL-17A (24, 29, 38, 74), knowledge of the factors involved in the cellular and molecular regulation of IL-17A remains limited.
The principle function of the active form of vitamin D, 1,25-dihydroxyvitamin D3 [1,25(OH)2D3], is the maintenance of calcium and phosphate homeostasis (13). However, vitamin D has numerous other functions, including downregulation of autoimmunity (7, 8, 25, 55). 1,25(OH)2D3 has been reported to at least partially protect against a number of experimental autoimmune diseases, including EAE (7, 8, 11, 35, 40, 53). In addition, numerous epidemiological studies have indicated a negative correlation between increased sun exposure, which would result in a higher vitamin D synthetic rate, and diets rich in vitamin D and MS prevalence (34, 55, 70). It has been reported that there is a high prevalence of vitamin D deficiency in MS and that early maintenance of vitamin D sufficiency is associated with a decreased risk of MS (42, 47). These findings strongly suggest a role for vitamin D in reducing the risk of MS and support that vitamin D or metabolites of vitamin D may prevent the progression of MS. However, the mechanisms by which vitamin D protects against MS, which have been suggested to involve its immune suppressive actions, have not been clearly defined.
The actions of 1,25(OH)2D3 are mediated through the vitamin D receptor (VDR). The ligand-occupied VDR heterodimerizes with the retinoid X receptor (RXR) and, together with coregulatory proteins, interacts with specific DNA sequences (vitamin D response elements [VDREs]) in the promoter regions of target genes, and modulates their transcription (6, 14). 1,25(OH)2D3 manifests a clear suppressive effect on immunity. Previous studies have shown that 1,25(OH)2D3 represses mRNA and transcription of several proinflammatory cytokines, including IL-2 and granulocyte-macrophage colony-stimulating factor (GM-CSF) (2, 55, 67, 69). Here we report, for the first time, that the negative effect of 1,25(OH)2D3 on IL-17A involves transcriptional repression [blocking of NFAT, recruitment of histone deacetylase (HDAC), sequestration of Runx1 by 1,25(OH)2D3/VDR, and a direct effect of 1,25(OH)2D3 on induction of Foxp3 (which associates with NFAT and Runx1 for transcriptional repression)]. Our results suggest that interaction among NFAT, VDR, Foxp3, and Runx1 is involved, at least in part, in suppression of proinflammatory Th17 responses by 1,25(OH)2D3.
MATERIALS AND METHODS
Materials.
Deoxy-[γ-32P]ATP (3,000 Ci/mmol) was obtained from PerkinElmer Life Sciences. A Random Primers DNA labeling kit was purchased from Invitrogen. Prestained protein molecular weight markers and an electrochemiluminescent detection system were obtained from PerkinElmer Life Sciences. VDR, NFATc1, Runx1, and β-actin antisera were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). 1,25(OH)2D3 was a generous gift from Milan Uskokovic (Hoffmann-LaRoche, Nutley, NJ).
Cell culture.
Jurkat (human T cell leukemia cell line), HUT102 (human T cell lymphoma cell line), and EL-4 (mouse lymphoma cell line) (HUT102 cells constitutively express IL-17A, and IL-17A mRNA is induced with stimulation in EL-4 cells) (28, 38) cells were maintained in RPMI media (Life Technologies, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gemini Biological Products, Calabasas, CA), 10% Dulbecco modified Eagle medium (DMEM; Life Technologies, Inc.), and antibiotics in a humidified atmosphere of 95% air-5% CO2 at 37°C. HEK293 cells were cultured in DMEM supplemented with 5% FBS and antibiotics in a humidified atmosphere of 95% air-5% CO2 at 37°C. Cells were grown to 70 to 80% confluence and changed to medium supplemented with 2% charcoal-dextran-treated FBS before activation and 1,25(OH)2D3 treatment. Cells were treated with the vehicle or the compounds noted at the indicated times and concentrations.
Transient-transfection and luciferase assay.
Human IL-17A (hIL-17A) promoter and deletion constructs, and the mouse Foxp3 constructs [including the construct with point mutations in the VDRE/retinoic responsive element (RARE) half-site (AGGTCA to AaGaCA; changes shown with lowercase letters) at position +2156 in the conserved noncoding sequence] were previously described (38, 64). The 2-kb mouse IL-17A (mIL-17A) promoter construct, from Strober's laboratory (74), was obtained from Addgene Inc. (Cambridge, MA). pAV-hVDR was a gift of Wesley Pike, University of Wisconsin. pSRα-NFATc1 was provided by G. R. Crabtree, Stanford University, Stanford, CA. pCMV-c-fos and pCMV-c-jun were provide by T. Curran, Children's Hospital of Philadelphia. The VDR plasmid lacking the DNA binding domain (amino acids [aa] 116 to 427) was provided by P. MacDonald, Case Western Reserve, Cleveland, OH. The mutant human VDR (hVDR) F251C expression vector, in which heterodimerization to RXR is defective, was from D. Feldman, Stanford University. pCMV-Runx1, pCMV-Runx2, and pCMV-Runx3 were provided by H. Drissi, University of Connecticut. Jurkat and EL-4 cells (1 × 106) were seeded in a 24-well culture dish 24 h prior to transfection. Cells were transfected using jetPEI (Polyplus-transfection Inc., New York, NY), according to the manufacturer's instructions. Cells (24 h posttransfection) were treated for 9 h with the following: (i) vehicle, (ii) 1,25(OH)2D3 (0.1 nM to 10 nM), (iii) activation with PMA (10−7 M) plus ionomycin (200 ng/ml), and (iv) 1,25(OH)2D3 plus PMA plus ionomycin. A luciferase assay (dual-luciferase reporter assay kit; Promega, Madison, WI) was performed as described previously (59).
Electrophoretic mobility shift assay (EMSA).
36-Mer complementary oligonucleotides spanning NFAT site 1 and a 34-mer spanning the NFAT site 2 binding site of the human IL-17A promoter or mutated NFAT site 1 and NFAT site 2 were used for the gel shift assays (prepared by the UMD Molecular Resource Facility, Newark, NJ). The wild-type sequence of the oligonucleotide used for NFAT site 1 was 5′-TTCCCATTTTCCTTCAGAAGGAGAGATTCTTCTATG-3′ and for NFAT site 2 was 5′-CTTCTATGACCTCATTGGGGGCGGAAATTTTAAC-3′. For mFoxp3, the VDRE wild-type sequence used was 5′-CCATTTACTGCAGAGGTCAAAAGTGTGGGTA-3′ (underlining shows the putative VDRE). The mutated sequence (shown in lowercase letters) used for mFoxp3 was 5′-CCATTTACTGCAGAaGatgAAAGTGTGGGTA-3′. Human full-length VDR was purified as previously described (68). Glutathione S-transferase (GST)–RXRα was obtained from D. Feldman, Stanford University. Nuclear preparations from activated Jurkat cells (source for NFAT) were used at concentrations as indicated in the figure legends. Gel shift analysis was performed as described previously (19).
ChIP assay.
HUT102 and EL-4 cells were cultured to 95% confluence prior to the experiment and treated with the following: (i) vehicle treatment, (ii) 1,25(OH)2D3 treatment (10 nM), (iii) activation with phorbol myristate acetate (PMA) (10−7 M) plus ionomycin (200 ng/ml), and (iv) 1,25(OH)2D3 plus PMA plus ionomycin for the indicated times. Human and mouse primary CD4+ T cells were isolated and polarized under Th17 or Treg conditions as described under mouse T cell cultures and human CD4+ T cell isolation. The chromatin immunoprecipitation (ChIP) assay was performed as previously described (59). Immunoprecipitations were performed with anti-NFATc1 (H-10; Santa Cruz Biotechnology), anti-VDR (C-20; Santa Cruz Biotechnology), anti-RXR (D20; Santa Cruz Biotechnology), anti-HDAC2 (F6; Santa Cruz Biotechnology), anti-AcH4 (Santa Cruz Biotechnology), and anti-Runx1 (DW71; Santa Cruz Biotechnology). DNA precipitates were isolated and subjected to PCR using the following primers designed to amplify the fragment of the hIL-17A promoter containing the NFAT binding sites: 5′AAAAGAGGACATGGTCTTTAGGAA3′ and 3′GCAACAAAGAAGGTTAGTTAC5′. For the murine IL-17A (mIL-17A) promoter, PCR was performed using the following primers designed to amplify the −1860-to-−1560 (−1860/−1560) region containing the Runx1 binding sites: 5′-GAGATGGCTCAGCAGTTAAGA-3′ and 5′-ATTCCCCGATAGAATAACTATTTGA-3′. The +2216/+2350 region of the mFoxp3 gene containing the putative VDRE was amplified using the primers 5′-CCAGTCTCCTTTATGGCTTCA-3′ and 5′-TTTTTATTAATGATGGTAGGTGCTCA-3′. PCR products were resolved in 1% agarose gel and visualized using ethidium bromide staining. DNA acquired prior to precipitation was collected and used as the input. A total of 10% of input was used for PCR evaluation. PCRs using the primers designed to amplify the upstream region of respective promoters were used as a negative control to exclude nonspecific binding.
Mouse T cell cultures for ChIP and immunoprecipitation assays.
All experiments using mice were carried out in accordance with the guidelines established by the National Institutes of Health Guide for the Care and Use of Laboratory Animals, as well as those established by the Stanford University and UMDNJ-New Jersey Medical School Institutional Animal Care and Use Committees. CD4+ T cells from BALB/c mice mesenteric lymph nodes (LNs) or spleens, depleted by a MACS column (CD4+ T cell isolation kit II; Miltenyi Biotec) according to the manufacturer's protocol, were cultured in the presence or absence of 1,25(OH)2D3 (10 nM) for 3 days under either Th17 conditions (20 ng/ml IL-6, 1 ng/ml transforming growth factor β [TGF-β], 20 μg/ml anti-gamma interferon [anti-IFN-γ], and 10 μg/ml anti-IL-4) or Treg conditions (100 U/ml IL-2, 0.5 ng/ml TGF-β, 20 μg/ml anti-IFN-γ, and 10 μg/ml anti-IL-4).
Immunoprecipitation.
Nuclear extracts (NE) were prepared from mouse primary CD4+ T cells isolated and polarized under Th17 conditions or from VDR- and Runx1-transfected HEK293 cells as previously described (19). A total of 500 μg of each preparation was used for immunoprecipitation with the addition of 4 μg anti-VDR (C-20; Santa Cruz Biotechnology) or 4 μg anti-Runx1 (DW71; Santa Cruz Biotechnology) for 24 h at 4°C. The complex was separated by SDS-PAGE and probed with either VDR antibody or Runx1 antibody.
Western blot analysis.
Western blot analysis was performed as described previously (19). Membranes were blocked with indicated antibodies, including anti-c-Fos (6-2H-2F), anti-c-Jun (G-4), anti-NFATc1 (H-10), anti-Runx1 (DW71), or anti-VDR (C-20) (Santa Cruz Biotechnology) at a dilution of 1:1,000 in 1% milk and Tris-buffered saline overnight. Membranes were then rinsed and incubated at room temperature with these respective secondary antibodies: for anti-VDR, goat anti-rabbit IgG–horseradish peroxidase (HRP) antibody (sc-2004); for anti-Runx1, anti-NFATc1, anti-c-Fos, and anti-c-Jun, goat anti-mouse IgG–HRP (sc-2005) at 1:3,000 dilution in 1% milk and TBS for 2 h.
RNA-mediated interference.
For knockdown of Runx1, CD4+ T cells were purified as described above and were directly transfected by nucleofection with small interfering RNA (siRNA) specific for Runx1 (predesigned ON-TARGETplus SMARTpool siRNA; Dharmacon) or “scrambled” control siRNA (Dharmacon). For transfection, 2 × 106 cells in 100 μl mouse T cell Nucleofector solution (Amaxa) were transfected with a total of 300 pmol of siRNA with the X-001 program (Amaxa). After transfection, cells were incubated for 4 h at 37°C, were activated with anti-CD3 (2 μg/ml) and anti-CD28 (1 μg/ml), and were cultured under Th17-polarizing conditions as described above. After 48 h of activation, transfected cells were collected, and RNA was made. RNA was then analyzed by reverse transcriptase (RT)-PCR as described below.
Peptides.
Myelin oligodendrocyte glycoprotein (MOG) p35-55 (MEVGWYRSPFSRVVHLYRNGK), PLPp139-151 (HCLGKWLGHPDKF) were synthesized on a peptide synthesizer (model 9050; MilliGen) by standard 9-fluorenylmethoxycarbonyl chemistry and purified by high-performance liquid chromatography (HPLC). Amino acid sequences were confirmed by amino acid analysis and mass spectroscopy. The purity of each peptide was greater than 95%.
Mice.
Female SJL/J and C57BL/6 mice (8 to 12 weeks old) were purchased from the Jackson Laboratory (Bar Harbor, ME). MOG p35-55 TCR transgenic mice were obtained from V. Kuchroo (4). All animal protocols were approved by the Division of Comparative Medicine at Stanford University, in accordance with the National Institutes of Health guidelines.
EAE induction, treatment, and cell preparation.
SJL/J mice, C57BL/6 mice, or MOG-specific TCR transgenic mice were immunized as described previously (9). On day 0 and 48 h postimmunization, C57BL/6 mice or 2D2 transgenic mice were intravenously injected with 200 ng of Bordetella pertussis toxin (BPT) in phosphate-buffered saline (PBS). Mice were examined daily for clinical signs of EAE and scored. In studies involving long-term treatment with 1,25(OH)2D3, mice were fed a diet without vitamin D added beginning at 3 days before immunization. Treatment was administrated to mice either intraperitoneally (i.p.) or orally [50 ng/mouse; this dose of 1,25(OH)2D3 is consistent with what has been used previously to prevent EAE] (7, 35). Mice treated with vehicle served as controls. Infiltrating cells from spinal cords or the brain stems and cerebellums from three or four perfused mice were isolated. Central nervous system (CNS) homogenates were incubated with collagenase (Roche) and DNase (Sigma) for 1 h at 37°C, and the cells were purified by a Percoll gradient.
Th17 CD4+ T cell differentiation and Th17 adoptive transfer.
Splenocytes and lymph nodes cells were isolated from MOG p35-55-specific TCR Tg (2D2) female mice (6 weeks old; n = 25). CD4+ T cells were isolated using L3T4 micromagnetic beads (Miltenyi Biotec). Cells were activated with 1 μg/ml each of plate-bound anti-CD3 (clone 145-2C11) and anti-CD28 (clone 37.51) (BD Biosciences). Cells were plated at 2 × 106 cells/ml in complete RPMI 1640 and treated with either vehicle or 1,25(OH)2D3 (10 nM) under the following Th17 conditions: IL-6 (20 ng/ml), TGF-β (3 ng/ml) (R&D Systems), anti-IFN-γ (50 μg/ml; clone R4-6A2), and anti-IL-4 (10 μg/ml; clone 11B11) (eBioscience). Cells were activated for 3 days and then expanded for 4 more days with fresh media supplemented with IL-23 (20 ng/ml; R&D Systems) and 1,25(OH)2D3 (10 nM) or vehicle. Cells were restimulated with anti-CD3 and anti-CD28 (1 μg/ml), IL-23 (20 ng/ml), anti IFN-γ, and 1,25(OH)2D3 or vehicle for 48 h and expanded as described above for a total of 3 activation cycles. Cells were then restimulated for 48 h, washed, and then resuspended in PBS. A total of 100 μl of cells (20 × 106 cells/mouse) were injected intravenously (i.v.) into 8-week-old female C57BL6/J recipient mice (n = 15). On the day of the cell transfer and 48 h later, mice were injected i.v. with 200 ng/mouse of BPT in PBS. EAE mice were scored daily for clinical signs as previously described.
2D2 mouse T cell cultures.
Resting (Pan) CD4+ T cells were isolated from naïve 2D2 mice using CD4 magnetic beads (L3T4; Miltenyi Biotec). Naïve and memory CD4+ T cells were sorted from total CD4+ T cells using anti-CD26L and anti-CD44 antibodies with magnetic beads (Miltenyi Biotec). Cells were treated with 1,25(OH)2D3 at the indicated concentrations or vehicle and activated with plate-bound anti-CD3/anti-CD28 at 1 μg/ml each (BD Biosciences) (73).
Intracellular flow cytometry and ELISA.
For IL-17A staining after PMA plus ionomycin stimulation of the CD4+ T cell population, surface staining was performed with anti-CD3, anti-CD45RO, and anti-CD4 (all obtained from BD Biosciences). Following fixation, permeabilization, and staining with anti-IL-17A (phycoerythrin [PE]; eBioscience), cells were acquired on a fluorescence-activated cell sorter (FACS) analyzer (Becton Dickinson) and analyzed using FlowJo software. For Foxp3 analysis, cultured cells were harvested on day 5. Surface staining was then performed with fluorochrome-conjugated anti-CD4, anti-CD25, and anti-CD45RO (all obtained from BD Pharmingen). Following fixation, permeabilization and Foxp3 staining were performed using the Foxp3 staining buffer kit from eBioscience, according to the manufacturer's protocol. Data were acquired on a FACSCalibur cytoflorometer and analyzed using CellQuest software (BD). Enzyme-linked immunosorbent assay (ELISA) kits for IL-17A, IL-22 (R&D systems), and IL-10 (BD Bioscience) were used to analyze the supernatants from 2- to 4-day cultures.
Human CD4+ T cell isolation and activation.
Human peripheral blood studies were approved by the Institutional Review Board of Stanford University and UMDNJ-New Jersey Medical School in accordance with the regulations mandated by the Department of Health and Human Services. CD4+ T cells were isolated from peripheral blood mononuclear cells (PBMCs) of healthy donors (after obtaining informed consent) by Ficoll-Hypaque gradient centrifugation (GE Healthcare).
Th17.
Resting CD45RO+ CD4+ T cells were isolated by magnetic beads (CD4+ T cell isolation kit II; Miltenyi Biotec). For short activation (72 h), resting CD4+ T cells were activated with anti-CD3 and anti-CD28 beads (T Cell Expander; Invitrogen). For Th17 differentiation, resting CD4+ T cells were cultured in serum-free X-VIVO 15 media (Lonza). Naive CD45RA+ T cells were isolated by two rounds of depletion with anti-CD45RO magnetic beads (Miltenyi Biotec). For Th17 differentiation, cells were activated with anti-CD3 and anti-CD28 in the presence of recombinant human IL-23 (rhIL-23) at 50 ng/ml, rhIL-6 at 30 ng/ml, IL-1β at 30 ng/ml (R&D Systems), human TGF-β (hTGF-β; 0.1 ng/ml), anti-human IFN-γ (hIFN-γ; 50 μg/ml), and anti-hIL-4 (20 μg/ml) (eBioscience). Cells were cultured for 4 to 6 days and then rested for an additional 4 to 6 days (1 stimulation cycle). Vehicle or 1,25(OH)2D3 at the indicated concentrations was added throughout.
Treg cells.
CD4+ CD25− T cells were purified using magnetic beads and reagents from Miltenyi Biotec. CD4+ T cells were isolated from PBMCs by immunomagnetic depletion of non-T helper cells (CD4+ T cell isolation kit) according to the manufacturer's instructions. CD4+ CD25− T cells were purified from the CD4+ T cells by negative selection using CD25 beads, and the purity was >95% in all experiments, as assessed by immunofluorescence flow cytometry. CD4+ CD25− T cells (2 × 105/well) were resuspended in 10% heat-inactivated FCS-RPMI 1640 containing CD28 Ab (2 μg/ml) and rIL-2 (100 IU/ml), in the presence and absence of 1,25(OH)2D3 (10 nM), and cultured in 96-well U-bottom plates coated with anti-CD3 (5 μg/ml) for 5 days. The same human T cell cultures were used for both FACS analysis and ChIP assays.
In vivo Treg induction.
EAE was induced in C57BL6/J mice and were treated (i.p.) with either vehicle or 1,25(OH)2D3 for 15 days, starting on the day of EAE induction (as described above). At day 7, before disease onset, 5 mice per group were killed, and spleens were isolated. CD4+ CD25+ Foxp3+ cells were examined using a mouse regulatory T cell staining kit (eBioscience), according to the manufacturer's instructions. On day 15, at the peak of disease, spinal cords were isolated and pooled from the 5 remaining mice in each group. Mononuclear infiltrating cells were isolated, and CD4+ CD25+ Foxp3+ cells were examined.
In vitro T cell suppression assay.
Naïve 2D2 splenocytes were cultured with MOG p35-55 (20 μg/ml) in the presence of vehicle, 1,25(OH)2D3 (10 nM), or TGF-β (5 ng/ml) for 72 h. Treg differentiation was tested by induction of Foxp3 using FACS analysis. Once Treg induction was confirmed under both 1,25(OH)2D3 and TGF-β treatment conditions, CD4+ CD25hi effector cells were sorted using FACS Aria (BD Biosciences). Responder CD4+ CD25− T cells were also sorted from naïve 2D2 splenocytes. For the suppression assays, 50,000 responder T cells were cocultured with the indicated number of effector CD4+ CD25hi T cells (or Treg cells under some conditions) and 200,000 irradiated syngeneic splenocytes [serving as antigen-presenting cells (APCs)] per well. Cells were activated in the presence of MOG p35-55 (20 μg/ml) for 72 h. The suppression activity of Treg cells was tested via a proliferation assay. At 16 h before harvesting, cells were pulsed with 1 μCi/well of [3H]thymidine (PerkinElmer). [3H]Thymidine incorporation was tested in triplicate wells using the MicroBeta counter (PerkinElmer). Proliferation was measured as counts per minute, and this experiment was repeated 3 times.
RT-PCR.
Total RNA was isolated from mouse and human primary CD4+ T cells using RNA-Bee reagent (Tel-Text, Inc.) or the reverse transcriptase (RT)-PCR miniprep kit (Agilent Technologies). RT-PCR was performed using 2 μg total RNA and the SuperScript one-step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen). Primers used for human IL-17A were forward primer 5′-ATGACTCCTGGGAAGACCTCATTG-3′ and reverse primer 5′-TTAGGCCACATGGTGGACAATCGG-3′ (40 cycles). Primers used for mouse IL-17A were 5′-GGTCAACCTCAAAGTCTTTAACTC-3′ and 5′-TTAAAAATGCAAGTAAGTTTGCTG-3′ (36 cycles). Data were normalized for the expression of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA within the sample (GAPDH primers, 5′-TCACCATCTTCCAGGAGCG-3′ and 5′-CTGCTTCACCACCTTCTTGA-3′). The cycles were chosen so that the amplification was conducted in the linear range of amplification efficiency. The resulting PCR products were subjected to electrophoresis on a 1% agarose gel containing ethidium bromide, and bands were visualized under UV light. Gel data were recorded using the Gene Genius bioimaging system (Syngene, Frederick, MD), and the relative densities of the bands were determined using Gene Tool software (Syngene).
Statistical analysis.
Data are presented as means ± standard errors (SE). For clinical scores, significance between each two groups was examined by using the Mann-Whitney U test. A P value of <0.05 was considered significant. All other statistics were analyzed by using a one-way- or two-way multiple-range analysis of variance (ANOVA) test for multiple comparisons. A P value of <0.05 was considered significant.
RESULTS
1,25(OH)2D3 inhibits human IL-17A and suppresses mouse IL-17A in mouse models of MS.
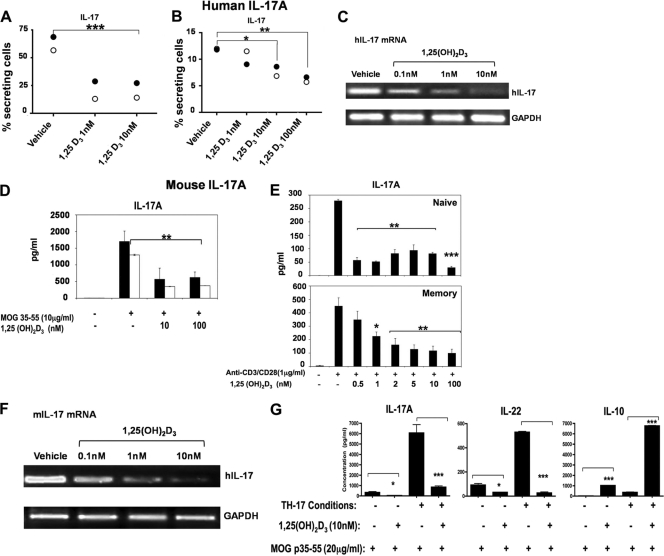
We first examined the effects of 1,25(OH)2D3 on human IL-17A (hIL-17A) using CD4+ T cells isolated from healthy donors. hIL-17A expression and mRNA are repressed in response to 1,25(OH)2D3 (Fig. 1A to C). Various mouse models for MS were also used to examine the effect of 1,25(OH)2D3 on mIL-17A. When splenocytes and lymph node (LN) cells isolated from naïve myelin oligodendrocyte glycoprotein (MOG) T cell receptor transgenic mice (2D2) were treated in vitro with either vehicle or 1,25(OH)2D3 in the presence of MOG p35-55, a significant reduction in mIL-17A secretion was observed in the presence of 1,25(OH)2D3 (Fig. 1D). Inhibitory effects of 1,25(OH)2D3 on both naïve and memory CD4+ T cells were observed (Fig. 1E). 1,25(OH)2D3 inhibition of mIL-17A secretion was also seen in CD8+ T cells, in γδ-T cells, and in NKT cells (data not shown), which also produce IL-17A (54, 60, 63). In addition, although it has been a matter of debate (12, 50), we found that 1,25(OH)2D3 inhibits mIL-17A mRNA in mouse CD4+ T cells polarized under Th17 conditions (Fig. 1F). To further investigate the effects of 1,25(OH)2D3, naïve 2D2 splenocytes were activated with MOG p35-55 and treated with vehicle or 1,25(OH)2D3 in the absence or presence of Th17 differentiation conditions. In the presence of 1,25(OH)2D3, mIL-17A and mIL-22 were significantly reduced, and mIL-10 was significantly increased (Fig. 1G).
Fig. 1.
Regulation of IL-17A expression and mRNA by 1,25(OH)2D3. (A to C) 1,25(OH)2D3 reduces the percentage of human IL-17A-secreting CD4+ T cells and hIL-17A mRNA levels. (A and B) Resting CD4+ T cells were isolated from PBMCs of healthy donors 1 (closed circles) and 2 (open circles), activated with anti-CD3 and anti-CD28 under Th17 conditions in the presence of 1, 10, or 100 nM 1,25(OH)2D3 or vehicle. In panel A, cells were passed through 3 cycles of activation and expansion and then stained to detect intracellular hIL-17A. In panel B, cells were activated for 72 h and then stained to detect intracellular hIL-17A. Data are presented as percentages of IL-17+ T cells. **, P < 0.01; ***, P < 0.001. (C) RT-PCR analysis of hIL-17A mRNA. Inhibition of hIL-17A mRNA by 1,25(OH)2D3 was observed using human primary CD4+ T cells polarized under Th17 conditions for 5 days in the presence or absence of 1,25(OH)2D3 (0.1 to 10 nM). Similar results were observed in two additional experiments. (D to G) 1,25(OH)2D3 treatment inhibits Th17 differentiation, mIL-17A mRNA, and mIL-17A secretion in mouse cells. (D) Lymph node cells (black bars) or splenocytes (white bars) isolated from T cell receptor transgenic mice (2D2) were activated with MOG p35-55 (10 μg/ml) in the presence or absence of 1,25(OH)2D3 (10 and 100 nM). A total of 48 h later, supernatants were used to measure IL-17A by ELISA. IL-17A was significantly reduced by 1,25(OH)2D3. **, P < 0.01 compared to MOG activation in the absence of 1,25(OH)2D3. (E) Naïve or memory CD4+ T cells were isolated from spleens of naïve 2D2 mice. Cells were activated and treated with 1,25(OH)2D3 (0.5 to 100 nM) 48 h later for memory cultures and 96 h later for naïve cultures. Supernatants were subjected to IL-17A ELISA. (F) RT-PCR analysis of mIL-17A mRNA. Inhibition of mIL-17A mRNA by 1,25(OH)2D3 was observed using mouse primary CD4+ T cells polarized under Th17 conditions for 3 days in the presence or absence of different concentrations of 1,25(OH)2D3. Similar results were observed in two additional experiments. (G) 1,25(OH)2D3 regulates Th17 cytokines during T cell activation under Th0 or Th17 differentiation conditions. Naïve 2D2 splenocytes were cultured with MOG p35-55 (20 μg/ml) under Th17 conditions or Th0 conditions. Cells were treated with either vehicle or 1,25(OH)2D3 (10 nM) for 72 h. The cytokines indicated in the figure were measured from supernatants using ELISA. Data are presented as the means of triplicate determinations ± SE. *, P < 0.05; ***, P < 0.001 (tested in two-way ANOVA).
Similar to what has previously been shown (8, 11, 35), we found that 1,25(OH)2D3, when given beginning on the day of immunization and then every other day, significantly inhibits the disease course in the EAE model of MS (not shown). When splenocytes and draining lymph node (DLN) cells were isolated on day 7 after immunization, before clinical symptoms appeared from 1,25(OH)2D3- or vehicle-treated mice, and then cultured in vitro in the presence of MOGp35-55, we found that mIL-17A was significantly reduced with 1,25(OH)2D3 treatment (data not shown). Parallel cultures from splenocytes and DLNs, activated with PMA and ionomycin for 5 h, also indicated a reduction in mIL-17A after in vivo 1,25(OH)2D3 treatment (data not shown).
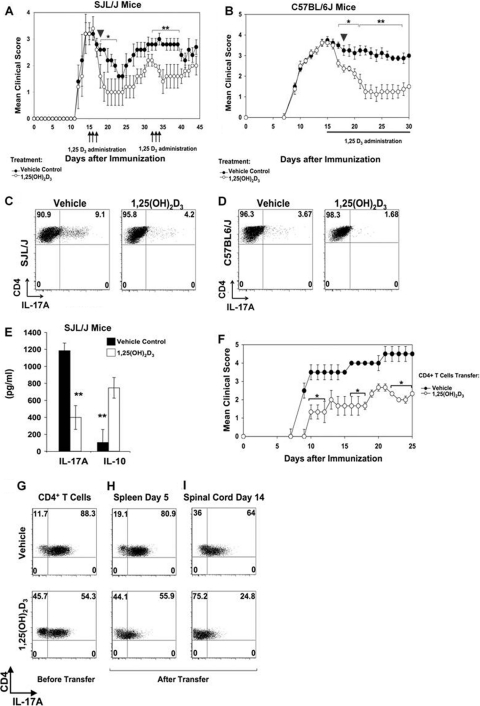
To determine whether 1,25(OH)2D3-mediated inhibition of mIL-17A is observed in protection against ongoing EAE and reversal of paralysis by 1,25(OH)2D3, EAE was induced in SJL/J mice with immunizations of PLPp139-151. After mice reached the peak clinical score with paralysis, they were treated with 1,25(OH)2D3 or vehicle for 3 consecutive days. The mice treated with 1,25(OH)2D3 showed a significant improvement in disease progression, which lasted for 8 days (P < 0.05). Retreatment with 1,25(OH)2D3 provided even further improvement (Fig. 2A). Next, C57BL6/J mice with ongoing MOG p35-55 EAE were treated with 1,25(OH)2D3 or vehicle every day starting from day 15 of disease induction. Treatment with 1,25(OH)2D3 caused marked reduction in the clinical scores, beginning 48 h posttreatment (P < 0.01), and this improvement was observed throughout the course of treatment (Fig. 2B). Infiltrating mononuclear cells were isolated from the spinal cord and brain of both the groups on day 18 (Fig. 2A and B, arrowheads) and stained for intracellular mIL-17A and cell surface CD4. mIL-17A was downregulated by 1,25(OH)2D3 treatment in the CD4+ infiltrating cells in the spinal cord and brain from both SJL/J mice (Fig. 2C) and C57BL6/J mice (Fig. 2D). Thus, the improvement in the clinical score after 1,25(OH)2D3 treatment is associated with inhibition of production of IL-17A. These findings are the first to correlate the reversal of paralysis in EAE mice by 1,25(OH)2D3 with reduced IL-17A-secreting CD4+ T cells. The mononuclear infiltrating cells isolated from the brain of the SJL/J treated group were activated with PLPp139-151 peptide in the presence or absence of 1,25(OH)2D3. Besides IL-17A inhibition, a significant upregulation of IL-10 was observed (Fig. 2E).
Fig. 2.
1,25(OH)2D3 treatment reverses ongoing EAE and ameliorates Th17-induced EAE in recipient mice. (A) PLPp139-151-EAE was induced in SJL/J mice and scored daily. At day 14 (peak of disease), mice were randomize into two groups (n = 12 per group). Mice were injected with either vehicle or 1,25(OH)2D3 for 3 consecutive days (arrows), days 15 to 18, and days 31 to 34. Mice showed reduced clinical scores after the transient treatments. *, P < 0.05; **, P < 0.01 (compared to vehicle-treated group). (B) MOG p35-55 EAE was induced in C57BL/6 mice and scored daily. At day 15 (peak of disease), mice were randomized into two groups (n = 15 per group). Treatment with either vehicle or 1,25(OH)2D3 was administered daily, starting on day 15 (days 15 to 30). Treated mice showed reduced clinical scores in the chronic phase. *, P < 0.05; **, P < 0.01 (compared to vehicle-treated group). (C and D) On day 18, as shown in panels A and B, 3 mice from each group were killed (arrowhead), and mononuclear cells were isolated from spinal cords for IL-17A analysis. (C) Lymphocytes isolated from a representative SJL/J mice (arrowhead in panel A). (D) Lymphocytes isolated from a representative C57BL/6 mice (arrowhead in panel B). Data are shown as representative FACS plots of 1 mouse out of 3. In each plot, CD45+ CD4+ IL-17A mononuclear infiltrated T cells are gated. (E) Mononuclear infiltrates isolated from brains of SJL/J mice killed as shown in panel C were activated with PLPp139-151 (50 ng/ml) for 72 h, and ELISAs were performed. Results shown are the means of triplicate determinations ± SE. **, P < 0.01 (1,25(OH)2D3 treatment versus that of vehicle). (F) Naïve CD4+ T cells were isolated from 2D2 splenocytes and were differentiated into Th17 cells in the presence of either vehicle or 1,25(OH)2D3 (10 nM). After 3 expansion cycles, differentiated into Th17 cells in the presence of either vehicle or 1,25(OH)2D3 (10 nM). After 3 expansion cycles, adoptive transfer into naïve recipient C57BL/6 mice was performed. A total of 20 ×106 Th17 cells treated by either 1,25(OH)2D3 or vehicle were injected i.v. on day 0 (n = 10 per group). Recipient C57BL/6 mice were scored daily. (G) Assessment of IL-17A-secreting cells after 1,25(OH)2D3 treatment (bottom) versus vehicle treatment (top) before adoptive transfer. (H) Assessment of IL-17A-secreting cells after 1,25(OH)2D3 treatment (bottom) versus vehicle treatment (top) in the spleen 5 days post-adoptive transfer. (I) Assessment of IL-17A-secreting cells after 1,25(OH)2D3 treatment (bottom) versus vehicle treatment (top) in the CNS. Assessment was performed by FACS analysis, and data are presented as dot plots of IL-17A versus CD4+ T cells. Each FACS plot is of a representative mouse.
Lastly, we tested whether 1,25(OH)2D3 treatment of Th17 cells was sufficient to give protection against EAE induction when adoptively transferred into naïve mice. Resting CD4+ T cells were isolated from naïve 2D2 mice activated under Th17 differentiation conditions in the presence or absence of 1,25(OH)2D3 (10 nM) and then injected into naïve C57BL6/J recipient mice. Recipients that received 1,25(OH)2D3-treated Th17 cells developed a milder EAE (Fig. 2F) [1,25(OH)2D3 treatment reduced the percentage of mIL-17A-producing cells (Fig. 2G)]. CD4+ T cells were isolated from representative mice at day 5 and at day 14 posttransfer to profile mIL-17A secretion in vivo in the periphery and CNS, respectively. The vehicle-treated Th17 cells showed high mIL-17A secretion in vivo, while 1,25(OH)2D3-treated Th17 cells produced significantly lower levels of IL-17A in both the spleen (Fig. 2H) and CNS (Fig. 2I). Collectively, these experiments indicate that the decrease in the severity of EAE in response to 1,25(OH)2D3 is associated with diminished secretion of mIL-17A.
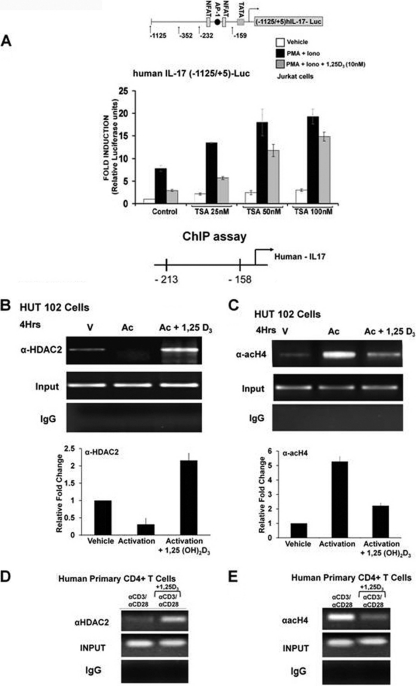
hIL-17A transcription is directly repressed by 1,25(OH)2D3, as VDR competes for DNA binding with NFATc1 and 1,25(OH)2D3 recruits HDAC to the hIL-17A promoter.
Although the experiments in both human and mouse cells and using various mouse models of MS indicate suppression of IL-17A by 1,25(OH)2D3, the molecular basis by which IL-17A production is regulated by 1,25(OH)2D3 remains poorly defined. IL-17A, similar to most T cell-derived cytokines, is regulated at least in part at the level of transcription (38, 74). Thus, to begin to define possible mechanisms involved in the repression of IL-17A by 1,25(OH)2D3, we examined the effects of 1,25(OH)2D3 on human and mouse IL-17A transcription.
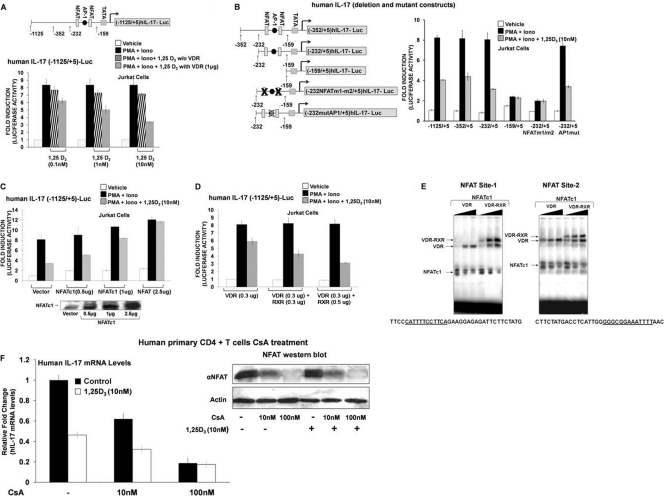
For studies related to 1,25(OH)2D3 suppression of hIL-17A, Jurkat cells were transfected with the hIL-17A promoter (−1125/+5) (Fig. 3A) or with different deletion constructs (Fig. 3B) and cotransfected with vector alone or VDR expression vector. Cells were stimulated with PMA and ionomycin for 9 h in the presence of vehicle or 1,25(OH)2D3. VDR was cotransfected since, as previously reported, T cells begin to express VDR only approximately 8 h postactivation (2, 5). A dose-dependent repression of activation of hIL-17A promoter (−1125/+5) activity was observed in the presence of 1,25(OH)2D3 (Fig. 3A). The repression was stronger in the presence of VDR expression vector. Thus, VDR is limiting for the suppression. When a VDR construct that lacked the DNA binding domain (DBD) was used, suppression of hIL-17A transcription above that attributed to endogenous VDR was not observed, indicating the importance of the DBD for the repression (data not shown). Using the −352/+5 and −232/+5 deletion constructs which contain two NFAT binding sites and an AP1 site, repression of activation by 1,25(OH)2D3 was also observed (Fig. 3B). The −159/+5 deletion construct which does not contain the NFAT and AP1 binding sites and the −232/+5 promoter construct with both NFAT sites mutated showed minimal activation and no repression by 1,25(OH)2D3 (Fig. 3B). These findings suggest that NFAT, which is an essential regulator of T cell-mediated hIL-17A gene transcription, is involved in mediating 1,25(OH)2D3 repression.
Fig. 3.
Regulation of hIL-17A transcription by 1,25(OH)2D3. NFATc1 relieves 1,25(OH)2D3-mediated repression of hIL-17A transcription, RXR with VDR contributes to transcriptional repression, and VDR/RXR bind to the NFAT sites. (A) Jurkat cells were cotransfected with the hIL-17A (−1125/+5) promoter and either vector alone (black bars and striped bars) or pAV-hVDR expression vector (1 μg) (gray bars). Cells were stimulated for 9 h with PMA and ionomycin (Iono) in the presence or absence of the indicated concentration of 1,25(OH)2D3. 1,25(OH)2D3 treatment (1 and 10 nM) resulted in a significant repression of hIL-17A transcriptional activation in the presence of transfected VDR (P < 0.01). In all subsequent transcription assays, 1 μg/well VDR expression plasmid was transfected (unless otherwise indicated). (B, left) Schematic of hIL-17A promoter deletion constructs or deletion constructs with specific mutations in the NFAT sites or the AP1 site (38). (Right) The human T cell line Jurkat was transfected with the hIL-17A promoter −1125/+5, deletion constructs −353/+5, −232/+5, and −159/+5, or deletion constructs with specific mutations in the NFAT sites (−232NFATm1/m2/+5) or the AP1 site (−232AP1mut/+5). Cells (24 h posttransfection) were stimulated for 9 h with PMA and ionomycin in the presence or absence of 10 nM 1,25(OH)2D3. (C) Overexpression of NFATc1 relieves 1,25(OH)2D3-mediated repression of hIL-17A transcription. Jurkat cells were cotransfected with hIL-17A promoter (−1125/+5) and pSRα-NFATc1 expression plasmid at the indicated concentrations. At 24 h posttransfection, cells were activated with PMA and ionomycin in the absence or presence of 1,25(OH)2D3 (10 nM) for 9 h. P < 0.05 [activation plus 1,25(OH)2D3, vector transfection versus activation plus 1,25(OH)2D3, and NFATc1 transfection (1 or 2.5 μg)]. (D) 1,25(OH)2D3-mediated repression is enhanced by RXR. Jurkat cells were transfected with the hIL-17A promoter (−1125/+5) together with either VDR (0.3 μg) or VDR plus RXR (0.3 μg or 0.5 μg). Cells were treated as described above. Transcriptional repression observed with 1,25(OH)2D3 treatment and VDR and RXR transfection (0.3 μg or 0.5 μg) is significantly different than repression observed with 1,25(OH)2D3 treatment and transfection of VDR alone (P < 0.01). (A, B, C, and D) Error bars represent the means ± SE from at least 3 to 5 independent experiments. (E) VDR displaces NFAT from its site. EMSA was performed using NFAT site 1 (left) and NFAT site 2 (right) present within the −232-to-−159 region of hIL-17A promoter as the radiolabeled probes (see Materials and Methods). Increasing amounts of recombinant VDR (15, 30, and 60 ng; lanes 1 to 3) or VDR-RXR (10, 20, and 40 ng each; lanes 4 to 6) and 5 μg of nuclear extract from activated Jurkat cells (source of NFAT) were used. (E) Similar results were observed in 3 to 5 independent experiments. (F) Calcineurin/activated NFAT is involved in hIL-17A gene regulation. Human CD4+ T cells were polarized under Th17 conditions and treated with cyclosporine A (CsA) in the presence or absence of 1,25(OH)2D3 (10 nM). RT-PCR analysis of hIL-17A mRNA was done 5 days post-CsA treatment. Similar results were observed in two additional experiments. (Inset) Western blot analysis of NFATc1 protein levels.
To further evaluate the involvement of NFAT in 1,25(OH)2D3-mediated repression, we assessed the ability of NFAT to rescue the 1,25(OH)2D3-mediated repression. Expression of NFATc1 relieved the effect of 1,25(OH)2D3 in a concentration-dependent manner, indicating that 1,25(OH)2D3 repression involved inhibition of NFAT-mediated activation (Fig. 3C). In contrast, overexpression of c-Fos and c-Jun (AP1 components) did not relieve the 1,25(OH)2D3 inhibition (data not shown). In addition, mutation of the AP1 site did not affect activation or 1,25(OH)2D3-mediated repression of hIL-17A transcription (Fig. 3B).
Next, the contribution of RXR to VDR-mediated transcriptional repression of hIL-17A was also assessed. 1,25(OH)2D3 repression was enhanced when RXR in addition to VDR was transfected (Fig. 3D). Lower levels of VDR were used in this assay [thus, the repression with 10 nM 1,25(OH)2D3 was less]. We also tested the VDR-RXR heterodimerization-defective mutant F251C. Using increasing concentrations of this hVDR mutant, repression was similar to 1,25(OH)2D3-mediated repression observed in the presence of endogenous VDR (not shown). These findings suggest that RXR plays a role in 1,25(OH)2D3-mediated repression of IL-17A.
To identify the mechanism by which 1,25(OH)2D3 inhibits NFAT-mediated activation of IL-17A transcription, we asked if the NFAT sites between positions −232 and −159 bind VDR and RXR. Electrophoretic mobility shift assays (EMSAs) indicated that VDR alone as well as VDR-RXR form protein-DNA complexes on the NFAT sites. RXR alone did not bind to either of the NFAT sites (data not shown). The mutated probes for both of the sites were unable to bind NFATc1, VDR, or VDR/RXR (data not shown). When increasing concentrations of recombinant VDR or VDR/RXR were added, two distinct complexes of NFAT and VDR or NFAT and the VDR-RXR heterodimer were observed on each NFAT site (Fig. 3E). Increasing concentrations of VDR or VDR-RXR resulted in a concomitant decrease in the amount of bound NFATc1 (Fig. 3E). These observations indicate that there is competition between NFAT and VDR/RXR to occupy the NFAT sites.
To confirm the role of NFAT in the production of hIL-17A (38), CD4+ T cells isolated from healthy donors were polarized under Th17 conditions and treated with cyclosporine A (CsA; acts by blocking the phosphatase calcineurin, which is responsible for activating NFAT) in the presence or absence of 1,25(OH)2D3 (10 nM). A dose-dependent inhibition of hIL-17A mRNA levels by CsA was observed (Fig. 3F). At the higher concentration of CsA, NFAT was not observed on the Western blot, hIL-17A mRNA levels were markedly decreased, and no inhibition by 1,25(OH)2D3 was observed in the presence of CsA (100 nM) (Fig. 3F). These findings provide evidence of the importance of NFAT in the regulation of endogenous hIL-17A.
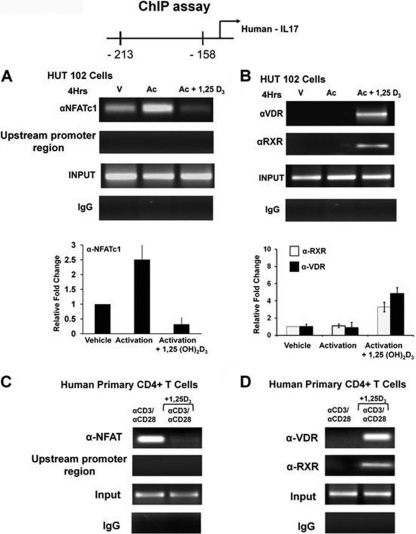
To determine that VDR or VDR-RXR can cooccupy the NFAT site in the hIL-17A promoter in vivo, chromatin immunoprecipitation (ChIP) assays were performed in HUT102 cells (a human cell line constitutively expressing IL-17A [38]) and in primary human CD4+ T cells isolated from healthy donors. Recruitment of NFATc1 to the NFAT sites under activation conditions was blocked in the presence of 1,25(OH)2D3 (Fig. 4A and C). Both in HUT102 cells and in primary human CD4+ T cells, VDR and RXR recruitment to the NFAT site was 1,25(OH)2D3 dependent (Fig. 4B and D). These findings indicate that VDR/RXR mediates hIL-17A repression by blocking NFATc1 from binding to its site on the hIL-17A promoter and by associating with the NFAT element.
Fig. 4.
VDR-RXR can occupy NFAT sites in the hIL-17A promoter in vivo. (A and B) ChIP assays were performed using HUT102 cells. HUT102 cells were transfected with VDR (1 μg) and stimulated with PMA and ionomycin in the presence or absence of 1,25(OH)2D3 (10 nM) for 4 h and cross-linked by 1% formaldehyde for 15 min. Cross-linked lysates were subjected to immunoprecipitation with NFATc1 antibody (A), VDR antibody (B), RXR antibody (B), or control rabbit IgG (A and B). DNA precipitates were isolated and then subjected to PCR using specific primers designed to amplify the −232/−159 region of the hIL-17A promoter, which contains NFAT sites. Analysis of input DNA (0.2%) was done prior to precipitation (input). For all ChIP assays in cells lines (but not in primary cells), VDR was transfected since T cells began to express VDR only 8 h postactivation (2). The bar graph represents quantification for HUT102 cells of three independent ChIP analyses (±SE). (C and D) ChIP assays were performed using human CD4+ T cells polarized under Th17 conditions. Human CD4+ T cells were activated in the presence or absence of 10 nM 1,25(OH)2D3 for 4 days. Formaldehyde cross-linked lysates were subjected to immunoprecipitation with NFAT antibody (C), VDR antibody (D), RXR antibody (D), or control rabbit IgG (C and D). DNA precipitates were isolated and subjected to PCR as described above. Similar results were observed in two additional experiments. In both HUT102 cells and primary human CD4+ T cells, PCR using the primers designed to amplify the upstream region of hIL-17A promoter −2800/−2500 (shown in panels A and C) was done as a negative control to exclude nonspecific binding. Similar results were also observed for the −3000/−2700 promoter region (not shown).
Previous studies have shown that NFAT binds to p300/CBP and recruits histone acetyltransferase (HAT) activity (20). It has been reported that 1,25(OH)2D3 negatively regulates 1α(OH)ase [the key enzyme involved in the synthesis of 1,25(OH)2D3] and PTH by p300 HAT dissociation and histone deacetylase (HDAC) association (32, 44). Whether this model of transrepression is applicable to other genes negatively regulated by 1,25(OH)2D3 is not known. Therefore, we investigated the involvement of HDAC in 1,25(OH)2D3-mediated inhibition of the hIL-17A transcription. Addition of trichostatin A (TSA), a histone deacetylase inhibitor, enhanced PMA- and ionomycin-induced hIL-17A transcription and reversed the 1,25(OH)2D3-mediated repression (Fig. 5A). To further understand the mechanism of 1,25(OH)2D3-mediated repression of hIL-17A in vivo, we examined the recruitment of HDAC and acetylated histone 4 to the hIL-17A promoter using HUT102 cells, primary human CD4+ T cells, and ChIP assay. ChIP analysis showed that in the presence of 1,25(OH)2D3 HDAC2 is recruited to the hIL-17A promoter (Fig. 5B and D). In addition, there is a decrease in acetylated histone 4 in response to 1,25(OH)2D3 (Fig. 5C and E). Together, these findings demonstrate that 1,25(OH)2D3 repression of hIL-17A transcription is mediated, at least in part, by HDAC recruitment, coupled to a decrease in HAT activity presumably via HAT coactivator dissociation.
Fig. 5.
1,25(OH)2D3 recruits histone deacetylase to the hIL-17A promoter. (A) Jurkat cells were transfected with the −1125/+5 hIL-17A promoter. Posttransfection cells were rested overnight and then stimulated with PMA and ionomycin in the presence or absence of 1,25(OH)2D3 (10 nM) for 9 h with the indicated concentrations of HDAC inhibitor, trichostatin A (TSA). P < 0.01 [activation plus 1,25(OH)2D3, control versus activation plus 1,25(OH)2D3, TSA (50 and 100 nM)]. Data represent the means ± SE from 3 separate experiments. (B to E) 1,25(OH)2D3 recruits histone deacetylase to the hIL-17A. (B and C) ChIP assays were performed using HUT102 cells. HUT102 cells were stimulated with PMA and ionomycin in the presence or absence of 1,25(OH)2D3 (10 nM) for 4 h and cross-linked with 1% formaldehyde for 15 min. Cross-linked lysates were prepared and subjected to immunoprecipitation with HDAC-2 antibody (B), AcH4 antibody (C), or control rabbit IgG (B and C). DNA precipitates were isolated and then subjected to PCR using specific primers designed to amplify the −213/−158 region of the hIL-17A promoter, which contains NFAT sites. Analysis of input DNA (0.2%) was taken prior to precipitation. The bar graph represents quantification for HUT102 cells of three independent ChIP analyses (±SE). (D and E) ChIP assays were performed using CD4+ T cells polarized under Th17 conditions. Human CD4+ T cells were activated in the presence or absence of 10 nM 1,25(OH)2D3 for 4 days. Formaldehyde cross-linked lysates were subjected to immunoprecipitation with HDAC2 antibody (D), AcH4 antibody (E), or control rabbit IgG (D and E). DNA precipitates were isolated and subjected to PCR as described above. Similar results were observed in two additional experiments.
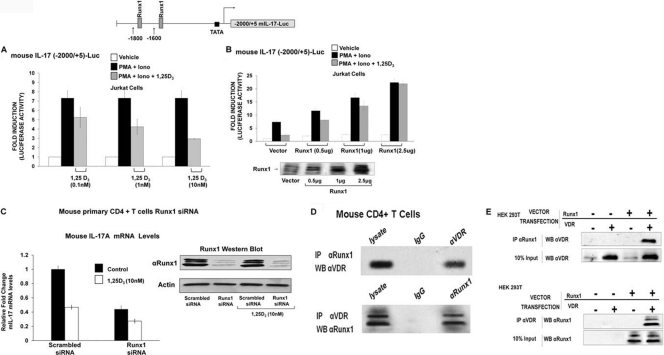
Mouse IL-17A transcription is directly repressed by 1,25(OH)2D3, as 1,25(OH)2D3 decreases activation-induced recruitment of Runx1 to the mIL-17A promoter.
To determine whether the mechanism of inhibition of mouse IL-17A secretion and mRNA also involves an effect on mIL-17A transcription, studies were done using the 2000/+5 mouse IL-17A promoter construct (74). Activation of the mIL-17A transcription was significantly repressed in response to 1,25(OH)2D3 (Fig. 6A). Since previous studies showed that expression and transcription of mIL-17A is dependent on Runx1 (74), we assessed the ability of Runx1 to rescue 1,25(OH)2D3-mediated inhibition of mIL-17A transcription. Runx1 overexpression relieved the repression by 1,25(OH)2D3 and enhanced activation of mIL-17A transcription (Fig. 6B). Runx2 was also able to enhance mIL-17A transcription and relieve 1,25(OH)2D3-mediated inhibition, albeit less efficiently (data not shown). Runx3 was the least efficient in enhancing activation of mIL-17A transcription (data not shown). These findings suggest that the repression by 1,25(OH)2D3 of mIL-17A transcription is at least partially dependent on Runx. We confirmed the dependence of mIL-17A expression on Runx1 by knockdown of endogenous Runx1. Runx1 siRNA transfection reduced expression of Runx1 (Fig. 6C, Western blot) as well as mIL-17A mRNA, and inhibition by 1,25(OH)2D3 of IL-17A mRNA was attenuated in the presence of Runx1 siRNA (Fig. 6C).
Fig. 6.
mIL-17A transcription is repressed by 1,25(OH)2D3, Runx1 relieves 1,25(OH)2D3-mediated repression, and VDR and Runx1 interact. (A) 1,25(OH)2D3 represses mIL-17A transcription. Jurkat cells transfected with the mIL-17A promoter (−2000/+5) were activated for 9 h with PMA and ionomycin in the presence of increasing concentrations of 1,25(OH)2D3. P < 0.05 [activation versus activation plus 1,25(OH)2D3 (1 and 10 nM)]. (B) Runx1 relieves 1,25(OH)2D3-mediated repression of mIL-17A transcription and enhances activation. Jurkat cells were cotransfected with the 2-kb mIL17A promoter and increasing concentrations of Runx1 expression vector or empty control vector. Cells were activated for 9 h with PMA and ionomycin in the presence or absence of 10 nM 1,25(OH)2D3. P < 0.05 [activation plus 1,25(OH)2D3, vector transfection versus activation plus 1,25(OH)2D3 Runx1 transfected (0.5, 1 and 2.5 μg)]. (A and B) The data represent the means ± SE from 3 to 6 separate experiments. (C) Runx1 is required for IL-17A expression in mouse CD4+ T cells. Mouse CD4+ T cells were isolated as described in Materials and Methods and were transfected with Runx1-specific siRNA or scrambled siRNA. Cells posttransfection were polarized under Th17 conditions in the presence or absence of 1,25(OH)2D3 (10 nM). RT-PCR analysis of mIL-17A mRNA was done 5 days post-siRNA transfection. Inhibition of mIL-17A mRNA levels by Runx1-specific siRNA was observed. (Inset) Western blot analysis of Runx1 protein levels was done to confirm knockdown of Runx1 by Runx1-specific siRNA. Similar results were observed in two additional experiments. (D and E) VDR and Runx1 interact. (D) Western blot (WB) analysis of Runx1 and VDR performed using mouse primary CD4+ T cells polarized under Th17 conditions, detected in nuclear extracts (NE) immunoprecipitated (IP) with Runx1 antibody or VDR antibody. Similar results were observed in 3 independent experiments. (E) Western blot analysis of Runx1 and VDR in HEK293 cells transfected with Runx1 and/or VDR expression vectors detected in NE immunoprecipitated with Runx1 antibody or VDR antibody. Similar results were observed in at least 3 independent experiments.
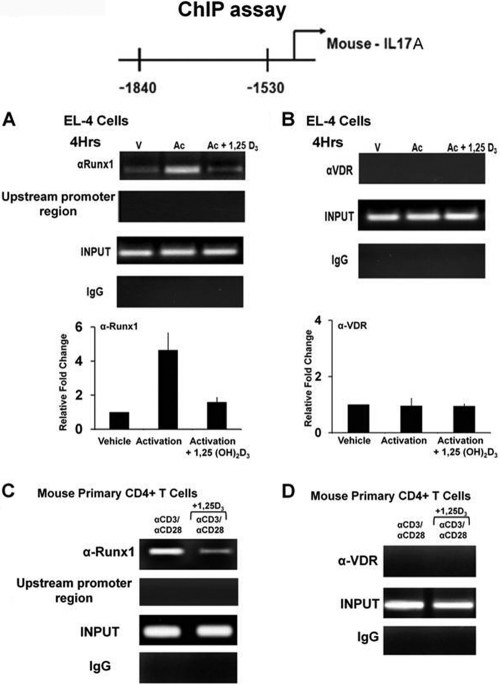
Since the Runx site is not homologous to VDRE half-sites, we hypothesized that VDR inhibits Runx function by direct physical interaction with Runx1, preventing Runx1 from binding to its sites. We assessed the interaction between Runx1 and VDR by coimmunoprecipitation using nuclear extracts from the mouse CD4+ T cells (Fig. 6D) or in HEK293T cells cotransfected with VDR and Runx1 (Fig. 6E). Runx1 and VDR were coimmunoprecipitated, indicating an interaction between the two proteins. ChIP assays were performed using mouse EL-4 cells [a tumor cell line that retains many T cell properties (28, 64, 66)] and mouse primary CD4+ T cells. Runx1 was recruited to the Runx1 sites in the mIL-17A promoter, but this recruitment was markedly decreased in the presence of 1,25(OH)2D3 (Fig. 7A and C). VDR recruitment to the Runx sites was not observed in either the presence or absence of 1,25(OH)2D3 (Fig. 7B and D), suggesting that repression of mIL-17A transcription by 1,25(OH)2D3 occurs through sequestration of Runx1 by VDR. Similar studies were also done using mouse IL-2 promoter (49). Runx1 also enhanced IL-2 transcription and relieved the repression of IL-2 transcription by 1,25(OH)2D3 (not shown). In addition, Runx1 enhanced hIL-17A promoter activity (data not shown). However, Runx1 only partially reversed the repressive effect of 1,25(OH)2D3 on hIL-17A transcription, even at higher concentrations of Runx1 (2.5 μg) (data not shown). Thus, control by 1,25(OH)2D3 of T cell function by affecting Runx1 is not specific for 1,25(OH)2D3 suppression of mIL-17A. Sequestration of Runx1 by 1,25(OH)2D3/VDR may be an additional control mechanism of 1,25(OH)2D3-mediated repression of inflammatory cytokines.
Fig. 7.
Activation-induced recruitment of Runx1 to the mIL-17A promoter is decreased in the presence of 1,25(OH)2D3. (A and B) ChIP assays were performed in EL-4 cells. EL-4 cells were transfected with VDR (1 μg), activated with PMA and ionomycin in the presence or absence of 1,25(OH)2D3 (10 nM) for 4 h, and cross-linked by 1% formaldehyde for 15 min. Cross-linked lysates were then subjected to immunoprecipitation with Runx1 antibody (A), VDR antibody (B), or control rabbit IgG (A and B). A total of 0.2% of chromatin obtained before (input) or after immunoprecipitation (IP) and were analyzed with primers specific to amplify the −1560/−1860 region of the mIL-17A promoter containing the Runx sites. The bar graph represents quantification of ChIP analyses for EL-4 cells. Data are presented as the means ± SE from three independent experiments. (C and D) ChIP assays were performed in mouse CD4+ T cells polarized under Th17 conditions. Mouse CD4+ T cells were activated in the presence or absence of 10 nM 1,25(OH)2D3 for 3 days. Formaldehyde cross-linked lysates were then subjected to immunoprecipitation with Runx1 antibody (C), VDR antibody (D), or control rabbit IgG (C and D). DNA precipitates were isolated and subjected to PCR as described above. Similar results were observed in two additional experiments. In both EL-4 cells and primary mouse CD4+ T cells, PCR using the primers designed to amplify the upstream region of mIL-17A promoter −2800/−2500 (shown in panels A and C) was done as a negative control to exclude nonspecific binding. Similar results were also observed for the −500/−200 promoter region (not shown).
Since sequence analysis indicated that the 2-kb mouse IL-17A promoter lacked a NFAT binding site, studies were done to examine if there was a cryptic but functional NFAT site within this region of the mouse IL-17A promoter. Overexpression of NFAT did not reverse the 1,25(OH)2D3-mediated inhibition of −2000/+5 mIL-17A promoter activity (data not shown). EMSAs and ChIP assays were performed using synthetic oligonucleotides corresponding to the recently reported NFAT site present at −3085/−3077 in the mouse IL-17A promoter (24). The sequence of the mIL-17A NFAT site (CCCTTATTT) was not identical to the sequences of the NFAT sites present in the hIL-17A promoter (CATTTTCCTTCA and GGGGCGGAAATTTT). Although NFAT bound to the site, VDR alone or the VDR-RXR heterodimer did not bind to it and was not recruited to this site (data not shown). Thus, the 1,25(OH)2D3-mediated inhibition of mIL-17A promoter activity is not reliant on this NFAT site.
Induction of Foxp3 by 1,25(OH)2D3.
Foxp3, a transcription factor involved in the development and function of regulatory T cells, a subset of CD4+ T cells that are important in inhibiting inflammation and in suppressing autoimmune processes (48), has recently been reported to negatively regulate both IL-17A and IL-2 transcription, at least in part by interacting with Runx1 (49, 74). Retinoic acid (RA), similar to 1,25(OH)2D3, has been reported to result in an enhancement of Treg cells and an inhibition of Th17 cells (57). Since RA has recently been reported to induce Foxp3 (64), we asked whether induction of Foxp3 may be an additional mechanism involved in the suppression of IL-17A by 1,25(OH)2D3.
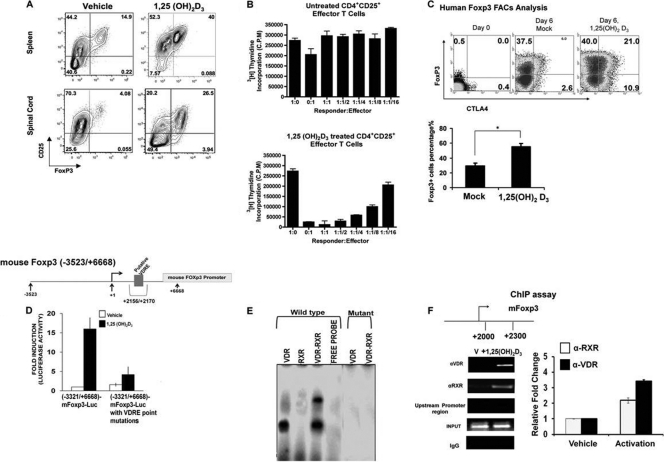
To examine whether 1,25(OH)2D3 induces Foxp3 and regulatory CD4+ T cells in vivo, MOG p35-55 EAE induced C57BL6/J mice were treated with either vehicle or 1,25(OH)2D3 starting at day of immunization. At day 7 (before disease onset), mice were sacrificed, and splenocytes were examined for the presence of Treg cells (Fig. 8A). We found that the percentage of Foxp3+ CD4+ CD25hi T cells increased from 14.9% in the vehicle-treated group to 40% in the 1,25(OH)2D3-treated group (Fig. 8A, top). At day 15 (peak of the disease), mononuclear infiltrating cells from the spinal cords (Fig. 8A, bottom) and brains (data not shown) were examined for the presence of Treg cells. As in the periphery, 1,25(OH)2D3 treatment induced a higher percentage of Treg cells in the spinal cords (4% in the vehicle-treated group versus 26.5% in the 1,25(OH)2D3-treated group). Similar data from the brains of these mice were observed (data not shown).
Fig. 8.
1,25(OH)2D3 induces functional Treg cells, regulation of Foxp3 transcription by 1,25(OH)2D3, and identification of a VDRE in the Foxp3 promoter. 1,25(OH)2D3 induces Treg cells in vivo. EAE was induced in C57BL6/J mice, and mice were treated with either vehicle or 1,25(OH)2D3 (50 ng/mouse) for 15 days, starting on the day of EAE induction. At day 7, before disease onset, and at day 15 (peak of disease), CD4+ CD25+ Foxp3+ cells in the spleens and spinal cords were examined, respectively. (Top) CD4-gated CD25+ Foxp3+ cells induced by either vehicle or 1,25(OH)2D3 in the spleen. (Bottom) CD4-gated CD25+ Foxp3+ cells induced by either vehicle or 1,25(OH)2D3 in the spinal cord. FACS plots are representative analyses of 1 mouse out of 5 tested. (B) 1,25(OH)2D3 induces functional T effector cells. Naïve spleens were isolated from 2D2 mice and activated with MOG p35-55 (20 μg/ml) in the presence of either vehicle or 1,25(OH)2D3 (10 nM). CD4+ CD25hi T cells were then sorted from these cultures to test for their suppressive capacity in vitro. Vehicle- or 1,25(OH)2D3-treated CD4+ CD25+ T (effector) cells were cocultured with (5 × 104 cells/well) naïve CD4+ CD25− T (responder) cells, sorted from naïve 2D2 spleens. T effector cells were added in titrated numbers at the T responder/T effector ratios indicated on the x axis. Cells were cultured in the presence of irradiated syngeneic APCs (2 × 105 cells/well) and MOG p35-55 (20 μg/ml) for 72 h. Mean [3H]thymidine incorporation was indicated as cpm ± SE. Data are representative of 3 independent experiments. (C) 1,25(OH)2D3 induces Foxp3 expression in purified human CD4+ CD25− cells. Cells were activated with immobilized CD3 and soluble CD28 monoclonal antibodies (MAbs) in the presence or absence of 1,25(OH)2D3 (10 nM) for 6 days in RPMI 1640 medium containing rhIL-2. Intracytoplasmic analysis of Foxp3, CTLA4, and CD25 was performed on day 0 and day 6 cultures. Analysis shows the percentage of Foxp3+ and CTLA4 cells of gated CD4+ CD45RO+ CD25+ T cells. One of five representative donors is illustrated. The expression of Foxp3 was measured by flow cytometry. The bar graph represents the means ± SE of results obtained from four donors analyzed in two independent experiments. *, P < 0.05. (D) Regulation of mFoxp3 transcription by 1,25(OH)2D3. HEK-293T cells were transfected with the mFoxp3-Luc promoter −3523/+6668 or −3523/+6668 mFoxp3-Luc with the VDRE mutated at positions +2156/+2170. Cells (24 h posttransfection) were treated with 10 nM 1,25(OH)2D3 for another 24 h. Results represent the means ± SE from at least 4 separate experiments. Note the significant activation of Foxp3 transcription by 1,25(OH)2D3 (P < 0.05 [vehicle versus 1,25(OH)2D3] [mutation in the VDRE blocked the response to 1,25(OH)2D3]. (E) VDR and VDR-RXR form protein-DNA complexes on the Foxp3 core enhancer region. EMSA was performed with recombinant VDR (15 ng), RXRα (20 ng), or VDR/RXRα (15 ng each) and 32P-labeled oligonucleotides containing the putative VDRE present at positions +2156/+2170 or mutated VDRE (see Materials and Methods). Similar results were observed in at least 3 independent experiments. (F) ChIP assays were performed using mouse CD4+ T cells polarized under Treg cell conditions. Mouse CD4+ T cells were activated in the presence or absence of 10 nM 1,25(OH)2D3 for 3 days and cross-linked with 1% formaldehyde. Cross-linked cell lysates were subjected to immunoprecipitation with VDR antibody, RXR antibody, or control rabbit IgG. A total of 0.2% of chromatin obtained before (input) or after immunoprecipitation (IP) was analyzed with primers specific to amplify the +2216/+2350 region of the mFoxp3 promoter. The bar graph represents the quantification of three independent ChIP analyses (±SE). PCR using the primers designed to amplify the other regions of Foxp3 promoter −800/−500 (shown in panel F) was done as a negative control to exclude nonspecific binding. Similar results were also observed for the +1500/+1200 region (not shown).
To prove the functionality of these Treg cells in suppressing proinflammatory CD4+ T cells, an in vitro suppression assay was performed. Naïve 2D2 splenocytes were activated with MOG p35-55 (20 μg/ml) for 72 h in the presence of vehicle or 1,25(OH)2D3 (10 nM). Treg differentiation was tested by the induction of Foxp3 using flow cytometry. Effector CD4+ T cells treated with 1,25(OH)2D3 induced a high percentage of Foxp3-positive T cells (data not shown). CD4+ CD25hi T cells from these activated cultures were then sorted out using CD4 and CD25 markers and added to responder CD4+ CD25− T cells, isolated from naïve 2D2 splenocytes (Fig. 8B). In the suppression assays, 50,000 responder T cells were cocultured with the indicated number of effector CD4+ CD25hi T cells and 200,000 irradiated syngeneic splenocytes (serving as APCs) per well. Cells were activated in the presence of MOG p35-55 (20 μg/ml) for 72 h. Our results show that 1,25(OH)2D3 induces functional Treg cells that can suppress the activity of proinflammatory CD4+ T cells, indicated by proliferation inhibition via thymidine incorporation (Fig. 8B). We also determined whether 1,25(OH)2D3 was able to induce human Foxp3. 1,25(OH)2D3 stimulated expression of Foxp3 and CTLA-4 in human CD4+ CD25− T cells (Fig. 8C).
Through in silico analysis we were able to identify a putative vitamin D response element (VDRE) present within the core enhancer region (positions +2079/+2198) at positions +2156/+2170 of the mouse Foxp3 gene. The core enhancer region is a highly conserved noncoding sequence (it is highly homologous between human and mouse) that contains a number of important regulatory sites important for Foxp3 transcriptional activation (66). To determine if 1,25(OH)2D3 affects mFoxp3 transcription, the mFoxp3-Luc construct (positions −3523/+6668) or the −3523/+6668 construct with the putative VDRE that was mutated was cotransfected with VDR in HEK293 cells. 1,25(OH)2D3 (10 nM) significantly induced mFoxp3 transcription (16-fold; P < 0.05 compared to vehicle-treated group) (Fig. 8D). Mutation of the putative VDRE blocked the response to 1,25(OH)2D3 (Fig. 8D), suggesting that the 1,25(OH)2D3 response is dependent on the VDRE present at positions +2156/+2170. EMSA indicated that VDR alone as well as VDR-RXR form protein-DNA complexes with the synthetic oligonucleotide corresponding to the VDRE within the core enhancer region (Fig. 8E). The oligonucleotide containing the mutated VDRE sequences failed to bind either VDR or VDR-RXR (Fig. 8E). In addition, ChIP assays performed with primary mouse CD4+ T cells using primers designed to amplify the region of the Foxp3 gene containing the VDRE site indicated 1,25(OH)2D3 recruitment of VDR and RXR to the VDRE site (Fig. 8F). Thus, 1,25(OH)2D3 may act by indirect and direct mechanisms to downregulate IL-17A. These findings suggest that induction of Foxp3 (which interacts with and inhibits Runx1 and NFAT function [71, 74]) is an additional mechanism involved in the suppression of IL-17A by 1,25(OH)2D3.
DISCUSSION
Previous studies have suggested a supporting role for vitamin D in reducing the risk of MS (43, 55, 61). In addition, 1,25(OH)2D3 is known to inhibit the development of EAE (8, 35), and recent studies have correlated a suppressive effect of 1,25(OH)2D3 on Th17 with the prevention of EAE (11). 1,25(OH)2D3 has also been shown to inhibit IL-17A production in T cells from multiple sclerosis patients (17) and from patients with early rheumatoid arthritis (15). The ability of 1,25(OH)2D3 to prevent experimental autoimmune uveitis and to reduce colitis in a mouse model has also been correlated to a suppression of IL-17A induction (18, 65). Although it is known that 1,25(OH)2D3 modulates inflammation and Th17-mediated autoimmunity, little is known about the molecular mechanisms mediating the effects of 1,25(OH)2D3. In addition, compared to what is known about VDR-mediated gene activation, little is known about the mechanisms and cofactors involved in 1,25(OH)2D3-induced gene repression. Our findings are the first to correlate the reversal of paralysis in EAE mice by 1,25(OH)2D3 with reduced IL-17A-secreting CD4+ T cells. In this study we also show by adaptive transfer that 1,25(OH)2D3 treatment of Th17 cells downregulates progression of EAE and that the mechanism involved in 1,25(OH)2D3-mediated suppression of IL-17A induction involves transcriptional repression. 1,25(OH)2D3 mediates this repression by dissociation of histone acetylase activity from the IL-17A promoter and recruitment of HDAC and VDR/RXR binding to the NFAT sites. In addition, our findings suggest that the suppressive effect of 1,25(OH)2D3 involves sequestration of Runx1 by 1,25(OH)2D3/VDR as well as induction by 1,25(OH)2D3 of Foxp3 (which is known to interact with Runx1 and NFAT and thus can negatively regulate IL-17A transcription) (27, 49, 74).
Our study shows that VDR and VDR/RXR compete with NFATc1 for common binding sites on the hIL-17A promoter. The two NFAT sites in the −232-to-−159 region have been shown to be crucial sensors of T cell receptor signaling on the hIL-17A promoter (38). Mutation of both NFAT sites was needed in order to reduce promoter activity to levels observed in the minimal IL-17A promoter (38). In previous studies examining the regulation of IL-2 and GM-CSF, a similar mechanism involving VDR-mediated repression of transcriptional activity by blocking NFAT binding on DNA has been reported (2, 67, 69). Similar to our findings, RXR enhanced the inhibition conferred by VDR on activated hIL-2 transcription (2). However, RXR is not involved in VDR-mediated transrepression of GM-CSF and thus is distinct from the negative regulation of IL-2 and hIL-17A by 1,25(OH)2D3. Although AP1 is known to cooperate with NFAT in the regulation of numerous cytokine genes (2, 67, 69), mutation of the AP1 site in the −232-to-−159 region of the hIL-17A promoter did not affect activation or 1,25(OH)2D3-mediated repression of hIL-17A transcription. Thus, in contrast to the IL-2 gene and the GM-CSF gene, suppression of NFAT activation of hIL-17A transcription, at least in the −232/−159 region of the promoter, does not require AP1 cooperativity, and the AP1 site adjacent to these NFAT sites does not demonstrate activation-inducible AP1 DNA binding activity. Thus, the lack of dependence on AP1 in the −232/−159 region of the hIL-17A promoter differentiates 1,25(OH)2D3 regulation of hIL-17A from the other known mechanisms of repression by 1,25(OH)2D3 of other cytokines regulated by NFAT. The lack of cooperation of NFAT with AP1 is not without precedence. Recent studies have found that NFAT binding to the promoter of CTLA-4 (a surface molecule which shares homology to CD28) does not require AP1 cooperativity (23).
This study is the first to show the involvement of HDAC in repression of a proinflammatory cytokine by 1,25(OH)2D3. 1,25(OH)2D3-dependent VDR/RXR binding to the NFAT sites resulted in the dissociation of acetylated histone H4 from the hIL-17A promoter and recruitment of HDAC2 to the NFAT sites. Previous studies have shown that NFAT proteins recruit histone acetyltransferases (HATs) and that these coactivators play an important role in NFAT-dependent transactivation (20). 1,25(OH)2D3 has been reported to induce transcriptional repression of PTH and 1α(OH)ase in part by recruitment of VDR/RXR and HDAC2 to the PTH or 1α(OH)ase promoter (31, 32, 45). Similar to our findings, a VDR mutant which is defective in heterodimerization with RXR was unable to mediate 1,25(OH)2D3 repression of PTH and 1α(OH)ase transcription, indicating that RXR plays a role in VDR-mediated repression of PTH and 1α(OH)ase. Unlike our findings, direct DNA binding of VDR or VDR/RXR on the PTH or 1α(OH)ase promoter was not detected (32, 45). Rather, a helix-loop-helix-type factor, designated vitamin D-interacting repressor (VDIR), was the direct binding factor for the negative vitamin D response element (composed of 2 E box-like motifs), and VDR/RXR binds to the VDIR in a 1,25(OH)2D3-dependent manner. Thus, the 1,25(OH)2D3 recruitment of VDR/RXR as well as HDAC2 to the NFAT sites represents a novel mechanism of repression by 1,25(OH)2D3. Negative feedback regulation of the thyrotropin β gene by thyroid hormone (T3) has been reported to involve ligand-dependent recruitment of thyroid hormone receptor (TR) and HDAC2 to a negative regulatory element. For repression of the thyrotropin gene by T3, RXR was not required (56). Thus, although recruitment of HDAC may be a mechanism of repression shared by these genes, the differences involved in hormone-induced transrepression may be due to cell- and promoter-specific functions of the steroid receptor. Our findings of recruitment of HDAC as a mechanism of repression of activation of IL-17A suggest that pharmacological modulation of histone acetylation is a potentially useful therapeutic strategy for Th17-mediated autoimmune diseases.
Although both human and mouse IL-17A expression is linked to NFAT activation (24, 38), unlike the NFAT sites in the hIL-17A promoter, VDR alone and VDR/RXR did not bind to and were not recruited by 1,25(OH)2D3 to the recently identified NFAT site present at position −3085 in the mIL-17A promoter (−3500/+1 of mIL-17-luc). In addition, using the −3,500/+1 mIL-17A promoter construct (from P. L. Schwartzberg, NIH, Bethesda, MD), NFAT did not reverse 1,25(OH)2D3-mediated repression of mIL-17A transcription (S. Joshi and S. Christakos, unpublished observation). When 20 kb upstream of the mIL-17A transcription start site was examined, the only NFAT site that was conserved between human and mouse was located at position −3085 (24). The sequence of the mIL-17A NFAT site, CCCTTATTT [position −3085; homologous NFAT site on the reverse strand (24)] is not identical to the sequences of the NFAT sites present in the hIL-17A promoter between positions −232 and −159 [distal site, CATTTTCCTTCA, homologous NFAT site on the reverse strand; proximal site, GGGCGGAAATTT (38)] [NFAT consensus sequence, (A/T)GGAAA(A/N)(A/T/C)N]. Although we cannot exclude the possibility that other NFAT sites in the mIL-17A gene not included in the constructs used in this study may be involved in the inhibition of mIL-17A by 1,25(OH)2D3 (in addition to sequestration of Runx1 by VDR), the species differences that we observed in IL-17A regulation are consistent with differences reported between the promoters of the mouse and human IL-17A genes (37).
This report also indicates that the suppressive effect of 1,25(OH)2D3 can involve sequestration of Runx1 by VDR. Recent studies have shown that Runx1 is important for the transcriptional activation of mIL-17A and mIL-2 (49, 74). The Runx1 family of transcription factors is comprised of the following 3 highly homologous Runx proteins, which have distinct roles in development: Runx1 (also known as AML1/CBFA2/PEBP2αB), which regulates normal hematopoiesis, Runx2 (also known as AML3/CBFA1/PEBP2αA), which is important for bone development and osteoblast differentiation, and Runx3 (also known as AML2/CBFA3/PEBP2αC), which has an important role in thymogenesis and neurogenesis (16, 72). We found that Runx1 was the most efficient in reversing the inhibitory effects of 1,25(OH)2D3, followed by Runx2, and Runx3 was the least efficient. This is consistent with the findings of Zhang et al. (74), who found that silencing of Runx1 (by siRNA) had the greatest effect on mIL-17A expression, silencing of Runx2 had only a modest effect, and silencing of Runx3 had no effect and that Runx1 maximally induced transcription of mIL-17A. Thus, although all 3 Runx family members are expressed in Th17 cells, Runx1 is most likely the main family member involved in Runx1 activation and repression by 1,25(OH)2D3/VDR. In our studies, Runx1 recruitment to its sites in the mIL-17A promoter was markedly decreased in the presence of 1,25(OH)2D3. We did not observe recruitment of VDR to the Runx sites (the Runx site is not homologous to the VDRE half-site). Similar results were observed using the mIL-2 promoter. Through immunoprecipitation studies, we found that Runx1 and VDR interact. Thus, Runx1 sequestration by 1,25(OH)2D3/VDR may be a common mechanism for downregulation of inflammatory cytokines positively regulated by Runx1. Previous studies have indicated an interrelationship between VDR and Runx in gene regulation. Runx2 and VDR have been shown to cooperate in the 1,25(OH)2D3 stimulation of osteopontin and osteocalcin in osteoblastic cells (51, 59). Our findings related to IL-17A regulation by 1,25(OH)2D3 are the first report of VDR interacting with Runx1 to inhibit Runx1 activation. Steroid hormones and thyroid hormone repress transcription by binding of the liganded receptors to DNA as well as by protein/protein interaction (36, 75). Although VDR binds to the NFAT sites in the hIL-17A promoter, our findings show that VDR can also suppress mIL-17A through interaction with Runx1, resulting in Runx1 sequestration and prevention of Runx1 activation. Since recent studies have revealed a potential role for Runx1 in autoimmune susceptibility, interaction between VDR and Runx1 may be a therapeutic strategy for control of pathological immune responses (1).
Runx1 has also been shown to interact with Foxp3 (49, 74). Foxp3 is a lineage-specific transcription factor for Treg cells and is involved in the development and function of this CD4+ T subset. The interaction of Foxp3 with Runx1 is needed for the negative effect of Foxp3 on IL-17A and IL-2 production (49, 74). In this study we show that 1,25(OH)2D3 upregulates Foxp3 at the transcriptional level. We identified a VDRE at positions +2156/+2170 in the highly conserved noncoding sequence of the mouse Foxp3 gene (66). In previous studies, a retinoic responsive element (RARE) was identified within this region of the Foxp3 promoter (64). Previous studies have shown that RA or 1,25(OH)2D3 can upregulate 25-hydroxyvitamin D3-24-hydroxylase (24-hydroxylase) and that VDREs in the 24(OH)ase promoter can also act as retinoic response elements (77). In addition, the human osteocalcin promoter has been shown to be stimulated by retinoids through a sequence containing a VDRE (58). Thus, Foxp3, similar to these other vitamin D target genes, is regulated by RA and 1,25(OH)2D3 though a common response element. We found that VDR alone as well as VDR/RXR can bind to the VDRE in the Foxp3 promoter. This is similar to what was observed for VDR binding to the VDRE in the osteopontin promoter but not for VDR binding to VDREs of other genes which are induced by 1,25(OH)2D3 [for example, 24(OH)ase and osteocalcin], which require the VDR/RXR heterodimer. It is likely that, although VDR can bind the Foxp3 VDRE, the interaction of VDR with RXR is needed for activation of Foxp3 transcription. Previous studies have shown that, unlike 1,25(OH)2D3-mediated transcriptional repression of GM-CSF, VDR mutants without the capability to form heterodimers are unable to activate transcription (46). In addition to activation of Foxp3 transcription, we also found, similar to previous reports (22, 30), that 1,25(OH)2D3 stimulates the expression of Foxp3 in mouse and human CD4+ CD25− T cells Thus, these findings suggest that 1,25(OH)2D3/VDR, Foxp3, and Runx1 may act together to suppress proinflammatory Th17 responses in autoimmune processes.
Other transcription factors may also participate in 1,25(OH)2D3-mediated suppression of IL-17. Recent data have indicated that RORγt is required for the differentiation of Th17 cells (76). Runx1 binds to and acts together with RORγt during mIL-17A transcription (28, 74). Foxp3 can interact with RORγt as well as with Runx1 to inhibit the production of IL-17 (49, 74). Thus, by stimulating the expression of Foxp3, 1,25(OH)2D3 may also result in suppression of IL-17 by increased Foxp3 interaction with RORγt. In addition, VDR can sequester the RORγt coactivator, Runx1, and thereby inhibit the transcriptional activity of RORγt. Although the sequence homology between the RORγt site (TGACCT) and the VDRE half-site (TGAACC) is suggestive, further studies are needed to determine whether an additional mechanism of regulation of IL-17 transcription by 1,25(OH)2D3 may be by VDR binding to the RORγt site in the IL-17 promoter, thus blocking RORγt activation.
Our in vivo data also demonstrate that 1,25(OH)2D3 represses expression of IL-17A. Concomitant with downregulation of IL-17A is the upregulation of IL-10 (Fig. 1). Previous studies have indicated a role for IL-10 in the effect of 1,25(OH)2D3 on EAE (62). Thus, multiple mechanisms are involved in the inhibition of EAE by 1,25(OH)2D3. Downregulation of IL-17A and upregulation of IL-10 are associated with the beneficial effect of IFN-β in EAE (3). Concordant upregulation of IL-10 is also seen in human CD4+ T cells treated with IFN-β (3). Our findings correlate reduced IL-17A expression not only with inhibition of EAE when 1,25(OH)2D3 treatment is given beginning prior to immunization but also with the reversal of paralysis in EAE mice by 1,25(OH)2D3. Recent studies show that there may be two forms of relapsing remitting MS, a Th17 form resistant to therapy with beta interferon and a Th1 form that responds well to IFN-β (3). Other forms of demyelinating disease like neuromyelitis optica have a strong signature of IL-17-mediated pathology (3). Neuromyelitis optica may be particularly responsive to treatment with 1,25(OH)2D3. It is possible that the Th17 form of MS may be particularly suitable for intervention with 1,25(OH)2D3. The polarization of relapsing remitting multiple sclerosis along the Th1 and Th17 axes and the key role of vitamin D deficiency as a susceptibility factor for this disease add to the relevance of this study. These studies result in new concepts with regard to the mechanisms involved in the interaction of the vitamin D endocrine system and the immune system and provide a foundation for trials involving 1,25(OH)2D3 or 1,25(OH)2D3 analogs for targeting Th17 immunity.
ACKNOWLEDGMENTS
We thank Milan Uskokovic (Hoffmann-LaRoche, Nutley, NJ) for providing 1,25(OH)2D3 and Puneet Dhawan and Anita Antes (UMDNJ, Newark, NS) for their assistance in certain aspects of this investigation. We acknowledge the assistance of Jessica Magid-Bernstein (Department of Neurology and Neurosciences, UMDNJ, Newark, NJ) with the human CD4+ T cell isolation and with the Runx1 siRNA experiments.
This work was supported by National Institutes of Health (NIH) grant DK-38961-22 to S.C., NIH grant NS55997 to L.S., NIH grant AR054389 to S.G., and NIH grant AI095055 to S.C. and L.S.
Footnotes
Published ahead of print on 11 July 2011.
REFERENCES
- 1. Alarcon-Riquelme M. E. 2004. Role of RUNX in autoimmune diseases linking rheumatoid arthritis, psoriasis and lupus. Arthritis Res. Ther. 6:169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alroy I., Towers T. L., Freedman L. P. 1995. Transcriptional repression of the interleukin-2 gene by vitamin D3: direct inhibition of NFATp/AP-1 complex formation by a nuclear hormone receptor. Mol. Cell. Biol. 15:5789–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Axtell R. C., et al. 2010. T helper type 1 and 17 cells determine efficacy of interferon-beta in multiple sclerosis and experimental encephalomyelitis. Nat. Med. 16:406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bettelli E., et al. 2003. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J. Exp. Med. 197:1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhalla A. K., Amento E. P., Clemens T. L., Holick M. F., Krane S. M. 1983. Specific high-affinity receptors for 1,25-dihydroxyvitamin D3 in human peripheral blood mononuclear cells: presence in monocytes and induction in T lymphocytes following activation. J. Clin. Endocrinol. Metab. 57:1308–1310 [DOI] [PubMed] [Google Scholar]
- 6. Bikle D., Adams J., Christakos S. 2008. Vitamin D: production, metabolism, mechanism of action, and clinical requirements, p. 141–149 In Rosen C. (ed.), Primer on the metabolic bone diseases and disorders of mineral metabolism, 7th ed. American Society for Bone and Mineral Research, Washington, DC [Google Scholar]
- 7. Cantorna M. T., Hayes C. E., DeLuca H. F. 1998. 1,25-Dihydroxycholecalciferol inhibits the progression of arthritis in murine models of human arthritis. J. Nutr. 128:68–72 [DOI] [PubMed] [Google Scholar]
- 8. Cantorna M. T., Hayes C. E., DeLuca H. F. 1996. 1,25-Dihydroxyvitamin D3 reversibly blocks the progression of relapsing encephalomyelitis, a model of multiple sclerosis. Proc. Natl. Acad. Sci. U. S. A. 93:7861–7864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chabas D., et al. 2001. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science 294:1731–1735 [DOI] [PubMed] [Google Scholar]
- 10. Chabaud M., Fossiez F., Taupin J. L., Miossec P. 1998. Enhancing effect of IL-17 on IL-1-induced IL-6 and leukemia inhibitory factor production by rheumatoid arthritis synoviocytes and its regulation by Th2 cytokines. J. Immunol. 161:409–414 [PubMed] [Google Scholar]
- 11. Chang J. H., Cha H. R., Lee D. S., Seo K. Y., Kweon M. N. 2010. 1,25-Dihydroxyvitamin D3 inhibits the differentiation and migration of T(H)17 cells to protect against experimental autoimmune encephalomyelitis. PLoS One 5:e12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang S. H., Chung Y., Dong C. 2010. Vitamin D suppresses Th17 cytokine production by inducing C/EBP homologous protein (CHOP) expression. J. Biol. Chem. 285:38751–38755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Christakos S. 2008. Vitamin D gene regulation, p. 779–795 In Bilezikian J. P., Raisz L. G., Martin T. J. (ed.), Principles of bone biology. Academic Press, San Diego, CA [Google Scholar]
- 14. Christakos S., Dhawan P., Liu Y., Peng X., Porta A. 2003. New insights into the mechanisms of vitamin D action. J. Cell. Biochem. 88:695–705 [DOI] [PubMed] [Google Scholar]
- 15. Colin E. M., et al. 2010. 1,25-Dihydroxyvitamin D3 modulates Th17 polarization and interleukin-22 expression by memory T cells from patients with early rheumatoid arthritis. Arthritis Rheum. 62:132–142 [DOI] [PubMed] [Google Scholar]
- 16. Collins A., Littman D. R., Taniuchi I. 2009. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat. Rev. Immunol. 9:106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Correale J., Ysrraelit M. C., Gaitan M. I. 2009. Immunomodulatory effects of vitamin D in multiple sclerosis. Brain 132:1146–1160 [DOI] [PubMed] [Google Scholar]
- 18. Daniel C., Sartory N. A., Zahn N., Radeke H. H., Stein J. M. 2008. Immune modulatory treatment of trinitrobenzene sulfonic acid colitis with calcitriol is associated with a change of a T helper (Th) 1/Th17 to a Th2 and regulatory T cell profile. J. Pharmacol. Exp. Ther. 324:23–33 [DOI] [PubMed] [Google Scholar]
- 19. Dhawan P., et al. 2005. Functional cooperation between CCAAT/enhancer-binding proteins and the vitamin D receptor in regulation of 25-hydroxyvitamin D3 24-hydroxylase. Mol. Cell. Biol. 25:472–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia-Rodriguez C., Rao A. 1998. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 187:2031–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garrett-Sinha L. A., John S., Gaffen S. L. 2008. IL-17 and the Th17 lineage in systemic lupus erythematosus. Curr. Opin. Rheumatol. 20:519–525 [DOI] [PubMed] [Google Scholar]
- 22. Ghoreishi M., et al. 2009. Expansion of antigen-specific regulatory T cells with the topical vitamin d analog calcipotriol. J. Immunol. 182:6071–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gibson H. M., et al. 2007. Induction of the CTLA-4 gene in human lymphocytes is dependent on NFAT binding the proximal promoter. J. Immunol. 179:3831–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gomez-Rodriguez J., et al. 2009. Differential expression of interleukin-17A and -17F is coupled to T cell receptor signaling via inducible T cell kinase. Immunity 31:587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holick M. F. 2004. Sunlight and vitamin D for bone health and prevention of autoimmune diseases, cancers, and cardiovascular disease. Am. J. Clin. Nutr. 80:1678S–1688S [DOI] [PubMed] [Google Scholar]
- 26. Hsu H. C., et al. 2008. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat. Immunol. 9:166–175 [DOI] [PubMed] [Google Scholar]
- 27. Hu H., Djuretic I., Sundrud M. S., Rao A. 2007. Transcriptional partners in regulatory T cells: Foxp3, Runx and NFAT. Trends Immunol. 28:329–332 [DOI] [PubMed] [Google Scholar]
- 28. Ichiyama K., et al. 2008. Foxp3 inhibits ROR gamma t-mediated IL-17A mRNA transcription through direct interaction with ROR gamma t. J. Biol. Chem. 283:17003–17008 [DOI] [PubMed] [Google Scholar]
- 29. Ivanov I. I., et al. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126:1121–1133 [DOI] [PubMed] [Google Scholar]
- 30. Jeffery L. E., et al. 2009. 1,25-Dihydroxyvitamin D3 and IL-2 combine to inhibit T cell production of inflammatory cytokines and promote development of regulatory T cells expressing CTLA-4 and FoxP3. J. Immunol. 183:5458–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kato S., Kim M. S., Yamaoka K., Fujiki R. 2007. Mechanisms of transcriptional repression by 1,25(OH)2 vitamin D. Curr. Opin. Nephrol. Hypertens. 16:297–304 [DOI] [PubMed] [Google Scholar]
- 32. Kim M. S., et al. 2007. 1Alpha,25(OH)2D3-induced transrepression by vitamin D receptor through E-box-type elements in the human parathyroid hormone gene promoter. Mol. Endocrinol. 21:334–342 [DOI] [PubMed] [Google Scholar]
- 33. Komiyama Y., et al. 2006. IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J. Immunol. 177:566–573 [DOI] [PubMed] [Google Scholar]
- 34. Kurtzke J. F. 2000. Epidemiology of multiple sclerosis. Does this really point toward an etiology? Lectio Doctoralis. Neurol. Sci. 21:383–403 [DOI] [PubMed] [Google Scholar]
- 35. Lemire J. M., Archer D. C. 1991. 1,25-Dihydroxyvitamin D3 prevents the in vivo induction of murine experimental autoimmune encephalomyelitis. J. Clin. Invest. 87:1103–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liberman A. C., et al. 2007. The activated glucocorticoid receptor inhibits the transcription factor T-bet by direct protein-protein interaction. FASEB J. 21:1177–1188 [DOI] [PubMed] [Google Scholar]
- 37. Liu X. K., Clements J. L., Gaffen S. L. 2005. Signaling through the murine T cell receptor induces IL-17 production in the absence of costimulation, IL-23 or dendritic cells. Mol. Cells 20:339–347 [PubMed] [Google Scholar]
- 38. Liu X. K., Lin X., Gaffen S. L. 2004. Crucial role for nuclear factor of activated T cells in T cell receptor-mediated regulation of human interleukin-17. J. Biol. Chem. 279:52762–52771 [DOI] [PubMed] [Google Scholar]
- 39. Lock C., et al. 2002. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat. Med. 8:500–508 [DOI] [PubMed] [Google Scholar]
- 40. Mathieu C., Waer M., Laureys J., Rutgeerts O., Bouillon R. 1994. Prevention of autoimmune diabetes in NOD mice by 1,25 dihydroxyvitamin D3. Diabetologia 37:552–558 [DOI] [PubMed] [Google Scholar]
- 41. Matusevicius D., et al. 1999. Interleukin-17 mRNA expression in blood and CSF mononuclear cells is augmented in multiple sclerosis. Mult. Scler. 5:101–104 [DOI] [PubMed] [Google Scholar]
- 42. Munger K. L., Levin L. I., Hollis B. W., Howard N. S., Ascherio A. 2006. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA 296:2832–2838 [DOI] [PubMed] [Google Scholar]
- 43. Munger K. L., et al. 2004. Vitamin D intake and incidence of multiple sclerosis. Neurology 62:60–65 [DOI] [PubMed] [Google Scholar]
- 44. Murayama A., Kim M. S., Yanagisawa J., Takeyama K., Kato S. 2004. Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J. 23:1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45. Murayama A., et al. 1998. The promoter of the human 25-hydroxyvitamin D3 1 alpha-hydroxylase gene confers positive and negative responsiveness to PTH, calcitonin, and 1 alpha,25(OH)2D3. Biochem. Biophys. Res. Commun. 249:11–16 [DOI] [PubMed] [Google Scholar]
- 46. Nakajima S., et al. 1994. The C-terminal region of the vitamin D receptor is essential to form a complex with a receptor auxiliary factor required for high affinity binding to the vitamin D-responsive element. Mol. Endocrinol. 8:159–172 [DOI] [PubMed] [Google Scholar]
- 47. Nieves J., Cosman F., Herbert J., Shen V., Lindsay R. 1994. High prevalence of vitamin D deficiency and reduced bone mass in multiple sclerosis. Neurology 44:1687–1692 [DOI] [PubMed] [Google Scholar]
- 48. O'Garra A., Vieira P. 2003. Twenty-first century Foxp3. Nat. Immunol. 4:304–306 [DOI] [PubMed] [Google Scholar]
- 49. Ono M., et al. 2007. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446:685–689 [DOI] [PubMed] [Google Scholar]
- 50. Palmer M. T., et al. 2011. Lineage-specific effects of 1,25-dihydroxyvitamin D3 on the development of effector Cd4 T cells. J. Biol. Chem. 286:997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paredes R., et al. 2004. The Runx2 transcription factor plays a key role in the 1alpha,25-dihydroxy vitamin D3-dependent upregulation of the rat osteocalcin (OC) gene expression in osteoblastic cells. J. Steroid Biochem. Mol. Biol. 89–90:269–271 [DOI] [PubMed] [Google Scholar]
- 52. Park H., et al. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6:1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pedersen L. B., Nashold F. E., Spach K. M., Hayes C. E. 2007. 1,25-Dihydroxyvitamin D3 reverses experimental autoimmune encephalomyelitis by inhibiting chemokine synthesis and monocyte trafficking. J. Neurosci. Res. 85:2480–2490 [DOI] [PubMed] [Google Scholar]
- 54. Rachitskaya A. V., et al. 2008. Cutting edge: NKT cells constitutively express IL-23 receptor and RORgammat and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J. Immunol. 180:5167–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Raghuwanshi A., Joshi S. S., Christakos S. 2008. Vitamin D and multiple sclerosis. J. Cell. Biochem. 105:338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sasaki S., et al. 1999. Ligand-induced recruitment of a histone deacetylase in the negative-feedback regulation of the thyrotropin beta gene. EMBO J. 18:5389–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schambach F., Schupp M., Lazar M. A., Reiner S. L. 2007. Activation of retinoic acid receptor-alpha favours regulatory T cell induction at the expense of IL-17-secreting T helper cell differentiation. Eur. J. Immunol. 37:2396–2399 [DOI] [PubMed] [Google Scholar]
- 58. Schule R., et al. 1990. Jun-Fos and receptors for vitamins A and D recognize a common response element in the human osteocalcin gene. Cell 61:497–504 [DOI] [PubMed] [Google Scholar]
- 59. Shen Q., Christakos S. 2005. The vitamin D receptor, Runx2, and the Notch signaling pathway cooperate in the transcriptional regulation of osteopontin. J. Biol. Chem. 280:40589–40598 [DOI] [PubMed] [Google Scholar]
- 60. Shin H. C., Benbernou N., Fekkar H., Esnault S., Guenounou M. 1998. Regulation of IL-17, IFN-gamma and IL-10 in human CD8(+) T cells by cyclic AMP-dependent signal transduction pathway. Cytokine 10:841–850 [DOI] [PubMed] [Google Scholar]
- 61. Simon K. C., Munger K. L., Xing Y., Ascherio A. 2010. Polymorphisms in vitamin D metabolism related genes and risk of multiple sclerosis. Mult. Scler. 16:133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Spach K. M., Nashold F. E., Dittel B. N., Hayes C. E. 2006. IL-10 signaling is essential for 1,25-dihydroxyvitamin D3-mediated inhibition of experimental autoimmune encephalomyelitis. J. Immunol. 177:6030–6037 [DOI] [PubMed] [Google Scholar]
- 63. Stark M. A., et al. 2005. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity 22:285–294 [DOI] [PubMed] [Google Scholar]
- 64. Takaki H., et al. 2008. STAT6 inhibits TGF-beta1-mediated Foxp3 induction through direct binding to the Foxp3 promoter, which is reverted by retinoic acid receptor. J. Biol. Chem. 283:14955–14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tang J., et al. 2009. Calcitriol suppresses antiretinal autoimmunity through inhibitory effects on the Th17 effector response. J. Immunol. 182:4624–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tone Y., et al. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202 [DOI] [PubMed] [Google Scholar]
- 67. Towers T. L., Freedman L. P. 1998. Granulocyte-macrophage colony-stimulating factor gene transcription is directly repressed by the vitamin D3 receptor. Implications for allosteric influences on nuclear receptor structure and function by a DNA element. J. Biol. Chem. 273:10338–10348 [DOI] [PubMed] [Google Scholar]
- 68. Towers T. L., Luisi B. F., Asianov A., Freedman L. P. 1993. DNA target selectivity by the vitamin D3 receptor: mechanism of dimer binding to an asymmetric repeat element. Proc. Natl. Acad. Sci. U. S. A. 90:6310–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Towers T. L., Staeva T. P., Freedman L. P. 1999. A two-hit mechanism for vitamin D3-mediated transcriptional repression of the granulocyte-macrophage colony-stimulating factor gene: vitamin D receptor competes for DNA binding with NFAT1 and stabilizes c-Jun. Mol. Cell. Biol. 19:4191–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. VanAmerongen B. M., Dijkstra C. D., Lips P., Polman C. H. 2004. Multiple sclerosis and vitamin D: an update. Eur. J. Clin. Nutr. 58:1095–1109 [DOI] [PubMed] [Google Scholar]
- 71. Wu Y., et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126:375–387 [DOI] [PubMed] [Google Scholar]
- 72. Yang X., Karsenty G. 2002. Transcription factors in bone: developmental and pathological aspects. Trends Mol. Med. 8:340–345 [DOI] [PubMed] [Google Scholar]
- 73. Youssef S., et al. 2002. The HMG-CoA reductase inhibitor, atorvastatin, promotes a Th2 bias and reverses paralysis in central nervous system autoimmune disease. Nature 420:78–84 [DOI] [PubMed] [Google Scholar]
- 74. Zhang F., Meng G., Strober W. 2008. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 9:1297–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang X. K., Wills K. N., Husmann M., Hermann T., Pfahl M. 1991. Novel pathway for thyroid hormone receptor action through interaction with jun and fos oncogene activities. Mol. Cell. Biol. 11:6016–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhou L., et al. 2008. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453:236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zou A., Elgort M. G., Allegretto E. A. 1997. Retinoid X receptor (RXR) ligands activate the human 25-hydroxyvitamin D3-24-hydroxylase promoter via RXR heterodimer binding to two vitamin D-responsive elements and elicit additive effects with 1,25-dihydroxyvitamin D3. J. Biol. Chem. 272:19027–19034 [DOI] [PubMed] [Google Scholar]