Abstract
Induction of heat shock proteins in Escherichia coli is primarily caused by increased cellular levels of the heat shock σ-factor σ32 encoded by the rpoH gene. Increased σ32 levels result from both enhanced synthesis and stabilization. Previous work indicated that σ32 synthesis is induced at the translational level and is mediated by the mRNA secondary structure formed within the 5′-coding sequence of rpoH, including the translation initiation region. To understand the mechanism of heat induction of σ32 synthesis further, we analyzed expression of rpoH–lacZ gene fusions with altered stability of mRNA structure before and after heat shock. A clear correlation was found between the stability and expression or the extent of heat induction. Temperature-melting profiles of mRNAs with or without mutations correlated well with the expression patterns of fusion genes carrying the corresponding mutations in vivo. Furthermore, temperature dependence of mRNA–30S ribosome–tRNAfMet complex formation with wild-type or mutant mRNAs in vitro agreed well with that of the expression of gene fusions in vivo. Our results support a novel mechanism in which partial melting of mRNA secondary structure at high temperature enhances ribosome entry and translational initiation without involvement of other cellular components, that is, intrinsic mRNA stability controls synthesis of a transcriptional regulator.
Keywords: rpoH, heat shock response, σ32, thermosensor, translational regulation
When cells are exposed to stresses such as heat or ethanol, a set of well-conserved heat shock proteins (HSPs) are rapidly induced. Many HSPs are molecular chaperones or proteases and play central roles in modulating protein folding, translocation, and degradation (Hendrick and Hartl 1993; Georgopoulos et al. 1994). Induction of HSPs is controlled primarily at the level of transcription (Morimoto et al. 1994).
In Escherichia coli, expression of HSPs is positively regulated by a heat shock transcription factor σ32 encoded by the rpoH gene (Yura et al. 1993; Gross 1996). Induction of HSP synthesis results from a transient increase in the level of σ32 caused by both increased synthesis and stabilization of σ32, which is normally unstable (t1/2 ∼ 1 min) (Grossman et al. 1987; Straus et al. 1987). On the other hand, some of the HSPs, particularly the DnaK, DnaJ, and GrpE chaperones, negatively regulate HSP synthesis by reducing the amounts of active σ32 (Tilly et al. 1983; Grossman et al. 1987; Straus et al. 1990). Thus, the initial accumulation of misfolded proteins on heat shock is thought to titrate free DnaK/DnaJ chaperones resulting in stabilization of σ32, whereas subsequent buildup of HSP, including DnaK/DnaJ, destabilizes σ32 and represses its synthesis (Tilly et al. 1989; Straus et al. 1990) forming an autogenous regulatory circuit (Craig and Gross 1991; Bukau 1993). Production of abnormal proteins without a temperature upshift can induce HSP synthesis (Goff and Goldberg 1985), and stabilization of σ32 (Wild et al. 1993), but not increased synthesis of σ32 (Kanemori et al. 1994), was observed under these conditions. In the feedback control mediated by the DnaK/DnaJ chaperones, a segment of σ32 polypeptide called region C (nucleotides 364–432) appears to be critical: Expression of certain rpoH–lacZ gene fusions lacking region C can be induced normally but not shut off on heat shock (Nagai et al. 1994). Moreover, apparently normal heat induction takes place in the dnaK, dnaJ mutants despite the constitutive synthesis of stable σ32 in these strains (Straus et al. 1990; Nagai et al. 1994). Thus, heat-induced synthesis of σ32 occurs independently of the chaperone-mediated regulatory circuits.
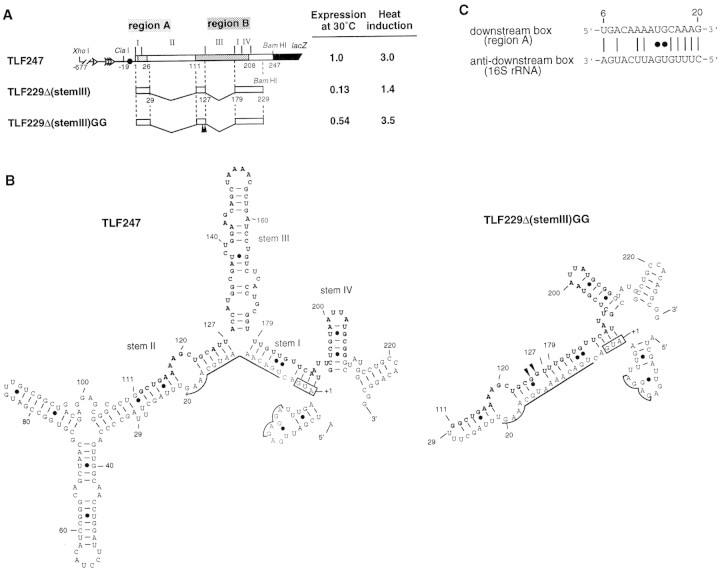
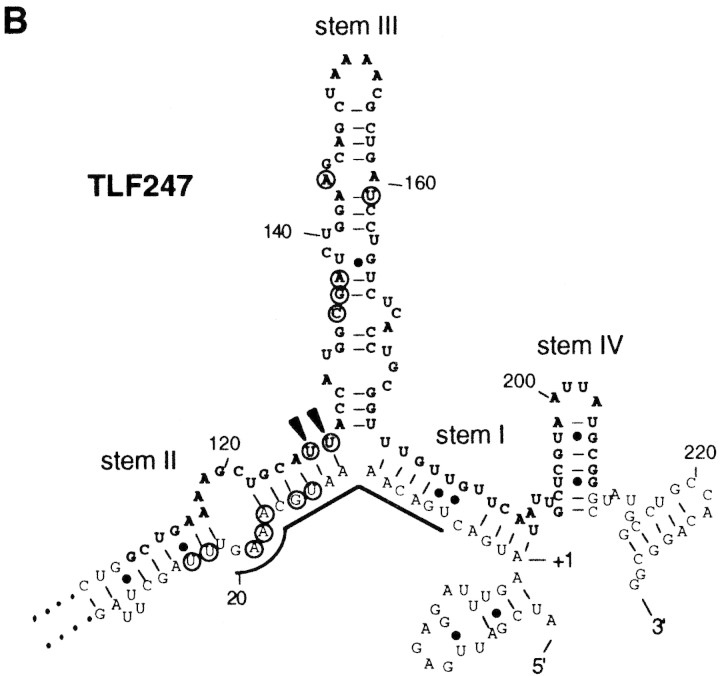
Induction of σ32 synthesis on typical heat shock (shift from 30°C to 42°C) occurs at the level of translation (Straus et al. 1987; Kamath-Loeb and Gross 1991; Nagai et al. 1991). Extensive deletion and mutation analyses of the rpoH–lacZ gene fusion led us to identify positive (region A) and negative (region B) elements of the rpoH mRNA required for translational induction (Nagai et al. 1991). Region A is located near the initiation codon (nucleotides 6–20) and represents a “downstream box” that is complementary to the 3′-region of 16S rRNA and that presumably acts as a translational enhancer (Fig. 1C; Sprengart et al. 1990). Region B is an internal coding segment of ∼100 nucleotides, located ∼100 nucleotides downstream of region A. The 5′ portion (−19 to +247) of rpoH mRNA was predicted to form base-pairings involving a part of the translational initiation region and region B (Fig. 1B; Nagai et al. 1991). Such a secondary structure model was supported through analyses of mutations (Yuzawa et al. 1993), by evolutionary conservation (Nakahigashi et al. 1995), and by direct chemical probing (Morita et al. 1999).
Figure 1.
Structure and expression of rpoH–lacZ gene fusion TLF247 and its derivatives. (A) Schematic diagram of TLF247 and its deletion derivatives. The locations of regions A and B are indicated by hatches. Segments of mRNA that correspond to each of the stem structures (I–IV; Fig. 1B) are shown above the diagram, and nucleotide numbers are shown below. Pulse-labeling (2 min) with [35S]methionine was performed before or 3 min after temperature upshift (30°C to 42°C), and immunoprecipitates were analyzed by SDS-PAGE as described in Materials and Methods. Synthesis rates were normalized to that of TLF247 labeled at 30°C. (Solid arrowheads) Positions of 2G substitutions. (B) mRNA secondary structures predicted for rpoH regions of TLF247 and TLF229Δ(stemIII)GG by mfold (Morita et al. 1999); the structures with minimum free energy are shown. The initiation codon, region A (nucleotides +6–20), SD sequence, and major stems (I–IV) are indicated. Region B (nucleotides +112–208) is shown as boldface letter. (●) G–U pairing. (C) Putative base-pairings between the downstream box (region A) of rpoH and the anti-downstream box of 16S rRNA (spanning 1469–1483 nucleotides).
The efficiency of translational initiation in bacteria can be markedly affected by the mRNA secondary structure involving the ribosome-binding site (from approximately −20 to +15 nucleotides), which includes the Shine–Dalgarno (SD) sequence and initiation codon (de Smit and van Duin 1990b). Thus, a part of the ribosome-binding site (+1–20), including the initiation codon and region A of rpoH, was thought to be masked by pairing with region B, restricting ribosome entry under nonstress conditions. Such mRNA structures were supposed to be disrupted on heat shock to enhance translation, although the mechanism remained unknown. Recently, we obtained a minimal rpoH–lacZ gene fusion containing 97 nucleotides of the rpoH coding region, which exhibits essentially normal thermoregulation (Morita et al. 1999; see Fig. 1). In addition, the appropriate stability of the mRNA secondary structure, rather than fine structure or specific sequences, appeared to be important for repression at low temperature and induction on temperature upshift.
In this study we systematically examined mutant derivatives of the minimal rpoH–lacZ gene fusion and found a clear correlation between expression or heat inducibility and stability of mRNA secondary structure involving the translational initiation region. The melting curve of rpoH mRNA fragments and accessibility of 30S ribosomes to mRNA examined in vitro gave results that were consistent with the expression patterns of the fusion constructs in vivo. These combined results suggest a novel mechanism in which melting of the mRNA secondary structure at high temperature facilitated ribosome binding and enhances σ32 synthesis, that is, a mechanism involving direct temperature sensing by mRNA structure.
Results
Expression of the minimal gene fusion is correlated with the predicted mRNA stability
Previously, we constructed several internal deletions of the rpoH–lacZ gene fusion to define the regions critical for heat induction of σ32 synthesis. Among them, a construct lacking the apical half of stem II (+30 to +110) and the entire stem III (+128 to +178) of the parental fusion TLF247 exhibited essentially normal thermoregulation provided that the appropriate instability of the mRNA secondary structure was maintained (Fig. 1A; Morita et al. 1999). The fusion TLF229Δ(stem III)GG carrying two base changes at +125 (A to G) and +126 (U to G) showed normal induction, whereas the same fusion without these changes showed very low expression at both 30°C and 42°C, despite retention of the native sequence. To substantiate further the relationship between the stability of the mRNA secondary structure and thermoregulation, a series of mutations was introduced into the minimal fusion that exhibits normal regulation and their effects on the synthesis of the fusion protein before and after temperature upshift were examined (Table 1 ). The secondary structure of the rpoH portion of mRNA was predicted, and stability was calculated for each mutant by the mfold program (Zuker 1989). As expected, most mutations predicted to decrease stability (16G, 123C, 124G, 181U, 184C) caused increased expression before and after temperature upshift (Table 1). In contract, those predicted to increase stability (5A, 14C, 118C, 121U, 126U, 125A, 125A/126U, 125A/126U/127C, 185G) resulted in reduced expression at both temperatures. These results suggested that the appropriate instability of the mRNA secondary structure, rather than a specific nucleotide sequence, is a primary requirement for the thermoregulation characteristic of rpoH translation.
Table 1.
Effects of base substitutions in minimal gene fusion TLF229Δ(stemlll)GG on expression of fusion protein
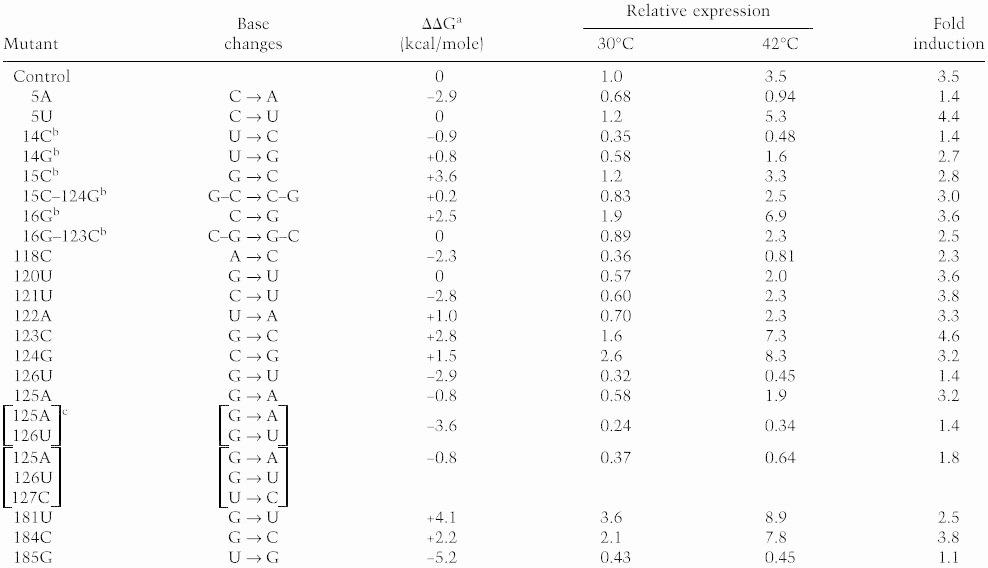 |
Nucleotide numbering is as in Fig. 1B. Synthesis rate of fusion protein was determined as in Fig. 1A and was normalized to that for the parental control labeled at 30°C.
Stability (ΔG) of each predicted mRNA structure was calculated as described in Materials and Methods. ΔΔG represents difference in ΔG at 30°C between each mutant and control RNA.
Mutants carrying base substitution(s) in region A.
This construct was identical with TLF229Δ(stemIII) shown in Fig. 1A.
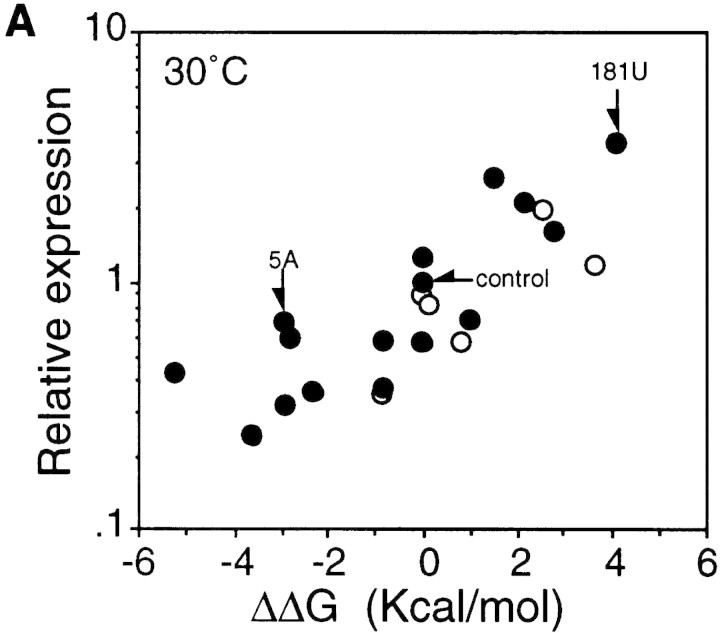
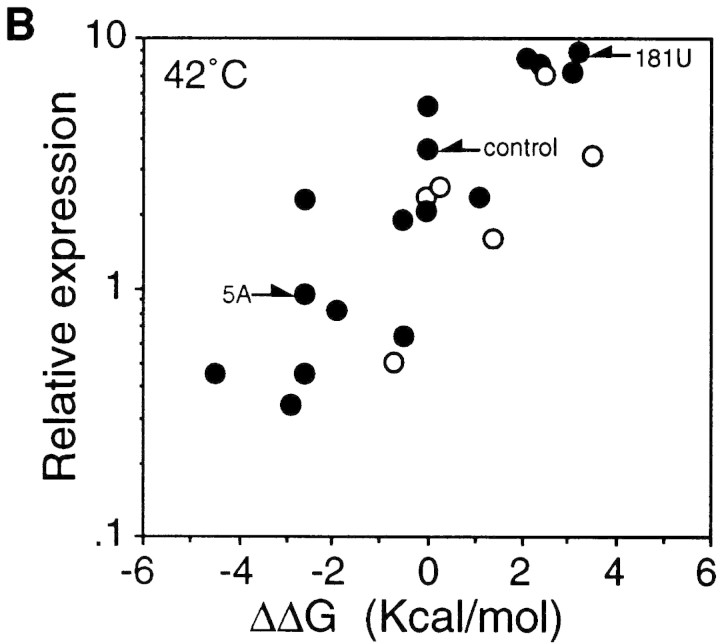
When relative synthesis rates of fusion protein for these mutants were plotted as a function of the difference in predicted stability (ΔΔG) between the mutant and control RNA, a highly significant correlation was found at both 30°C and 42°C (Fig. 2). A less clear but significant correlation was observed between the extent of heat induction and RNA stability (Table 1). These results suggest that the translational efficiency of the minimal gene fusion at both temperatures and the extent of heat induction are greatly affected by the stability of the mRNA secondary structure presumably formed between the translation initiation region and the internal region.
Figure 2.
Correlation between the predicted stability of mRNA secondary structure and synthesis rates of fusion proteins from the minimal fusion constructs at 30°C (A) and 42°C (B) based on the results shown in Table 1. The log of relative synthesis rate was plotted against differences in the predicted stability of the rpoH portion of the mRNA between each mutant and control. (Arrows) Control or mutant constructs used in Fig. 3. ΔΔG values were as described in footnote a to Table 1; the values at 42°C used in B were slightly different from those at 30°C. (○) Expression from fusions carrying mutation(s) in region A (see footnote b in Table 1).
Correlation between the temperature profile (melting curve) of RNA and expression in vivo
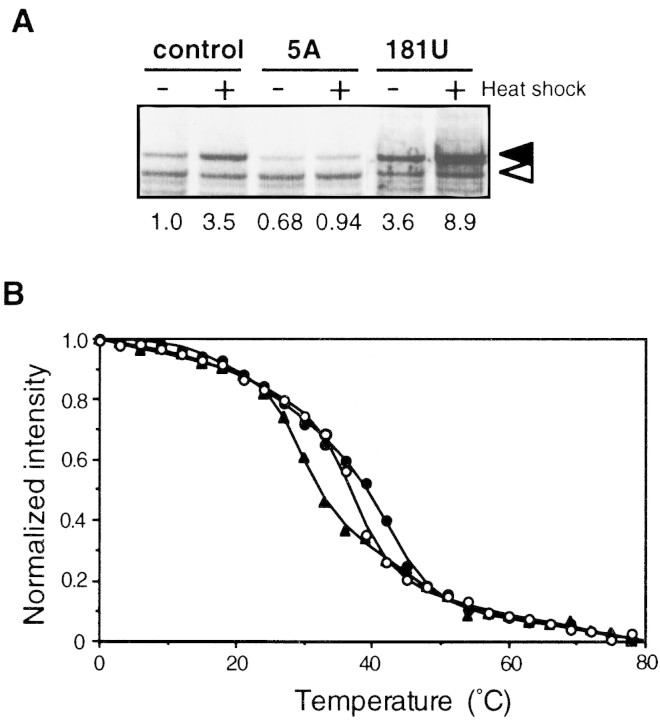
To examine whether expression levels in vivo are correlated with the actual thermostability of the mRNA, we measured the temperature profile of circular dichroism (CD) spectra using mRNA fragments derived from three representative constructs (control, 5A and 181U mutants). RNAs (∼160 nucleotides of rpoH) were prepared by in vitro transcription as described in Materials and Methods, and CD spectra were measured at various temperatures. The CD spectral patterns obtained below 25°C were similar to those of A-form RNA duplex indicating a certain folded structure, whereas the patterns at 81°C indicated denatured structure (data not shown). The melting curves based on the normalized intensities of CD spectra at 255 nm are presented in Figure 3B. The two mutant RNAs revealed significant differences from the control RNA and from each other that should have functional relevance. Whereas the 5A mutation, which reduced expression before and after temperature upshift, was expected to increase the stability of the mRNA structure, 181U, which enhanced expression at both 30°C and 42°C, was predicted to reduce the stability (Table 1 and Fig. 3A). Indeed, 5A-RNA was denatured at higher temperatures than the control RNA, whereas 181U-RNA was denatured at lower temperatures (Fig. 3B). Thus, the actual thermostability of these RNAs determined in vitro correlated well with the synthesis rate of fusion proteins from respective RNA under the conditions used.
Figure 3.
Expression and thermoregulation in vivo and mRNA thermostability in vitro of the minimal fusion constructs. (A) SDS-PAGE patterns of fusion proteins expressed from TLF229Δ(stemII)GG (control) and its derivatives with the super-repressed (5A) or derepressed (181U) phenotype (see Table 1). Cells were grown at 30°C, shifted to 42°C, and synthesis rates of fusion protein were analyzed as in Fig. 1. (Solid and open arrows) Fusion protein and β-galactosidase ω protein (internal reference), respectively. (B) Temperature profiles of CD spectra at 255 nm for the 5′ portion (nucleotides −20 to +229) of mRNA from each fusion. Higher intensity reflects folded structure, and lower intensity indicates melting of RNA duplexes. (○, ●, ▴) Control, 5A, and 181U, respectively. CD spectra were determined, and data were processed as described in Materials and Methods.
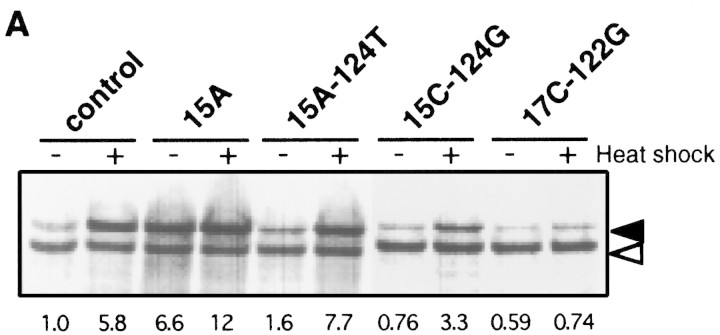
Because the structure of the minimal gene fusion used here was markedly different from the parental TLF247, we also examined thermostability of mRNA from TLF247 and its mutant derivatives. For the sake of clarity, heat-induced synthesis of fusion proteins from the representative gene fusions together with structural features of their RNAs (Morita et al. 1999) are indicated in Figure 4, A and B. The 15A mutation predicted to disrupt the 15G–124C base-pairing (Fig. 1B) enhanced expression at 30°C and reduced heat induction, whereas a pair of compensatory mutations (15A–124T) expected to restore the base-pairing showed recovery of almost normal thermoregulation (Fig. 4A). The somewhat higher expression of 15A–124T was presumably due in part to the slight instability of A–U pairing as compared with G–C (wild type) pairing. In addition, it should be noted that a mutation within region A can affect expression by altering complementarity to 16S rRNA (Fig. 1C; Nagai et al. 1991). Thus, the higher or the lower expression relative to wild type observed in the 15A–124T or 15C–124G double mutant, respectively, correlated well with the increased or decreased complementarity to 16S rRNA, respectively. The lower expression of 17C–122G may also be partially explained on the same basis. Consistent with the expression patterns observed in vivo, striking alterations in mRNA secondary structure of 15A RNA were found by means of chemical probing (Fig. 4B). In sharp contrast, no detectable alterations were found with 15A–124T RNA containing the compensatory mutations. Moreover, the 17C–122G double mutant predicted to form more stable C–G pairing (than the parental A–U pairing) exhibited reduced expression at 30°C and little or no induction on shift to 42°C. In agreement with the latter result, reverse transcriptase often stalled at the branchpoint of stems II and III even without chemical modification, indicating that stem II in this mutant RNA became more stable than in the parental TLF247 (Fig. 4B).
Figure 4.
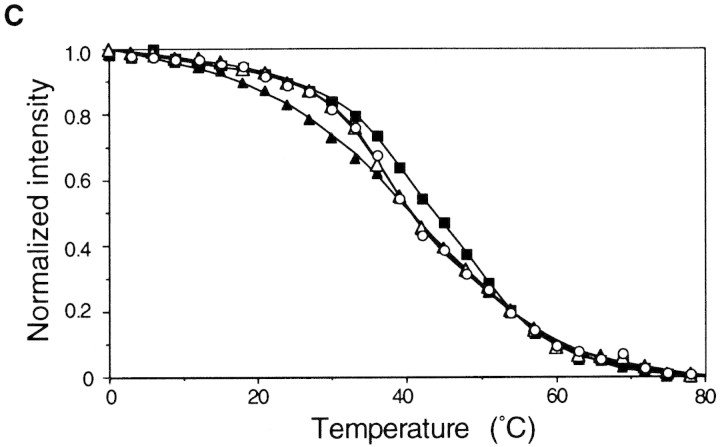
Expression, thermoregulation, and mRNA thermastability of native TLF247 fusion constructs. (A) SDS-PAGE patterns of fusion protein expressed from TLF247 or its mutant derivatives. Cells were grown at 30°C, shifted to 42°C, and synthesis rates of fusion protein were analyzed as in Fig. 1. Fusion proteins and internal references are indicated as in Fig. 3A. The values were normalized to that of the control at 30°C. (B) Summary of structural alterations detected for 15A and 17C–122G RNAs by chemical probing (Morita et al. 1999). Circled nucleotides indicate those modified more strongly in 15A than the control RNA by CMCT or DEP, which specifically modify unpaired nucleotides U/G or A, respectively. (Arrowheads) Nucleotides at which reverse transcriptase was often arrested in 17C–122G RNA. Initiation codon, region A and B, and SD sequence are indicated as in Fig. 1B. (C) Temperature profiles of CD spectra at 255 nm for the 5′ portion of the mRNA (∼290 nucleotides) of each fusion. (○) Control; (▴) 15A; (▵) 15A–124T; (█) 17C–122G. CD spectra were determined, and data were processed as in Fig. 3B.
To examine the possible correlation between the thermostability of TLF247 mRNAs determined in vitro and the results of chemical probing and in vivo expression analyses, we measured the thermostability of mRNA fragments (nucleotides −20 to +247) from wild-type control and mutant derivatives 15A, 15A–124T, and 17C–122G. As shown in Figure 4C, 15A RNA was denatured at lower temperatures (between 9°C and 39°C) as compared with the control, whereas 17C–122G RNA was denatured at significantly higher temperatures throughout the transition from folded to unfolded structures. On the other hand, the melting curve of 15A–124T RNA with compensatory mutations could be superimposed on that of the control. Thus, the relative thermostability of the mRNA of both the native and the minimal fusion constructs correlated well with the characteristic expression patterns of fusion proteins. In addition, the results were in good agreement with the mutational alterations of the mRNA secondary structure as detected by chemical probing analyses (Fig. 4B). Taken together, these results strongly support the notion that translational thermoregulation of the rpoH or rpoH–lacZ gene fusion depends primarily on the thermostability of the secondary structure of mRNA involving the translational initiation region.
The binding of 30S ribosomes to rpoH mRNA depends on high temperature
Given that the regulation of TLF247 expression is primarily dependent on the thermostability of the 5′ portion of rpoH mRNA, we addressed whether the stability of the RNA structure as well as in vivo expression are correlated with formation of translational initiation complexes. It was also of interest to determine whether any cellular factors such as RNA-binding proteins are involved in modulating translation. To examine the effects of mRNA secondary structure on ribosome binding, we analyzed the accessibility of 30S ribosomes to rpoH mRNA by means of primer extension inhibition, or toeprinting analysis (Hartz to al. 1988). A sample of wild-type rpoH mRNA fragment (−60 to +247) was first annealed to a fluorescent 5′-labeled primer and was mixed with 30S ribosomes and initiator tRNAfMet to allow formation of an mRNA–ribosome–rRNAfMet ternary complex. The extent of complex formation was estimated by reverse transcription reaction initiating from the primer hybridized downstream of the ribosome-binding site (nucleotides +227 to +247); the reaction was arrested effectively at or near the edge of the bound ribosome complex, producing a toeprint at position +16 (Fig. 5A).
Figure 5.
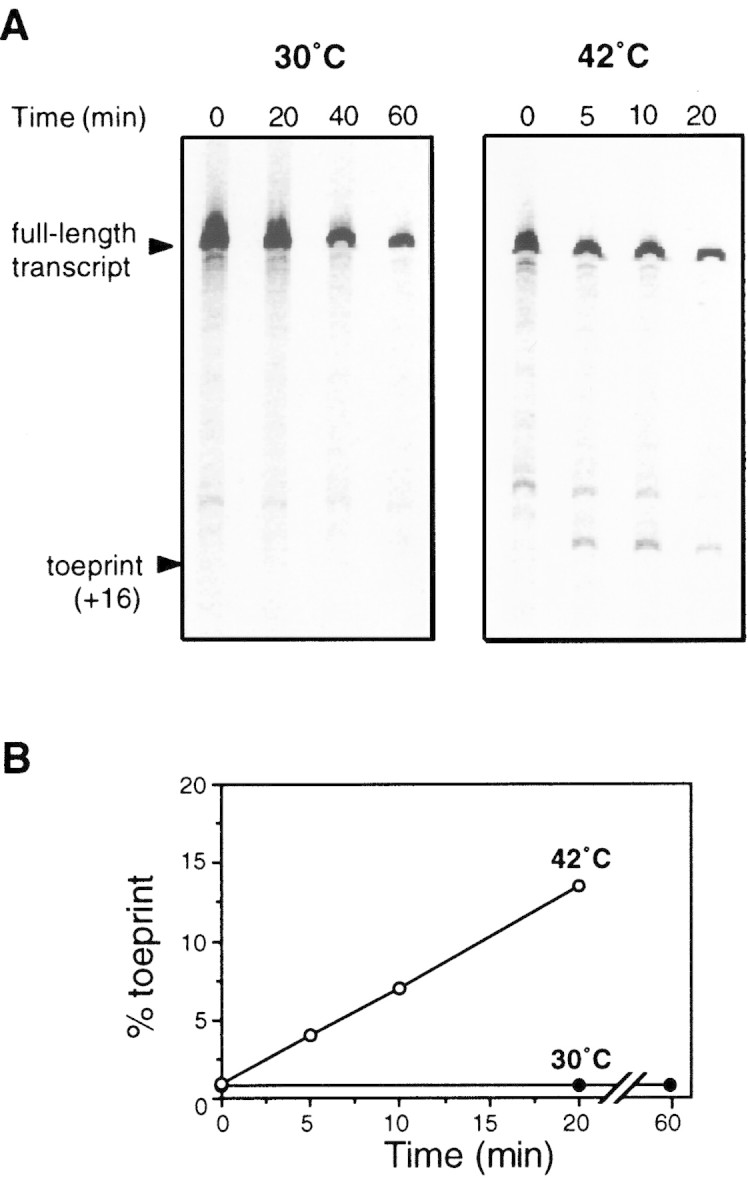
Ternary complex formation with 5′ portion of rpoH mRNA as detected by toeprint analysis. (A) Urea–PAGE patterns of products of primer extension analysis. Toeprint assays with wild-type rpoH mRNA (nucleotides −60–+247) were carried out at 30°C or 42°C as described in Materials and Methods. The positions of full-length transcript and toeprint at +16 (as judged by sequence ladder run simultaneously) are indicated. Incubation times indicated (top) were different at the two temperatures. (B) Toeprint and full-length signals were quantified as described in Materials and Methods. (% toeprint) Calculated as the intensity of the toeprint signal divided by the sum of toeprint and full-length signals multiplied by 100.
If a hypothetical cellular factor is required to repress rpoH translation at 30°C, toeprints may be expected to emerge at both 30°C and 42°C because the toeprint assay system should be free of such cellular components except for 30S ribosomes. On the other hand, if an activator is required for translation, little or no toeprint would be expected at either temperature. Neither of these predictions was supported by our experiments. Whereas no significant toeprint was detected at 30°C even on prolonged incubation (60 min), significant levels of toeprint appeared within 5 min at 42°C and increased steadily for at least 20 min (Fig. 5A,B). Thus, the formation of a ternary complex under these conditions was strongly prevented at 30°C apparently in the absence of any cellular factors other than purified 30S ribosomes. In marked contrast, ternary complexes formed rapidly and effectively at 42°C under the same conditions.
Mutations altering rpoH thermoregulation affect ribosome binding
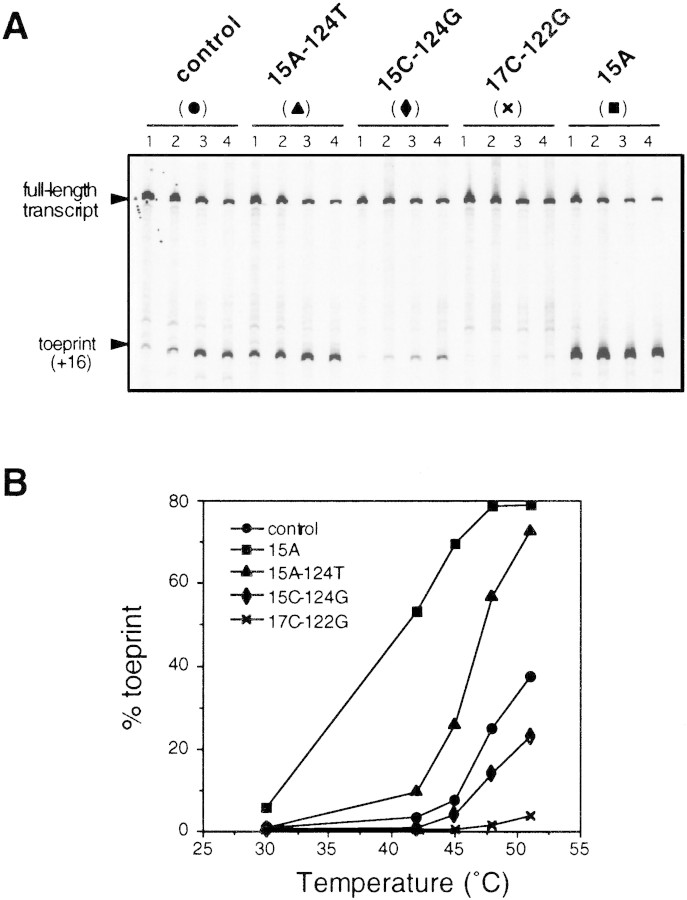
To examine the effects of mutations on ternary complex formation in vitro, toeprint analyses were carried out with RNA containing mutations altering expression or thermoregulation and thermostability (Fig. 4). In contrast to the results with wild-type RNA, 15A RNA with a less stable secondary structure rapidly yielded toeprints even at 30°C, and complex formation occurred much faster at 42°C (Fig. 6). The RNA containing two compensatory mutations (15A–124T) failed to form ternary complexes at 30°C as in the wild-type RNA, and toeprints emerged only at 42°C; complex formation was significantly faster with 15A–124T than with wild-type RNA, presumably reflecting both reduced thermostability and increased complementarity to 16S rRNA (Fig. 1C). In 15C–124G RNA predicted to form C–G instead of G–C pairing, complex formation occurred much more slowly than with wild-type RNA despite the equivalent base pairings involved. This result was not unexpected because the 15C–124G RNA has decreased complementarity to 16S rRNA and may affect the affinity to 30S ribosomes in toeprint analysis. No significant toeprint was detected with 17C–122G RNA at either temperature. Thus, the effects of these mutations on ternary complex formation correlated well with those on expression and thermoregulation of the rpoH–lacZ gene fusion in vivo. In addition, the relative rates of ternary complex formation with wild-type, 15A–124T, and 15C–124G RNA correlated closely with the synthesis rates of fusion protein from these constructs (Fig. 4A). These results indicate that the ternary complex formation observed here reflects, at least in part, translational initiation events in vivo. The results suggest that thermoregulation of σ32 synthesis may not require any specific cellular factors other than 30S ribosomes, although possible roles of factors contaminating the 30S ribosome preparation used cannot be excluded.
Figure 6.
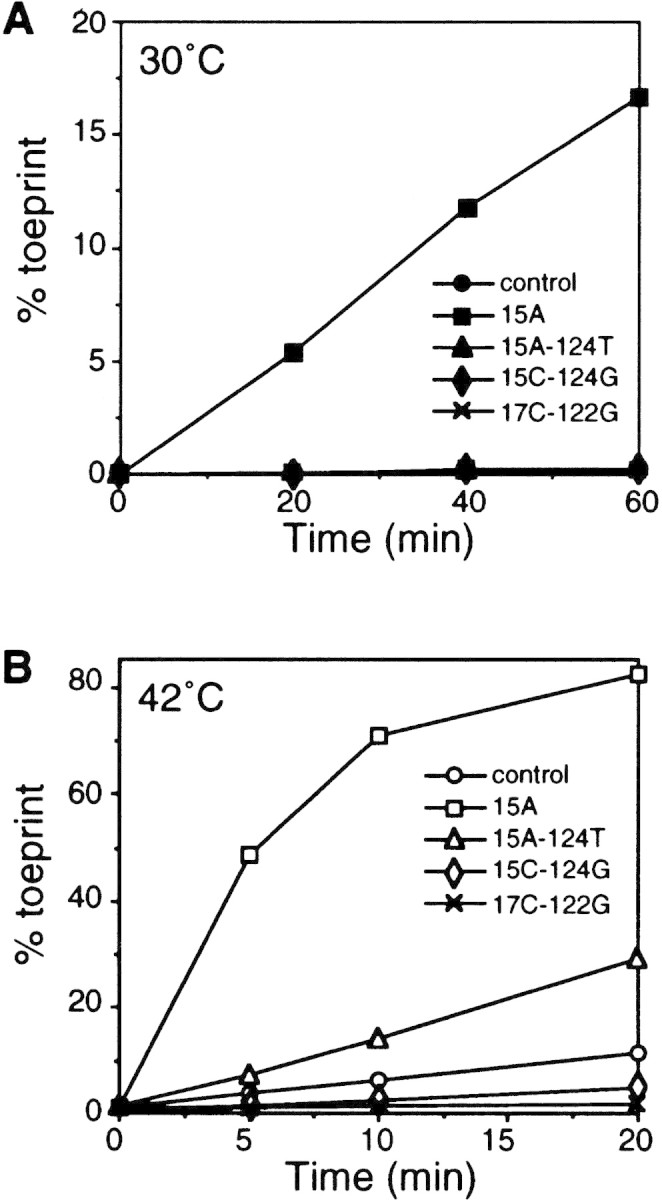
Mutations known to affect expression and thermoregulation in vivo also affect formation of the toeprint complex in vitro. Ternary complex formation with mutant mRNAs was performed at 30°C (A) or at 42°C (B). Toeprint and full-length signals were quantified. (% toeprint) Calculated as in Fig. 5B. Note that the values obtained with mutants other than 15A at 30°C are superimposed to that of the control in A.
Next, we carried out toeprint assays (5-min reaction) at even higher temperatures with wild-type and the same mutant RNAs (Fig. 7). As expected, ternary complex formation depended strongly on temperature, yielding “RNA melting curves” for the temperature range examined. The curves obtained for 15A and 15A–124T RNA were significantly shifted toward lower temperatures than with wild-type RNA, whereas that for 15C–124G RNA was shifted toward higher temperatures. Toeprints with 17C–122G RNA were barely seen at temperatures above 45°C. The temperature dependency of ternary complex formation in vitro correlated highly with that of expression observed in vivo (Figs. 4A and 7). These results strongly suggest that translational initiation complex formation is primarily controlled by the thermostability of the mRNA secondary structure formed at the translational initiation region. When such a folded structure becomes loose or unfolded on heat shock, ribosome entry and translational initiation would be increased, resulting in induction of σ32 synthesis. Single base substitutions affecting the thermostability of RNAs containing the translational initiation region can markedly affect the temperature sensitivity of the mRNA with respect to ribosome entry.
Figure 7.
Effects of temperature on ternary complex formation with control and mutant mRNAs. (A) Urea–PAGE patterns of products of primer extension analysis. Toeprint assays were carried out at 30°C (not shown), 42°C (lanes 1), 45°C (lanes 2), 48°C (lanes 3), or 51°C (lanes 4) for 5 min under standard conditions. The position of full-length transcript and toeprint at +16 are indicated. (B) Toeprint and full-length signals were quantified and the percent toeprint was calculated as in Fig. 5B. The values are plotted as a function of temperature.
Discussion
Here, we show that expression and heat induction of rpoH–lacZ gene fusions in vivo correlate well with both the predicted and actual thermostability of rpoH mRNA secondary structure involving part of the ribosome-binding site. Furthermore, accessibility of 30S ribosomes to RNA fragments carrying a specific mutation analyzed by toeprint assays was correlated well with the expression pattern of corresponding fusion constructs in vivo. In addition, distinct temperature dependency of ribosome binding to the wild-type RNA fragment was observed, suggesting that cellular factor(s) other than 30S ribosomes and initiator tRNA are not required for thermoregulation of σ32 synthesis, although the possible involvement of some other factors contaminating the 30S ribosome fraction cannot be excluded. These results, together with other considerations (see below), led us to propose that σ32 is a heat shock transcription factor with a unique thermosensing capacity built into its own mRNA.
The efficiency of translation in E. coli can be markedly affected at the stage of initiation by the secondary structure of mRNA containing the ribosome-binding site (Gold 1988; de Smit and van Duin 1990b). Such secondary structure can exert a regulatory role in translation by a variety of mechanisms (de Smit and van Duin 1990b; Springer 1996). In the case of rpoH, the initiation codon and region A are masked by region B located far downstream, forming a complex structure that consists of several stem–loops (Fig. 1B). de Smit and van Duin (1990a) observed a linear relationship between the log of synthesis rate and change in stability of a short RNA hairpin formed at the ribosome-binding site of the MS2 phage coat gene; expression decreases by 10-fold for each 1.4 kcal/mole increment in stability. We observed a similar relationship for expression of the minimal fusion construct (Fig. 2), although the effect of stability on expression was not as marked as that observed in the MS2 coat gene. Because the regulatory regions of rpoH are separated by ∼100 nucleotides of intervening coding sequence, RNA folding kinetics may also play some role in thermoregulation, as in the case of MS2 gene A whose regulatory regions (SD and its complement) are separated by 80 nucleotides, forming three stem–loop structures (Groeneveld et al. 1995; Poot et al. 1997). Consistent with this expectation, when mRNA folding was supposed to be completed as in toeprint assays, no binding of 30S ribosomes to RNA was detected at low temperature (Fig. 5). In living cells, transcription–translation coupling may facilitate a productive interaction between rpoH mRNA and ribosomes because of the expected delay in forming stem I structure due to the distance (∼180 nucleotides) separating the AUG codon from the inhibitory sequence. Such a time lag may ensure some basal level of expression at certain temperatures (e.g., 30°C) where σ32 is required for survival and growth.
Translational regulatory elements, that is, the downstream box and the mRNA secondary structure, are widely conserved among RpoH homologs from γ-proteobacteria (Nakahigashi et al. 1995). Moreover, several base-pairings in stem II (15–124, 16–123, 17–122) that are critical for thermoregulation in E coli (Yuzawa et al. 1993) are particularly well conserved. The possible involvement of trans-acting factor(s) that can bind to rpoH mRNA in a sequence-specific manner was suspected initially because of the unexpected phenotypes found with double mutants carrying compensatory mutations in this region; no heat induction in the 15C–124G mutant and little induction in the 16G–123C mutant (Yuzawa et al. 1993). It should be noted, however, that the mutations in this conserved segment of region A can markedly affect translation efficiency by altering complementarity to the antidownstream box of 16S rRNA (Fig. 1C). In addition, the 15C–124G mutant actually exhibited slightly reduced but appreciable heat induction (Morita et al. 1999). Meanwhile we performed extensive screening for factor(s) that could control rpoH translation and found several candidate genes that can reduce expression of the rpoH–lacZ gene fusion when expressed from multicopy plasmids. However, none of them affected authentic σ32 synthesis when the chromosomal genes were individually disrupted (H. Yuzawa and M.T. Morita, unpubl.). Recent results of chemical probing analyses supported the importance of 15–124 and 17–122 base pairings not only for complementarity to 16S rRNA but for forming RNA secondary structure with the appropriate stability (see Fig. 4B; Morita et al. 1999). Although heat-induced translation of rpoH mRNA is conserved within γ-proteobacteria, the extents of heat induction varied between three- and eightfold among different species (Nakahigashi et al. 1998). Interestingly, quantitatively similar extents of induction were observed when these homologs were expressed in E. coli, suggesting that the differential extents of induction might reflect differential stability of their rpoH mRNA secondary structures (Nakahigashi et al. 1998). Thus, the critical roles and strong conservation of the above base pairings appear to be explained by their involvement in forming mRNA secondary structure and in interaction with ribosomes. This should also provide a basis for the potentially high efficiency of translation required for the rapid response to heat shock stress.
Ethanol and nalidixic acid are known to increase σ32 levels, resulting in the induction of HSPs similar to those induced by heat shock (Straus et al. 1987; Mizushima et al. 1996). Ethanol causes both induction of synthesis and stabilization of σ32 (Straus et al. 1987 and unpublished results cited therein), but the mechanism of these effects remains unclear. Synthesis of TLF247 fusion protein was induced by 4% ethanol, although the transcriptional fusion OF-0 lacking the translational regulatory region of rpoH showed similar induction (M.T. Morita, unpubl.). When the promoter region of translational fusions was replaced by the trc or ara promoter (chimeric translational fusion), no induction by ethanol was observed despite the presence of a translational regulatory region. Therefore, induction of σ32 synthesis by ethanol appears to occur primarily at the transcriptional level. Essentially similar results were obtained with the heat shock response following treatment with nalidixic acid, using chimeric translational fusions (M.T. Morita unpubl.). Neither ethanol nor nalidixic acid seems to elicit translational induction: Heat shock appears to be the only stress condition that clearly causes translational induction of σ32 synthesis.
To our knowledge, two transcriptional regulators that can respond directly to high temperatures have been described: TlpA, an autoregulatory repressor protein of Salmonella typhimurium, and heat shock factor 1 (HSF1) of Drosophila (Hurme et al. 1997; Zhong et al. 1998). Both proteins exhibited temperature- and concentration-dependent conformational changes required for both multimerization and DNA binding. In addition, possible temperature sensing by structural changes in mRNA was suggested for the lcrF gene of Yersinia pestis, which encodes a transcription activator for induction of several virulence-related proteins (Hoe and Goguen 1993), although this conclusion was based solely on the calculated stability at various temperature of predicted hairpin structures.
In view of the known characteristics of translational initiation in prokaryotes, mRNA melting at higher temperature followed by translational initiation appears to provide a simple mechanism for rapid induction of critical regulators of the cell. There are other examples in which RNA structures containing SD were reported to be unfolded at higher temperature resulting in faster toeprint complex formation (Spedding and Draper 1993; Brunel et al. 1995). In the present system, melting of mRNA structure by heat was directly linked with the biological function of its product protein σ32. The results reported here do not exclude the involvement of trans-acting factor(s) that could regulate rpoH translation by interacting with mRNA only when it forms a specific structure. Even though such factors might modulate translation, RNA structure formation would be regarded as a primary determinant of this regulation. For bacteria or other unicellular organisms that often experience drastic changes in environmental temperature, thermoregulation of expression or activity of gene regulators by modulating the conformation of mRNA or protein should provide simple, rapid, and versatile mechanisms that should be of great advantage for their survival.
Materials and methods
Strains, phage, and media
E. coli K-12, strain MC4100 [araD Δ(argF-lac)U169 rpsL relA flbB deoC ptsF rbsR] (Casadaban 1976) was used for all experiments in vivo. The λTLF97-3 vector (St. Pierre and Linn 1996) was used to construct rpoH–lacZ gene fusions. Minimal medium M9 (Nagai et al. 1991) supplemented with 0.2% glucose, thiamine (2 μg/ml) and all amino acids except for methionine (20 μg/ml each) was used for pulse-labeling experiments. MacConkey lactose agar (DIFCO) and L agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; 30 μg/ml) were used to isolate λ lysogens containing rpoH–lacZ gene fusions. Recombinant DNA and other general techniques were performed as described by Sambrook et al. (1989) and by Miller (1972).
Chemical, enzymes, and buffers
E. coli uncharged tRNAfMet was purchased from Sigma, and avian myeloblastosis virus (AMV) reverse transcriptase was from Life Sciences. Buffer A was comprised of 10 mm Tris-acetate (pH 7.4), 60 mm NH4Cl, 6 mm β-mercaptoethanol; buffer B was comprised of buffer A to which 10 mm Mg-acetate was added. Reverse transcription buffer was buffer B containing 0.375 mm dNTPs and 0.1 U/μl of AMV reverse transcriptase.
Construction of rpoH–lacZ gene fusions
The gene fusion (translational fusion) TLF247, its derivatives carrying base substitutions, minimal gene fusion TLF229Δ(stemIII)GG, and plasmids used to construct these fusions [pBSK247 and derivatives; pBSK229Δ(stemIII)GG] have been described (Morita et al. 1999). A set of base substitutions of TLF229Δ(stemIII)GG was constructed by use of the ExSite PCR-based site-directed mutagenesis kit (Stratagene) with synthetic oligonucleotide primers carrying base substitutions and pBSK229Δ(stemIII)GG as the DNA template. ClaI–BamHI fragments of PCR-amplified DNAs were cloned into pBSK229Δ(stemIII)GG by replacing the corresponding region. Nucleotide sequences of PCR amplified regions were confirmed by dideoxy sequencing. The XhoI–BamHI fragments of the resulting plasmids containing rpoH promoters and the 5′ portion of the coding region were then inserted into the λTLF97-3 vector to make a fusion in-frame to lacZ. The resulting gene fusion constructs were transferred to MC4100 by in vitro packaging followed by infection. After repeated single colony isolation, monolysogeny of each construct was confirmed by PCR as described by Powell et al. (1994).
Prediction of RNA secondary structure
RNA secondary structures and their stability (ΔG) at 30°C and 42°C were predicted by use of the mfold program provided by Zuker and Turner on the internet (http://www.ibc.wustl.edu/zuker/rna).
Determination of synthesis rates of fusion proteins
The procedure used was essentially as described previously (Nagai et al. 1994). Portions (0.1 ml) of log phase cultures were pulse-labeled with l-[35S]methionine (1200 Ci/mmole, 200 μCi/ml) for 2 min. Extracts were prepared after TCA precipitation and suspension in SDS buffer, and portions with equal radioactivity were mixed with a fixed amount of JM103 cell extract (labeled with [35S]methionine) containing β-galactosidase θ protein, and treated with antibody against β-galactosidase (Organon Teknika Cappel). The immunoprecipitates were subjected to SDS-PAGE (7.5% gel), and intensities of radioactive bands were quantified by use of a Fujix BAS2000 imaging analyzer to determine synthesis rates of fusion protein after correcting for recovery with θ protein as a reference.
Measurement of temperature profiles of mRNA CD spectra (melting curve)
RNAs containing the upstream and part of the rpoH coding region were prepared in vitro by T7 RNA polymerase and a MEGAscript Kit (Ambion). The EcoRV (positioned at −22)–BamHI fragments of pBSK247, pBSK229Δ(stemIII)GG, or their derivatives carrying mutations were placed under the control of the T7 promoter of the pSP72 vector. The resulting plasmids were digested with BamHI and used as templates for RNA synthesis. All samples for CD measurements were desalted by use of a reverse-phase column (COSMOSIL, Nacalai Tesque) on an HPLC apparatus and quantified by UV absorbance at 260 nm after P1 nuclease digestion. The pH of solution for CD spectra was buffered with 25 mm sodium phosphate (pH 7.0). The concentration of RNA varied from 0.05 to 9 μm for the control RNA derived from pSP247, and from 0.3 to 3 μm for other RNAs. Temperature profiles of CD spectra were measured by use of a Jasco J-720WI spectropolarimeter equipped with a Pertier type temperature controller, PTC-348WI. CD spectra were acquired from 220 to 320 nm at 0°C–81°C (every 3°C) with a scanning speed of 50 nm/min, time constant of 1 sec, spectral band width of 1.0 nm and seven scans for each temperature. Temperature varied with heating (up-scan) or cooling (down-scan). The CD spectra at each temperature were found to be reproducible in several experiments. Temperature profiles of CD spectra were recorded for solutions at different concentrations to determine whether the denaturation processes of all the samples were unimolecular processes. The results were consistent with each other within a range of noise level as assumed as an unimolecular process. The CD spectra at an RNA concentration of 3 μm (down-scan) were processed further as described (Kodama et al. 1998). Each signal intensity at 255 nm was plotted against temperature, and curve fitting for the above data was done as described (Petersheim and Turner 1983; Kodama et al. 1997) assuming a three-state equilibrium, as two transitions were observed (data not shown). Then, the CD signal intensity at 255 nm for each temperature was extracted and normalized by use of the signal intensities at 0°C and 81°C.
Primer extension inhibition (toeprint) assays
RNA containing the upstream and part of the rpoH coding region (nucleotides −60 to +247) was prepared in vitro by T7 RNA polymerase with an RNA transcription kit (Stratagene). The AflII–BamHI fragments of pBSK247 were placed under the control of the T7 promoter of the pSP72 vector, and the resulting plasmids were digested with BamHI and used as a template for RNA synthesis. The formation of a ternary complex composed of the 30S ribosomal subunit, target mRNA and tRNAfMet, and extension inhibition analyses were carried out basically according to Hartz to al. (1988). Purified samples of 30S ribosomes were kindly provided by Dr. A. Wada (Osaka Medical College; Wada 1986). RNAs (100 nm) were renatured in the presence of a fluorescent 5′ labeled primer FAM1p (1 μm) complementary to nucleotides from +227 to +247 by heating the mixture at 65°C for 3 min followed by slow cooling to room temperature in 10 μl of buffer A. Then, Mg-acetate was added to the renatured RNA mixture at a final concentration of 10 mm. The reaction mixture (10 μl) contained 4 μl of renatured RNA, 0.375 mm dNTPs, 500 nm 30S ribosome, and 1 μm uncharged initiator tRNA in buffer B. After incubation for 5–60 min at the indicated temperatures, the reactions were terminated by addition of 190 μl of reverse transcription buffer (20-fold dilution; see below). Ternary complex formation was estimated quantitatively by performing cDNA synthesis at 42°C for 30 min. Samples were treated with proteinase K and ribonuclease A, and the product DNA was extracted with phenol/chloroform followed by ethanol precipitation. Portions of primer extension products were loaded onto a 5% polyacrylamide/8 m urea sequencing gel, and electrophoresed at 1400 V for 2.5 hr. Primer extension products were quantified with a Fluorescence BioImage Analyzer FMBIO II Multi-View (Hitachi). The positions of toeprints were determined by comparison with sequence ladder run simultaneously with the same end-labeled primers.
As region B of rpoH mRNA involved in repressing translation is located far downstream from the toeprint position, we wished to avoid possible complications arising from toeprint complexes formed during incubation for reverse transcription. Thus, the toeprint assay was carried out in two steps; complex formation and reverse transcription. After complex formation, the reaction mixture was diluted 20-fold, which effectively prevented formation of further complexes. We detected no ternary complex formation when mRNA and 30S subunits were mixed at such low concentration (data not shown). Once ternary complex was formed during the first incubation, the complex remained stable and was hardly dissociated during the second incubation (data not shown).
Acknowledgments
We are grateful to M. Springer for helpful discussion, to A. Wada for kindly donating 30S ribosome, and K. Watanabe for tRNAfMet. We also thank H. Kubota and K. Nakahigashi for kind advice, and M. Nakayama, and H. Kanazawa for technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tyura@hsp.co.jp; FAX (81)-75-315-8659.
References
- Brunel C, Romby P, Sacerdot C, de Smit M, Graffe M, Dondon J, van Duin J, Ehresmann B, Ehresmann C, Springer M. Stabilised secondary structure at a ribosomal binding site enhances translational repression in E. coli. J Mol Biol. 1995;253:277–290. doi: 10.1006/jmbi.1995.0552. [DOI] [PubMed] [Google Scholar]
- Bukau B. Regulation of the Escherichia coliheat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coliusing bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Craig EA, Gross CA. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: A quantitative analysis. Proc Natl Acad Sci. 1990a;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit MH, van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990b;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coliand the autoregulation of the heat shock response. In: Morimoto RI, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 209–249. [Google Scholar]
- Goff SA, Goldberg AL. Production of abnormal proteins in E. coli stimulates transcription of Ionand other heat shock genes. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Groeneveld H, Thimon K, Van Duin J. Translational control of maturation-protein synthesis in phage MS2: A role for the kinetics of RNA folding? RNA. 1995;1:79–88. [PMC free article] [PubMed] [Google Scholar]
- Gross CA. Function and regulation of the heat shock proteins. In: Neidhardt FC, Curtis III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- Grossman AD, Straus DB, Walter WA, Gross CA. σ32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes & Dev. 1987;1:179–184. doi: 10.1101/gad.1.2.179. [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Hoe NP, Goguen JD. Temperature sensing in Yersinia pestis: Translation of the LcrF activator protein is thermally regulated. J Bacteriol. 1993;175:7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurme R, Berndt KD, Normark SJ, Rhen M. A proteinaceous gene regulatory thermometer in Salmonella. Cell. 1997;90:55–64. doi: 10.1016/s0092-8674(00)80313-x. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Gross CA. Translational regulation of σ32synthesis: Requirement for an internal control element. J Bacteriol. 1991;173:3904–3906. doi: 10.1128/jb.173.12.3904-3906.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemori M, Mori H, Yura T. Induction of heat shock proteins by abnormal proteins results from stabilization and not increased synthesis of σ32 in Escherichia coli. J Bacteriol. 1994;176:5648–5653. doi: 10.1128/jb.176.18.5648-5653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama TS, Kyogoku Y, Sugeta H. A novel method for estimating thermodynamic parameters in multi-equilibrium system by global analysis of spectrophotometric data. In: Carmona P, Navarro R, Hernanz A, editors. Spectroscopy of biological molecules: Modern trends. The Netherlands: Kluwer Academic Publishers; 1997. pp. 577–578. [Google Scholar]
- Kodama TS, Tanaka Y, Yasuno K, Teruya K, Oda N, Sugeta H, Kyogoku Y. Systematic errors of CD spectra in thermodynamic analysis of nucleic acids. Nucleic Acids Symp Ser. 1998;39:61–62. [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mizushima T, Ohtsuka Y, Mori H, Miki T, Sekimizu K. Increase in synthesis and stability of σ32 on treatment with inhibitors of DNA gyrase in Escherichia coli. Mol & Gen Genet. 1996;253:297–302. doi: 10.1007/pl00008596. [DOI] [PubMed] [Google Scholar]
- Morimoto RI, Jurivich DA, Kroeger PE, Msthr SK, Murphy SP, Nakai A, Sarge K, Abravaya K, Sistonen LT. Regulation of heat shock gene transcription by a family of heat shock factors. In: Morimoto RI, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Morita M, Yanagi H, Yura T. Heat induced synthesis of σ32 in Escherichia coli: Structural and functional dissection of rpoHmRNA secondary structure. J Bacteriol. 1999;181:401–410. doi: 10.1128/jb.181.2.401-410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Yuzawa H, Yura T. Interplay of two cis-acting mRNA regions in translational control of σ32 synthesis during the heat shock response of Escherichia coli. Proc Natl Acad Sci. 1991;88:10515–10519. doi: 10.1073/pnas.88.23.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Yuzawa H, Kanemori M, Yura T. A distinct segment of the σ32 polypeptide is involved in DnaK-mediated negative control of the heat shock response in Escherichia coli. Proc Natl Acad Sci. 1994;91:10280–10284. doi: 10.1073/pnas.91.22.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding σ32homologs from gram negative bacteria; Conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 1995;23:4383–4390. [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi K, Yanagi H, Yura T. Regulatory conservation and divergence of σ32 homologs from gram-negative bacteria: Serratia marcescens, Proteus mirabils, Pesudomonas aeruginosa, and Agrobacterium tumefaciens. J Bacteriol. 1998;180:2402–2408. doi: 10.1128/jb.180.9.2402-2408.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersheim M, Turner DH. Base-stacking and base-pairing contributions to helix stability: Thermodynamics of double-helix formation with CCGC, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- Poot RA, Tsareva NV, Boni IB, Van Duin J. RNA folding kinetics regulate translation of phage MS2 maturation gene. Proc Natl Acad Sci. 1997;94:10110–10115. doi: 10.1073/pnas.94.19.10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell BS, Court DL, Nakamura Y, Rivas MP, Turnbough CL., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Spedding G, Draper D. Allosteric mechanism for translational repression in the E. coliα operon. Proc Natl Acad Sci. 1993;90:4399–4403. doi: 10.1073/pnas.90.10.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengart ML, Fatscher HP, Fuchs E. The initiation of translation in E. coli: Apparent base pairing between the 16S rRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990;18:1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. Translational control of gene expression in E. coli and bacteriophage. In: In: Lin ECC, Simon Lynch A, editors. Regulation of gene expression in E. coli and Bacterophage. Austin, TX: R.G. Landes Company; 1996. pp. 85–126. [Google Scholar]
- St. Pierre R, Linn T. A refined vector system for the in vitro construction of single-copy transcriptional or translational fusions to lacZ. Gene. 1996;169:65–68. doi: 10.1016/0378-1119(95)00787-3. [DOI] [PubMed] [Google Scholar]
- Straus DB, Walter WA, Gross CA. The heat shock response of E. coli is regulated by changes in the concentration of σ32. Nature. 1987;329:348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- Straus D, Walter W, Gross CA. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of σ32. Genes & Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- Tilly K, McKittrick N, Zylicz M, Georgopoulos C. The dnaK protein modulates the heat-shock response of Escherichia coli. Cell. 1983;34:641–646. doi: 10.1016/0092-8674(83)90396-3. [DOI] [PubMed] [Google Scholar]
- Tilly K, Spence J, Georgopoulos C. Modulation of stability of the Escherichia coliheat shock regulatory factor sigma 32. J Bacteriol. 1989;171:1585–1589. doi: 10.1128/jb.171.3.1585-1589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A. Analysis of Escherichia coliribosomal proteins by an improved two dimensional gel eletrophoresis. I. Detection of four new proteins. J Biochem. 1989;100:1583–1594. doi: 10.1093/oxfordjournals.jbchem.a121866. [DOI] [PubMed] [Google Scholar]
- Wild J, Walter WA, Gross CA, Altman E. Accumulation of secretory protein precursors in Escherichia coliinduces the heat shock response. J Bacteriol. 1993;175:3992–3997. doi: 10.1128/jb.175.13.3992-3997.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- Yuzawa H, Nagai H, Mori H, Yura T. Heat induction of σ32 synthesis mediated by mRNA secondary structure: A primary step of the heat shock response in Escherichia coli. Nucleic Acids Res. 1993;21:5449–5455. doi: 10.1093/nar/21.23.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by Drosophilaheat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]