Abstract
Enterotoxigenic Escherichia coli (ETEC) is an important cause of diarrhea. Three adhesins (Tia, TibA, EtpA), an iron acquisition system (Irp1, Irp2, and FyuA), a GTPase (LeoA), and an autotransporter (EatA) are ETEC virulence-related proteins that, in contrast to the classical virulence factors (enterotoxins and fimbrial colonization factors) have not heretofore been targets in characterizing isolates from epidemiological studies. Here, we determined the occurrence of these nonclassical virulence genes in 103 ETEC isolates from Chilean children with diarrhea and described their association with O serogroups and classical virulence determinants. Because tia, leoA, irp2, and fyuA are harbored by pathogenicity islands inserted into the selC and asnT tRNA genes (tDNAs), we analyzed the regions flanking these loci. Ten additional tDNAs were also screened to identify hot spots for genetic insertions. Associations between the most frequent serogroups and classical colonization factor (CF)-toxin profiles included O6/LT-STh/CS1-CS3-CS21 (i.e., O6 serogroup, heat-labile [LT] and human heat-stable [STh] enterotoxins, and CFs CS1, -3 and -21), O6/LT-STh/CS2-CS3-CS21, and O104-O127/STh/CFAI-CS21. The eatA and etpA genes were detected in more than 70% of the collection, including diverse serogroups and virulence profiles. Sixteen percent of the ETEC strains were negative for classical and nonclassical adhesins, suggesting the presence of unknown determinants of adhesion. The leuX, thrW, and asnT tDNAs were disrupted in more than 65% of strains, suggesting they are hot spots for the insertion of mobile elements. Sequences similar to integrase genes were identified next to the thrW, asnT, pheV, and selC tDNAs. We propose that the eatA and etpA genes should be included in characterizations of ETEC isolates in future epidemiological studies to determine their prevalence in other geographical regions. Sequencing of tDNA-associated genetic insertions might identify new ETEC virulence determinants.
INTRODUCTION
Enterotoxigenic Escherichia coli (ETEC) causes nearly 400 million diarrhea episodes every year in children younger than 5 years of age and is responsible for approximately 50% of all traveler's diarrhea episodes (35). ETEC colonizes the small-bowel epithelium, producing heat-labile (LT) and/or heat-stable (ST) enterotoxins (7) that cause secretion of water and electrolytes. Toxin variants LT-I and STh are produced only by strains that infect humans. LT-II and STp are expressed by strains that infect mostly piglets, although they occasionally also infect humans (20).
Other classical ETEC virulence determinants are the colonization factors (CFs), also referred to as coli surface antigens (20), adhesins that direct colonization of the small bowel epithelium. Currently, 22 different putative ETEC CFs have been identified, designated CS followed by a number depending on their order of discovery, with the exception of CFAI, which maintains its original denomination (10, 31). The most prevalent CFs associated with the occurrence of diarrhea worldwide are CFAI, CS1, CS2, CS3, and CS6; more recently CS8, CS14, and CS21 have been detected with relatively high frequencies (30). Nevertheless, a significant proportion of ETEC isolates obtained from individuals with diarrhea are CF negative, suggesting that additional, yet-unknown adhesion proteins remain to be identified (31, 36).
Inactivated enterotoxins, CFs such as CFAI and CS1 to -6, and attenuated and inactivated strains harboring one or more of these virulence traits have been used as vaccine candidates in clinical studies (30, 35) but, heretofore, none of them has yielded results in clinical trials sufficiently robust to foster completion of clinical development. An alternative approach to produce a vaccine would be to focus on ETEC antigens other than enterotoxins.
Other virulence-related proteins have been identified from the prototype strain ETEC H10407 (O78:H11/LTI-STh-STp/CFAI). These “nonclassical” virulence factors include adhesins and others proteins with diverse functions. Tia and TibA, two outer-membrane proteins of 26 and 104 kDa, respectively, have been shown to mediate adhesion to and invasion of the colonic epithelial cell line HCT-8 (9, 18). Isogenic mutants of both tia and tibA genes show reduced ability to adhere to and enter cells (8). EtpA is a 177-kDa protein involved in adhesion, secreted by the two-partner apparatus EtpBC encoded by the same plasmid that harbors genes for toxins and CFAI in strain H10407 (12). EtpA binds to the flagellum tip, and this complex mediates adherence to HCT-8 and Caco-2 epithelial cells (26). EatA is a 110-kDa autotransporter belonging to the serine protease autotransporters of Enterobacteriaceae (SPATE family). Although its role in ETEC pathogenesis remains unclear, rabbits infected with an isogenic H10407 eatA mutant show less fluid accumulation compared to wild-type-infected controls (22). LeoA is a cytoplasmic protein with GTPase activity, required for maximal LT secretion. An isogenic H10407 leoA mutant strain produces smaller amounts of LT membrane vesicles and shows less fluid accumulation in the rabbit ileal loop model than those of wild-type controls (5, 11). The gene coding for LeoA is located in a 46-kb pathogenicity island (PAI) that also carries the tia gene. This PAI, known as Tia-PAI, is inserted next to the 3′ end of selC, the tRNA gene (tDNA) coding for selenocysteine tRNA (11). Finally, the high-pathogenicity island (HPI) which carries genes encoding the yersiniabactin iron scavenging system has been detected in ETEC isolates. HPI is inserted next to asnT tDNA. The irp2 and fyuA genes that encode proteins involved in the synthesis of yersiniabactin and its receptor, respectively, have been detected in some diarrheagenic E. coli, including one ETEC clinical isolate (28).
The above-described proteins and coding sequences have been detected with different frequencies in collections of ETEC strains but mainly in works that reported their discovery or their functional analysis (except for the HPI genes that were discovered in other bacterial species). To our knowledge, this broad group of genes associated with ETEC virulence has not been systematically evaluated in epidemiological studies.
Our aim was to detect these nonclassical virulence genes among ETEC isolates from clinical cases obtained in Chile during a large epidemiologic surveillance study performed during the 1980s. The presence of these nonclassical virulence genes was correlated with O-antigen serogroup and classical virulence genes (enterotoxins and CFs); genetic relationships among isolates were established by multilocus variable number of tandem repeats (VNTR) analysis (MLVA). Furthermore, as an approach to identify new potential loci encoding ETEC virulence factors, we performed a screening of hot spots for insertion of genetic elements by analyzing the integrity of 12 tDNA genetic contexts frequently used as insertion sites for known genomic islands in E. coli strains. Different categories of pathogenic E. coli strains bear important virulence genes in genetic chromosomal insertions as PAIs and prophages associated with tDNAs. To date, Tia-PAI and HPI are the only PAIs in ETEC strains, but it is possible that other unknown loci could be contributing to their virulence. We therefore intended to identify tDNA hot spots for insertion of genetic elements in ETEC clinical strains as the first step in the identification of new chromosomal virulence genes.
MATERIALS AND METHODS
Bacterial isolates.
One hundred three ETEC isolates collected from children with diarrhea during an epidemiologic surveillance study performed in 1985 in a peri-urban community of Santiago, Chile, were studied (16). Isolates were identified as E. coli by standard biochemical tests and as ETEC by detection of genes encoding LT (corresponding to LTI), STp, and STh.
Serogroup and MLVA.
The O antigen was determined as described by Guinée et al. (14) using antisera that identify O antigen serogroups O1 to O185. Isolates that did not react with O antisera were classified as nontypeable (ONT). All antisera were obtained and adsorbed with the corresponding cross-reacting antigens to remove nonspecific agglutinins. For MLVA, seven different polymorphic chromosomal repeats (CVN001, -002, -003, -004, -007, -014, and -015) were amplified by PCR (17). Products were analyzed by gel electrophoresis with 3% agarose, and sizes were determined with Gel Compar II software (Applied Maths). Allele numbers were established according to product size ranges, including 11 different alleles for CVN001, 18 for CVN002, 13 for CVN003, 15 for CVN004, six for CVN007, 20 two for CVN014, and 4 for CVN015. In addition, seven nonreported alleles assigned to CVN004 (ranging between 404 and 506 bp), one to CVN007 (419 to 435 bp), one to CVN0014 (114 to 118 bp), and one to CVN0015 (243 to 253 bp) were included. Different alleles for each isolate were represented as a numerical matrix, and a dendrogram was constructed using the unweighted-pair group method using average linkages (UPGMA) with Treecon software version 1.3b (33).
Classical virulence genes.
Multiplex PCR was performed for detection of classical virulence genes, using previously described primers: LT, STh, STp, CFAI, CS1, CS2, CS3, CS4, and CS6 (34); CS5, CS20, and CS21 (24). New primers were designed for CS7, CS12, CS17, and CS19 (see Table S1 in the supplemental material). The following reference strains were used as positive controls: ETEC H10407 (CFAI), E24377A (CS1, CS3), C91F (CS2), 1181c (CS4, CS6), and 75688 (CS5, CS6). For detection of CS7, CS12, CS17, CS19, CS20, and CS21, Chilean ETEC isolates 15758a, 14154a, 3148a, 3232c, E275a, and 3382a1 were used, respectively, as positive controls after confirmation by PCR product sequencing.
Nonclassical virulence genes.
Genes encoding Tia (GenBank accession number U20318), LeoA (AF170971), TibA (AF109215), EatA (AY163491), EtpB (AY920525, nucleotides 817 to 2628), EtpA (AY920525, nucleotides 2718 to 8021), Irp2 (NC_003143, nucleotides 2156049 to 2162156), and FyuA (NC_003143.1, nucleotides 2140840 to 2142861) were detected by PCR using primers listed in Table S1 in the supplemental material. Primers were designed using Primer3 software (version 0.4.0; available at http://frodo.wi.mit.edu/primer3/) and evaluated with OligoAnalyzer version 1.0.2. PCR mixes were composed of 1× reaction buffer, 1.5 mM MgCl2, 0.8 mM dNTPs, 10 pmol primers, and 0.6 U Taq DNA polymerase. All reactions were performed with purified genomic DNA as template. Cycling conditions included denaturation for 30 s at 94°C, annealing for 30 s at 55°C, and extension for 60 s at 72°C. ETEC H10407 was included as a positive control. Products were analyzed by agarose gel electrophoresis and stained with ethidium bromide.
Genomic island insertion site screening.
The integrity of the selC, pheU, pheV, thrW, leuX, asnT, asnU, asnV, ansW, argU, metV, and aspV tDNAs was analyzed by PCR using primers listed in Table S2 in the supplemental material. Primers were directed against genes flanking each tDNA using the genomic sequence of Escherichia coli K-12 in coliBase as a reference (http://xbase.bham.ac.uk/colibase/). The sequences were aligned using the ClustalW2 server to design primers for conserved regions. Products were analyzed by agarose gel electrophoresis, and results were interpreted as follows. A tDNA was considered intact if a PCR product of a size similar to that predicted for E. coli K-12 was obtained. A tDNA was considered disrupted when no such product was obtained. The same analysis was carried out with the ETEC E24377A genome sequence (J. Craig Venter Institute, MD).
Primer walking.
Sequences downstream of tDNAs were determined by primer walking using a Genome walker universal kit (Clontech), according to the manufacturer's instructions. Final products were purified, cloned in pGEM-T Easy vector and sequenced using T7 and SP6 universal primers that flank the multiple cloning site. Sequences were compared with genomic databases using the blastn algorithm of BLAST.
Nucleotide sequence accession numbers.
Sequences from this study were deposited in GenBank under accession numbers JF421572 (thrW), JF421569 (asnT), JF421570 (pheV), and JF421571 (selC).
RESULTS
O serogroups, MLVA clusters, and classical virulence genes of ETEC isolates.
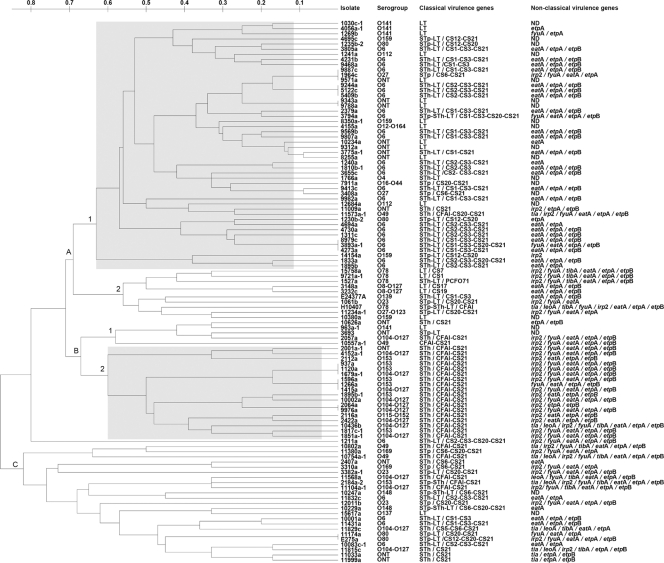
The 103 ETEC isolates belonged to 21 different O serogroups. However, 51 (50%) belonged to one of three serogroups: O6 (28.2%), O104/O127 (13.6%) (these isolates appeared to express both O104 and O127 antigens), or O153 (7.8%). Fifteen less-common serogroups were identified, and 14 isolates were ONT (13.6%). Strains harboring the most prevalent O antigens and classical virulence factors fell mainly into two major clusters, according to MLVA; each cluster included strains sharing about 40% similarity (Fig. 1). Cluster A1 contained 19 O6/ST1h-LT+ strains associated with CS1-CS3-CS21 or CS2-CS3-CS21 profiles, whereas cluster B contained 17 O104-O127/STh+ or O153/STh+ strains associated with a CFAI-CS21 profile. The STh gene, carried by 65 strains (63.1%), constituted the most common enterotoxin type among these ETEC isolated from cases of diarrhea. In this study, we included the strain 10557a-1 that was negative for all three enterotoxin genes by PCR, because it had been classified as ETEC according to enterotoxin genes detection by DNA hybridization in a previous study (16) and harbors CFAI and CS21. It is not uncommon that ETEC spontaneously loses these enterotoxin genes upon storage and subculture (29).
Fig. 1.
Characterization of ETEC isolates according to O serogroup and classical and nonclassical virulence genes. All isolates were named, and their serogroup and profiles of virulence genes are shown beside them. A dendrogram showing genetic relationships between strains was built based on MLVA. Genetic distance is shown above the tree. Reference ETEC strains H10407 (O78, STh-STp/CFAI) and E24377A (O139, STh-LT/CS1-CS3) carrying known features were included. Shaded squares show the isolates carrying the most prevalent features. ONT, nontypeable O serogroup; ND, not detected.
By far the most common CF detected was CS21, carried by 74 strains (71.8%); while 18 (17.5%) of the isolates that carried enterotoxin genes were negative for CFs. Strains from less frequently encountered serogroups and classical virulence gene profiles were grouped mainly in clusters A2, B1, and C.
Detection of nonclassical virulence genes.
The nonclassical ETEC virulence genes were detected with different frequencies among 83 strains (80.6%) within the collection (Fig. 1). Among the remaining 20 strains, 15 were CF negative (14.6%). Strains carrying Tia-PAI-associated genes were uncommon. Only 9 strains (8.7%) were positive for the tia gene and 6 for the leoA gene (5.8%), the majority grouped in branch C, which carried the STh/CFAI-CS21 profile (Fig. 1). The irp2 and fyuA HPI genes were detected in 39 (37.9%) and 37 strains (35.9%), respectively, including serogroups O104-O127 and O153 and the STh/CFAI-CS21 profile from branches B2 and C. The adhesin/invasin encoded by the tibA gene was detected in 11 strains (10.7%) from three different O serogroups (O78, O49, and O104-O127), 7 of them carrying the STh/CFAI-CS21 profile. In contrast, the eatA (70.9%), etpA (74.8%), and etpB (62.1%) genes were widely distributed and were present in strains from 13 different O serogroups and from strains with almost all of the identified virulence factor profiles. Only 2 strains negative for CF genes were positive for nonclassical adhesin-encoding genes, whereas 16 strains were negative for all the adhesin genes tested for in this study (15.5%). Overall, there were 15 strains (14.6%) positive for enterotoxin genes but negative for any other ETEC virulence gene, including CFs and nonclassical virulence genes. The majority of them were associated with the LT profile in cluster A1 (Fig. 1).
Screening of tDNA context integrity.
In contrast to the E. coli K-12 genome, every ETEC isolate in our collection showed disruption of at least 1 of the 12 tDNAs included in this study, with typically from 4 to 5 disrupted genes in each strain. The leuX, thrW, and asnT tDNAs were disrupted in more than 65% of the isolates (Table 1). The pheV and selC tDNAs were changed mainly in strains from cluster A1 (30 strains for the pheV gene and 23 for the selC gene), whereas the argU gene was disrupted in cluster B2 (14 strains) and non-O6 strains from cluster A1 (12 strains). In the case of the asnV gene, disruption was associated with clusters B2 (11 strains) and C (9 strains). In representative strains, we were able to identify sequences with high identity to integrase genes, next to the 3′ end of the thrW, asnT, pheV, and selC genes (see Table S3 in the supplemental material). We were unable to obtain products suitable for sequencing from leuX using genome walking. tDNAs pheU, asnU, asnW, metV, and aspV were intact in more than 85% of the strains in the collection (Table 1).
Table 1.
Analysis of tDNA context integrity
| tDNA | No. (%) of strains with disrupted contexta | Associated features |
|||
|---|---|---|---|---|---|
| Serogroup(s)b | Toxin type(s) | Colonization factorsc | Nonclassical virulence genesd | ||
| selC | 40 (38.8) | O6, 8–127, 12–164, 27–123, 49, 78, 102, 104–127, 141, 148, 159, ONT | STh, STp, LT, ND | CFAI, CS1, 2, 3, 5, 6, 7, 19, 20, 21, PCFO71, ND | tia, leoA, irp2, fyuA, tibA, eatA, etpA, etpB, ND |
| pheU | 12 (11.7) | O6, 27, 49, 78, 137 | STh, STp, LT | CFAI, CS1, 2, 3, 6, 20, 21, ND | tia, irp2, fyuA, tibA, eatA, etpA, etpB, ND |
| pheV | 40 (38.8) | O6, 8–127, 16–44, 23, 27, 78, ONT | STh, STp, LT | CS1, 2, 3, 6, 19, 20, 21, ND | tia, leoA, irp2, tibA, fyuA, eatA, etpA, etpB, ND |
| thrW | 85 (82.5) | O6, 8–127, 12–164, 16–44, 23, 27, 27–123, 49, 78, 80, 102, 104–127, 112, 115–152, 137, 141, 148, 153, 159, 169, ONT | STh, STp, LT | CFAI, CS1, 2, 3, 6, 5, 7, 12, 17, 19, 20, 21, PCFO71, ND | tia, leoA, irp2, fyuA, tibA, eatA, etpA, etpB, ND |
| leuX | 92 (89.3) | O6, 8–127, 12–164, 16–44, 23, 27, 27–123, 49, 78, 80, 102, 104–127, 112, 115–152, 137, 141, 148, 153, 159, 169, ONT | STh, STp, LT, ND | CFAI, CS1, 2, 3, 6, 5, 7, 12, 17, 19, 20, 21, PCFO71, ND | tia, leoA, irp2, fyuA, tibA, eatA, etpA, etpB, ND |
| asnT | 68 (66) | O6, 8–127, 23, 27, 49, 78, 102, 104–127, 137, 153, 169, ONT | STh, STp, LT, ND | CFAI, CS1, 2, 3, 6, 7, 17, 19, 20, 21, PCFO71, ND | tia, leoA, irp2, fyuA, tibA, eatA, etpA, etpB, ND |
| asnU | 1 (1) | O49 | STh | CFAI, CS21 | tia, irp2, fyuA, tibA, eatA, etpA, etpB |
| asnV | 24 (23.3) | O49, 80, 104–127, 153, ONT | STh, ND | CFAI, CS5, 6, CS20, 21 | tia, leoA, irp2, fyuA, tibA, eatA, etpA, etpB |
| asnW | 14 (13.6) | O6, 49, 78, 104–127, 115–152, 153, ONT | STh, LT | CFAI, CS1, 3, 5, 6, 20, 21, PCFO71 | tia, leoA, irp2, fyuA, tibA, eatA, etpA, etpB |
| argU | 35 (34) | O4, 27, 49, 78, 80, 104–127, 112, 115–152, 137, 148, 153, 159, ONT | STh, STp, LT | CFAI, CS6, 12, 19, 20, 21, PCFO71, ND | tia, irp2, fyuA, tibA, eatA, etpA, etpB, ND |
| metV | 14 (13.6) | O6, 23, 78, 137, 141, 148, 159 | STh, STp, LT | CS1, 2, 3, 6, 7, 20, 21, PCFO71, ND | irp2, fyuA, tibA, eatA, etpA, etpB, ND |
| aspV | 8 (7.8) | O6, 78, 141, ONT | STh, STp, LT | CS1, 3, 21, PCFO71, ND | irp2, fyuA, tibA, eatA, etpA, etpB, ND |
Percentage of 103 strains is given in parentheses.
Numbers without prefixes are O numbers. ONT, nontypeable O serogroup.
Numbers without prefixes are CS numbers.
ND, not detected.
For selC and asnT, there were 7 and 8 strains, respectively, for which tDNAs were intact but strains were positive for at least one gene from associated PAIs (Tia-PAI for selC and HPI for asnT) (Table 2). In contrast, there were several strains with disrupted selC and/or asnT genes that carried neither Tia-PAI nor HPI genes (Table 2), but the vast majority of them were positive for tDNA-associated integrase genes.
Table 2.
Association between detection of PAI-associated genes and tDNA integrity
| PAI | Genea | No. of strains with: |
|
|---|---|---|---|
| Intact tDNAb | Disrupted tDNAb | ||
| Tia-PAI | tia | 6 | 3 |
| leoA | 3 | 3 | |
| selC-int | 1 | 21 | |
| HPI | irp2 | 5 | 34 |
| fyuA | 6 | 31 | |
| asnT-int | 4 | 62 | |
selC-int, selC-associated integrase; asnT-int, asnT-associated integrase.
selC tDNA for Tia-PAI genes (tia, leoA, and selC-associated integrase). asnT tDNA for HPI genes (irp2, fyuA, and asnT-associated integrase).
DISCUSSION
Among 103 ETEC isolates obtained from Chilean children with acute diarrhea, the eatA and etpA nonclassical virulence genes were frequently present and were more common than the classical virulence genes. Thirteen different O serogroups, 7 toxigenic profiles, and 22 CFs profiles were identified. MLVA allowed us to discern genetic relationships and demonstrated that within this collection obtained from a localized geographical niche, isolates from the same serogroup and sharing virulence profiles were not always clonally related. The MLVA carried out in this work has a high discriminatory power which proved to be particularly useful in epidemiological studies (17). Serogroups O6, O153, O78, and O159 detected in our collection have been reported for ETEC isolates from other locations (3, 4, 36). However, strains reacting simultaneously with anti-O104 and anti-O127 sera are not commonly isolated and may represent a geographically confined group of strains. STh was the toxin type most frequently found in Chilean diarrhea-associated ETEC, being detected in more than 60% of strains, whether alone or in combination with other variants. In agreement with previous studies, 75% of ETEC strains produced ST, whereas 53% produced LT, so the former might be a promising vaccine candidate if it could be rendered nontoxic but immunogenic (35, 36). Unfortunately, because ST is a short peptide, it has low immunogenic activity and an efficient carrier-conjugated formulation has not been developed to date.
The most common CF detected in our strains, CS21 (also called longus), is a long rod-like fimbria that directs adhesion to intestinal epithelial cells (19). CS21 is common in ETEC strains from Argentina, Brazil, Bolivia, Egypt, and Bangladesh (13, 21, 23, 24). In a previous study carried out with Chilean ETEC isolates, CS21 was detected by probe hybridization in a lower percentage of strains (28%) than that in our current study (71.8%) (13). The higher proportion of isolates with CS21 in this study might be due to the fact that we included ETEC strains only from children with diarrhea, in which CS21 ETEC seems to be more common (21). To date the immunogenic potential of CS21 is unknown. Nevertheless, taking into account its high prevalence and association with numerous serogroups, toxigenic profiles, and other CFs, it might be a useful candidate for inclusion in an ETEC vaccine.
The nonclassical ETEC virulence genes were detected in 81% of the isolates, and within the remaining 19%, 15 strains were also negative for known CFs. This indicates that some ETEC strains are negative for all the virulence genes identified to date, with the exception of ST and LT. There were only two CF-negative strains that were positive for nonclassical adhesins. So, despite identifying nonroutinely studied genes, such as the tia, tibA, and etpA genes, a surprisingly high percentage of ETEC strains were negative for adhesin genes; this suggests that there may exist additional colonization determinants yet to be identified. Genes of the Tia-PAI, the tia and leoA genes, were detected with low frequency in our collection, almost exclusively in STh/CFAI-CS21+ strains. This distribution coincides with a previous report showing a low frequency of the tia and leoA genes (32). In contrast, irp2 and fyuA genes were detected in about a third of the strains and were associated predominantly with the STh/CFAI-CS21 profile. In a previous report, these genes were detected in only one ETEC strain out of 20 (28). That study included other diarrheagenic E. coli categories of which enteroaggregative E. coli (EAEC) were the most prevalent. The yersiniabactin iron capture system may be used by STh/CFAI-CS21+ ETEC strains for survival within the host. However, more data are needed to test this hypothesis.
The adhesin/invasin-encoding tibA gene was also uncommon in our collection and was mostly associated with the STh/CFAI-CS21 profile. Previously, the tibC gene (residing within an operon along with the tibA gene) has been detected in low frequency (32). In contrast, eatA, etpA, and etpB genes were frequently detected within every serogroup and were associated with all the classical virulence gene profiles found in our isolates. The eatA gene has been previously detected in different frequencies (26% of 209 strains and 61% of 41) (22, 32), while the etpA and etpB genes were detected in high frequency in the report describing its discovery (58% and 42% of 19 strains, respectively) (12). The difference between the numbers of strains positive for the etpA gene compared with strains positive for the etpB gene was unexpected, because these two genes, in addition to the etpC gene, are cotranscribed and are required for EtpA secretion (12). However, three of the etpB-negative and etpA-positive strains proved to be positive by PCR using an etpB forward primer and an etpA reverse primer (data not shown), suggesting some variability in the etpB sequence.
It is well known that tDNAs are frequent insertion sites for mobile genetic elements, such as genomic islands and, specifically, PAIs (15). Our results suggest that the leuX, thrW, and asnT genes could be hot spots for insertion of genetic elements in ETEC strains. The tDNAs selC, pheV, argU, and asnV were disrupted in more than 20% of isolates and could also be insertion sites for mobile DNA. In some of those cases, it was possible to establish an association between the serogroup/virulence profile and tDNA disruption. For example, the pheV gene was disrupted in most O6 strains (27 strains), but it was always intact in strains from cluster B2 (carrying STh/CFAI-CS21 profile). The opposite association was found for the argU and asnV tDNAs, which were disrupted mainly in isolates from cluster B2 and non-O6 isolates from cluster A1. Further sequencing of the insertions in these tDNAs will allow a more accurate determination of relevant associations. As mentioned, some strains with intact selC and/or asnT tDNAs were positive for Tia-PAI and/or HPI genes, suggesting that these gene clusters also occur at different insertion sites. This is to be expected, given that alternative locations of PAIs have been reported. The locus of enterocyte effacement (LEE), the PAI of enterohemorrhagic E. coli (EHEC) that encodes a type III secretion system crucial for pathogenicity, has been shown to be inserted at selC, pheU, or pheV tDNAs in different strains (2). Therefore, in clinical ETEC isolates, Tia-PAI and HPI could also be inserted at sites different than those reported for ETEC H10407 (6). Unexpectedly, there were strains with intact selC and asnT contexts that were positive for adjacent integrase gene detection (Table 2). This could be explained by an equilibrium between excision and insertion of active mobile genetic elements next to tDNAs, as has been recently reported for a Salmonella enterica serovar Enteritidis prophage-like element (27).
The widespread distribution of the etpA and eatA genes fulfills one of the main prerequisites for being included as a candidate for use in an ETEC vaccine. Furthermore, there are reports suggesting that these proteins could be used as antigens to stimulate protective immunity against ETEC infection. Intranasal administration of purified EtpA conferred protection against subsequent ETEC H10407 infection in mice (25). Although the immunogenic potential of EatA has not been determined, proteins of the same family of autotransporters, such as Pic and Pet, are recognized by human sera obtained from infected patients (1). Future epidemiological studies should look for the presence of these genes in ETEC obtained from other regions to determinate their global prevalence and importance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant FONDECYT 1061088, MECESUP higher education program project UCH407, doctoral thesis support fellowship 24091103 (CONICYT, Ministerio de Educación, Gobierno de Chile), “Diarrheal disease in infants and young children in developing countries” (grant 38874) from the Bill and Melinda Gates Foundation, Red Española de Investigación en Patología Infecciosa (REIPI RD06/0008/1018-1016-0026), grant PS09/01273 (Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III, Fondo de Investigación Sanitaria, Gobierno de España), and grants 09TAL007261PR and 2007/000044-0 (Xunta de Galicia and The European Regional Development Fund [ERDF], respectively).
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Bellini E. M., et al. 2005. Antibody response against plasmid-encoded toxin (Pet) and protein involved in intestinal colonization (Pic) in children with diarrhea produced by enteroaggregative Escherichia coli. FEMS Immunol. Med. Microbiol. 43: 259–264 [DOI] [PubMed] [Google Scholar]
- 2. Bertin Y., Boukhors K., Livrelli V., Martin C. 2004. Localization of the insertion site and pathotype determination of the locus of enterocyte effacement of shiga-toxin producing Escherichia coli strains. Appl. Environ. Microbiol. 70: 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blanco J., et al. 1991. Enterotoxigenic Escherichia coli associated with infant diarrhoea in Galicia, north-western Spain. J. Med. Microbiol. 35: 162–167 [DOI] [PubMed] [Google Scholar]
- 4. Blanco J., et al. 1993. Serotypes and colonization factors of enterotoxigenic Escherichia coli isolated in various countries. Eur. J. Epidemiol. 9: 489–496 [DOI] [PubMed] [Google Scholar]
- 5. Brown E. A., Hardwidge P. R. 2007. Biochemical characterization of the enterotoxigenic Escherichia coli LeoA protein. Microbiology 153: 3776–3784 [DOI] [PubMed] [Google Scholar]
- 6. Crossman L. C., et al. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192: 5822–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Croxen M. A., Finlay B. B. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8: 26–38 [DOI] [PubMed] [Google Scholar]
- 8. Elsinghorst E. A., Kopecko D. J. 1992. Molecular cloning of epithelial cell invasion determinants from enterotoxigenic Escherichia coli. Infect. Immun. 60: 2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elsinghorst E. A., Weitz J. A. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62: 3463–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Elsinghorst E. A. 2002. Enterotoxigenic Escherichia coli, p. 155–187 In Donnenberg M. S. (ed.), Escherichia coli virulence mechanisms of a versatile pathogen, 1st ed. Academic Press, San Diego, CA [Google Scholar]
- 11. Fleckenstein J. M., Lindler L. E., Elsinghorst E. A., Dale J. B. 2000. Identification of a gene within a pathogenicity island of enterotoxigenic Escherichia coli H10407 required for maximal secretion of the heat-labile enterotoxin. Infect. Immun. 68: 2766–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fleckenstein J. M., Roy K., Fisher J. F., Burkit M. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74: 2245–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Girón J. A., et al. 1995. Prevalence and association of the longus pilus structural gene (lngA) with colonization factor antigens, enterotoxin types, and serotypes of enterotoxigenic Escherichia coli. Infect. Immun. 63: 4195–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guinée P. A. M., Agterberg C. M., Jansen W. H. 1972. Escherichia coli O antigen typing by means of a mechanized microtechnique. Appl. Microbiol. 24: 127–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hou Y. M. 1999. Transfer RNAs and pathogenicity islands. Trends Biochem. Sci. 24: 295–298 [DOI] [PubMed] [Google Scholar]
- 16. Levine M. M., et al. 1993. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am. J. Epidemiol. 138: 849–869 [DOI] [PubMed] [Google Scholar]
- 17. Lindstedt B., Brandal L. T., Aas L., Vardund T., Kapperud G. 2007. Study of polymorphic variable-number of tandem repeats loci in the ECOR collection and in set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. J. Microbiol. Methods 69: 197–205 [DOI] [PubMed] [Google Scholar]
- 18. Mammarappallil J. G., Elsinghorst E. A. 2000. Epithelial cell adherence mediated by the enterotoxigenic Escherichia coli Tia protein. Infect. Immun. 68: 6595–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mazariego-Espinosa K., Cruz A., Ledesma M. A., Ochoa S. A., Xicohtencatl-Cortes J. 2010. Longus, a type IV pilus of enterotoxigenic Escherichia coli, is involved in adherence to intestinal epithelial cells. J. Bacteriol. 192: 2791–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nataro J. P., Kaper J. B. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11: 142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nishimura L. S., Girón J. A., Nunes S. L., Guth B. E. C. 2002. Prevalence of enterotoxigenic Escherichia coli strains harboring the longus pilus gene in Brazil. J. Clin. Microbiol. 40: 2606–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patel S. K., Dotson J., Allen K. P., Fleckenstein J. M. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72: 1786–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pichel M. G., Binsztein N., Qadri F., Girón J. A. 2002. Type IV longus pilus of enterotoxigenic Escherichia coli: occurrence and association with toxin types and colonization factors among strains isolated in Argentina. J. Clin. Microbiol. 40: 694–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodas C., et al. 2009. Development of multiplex PCR assays for detection of enterotoxigenic Escherichia coli colonization factors and toxins. J. Clin. Microbiol. 47: 1218–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roy K., Hamilton D., Allen K. P., Randolph M. P., Fleckenstein J. M. 2008. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect. Immun. 76: 2106–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roy K., et al. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457: 594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santiviago C. A., et al. 2010. Spontaneous excision of the Salmonella enterica serovar Enteritidis-specific defective prophage-like element φSE14. J. Bacteriol. 192: 2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schubert S., Rakin A., Karch H., Carniel E., Heesemann J. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sommerfelt H., et al. 1989. Mechanism of spontaneous loss of heat-stable toxin (STa) production in enterotoxigenic Escherichia coli. APMIS 97: 436–440 [DOI] [PubMed] [Google Scholar]
- 30. Svennerholm A. M., Tobias J. 2008. Vaccines against enterotoxigenic Escherichia coli. Expert Rev. Vaccines 7: 795–804 [DOI] [PubMed] [Google Scholar]
- 31. Turner S. M., Scott-Tucker A., Cooper L. M., Henderson I. R. 2006. Weapons of mass destruction: virulence factors of the global killer enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 263: 10–20 [DOI] [PubMed] [Google Scholar]
- 32. Turner S. M., et al. 2006. Phylogenetic comparisons reveal multiple acquisitions of the toxin genes by enterotoxin Escherichia coli strains of different evolutionary lineages. J. Clin. Microbiol. 44: 4528–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van de Peer Y., De Wachter R. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10: 569–570 [DOI] [PubMed] [Google Scholar]
- 34. Vidal R., et al. 2009. New multiplex PCRs for characterization of enterotoxigenic Escherichia coli (ETEC) colonization factor antigen genes. Diagn. Microbiol. Infect. Dis. 65: 217–223 [DOI] [PubMed] [Google Scholar]
- 35. Walker R. I., Steele D., Aguado T., et al. 2007. Analysis of strategies to successfully vaccinate infants in developing countries against enterotoxigenic Escherichia coli disease. Vaccine 25: 2545–2566 [DOI] [PubMed] [Google Scholar]
- 36. Wolf M. K. 1997. Occurrence, distribution and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10: 569–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.