Abstract
High-throughput sequencing furnishes a large number of short sequence reads from uncloned DNA and has rapidly become a major tool for identifying viruses in biological samples, and in particular when the target sequence is undefined. In this study, we assessed the analytical sensitivity of a pipeline for detection of viruses in biological samples based on either the Roche-454 genome sequencer or Illumina genome analyzer platforms. We sequenced biological samples artificially spiked with a wide range of viruses with genomes composed of single or double-stranded DNA or RNA, including linear or circular single-stranded DNA. Viruses were added at a very low concentration most often corresponding to 3 or 0.8 times the validated level of detection of quantitative reverse transcriptase PCRs (RT-PCRs). For the viruses represented, or resembling those represented, in public nucleotide sequence databases, we show that the higher output of Illumina is associated with a much greater sensitivity, approaching that of optimized quantitative (RT-)PCRs. In this blind study, identification of viruses was achieved without incorrect identification. Nevertheless, at these low concentrations, the number of reads generated by the Illumina platform was too small to facilitate assembly of contigs without the use of a reference sequence, thus precluding detection of unknown viruses. When the virus load was sufficiently high, de novo assembly permitted the generation of long contigs corresponding to nearly full-length genomes and thus should facilitate the identification of novel viruses.
INTRODUCTION
Identification of new viruses, i.e., phylogenetically distant from those already described, has been in the past an arduous task. Identification of viruses in biological samples has been dominated by the use of cell culture and, more recently, by molecular characterization (for a review, see reference 33). Both of these methodologies suffer from two major limitations: (i) isolation in cell culture is unsuitable for viruses for which no permissive cell line is known (like all hepatitis viruses), and (ii) molecular identification of known viruses is mostly based on (reverse transcriptase [RT]-)PCRs, which for practical reasons can generally be conducted only for a selected number of known viruses. Although DNA arrays can substantially increase the number of targets that can be explored simultaneously (24, 26, 36), currently only resequencing microarrays (RMA; Affymetrix) are able to identify divergent viral strains, by virtue of their large set of probes with mismatches to account for strain diversity. Both technologies, however, still suffer from a sensitivity lower than that of optimized (quantitative) (RT-)PCRs (3), even if this sensitivity does allow the detection of viruses present at high load in biological fluids and tissues (8, 9, 11). Some subtractive techniques, such as representational difference analysis (RDA), have allowed identification of previously unknown viruses, such as human herpesvirus type 8 (HHV8) (7), human GB virus (31), torque teno virus (TTV) (22), and bocavirus (2). Random amplification of nuclease resistant (capsid protected) viral genomes, such as by sequence-independent single primer amplification, has led to the identification of other viruses, such as GB virus C (18), bovine parvoviruses 2 and 3 (1), and human parvovirus 4 (14). All of these techniques are generally poorly sensitive and too time-consuming to be used on numerous biological samples and thus are unsuited to large-scale analysis.
Several studies have now demonstrated that high-throughput sequencing (HTS) is a very useful new research tool for detecting viruses present in biological fluids if their sequence is already known or close to that of viruses that have already been described (10, 19, 24). Although these techniques are still time-consuming and costly, their increasing affordability should enable their application for research purposes and probably in diagnostic settings. Nevertheless, the sensitivity of such techniques has not been evaluated with regard to the detection of low levels of viruses and/or viruses that are genetically distant from known viruses, which is indeed of paramount importance both in research and clinical settings. Also, Roche-454 pyrosequencing has been widely used because of the longer length of the sequences (“reads”) that are generated (up to 500 nucleotides [nt]), compared to Illumina Solexa reads (currently 100 nt). The output of Illumina Solexa is nevertheless much higher than that of Roche-454 (0.6 versus 3 to 20 Gb per run, respectively [for a recent review, see reference 23]). Using a set of samples that were artificially spiked with 11 different viruses, selected so as to represent the diversity of genomic composition found in viruses, we have explored the analytical sensitivity of current HTS using a specifically devoted pipeline.
MATERIALS AND METHODS
Cells and viruses.
HeLa (ATCC CCL-2) and Hep2 cells were propagated in Dulbecco modified Eagle medium. MRC5 cells were purchased from the American Type Culture Collection (ATCC; CCL-171) and propagated in Eagle minimum essential medium supplemented with 10% fetal bovine serum, 4 mM glutamine, and 1% nonessential amino acids. The CVS-11 strain of fixed rabies virus (RABV; reference GI:299542131) was amplified in HeLa cells and titrated in BSR cells, a cloned cell line derived from baby hamster kidney (BHK-21) cells (29). A poliovirus Sabin (PV; reference sequence GI:260907738) strain and a clinical isolate of human herpesvirus type 1 (HHV-1; reference sequence GI:9629378) were amplified and titrated in Hep2 cells (PFU or PFU/ml). Human adenovirus type 5 (HADV-5; ATCC VR-5) was produced and titrated in A549 cells. The NADL strain of bovine viral diarrhea virus (BVDV-1-NADL; ATCC VR-534) was produced and titrated in MDBK cells. Equine influenza virus A/Equine/2/Miami/1/63 (H3N8) (ATCC VR-317) was produced in MDCK cells and titrated in genome copies (gc) per ml by quantitative RT-PCR. The influenza virus B/Lee/40 (FLUBV-B/LEE/40; ATCC VR-1535) was produced in specific-pathogen-free eggs and titrated in gc per ml by quantitative RT-PCR. Strain 68-1 of rhesus cytomegalovirus (RhCMV) was purchased from the ATCC (VR-677) and titrated for 50% tissue culture infective doses (TCID50)/ml. Standard titrated sources of parvovirus B19 (B19), hepatitis B virus (HBV), and hepatitis C virus (HCV) were World Health Organization International Standard virus stocks (NIBSC 99/800, 97/750, and 06/100, respectively [www.nibsc.ac.uk]) and were expressed in international units (IU).
Samples.
To evaluate the analytical sensitivity of Roche-454 pyrosequencing, a set of plasma (taken from a healthy volunteer) and cerebrospinal fluid (CSF; used for diagnosis in patients suspected to have Epstein-Barr [HHV-4]-associated lymphoma) samples were spiked with known viruses at different concentrations. A spiked CSF sample, chosen to be at the limit of detection (LOD) with Roche-454 equipment (see Table 1, CSF 1000), was also sequenced using the Illumina platform. Because the Illumina technology appeared to be more sensitive, this technique was further evaluated using another set of samples corresponding to human MRC5 cell supernatant spiked with a wide range of viruses at two concentrations, namely, 0.8- and 3-fold the LOD of the corresponding quantitative (RT-)PCRs except for B19, for which the spike concentration was expressed on the basis of the known concentration of the international NIBSC 99/800 standard.
Table 1.
Sample spiking schemea
| Virusb | Titration method | Genome structure | Sample identification (titer) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma |
CSF |
Blood |
Cell supernatant |
||||||||||
| P10 | P1000 | P100000 | CSF10 | CSF100 | CSF1000 | B100 | B1000 | B10000 | S3LODx | S0.8LODx | |||
| RABV | FFU or cc | ssRNA | 10 | 103.0 | 105.0 | 10 | 102 | 103 | 102 | 103 | 104 | ||
| PV | PFU or cc | 10 | 103.0 | 105.0 | 10 | 102 | 103 | 102 | 103 | 104 | |||
| HHV-1 | PFU or cc | dsDNA | 10 | 103.0 | 105.0 | 102 | 103 | 104 | |||||
| HHV-4 | gc | 103.8 | 104.3 | 104.7 | |||||||||
| RhCMV | TCID50 | 2 | 8 | ||||||||||
| HADV-5 | TCID50 | 103.0 | 102.4 | ||||||||||
| HBV | IU | 104.0 | 103.4 | ||||||||||
| B19 | IU | ssDNA | 103.0 | 102.4 | |||||||||
| BVDV-1-NADL | TCID50 | ssRNA | 21 | 6 | |||||||||
| H3N8 | gc | 103.0 | 102.4 | ||||||||||
| FLUBV-B/LEE/40 | gc | 103.0 | 102.4 | ||||||||||
| HCV | IU | 103.5 | 102.9 | ||||||||||
The titers of viruses spiked in plasma, CSF, or in MRC5 cell supernatants are expressed per ml (FFU, focus-forming unit; gc, genome copies; IU, international units roughly equivalent to gc; TCID50: 50% tissue culture infectious dose). Virus-infected cells were spiked in blood, and these concentrations are given as the number of virus-infected cells per ml of blood (cc). LOD, limit of detection of quantitative (RT-)PCR tests.
Abbreviations: BVDV-1-NADL, bovine viral diarrhea virus; B19, parvovirus B19 (B19) (NIBSC 99/800); FLUBV-B/LEE/40, influenza B virus; HADV-5, adenovirus type 5; H3N8, equine influenza virus A/Equine/2/Miami/1/63; HBV, hepatitis B virus; HCV, hepatitis C virus; HHV-1, human herpesvirus type 1; PV, poliovirus Sabin; RABV, rabies virus; RhCMV, rhesus cytomegalovirus.
A total of 11 biological samples were used for the present study as described in Table 1. Three plasma samples were spiked with 10, 103, or 105 PFU or focus-forming units (FFU) of PV, RABV, or HHV-1/ml. For the CSF, the HHV-1 was replaced by HHV-4 as a model of herpesvirus. Three CSF samples containing increasing amounts of HHV-4 (6,400, 21,500, and 51,000 gc/ml) were generated by mixing appropriate volumes of single CSF samples with different HHV-4 loads. The resulting CSF samples were spiked, respectively, with 10, 102, or 103 FFU or PFU of RABV or PV/ml. Three blood samples were spiked with increasing concentrations (102 to 104 cells/ml) of HeLa cells infected with RABV and Hep2 cells infected with PV or HHV-1.
Sample preparation. (i) Plasma, CSF, and cell supernatant samples.
The extraction procedure was optimized for the isolation of viral DNA or RNA genomes without precluding the identification of bacterial and fungal nucleic acids. A volume of 150 μl of each sample was extracted using a Nucleospin RNA virus kit (Macherey-Nagel), which allows recovery of both DNA and RNA, and then amplified by the bacteriophage φ29 polymerase based multiple-displacement-amplification (MDA) assay using random primers. This technique allows DNA synthesis from DNA samples and, according to a recently developed procedure, also from cDNA fragments from viral genomes previously colligated prior to φ29 polymerase-MDA (4). Briefly, the protocol of a QuantiTect whole transcriptome kit (Qiagen) was followed, except that the cDNA synthesis step was performed with random hexamer primers. A mix with 8 μl of RNA, 1 μl of primer (50 μM), and 1 μl of deoxynucleoside triphosphates (10 mM) was incubated at 75°C for 5 min and cooled on ice for 5 min. Then, 10 μl of 2× enzyme mix was added. This enzyme mix was composed of 2 μl of 10× RT buffer for SSIII (Invitrogen, Inc.), 4 μl of 25 mM MgCl2, 2 μl of 0.1 M dithiothreitol, 1 μl of a 40-U/μl mixture of RNaseOUT (Invitrogen, Inc.), 1 μl of SuperScript III reverse transcriptase (Invitrogen, Inc.), and 0.5 μl of dimethyl sulfoxide (Sigma-Aldrich). The final mix was incubated at 25°C for 10 min, then at 45°C for 90 min, and finally at 95°C for 5 min. All cDNAs were stored at −20°C or immediately used. The two following steps (ligation and WGA) were performed with the QuantiTect whole transcriptome kit (Qiagen) according to the manufacturer's instructions. This provides concatemers of high-molecular-weight DNA.
(ii) Blood cell samples.
HeLa cells were infected with RABV at a multiplicity of 10 FFU/cell as described previously (15). Hep2 cells were infected under the same conditions with PV or HHV-1 at 10 PFU/cell. When a cytopathic effect was observed (for PV or HHV-1) or when ca. 30% of the cells scored positive in a rabies-specific immunofluorescence assay, the cells were recovered by scraping and mixed with total human blood at the concentrations indicated in Table 1 as virus-infected cells per ml. Total RNAs were then extracted from the resulting blood cells with the QIAamp RNA Blood Easy kit (Qiagen).
High-throughput sequencing.
Illumina and Roche-454 sequencing were subcontracted to GATC Biotech AG (Constance, Germany). For blood cell samples, standard full-length cDNA libraries were prepared by GATC, and 5 μg of amplified cDNA was used for Roche-454 library construction and sequencing. In all other cases, high-molecular-weight DNA (5 μg), resulting from isothermal amplification of the pool of genomic DNAs and cDNAs made from genomic RNAs as described above, was fragmented into 200- to 350-nt fragments, to which adapters were ligated. Adapters included a nucleotide tag allowing for multiplexing several samples per lane or channel. Sequencing was conducted with a mean depth per sample of 5 × 104 reads (range, 2 × 104 to 8 × 104) (Roche-454 sequencing, together with the GS FLX Titanium series reagents) or 5 × 106 sequences per sample (range, 3 × 106 to 10 × 106) (Illumina GAII sequencer).
Bioinformatics.
Sequencing and bioinformatics analysis were conducted in a blinded fashion. Sorting out the flow of Illumina sequences was first performed by a subtractive database comparison procedure. To this end, the whole host genome sequence (mostly human: NCBI build 37.1/assembly hg19) was scanned with the reads using SOAPaligner. A quick and very restrictive BLASTN study was also performed to eliminate additional host reads. The best parameters to be used were determined in a pilot experiment. A number of assembly programs dedicated to short or medium-sized reads (Velvet [http://www.ebi.ac.uk/∼zerbino/velvet/], SOAPdenovo [http://soap.genomics.org.cn/], and CLC Genomics Workbench) have been tested for their efficiency in our pipeline. Optimal parameters have been set. Comparison of the single reads and contigs to known genomic and taxonomic data was performed using dedicated specialized viral, bacterial, and generalist databases maintained locally (GenBank viral and bacterial databases, nr). The aforementioned databases were scanned using BLASTN and BLASTX. We used Paracel BLAST, a software that distributes the BLAST tasks on multiple processors. Binning (or taxonomic assignment) was based on the lowest common ancestor from the best hits among reads with a significant e-value (generally 10−4).
The previous 454 pyrosequencing analysis was partly performed by GATC Biotech AG. Filtering of host reads was performed by using BLASTN and assembly of the remaining data by using Newbler. Subsequent analysis was similar to that described above for Illumina reads.
PathogenID resequencing microarray.
An Affymetrix PathogenID RMA, optimized for the detection and sequence determination of rhabdoviruses, was used as described previously (11).
Quantification of virus targets or genomes using quantitative (RT-)PCRs.
Detection and quantification of RABV in samples were performed by using SYBR green real-time PCR (7500 real-time PCR system; Applied Biosystems) using two sets of primers targeting conserved regions of the viral polymerase gene (TaqCVSc_S1 [5′-GCAAGGGCTTTTGGACTATTCTAG-3′] and TaqCVSc_AS1 [5′-TCTTTGATGATTGTTCCATTTCTCA-3′] or TaqCVSc_S2 [5′-ACCTGGACTATGAGAAATGGAACAA-3′] and TaqCVSc_AS2 [5′-TCAATCCAAACACCTGATCTAGGA-3′]) and designed using PrimerExpress 3.0 software (Applied Biosystems). Quantification was performed using a standard dilution curve using a plasmid encompassing the nucleotide target, with limits of quantification and detection for both primer sets of >20 and 0.5 copies/μl, respectively. Other quantitative (RT-)PCR have been described previously (RhCMV [16], HADV-5 [34a], BVDV-1-NADL [5], H3N8 [37], FLUBV-B/LEE/40 [37], and HCV [21]) and have been conducted and validated by Texcell (Evry, France), a company specialized in providing services for the biological products manufacturing industry. Validations of the viral spikes detection limit (LOD) are provided (see Appendix SA1 in the supplemental material), and the results are summarized in Table 5. Validation of the limits of detection and experiments were conducted according to good laboratory procedures.
Table 5.
Illumina hits from two cell culture supernatants spiked with eight different virusesa
| Virus | Sensitivity of quantitative (RT)-PCRs |
Supernatant 3LODxa (no. of reads = 7,891,140) |
Supernatant 0.8LODx (no. of reads = 8,342,364) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Validated LOD (per ml) | CT for 10/100/1,000 plasmid copies (per 5 μl) | Quantitative (RT)-PCR | CT | No. of hits | Identity (%) | Length (hits) | Quantitative (RT)-PCR | CT | No. of hits | Identity (%) | Length (hits) | |
| RhCMV | 3 TCID50 | ND/35/32 | +/+ | 31/30 | 2,142 | 99 | 65 | +/+ | 32/33 | 662 | 99 | 68 |
| HADV-5 | 357 TCID50 | ND/37/33 | +/+ | 38/39 | 13 | 99 | 58 | −/− | 4 | 99 | 59 | |
| HBV | 3,571 IU | ND/40/35 | +/+ | 35/35 | 2 | 99 | 68 | +/+ | 37/37 | 0 | ||
| B19 | 2 | 99 | 70 | 6 | 97 | 70 | ||||||
| BVDV-1-NADL | 10 TCID50 | ND/42/36 | +/+ | 38/38 | 2 | 100 | 71 | +/+ | 45/45 | 1 | 95 | 58 |
| H3N8 | 357 cp | ND/39/36 | +/+ | 33/34 | 10 | 100 | 70 | −/− | 0 | |||
| FLUBV-B/LEE/40 | 357 cp | ND/35/33 | −/− | 4 | 99 | 66 | −/− | 0 | ||||
| HCV | 1,071 IU | ND/>40/ND | +/− | 40/ | 0 | +/− | >40/ | 0 | ||||
MRC5 cell supernatants were spiked with viruses at a concentration corresponding to 3× or 0.8× the limit of detection (LOD) of the validated threshold of detection of corresponding quantitative (RT-)PCRs, as detailed in Table 1. The number of reads generated from the samples and the number of virus-specific hits are given, together with the results from two runs of quantitative (RT-)PCRs (except for B19): validated limits of detection of virus spikes, cycle thresholds (CT) corresponding to a given concentration of positive controls plasmids, and CT corresponding to the samples are shown for comparison. The percent identities were obtained after comparison with the reference sequences (see the text). The length of the hits and the percent identities correspond to averages. Abbreviations: IU, international unit; TCID50, tissue culture infective dose 50%; cp, DNA copies; ND, not determined; LOD, limit of detection of quantitative (RT-)PCR. +/+, +/−, and −/−, results of two independent runs of quantitative (RT-)PCRs.
Ethical approval.
The studies were approved by the Institut Pasteur Comité de Recherche Clinique and the French Commission Nationale Informatique et Libertés (09.465). Written consent was sought for human samples according to French regulations.
RESULTS
Analytical sensitivity of Roche-454 using spiked samples.
To estimate how the results of pyrosequencing correlated with those of infectivity assays, human plasma and CSF samples were spiked with increasing amounts of RABV, HHV-1, and PV. CSF samples contained increasing concentrations of HHV-4 as a test for herpesvirus detection (Table 1). Nucleic acids, including DNA and RNA, were isolated, amplified, and sequenced as previously described. As shown in Table 2, RABV, PV, and HHV-1 were detected at a concentration of 105 PFU or FFU/ml. Rare positive hits were detected for HHV-1 or RABV at a concentration of 103 PFU/ml, whereas HHV-4 was not detected even at a concentration of 51,000 genomes/ml. Thus, although some viruses could be detected at the lowest concentration tested (150 PFU or FFU), the threshold for consistent detection appeared to be between 103 and 105 PFU or FFU/ml. Concurrent analysis of the same batch of amplified nucleic acids with the recently described PathogenID RMA (11) gave a positive response only for the concentration of 105 FFU/ml, whereas Roche-454 and quantitative (RT-)PCR were positive at concentrations of 103 FFU/ml (5,600 target copies/ml) and above (not shown).
Table 2.
Roche-454-specific hits in plasma and CSF spiked with known virusesa
| Sample | No. of reads per sample | Avg length (nt) | No. of virus-specific hits per sample |
Length (no. of virus-specific hits)b | Identity (%) | |||
|---|---|---|---|---|---|---|---|---|
| RABV | PV | HHV-1 | HHV-4 | |||||
| Plasma 10 | 27,516 | 247 | 0 | 0 | 0 | NDc | ND | ND |
| Plasma 1000 | 23,009 | 264 | 5 | 0 | 4 | ND | 443 | 98 |
| Plasma 100000 | 44,780 | 324 | 34 | 29 | 591 | ND | 403 | 99 |
| CSF 10 | 33,453 | 352 | 0 | 0 | 0 | ND | ND | ND |
| CSF 100 | 61,161 | 360 | 0 | 0 | 0 | ND | ND | ND |
| CSF 1000 | 27,730 | 358 | 1 | 0 | 0 | ND | 438 | 99 |
For each virus, the number of hits per sample is shown. The average length (nt) is given for all reads in a given sample and for the virus-specific hits. Percentages of identity were obtained after comparison to the reference sequences (for RABV and HHV-1 see text, for HHV-4, reference sequence GI:139424470).
When several viruses were detected in a sample, the length of the hits and the percent identity correspond to averages.
ND, not determined.
We also tested whether we would be able to detect viral RNAs in actively infected cells as a correlate of infection. To this end, we spiked whole blood samples with decreasing concentration of cells infected with HHV-1, PV, and RABV. Blood cells were isolated and the bulk of RNAs was extracted, reverse transcribed, and sequenced by Roche-454 as previously described. The three viruses could be detected at a concentration of 10,000 infected cells per ml of blood (10,500 target copies/ml for RABV), which would correspond to a frequency of infected cells of between 1/100 and 1/1,000. The numbers of hits were 2, 150, and 4 for RABV, PV, and HHV-1, respectively. The mean length of hits was 404 nt, and on average they displayed 99.1% identity with the reference sequences (data not shown).
Comparison of Illumina to Roche-454 sequencing.
We performed Illumina sequencing on samples containing quantities of virus at the LOD for 454 or quantitative (RT-)PCR. We chose a sample (CSF 1000) at or below the LOD of pyrosequencing for an initial evaluation of the sensitivity of Illumina technology. To this end, we used an aliquot of the same preparation of nucleic acids that had been amplified and subjected to pyrosequencing. As shown in Tables 3 and 4, hits assignable to HHV-4, PV, and RABV were clearly identified. Moreover, we also detected additional viruses naturally present in this mixture of CSF. These included a very high number of hits (n = 16,793) for torque teno virus (TTV), which is a ubiquitous anellovirus present in most biological samples (13), and 7 hits from HHV-1, a neurotropic virus responsible of a high prevalence of latent infections. It is of note that only 176 hits for TTV and no HHV-1 sequences were found using the Roche-454 platform. We also detected 1 hit for human papillomavirus type 18 (HPV18), which likely came from the RABV spike, since its single read exactly matched the HPV18 DNA previously described in HeLa cells (i.e., the cells used to grow the strain of RABV used for spiking) (20). Finally, we detected 3 hits from a bunyavirus (Rift Valley fever virus) which were identical to the sequence of a plasmid amplified several months before in the same laboratory room that served for the present extraction and was thus considered to be a contaminant from the environment. These results suggested that Illumina technology was more sensitive than pyrosequencing and capable of detecting even traces of contaminants.
Table 3.
Comparison of Roche-454 and Illumina hits from CSF 1000: basic parametersa
| Parameter | Roche-454 | Illumina |
|---|---|---|
| No. of reads | 27,730 | 5,073,149 |
| Avg length (nt) | 358 | 72 |
| Avg length (no. of hits) | 334a | 72 |
This value includes TTV reads.
Table 4.
Comparison of Roche-454 and Illumina hits from CSF 1000: viruses identifieda
| Virus identified | Roche-454 |
Illumina |
||
|---|---|---|---|---|
| No. of hits | Identity (%) | No. of hits | Identity (%) | |
| RABV | 1 | 99 | 569 | 99 |
| PV | 0 | 37 | 99 | |
| HHV-4 | 0 | 95 | 99 | |
| TTV | 176 | 78 | 16,793 | 87 |
| HHV-1 | 0 | 7 | 100 | |
| HPV-18 | 0 | 1 | 100 | |
| Rift Valley fever virus | 0 | 3 | 99 | |
The percent identity values correspond to averages. The percent identities for TTV, Rift Valley fever virus, and HPV-18 were obtained after comparison to the closest reference sequences identified by BLAST in the database nr. Other viruses are as described in Table 2.
Analytical sensitivity of Illumina.
Since the study described above suggested that Illumina was highly sensitive, we studied its analytical sensitivity on a range of different viruses representing all possible genome compositions, as shown in Table 1. To this end, two samples of supernatant from MRC5 cells were spiked with eight viruses, including seven viruses at theoretical genome concentrations 0.8- and 3-fold the level of detection of the corresponding quantitative (RT-)PCRs and another virus (B19) at, respectively, 103.0 and 102.4 gc/ml (with 102.4 gc/ml being the lower detection limit of the LightCycler parvovirus B19 quantification kit [Roche Diagnostics] at a 95% hit rate [30]). These low concentrations were chosen so as to bracket the threshold of detection of the Illumina pipeline on the basis of our previous experience. The results of two independent runs of quantitative (RT-)PCRs are shown in Table 5 , together with those of sequencing. For B19, no quantitative PCR using a published technique was performed with the samples. For the tube spiked with the highest concentration, seven of eight viruses were detected by Illumina. It can nevertheless be noted that, for RhCMV, quantitative PCR results from the 0.8× and 2× LOD samples suggest that the spike could have been done at higher concentrations than anticipated. As shown in Table 5, the undetected virus corresponds to HCV (detected in one of the two duplicates by quantitative RT-PCR). For the tube spiked at 102.4 gc/ml (B19) or below the validated LOD of quantitative (RT-)PCR, four of eight viruses were still identified, i.e., approximately the same number of viruses that were detected for both of duplicate runs of quantitative (RT-)PCRs (four of seven). Interestingly, in two cases [FLUBV-B/LEE/40 and HADV-5 at, respectively, 3- and 0.8-fold the level of detection of quantitative (RT-)PCRs], the viruses could be detected by sequencing but not by quantitative (RT-)PCR. When the number of hits was high enough, the ratio of the number of hits between the two samples was roughly proportional to concentrations (see the RhCMV results). At very low concentrations, this proportionality was inconsistent, likely reflecting the imprecision of target concentration near the LOD in the 150 μl of sample subjected to amplification, conforming to Poisson's distribution.
De novo assembling of nonhuman reads in viral contigs.
We wondered whether we would have been able to detect unknown viruses, whose sequences would not be present in the nucleotide sequence databases. To address this question, we conducted de novo assembling of nonhuman reads using three software programs (CLC, SOAPdenovo, and Velvet), that is, without mapping the reads on a reference sequence. Once the contigs were assembled, for the purpose of comparison, we checked by BLASTN on general databank (nr) and taxonomic assignment those corresponding to known viruses. As can be seen in Table 6, when the level of spiking was high enough, as was the case for sample CSF 1000, which was naturally contaminated by an anellovirus (TTV), both Roche-454 and Illumina sequencing gave equivalent results: a contig of more than 2,600 nt was identified, whose genetic organization (not shown) would have strongly suggested the presence of a virus. The contig represented more than 85% of the whole viral genome. Regarding Roche-454, contigs in the range of 1,164 to 3,785 nt were identified for a virus load of 105 PFU or FFU/ml for model viruses (PV, HHV-1, and RABV), which could have sufficed to suggest the presence of a new virus, particularly if an open reading frame was present (Table 6). We have not tested Illumina sequencing for a virus concentration of 105 PFU or FFU/ml. At a much lower concentration of 103 FFU/ml (Table 6), no contig could be assembled from the Roche-454 reads, while the Illumina reads could be assembled into a contig of 1,121 nt for RABV, which allowed identification of a viral signature. No contigs of more than 200 nt were identified for the two other viruses (PV and HHV-4).
Table 6.
De novo assembling and mapping on reference sequences of Roche-454 and Illumina reads a
| Method, sample, and virus | De novo assembly |
Mapping on a reference sequence |
|||||
|---|---|---|---|---|---|---|---|
| Largest contig (CLC or Velvet) | Reference sequence assembled from the largest contig (%) | Depth coverage (fold) | Length of reference sequence (nt) | No. of mapped hits | Reference sequence mapped (%) | Depth coverage (fold) | |
| Roche-454 | |||||||
| P100000 | |||||||
| HSV 1 | 3,785 | 2 | 5 | 152,261 | 583 | 42 | 1.5 |
| RABV | 1,164 | 10 | 1.7 | 11,927 | 34 | 51 | 1.1 |
| PV | 1,619 | 22 | 1.9 | 7,439 | 29 | 77 | 1.7 |
| CSF 1000, TTV | 2,885 | 94 | 25 | 3,080 | 35 | 54 | 4.9 |
| Illumina | |||||||
| CSF 1000 | |||||||
| HHV-4 | 150 | 0.1 | 1.5 | 171,823 | 156 | 3 | 0.05 |
| RABV | 1,121 | 9 | 4.5 | 11,927 | 571 | 90 | 3.3 |
| PV | <100 | 7,439 | 38 | 28 | 0.3 | ||
| Anellovirus | 2,624 | 85 | 683.7 | 3,080 | 5,592 | 61 | 118.7 |
For the de novo assembling process, the nonhuman reads were assembled, and the resulting largest contig was mapped on the corresponding reference sequence for confirmation. Mapping of reads identified as virus hits on the reference sequences (see Table 2) are also presented for comparison. Abbreviations: CLC, CLC genomics.
Recovery of whole viral genomes.
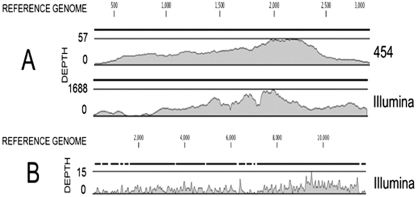
We evaluated the total length of the hits that could be mapped on a reference sequence and the coverage or depth of the sequencing (corresponding to the number of hits obtained for a given nucleotide position). As can be seen, using Roche-454 sequencing (Table 6) for a concentration of 105 PFU or FFU/ml (sample Plasma 100000), 42 to 77% of the genomes of PV, RABV, and HHV-1 were sequenced with a mean depth of 1.1 to 1.7. At a virus concentration 100 times lower (103 PFU or FFU/ml), Illumina reads (Table 6) covered 28% (PV) to 90% (RABV) of the length of the genome but only 3% of that of the much larger HHV-4 genome present at a concentration of 51,000 copies/ml. Interestingly, the coverage of the highly variable single-stranded DNA (ssDNA) genome of TTV was much higher with the contig generated from de novo assembling than by mapping the single hits on a reference sequence. This was expected because the mapping process tolerates very few mismatches for a given read and reflects the absence of sequence data for genetically similar isolates in nucleotide public sequence databases. The distribution of the depth of Illumina reads along the genomes of CVS strain of RABV and anellovirus is shown in Fig. 1. Almost the entire length of the genomes was sequenced.
Fig. 1.
Distribution of the coverage resulting from de novo assembling or mapping on reference sequences of Roche-454 and Illumina reads. (A) Coverage of the anellovirus sequence by the contigs resulting from de novo assembling of Roche-454 and Illumina reads. Depth (y axis) corresponds to the number of reads representing a given nucleotide in the reconstructed sequence. The x axis corresponds to nucleotide positions in the whole genome. One contig from the Roche-454 covered 93.7% of the genome. Two contigs from the Illumina reads covered 92.6% of the genome. (B) Resequencing coverage of the genome of CVS strain of RABV from the hits directly mapped on the reference sequence. Note the difference of scale for the depth between the three graphs.
DISCUSSION
In this study, we used artificially spiked and naturally infected biological samples to evaluate the analytical sensitivity of virus detection using HTS based on Roche-454 and Illumina platforms, defined by the lowest genome copy number and/or the PFU or FFU of known viruses that could be detected. We also evaluated the ability to detect the signatures of unknown viruses (defined as the capacity to identify large contigs from de novo assembly).
Regarding the analytical sensitivity, the study design took into account the rapid evolution of sequencing techniques and the existence of two current major technologies; namely, Illumina/Solexa and Roche-454. Indeed, these two techniques differ in several respects. The length of Roche-454 reads is higher (350 nt on average at the time of our study) than that of Illumina (76 nt for the present study and, more recently, 100 nt [Illumina]). On the other hand, the output of Illumina per run is much higher (35). Using the same number of reads for each sample would have been meaningless, as in practice the cost associated with sequencing and the daily outputs of the sequencers determine the choice of a given depth. Illumina GAIIx generates 160 to 250 million reads per run. For Roche-454's GS FLX system, the average number of reads per run is ∼1 million. Both techniques allow for multiplexing the samples, which permits use of a part of a lane or run for each sample. We have considered that a fair comparison between the two techniques corresponded to a number of reads corresponding to around 1/20 of a run; that is, a range of 2 × 104 to 8 × 104 Roche-454 and 3 × 106 to 10 × 106 Illumina single reads. In principle, a higher number of reads per sample should be an advantage regarding the sensitivity, and longer reads should allow easier de novo assembling and a better chance for the detection of viruses that are completely unknown or whose sequences are distant from those in the databases (32).
The sensitivity and the range of detection are also a function of the preparation of the sample. Our goal was to set up a procedure compatible with the detection of bacteria and fungi, so we avoided any step that would have counterselected such microorganisms. For example, we used random rather than oligo(dT) priming for all cDNA synthesis. Detection of virus infection in cells (like that present in blood samples spiked with virus-associated cells) used standard full-length cDNA libraries made from total RNAs. For biological fluids (such as CSF or plasma) or cell culture supernatants, for which the goal was to identify cell-free viral particles (or bacteria and fungi), we used a previously described procedure able to amplify minute amounts of any RNA or DNA present in the sample, based on a nonspecific and unbiased isothermal amplification (4). Most of the results presented here were obtained under these conditions. This means that results cannot be strictly correlated with the number of copies of viral genomes, since free viral (m)RNAs could also be amplified. Also, for a given genome copy number, a larger genome increases the probability of detection, whereas this is not the case for PCR.
We show here that the Illumina technology is more efficient at detecting and characterizing viruses whose sequence is already present in the databases. For example, the PV and HHV-4 viruses that were not detected by the Roche-454 were clearly evidenced by Illumina (Tables 3 and 4). A more complete validation was conducted on eight different viruses. Of the seven viruses spiked at 3-fold the LOD of quantitative (RT-)PCRs (RhCMV may have been spiked at a higher concentration), five gave consistently reproducible positive results for the two runs of (RT-)PCR, and all of these were detected by HTS. A parvovirus (B19) was identified at a concentration of 103 gc/ml. On the other hand, one virus (FLUBV-B/LEE/40) was detected by HTS but not (RT-)PCR. One virus (HCV) did not give any identifiable read but the quantitative (RT-)PCR was positive in 50% of the assays (one run out of two). When the concentration of the seven targets was decreased to 102.4 gc/ml (B19) or 0.8-fold the level of detection of quantitative (RT-)PCRs, three of six viruses remained consistently detected by quantitative (RT-)PCRs, and two gave identifiable Illumina reads. B19 gave also identifiable Illumina reads. It is noteworthy that one virus not detected by quantitative PCR (HADV-5) was identified by HTS. Thus, Illumina sequencing with a depth of ∼7 millions reads per sample allowed identification of viruses at a level close to dedicated and optimized quantitative (RT-)PCRs. Moreover, the identification of viruses was highly specific. First, we recorded no incorrect result during this blinded study (i.e., reads falsely attributed to a virus), despite the fact that our taxonomic assignment was based on the best BLAST hits, which was previously shown to give frequent misclassification (17). Moreover, in all cases, we were able to identify the viruses at the level of the virus species and even, for the equine influenza A virus, at the level of the subtype (H3N8) (data not shown).
Regarding the capacity to identify unknown viruses, we evaluated the potential of de novo assembly for the identification of contigs of sufficient size to suggest the presence of a new virus. The probability of obtaining a contig of a given size of an unknown species from a few reads is a function of the complexity and size distribution of the genomes within the pool (32). When the virus load was high enough to give a sufficient number of reads, as was the case for the TTV present in sample CSF 1000, both Illumina and Roche-454 gave a contig almost as large as a full-length genome (≥85%). The number of reads that could have been assembled de novo was higher than the number of reads that could be mapped on the closest known sequence (Tables 4 and 6). This reflects the capacity of assembling de novo reads generated by both techniques for the identification of viral genomes whose sequence is distant from known sequences.
Comparing the sequence of a new isolate to a standard reference is a classical task in virology. One of the advantages of HTS over the Sanger method is that it is easier to generate full-length genome sequences directly from biological samples. Indeed, this is of incalculable advantage for uncultivable viruses, but it also avoids any bias in virus populations associated with the procedure of isolation in cell culture. Once major parts of the genome have been characterized, it remains possible to complete the gaps by Sanger sequencing. We present here two examples of such approaches. In the first, from a CSF sample in which RABV had been spiked at a very low concentration (103 FFU/ml corresponding to 5,600 targets/ml), we have mapped the available hits on the reference sequence and were able to resequence 90% of its 11,927-nt single-strain RNA genome with an average coverage of 3.3×. In the second example, the single strain circular DNA TTV genome naturally present in the same CSF was sequenced with an average coverage of 118×. For this purpose, we used the contig obtained after de novo assembling from the Illumina reads, which incorporated many more reads than those that could be mapped on the known but more distant reference sequences. It was then possible to analyze with precision the distribution of the quasispecies, as already shown by others with the Roche-454 technology (28). Curiously, the two virtual extremities of the circular genome were underrepresented, possibly because sequencing was performed on both genomic DNA and mRNAs and/or the presence of nonencapsidated linear DNA molecules.
Finally, it should be underscored that these results were obtained without optimizing the nucleic acid extraction for the identification of viral genomes, since the identification of bacteria and fungi was also targeted. Indeed, enrichment of viral sequences by DNase and/or RNase treatments or physical separation would probably increase the performance of viral detection (6, 34). Also, the comparison of Roche-454 and Illumina has taken into account the different outputs of the two sequencers. For each technique, we used approximately 1/20 of a run with a similar cost. Indeed, as the cost per base of HTS will certainly continue to decrease with improvements of current technologies and the appearance of third-generation techniques, the sensitivity (for a given cost) of HTS will further increase. As it stands now, we show here that HTS is able to identify up to seven different viruses simultaneously in a given biological sample with a level of sensitivity close to that of optimized quantitative (RT-)PCRs. Indeed, like PCR, this technique is prone to reveal subtle cross-contamination of samples and must be used with the same degree of caution as PCR. Currently, the time to results is at least 3 weeks, including the bioinformatics analysis with the Illumina technology. Nevertheless, there is little doubt that HTS, which is already a method of choice for virus discovery as exemplified by others (12, 24, 33), could also become a tool for in depth or even routine diagnosis as soon as the cost and delays decrease significantly.
Supplementary Material
ACKNOWLEDGMENTS
We thank the group Logiciels et Banques de Données of the Institut Pasteur and, more particularly, Louis Jones for the creation and upgrade of the local nucleotide databases and blast machines, and we thank Nicolas Joly for the development of “Golden,” a program retrieving databank entries, including taxonomy. We also thank Muriel Eliaszewicz for her constant support for this program. We thank Jennifer Richardson for help in the redaction of the manuscript.
The platform Genotyping of Pathogens and Public Health is supported in part by the Institut de Veille Sanitaire (Saint-Maurice, France). This study was mainly supported by Programme Transversal de Recherche (PATHODISC 301) from the Institut Pasteur (France) and by grants from region Ile de France.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 29 June 2011.
REFERENCES
- 1. Allander T., Emerson S. U., Engle R. E., Purcell R. H., Bukh J. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. U. S. A. 98: 11609–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allander T., et al. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102: 12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berthet N., et al. 2010. High-density resequencing DNA microarrays in public health emergencies. Nat. Biotechnol. 28: 25–27 [DOI] [PubMed] [Google Scholar]
- 4. Berthet N., et al. 2008. Phi29 polymerase-based random amplification of viral RNA as an alternative to random RT-PCR. BMC Mol. Biol. 9: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhudevi B., Weinstock D. 2001. Fluorogenic RT-PCR assay (TaqMan) for detection and classification of bovine viral diarrhea virus. Vet. Microbiol. 83: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breitbart M., Rohwer F. 2005. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 39: 729–736 [DOI] [PubMed] [Google Scholar]
- 7. Chang Y., et al. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266: 1865–1869 [DOI] [PubMed] [Google Scholar]
- 8. Chiu C. Y., et al. 2007. Diagnosis of a critical respiratory illness caused by human metapneumovirus by use of a pan-virus microarray. J. Clin. Microbiol. 45: 2340–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiu C. Y., et al. 2008. Utility of DNA microarrays for detection of viruses in acute respiratory tract infections in children. J. Pediatr. 153: 76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cox-Foster D. L., et al. 2007. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318: 283–287 [DOI] [PubMed] [Google Scholar]
- 11. Dacheux L., et al. 2010. Application of broad-spectrum resequencing microarray for genotyping rhabdoviruses. J. Virol. 84: 9557–9574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng H., Shuda M., Chang Y., Moore P. S. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hino S., Miyata H. 2007. Torque Teno virus (TTV): current status. Rev. Med. Virol. 17: 45–57 [DOI] [PubMed] [Google Scholar]
- 14. Jones M. S., et al. 2005. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 79: 8230–8236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kassis R., Larrous F., Estaquier J., Bourhy H. 2004. Lyssavirus matrix protein induces apoptosis by a TRAIL-dependent mechanism involving caspase-8 activation. J. Virol. 78: 6543–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaur A., et al. 2002. Decreased frequency of cytomegalovirus (CMV)-specific CD4+ T lymphocytes in simian immunodeficiency virus-infected rhesus macaques: inverse relationship with CMV viremia. J. Virol. 76: 3646–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koski L. B., Golding G. B. 2001. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 52: 540–542 [DOI] [PubMed] [Google Scholar]
- 18. Linnen J., et al. 1996. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271: 505–508 [DOI] [PubMed] [Google Scholar]
- 19. Loh J., et al. 2009. Detection of novel sequences related to African swine fever virus in human serum and sewage. J. Virol. 83: 13019–13025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meissner J. D. 1999. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa, and HeLa cervical carcinoma cell lines. J. Gen. Virol. 80: 1725. [DOI] [PubMed] [Google Scholar]
- 21. Mercier B., Burlot L., Férec C. 1999. Simultaneous screening for HBV DNA and HCV RNA genomes in blood donations using a novel TaqMan PCR assay. J. Virol. Methods 77: 1–9 [DOI] [PubMed] [Google Scholar]
- 22. Nishizawa T., et al. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241: 92–97 [DOI] [PubMed] [Google Scholar]
- 23. Nowrousian M. 2010. Next-generation sequencing techniques for eukaryotic microorganisms: sequencing-based solutions to biological problems. Eukaryot. Cell 9: 1300–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Palacios G., et al. 2008. A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 358: 991–998 [DOI] [PubMed] [Google Scholar]
- 25. Reference deleted.
- 26. Palacios G., et al. 2007. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 13: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reference deleted.
- 28. Rozera G., et al. 2009. Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte subpopulations. Retrovirology 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sato M., Tanaka H., Yamada T., Yamamoto N. 1977. Persistent infection of BHK21/WI-2 cells with rubella virus and characterization of rubella variants. Arch. Virol. 54: 333–343 [DOI] [PubMed] [Google Scholar]
- 30. Schorling S., Schalasta G., Enders G., Zauke M. 2004. Quantification of parvovirus B19 DNA using COBAS AmpliPrep automated sample preparation and LightCycler real-time PCR. J. Mol. Diagn. 6: 37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simons J. N., et al. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. U. S. A. 92: 3401–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stanhope S. A. 2010. Occupancy modeling, maximum contig size probabilities and designing metagenomics experiments. PLoS One 5: e11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tang P., Chiu C. 2010. Metagenomics for the discovery of novel human viruses. Future Microbiol. 5: 177–189 [DOI] [PubMed] [Google Scholar]
- 34. Thurber R. V., Haynes M., Breitbart M., Wegley L., Rohwer F. 2009. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4: 470–483 [DOI] [PubMed] [Google Scholar]
- 34a. van de Pol A. C., et al. 2007. Increased detection of respiratory syncytial virus, influenza viruses, parainfluenza viruses, and adenoviruses with real-time PCR in samples from patients with respiratory symptoms. J. Clin. Microbiol. 45: 2260–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voelkerding K. V., Dames S. A., Durtschi J. D. 2009. Next-generation sequencing: from basic research to diagnostics. Clin. Chem. 55: 641–658 [DOI] [PubMed] [Google Scholar]
- 36. Wang D., et al. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. U. S. A. 99: 15687–15692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watzinger F., et al. 2004. Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J. Clin. Microbiol. 42: 5189–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.