Abstract
Phoenix 100 and Vitek 2 (operating with the current colorimetric cards) are commonly used in hospital laboratories for rapid identification of microorganisms. The present meta-analysis aims to evaluate and compare their performance on Gram-positive and Gram-negative bacteria. The MEDLINE database was searched up to October 2010 for the retrieval of relevant articles. Pooled correct identification rates were derived from random-effects models, using the arcsine transformation. Separate analyses were conducted at the genus and species levels; subanalyses and meta-regression were undertaken to reveal meaningful system- and study-related modifiers. A total of 29 (6,635 isolates) and 19 (4,363 isolates) articles were eligible for Phoenix and colorimetric Vitek 2, respectively. No significant differences were observed between Phoenix and Vitek 2 either at the genus (97.70% versus 97.59%, P = 0.919) or the species (92.51% versus 88.77%, P = 0.149) level. Studies conducted with conventional comparator methods tended to report significantly better results compared to those using molecular reference techniques. Speciation of Staphylococcus aureus was significantly more accurate in comparison to coagulase-negative staphylococci by both Phoenix (99.78% versus 88.42%, P < 0.00001) and Vitek 2 (98.22% versus 91.89%, P = 0.043). Vitek 2 also reached higher correct identification rates for Gram-negative fermenters versus nonfermenters at the genus (99.60% versus 95.90%, P = 0.004) and the species (97.42% versus 84.85%, P = 0.003) level. In conclusion, the accuracy of both systems seems modified by underlying sample- and comparator method-related parameters. Future simultaneous assessment of the instruments against molecular comparator procedures may facilitate interpretation of the current observations.
INTRODUCTION
Early provision of microorganism identification and susceptibility data permits efficient management of patients with infectious diseases and is associated with significant clinical and financial benefits, via the reduction of mortality rates and overall hospitalization costs (17). In view of this assumption, identification and antimicrobial susceptibility testing (AST) of clinical isolates is mainly achieved by means of fully automated systems in most medium- to high-throughput microbiology laboratories. Apart from shortened turnaround times, improved specimen handling, enhanced quality control, reproducibility, accuracy, and the ability to track results are further advantages prompting routine laboratories to adopt automated technologies for bacterial processing (18). Since the release of the AutoMicrobic System, designed in the late 1960s by McDonnell Douglas at the request of NASA, a plethora of products have made their appearances in this extremely demanding marketplace. Two of the major competitors in the field are the Phoenix 100, launched by Becton Dickinson in 2003, and the Vitek 2 system, introduced by bioMérieux in 1997.
With regard to identification, Phoenix utilizes a series of modified conventional, fluorogenic and chromogenic, substrates to cover a total of 145 Gram-positive and 161 Gram-negative taxa within 3 to 4 h (4). Vitek 2, combined with the original (Gram-positive) ID-GPC and (Gram-negative) ID-GNB identification cards, using fluorescence reading technology, required up to 3 h to identify 52 Gram-positive and 98 Gram-negative taxa; with the redesigned (Gram-positive) ID-GP and (Gram-negative) ID-GN formats, based on colorimetric detection, the system covers a broadened database of 115 Gram-positive and 135 Gram-negative taxa in an approximate turnaround time of 10 h (6).
Speciation of an isolate provides essential information on its pathogenic potential and is of utmost importance for the correct interpretation of AST results; therefore, the identification performance of both Phoenix and Vitek 2 has already undergone numerous evaluations by laboratories having them integrated in the routine diagnostic workflow. The design of individual studies, with regard to the system being assessed, the identification procedure used as the comparator method, and the composition of the strain battery under investigation may account for the heterogeneous conclusions inferred by various researchers. A comprehensive quantitative synthesis of all published articles is necessary to shed light on the controversies of the literature. This meta-analysis aims to estimate and compare the accuracy of Phoenix and Vitek 2 for the identification of Gram-positive and Gram-negative species.
MATERIALS AND METHODS
Search strategy.
The present meta-analysis was performed in accordance with the PRISMA guidelines (42). A systematic computerized search of MEDLINE bibliographical database was performed to identify relevant studies (end-of-search date: 30 October 2010), using the search string “[Phoenix OR Vitek] AND identification”. Language restrictions were not applied, while the references of eligible articles were also checked. Two investigators (K.-S.C. and T.N.S.) working independently searched the literature and extracted data from eligible studies. Disagreements were resolved by discussion and consensus.
Eligible studies and data abstraction.
Articles evaluating Phoenix, Vitek 2, or both systems concomitantly for their ability to identify Gram-positive and/or Gram-negative bacteria were considered eligible for the meta-analysis; the use of a comparator method had to be clearly stated by the authors.
The following exclusion criteria were adopted during selection of eligible trials and data abstraction.
(i) The present meta-analysis focused on common aerobic and facultative anaerobic pathogens, to which Phoenix panels for Gram-positive, Gram-negative, and streptococcal taxa and Vitek 2 cards for Gram-positive (fluorescent ID-GPC and colorimetric ID-GP) and Gram-negative (fluorescent ID-GNB and colorimetric ID-GN) taxa are dedicated. Studies assessing the performance of NH (for Neisseria and Haemophilus species), ANC (for anaerobes and corynebacteria), ID-YST, and YST (for yeasts) cards were excluded.
(ii) Both Phoenix and Vitek 2 are designed for inoculation with pure colonies grown on appropriate solid media. Studies performing direct inoculation of the systems with positive blood cultures were excluded from the analysis.
(iii) Only results of clinical isolates were of concern. Therefore, strains of environmental, veterinary, or unspecified origin were also excluded.
(iv) Strains representing species not included in the database of the system under evaluation were not considered for the calculations.
(v) Reference or type strains were also excluded from the calculations, to circumvent their inevitable overlapping and repetition among relevant studies.
(vi) Enough evidence has already accumulated in the literature regarding the suboptimal performance of Vitek 2 fluorescent cards to accurately identify bacteria; since 2004, bioMérieux exclusively markets the new colorimetric formats designed to improve the accuracy and broaden the database of the previous fluorescent versions. In this context, studies evaluating Vitek 2 fluorescent identification cards for Gram-positive (ID-GPC) and Gram-negative (ID-GNB) bacteria were also excluded; nevertheless, a grand-total analysis on all studies (using either fluorescent or colorimetric cards) is secondarily presented in the supplemental material to ensure the comprehensiveness of the meta-analytical approach.
Data abstraction was conducted for the total number of isolates, Gram-positive, and Gram-negative strains under investigation; Staphylococcus aureus strains, coagulase-negative staphylococci (CoNS), enterococci, streptococci, Gram-negative glucose fermenters, and nonfermenters were also considered separately. In addition, the following data were abstracted: first author name, publication year, type of comparator method, type of identification card (fluorescent or colorimetric) for Vitek 2, the genera and species under investigation, and the proportions of Gram-positive, Gram-negative, S. aureus isolates, CoNS, enterococci, streptococci, fermenters, and nonfermenters included in individual studies.
Definitions.
The Phoenix system leads to an identification result when a species or group of species is identified with a >90% confidence level. For Vitek 2, the confidence value is expressed by seven different categories of results: excellent, very good, good, acceptable identification (only one result is provided), low discrimination (more than one result is given, whereupon the software suggests additional tests), inconclusive identification, and unidentified.
Therefore, each identification result obtained by Phoenix and/or Vitek 2 in comparison to the reference method was classified as follows. (i) The first classification was “correct identification” at the species and/or genus level. For Phoenix, correct identification was defined as any result concordant with the reference method at the species and/or genus level; accordingly for Vitek 2 any excellent/very good/good/acceptable identification concordant with the reference method was referred to as correct identification at the species and/or genus level. A low discrimination result between species of the correct genus (including or not the correct species) was considered correct identification at the genus level. (ii) The second classification was “misidentification.” For either system, misidentification was defined as any result discordant with the reference method at the genus and species level. (iii) The third classification was “no identification.” This category was allocated when either system was unable to provide any identification or yielded an “inconclusive” (in the case of Vitek 2) result. The “misidentification” and “no identification” categories were merged for the purposes of this meta-analysis to establish a binary “correct/not correct” conceptual framework.
For low discrimination results suggesting the correct species among viable choices, an alternative (species level) analysis was undertaken, counting these results as correct identifications at the genus and species level. Of note, the resolution of multiple-choice identifications required the performance of supplemental tests, implying a significant delay in the definitive speciation of the respective isolates.
Of note, from a biostatistical point of view, correct identification rates should be interpreted as sensitivity rates, since they conceptually correspond to the number of true positives divided by the sum of true-positive and false-negative results.
Statistical analysis.
For both Phoenix and Vitek 2, separate analyses were performed regarding their identification performance at the genus and species levels.
Based on the appropriate numbers in each study, the correct identification rates at the genus and species levels (isolates with correct identification/total number of isolates) were calculated; importantly, given that in numerous studies these rates were very close or equal to unity the arcsine (Freeman-Tukey), transformation was implemented (25) and preferred over logistic regression (53). This transformation results in a roughly normally distributed variable (54, 63) and exhibits satisfactory properties in terms of variance stabilization, the latter representing a problem during the meta-analysis of rates (53).
The arcsine-transformed rates were subsequently pooled through random-effects models (16). Pooled correct identification rates were derived after back-transformation of the pooled arcsine-transformed summary estimates. Apart from the overall meta-analysis, subanalyses were undertaken in Gram-positive (total, S. aureus, CoNS, Enterococcus spp., and Streptococcus spp.) and Gram-negative (total, fermenters, and nonfermenters) bacteria. The appropriate z-tests were performed to estimate the level of statistical significance regarding (i) the difference in the respective correct identification rates between Phoenix and Vitek 2, as well as (ii) within-system differences (molecular versus conventional comparator methods, Gram-positive versus Gram-negative isolates, S. aureus versus CoNS strains and Gram-negative fermenters versus nonfermenters). Z-tests are univariate tests, which were performed given that arcsine-transformed rates are normally distributed (3, 54, 63). Each z-value was appropriately calculated as the difference in pooled arcsine-transformed rates divided by the standard error of the difference. The level of statistical significance for z-tests was set at P < 0.05. Of note, a subanalysis on studies directly comparing Phoenix and Vitek 2 on the same isolates was performed in order to minimize any confounding.
To circumvent the effect of within-system multiple comparisons, as well as possible confounding at the numerous subanalyses, a multiple (multivariate) meta-regression adjustment algorithm (29) was performed for the items selected as significant at the univariate analysis (the P value for entry was fixed at < 0.05) for both Phoenix and Vitek 2.
Meta-regression was performed to evaluate whether correct (arcsine-transformed) identification rates were modified by the proportion of Gram-positive, Gram-negative, S. aureus, CoNS, Enterococcus spp., Streptococcus spp., Gram-negative fermenters, and nonfermenters included in each study; the increment was set at increase by 1% in the proportion of each pathogen.
Between-study heterogeneity was quantified using the I2 measure (32). Publication bias was evaluated by using the rank correlation method of Begg (5), the Egger's regression method (20), and its random-effects analogue (61); for the interpretation of publication bias, statistical significance was defined as P < 0.1.
Statistical analyses were conducted with STATA 11.1 Intercooled (STATA Corp., College Station, TX). Forest plots were generated by StatsDirect statistical software Version 2.7.2 (StatsDirect, Ltd., Altrincham, Cheshire, United Kingdom).
RESULTS
Eligible studies.
Of the 624 abstracts retrieved through the search criteria, 527 articles were excluded from the meta-analysis as irrelevant; these articles included studies assessing the AST performance of Phoenix or Vitek 2, reports evaluating the AutoMicrobic or the first-generation Vitek system, other types of publications employing Phoenix or any of the Vitek versions for the processing of clinical isolates outside the context of an evaluation (e.g., case reports, surveillance studies, etc.) and nonmicrobiological papers. Among relevant articles, 19 studies were excluded given that they reported on the accuracy of Vitek 2 NH, ANC, ID-YST, and YST cards, nine studies were excluded because inoculation of Phoenix or Vitek 2 was performed directly from positive blood cultures, one study (using the colorimetric Vitek 2 ID-GN card) was excluded since it was carried out exclusively on environmental strains, four studies (one using the colorimetric ID-GP and three using the fluorescent ID-GPC Vitek 2 cards) were excluded due to reporting reasons (clinical isolates were tested together with reference and/or animal strains, while identification results for the former were not provided separately), two studies were excluded because the type of Vitek 2 card was not specified, and 18 studies were excluded since they assessed exclusively the identification performance of Vitek 2 fluorescent ID-GPC and ID-GNB cards. As a result, a total of 44 publications were included in the meta-analysis: 29 (8–11, 13, 14, 18, 19, 21–24, 28, 31, 33, 35, 36, 39–41, 43–46, 48, 50, 56, 59, 60) reported on the identification performance of Phoenix, and 19 (1, 2, 15, 19, 26, 27, 30, 34, 37, 38, 40, 41, 46, 47, 49, 51, 55, 62, 64) reported on the identification performance of Vitek 2 operating with the colorimetric ID-GP and ID-GN cards; among them four (19, 40, 41, 46) were direct-comparison studies (see Fig. S1 in the supplemental material for the trial flow chart and Table S1 in the supplemental material for the characteristics of eligible studies).
Comparison between Phoenix and Vitek 2.
Table 1, Table 2, and Fig. 1 depict the results of the meta-analysis at the genus and species levels (see Table S2 in the supplemental material for the results of the alternative analysis).
Table 1.
Results of the meta-analysis at the genus levela
| Genus level | Phoenix |
Vitek 2 |
P between systems (z-value) | ||||
|---|---|---|---|---|---|---|---|
| No. of isolates (no. of studies) | Correct identification rate (95% CI) | P within system (z-value) | No. of isolates (no. of studies) | Correct identification rate (95% CI) | P within system (z-value) | ||
| Overall analysis | 4,763 (21) | 97.70 (96.22-98.81) | 3,423 (15) | 97.59 (95.76-98.92) | 0.919 (0.102) | ||
| Comparator method | 0.013 (2.481) | 0.115 (1.576) | |||||
| Molecular | 387 (7) | 94.56 (90.82-97.39) | 543 (6) | 94.34 (85.31-99.25) | 0.954 (0.058) | ||
| Conventional | 4,376 (14) | 98.44 (97.04-99.40) | 2,880 (9) | 98.71 (97.79-99.39) | 0.704 (0.380) | ||
| Gram stain | 0.258 (1.131) | 0.518 (0.647) | |||||
| Positive | 2,626 (15) | 98.32 (96.96-99.30) | 1,523 (8) | 98.22 (95.48-99.72) | 0.933 (0.084) | ||
| Negative | 2,588 (11) | 97.13 (95.10-98.64) | 1,900 (8) | 97.16 (94.37-99.04) | 0.982 (0.022) | ||
| Subanalysis on Gram-positive bacteria | |||||||
| Comparator method | 0.024 (2.260) | 0.663 (0.436) | |||||
| Molecular | 193 (4) | 95.52 (91.83-98.13) | 330 (3) | 97.16 (84.09-99.48) | 0.733 (0.341) | ||
| Conventional | 2,433 (11) | 98.81 (97.55-99.63) | 1,193 (5) | 98.72 (96.61-99.83) | 0.365 (0.907) | ||
| Staphylococcus spp. | 0.053 (1.932) | 0.498 (0.677) | |||||
| Staphylococcus aureus | 791 (9) | 99.78 (99.33-99.98) | 69 (3) | 99.09 (95.56-99.96) | 0.435 (0.780) | ||
| Coagulase-negative staphylococci | 670 (8) | 98.70 (97.03-99.70) | 372 (4) | 99.74 (98.96-99.99) | 0.113 (1.587) | ||
| Enterococcus spp. | 526 (8) | 98.27 (95.39-99.78) | 239 (3) | 99.70 (98.61-99.99) | 0.153 (1.430) | ||
| Streptococcus spp. | 778 (7) | 96.70 (94.83-98.14) | 841 (5) | 96.10 (91.39-99.01) | 0.774 (0.287) | ||
| Subanalysis on Gram-negative bacteria | |||||||
| Comparator method | 0.077 (1.769) | 0.050 (1.957) | |||||
| Molecular | 192 (3) | 92.40 (83.24-98.14) | 213 (3) | 90.30 (74.85-98.84) | 0.771 (0.291) | ||
| Conventional | 2,396 (8) | 98.02 (96.16-99.27) | 1,687 (5) | 98.91 (97.95-99.56) | 0.293 (1.051) | ||
| Glucose fermentation | 0.811 (0.239) | 0.004 (2.898) | |||||
| Fermenters | 1,907 (9) | 97.62 (95.56-99.05) | 1,028 (4) | 99.60 (99.12-99.89) | 0.006 (2.765) | ||
| Nonfermenters | 353 (7) | 97.93 (95.70-99.36) | 872 (7) | 95.90 (91.61-98.71) | 0.292 (1.055) | ||
| Subanalysis on direct comparison studies | 480 (3) | 97.31 (95.68-98.56) | 480 (3) | 98.11 (94.90-99.77) | 0.598 (0.527) | ||
CI, confidence interval.
Table 2.
Results of the meta-analysis at the species levela
| Species level | Phoenix |
Vitek 2 |
P between systems (z-value) | ||||
|---|---|---|---|---|---|---|---|
| No. of isolates (no. of studies) | Correct identification rate (95% CI) | P within system (z-value) | No. of isolates (no. of studies) | Correct identification rate (95% CI) | P within system (z-value) | ||
| Overall analysis | 6,635 (29) | 92.51 (89.54-94.99) | 4,363 (19) | 88.77 (83.91-92.82) | 0.149 (1.442) | ||
| Comparator method | 0.00001 (4.358) | 0.00003 (4.158) | |||||
| Molecular | 697 (9) | 80.07 (70.94-87.80) | 903 (9) | 78.12 (67.39-87.21) | 0.770 (0.292) | ||
| Conventional | 5,938 (20) | 95.76 (93.84-97.36) | 3,460 (10) | 95.22 (93.67-96.55) | 0.645 (0.461) | ||
| Gram stain | 0.652 (0.451) | 0.712 (0.369) | |||||
| Positive | 3,152 (18) | 93.60 (89.82-96.55) | 2,056 (11) | 90.18 (84.71-94.56) | 0.253 (1.142) | ||
| Negative | 3,481 (16) | 92.45 (88.38-95.68) | 2,307 (10) | 88.58 (80.70-94.61) | 0.317 (1.001) | ||
| Subanalysis on Gram-positive bacteria | |||||||
| Comparator method | 0.003 (2.989) | 0.006 (2.734) | |||||
| Molecular | 418 (5) | 82.45 (69.94-92.11) | 614 (5) | 83.01 (72.65-91.14) | 0.939 (0.077) | ||
| Conventional | 2,734 (13) | 96.35 (94.06-98.11) | 1,442 (6) | 94.56 (91.61-96.92) | 0.292 (1.054) | ||
| Staphylococcus spp. | <0.00001 (4.565) | 0.043 (2.028) | |||||
| Staphylococcus aureus | 791 (9) | 99.78 (99.33-99.98) | 90 (4) | 98.22 (94.52-99.90) | 0.114 (1.579) | ||
| Coagulase-negative staphylococci | 895 (9) | 88.42 (79.38-95.12) | 740 (7) | 91.89 (84.96-96.82) | 0.489 (0.692) | ||
| Enterococcus spp. | 570 (9) | 96.91 (93.38-99.13) | 275 (4) | 95.84 (91.39-98.73) | 0.654 (0.448) | ||
| Streptococcus spp. | 778 (7) | 93.18 (89.57-96.04) | 930 (6) | 88.92 (79.55-95.68) | 0.311 (1.014) | ||
| Subanalysis on Gram-negative bacteria | |||||||
| Comparator method | 0.001 (3.236) | 0.003 (2.996) | |||||
| Molecular | 277 (4) | 76.91 (61.90-89.05) | 289 (4) | 70.90 (48.06-89.23) | 0.640 (0.468) | ||
| Conventional | 3,204 (12) | 95.54 (92.74-97.70) | 2,018 (6) | 95.80 (94.31-97.08) | 0.854 (0.183) | ||
| Glucose fermentation | 0.546 (0.604) | 0.003 (3.005) | |||||
| Fermenters | 2,446 (11) | 94.94 (91.11-97.74) | 1,271 (5) | 97.42 (94.66-99.21) | 0.221 (1.224) | ||
| Nonfermenters | 686 (10) | 92.82 (85.28-97.82) | 1,036 (9) | 84.85 (73.76-93.28) | 0.172 (1.365) | ||
| Subanalysis on direct comparison studies | 705 (4) | 83.61 (72.03-92.53) | 705 (4) | 88.77 (78.61-95.94) | 0.457 (0.744) | ||
CI, confidence interval.
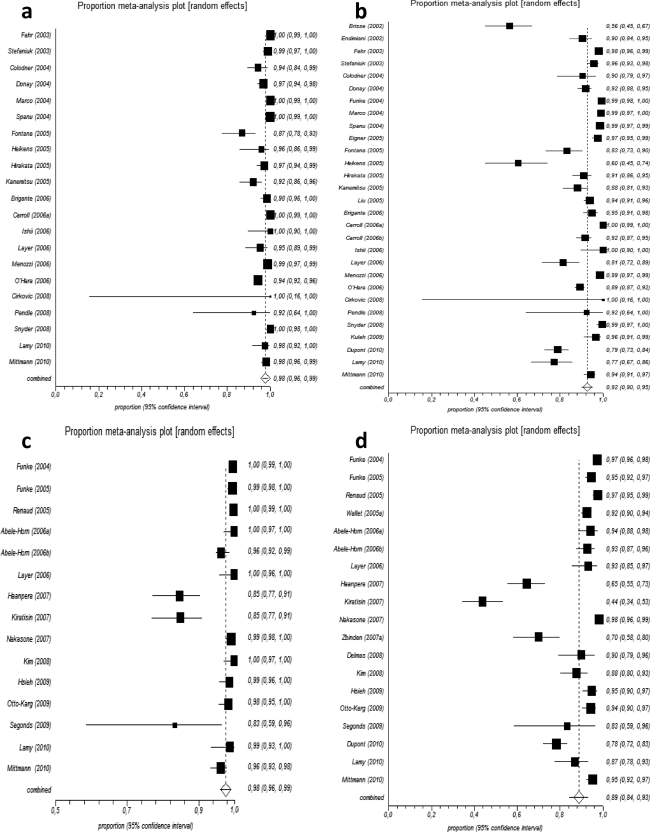
Fig. 1.
Results of the meta-analysis for Phoenix and Vitek 2 at the genus (a and c, respectively) and species (b and d, respectively) levels. Each study is shown by a point estimate of the effect size (correct identification rate) and its 95% confidence intervals derived from the arcsine (Freeman-Tukey) transformation algorithm. The diamond represents the summary random-effects estimate from the meta-analysis.
Regarding the overall analyses, no significant differences were observed between Phoenix and Vitek 2 either at the genus (97.70% versus 97.59%, P = 0.919) or the species (92.51% versus 88.77%, P = 0.149) level. The lack of difference persisted at the overall subanalyses on studies conducted with conventional (P = 0.704 for genus and P = 0.645 for species) or molecular reference methods (P = 0.954 and P = 0.770, respectively), as well as on Gram-positive (P = 0.933 and P = 0.253, respectively) and Gram-negative (P = 0.982 and P = 0.317, respectively) bacteria.
Accordingly, the subanalyses on S. aureus, CoNS, Enterococcus spp, Streptococcus spp., and Gram-negative nonfermenters did not reveal any significant finding at both genus and species levels. The superiority of Vitek 2 over Phoenix in the identification of Gram-negative fermenters was demonstrated at the genus level (97.62% versus 99.60%, P = 0.006), albeit not replicated at the species level.
The subanalysis on studies directly comparing the systems did not demonstrate any significant difference (97.31% versus 98.11%, P = 0.598 for genus and 83.61% versus 88.77%, P = 0.457 for species identification).
Within-systems analyses.
With respect to Phoenix, correct identification rates were higher in conventional compared to molecular-based studies, as a rule. No significant difference was observed between Gram-positive and Gram-negative bacteria at both genus and species levels. Species identification of S. aureus was significantly more accurate in comparison to CoNS (99.78% versus 88.42%, P < 0.00001), with a borderline significance obtained at the genus level (P = 0.053).
Regarding Vitek 2, studies conducted with conventional comparator methods yielded significantly better results than those using molecular techniques at the overall species analysis (P = 0.00003), as well as at the subanalyses on Gram-positive (P = 0.006) and Gram-negative (P = 0.003) bacteria; at the genus level, this finding was confined to the latter (P = 0.050). No significant difference was observed between Gram-positive and Gram-negative bacteria regarding genus and species identification. Vitek 2 was significantly more accurate in the identification of S. aureus versus CoNS at the species level (98.22% versus 91.89%, P = 0.043), as well as in the identification of fermenters versus nonfermenters at both genus (99.60% versus 95.90%, P = 0.004) and species (97.42% versus 84.85%, P = 0.003) levels.
Multiple meta-regression analysis, where applicable, confirmed the univariate associations on all occasions except for one (see Table S3 in the supplemental material). Specifically, the superior performance of Vitek 2 for species level identification of S. aureus did not persist at the multiple meta-regression approach (P = 0.524 for S. aureus and P = 0.023 for the comparator method). Importantly, it should be stressed that only one study on S. aureus had used a molecular comparator method (15); the accumulation of further studies using molecular reference procedures for S. aureus speciation seems mandatory, so as to elucidate the independence of the effects mediated by the comparator method and staphylococcal species per se.
Assessment of sample-related modifiers and publication bias.
Regarding Phoenix (see Fig. S2 in the supplemental material), meta-regression revealed that correct identification rates correlated positively with the proportion of S. aureus isolates in individual studies at the genus (b = +0.004, P = 0.024) and species (b [regression coefficient] = +0.0069, P = 0.012) levels. Furthermore, the accuracy of the system at the species level correlated marginally negatively with the proportion of CoNS (b = −0.0039, P = 0.053). No modifying effects were observed regarding the proportions of Gram-positive, Gram-negative bacteria, enterococci, streptococci, fermenters, and nonfermenters.
Concerning Vitek 2, meta-regression did not reveal any significant effects mediated by the potential modifiers.
Significant publication bias emerged on numerous occasions (see Tables S4 and S5 in the supplemental material). Close inspection of Phoenix and Vitek 2 Begg's plots (genus and species level overall analyses) revealed that the missing, hypothetically existing, studies were located in the upper right quadrant of the funnel plots in all cases, i.e., underestimation of performance tended to emerge in smaller studies (see Fig. S3 and S4 in the supplemental material).
DISCUSSION
The present meta-analysis highlights the comparability in the identification performance of Phoenix and Vitek 2 at both genus and species levels. Subanalyses on Gram-positive and Gram-negative bacteria, as well as on studies using conventional or molecular comparator methods reproduced the lack of significant differences between the instruments. The analysis conducted exclusively on direct comparison studies, as well as the alternative analysis for Vitek 2 (counting low discrimination results as correct species identifications) lent further support to the above observations, yielding no significant differences between Phoenix and Vitek 2.
Within-system analyses revealed that accuracy rates of both instruments ranged widely depending on the type of method adopted for reference identification; studies conducted with conventional reference methods tended to report significantly better results compared to those using molecular comparator techniques, as a rule. Molecular evaluations of Phoenix and Vitek 2 may be considered of higher quality than conventional investigations. The majority of the latter reports, published to date, have not used tedious and time-consuming reference identification schemes; instead, the instruments were compared to other phenotypic systems, most often the API galleries and molecular confirmation was not undertaken in case of concordant results. Theoretically, some of these “correctly identified” strains might actually have been misidentified by both the reference phenotypic system and the system under evaluation (41), calling into question the accuracy of the results furnished by such evaluations.
Apart from comparator method-related parameters, the present meta-analysis uncovered meaningful sample-related modifiers of the systems' performance. Species identification of S. aureus was significantly more accurate in comparison to CoNS by both Phoenix and Vitek 2, this difference being well explained by the variable phenotypic expression of the coagulase negative species, as well as by their slow metabolic rates, leading to ambiguous reactions within the short incubation times used by automated instruments (31, 58). Furthermore, Vitek 2 reached higher correct identification rates for Gram-negative fermenters versus nonfermenters at both genus and species levels, with the phenotypic variation, atypical biochemical characteristics and slow growth rates of the latter most probably accounting for this difference (7). Interestingly, the results of the meta-regression confirmed the findings derived from within-system analysis for Phoenix, whose accuracy correlated positively with S. aureus and negatively with CoNS relative frequency. The smaller number of studies on Vitek 2 may have precluded the reproduction of within-system findings by meta-regression, the latter yielding null associations.
An important measure of the value of a highly standardized commercial system is the capability of the manufacturer to maintain or even improve its performance over time. In this context, bioMérieux has converted fluorescent biochemicals and optics to colorimetric biochemicals and optics to broaden the database and boost the accuracy of the system, particularly for streptococci and Gram-negative nonfermenters (62). The same meta-analytical approach encompassing all published studies on Vitek 2 (see Tables S6 and S7 in the supplemental material) corroborated the superiority of the current colorimetric over the previous fluorescent identification cards.
Finally, several meaningful limitations of the present meta-analysis, which nevertheless seem quite inherent in the current literature, should be acknowledged. First, conference proceedings were not included to ensure detailed reporting of data. Moreover, the findings of the meta-analysis may have been distorted, at a certain extent, due to the existence of significant publication bias, which seems fairly common in the context of meta-analyses on diagnostic accuracy (57). Nevertheless, visual inspection of the relevant funnel plots revealed that the pattern of asymmetry was essentially the same for both Phoenix and Vitek 2, possibly affecting the results at the same direction for each system and thus not substantially interfering with potential underlying differences.
At present, the application of more elaborate, bivariate meta-analysis models was not feasible (52), as the number of direct comparison studies was less than five (i.e., only four). Forty of forty-four eligible articles evaluated separately Phoenix or Vitek 2 in various laboratories, by different researchers, on a wide range of bacterial species, recovered from variable patient populations and clinical conditions; the synthesis of such reports implied the existence of underlying confounding and sizeable heterogeneity. In an attempt to overcome this drawback, a subanalysis on the four direct comparison studies was undertaken, although the generalization of the relevant findings was precluded by the fact that these publications focused on particular genera, namely, Staphylococcus spp. (19, 41), Streptococcus spp. (46), and Aeromonas spp. (40).
According to the Manual of Clinical Microbiology, automated identification systems should ideally achieve an accuracy rate of no less than 90% in comparison to reference methods (12). Evidently, the present meta-analysis points to the potential for further improvement in the performance of both Phoenix and Vitek 2. When interpreting individual studies, the scientific audience should be aware of the underlying meaningful system-, sample-, and comparator method-related parameters affecting the reported results. Future studies to evaluate the instruments should preferably use molecular methods for reference identification, directly compare both systems wherever feasible, and provide data for the less common species, whose separate synthesis was not possible in this meta-analysis.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Abele-Horn M., Hommers L., Trabold R., Frosch M. 2006. Validation of Vitek 2 version 4.01 software for detection, identification, and classification of glycopeptide-resistant enterococci. J. Clin. Microbiol. 44: 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abele-Horn M., Stoy K., Frosch M., Reinert R. R. 2006. Comparative evaluation of a new Vitek 2 system for identification and antimicrobial susceptibility testing of Streptococcus pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 25: 55–57 [DOI] [PubMed] [Google Scholar]
- 3. Altman D. G., Bland J. M. 2003. Interaction revisited: the difference between two estimates. BMJ 326: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dickinson Becton. 2005. BD Phoenix system user's manual, document L003342(M). Becton Dickinson and Company, Sparks, MD [Google Scholar]
- 5. Begg C. B., Mazumdar M. 1994. Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101 [PubMed] [Google Scholar]
- 6.bioMérieux 2006. Vitek 2 product information, document 510769-4EN1. bioMérieux, Inc., Durham, NC [Google Scholar]
- 7. Bosshard P. P., et al. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the Vitek 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 44: 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brigante G., et al. 2006. Use of the Phoenix automated system for identification of Streptococcus and Enterococcus spp. J. Clin. Microbiol. 44: 3263–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brisse S., et al. 2002. Comparative evaluation of the BD Phoenix and Vitek 2 automated instruments for identification of isolates of the Burkholderia cepacia complex. J. Clin. Microbiol. 40: 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carroll K. C., et al. 2006. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of staphylococci and enterococci. J. Clin. Microbiol. 44: 2072–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carroll K. C., et al. 2006. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterobacteriaceae. J. Clin. Microbiol. 44: 3506–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carroll K. C., Weinstein M. P. 2007. Manual and automated systems for detection and identification of microorganisms, p. 192–217 In Murray P. R. (ed.), Manual of clinical microbiology, 9th ed., vol. 1 ASM Press, Washington, DC [Google Scholar]
- 13. Cirkovic I., et al. 2008. Identification and antimicrobial susceptibility testing of Staphylococcus vitulinus by the BD Phoenix automated microbiology system. Curr. Microbiol. 57: 158–160 [DOI] [PubMed] [Google Scholar]
- 14. Colodner R., et al. 2004. Identification of the emerging pathogen Vibrio vulnificus biotype 3 by commercially available phenotypic methods. J. Clin. Microbiol. 42: 4137–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delmas J., et al. 2008. Evaluation of the Vitek 2 system with a variety of Staphylococcus species. J. Clin. Microbiol. 46: 311–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DerSimonian R., Laird N. 1986. Meta-analysis in clinical trials. Control Clin. Trials 7: 177–188 [DOI] [PubMed] [Google Scholar]
- 17. Doern G. V., Vautour R., Gaudet M., Levy B. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 32: 1757–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Donay J. L., et al. 2004. Evaluation of the automated phoenix system for potential routine use in the clinical microbiology laboratory. J. Clin. Microbiol. 42: 1542–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dupont C., et al. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin. Microbiol. Infect. 16: 998–1004 [DOI] [PubMed] [Google Scholar]
- 20. Egger M., Davey Smith G., Schneider M., Minder C. 1997. Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eigner U., Schmid A., Wild U., Bertsch D., Fahr A. M. 2005. Analysis of the comparative workflow and performance characteristics of the Vitek 2 and Phoenix systems. J. Clin. Microbiol. 43: 3829–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Endimiani A., et al. 2002. Identification and antimicrobial susceptibility testing of clinical isolates of nonfermenting gram-negative bacteria by the Phoenix automated microbiology system. New Microbiol. 25: 323–329 [PubMed] [Google Scholar]
- 23. Fahr A. M., et al. 2003. Two-center collaborative evaluation of the performance of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterococcus spp. and Staphylococcus spp. J. Clin. Microbiol. 41: 1135–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fontana C., Favaro M., Pelliccioni M., Pistoia E. S., Favalli C. 2005. Use of the MicroSeq 500 16S rRNA gene-based sequencing for identification of bacterial isolates that commercial automated systems failed to identify correctly. J. Clin. Microbiol. 43: 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Freeman M., Tukey J. 1950. Transformations related to the angular and the square root. Ann. Math. Statist. 21: 607–611 [Google Scholar]
- 26. Funke G., Funke-Kissling P. 2004. Evaluation of the new Vitek 2 card for identification of clinically relevant gram-negative rods. J. Clin. Microbiol. 42: 4067–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Funke G., Funke-Kissling P. 2005. Performance of the new Vitek 2 GP card for identification of medically relevant gram-positive cocci in a routine clinical laboratory. J. Clin. Microbiol. 43: 84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Funke G., Funke-Kissling P. 2004. Use of the BD Phoenix automated microbiology system for direct identification and susceptibility testing of gram-negative rods from positive blood cultures in a three-phase trial. J. Clin. Microbiol. 42: 1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Greco S., Rulli M., Girardi E., Piersimoni C., Saltini C. 2009. Diagnostic accuracy of in-house PCR for pulmonary tuberculosis in smear-positive patients: meta-analysis and meta-regression. J. Clin. Microbiol. 47: 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haanpera M., Jalava J., Huovinen P., Meurman O., Rantakokko-Jalava K. 2007. Identification of alpha-hemolytic streptococci by pyrosequencing the 16S rRNA gene and by use of Vitek 2. J. Clin. Microbiol. 45: 762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heikens E., Fleer A., Paauw A., Florijn A., Fluit A. C. 2005. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 43: 2286–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Higgins J. P., Thompson S. G., Deeks J. J., Altman D. G. 2003. Measuring inconsistency in meta-analyses. BMJ 327: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirakata Y., et al. 2005. Evaluation of the BD Phoenix automated microbiology system SMIC/ID panel for identification and antimicrobial susceptibility testing of Streptococcus spp. Diagn. Microbiol. Infect. Dis. 53: 169–173 [DOI] [PubMed] [Google Scholar]
- 34. Hsieh W. S., Sung L. L., Tsai K. C., Ho H. T. 2009. Evaluation of the Vitek 2 cards for identification and antimicrobial susceptibility testing of non-glucose-fermenting Gram-negative bacilli. APMIS 117: 241–247 [DOI] [PubMed] [Google Scholar]
- 35. Ishii Y., et al. 2006. Identification of biochemically atypical Staphylococcus aureus clinical isolates with three automated identification systems. J. Med. Microbiol. 55: 387–392 [DOI] [PubMed] [Google Scholar]
- 36. Kanemitsu K., et al. 2005. Evaluation of the BD Phoenix SMIC/ID, a new streptococci identification and antimicrobial susceptibility panel, for potential routine use in a university-based clinical microbiology laboratory. Diagn. Microbiol. Infect. Dis. 53: 101–105 [DOI] [PubMed] [Google Scholar]
- 37. Kim M., et al. 2008. Comparison of the MicroScan, Vitek 2, and Crystal GP with 16S rRNA sequencing and MicroSeq 500 v2.0 analysis for coagulase-negative staphylococci. BMC Microbiol. 8: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kiratisin P., Santanirand P., Chantratita N., Kaewdaeng S. 2007. Accuracy of commercial systems for identification of Burkholderia pseudomallei versus Burkholderia cepacia. Diagn. Microbiol. Infect. Dis. 59: 277–281 [DOI] [PubMed] [Google Scholar]
- 39. Kulah C., et al. 2009. Detecting imipenem resistance in Acinetobacter baumannii by automated systems (BD Phoenix, Microscan WalkAway, Vitek 2): high error rates with Microscan WalkAway. BMC Infect. Dis. 9: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lamy B., et al. 2010. Accuracy of 6 commercial systems for identifying clinical Aeromonas isolates. Diagn. Microbiol. Infect. Dis. 67: 9–14 [DOI] [PubMed] [Google Scholar]
- 41. Layer F., Ghebremedhin B., Moder K. A., Konig W., Konig B. 2006. Comparative study using various methods for identification of Staphylococcus species in clinical specimens. J. Clin. Microbiol. 44: 2824–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liberati A., et al. 2009. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J. Clin. Epidemiol. 62: e1–e34 [DOI] [PubMed] [Google Scholar]
- 43. Liu Z. K., Ling T. K., Cheng A. F. 2005. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of common clinical isolates. Med. Principles Pract. 14: 250–254 [DOI] [PubMed] [Google Scholar]
- 44. Marco F., Jurado A., Jimenez de Anta M. T. 2004. Evaluation of the Phoenix system for identifying and determining the susceptibility of clinical isolates: comparative study with the Microscan system. Rev. Esp. Quimioter. 17: 169–176 (In Spanish.) [PubMed] [Google Scholar]
- 45. Menozzi M. G., et al. 2006. Two-center collaborative evaluation of performance of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of gram-negative bacteria. J. Clin. Microbiol. 44: 4085–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mittman S. A., Huard R. C., Della-Latta P., Whittier S. 2010. Comparison of the automated Phoenix with the Vitek 2 for the identification of Streptococcus pneumoniae. Can. J. Microbiol. 56: 326–332 [DOI] [PubMed] [Google Scholar]
- 47. Nakasone I., Kinjo T., Yamane N., Kisanuki K., Shiohira C. M. 2007. Laboratory-based evaluation of the colorimetric Vitek-2 compact system for species identification and of the advanced expert system for detection of antimicrobial resistances: Vitek-2 compact system identification and antimicrobial susceptibility testing. Diagn. Microbiol. Infect. Dis. 58: 191–198 [DOI] [PubMed] [Google Scholar]
- 48. O'Hara C. M. 2006. Evaluation of the Phoenix 100 ID/AST system and NID panel for identification of Enterobacteriaceae, Vibrionaceae, and commonly isolated nonenteric gram-negative bacilli. J. Clin. Microbiol. 44: 928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Otto-Karg I., et al. 2009. Validation of Vitek 2 nonfermenting gram-negative cards and Vitek 2 version 4.02 software for identification and antimicrobial susceptibility testing of nonfermenting gram-negative rods from patients with cystic fibrosis. J. Clin. Microbiol. 47: 3283–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pendle S., et al. 2008. Difficulties in detection and identification of Enterococcus faecium with low-level inducible resistance to vancomycin, during a hospital outbreak. Clin. Microbiol. Infect. 14: 853–857 [DOI] [PubMed] [Google Scholar]
- 51. Renaud F. N., et al. 2005. Evaluation of the new Vitek 2 GN card for the identification of gram-negative bacilli frequently encountered in clinical laboratories. Eur. J. Clin. Microbiol. Infect. Dis. 24: 671–676 [DOI] [PubMed] [Google Scholar]
- 52. Riley R. D., Thompson J. R., Abrams K. R. 2008. An alternative model for bivariate random-effects meta-analysis when the within-study correlations are unknown. Biostatistics 9: 172–186 [DOI] [PubMed] [Google Scholar]
- 53. Rucker G., Schwarzer G., Carpenter J., Olkin I. 2009. Why add anything to nothing? The arcsine difference as a measure of treatment effect in meta-analysis with zero cells. Stat. Med. 28: 721–738 [DOI] [PubMed] [Google Scholar]
- 54. Ryu T., Mavromatis C. H., Bayer T., Voolstra C. R., Ravasi T. 2011. Unexpected complexity of the reef-building coral Acropora millepora transcription factor network. BMC Syst. Biol. 5: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Segonds C., et al. 2009. Microbiological and epidemiological features of clinical respiratory isolates of Burkholderia gladioli. J. Clin. Microbiol. 47: 1510–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Snyder J. W., Munier G. K., Johnson C. L. 2008. Direct comparison of the BD Phoenix system with the MicroScan WalkAway system for identification and antimicrobial susceptibility testing of Enterobacteriaceae and nonfermentative gram-negative organisms. J. Clin. Microbiol. 46: 2327–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Song F., Khan K. S., Dinnes J., Sutton A. J. 2002. Asymmetric funnel plots and publication bias in meta-analyses of diagnostic accuracy. Int. J. Epidemiol. 31: 88–95 [DOI] [PubMed] [Google Scholar]
- 58. Spanu T., et al. 2003. Use of the Vitek 2 system for rapid identification of clinical isolates of staphylococci from bloodstream infections. J. Clin. Microbiol. 41: 4259–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Spanu T., et al. 2004. Identification of methicillin-resistant isolates of Staphylococcus aureus and coagulase-negative staphylococci responsible for bloodstream infections with the Phoenix system. Diagn. Microbiol. Infect. Dis. 48: 221–227 [DOI] [PubMed] [Google Scholar]
- 60. Stefaniuk E., Baraniak A., Gniadkowski M., Hryniewicz W. 2003. Evaluation of the BD Phoenix automated identification and susceptibility testing system in clinical microbiology laboratory practice. Eur. J. Clin. Microbiol. Infect. Dis. 22: 479–485 [DOI] [PubMed] [Google Scholar]
- 61. Thompson S. G., Sharp S. J. 1999. Explaining heterogeneity in meta-analysis: a comparison of methods. Stat Med. 18: 2693–2708 [DOI] [PubMed] [Google Scholar]
- 62. Wallet F., Loiez C., Renaux E., Lemaitre N., Courcol R. J. 2005. Performances of Vitek 2 colorimetric cards for identification of gram-positive and gram-negative bacteria. J. Clin. Microbiol. 43: 4402–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Warton D. I., Hui F. K. 2011. The arcsine is asinine: the analysis of proportions in ecology. Ecology 92: 3–10 [DOI] [PubMed] [Google Scholar]
- 64. Zbinden A., Bottger E. C., Bosshard P. P., Zbinden R. 2007. Evaluation of the colorimetric Vitek 2 card for identification of gram-negative nonfermentative rods: comparison to 16S rRNA gene sequencing. J. Clin. Microbiol. 45: 2270–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.