Abstract
Norovirus is a major cause of acute gastroenteritis in humans. A norovirus outbreak occurred in Ohio in January 2010. Stool and saliva samples were obtained from six infected individuals. The full-length genomes of two representative strains (HS206 and HS210) were characterized. They belonged to GII.12 in the capsid but GII.g in the RNA polymerase region. Interestingly, an immunocompetent 2-year-old male shed virus for up to 30 days, as detected by real-time reverse transcription (RT)-PCR. Histo-blood group antigen (HBGA) typing of saliva showed that the norovirus strains infected various types of secretor-positive individuals (types A, B, and O). The viruslike particles of strain HS206 did not bind substantially to A/B/O antigens by synthetic HBGA binding, hemagglutination, or saliva binding assays. These results suggest that infection by this strain may not be A/B/O antigen dependent or that in vitro binding patterns do not always accurately reflect in vivo HBGA usage. This is different from the HBGA binding pattern of the previously reported GII.12/Aichi76 strain. Structural analysis of the predicted capsid of these GII.12 strains revealed two amino acid mismatches located near the HBGA binding sites. Four gnotobiotic pigs were inoculated orally with HS206 (6 × 1010 genomic equivalents [GE]/pig). Virus shedding began at postinoculation days (PID) 1 to 3 and continued up to PID 16 (1 × 105 to 2 × 107 GE/ml). Gastroenteritis cases caused by GII.12 noroviruses have been recently reported worldwide. We observed that this emerging GII.12 norovirus infected humans regardless of A/B/O blood type. The infection of pigs by strain HS206 suggests that interspecies transmission of this strain is possible under experimental conditions.
INTRODUCTION
Norovirus (NoV) is a major cause of nonbacterial acute gastroenteritis in both children and adults (17, 52, 54). Outbreaks are common in confined environments such as families, hospitals, and schools, and secondary attack rates are as high as 30% in these settings (22). Small numbers of infectious particles can cause infection (67). Virus shedding from patients recovered from illness (2, 46, 57, 63) or from asymptomatic individuals (1, 2, 21, 51) has been well documented, rendering control more difficult.
Norovirus, a member of the family Caliciviridae, has a single-stranded, positive-sense RNA genome of 7.3 to 7.7 kb, which encodes 3 open reading frames (ORFs) (23). ORF1 encodes a polyprotein that is cleaved by the virus-encoded protease to produce several nonstructural proteins, including the RNA-dependent RNA polymerase (RdRp), while ORF2 encodes a major capsid protein (VP1), and ORF3 encodes a minor structural protein (VP2) (3, 26). The capsid protein consists of a conserved shell (S) domain and a variable protruding (P) domain (55). Noroviruses are genetically diverse and are currently classified into five genogroups (GI to GV), with each genogroup being further divided into multiple genotypes (75). Among them, GI, GII, and GIV strains infect humans, with GII being the most prevalent and genetically diverse, including some GII NoV strains of swine origin (71, 75). Genotyping based on the complete capsid region remains the gold standard (32, 70, 75), and more than 30 genotypes have been designated, with the GII.4 genotype being dominant in humans. However, RNA recombination complicates genetic classification, since recombinant strains cluster within two distinct genotypes when different genomic regions (mainly before and after the ORF1-ORF2 junction region) are subjected to phylogenetic analysis (7, 8, 53). Consequently, genotyping based on the ORF1 (RdRp) region has been proposed (5, 8, 21), and numerous recombinant strains designated by RdRp genotype/capsid genotype have been reported, such as GI.2/GI.6 (33), GII.b/GII.3 (4, 13, 16), GII.b/GII.16 (15), GII.3/GII.13 (14), GII.4/GII.3 (15), GII.4/GII.12 (18), GII7/GII13 (47), and GIII.2/GIII.1 (8).
Norovirus recognizes histo-blood group antigens (HBGAs) as receptors or coreceptors (29, 39). In humans, HBGAs are complex carbohydrates present on red blood cells, and, in most of the population who have an active FUT2 gene, also on the surface of mucosal epithelial cells, as well as in body fluids such as saliva (secretor positive [Se+]) (68). Associations between HBGA phenotypes, secretor status, and susceptibility to certain NoV strains have been reported. In studies with human volunteers, individuals with blood type B had a decreased risk of infection, and nonsecretors were resistant to Norwalk virus (GI.1) infection (29, 39), whereas individuals were equally susceptible to Snow Mountain virus (SMV) (GII.2) regardless of their HBGA or secretor status (38). Nonsecretors were resistant to GII.3 and GII.4 NoVs, and individuals with blood type A were more susceptible to GII.4 strains in outbreak cases (65). There have been few reports on HBGA types and secretor statuses for uncommon NoV genotype infections (50).
Gnotobiotic (Gn) pigs are susceptible to strains of the dominant human NoV genotype, GII.4 (strain HS66) (12, 61, 62), and are an important animal model for human NoV infection. The detection of human NoV RNA in swine (48) and the high prevalence of antibodies against human GI and GII NoVs in porcine sera (19) raise questions as to the possible interspecies transmission of NoVs between humans and pigs. It is of interest, therefore, to expand previous studies to determine if other emerging new strains infect Gn pigs.
In this study, we delineate the clinical course of recent NoV outbreak cases in Ohio and characterize the full-length genomes of two representative strains. Viruslike particles (VLPs) of one strain were expressed in the baculovirus expression system and were used to examine its HBGA binding pattern. Three-dimensional (3D) structural analysis was performed to investigate the differences between current and previously reported GII.12 strains. Finally, Gn pigs were inoculated with one strain (HS206) to investigate whether it infects pigs and, thus, its potential for interspecies transmission between humans and pigs.
MATERIALS AND METHODS
Outbreak profile and sample collection.
From 9 January to 5 February 2010, 14 people from eight families in the departmental staff or their family members reported symptoms of acute gastroenteritis (vomiting and/or diarrhea) to the calicivirus research team at the Food Animal Health Research Program of the Ohio Agricultural Research Development Center, The Ohio State University, Wooster, OH. Fresh stool samples (total, 37) were provided voluntarily from six persons (patients A to F) from two families (families 1 and 2) (Table 1). None of them suffered from any underlying disease. An additional sample (strain HS239) was provided by patient A in July 2010 as a potential negative control when patient A had been free of symptoms for 4 months. Saliva samples were also provided by each person for HBGA typing (56). All the samples were immediately deidentified by the investigators so that only coded-log-number samples were used.
Table 1.
Profiles of individuals in the NoV outbreak and virus shedding in stool samples
| Family | Patient | Gender | Age | HBGA typea | Symptom(s) | No. of stool samples tested | Virus isolates | Range of NoV titers (GE/ml) | Duration of asymptomatic viral shedding/total duration of viral shedding (days) | Description |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A | Male | 2 yr | O, Le(a+b+) | Diarrhea, fever, appetite loss | 16 | HS200, HS207, HS234, HS239, HS255 | 2.6 × 105–2.0 × 1012 | 20/30 | Subsequent asymptomatic NoV GII.6 infection in July 2010 and symptomatic NoV GII.2 infection in January 2011 |
| B | Female | 34 yr | A, Le(a−b−) | Diarrhea, vomiting, headache | 7 | HS206, HS256 | 1.8 × 105–2.7 × 1011 | 12/15 | Subsequent symptomatic NoV GII.2 infection in January 2011 | |
| 2 | C | Male | 2 yr | O, Le(a−b−) | Vomiting | 3 | HS194, HS210, HS245 | 7.6 × 109–1.2 × 1012 | 7/8 | Prior symptomatic NoV GII.4 infection in January 2009 and subsequent symptomatic NoV GII.6 infection in December 2010 |
| D | Female | 9 mo | B, Le(a−b−) | None | 6 | HS209, HS246 | 1.8 × 1011–2.5 × 1012 | 9/9b | Subsequent symptomatic NoV GII.6 infection in December 2010 | |
| E | Male | 35 yr | O, Le(a−b+) | Diarrhea | 3 | HS224, HS244 | 8.0 × 108–9.7 × 109 | 1/3 | Subsequent symptomatic NoV GII.6 infection in December 2010 | |
| F | Female | 39 yr | B, Le(a−b+) | None | 2 | HS219, HS247 | 4.5 × 107 | 6/6b | Subsequent symptomatic NoV GII.6 infection in December 2010 |
Determined by using saliva samples in an ELISA. Le, Lewis.
Days after the onset of vomiting for patient C.
RNA extraction and viral detection/titration from stool samples.
Sample RNA was extracted from 10% (wt/vol) stool suspensions in sterile minimal essential medium (MEM; Invitrogen, Carlsbad, CA) by using a MagMAX viral RNA isolation kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Subsequently, RNA was screened for common diarrheal viruses, including GI and GII NoVs; human rotavirus groups A, B, and C; sapovirus; astrovirus; and adenovirus, by reverse transcription (RT)-PCR or PCR assays as described previously (31, 72, 73). All the stool samples were titrated in duplicate for GII NoV by real-time RT-PCR (31), with a standard curve generated from serial dilutions of plasmid DNA carrying the GII.4/HS66-specific COG2F/2R amplicon.
Sequence analysis of NoV strains.
The full-length genomes of two representative strains (HS206 from family 1 and HS210 from family 2) were characterized. The primers were designed based on the conserved regions among GII NoV genomes reported to date (Table 2). The cDNA was generated by using a SuperScript III first-strand cDNA synthesis kit (Invitrogen) and amplified with Takara Ex Taq polymerase (TaKaRa Mirus Bio, Madison, WI) according to the manufacturer's instructions. Three independent RT-PCR assays were conducted to determine the consensus genome sequences. First, two overlapping cDNAs encompassing the near-full-length NoV genome were synthesized with antisense primers Noro-F1-R1 and Noro-F2-R, respectively. The first-round PCRs were carried out with primer sets Noro-F1-F/Noro-F1-R1 and Noro-F2-F1/Noro-F2-R, respectively. Seminested secondary PCRs were performed with primer sets Noro-F1-F/Noro-F1-R2 and Noro-F2-F2/Noro-F2-R, respectively. The 5′ end of the NoV genome was determined by the 5′ rapid amplification of cDNA ends (5′-RACE) system according to the manufacturer's instructions (Invitrogen). The 3′ end of the norovirus genome was characterized according to a 3′-end cDNA amplification protocol described previously (58).
Table 2.
Primers designed in this study
| Primer | Nucleotide sequence (5′-3′) | Polarity | Positionsa |
|---|---|---|---|
| Noro-F1-F | GTGAATGAAGATGGCGTCTAACGAC | Sense | 1–25 |
| Noro-F1-R1 | CTCCAGAATTTGGTTTTAGTAC | Antisense | 3645–3666 |
| Noro-F1-R2 | GTCATTTCCTCTCTTGTAGATGTATGG | Antisense | 3458–3484 |
| Noro-F2-F1 | GGTCTACTGGAGAGCTGATGCC | Sense | 3300–3321 |
| Noro-F2-F2 | GCGCACTGTTGGAGGGCAGATG | Sense | 3367–3388 |
| Noro-F2-R | GCGCCAGTCCAGGAGTCCAAAAC | Antisense | 7391–7413 |
Location based on HS206.
For patient A, the complete capsid was amplified for samples from different time points in his infection course (HS200 at 3 days, HS207 at 7 days, and HS234 at 30 days after the onset of disease) to investigate if mutations occurred during the month-long infection. First-round PCR was performed by using primer pairs p290 and QO, followed by second-round PCR with primer pair Mon431 and QI (27, 49, 51). To obtain the consensus sequence of capsid sequences of HS200, HS207, and HS234, five independent clones of each strain were selected for sequence analysis. Capsid region C was amplified for samples from the remaining patients (patient D [HS209], patient E [HS224], and patient F [HS219]) and from HS239 from patient A, collected in July 2010, by using primer set COG2F and G2-SKR for genotyping (31, 32, 70).
The PCR products were purified by use of the QIAquick gel extraction kit (Qiagen, Inc.) before direct sequencing or cloning into the PCR XL vector by using a TOPO XL PCR cloning kit for sequencing (Invitrogen). BigDye Terminator cycle sequencing and an ABI prism 3100 Genetic Analyzer (Applied Biosystems) were used to obtain DNA sequence data. Sequence editing was carried out by using the Lasergene software package (version 8; DNASTAR Inc., Madison, WI). The Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST) was used to find homologous hits. Multiple-sequence alignment was performed by using CLUSTAL W software (version 2) (36). A phylogenetic tree with 1,000 bootstrap replicates of the sequence alignment data sets was generated by using the neighbor-joining method with MEGA (version 4) (64). Recombination analysis was performed by using Simplot software (version 1.3) (http://sray.med.som.jhmi.edu/SCRoftware/simplot/). NoV reference strains used for analysis are summarized in the table in the supplemental material. Genotype (capsid) reference strains whose complete ORF1 to ORF3 sequences were available in the GenBank database were used. Representative GII.4 NoVs were selected to represent each branch in a GII.4 phylogenetic tree (45). Reference strains for RdRp genotypes are included only when the assigned genotype numbers differ from the capsid genotypes according to the nomenclature proposed by the National Institute for Public Health and the Environment, Netherlands (35).
Expression and purification of HS206 VLPs.
The recombinant baculovirus carrying the ORF2 and ORF3 genes of HS206 was generated by using the BaculoDirect baculovirus expression system (Invitrogen) according to the manufacturer's instructions. Briefly, a Gateway entry clone containing the ORF2 and ORF3 genes of HS206 (pENTR/SD/D-TOPO-HS206) was generated and used with BaculoDirect linear DNA to perform an LR recombination reaction to generate recombinant baculovirus DNA carrying the HS206 genes. Insect Sf9 cells were transfected by the recombination reaction products, and the cells containing recombinant baculovirus DNA were positively selected by use of ganciclovir. The expression of HS206 capsid proteins in Sf9 cells and culture supernatants was examined by immunoblotting using guinea pig antiserum against Hu/NoV/GII.4/HS191. The recombinant baculoviruses (rBac-HS206) were propagated to prepare virus stocks with high titers for routine VLP expressions.
The production and purification of HS206 VLPs were performed as described previously (24). Briefly, Sf9 cells were infected with rBac-HS206 at a multiplicity of infection of 5 to 10. The cell culture supernatants were harvested at postinoculation day (PID) 7. The VLPs were purified by ultracentrifugation through a 40% (wt/vol) sucrose cushion, followed by CsCl isopycnic gradient (0.39 g/ml) ultracentrifugation. The protein concentration of the VLPs was measured with Bradford reagent (Sigma-Aldrich, St. Louis, MO). The VLPs were negatively stained with 3% phosphotungstic acid (pH 7.0) and examined by transmission electron microscopy (EM) as described previously (24). The HS206 VLPs that were confirmed by EM to contain the correct morphologies of particles were used for hemagglutination (HA), saliva-VLP binding, and synthetic HBGA-VLP binding assays.
Screening of rabbit antisera against NoV VLPs.
GII.12/HS206 VLPs were characterized by an enzyme-linked immunosorbent assay (ELISA), as described previously (41). Plates were coated at 1 μg/ml VLP in phosphate-buffered saline (PBS) for 4 h at room temperature and blocked overnight at 4°C in 5% Carnation dry milk in PBS-0.05% Tween 20 before the addition of rabbit anti-NoV polyclonal sera and incubation for 45 min, followed by anti-rabbit IgG-horseradish peroxidase (HRP) (Sigma-Aldrich) for 30 min and color development with 1-Step Ultra TMB ELISA HRP substrate solution (Thermo Fischer Scientific Inc., Rockford, IL). All incubations were completed at room temperature. Each step was followed by washing with PBS-0.05% Tween 20, and all antibodies were diluted in 5% Carnation dry milk in PBS-0.05% Tween 20. All samples were assayed in triplicate. Antibodies were considered positive for reactivity if the mean optical density for VLP-coated wells was greater than three times the mean optical density for PBS-coated wells.
Carbohydrate binding assays.
Viruslike particle binding to HBGA-phenotyped boiled salivary samples and synthetic biotinylated HBGAs (GlycoTech, Gaithersburg, MD) was determined as previously reported (40, 42), except as noted. Saliva samples were diluted 1:200 for plate coating, and bound VLPs were detected by rabbit anti-GI and anti-GII NoV pooled polyclonal serum followed by anti-rabbit IgG-HRP and 1-Step Ultra TMB ELISA HRP substrate solution. All assays were performed at room temperature. Binding was considered positive for ligand reactivity if the mean optical density for carbohydrate-coated wells was greater than three times the mean optical density for PBS-coated wells.
HA assay.
An HA assay was performed as described previously (30). Briefly, 50 μl of 0.5% packed human red blood cells (RBCs) (Immucor, Norcross, GA) was combined with an equal volume of serially diluted VLPs (starting concentration of 4 μg/ml) at three different pHs (0.01 M PBS for pH 6.8 and pH 7.4 and citric acid-Na2HPO4-buffered saline [5 mM citric acid, 0.01 M Na2HPO4, and 0.85% NaCl] for pH 5.5) at three different temperatures (4°C, room temperature, and 37°C). Inactivated human influenza virus (strain Hum/OH/K1130/06) and the Sf9 cell protein were used as positive and negative controls, respectively. The HA titer was the reciprocal of the highest sample dilution that completely blocked the sedimentation of the RBCs.
Structure analysis.
Three-dimensional structural models of the capsid P-domain dimers were constructed by MOE-Align and MOE-Homology in the Molecular Operating Environment (MOE; Chemical Computing Group, Inc., Montreal, Quebec, Canada), as described previously (44, 45), for strains HS206, HS210, and GII.12/Aichi76 (59). Briefly, the P-domain monomer models were first constructed by using the crystal structure of the capsid P domain of strain GII.4/VA387 at a resolution of 2.00 Å (Protein Data Bank [PDB] accession number 2OBS [9]) as the template. P-domain dimer models were constructed based on the P-domain monomer models by superimposing chains A and B using the crystal structure of the GI.1/Norwalk virus capsid dimer as a template (PDB accession number 1IHM [55]).
Inoculation of Gn pigs.
Near-term pigs were derived by surgery and maintained in sterile isolator units as previously described (43). Six-day-old Gn pigs were allocated into the HS206-inoculated (pigs 1 to 4) or mock-inoculated control (pigs 5 and 6) group. A single aliquoted pool of the original human fecal sample obtained from patient B (strain HS206) was used for the oral inoculation of the Gn pigs. Each Gn pig received one oral dose of the NoV inoculum, consisting of the original HS206 strain diluted 10% (wt/vol) in MEM, which was processed by vortexing, centrifugation at 1,800 × g for 30 min, and filtration through 0.2-μm-pore-size filters. The viral titer of the inoculum was 2.4 × 1010 genomic equivalents (GE)/ml, and 6.0 × 1010 GE was used per pig. The mock inoculum was the same volume of MEM. Two HS206-inoculated pigs (pigs 1and 3) and 1 mock-inoculated control pig (pig 5) were euthanized at the acute phase (PID 4), and the rest were euthanized at the convalescent phase (PID 22). Rectal swabs were collected daily for the detection of diarrhea and virus shedding. Fecal consistency was scored as follows, with scores of 2 or higher being considered diarrheic: 0, solid; 1, pasty; 2, semiliquid; 3, liquid (12). HBGA typing of experimental pigs was done on formalin-fixed, paraffin-embedded intestinal and buccal tissues by immunofluorescent staining as described previously (11). The pigs were maintained in accordance with the guidelines of the Institutional Animal Care and Use Committee of The Ohio State University.
Nucleotide sequence accession numbers.
The sequences determined in this study were submitted to the GenBank database under the following accession numbers: GU325839 (Hu/NoV/GII.4/HS194/09/US), HQ664990 (Hu/NoV/GII.12/HS206/10/US), HQ401025 (Hu/NoV/GII.12/HS207/10/US), HQ449728 (Hu/NoV/GII.12/HS210/10/US), HQ880574 (Hu/N0V/GII.6/HS239/l0/US), JN022612 (Hu/NoV/GII.6/HS245/10/US), and JN022613 (Hu/NoV/GII.2/HS255/11/US).
RESULTS
GII.12 NoV (capsid) was identified from the gastroenteritis outbreak.
The screening of the first 6 stool samples collected from each of the six individuals of the two families for diarrheal viruses by RT-PCR or PCR resulted in the exclusive detection of GII NoV in all of the samples. Sequence analysis of the samples provided by all the patients revealed that strains detected from the same family were identical in capsid region C (282 nucleotides [nt]) and belonged to GII.12 (32, 70). Clinical symptoms, HBGA typing of saliva, and NoV GII viral shedding of each individual are summarized in Table 1. A 9-month-old girl (patient D, saliva type B) shed high titers of NoV (2.5 × 1012 GE/ml) but without any clinical symptoms. The titer remained high (≥1.8 × 1011 GE/ml) for at least 6 days. The asymptomatic excretion of NoV was recorded for all 6 patients. The complete course of virus shedding was examined for patient A (Fig. 1), who shed virus up to 30 days after the onset of diarrhea but for 20 days without symptoms. Interestingly, all infected individuals were secretor positive and had diverse HBGA types (types A, B, and O), while the two B phenotype individuals did not show clinical symptoms.
Fig. 1.
Fecal virus shedding of GII NoV and clinical symptoms of patient A (2-year-old male). Samples analyzed for complete capsid sequences are marked by circles.
Because patient A shed NoV for 1 month, we analyzed the complete capsid region of the viral genomes from three fecal samples from this patient, HS200, HS207, and HS234, at 3, 7, and 30 days after onset of illness, respectively, to examine if mutations occurred in the viral genome in this immunocompetent boy. We found no mutations during the course of this 1-month infection (Fig. 2).The sample provided from this patient as a control when he was free of symptoms in July 2010 (HS239) turned out to be NoV positive and was of GII.6 by sequence analysis of capsid region C.
Fig. 2.
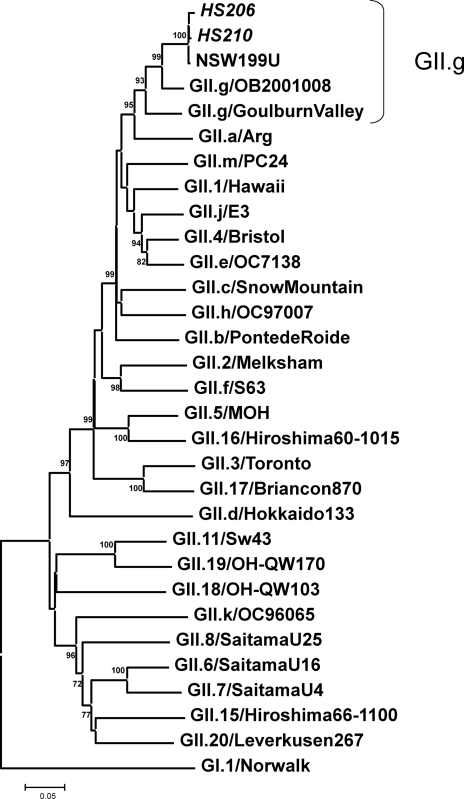
Neighbor-joining phylogenetic analysis of GII NoVs based on the complete capsid region (535 to 557 amino acids). The newly sequenced strains in this study are in italic type. Bootstrap values are provided as percentages of 1,000 replicates for values of ≥70%.
Full-length genome analyses of two representative strains revealed that they were potentially recombinant strains (GII.g [RdRp]/GII.12 [capsid]).
Strain HS206 from family 1 and strain HS210 from family 2 were subjected to full-length genome sequence analysis. Both strains shared high genomic identity (98.7% nucleotide identity). A BLAST search showed that they shared the highest genomic identity (99.0 to 99.2% nucleotide identity) with strain NSW199U, detected in a sporadic case in Australia in 2008 (16). Strain HS206 was compared for each ORF region with capsid (ORF2) genotype reference strains that have been completely sequenced (total, 36) (Table 3). Complete genome sequences were not available in GenBank for certain genotypes (GII.5, GII.7, GII.9, GII.13, GII.14, GII.15, GII.17, GII.18, GII.19, GII.20, and GII.21). For our analysis, previously reported recombinant strains (GII.c/GII.2 Snow Mountain virus [27], GII.4/GII.3 CBNU1 [74], GII.4/GII.10 Mc37 [25], GII.4/GII.12 Gifu′96, and GII.4/GII.12 Saitama U1 [18]) were excluded from the ORF1 analysis to avoid confusion. Strikingly higher identities were found for all ORFs (88.3 to 99.2% nucleotide identity and 94.4 to 100.0% amino acid identity) between HS206 and NSW199U than with the other genotypes of NoV strains. As expected, the lowest genomic identity (53.4 to 62.6% nucleotide identity and 50.4 to 67.5% amino acid identity) was with the porcine GII.11 NoV.
Table 3.
Sequence identity between HS206 and other completely sequenced reference strains based on available genotypes (capsid) in genogroup II
| Genogroup and genotype (capsid) (no. of strains analyzed) | % sequence identitya |
|||||
|---|---|---|---|---|---|---|
| ORF1 |
ORF2 |
ORF3 |
||||
| Nucleotide | Amino acid | Nucleotide | Amino acid | Nucleotide | Amino acid | |
| GII.1 (1) | 82.7 | 94.2 | 74.8 | 86.4 | 88.1 | 94.4 |
| GII.2 (2) | 77.1b | 90.9b | 68.7–69.0 | 74.9–76.2 | 67.2–68.4 | 69.8–71.2 |
| GII.3 (4) | 73.7–81.7c | 87.0–93.8c | 62.3–62.8 | 69.2–70.5 | 66.5–67.7 | 74.1–76.9 |
| GII.4 (16) | 81.5–82.9 | 94.2–94.4 | 59.8–60.1 | 60.9–62.8 | 59.1–61.4 | 53.0–54.7 |
| GII.6 (5) | 62.7–63.4 | 71.3–71.5 | 62.9–64.3 | 69.2–71.0 | 62.0–65.5 | 66.0–69.0 |
| GII.8 (1) | 64.3 | 71.3 | 60.9 | 65.6 | 65.2 | 65.3 |
| GII.10 (2) | 71.6d | 93.9d | 67.8–68.2 | 78.2–78.4 | 70.1 | 77.5–78.9 |
| GII.11 (1) | 62.6 | 67.5 | 59.8 | 64.5 | 53.4 | 50.4 |
| GII.12 (3) | 99.2e | 99.6e | 90.5–99.1 | 97.6–99.6 | 88.3–99.1 | 94.4–100.0 |
| GII.16 (1) | 72.6 | 87.2 | 70.0 | 80.6 | 80.8 | 84.8 |
The highest identities for each ORF are highlighted in boldface type. Recombinant strains were excluded for the ORF1 analysis.
Snow Mountain (GII.c/GII.2) was excluded from the ORF1 analysis.
CBNU1 (GII.4/GII.3) was excluded from the ORF1 analysis.
Mc37 (GII.4/GII.10) was excluded from the ORF1 analysis.
Gifu′96 and SaitamaU1 (GII.4/GII.12) were excluded from the ORF1 analysis.
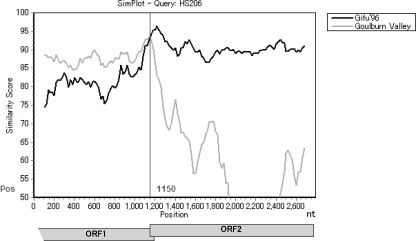
Based on the capsid sequences, HS206 and HS210 clearly clustered with GII.12 together with NSW199U and other GII.12 reference strains (Fig. 2) (75). In contrast, our strains and NSW199U clustered in the distinct GII.g lineage in the partial RdRp region (780 nt), according to the nomenclature proposed by the National Institute for Public Health and the Environment, Netherlands (Fig. 3) (35). Because no ORF1 of GII.12 strains other than strain NSW199U has been identified to date, the GII.g ORF1 could be the corresponding GII.12 ORF1, or a recombination event may have occurred between two distinct genotypes of NoVs, GII.g (RdRp) and GII.12 (capsid). Simplot analysis was carried out by placing the 3′-end ORF1 (RdRp) and ORF2 of HS206 as a query sequence and the corresponding sequences of GII.12 (capsid) reference strain Gifu′96 and GII.g (RdRp) reference strain Goulburn Valley as background sequences (Fig. 4). The HS206 strain shared a high level of identity with strain Goulburn Valley in the RdRp, but the identity decreased in the capsid region, where it shared a high level of identity with the Gifu′96 strain. The breakpoint of recombination was predicted where these two parental strains shared the same identity (i.e., where the curves cross) at the ORF1-ORF2 overlap region (around nucleotide position 1150 in Fig. 4).
Fig. 3.
Neighbor-joining phylogenetic analysis of GII NoVs based on the 3′ end of the RNA-dependent RNA polymerase region (780 nt). The newly sequenced strains in this study are in italic type. Bootstrap values are provided as percentages of 1,000 replicates for values of ≥70%.
Fig. 4.
Simplot analysis of strain HS206. The partial genome of HS206 covering the ORF1/ORF2 overlap region was analyzed for recombination events. The window size was 200 nt, and the step size was 20 nt. At each position of the window, the query sequence (HS206) was compared to each of the background genotype representatives (GII.12 Gifu′96 and GII.d Goulburn Valley). The nucleotide positions of ORF1 and ORF2 are indicated beneath the diagram.
HS206 VLPs did not bind substantially to any HBGA types tested.
To characterize the new strain more completely, we constructed VLPs of a representative outbreak strain (HS206) to investigate their HBGA binding patterns and compare them with those of other well-characterized NoV strains. First, to determine the antigenic properties of HS206 relative to those of previously characterized strains, VLPs were screened for reactivity to a panel of rabbit anti-NoV sera by ELISA. As expected, GI.1/Norwalk VLPs reacted with the anti-GI NoV sera but with none of the anti-GII sera, while GII.4/1997, GII.1/Hawaii, and GII.2/Snow Mountain virus (SMV) reacted with antisera directed against genogroup II VLPs. HS206 VLPs reacted with all the anti-NoV sera similarly to the GII.1/Hawaii VLPs (Fig. 5).
Fig. 5.
Screening of rabbit antisera for antibody reactivity against NoV VLPs. Plates were coated with VLPs before the addition of rabbit anti-norovirus polyclonal sera. Anti-rabbit IgG-HRP was added, and color was developed with 1-Step Ultra TMB ELISA HRP substrate solution. OD, optical density.
By using pooled NoV antisera as detecting antibodies, the profiles of binding of HS206 VLPs to synthetic HBGAs was tested (Fig. 6A). While GI.1/Norwalk VLPs bound to H type 1, H type 3, and the Lewis B and A trimer and GII.4/1997 bound to H type 3 and the Lewis Y, A, and B trimer, as reported previously (40, 42), HS206 VLPs did not bind to any of the tested HBGAs under the assay conditions used.
Fig. 6.
Binding assay of VLPs of HS206. (A) Binding between VLPs and synthetic histo-blood group carbohydrates. A neutravidin-coated plate was incubated with biotin-labeled synthetic HBGA carbohydrates. The binding of VLPs to HBGAs was detected by rabbit anti-GI and anti-GII NoV pooled polyclonal sera. (B) Binding between VLPs and saliva from HS206- or HS210-infected individuals. Plates were coated with boiled saliva at a 1:200 dilution. The binding of VLPs to saliva was detected by rabbit anti-GI and anti-GII NoV pooled polyclonal sera.
To investigate the properties of the binding of HS206 VLPs to biological HBGAs, we performed a VLP-saliva binding assay using saliva from GII.12-infected patients in this outbreak and an HA assay using commercial human pooled RBCs. In the VLP-saliva binding assay, GI.1/Norwalk VLPs bound to saliva types O and A at high levels but bound to type B only at room temperature and at a lower level, as reported previously (40, 42). GII.4/1997 VLPs bound to saliva types O, A, and B at low levels. GII.2/SMV VLPs bound to type B saliva at a low level (28), and GII.1/Hawaii VLPs bound weakly to type B saliva, just above the cutoff value. HS206 VLPs did not bind to any saliva HBGA type when 1 μg/ml of VLPs was used but bound to type B saliva weakly when 5 μg/ml of VLPs was used (Fig. 6B).
In the HA assays, the negative-control Sf9 cell proteins did not hemagglutinate any RBCs, whereas the positive influenza virus control gave HA titers of 128 to 256 under all the pHs (pH 5.5, 6.8, and 7.4) and temperature (4°C, room temperature, and 37°C) conditions tested. HS206 VLPs did not hemagglutinate any RBC types at any pHs and incubation temperatures tested.
Mismatches between our strains and Aichi76 within the P2 domain are located near the HBGA binding sites.
Since previously reported GII.12 (capsid) strain Aichi76 bound to saliva from type B individuals (6 out of 6), we further analyzed if the sequences of these strains are different at critical HBGA binding sites. When the GII.12 NoV strains in this study (HS206 and HS210) and Aichi76 were aligned at the capsid region, 9 and 11 amino acid mismatches were noted for HS206 and HS210, respectively (data not shown). Among the 8 amino acid mismatches that were common for both strains, 2 amino acid mismatches were in the most variable P2 domain, E373Q and N392S. By 3D structural analysis, the two amino acids were located in the vicinity of the HBGA binding interfaces (site I, positions 343 to 347; site II, position 374; site III, positions 440 to 443 [in the GII.4/VA387 strain]) (66) in the P-domain dimer structural models generated by homology modeling (Fig. 7). Although the shape of the binding pocket was not predicted to have changed, the negatively charged amino acid residue (E) at position 373 became a neutral amino acid (Q).
Fig. 7.
Location of amino acid mismatches in the P2 domain among the GII.12 NoV strains from a study described previously by Shirato et. al (59) (Aichi76 strain) and the strains in this outbreak (HS206 and HS210). Shown are P-domain dimer structures of the three strains. HBGA binding interfaces (sites I to III) and two different amino acid changes (E373Q and N392S) from the Aichi76 strain are shown in orange and aqua, respectively. A to C are top views and D to F are magnified views of HBGA binding interfaces of Aichi76, HS206, and HS210, respectively.
HS206 infected Gn pigs.
To investigate whether this emerging new strain infects pigs and, thus, its potential for interspecies transmission between humans and pigs, Gn pigs were inoculated with HS206. HS206-inoculated Gn pigs shed virus from PID 1 to 3, which coincided with mild diarrhea (fecal consistency score of 2) in pig 1 (Fig. 8). The virus titer peaked (1.8 × 106 to 1.7 × 107 GE/ml) from PID 2 to 5. The longest duration of virus shedding, up to PID 16, was observed for pig 4, which was euthanized at PID 16. Neither of the two mock-inoculated pigs shed virus or had diarrhea. All the HBGA phenotypes of the experimental pigs were determined to be A+.
Fig. 8.
Fecal virus shedding in four Gn pigs inoculated with NoV HS206 determined by real-time RT-PCR. Arrows indicate diarrhea in a pig. PID, postinoculation day.
DISCUSSION
For decades, GII.4 was the predominant genotype among NoV gastroenteritis outbreaks. In this study, we identified new GII.g/GII.12 strains from a gastroenteritis outbreak in Wooster, OH, in January 2010. Similar strains caused sporadic gastroenteritis cases in Australia in 2008 (16) and food-borne outbreaks in Hungary, France, and the Netherlands from 2009 to 2010 (20). GII.12 (capsid) strains were also reported to have caused 32 outbreaks from October 2009 to March 2010 in the United States (69). These findings suggest the emergence of GII.12 (capsid) NoV strains worldwide in 2008 to 2010. This was probably due to their escape from existing herd immunity to previously prevalent genotypes, such as GII.4 (22). Further monitoring of the epidemiological trends for NoV infections is warranted.
Among the 6 individuals involved in this study, patient A was asymptomatically infected with GII.6 NoV (strain HS239) 4 months later and was infected together with his mother (patient B, strain HS256) with NoV GII.2 (strain HS255), with vomiting and diarrhea, 1 year after the GII.12 infection, respectively. Patient C had been symptomatically infected with NoV GII.4 (strain HS194), with diarrhea and vomiting, 1 year prior and was infected together with his sister (patient D, stain HS246), his father (patient E, strain HS244), and his mother (patient F, strain HS247) with NoV GII.6 (HS245), with vomiting and diarrhea, 11 months after the GII.12 infection, respectively (our unpublished data). Except for strain HS194, for which the genome has been sequenced, the other strain genotypes were characterized based on region C sequence analysis. These results suggest that intergenotype protection from NoV infection is lacking.
The GII.12 strains from the two families were very similar, falling into the same lineage in phylogenetic trees based on both the capsid and RdRp regions, but are apparently different strains showing 1.3% nucleotide diversity in the complete genome, suggesting that the sources of infection of the 2 families differed.
There are increasing reports of prolonged NoV shedding detected by RT-PCR even in immunocompetent patients. Rockx et al. (57) reported previously that 26 of 99 NoV-infected children shed NoV for up to 22 days after the onset of illness. Murata et al. (46) found previously that infants less than 6 months old shed NoV for up to 47 days. Kirkwood and Streitberg (34) reported the shedding of a GII.4 strain in a 21-month-old child for 100 days, and Siebenga et al. (60) described the shedding of a GII.3 strain in a 4-month-old infant for 34 days. These findings may reflect the increased sensitivity of detection by RT-PCR and real-time RT-PCR assays. However, little sequence information on the NoVs detected during prolonged virus shedding has been reported. In our study, the complete capsids of three samples (collected at 3, 7, and 30 days after the onset of illness) from patient A, who is immunocompetent and shed virus for 1 month, were sequenced. Surprisingly, no mutations were noted among the three samples in the complete capsid region. In studies of immunocompromised patients, a NoV GII.3 strain accumulated 32 amino acid mutations in the complete capsid region during 1 year of infection (49), and a GII.4 NoV strain accumulated 11 amino acid mutations during 119 days of infection (60). In the latter study, those authors reported that a GII.3 NoV mutated more in an immunocompetent patient than in immunocompromised patients, suggesting an immune-driven selection of mutants. Those authors also noted lower mutation rates in GII.3 than in GII.4 NoV strains in their patients. Recently, Bull et al. (6) confirmed by in vitro experiments that GII.4 strains had a higher substitution rate than did GII.3 and GII.7 strains. These results suggest that genotype/strain differences play an important role in NoV evolution. To date, our study is the first to depict the prolonged shedding of a NoV GII.12 strain in an immunocompetent child up to 30 days after the onset of gastroenteritis. Whether the viruses in asymptomatically infected individuals detected by real-time RT-PCR are infectious or not remains uncertain (22, 63), and until an effective cell culture system for NoV is established to examine infectivity, hygienic precautions should be implemented even after the symptoms have subsided.
Another interesting finding of the current study was that individuals of various HBGA types (types A, B, and O) were infected with GII.12 strains, although the two type B individuals remained asymptomatic. Whether the two type B individuals were infected previously with a NoV that provided partial protection is unknown. When we analyzed the binding pattern of VLPs of one representative strain from this outbreak (HS206), we found little if any binding to any synthetic or biological (those expressed on RBCs and those secreted in saliva) HBGAs that were evaluated. All these assays were performed in vitro at room temperature and might not represent the binding properties of NoV in vivo. Nevertheless, these in vitro results were consistent with the clinical profile that HS206 infected individuals regardless of A, B, or O phenotype. This clinical pattern is similar to that observed previously for GII.2/SMV infection in a volunteer study, which was not type A/B/O dependent (38). A seroprevalence study using donated blood also revealed that the presence of IgG titers against GII.4 strain was type A/B/O independent but correlated with the secretor phenotype (37). In another report, symptomatic GII.4 infections were also strongly associated with the Se+ phenotype (10). Although all the individuals in the current outbreak were Se+, it is hard to conclude that GII.12 infection is Se+ dependent without Se-negative (Se−) individuals exposed to these GII.12 strains. Higher concentrations of HS206 VLPs did not give positive results in the HA assay (40 μg/ml) or in VLP-synthetic HBGA binding assays (5 μg/ml), whereas they bound weakly to type B saliva at a concentration of 5 μg/ml, suggesting that they have low-affinity binding to the B antigen. Of note, this pattern is very similar to that of GII.1/Hawaii, which lacks synthetic HBGA binding (28) yet weakly binds to type B saliva (Fig. 6B). In addition, GII.12/HS206 and GII.1/Hawaii VLPs react similarly to rabbit anti-NoV sera (Fig. 5). These VLPs also reacted with mouse anti-NoV sera in a similar pattern: among mouse anti-GI.1/Norwalk virus, anti-GII.1/Hawaii, anti-GII.2/SMV, and anti GII.4/1997 antisera, they reacted with anti-GII.1/Hawaii and anti-GII.2/SMV sera but with not anti-GI.1/Norwalk virus and anti-GII.4/1997 sera (data not shown). These similarities may be explained by the high sequence identity in the ORF2 region (Table 3) (74.8% nucleotide identity and 86.4% amino acid identity) between these two strains among GII NoVs.
The GII.12/Aichi76 strain bound to saliva from type B individuals (6 out of 6) (59). The authors of that study used the same concentration of VLPs (1 μg/ml) as we did but more-diluted saliva (1:1,600) in the VLP-saliva binding assay. However, the current GII.12/HS206 VLPs did not bind to saliva from type B individual at 1 μg/ml, but the VLPs bound at a higher concentration (5 μg/ml). These results suggest that HS206 was a weak binder to the B antigen. These differences between the two GII NoV strains (Aichi76 and HS206) could be due to their different capsid sequences, especially at the P2 domain that is exposed on the surface of virus particles. To determine if the sequences of these GII.12 strains are different at critical HBGA binding sites of the P2 domain, we analyzed the P-domain dimer structural models generated by homology modeling. Two amino acid mismatches between our strains and strain Aichi76, E373Q and N392S, were predicted to be located in the vicinity of the HBGA binding interfaces. These changes did not induce any detectable changes in the structural shape. However, the change at position 373 alters a negatively charged residue to a neutral amino acid. In many cases, the electrostatic interaction affects the binding affinity. Therefore, we speculate that E373Q would impact the affinity of binding between virus particles and ligands and might have contributed to the difference of binding patterns between the two GII.12 human NoV strains HS206 and Aichi76.
The assigned nomenclature for RdRp genotypes is unclear and has resulted in the naming of almost identical sequence strains as different genotypes, such as the identification of NSW199U as being of GII.e/GII.12 in one publication (16) but GII.g/GII.12 in our analysis. In our study, we followed the nomenclature developed by the National Institute for Public Health and the Environment, Netherlands, as it includes the widest range of reference strains in the database (35). In this classification system for the RdRp region (780 nt), within-genotype identities (amino acids) ranged from 89.9% (GII.8) to 99.6% (GII.18), whereas intergenotype identities ranged from 61.3% (between GII.a and GII.3) to 91.0% (between GII.4 and GII.e), which indicates the difficulty of assigning a clear cutoff percent identity to denote a genotype in this region.
The HS206 strain infected all four inoculated Gn pigs, as documented by real-time RT-PCR. Three of the four pigs began fecal virus shedding at PID 2 or 3, and in all four pigs, virus titers first increased and then decreased after the onset of virus shedding. This fecal virus shedding pattern was similar to that of human volunteers infected with GI.1/Norwalk virus, who showed peak fecal virus titers at 2 to 4 days after oral inoculation, which decreased progressively thereafter (2). Although the viral titers in the feces were lower than that in the original inocula, we were unable to accurately calculate the total amount of virus shed due to difficulties in collecting all the feces separately from isolator units. The Gn pigs infected with human rotavirus presented similar fecal shedding patterns. Rotavirus-inoculated pigs displayed obvious diarrhea, but the rotavirus titers in the feces were 100- to 1,000-fold lower than the titers of the inocula, as determined by a fluorescent-focus assay (A. Vlasova and L. J. Saif, unpublished data). Moreover, in Gn pigs inoculated with a heat-inactivated NoV GII.4 strain, no viral RNA was detected in feces, suggesting that infectious virus was detected by our real-time RT-PCR (K. Jung and L. J. Saif, unpublished data). Collectively, these data support our assertion that the viral RNA detected was not from pass-through inocula, in which case initially high and progressively decreasing titers would occur. Our data suggest that the interspecies transmission of a human GII.12 strain to pigs is possible under experimental conditions, as was previously observed for a human GII.4 NoV strain (12).
In summary, we detected and characterized GII.g (RdRp)/GII.12 (capsid) NoVs from a gastroenteritis outbreak in Wooster, OH. Prolonged virus shedding (1 month) was documented in one immunocompetent child in this outbreak, reemphasizing the importance of hygienic precautions. The results of HBGA typing of saliva of infected individuals confirmed that A, B, and O HBGA Se+ types were all susceptible to this strain, and the finding that HS206 VLPs did not bind substantially to any HBGA in vitro supports the speculation that infection by this strain is not A/B/O antigen dependent. The new GII.12 strain infected young Gn piglets, confirming the interspecies transmission of human NoVs to pigs under experimental conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Atmar and Frederick Neill at Baylor College of Medicine for performing HBGA typing ELISAs and Marion Koopmans and Harry Vennema at the National Institute for Public Health and the Environment, Netherlands, for assistance with NoV nomenclature. We also acknowledge Juliette Hanson and Rich McCormick for assistance with animal care, Kelly Scheuer for assistance with EM examination, Chang-Won Lee for providing inactivated influenza virus for HA assays, and Alexander Rodriguez-Palacios and Sanja Ilic for helpful discussion.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (R21 AI081009-2, R01 AI056351, and U01AI08001). We have no potential conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Akihara S., et al. 2005. Existence of multiple outbreaks of viral gastroenteritis among infants in a day care center in Japan. Arch. Virol. 150: 2061–2075 [DOI] [PubMed] [Google Scholar]
- 2. Atmar R. L., et al. 2008. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 14: 1553–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belliot G., et al. 2003. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 77: 10957–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruggink L. D., Marshall J. A. 2009. Molecular and epidemiological features of GIIb norovirus outbreaks in Victoria, Australia, 2002-2005. J. Med. Virol. 81: 1652–1660 [DOI] [PubMed] [Google Scholar]
- 5. Buesa J., et al. 2002. Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J. Clin. Microbiol. 40: 2854–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bull R. A., Eden J. S., Rawlinson W. D., White P. A. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6: e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bull R. A., et al. 2005. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 11: 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bull R. A., Tanaka M. M., White P. A. 2007. Norovirus recombination. J. Gen. Virol. 88: 3347–3359 [DOI] [PubMed] [Google Scholar]
- 9. Cao S., et al. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81: 5949–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlsson B., et al. 2009. The G428A nonsense mutation in FUT2 provides strong but not absolute protection against symptomatic GII.4 norovirus infection. PLoS One 4: e5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheetham S., et al. 2007. Binding patterns of human norovirus-like particles to buccal and intestinal tissues of gnotobiotic pigs in relation to A/H histo-blood group antigen expression. J. Virol. 81: 3535–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheetham S., et al. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 80: 10372–10381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chhabra P., Walimbe A. M., Chitambar S. D. 2010. Complete genome characterization of genogroup II norovirus strains from India: evidence of recombination in ORF2/3 overlap. Infect. Genet. Evol. 10: 1101–1109 [DOI] [PubMed] [Google Scholar]
- 14. Chhabra P., Walimbe A. M., Chitambar S. D. 2010. Molecular characterization of three novel intergenotype norovirus GII recombinant strains from western India. Virus Res. 147: 242–246 [DOI] [PubMed] [Google Scholar]
- 15. Chung J. Y., Han T. H., Park S. H., Kim S. W., Hwang E. S. 2010. Detection of GII-4/2006b variant and recombinant noroviruses in children with acute gastroenteritis, South Korea. J. Med. Virol. 82: 146–152 [DOI] [PubMed] [Google Scholar]
- 16. Eden J. S., et al. 2010. Norovirus GII.4 variant 2006b caused epidemics of acute gastroenteritis in Australia during 2007 and 2008. J. Clin. Virol. 49: 265–271 [DOI] [PubMed] [Google Scholar]
- 17. Estes M. K., Prasad B. V., Atmar R. L. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19: 467–474 [DOI] [PubMed] [Google Scholar]
- 18. Etherington G. J., Dicks J., Roberts I. N. 2006. High throughput sequence analysis reveals hitherto unreported recombination in the genus Norovirus. Virology 345: 88–95 [DOI] [PubMed] [Google Scholar]
- 19. Farkas T., et al. 2005. Seroprevalence of noroviruses in swine. J. Clin. Microbiol. 43: 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. FBVE. 2010. NoroNet report May 2010. RIVM, Bilthoven, The Netherlands [Google Scholar]
- 21. Gallimore C. I., Cubitt D., du Plessis N., Gray J. J. 2004. Asymptomatic and symptomatic excretion of noroviruses during a hospital outbreak of gastroenteritis. J. Clin. Microbiol. 42: 2271–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Glass R. I., Parashar U. D., Estes M. K. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361: 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Green K. (ed.) 2007. The noroviruses, p. 949–980 In Knipe D. M. (ed.), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 24. Han M. G., Wang Q., Smiley J. R., Chang K. O., Saif L. J. 2005. Self-assembly of the recombinant capsid protein of a bovine norovirus (BoNV) into virus-like particles and evaluation of cross-reactivity of BoNV with human noroviruses. J. Clin. Microbiol. 43: 778–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hansman G. S., et al. 2004. Genetic diversity of norovirus and sapovirus in hospitalized infants with sporadic cases of acute gastroenteritis in Chiang Mai, Thailand. J. Clin. Microbiol. 42: 1305–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hardy M. E. 2005. Norovirus protein structure and function. FEMS Microbiol. Lett. 253: 1–8 [DOI] [PubMed] [Google Scholar]
- 27. Hardy M. E., Kramer S. F., Treanor J. J., Estes M. K. 1997. Human calicivirus genogroup II capsid sequence diversity revealed by analyses of the prototype Snow Mountain agent. Arch. Virol. 142: 1469–1479 [DOI] [PubMed] [Google Scholar]
- 28. Harrington P. R., Lindesmith L., Yount B., Moe C. L., Baric R. S. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76: 12335–12343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hutson A. M., Atmar R. L., Graham D. Y., Estes M. K. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 185: 1335–1337 [DOI] [PubMed] [Google Scholar]
- 30. Hutson A. M., Atmar R. L., Marcus D. M., Estes M. K. 2003. Norwalk virus-like particle hemagglutination by binding to H histo-blood group antigens. J. Virol. 77: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kageyama T., et al. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41: 1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kageyama T., et al. 2004. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J. Clin. Microbiol. 42: 2988–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Katayama K., et al. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299: 225–239 [DOI] [PubMed] [Google Scholar]
- 34. Kirkwood C. D., Streitberg R. 2008. Calicivirus shedding in children after recovery from diarrhoeal disease. J. Clin. Virol. 43: 346–348 [DOI] [PubMed] [Google Scholar]
- 35. Kroneman A., et al. 2011. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 51: 121–125 [DOI] [PubMed] [Google Scholar]
- 36. Larkin M. A., et al. 2007. ClustalW and ClustalX version 2. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- 37. Larsson M. M., et al. 2006. Antibody prevalence and titer to norovirus (genogroup II) correlate with secretor (FUT2) but not with ABO phenotype or Lewis (FUT3) genotype. J. Infect. Dis. 194: 1422–1427 [DOI] [PubMed] [Google Scholar]
- 38. Lindesmith L., et al. 2005. Cellular and humoral immunity following Snow Mountain virus challenge. J. Virol. 79: 2900–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lindesmith L., et al. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9: 548–553 [DOI] [PubMed] [Google Scholar]
- 40. Lindesmith L. C., et al. 2010. Heterotypic humoral and cellular immune responses following Norwalk virus infection. J. Virol. 84: 1800–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lindesmith L. C., Donaldson E. F., Baric R. S. 2011. Norovirus GII.4 strain antigenic variation. J. Virol. 85: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindesmith L. C., et al. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meyer R. C., Bohl E. H., Kohler E. M. 1964. Procurement and maintenance of germ-free swine for microbiological investigations. Appl. Microbiol. 12: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Motomura K., et al. 2008. Identification of monomorphic and divergent haplotypes in the 2006-2007 norovirus GII/4 epidemic population by genomewide tracing of evolutionary history. J. Virol. 82: 11247–11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Motomura K., et al. 2010. Divergent evolution of norovirus GII/4 by genome recombination from May 2006 to February 2009 in Japan. J. Virol. 84: 8085–8097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murata T., et al. 2007. Prolonged norovirus shedding in infants < or =6 months of age with gastroenteritis. Pediatr. Infect. Dis. J. 26: 46–49 [DOI] [PubMed] [Google Scholar]
- 47. Nakamura K., et al. 2009. Detection of a novel recombinant norovirus from sewage water in Toyama prefecture, Japan. Jpn. J. Infect. Dis. 62: 394–398 [PubMed] [Google Scholar]
- 48. Nakamura K., et al. 2010. Frequent detection of noroviruses and sapoviruses in swine and high genetic diversity of porcine sapovirus in Japan during Fiscal Year 2008. J. Clin. Microbiol. 48: 1215–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nilsson M., et al. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 77: 13117–13124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nordgren J., Kindberg E., Lindgren P. E., Matussek A., Svensson L. 2010. Norovirus gastroenteritis outbreak with a secretor-independent susceptibility pattern, Sweden. Emerg. Infect. Dis. 16: 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ozawa K., Oka T., Takeda N., Hansman G. S. 2007. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J. Clin. Microbiol. 45: 3996–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Patel M. M., et al. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14: 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Phan T. G., et al. 2007. Genetic heterogeneity, evolution, and recombination in noroviruses. J. Med. Virol. 79: 1388–1400 [DOI] [PubMed] [Google Scholar]
- 54. Phan T. G., et al. 2006. Detection and genetic characterization of norovirus strains circulating among infants and children with acute gastroenteritis in Japan during 2004-2005. Clin. Lab. 52: 519–525 [PubMed] [Google Scholar]
- 55. Prasad B. V., et al. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286: 287–290 [DOI] [PubMed] [Google Scholar]
- 56. Reeck A., et al. 2010. Serological correlate of protection against norovirus-induced gastroenteritis. J. Infect. Dis. 202: 1212–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rockx B., et al. 2002. Natural history of human calicivirus infection: a prospective cohort study. Clin. Infect. Dis. 35: 246–253 [DOI] [PubMed] [Google Scholar]
- 58. Scotto-Lavino E., Du G., Frohman M. A. 2006. 3′ end cDNA amplification using classic RACE. Nat. Protoc. 1: 2742–2745 [DOI] [PubMed] [Google Scholar]
- 59. Shirato H., et al. 2008. Noroviruses distinguish between type 1 and type 2 histo-blood group antigens for binding. J. Virol. 82: 10756–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Siebenga J. J., et al. 2008. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: a model for in vivo molecular evolution. J. Infect. Dis. 198: 994–1001 [DOI] [PubMed] [Google Scholar]
- 61. Souza M., Cheetham S. M., Azevedo M. S., Costantini V., Saif L. J. 2007. Cytokine and antibody responses in gnotobiotic pigs after infection with human norovirus genogroup II.4 (HS66 strain). J. Virol. 81: 9183–9192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Souza M., Costantini V., Azevedo M. S., Saif L. J. 2007. A human norovirus-like particle vaccine adjuvanted with ISCOM or mLT induces cytokine and antibody responses and protection to the homologous GII.4 human norovirus in a gnotobiotic pig disease model. Vaccine 25: 8448–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sukhrie F. H., Siebenga J. J., Beersma M. F., Koopmans M. 2010. Chronic shedders as reservoir for nosocomial transmission of norovirus. J. Clin. Microbiol. 48: 4303–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tamura K., Nei D. J. M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 65. Tan M., et al. 2008. Outbreak studies of a GII-3 and a GII-4 norovirus revealed an association between HBGA phenotypes and viral infection. J. Med. Virol. 80: 1296–1301 [DOI] [PubMed] [Google Scholar]
- 66. Tan M., et al. 2009. Conservation of carbohydrate binding interfaces: evidence of human HBGA selection in norovirus evolution. PLoS One 4: e5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Teunis P. F., et al. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80: 1468–1476 [DOI] [PubMed] [Google Scholar]
- 68. Thorven M., et al. 2005. A homozygous nonsense mutation (428G→A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J. Virol. 79: 15351–15355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vega E., et al. 2010. A paradigm shift in norovirus outbreaks: the emergence of II.12 strains as a major contributor to disease, abstr. VII-4, p. 59. Abstr. 4th Int. Conf. Caliciviruses. [Google Scholar]
- 70. Vinje J., Hamidjaja R. A., Sobsey M. D. 2004. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods 116: 109–117 [DOI] [PubMed] [Google Scholar]
- 71. Wang Q. H., et al. 2005. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 11: 1874–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yan H., et al. 2004. Development of RT-multiplex PCR assay for detection of adenovirus and group A and C rotaviruses in diarrheal fecal specimens from children in China. Kansenshogaku Zasshi 78: 699–709 [DOI] [PubMed] [Google Scholar]
- 73. Yan H., Yagyu F., Okitsu S., Nishio O., Ushijima H. 2003. Detection of norovirus (GI, GII), sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J. Virol. Methods 114: 37–44 [DOI] [PubMed] [Google Scholar]
- 74. Yun S. I., et al. 2010. Complete genome sequence and phylogenetic analysis of a recombinant Korean norovirus, CBNU1, recovered from a 2006 outbreak. Virus Res. 152: 137–152 [DOI] [PubMed] [Google Scholar]
- 75. Zheng D. P., et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346: 312–323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.