Abstract
The yeast Candida albicans causes life-threatening candidemia. A general-purpose genotype (GPG; corresponds to clade 1) causes more infections than other C. albicans genotypes. To investigate if GPG strains also cause higher mortality, we developed a duplex PCR assay which was 98% accurate in identifying GPG strains in an international collection of strains typed with probe Ca3. We applied the assay to 635 European C. albicans candidemia isolates. Of these, 18% conformed to the GPG genotype, 4% were of a borderline genotype, and 78% were of a non-GPG genotype, broadly consistent with genotype distributions in earlier studies. The prevalence of GPG strains was increased in females and in younger patients, exceeding 40% in infants aged ≤1 year. Logistic regression confirmed sex and age as significant determinants of GPG prevalence. Across the entire patient cohort, there was no difference in mortality for patients infected with GPG strains or other strains (36% versus 37%). However, mortality in patients aged ≤48 years was 33% for infection with GPG strains but only 15% for infection with other strains (z test; P < 0.01). Mortality rates associated with GPG and non-GPG strains were comparable in older patients (39% versus 46%). A logistic regression analysis confirmed the age-dependent impact of genotype on mortality. Thus, GPG strains may be more virulent than other strains in younger patients. Because candidemia is usually caused by endogenous strains, our PCR assay could potentially be used as a risk assessment tool for identifying younger patients most at risk of death from candidemia.
INTRODUCTION
The yeast Candida albicans is a major opportunistic pathogen causing life-threatening systemic disease (11, 19, 40). We previously identified a cluster of genotypically similar strains which comprised 37% of 266 infection-causing isolates in a global collection (28). The cluster had a prevalence that was 10 to 100 times higher than that of each of the remaining 37 groups (prevalences of 0.3 to 4.5%). Because the cluster was highly prevalent in all patient types, we proposed that its members have a widely adapted general-purpose genotype (GPG) (28). Multilocus sequence typing confirmed the global high prevalence of this group, designated clade 1 by Odds and coworkers (22). GPG strains are more frequent as agents of superficial disease than expected on the basis of their frequency as commensal colonizers (22, 28). They also cause candidemia more frequently than any other type of strains (although the percentage of all clade 1 isolates that cause bloodstream infections is lower than the percentage of blood isolates encountered in other clades) (22). Some of the features specific to GPG strains could conceivably be virulence determinants, such as increased resilience to chemicals, increased adhesion, GPG-specific alleles of DNA tandem repeat-containing genes, and genes involved in dimorphism (15, 29, 41–43).
The high prevalence of GPG strains as pathogens suggests that they could be more virulent than other strains, at least under some circumstances. GPG (clade 1) strains are not more virulent than those of other clades in an intravenous mouse model (15). However, aside from physiological differences between mice and humans, in the mouse model large numbers of C. albicans cells are introduced directly into the bloodstream and may thus not give a complete assessment of virulence, since in human disease C. albicans must penetrate epithelia and must multiply in the host to reach high cell concentrations in sterile sites (16, 20). We undertook the current study to investigate if the presence of GPG strains was associated with higher mortality of candidemia patients, which would be an indication that GPG strains may be more virulent in the human host.
MATERIALS AND METHODS
Isolates and patient data.
Isolates for the development of a duplex PCR assay for GPG strain identification were from Britain, Fiji, Colombia, Malaysia, New Zealand, and the United States (see Table S1 in the supplemental material) and were part of an international collection that was DNA fingerprinted with the repetitive sequence Ca3 (28).
Isolates for analysis of the impact of genotype on mortality had been collected as part of a survey conducted in the Paris area in France in 2005 to 2006 (5) and as part of two European Confederation of Medical Mycology (ECMM) surveys, one held in 1997 to 1999 (36) and one in 2006 to 2008 (http://ecmm.eu/node/81). See Table S2 in the supplemental material for detailed information on isolates. The yeasts had been identified as C. albicans on the basis of germ tube production in serum at 37°C and chlamydospore production on potato-carrot-ox gall agar at 28°C. Outcome (survival or death) had been assessed 30 days after the initial diagnosis of candidemia. Where APACHE II and SAPS scores had been recorded at admission, these were converted into predicted mortality (10, 12); for APACHE II scores, the conversion was based on Fig. 3 in reference 10.
Fig. 3.
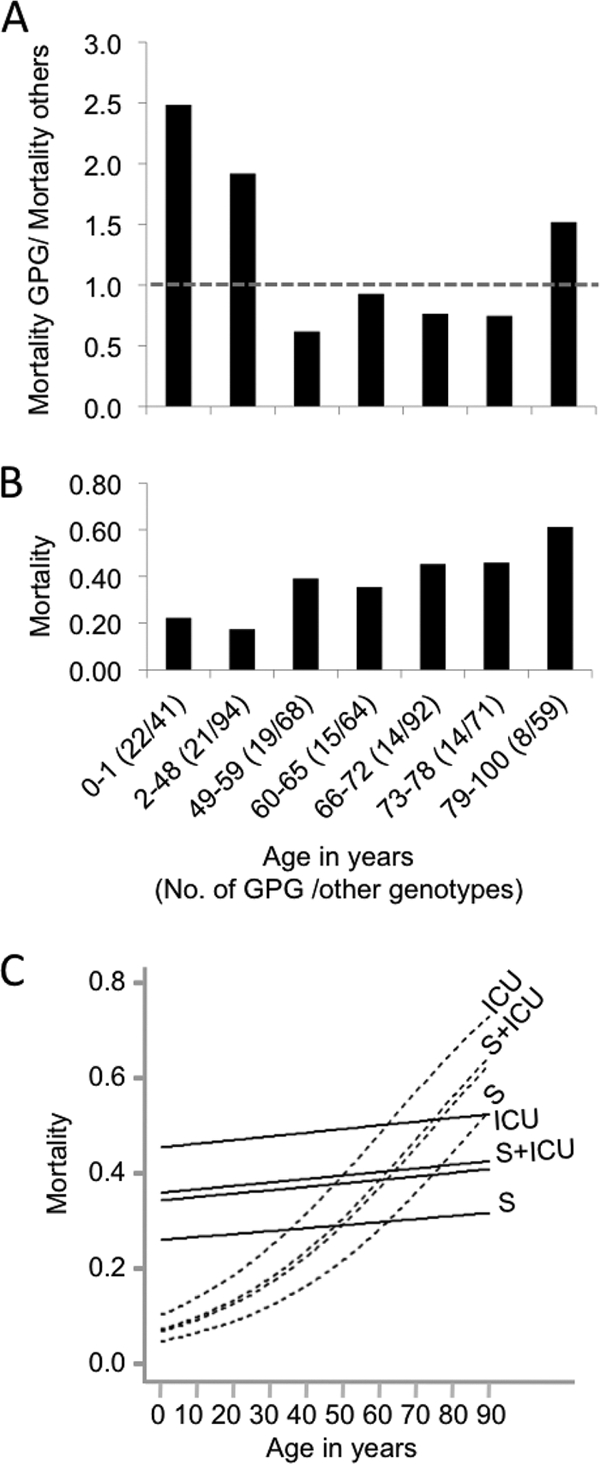
Higher mortality is associated with GPG isolates in young patients. (A) Mortality in different age groups of patients with GPG isolates relative to that of patients with non-GPG strains. The dashed line indicates a ratio of 1.0. (B) Overall mortality of patients of different ages. Numbers in parentheses after the age range are the numbers of GPG isolates and non-GPG isolates in the cohort. Age ranges were set so that at least 5 deaths of patients with GPG strains occurred in each cohort. (C) Plot of dependence of mortality on age, as deduced from logistic regression, for ICU patients, ICU patients who had undergone surgery (S+ICU), non-ICU patients who had undergone surgery (S), and non-ICU patients who had not undergone surgery (unlabeled). Solid lines indicate the mortality of patients with GPG strains, and dashed lines indicate the mortality of patients with non-GPG strains. Patients with borderline strains were excluded from these analyses.
Duplex PCR assay.
A portion of a C. albicans colony from a yeast extract-peptone-dextrose (YPD) plate was picked with a 10-μl pipette tip and mixed with a 20-μl PCR mixture containing 4 μl Q solution, 2 μl 10× CoralLoad buffer (Qiagen), 5 nmol of each deoxynucleoside triphosphate (dNTP), 2.25 mM MgCl2, 16 pmol each of primers MG1pf (CCTCCCTTCTCTTAAGAG) and MG1pr (AACAGGAGAGGTTAAGAG), 2 pmol each of primers YW1pf (TCAAGTTCTGCTTCCCCATCG) and YWP1pr (CGTGGACCGTAGTGACACCAATAC), and 1 unit Taq polymerase (Qiagen). Cycling conditions included an initial incubation for 5 min at 96°C followed by 30 cycles of 45 s at 94°C, 1 min at 50°C, and 1 min at 72°C, with the last cycle being followed by an additional 7 min at 72°C. Samples were loaded onto a 2.5% Tris-acetate-EDTA (TAE) gel (3) and run at 30 V for 20 h, stained in TAE buffer with 0.5 mg/ml ethidium bromide for 45 min, and destained in water for 30 min. Strains of known genotype were included as quality controls and molecular weight markers in every set of unknown samples that were typed (see Fig. 1B) (we initially used 6 strains of known genotype from our DNA fingerprinted collection [28] as controls and then gradually replaced them with 7 isolates typed as part of the current survey, to gain a more realistic assessment of the reproducibility of the colony PCR with survey isolates; the patterns of these strains were highly reproducible). Isolates producing an MG1 product and a single 276-bp YWP1 product were classified as GPG strains, and isolates producing no MG1 products and YWP1 products of sizes unequal to 276 bp were classified as non-GPG strains. Strains producing other banding patterns were retested, and if these patterns were reproducible, the strains were considered borderline strains. Isolates producing no PCR products were also retested, first by PCR and then by species identification through chlamydospore and germ tube formation (all isolates that reproducibly did not produce PCR products were not C. albicans).
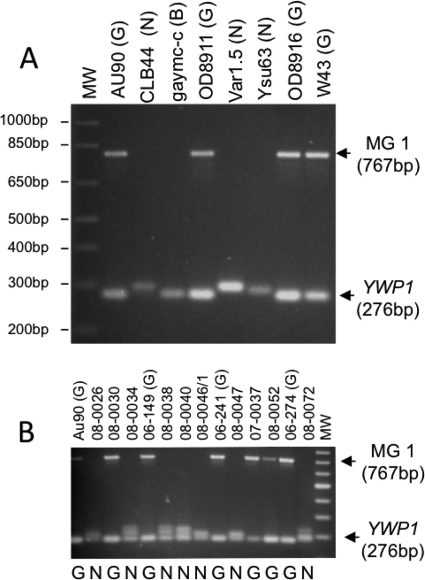
Fig. 1.
Duplex PCR assay. (A) PCR products for isolates from an international collection of isolates of known genotype (G=GPG, N=non-GPG, and B=borderline) (see Table S1 in the supplemental material) on which development of the assay was based. The GPG-specific MG1 and YWP1 products and their molecular weights are marked on the right, and molecular weights of bands of a 1-kb Plus ladder (MW) are shown on the left. (B) Example of a gel used to type isolates of unknown genotype, in this case a set of Italian isolates (see Table S2 in the supplemental material). Coamplified samples from previously typed GPG strains were included in every 4 to 5 lanes as controls [marked by “(G)” after the strain name]. Genotypes deduced from the assay are shown below each lane. A 100-bp ladder was loaded in the right lane (MW).
Logistic regression for GPG mortality and prevalence.
The probability of patient death in the first 30 days was modeled on the logistic scale as a linear function of the covariates. To allow for possible correlations in the experiences of patients at the same hospital in the same year, the random effect term “hospital year” was included. Thus, the model is log (πij)/(1−πij)=xijtβ+ui, where πij is the probability of death in the first 30 days for patient j in hospital year i, xij is a vector of covariates for that patient, β is a vector of fixed covariate effects to be estimated from the data, and ui is the random effect associated with hospital year i.
A similar analysis was carried out to model the probability that the infecting strain of C. albicans was a GPG strain.
The model can be fitted using the statistical software package R (25). Initially, a full set of covariates was used, including constant, age, sex, surgery, and ICU, plus GPG for mortality analyses. The effect of GPG on mortality was found to be significant only when an interaction with age was included. Because of the importance of age in assessing the effect of GPG, a generalized additive mixed model (13) was used to explore the functional relationship between age and mortality. This showed no significant departure from linearity for GPG or non-GPG strains, so linear functions were used. Interactions between GPG and other covariates were investigated and found to be nonsignificant, as was the effect of sex on mortality.
Survival analysis.
Actual death times (in days) were recorded for only 31% of patients; the death times for 62% of patients were right censored at 30 days, as these patients were still alive, and those for 7% of patients were left censored at 30 days, as these patients were known to have died but had no recorded death time. The presence of left censoring makes analysis of the survival times difficult. The Kaplan-Meier estimate of survival probability can accommodate right but not left censoring, and omitting the left-censored patients would result in bias. Estimates of the survival functions for GPG and non-GPG groups were calculated using Turnbull's estimator (37) separately for patients aged 48 or less and those above 48 years of age. Because of the heavy censoring and the difficulty introduced by left censoring, more sophisticated survival models with multiple covariate effects were not pursued.
RESULTS
Development of a PCR method for the identification of GPG strains.
Since the current study involved isolates from several hundred patients, we developed a duplex PCR method for rapid identification of GPG strains by combining two primer sets which had each been shown to amplify products highly specific to GPG strains. The first set (MG1pf and MG1pr) generates a 767-bp GPG-specific fragment caused by a GPG-specific point mutation upstream of YHB4 that is present in all of 40 GPG strains but none of 20 non-GPG strains from an international collection of Ca3-typed isolates, as well as in two borderline strains for which Ca3 typing and other molecular markers gave conflicting results (43). A second set of primers (YWP1pf and YWP1pr) amplifies a microsatellite-containing region within the YWP1 gene, producing a single 276-bp product in 50/50 GPG strains, 3/6 borderline strains, and 2/43 non-GPG strains from a representative set (8) chosen from an international collection (28) of infection-causing isolates. The remaining non-GPG strains gave products of different sizes instead of or in addition to the 276-bp product (S. Wattimena, M. Patchett, and J. Schmid, unpublished observations).
A duplex colony PCR assay utilizing both markers correctly identified 36/36 GPG isolates in a set of 59 isolates from our international collection (see Table S1 in the supplemental material), based on the presence of (i) a 767-bp MG1 product and (ii) a single, 276-bp YWP1 band. Twenty-one non-GPG strains in the set produced no MG1 product and produced YWP1 products that differed in size from the 276-bp GPG product (some formed two YWP1 products, one of which had a size of 276 bp). Of two strains classified as borderline by other methods, one produced the GPG-specific MG1 product but not the GPG-specific YWP1 product. The other had both the GPG-specific MG1 product and the single 276-bp YWP1 product, i.e., it would have been classified as a GPG strain by our assay (see Fig. 1A for examples of patterns). Thus, of 59 isolates, only 1 isolate (a borderline strain) was misclassified as a GPG strain, indicating that the method is 98% accurate in identifying GPG strains (binominal confidence interval, 91% to 100%).
The assay apparently does not produce PCR products with other species. When the assay was used in the survey described below, no PCR products were obtained for 19 isolates. While these had been submitted to us as C. albicans strains, all of them were shown to be different species when tested for germ tube and chlamydospore formation.
The prevalence of GPG strains is elevated in younger patients and in females.
We applied the duplex assay (Fig. 1B) to 635 C. albicans isolates from bloodstream infections of European patients, using samples collected between 1997 and 2008. The majority of the patients (437 patients) were Italian, 98 were French, 43 Swedish, 23 Austrian, 10 Greek, 10 Spanish, 5 German, 5 Finnish, and 4 Portuguese. The average age of the patients was 55 years, 40% were female, 61% had undergone surgery, and 68% had been admitted to an intensive care unit (ICU) (see Table S2 in the supplemental material for information on individual patients).
A total of 114 isolates had the typical GPG pattern, 496 were classified as non-GPG strains, and 25 gave PCR patterns that identified them as borderline strains. Thus, 18% of all C. albicans isolates were of the GPG type, 78% were of the non-GPG type, and 4% were of the borderline genotype, which is broadly consistent with the genotype distribution we observed earlier among isolates from a worldwide collection of predominantly superficial infections (28, 43) and with the frequency of 28% GPG strains among 327 European bloodstream isolates (22).
The prevalence of GPG strains appeared to be higher in younger patients, in particular those of less than 1 year of age, and in females (Fig. 2A) (age brackets were set to be consistent with those used in mortality analyses; see below). Logistic regression confirmed that both age (P=0.011) and sex (P=0.0005), but no other factors, significantly impacted GPG prevalence (Fig. 2B) (borderline strains were excluded from these analyses).
Fig. 2.
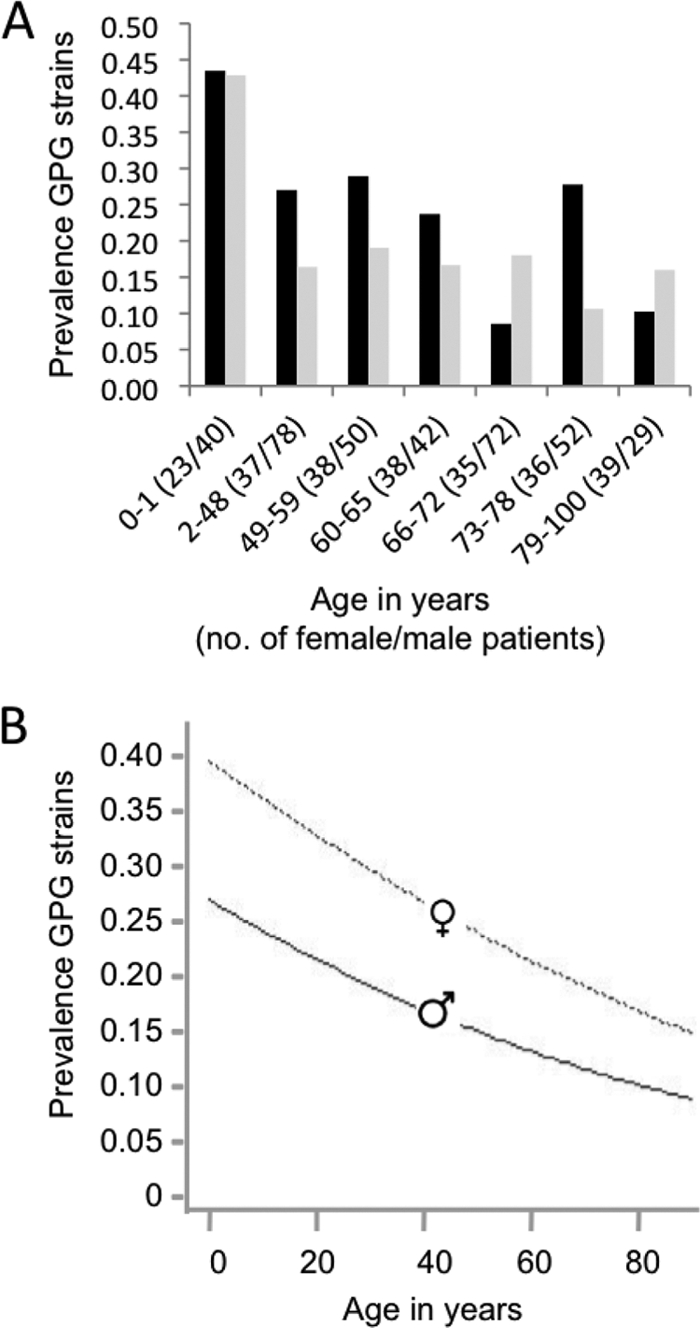
Prevalence of GPG strains. (A) Prevalence of GPG strains in female (black bars) and male (gray bars) patients in different age brackets. Age brackets were set to be consistent with those set for mortality analyses (see Fig. 4). Numbers in parentheses are the numbers of male and female patients in each age category. (B) Plot of dependence of prevalence on age for males and females, as deduced from logistic regression. Borderline strains were excluded from these analyses, as was one isolate where the age of the patient was unknown.
For 125 patients, the expected mortality at admission could be calculated based on SAPS, APACHE, or PIM2 scores. Because of the relatively small number of data points, we did not include predicted mortality in the regression analysis, but there was no indication that the severity of the underlying illness had an impact on GPG strain prevalence. The predicted mortality at admission for 21 patients from whom GPG strains were later isolated was 42% ± 27%, and the predicted mortality at admission for 104 patients from whom non-GPG strains were later isolated was 40% ± 29%.
Presence of GPG strains is associated with higher mortality in young patients.
Overall mortality (assessed 30 days after the diagnosis of candidemia) was 37%, and for those patients who died within the observation period, the average time between diagnosis of candidemia and death was 10 days (the latter average is based on 195 patients for whom the time interval between diagnosis and death had been recorded [see Table S2 in the supplemental material]).
For analysis of the impact of genotype on mortality, only GPG strains and non-GPG strains were considered (i.e., borderline strains were excluded). Across all patients, there was no apparent impact of genotype on mortality (36% for GPG and 37% for non-GPG strains) or the average time between diagnosis of candidemia and death (10 days for both types). Likewise, the ratio between actual and predicted mortality at admission (based on SAPS, APACHE, or PIM2 scores) was not significantly different for patients infected with GPG strains from that for patients infected with other strains. The actual mortality was 86% of predicted mortality for 21 patients with GPG isolates and 93% of predicted mortality for 104 patients with non-GPG isolates.
However, when mortality was assessed for different age brackets, GPG isolates were associated with increased mortality in younger patients (Fig. 3A and B) (age ranges were set so that at least 5 deaths of patients with GPG strains occurred in each cohort). For patients aged 48 years or less, mortality was >2-fold higher when GPG strains were present (33% versus 15%) (z test; P < 0.01). Mortality of patients aged 49 and above was 39% when GPG strains were present, versus 46% for non-GPG strains, but this difference was not significant in a z test.
We verified the age-dependent impact of genotype on mortality by using logistic regression (Fig. 3C; Table 1 ). To allow for variations between hospitals and between the years in which samples were collected, we used hospital year as a random effect. The results confirmed the higher mortality associated with GPG strains in younger patients, with the effect gradually reducing and possibly reversing in older patients. Among the other variables, surgery decreased mortality and admission to ICU and age increased mortality; sex had no effect (Table 1).
Table 1.
Logistic regression model results for mortality
| Effecta | Estimateb | SE | z value | P valuec |
|---|---|---|---|---|
| Interceptd | −2.65549 | 0.386138 | −6.877 | 6.11E−12*** |
| GPG | 1.996333 | 0.525864 | 3.796 | 0.000147*** |
| Age | 0.035276 | 0.005522 | 6.388 | 1.68E−10*** |
| Surgery | −0.397701 | 0.20292 | −1.96 | 0.050009† |
| ICU | 0.467444 | 0.207856 | 2.249 | 0.02452* |
| GPG-age | −0.032178 | 0.008941 | −3.599 | 0.00032*** |
Hospital year was fitted as a random effect to allow for differences in mortality between hospitals or in different years at the same hospital.
Estimates of model terms that, when added together, give the logit {log[P/ (1 − P)]} of patient mortality.
*, P < 0.05; ***, P < 0.001; †, 0.1.
Logit of probability of death of patients of age 0 infected with non-GPG strains.
The age-dependent effect of genotype on mortality was also apparent by a steeper decline of survival probability over 30 days for GPG strain-infected patients aged 48 or below than that for patients aged 48 or below infected with other strains (Fig. 4A). In older patients, the decline of survival probability was not significantly affected by strain genotype (Fig. 4B).
Fig. 4.
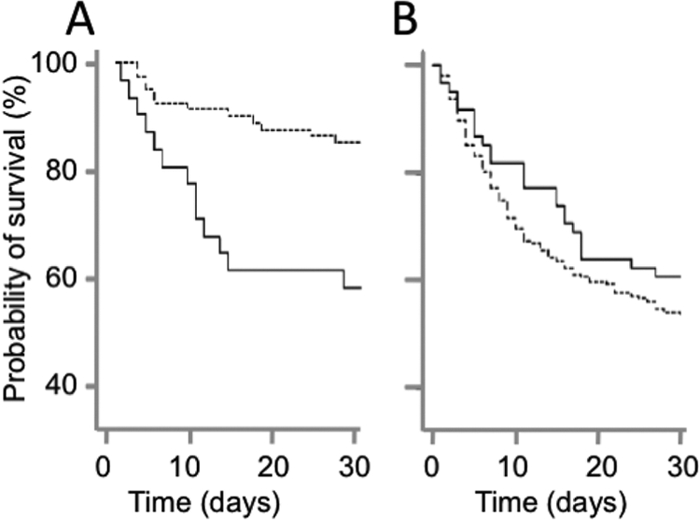
Time dependence of survival probability of patients. (A) Time dependence of the probability of survival of patients aged 48 years or less with GPG isolates (solid line) and non-GPG isolates (dashed line). (B) Time dependence of the probability of survival of patients older than 48 years with GPG isolates (solid line) and non-GPG isolates (dashed line).
DISCUSSION
This is the first report, to our knowledge, that a specific C. albicans genotype, namely, the GPG (clade 1) strain type, is associated with increased patient mortality—albeit only in younger patients. It is difficult to establish that the death of a patient is attributable to candidemia (7, 24), and we cannot exclude the possibility that unknown factors that increased the likelihood of death of young patients also increased the likelihood of the presence of GPG strains, even though we found no indications supporting such a scenario. However, while we cannot be absolutely certain, a causal link between the presence of GPG strains and increased mortality seems the more likely explanation for our data.
Such a causal link is not likely to involve susceptibility to antifungal agents. GPG (clade 1) strains do not differ from other strains in their susceptibility to antifungal agents commonly used to treat candidemia; the exception is an elevated frequency of flucytosine resistance (21, 22, 29), but this agent is not commonly used for monotherapy (26).
The most likely explanation for our results is that GPG strains are more virulent than non-GPG strains in younger patients. This would not be unexpected, since their high prevalence as commensals and pathogens (22, 28) and some of the traits that distinguish them from other strains (15, 29, 41–43) indicate that they are more apt at coping with the host's defense mechanisms than other strains. On the other hand, GPG strains are not more virulent than strains of other clades in the tail vein mouse model (15) (the age of the mice used was not provided, but presumably they were young, as also suggested by their weights [18]). This discrepancy could stem from the fact that for the mouse model, virulence comparisons are based on mortality differences observed after the introduction of equal (and large) numbers of C. albicans cells into the bloodstream, while in the human patient the ability to reach sterile sites in sufficiently large numbers to cause disease would be a prerequisite for causing candidemia and death. Our data might reflect differences between GPG strains and other strains in the ability to reach the bloodstream. This would also be consistent with our observation that the prevalence of GPG strains, i.e., their ability to cause candidemia (which should in part depend on their ability to cross into the bloodstream), and mortality follow very similar trends.
We note that the current study involved predominantly Italian patients, and we cannot assess to what degree our findings are applicable to patients regardless of ethnicity or geographical region. However, we did see a similar age-dependent impact of genotype on mortality and a similar age dependence of GPG prevalence in a preliminary study using 90 of the French patients (27).
An obvious question, to which we have no answer, is which key factors could make GPG strains more virulent. Indeed, there may be no small set of easily definable key factors. Comparative studies have identified numerous differences between GPG (clade 1) strains and strains from other clades (6, 14, 15, 22, 23, 29, 41–44). Most likely, hundreds of epistatically interacting DNA polymorphisms, rather than a few key mutations, are responsible for the greater success of GPG strains in the human host in general, and potentially also for their increased virulence (43). This assumption is also consistent with the large number of genes—over 200—that are C. albicans virulence determinants in animal models (34). An additional complication is that while our results indicate that the average GPG strain could be more virulent than the average non-GPG strain, non-GPG strains constitute a diverse collection of genotypes, some of which might very well be more virulent than GPG strains (22, 28).
C. albicans genotype was not the only factor impacting mortality. One other factor, an association of lower mortality with surgery, is a well-established phenomenon used in mortality predictions for critically ill patients (APACHE) (10). Another, the association of admission to an ICU with increased mortality, would be expected, since ICU admission indicates that the patient's state was already critical prior to the onset of candidemia.
More puzzling is the impact of age, namely, the observation that GPG strains are associated with higher mortality in younger patients, including neonates, but not in the elderly (where they may even cause less mortality). Neither neonates nor the elderly have a fully functional immune system (32, 39), and if GPG strains were causing higher mortality because they were better at overcoming existing immune defenses, they would be expected to cause higher mortality than other strains in both groups. Previous exposure to the pathogen has been used to explain reduced mortality among older patients in the 1918 influenza epidemic (1) and could also conceivably play a role in protecting older patients against GPG strains, given that acquired immunity does play some role in the host's defense against candidemia (2) and that older patients are likely to have encountered the common GPG strains at some stage during their life. Another factor contributing to age dependence of both mortality and prevalence could be the relatively recent increase in human life expectancy (9). GPG strains presumably evolved to be adapted optimally toward young individuals, which until the 19th century were the typical human hosts of C. albicans. Among non-GPG strains, which are less successful in these hosts, there may have been stronger selection for traits allowing them to expand into the novel cohort of older hosts.
One factor that did not influence mortality but did influence GPG strain prevalence was gender. Being female increased the probability that candidemia was caused by a GPG strain approximately 1.5-fold (Fig. 2B). We are not aware of other reports on this issue, but some of our own earlier work suggests that this gender effect could be specific to candidemia. We found a lower GPG prevalence in females than in males (33% versus 41%) among 266 isolates from mainly superficial infections (28). Commensal colonization data we retained from other studies (30, 31, 33) do not show a statistically significant impact on gender (28/69 [41%] isolates from females and 7/21 [33%] isolates from males were of the GPG type). However, the latter data are likely to underrepresent the frequency at which GPG strains are present as commensals on an individual. In 11 of 16 healthy females (69%) from whom C. albicans could be cultured from several body locations, GPG isolates were recovered from at least one location (33). More work is required to resolve this issue, especially since the frequent presence of GPG strains on females could be part of the explanation for why GPG strains are highly prevalent as infection-causing agents in infants. Infants are often colonized by maternal strains (4). If GPG strains are present on most mothers and if they are better colonizers than other strains (28), infants may initially be colonized predominantly by GPG strains (which later cause disease), even if a GPG strain is only one of several strains present in the mother.
Finally, our results have potential impact on antifungal prophylaxis. The prophylactic administration of antifungal drugs can reduce the incidence of fungemia, although its effect on mortality is less well established. However, prophylaxis is thought to accelerate the spread of antifungal resistance, is not always well tolerated, and should thus be targeted toward those patients at most risk (11, 19, 24, 35). Including the impact of genotype on mortality in evaluations of the efficacy of prophylaxis should assist in clarifying its benefits in terms of reducing mortality. Furthermore, since candidemia is caused predominantly by previously present colonizing strains (17, 38), our assay, if used for detection of GPG strains among colonizing isolates from young patients, could be a risk assessment tool to identify those patients who would benefit most from antifungal prophylaxis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Francoise Dromer and Stephane Bretagne for their assistance in setting up the initial part of this work and in interpreting its results and Richard Cannon for valuable comments on the manuscript.
This work was partially funded by a Marsden grant from the Royal Society of New Zealand and by a grant from the International Mobility Fund, administered by the Royal Society of New Zealand, to J.S.
The French Mycoses Study Group participants contributing isolates used in this study were Claire Bouges-Michel (Hôpital Avicenne, Bobigny), Isabelle Poilane (Hôpital Jean Verdier, Bondy), Marie-Elisabeth Bougnoux and Jean Dunand (Hôpital Ambroise Paré, Boulogne), Guy Galeazzi (Hôpital Louis Mourier, Colombes), Stéphane Bretagne (Hôpital Henri Mondor, Créteil), Nathalie Fauchet (Centre Hospitalier Intercommunal de Créteil, Créteil), Elisabeth Forget (Hôpital Beaujon, Clichy), Françoise Botterel and Christine Bonnal (Hôpital du Kremlin Bicêtre), Odile Eloy (Hôpital Mignot, Le Chesnay), Christine Lawrence (Hôpital Raymond Poincaré, Garches), Marie-Françoise David and Liliana Mihaila (Hôpital Paul Brousse, Villejuif), Elisabeth Chachaty and Olivier Adam (Institut Gustave Roussy, Villejuif), and the following participants in Paris: Christian Chochillon (Hôpital Bichat), André Paugam and Marie-Thérèse Baixench (Hôpital Cochin), Muriel Cornet (Hôpital de l'Hôtel Dieu), Marie-Christine Escande (Institut Curie), Svetlana Challier and Marie-Elisabeth Bougnoux (Hôpital Necker), Eric Dannaoui (Hôpital Européen Georges Pompidou), Annick Datry, Houria Laklache, Bader Lminmouni, and Sophie Brun (Hôpital de la Pitié-Salpétrière), Jean-Louis Poirot (Hôpital Saint Antoine), Claire Lacroix (Hôpital Saint Louis), Didier Moissenet (Hôpital Trousseau), Michel Develoux (Hôpital Tenon), and Stéphane Bonacorsi (Hôpital Robert Debré).
The ECMM survey participants contributing isolates used in this study were Anna Maria Tortorano, Giovanna Dho, Anna Prigitano, Anna Grancini, Cristina Ossi, Giuseppe Giuliani, Milvana Tejada, Chiara Vismara, Giovanna Viola, Caterina Cavanna, Marco Passera, Claudio Farina, Antonio Grossi, Gabriele Pinsi, Antonio Ceraminiello, Pietro Casella, Carlo Agrappi, Carla Sturla, Gianluigi Lombardi, Giuliana LoCascio, Maria Luisa Deiana and Maria Graziella Garau (Italy), Lean Klingspor (Sweden), Andrea Weigel, Barbara Graf, and Markus Ruhnke (Germany), Javier Peman (Spain), Juha H. Salonen, Olli Meurman, and Hannu Sarkkinen (Finland), Raquel Sabino and Zélia Videira (Portugal), Harald Kirschner, Cornelia Lass-Flörl, Matthias Maass, Regina Watschinger, Birgit Willinger, and Eva-Maria Zeitlberger (Austria), and Aristea Velegraki, Stavroula Antonopoulou, George Dimopoulos, Evangelia Kouskouni, Efthimia Petinaki, Eleftheria Trikka-Graphakos, A. Timoleon Vyzantiadis, and Loukia Zerva (Greece).
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Ahmed R., Oldstone M. B., Palese P. 2007. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat. Immunol. 8: 1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashman R. B., et al. 2004. Innate versus adaptive immunity in Candida albicans infection. Immunol. Cell Biol. 82: 196–204 [DOI] [PubMed] [Google Scholar]
- 3. Ausubel F. M., et al. 2006. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY [Google Scholar]
- 4. Bliss J. M., Basavegowda K. P., Watson W. J., Sheikh A. U., Ryan R. M. 2008. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr. Infect. Dis. J. 27: 231–235 [DOI] [PubMed] [Google Scholar]
- 5. Dannaoui E Comparison of antifungal MICs for yeasts obtained using the EUCAST method in a reference laboratory and the Etest in nine different hospital laboratories. Clin. Microbiol. Infect. 16: 863–869 [DOI] [PubMed] [Google Scholar]
- 6. Giblin L., et al. 2001. A DNA polymorphism specific to Candida albicans strains exceptionally successful as human pathogens. Gene 272: 157–164 [DOI] [PubMed] [Google Scholar]
- 7. Gudlaugsson O., et al. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37: 1172–1177 [DOI] [PubMed] [Google Scholar]
- 8. Holland B. R., Schmid J. 2005. Selecting representative model micro-organisms. BMC Microbiol. 5: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horiuchi S. 2000. Greater lifetime expectations. Nature 405: 744–745 [DOI] [PubMed] [Google Scholar]
- 10. Knaus W. A., Draper E. A., Wagner D. P., Zimmerman J. E. 1985. APACHE II: a severity of disease classification system. Crit. Care Med. 13: 818–829 [PubMed] [Google Scholar]
- 11. Lass-Flörl C. 2009. The changing face of epidemiology of invasive fungal disease in Europe. Mycoses 52: 197–205 [DOI] [PubMed] [Google Scholar]
- 12. Le Gall J. R., Lemeshow S., Saulnier F. 1993. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA 270: 2957–2963 [DOI] [PubMed] [Google Scholar]
- 13. Lin X., Zhang D. 1999. Inference in generalized additive mixed models by using smoothing splines. J. R. Stat. Soc. Series B Stat. Methodol. 55: 381–400 [Google Scholar]
- 14. Lott T. J., Holloway B. P., Logan D. A., Fundyga R., Arnold J. 1999. Towards understanding the evolution of the human commensal yeast Candida albicans. Microbiology 145: 1137–1143 [DOI] [PubMed] [Google Scholar]
- 15. MacCallum D. M., et al. 2009. Property differences among the four major Candida albicans strain clades. Eukaryot. Cell 8: 373–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Magee P. T. 2010. Fungal pathogenicity and morphological switches. Nat. Genet. 42: 560–561 [DOI] [PubMed] [Google Scholar]
- 17. Marco F., et al. 1999. Elucidating the origins of nosocomial infections with Candida albicans by DNA fingerprinting with the complex probe Ca3. J. Clin. Microbiol. 37: 2817–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matzel L. D., Grossman H., Light K., Townsend D., Kolata S. 2008. Age-related declines in general cognitive abilities of Balb/C mice are associated with disparities in working memory, body weight, and general activity. Learn. Mem. 15: 733–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morgan J. 2005. Global trends in candidemia: review of reports from 1995-2005. Curr. Infect. Dis. Rep. 7: 429–439 [DOI] [PubMed] [Google Scholar]
- 20. Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42: 590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Odds F. C. 2009. In Candida albicans, resistance to flucytosine and terbinafine is linked to MAT locus homozygosity and multilocus sequence typing clade 1. FEMS Yeast Res. 9: 1091–1101 [DOI] [PubMed] [Google Scholar]
- 22. Odds F. C., et al. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6: 1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oh S.-H., et al. 2005. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology 151: 673–681 [DOI] [PubMed] [Google Scholar]
- 24. Pfaller M. A., Diekema D. J. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20: 133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. R Development Core Team. 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 26. Sanglard D., Odds F. C. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2: 73–85 [DOI] [PubMed] [Google Scholar]
- 27. Schmid J., et al. 2008. Differences in mortality between patients infected with different Candida albicans genotypes, abstr. S2:1, p. 19. Abstr. 9th ASM Conf. Candida Candidiasis, Jersey City, NJ [Google Scholar]
- 28. Schmid J., et al. 1999. Evidence for a general-purpose genotype in Candida albicans, highly prevalent in multiple geographic regions, patient types and types of infection. Microbiology 145: 2405–2414 [DOI] [PubMed] [Google Scholar]
- 29. Schmid J., Hunter P. R., White G. C., Nand A. K., Cannon R. D. 1995. Physiological traits associated with success of Candida albicans strains as commensal colonisers and pathogens. J. Clin. Microbiol. 33: 2920–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmid J., Rotman M., Reed B., Pierson C. L., Soll D. R. 1993. Genetic similarity of Candida albicans strains from vaginitis patients and their partners. J. Clin. Microbiol. 31: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmid J., et al. 1995. Evidence for nosocomial transmission of Candida albicans obtained by Ca3 fingerprinting. J. Clin. Microbiol. 33: 1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Solana R., Pawelec G., Tarazona R. 2006. Aging and innate immunity. Immunity 24: 491–494 [DOI] [PubMed] [Google Scholar]
- 33. Soll D. R., et al. 1991. Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J. Clin. Microbiol. 29: 1702–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Szabo E. K., MacCallum D. M. 2011. The contribution of mouse models to our understanding of systemic candidiasis. FEMS Microbiol. Lett. 320: 1–8 [DOI] [PubMed] [Google Scholar]
- 35. Timmers G. J., et al. 2000. Amphotericin B colloidal dispersion (Amphocil) vs fluconazole for the prevention of fungal infections in neutropenic patients: data of a prematurely stopped clinical trial. Bone Marrow Transplant. 25: 879–884 [DOI] [PubMed] [Google Scholar]
- 36. Tortorano A. M., et al. 2004. Epidemiology of candidaemia in Europe: results of 28-month European Confederation of Medical Mycology (ECMM) hospital-based surveillance study. Eur. J. Clin. Microbiol. Infect. Dis. 23: 317–322 [DOI] [PubMed] [Google Scholar]
- 37. Turnbull B. W. 1976. The empirical distribution function with arbitrarily grouped censored and truncated data. J. R. Stat. Soc. Series B Stat. Methodol. 38: 290–295 [Google Scholar]
- 38. Vanhee L. M. E., Nelis H. J., Coenye T. . What can be learned from genotyping of fungi? Med. Mycol. 48: S60–S69 [DOI] [PubMed] [Google Scholar]
- 39. Weng N.-P. 2006. Aging of the immune system: how much can the adaptive immune system adapt? Immunity 24: 495–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zaoutis T. 2010. Candidemia in children. Curr. Med. Res. Opin. 26: 1761–1768 [DOI] [PubMed] [Google Scholar]
- 41. Zhang N., Cannon R. D., Holland B., Patchett M., Schmid J. 2010. Impact of genetic background on allele selection in a highly mutable Candida albicans gene, PNG2. PLoS One 5: e9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang N., et al. 2003. Sixty alleles of the ALS7 open reading frame in Candida albicans: ALS7 is a hypermutable contingency locus. Genome Res. 13: 2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang N., et al. 2009. Distribution of mutations distinguishing the most prevalent disease-causing Candida albicans genotype from other genotypes. Infect. Genet. Evol. 9: 493–500 [DOI] [PubMed] [Google Scholar]
- 44. Zhao X., et al. 2007. Analysis of ALS5 and ALS6 allelic variability in a geographically diverse collection of Candida albicans isolates. Fungal Genet. Biol. 44: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.