Abstract
A middle-aged woman developed fatal urosepsis due to a multidrug-resistant Escherichia coli strain representing sequence type ST131, a recently emerged, disseminated, multidrug-resistant extraintestinal pathogen, after presumably having acquired it from her extensively antibiotic-exposed sister with chronic recurrent cystitis. Susceptibility results (reported on day 4) showed resistance to the initially selected regimen.
CASE REPORT
The index patient was a middle-aged female with chronic, recurrent symptomatic urinary tract infections (UTIs) due to Escherichia coli. Her urine isolates had become increasingly antimicrobial resistant over a 5-year period, including having become, for the past several years, extended-spectrum beta-lactamase (ESBL) positive.
Both the index patient and a younger sister had alpha-1-anti-trypsin deficiency. Whereas the index patient was only mildly affected, the younger sister was severely affected, leading to long-term care facility placement and a referral for lung transplantation (which was denied, because of continued smoking). The index patient had cared for the younger sister in the index patient's home for several months during the summer prior to the younger sister's acute illness.
Shortly after the younger sister's temporary residence in the index patient's home, she began to experience mild dysuria when voiding (no indwelling or intermittent urinary catheter use) and low-grade fever. After a week of progressive symptoms, she was admitted to the hospital for presumed pyelonephritis.
On admission, the patient appeared to be in mild distress with right-sided flank pain. Her temperature was 37.6°C, pulse was 52 beats per minute, blood pressure was 153/82 mm Hg, and respirations were 24 per minute. Oxyhemoglobin saturation was 92% on 4 liters of oxygen. The right flank was tender. Laboratory findings included leukocytosis (17,800 cells/μl) with 88% neutrophils. Urinalysis showed marked pyuria and bacteriuria. Ciprofloxacin (400 mg intravenously every 12 h) and stress-dose corticosteroids were administered.
Over 3 days, her clinical status worsened, and she was transferred to the intensive care unit for septic shock, presumed to be secondary to pyelonephritis. Her white blood cell (WBC) count rose to 18,400 cells/μl (29% band forms), and her serum creatinine level increased to 2.7 mg/dl. The patient received mechanical ventilation and vasopressor therapy. Renal ultrasonography revealed moderate right hydronephrosis; a right percutaneous nephrostomy tube was placed. Blood and urine cultures grew E. coli.
Due to the patient's precipitous decline, antimicrobial therapy was broadened empirically by adding piperacillin-tazobactam, based on local antibiogram data. On hospital day 4, septic shock worsened, requiring additional vasopressor support. Susceptibility data, which were now reported, showed the E. coli isolates to be resistant to fluoroquinolones and extended-spectrum cephalosporins but susceptible to piperacillin-tazobactam, amikacin, carbapenems, and trimethoprim-sulfamethoxazole. Therapy was changed to meropenem, but the patient died within hours.
Molecular characterization of isolates.
Genomic profiling was performed by pulsed-field gel electrophoresis (PFGE) of XbaI-digested total DNA (5). Isolates were screened for blaCTX-M alleles by PCR with primers specific for various blaCTX-M groups using Taq polymerase and a genomic DNA template. Purified CTX-M PCR amplicons (QIAquick PCR purification kit; Qiagen, Valencia, CA) were cloned into plasmid pCR-XL-TOPO and transformed into competent E. coli TOP10 cells, as described by the manufacturer (Invitrogen, Carlsbad, CA). Isolated plasmid DNA (Qiagen plasmid minikit) was bidirectionally sequenced by ACGT, Inc. (Wheeling, IL). Seven-locus multilocus sequence typing (MLST) was done as specified using the MLST database (http://mlst.ucc.ie/mlst/dbs/Ecoli). The presence of 52 extraintestinal virulence-associated genes and the O25b rfb (O-lipopolysaccharide) variant was assessed using established PCR assays (5). Susceptibility testing was performed using MicroScan II (Siemens, Deerfield, IL).
Microbiology results.
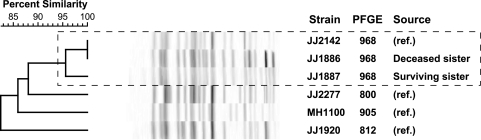
The E. coli isolates from both patients (surviving sister, urine; deceased sister, blood) exhibited identical susceptibility profiles and MLST allelic profiles corresponding with ST131. They also had highly (96%) similar PFGE profiles, which suggested that they represented the same strain (14), likely transmitted from one sister to the other. Their PFGE profiles corresponded with pulsotype 968, the most common pulsotype within a private PFGE profile reference library comprising 167 PFGE types from 566 ST131 isolates, as obtained from approximately 85 U.S. and international locales (J. R. Johnson, unpublished data) (Fig. 1). Both isolates exhibited blaCTX-M-15 (which encodes the ST131-associated CTX-M-15 ESBL variant), the (ST131-associated) O25b lipopolysaccharide-encoding rfb variant, and a typical ST131-like virulence profile, which included the F10 papA (P fimbria) allele but no other pap genes, fimH (type 1 fimbria), iha (siderophore-adhesin), fyuA (yersiniabactin system), iutA (aerobactin system), kpsMII (group 2 capsule), sat (secreted autotransporter toxin), usp (uropathogenic-specific protein), ompT (outer membrane protease), and malX (pathogenicity island marker). The only virulence profile difference detected involved traT (plasmid borne, serum resistance associated; present in the surviving sister's urine isolate but not the deceased sister's blood isolate). Notably absent, as for most ST131 strains, were certain other typical virulence genes of uropathogenic E. coli, such as papG (P fimbria adhesin), sfa/foc (S and F1C fimbriae), hlyA (hemolysin), and cnf1 (cytotoxic necrotizing factor 1).
Fig. 1.
Dendrogram of XbaI pulsed-field gel electrophoresis (PFGE) profiles. Included in the analysis is a blood isolate from the deceased sister (strain JJ1886), a urine isolate from the surviving sister (strain JJ1887), and four sequence type ST131 reference (ref.) strains. The reference strains are the index isolates from the four most prevalent pulsotypes in a large private PFGE library that comprises 566 ST131 isolates and their 167 distinct pulsotypes (as defined at a 94% Dice coefficient similarity threshold, corresponding with an approximately 3-band profile difference). The 2% band tolerance limit used explains the calculated 100% profile similarity between JJ2142 and JJ1886, despite visible differences. The dashed rectangle encloses the two case isolates and the index isolate for pulsotype 968 (24% of reference library isolates), which cluster at approximately 96% similarity, consistent with them all representing the same strain.
Escherichia coli strains of sequence type ST131, as defined by multilocus sequence typing (MLST), have emerged globally over the past decade both in hospitals and in the community, causing outbreaks of multidrug-resistant (MDR) infections in Europe, Canada, and Asia (12, 13). Such strains are almost uniformly resistant to fluoroquinolones and frequently produce ESBLs, specifically CTX-M-15, which is the most prevalent of the now-dominant CTX-M (cefotaximase) family of ESBLs. ST131 strains can cause serious or fatal disease and may be community acquired. ST131 was reported in the United States only recently, including in a surveillance study showing that ST131 was the main cause of significantly antimicrobial-resistant E. coli infections in 2007 (5), plus in several case reports and series documenting likely within-household transmission (2, 4, 6, 8). We report a fatal case of urosepsis that involved community-associated intrafamilial spread of a multiresistant strain of E. coli ST131.
The use of empirical therapy with fluoroquinolones for presumed pyelonephritis, as recommended by guidelines, is increasingly problematic due to emerging resistance among uropathogens (10). Many countries have experienced a disturbing decline in E. coli susceptibility to fluoroquinolones, likely due in part to both extensive drug usage and clonal spread (10). In this report, we document the presumed clonal spread in the outpatient setting of a virulent strain of fluoroquinolone-resistant, ESBL-producing E. coli from an extensively antimicrobial-exposed patient with chronic/recurrent UTIs to her antimicrobial-naïve sister without a history of recurrent UTIs.
Within-household sharing of E. coli strains, including members of sequence type ST131, among human and animal household members has been extensively documented (2, 4, 8). This process, which likely reflects host-to-host transmission, may contribute significantly to the community-wide dissemination of emerging antimicrobial-resistant uropathogenic E. coli lineages, such as clonal group A (trimethoprim-sulfamethoxazole resistance) (7, 9), the O15:K52:H1 clonal group (fluoroquinolone resistance) (7), and perhaps of greatest concern, ST131 (fluoroquinolone and extended-spectrum cephalosporin resistance) (5, 12, 13). To our knowledge, this is the first report of a fatal infection resulting from within-household transmission of extraintestinal pathogenic E. coli, including ST131.
This case also emphasizes that appropriate broad-spectrum antimicrobial therapy is critical for patients with sepsis (11). Ultimately, the design of empirical therapy regimens in patients presenting with urosepsis must consider local resistance patterns, including rising rates of fluoroquinolone resistance among E. coli strains and the emergence of community-associated ESBLs, plus patient-specific risk factors for resistance. Thus, elicitation of a good medical history, including antibiotic exposures (both direct and indirect), travel, and health care contact, is vital. A future rapid molecular test for relevant multidrug-resistant pathogens, or specific resistance elements, could supplement the careful clinical assessment, allowing more appropriately targeted initial therapy (1).
This case also provides an example of the “societal” effects of antimicrobials, since the deceased sister, who had not been directly exposed to outpatient antibiotics, presumably acquired a virulent MDR pathogen from an exposed household member, leading to her demise. This is consistent with other evidence that household-level antimicrobial use may be more predictive of colonization with a resistant E. coli strain than an individual's personal exposure history (3).
Summary.
A patient developed fatal urosepsis from an ESBL-positive, fluoroquinolone-resistant E. coli ST131 strain that she likely acquired in the community from a sister with chronic recurrent UTIs. Empirical antimicrobial therapy with ciprofloxacin and then piperacillin-tazobactam (to which the organism exhibited in vitro susceptibility) failed to prevent progressive septic shock and multisystem failure. Community dissemination of virulent MDR pathogens, including E. coli sequence type ST131, requires heightened vigilance on the part of clinicians, laboratories, and public health authorities for both prevention and appropriate management. In the future, rapid detection of ST131 (or other multidrug-resistant pathogens or their resistance elements) by the clinical microbiology laboratory might allow refined selection of empirical antimicrobial therapy, thereby improving clinical outcomes while avoiding the “collateral damage” resulting from routine widespread use of broad-spectrum agents.
Acknowledgments
This work was supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs.
Connie Clabots (VA Medical Center) and Brian Johnston (VA Medical Center and University of Minnesota) provided invaluable technical assistance. Dave Prentiss (Minneapolis VA Medical Center) prepared the figure image.
J. R. Johnson has received grants or contracts from Merck, Rochester Medical, and Syntiron. J. Quinn received financial support from the Chicago Infectious Diseases Research Institute and is currently an employee and shareholder in Pfizer Global Research. The other authors have no conflicts of interest with this work.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Colgan R., Johnson J. R., Kuskowski M., Gupta K. 2008. Risk factors for trimethoprim-sulfamethoxazole resistance in patients with acute uncomplicated cystitis. Antimicrob. Agents Chemother. 52:846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ender P. T., et al. 2009. Transmission of extended-spectrum beta-lactamase-producing Escherichia coli (sequence type ST131) between a father and daughter resulting in septic shock and emphysematous pyelonephritis. J. Clin. Microbiol. 47:3780–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hannah E. L., et al. 2005. Drug-resistant E. coli, rural Idaho. Emerg. Infect. Dis. 11:1614–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson J. R., Anderson J. T., Clabots C., Johnston B., Cooperstock M. 2010. Within-household sharing of a fluoroquinolone-resistant Escherichia coli sequence type ST131 strain causing pediatric osteoarticular infection. Pediatr. Infect. Dis. J. 29:474–475 [DOI] [PubMed] [Google Scholar]
- 5. Johnson J. R., Johnston B., Clabots C., Kuskowski M. A., Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 6. Johnson J. R., et al. 2010. Escherichia coli sequence type ST131 as an emerging fluoroquinolone-resistant uropathogen among renal transplant recipients Antimicrob. Agents Chemother. 54:546–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson J. R., et al. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002–2004. Antimicrob. Agents Chemother. 53:2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson J. R., Miller S., Johnston B., Clabots C., DebRoy C. 2009. Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and urovirulent E. coli strains among dogs and cats within a household. J. Clin. Microbiol. 47:3721–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson J. R., et al. 2005. Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11:141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson L., et al. 2008. Emergence of fluoroquinolone resistance in outpatient urinary Escherichia coli isolates. Am. J. Med. 121:876–884 [DOI] [PubMed] [Google Scholar]
- 11. Lodise T. P. J., et al. 2007. Predictors of 30-day mortality among patients with Pseudomonas aeruginosa bloodstream infections: impact of delayed appropriate antibiotic selection. Antimicrob. Agents Chemother. 51:3510–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peirano G., Pitout J. D. D. 2010. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents. 35:316–321 [DOI] [PubMed] [Google Scholar]
- 13. Rogers B. A., Sidjabat H. E., Paterson D. L. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 14. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]