Abstract
We report the first case of Segniliparus rotundus pneumonia in an adult with non-cystic fibrosis bronchiectasis. All isolates were identified as S. rotundus by 16S rRNA gene sequencing and rpoB PCR-restriction analysis. Antibiotic therapy with clarithromycin and ciprofloxacin for 2 months improved the patient's condition and achieved successful sputum conversion.
CASE REPORT
A 43-year-old woman was referred to Samsung Medical Center for chronic cough and sputum lasting more than 3 months. The patient's medical history included a tuberculous pleurisy at the age of 22 years. She lived in an urban area and had no history of smoking, alcoholism, or immunosuppressive drug use.
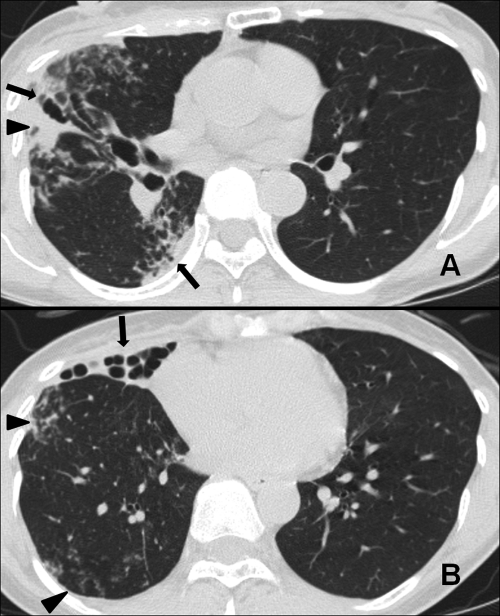
A human immunodeficiency virus (HIV) antibody test was negative. A high-resolution computed tomography (HRCT) chest scan revealed severe bronchiectasis, multiple small nodules, and branching centrilobular nodules. These findings suggested bronchiolitis, especially in the right lung (Fig. 1). There was no evidence indicating a specific cause of the bronchiectasis, such as cystic fibrosis. Multiple sputum specimens were examined for mycobacteria. The specimens were decontaminated and concentrated with NaOH plus N-acetyl-l-cysteine. Acid-fast bacilli (AFB) were detected more than five times on both auramine-rhodamine fluorescent stain and Ziehl-Neelsen methods, and liquid cultures of these multiple sputum specimens were incubated for 9 to 11 days with a Bactec MGIT 960 system (BD Diagnostics, Sparks, MD).
Fig. 1.
A 43-year-old woman with Segniliparus rotundus pneumonia. (A) A high-resolution computed tomography (HRCT) chest scan at the level of the proximal lower lobar bronchus shows extensive bronchiectasis and lobular consolidation. (B) An HRCT scan obtained at the lung base shows severe bronchiectasis in the right middle lobe. Multiple small nodules and branching centrilobular nodules form a tree-in-bud pattern in the right lower lobe.
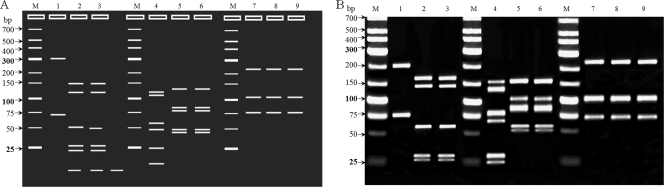
To diagnose the etiological agent, bacteria grown in the MGIT 960 culture system were initially propagated in 7H9 broth (Difco Laboratories, Detroit, MI) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC; Becton Dickinson) for 7 days at 37°C and genomic DNA was extracted from cultured bacteria. Initial species identification using a reverse line blot hybridization assay (REBA Myco-ID; M&D, Inc., Wonju, South Korea) was unsuccessful. Because this assay was designed to detect and identify Mycobacterium tuberculosis and 19 species of nontuberculous mycobacteria (NTM) (7), we considered our samples to be outside this detection range. We conducted a PCR-restriction fragment length polymorphism analysis (PRA) of the hsp65 and rpoB genes (Table 1) which has been used effectively for the simultaneous identification of many mycobacterial species (4, 8, 11). Digestion of the hsp65 gene at 37°C with MspI (New England BioLabs, MA) produced a 527-bp segment of the amplified PCR product, displaying a restriction pattern of 250-, 105-, and 75-bp DNA fragments (Fig. 2). This restriction pattern was identical to previously reported patterns of Segniliparus rotundus and S. rugosus (1). To confirm these results, reference strains of S. rotundus CIP 108380T and S. rugosus CIP 108378T (Institut Pasteur, Paris, France) were included in subsequent experiments. PRA of the hsp65 gene found no difference between a clinical isolate and the two reference strains. PRA of the rpoB gene was performed using MspI and HaeIII to further distinguish the two species. These restriction enzymes were selected from the rpoB genes of S. rotundus CIP 108380T (10) and S. rugosus (GenBank accession no. GL622756). The rpoB PRA using MspI and HaeIII restriction showed a unique digestion pattern for S. rotundus that distinguished it from S. rugosus and Mycobacterium spp. (Fig. 2) (11). Furthermore, the rpoB PRA of the clinical isolate using these two enzymes displayed a pattern identical to that of S. rotundus CIP 108380T.
Table 1.
Genes and oligonucleotide primers used and percent similarity in sequencing analysis with Segniliparus rotundus reference strain (GenBank accession no. CP001958)
| Gene | Primer sequence (5′–3′)c | Product size (bp) | Similarity (%) | Reference |
|---|---|---|---|---|
| hsp65 | (F) GAG GGC GTC ATC ACC GTC GAG G | 527 | NAd | 4 |
| (R) CGG CGA TGG CGT CGG AGT CAC C | ||||
| rpoBa | (F) TCA AGG AGA AGC GCT ACG A | 360 | 99 | 7 |
| (R) GGA TGT TGA TCA GGG TCT GC | ||||
| 16S–23S ITSb | (F) TTGTACACACCGCCCGTCA | 487 | 100 | 10 |
| (R) TCTCGATGCCAAGGCATCCACC | ||||
| 16S rRNA | (F) GAGTTTGATCCTGGCTCAGGA | 560 | 100 | 9 |
| (R) AAGGAGGTGATCCAGCCGCA |
The gene used in both molecular methods of PRA and sequence analysis.
16S–23S rRNA internal transcribed spacer of mycobacteria.
F, forward; R, reverse.
NA, not applicable, due to no information.
Fig. 2.
PCR restriction-enzyme polymorphism analysis (PRA). (A) Simulation of PRA of the hsp65 and rpoB genes. (B) PRA electrophoresis results. M, size marker; lanes 1 to 3, hsp65 amplicons digested by MspI from S. rugosus CIP 108378T, S. rotundus CIP 108380T, and the clinical isolate of this case, respectively; lines 4 to 6, rpoB amplicons digested by MspI from S. rugosus CIP 108378T, S. rotundus CIP 108380T, and the clinical isolate of this case, respectively; lines 7 to 9, rpoB amplicons digested by HaeIII from S. rugosus CIP 108378T, S. rotundus CIP 108380T, and the clinical isolate of this case, respectively.
To confirm the accuracy of this identification, sequencing analyses of rpoB (7), the 16S-23S rRNA internal transcribed spacer (ITS) sequence (5), and 16S rRNA were performed (9). The rpoB sequences showed 99% similarity, and the 16S-23S rRNA ITS and 16S rRNA sequences showed 100% similarity to that of the S. rotundus reference strain (GenBank accession no. CP001958 and FJ468343) (10).
Drug susceptibility testing was performed at the Korean Institute of Tuberculosis by using a broth microdilution method in the same manner as used for rapidly growing mycobacteria (RGM), according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (3, 12). MICs were determined after incubation at 30°C for 3 days. The Segniliparus species reference strains and the clinical isolate were resistant to amikacin, sulfamethoxazole, tobramycin, and ethambutol. The clinical isolate was more resistant than S. rotundus CIP 108380T to cefoxitin, doxycycline, and imipenem. The clinical isolate and S. rotundus CIP 108380T were susceptible to clarithromycin, ciprofloxacin, and moxifloxacin (Table 2).
Table 2.
Antimicrobial susceptibility patterns for S. rotundus and both reference strains of Segniliparus species
| Antibiotic | MIC (μg/ml)a |
||
|---|---|---|---|
| Clinical isolate of S. rotundus identified in this report | S. rotundus CIP 108380T | S. rugosus CIP 108378T | |
| Amikacin | >128 | >128 | >128 |
| Cefoxitin | >256 | 2 | 8 |
| Ciprofloxacin | 1 | ≤0.125 | >16 |
| Clarithromycin | 2 | 4 | 64 |
| Doxycycline | >32 | 2 | >32 |
| Imipenem | 16 | ≤0.5 | 4 |
| Moxifloxacin | 1 | ≤0.125 | ≤0.125 |
| Rifampin | >16 | ≤0.125 | >16 |
| Sulfamethoxazole | >128 | 16 | >128 |
| Tobramycin | >32 | >32 | >32 |
| Ethambutol | >32 | >32 | >32 |
MICs were interpreted using criteria for mycobacteria and aerobic actinomycetes.
The patient was treated with oral clarithromycin (1,000 mg/day) and ciprofloxacin (1,000 mg/day) for 2 months. The treatment outcome was favorable; the patient's symptoms resolved completely, radiographic findings improved, and sputum smear and culture were negative after 2 months of antibiotic therapy.
Segniliparus is a novel genus consisting of two species (S. rotundus and S. rugosus) that was first named in 2005 (1). Both reference strains of Segniliparus were isolated from human sputum (1, 10), and the characterization study demonstrated that they shared some phenotypical characteristics with rapidly growing mycobacteria (1).
In this article, we have described a case of S. rotundus pneumonia in a patient with non-cystic fibrosis bronchiectasis. To our knowledge, this is the first reported case of S. rotundus pneumonia. S. rugosus in patients with cystic fibrosis in the United States and Australia has recently been reported (2, 6). These cases suggest that Segniliparus species may be emerging respiratory pathogens that can cause pneumonia in patients with bronchiectasis.
The accurate identification of bacteria is important for the evaluation of the clinical implications of emerging pathogens in respiratory infections. However, Segniliparaceae may be confused with nonchromogenic rapidly growing mycobacteria in microscopic examination, due to the acid-fast staining properties of these species. In our case, molecular diagnostic methods allowed the accurate identification of S. rotundus and its differentiation from mycobacteria and S. rugosus, especially in the sequencing analyses of 16S rRNA genes and rpoB PRA using MspI and HaeIII.
Few studies of S. rotundus infection have been published, and reliable information about antibiotic therapy is limited. The S. rotundus reference strain and isolate were susceptible to several oral antibiotics, including clarithromycin, ciprofloxacin, and moxifloxacin. In contrast, the S. rugosus reference strain was highly resistant to these antibiotics. These results were consistent with those of previous studies (2, 6). Our patient showed symptomatic, radiographic, and microbiological improvements after 2 months of treatment with clarithromycin and ciprofloxacin. Further studies should seek to establish the optimal antibiotic therapy for S. rotundus infection.
Clinicians and physicians should be aware that AFB bacteria other than Mycobacterium spp. in respiratory infections may be S. rotundus, and further studies should investigate the significance and clinical importance of Segniliparus species.
Acknowledgments
This work was supported by National Research Foundation of Korea (NRF) grant no. 2011-0006228 funded by the Korean government (MEST).
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Butler W. R., et al. 2005. Novel mycolic acid-containing bacteria in the family Segniliparaceae fam. nov., including the genus Segniliparus gen. nov., with descriptions of Segniliparus rotundus sp. nov. and Segniliparus rugosus sp. nov. Int. J. Syst. Evol. Microbiol. 55:1615–1624 [DOI] [PubMed] [Google Scholar]
- 2. Butler W. R., et al. 2007. First isolations of Segniliparus rugosus from patients with cystic fibrosis. J. Clin. Microbiol. 45:3449–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. CLSI 2011. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes. M24-A2. CLSI, Wayne, PA: [PubMed] [Google Scholar]
- 4. Devallois A., Goh K. S., Rastogi N. 1997. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J. Clin. Microbiol. 35:2969–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frothingham R., Wilson K. H. 1993. Sequence-based differentiation of strains in the Mycobacterium avium complex. J. Bacteriol. 175:2818–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hansen T., et al. 2009. Segniliparus rugosus infection, Australia. Emerg. Infect. Dis. 15:611–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee H., Bang H. E., Bai G. H., Cho S. N. 2003. Novel polymorphic region of the rpoB gene containing Mycobacterium species-specific sequences and its use in identification of mycobacteria. J. Clin. Microbiol. 41:2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee H., Park H. J., Cho S. N., Bai G. H., Kim S. J. 2000. Species identification of mycobacteria by PCR-restriction fragment length polymorphism of the rpoB gene. J. Clin. Microbiol. 38:2966–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ninet B., et al. 1996. Two different 16S rRNA genes in a mycobacterial strain. J. Clin. Microbiol. 34:2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sikorski J., et al. 2010. Complete genome sequence of Segniliparus rotundus type strain (CDC 1076). Stand. Genomic Sci. 2:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whang J., et al. 2011. PCR-restriction fragment length polymorphism of the rpoB gene for identification of Mycobacterium avium subsp. paratuberculosis and differentiation of Mycobacterium avium subspecies. Diagn. Microbiol. Infect. Dis. 70:65–71 [DOI] [PubMed] [Google Scholar]
- 12. Woods G. L. 2000. Susceptibility testing for mycobacteria. Clin. Infect. Dis. 31:1209–1215 [DOI] [PubMed] [Google Scholar]