Abstract
Hepatitis C virus (HCV) RNA measurement has been facilitated by the introduction of real-time PCR-based assays with low limits of detection and broad dynamic ranges for quantification. In the present study, the performance of two second-version prototypes of the Cobas AmpliPrep/Cobas TaqMan HCV Quantitative Test (CAP/CTM v2) with decreased sample input volume and improved genotype inclusivity was investigated. A total of 232 serum and plasma samples derived from patients with chronic hepatitis C (genotype 1 [GT1], n = 108; GT2, n = 8; GT3, n = 24; GT4, n = 87; GT5, n = 3; and GT6, n = 2) were processed in parallel with the Cobas AmpliPrep/Cobas TaqMan HCV Test (CAP/CTM), Cobas Amplicor HCV Monitor Test v2.0 (CAM), and two second-version prototype formulations of CAP/CTM, Mastermix 1 (MMx1) and MMx2. In addition, three GT4 transcripts containing rare variant sequences were tested. The mean log10 HCV RNA differences for the best-performing CAP/CTM v2/MMx2 formulation in comparison to CAM were −0.05, 0.05, −0.12, −0.10, −0.44, and −0.29 for patients with GT1, GT2, GT3, GT4, GT5, and GT6 infections, respectively. GT1, GT2, and GT4 samples including isolates with known variants within the 5′ untranslated region (G145A, A165T) that were underquantified with CAP/CTM were correctly quantified with the second-version prototype. In addition, CAP/CTM v2 was able to accurately quantify the three transcripts with rare variant sequences. In conclusion, CAP/CTM v2 accurately quantifies HCV RNA across all HCV genotypes, including specimens with rare polymorphisms previously associated with underquantification.
INTRODUCTION
Chronic hepatitis C affects up to 170 million people worldwide, and more than 350,000 people die from hepatitis C virus (HCV)-related liver diseases each year (23). However, with the current standard of care, a combination of pegylated interferon alfa plus ribavirin, HCV can be permanently eradicated in ca. 50% of patients (6, 11).
The study of HCV replication and of its kinetics during antiviral therapy with pegylated interferon and ribavirin has led to the identification of distinct response patterns that serve to individualize treatment durations. Response-guided therapy durations are recommended by national and international treatment guidelines (5, 15). However, these require the use of HCV RNA assays that allow for precise measurement and monitoring of viral kinetics. Real-time PCR has increasingly gained in importance for the detection and quantification of HCV RNA because of its higher sensitivity and broader dynamic range compared to conventional, endpoint PCR (4, 19). Currently, there are two real-time PCR-based assays commercially available: the Cobas AmpliPrep/Cobas TaqMan HCV Test (CAP/CTM; Roche Molecular Systems, Pleasanton, CA) and the Abbott RealTime HCV Test (Abbott Molecular, Des Plaines, IL).
The TaqMan-based CAP/CTM test has been successfully used in clinical practice and in drug development trials and has been approved by European and U.S. authorities for the monitoring of anti-HCV therapy. However, CAP/CTM has been reported to occasionally underquantify certain genotype 4 (GT4) samples, as well as isolates of other HCV genotypes that may include nucleotide polymorphisms at the hybridization sites for the PCR primers and/or TaqMan probe in the 5′ untranslated region (5′UTR) of the HCV genome (1–3, 8, 10, 16, 17, 20, 22). This has prompted the development of an upgraded, second-generation assay that could provide an improved genotype inclusivity and workflow.
In the present study, we evaluated the genotype-specific performance of two formulations (Mastermix 1 [MMx1] and MMx2) of a second-version prototype of the Cobas AmpliPrep/Cobas TaqMan HCV Quantitative Test (CAP/CTM v2) in comparison to the already licensed CAP/CTM assay and the Cobas Amplicor HCV Monitor Test v2.0 (CAM; Roche Molecular Systems), the latter serving as our reference method.
MATERIALS AND METHODS
Clinical specimens and genotyping.
A total of 232 leftover clinical specimens, 121 serum samples, and 111 EDTA plasma samples from patients with chronic HCV infection were obtained at the J. W. Goethe University Hospital, Frankfurt, Germany, the German University in Cairo, Cairo, Egypt, and the Bioscientia Institut für Medizinische Diagnostik, Ingelheim, Germany. The ethics committee of the J. W. Goethe University Hospital approved the study. All samples were frozen immediately at −40 to −80°C and shipped to the testing site in Frankfurt. A nonblinded operator performed all testing at the J. W. Goethe University Hospital.
Due to the large volume required for testing with multiple assays, all of the clinical specimens were diluted with pools of HCV RNA-negative human serum or EDTA plasma at a concentration of 1:12 prior to testing. HCV genotypes and subtypes of each sample were determined by sequencing of the 5′UTR region of the HCV genome.
Technical system features.
The Cobas AmpliPrep/Cobas TaqMan HCV Test (CAP/CTM; Roche Molecular Systems) is based on reverse transcription-PCR technology. The system combines automated sample extraction (required sample input volume, 1 ml) with automated amplification and detection on the Cobas TaqMan HCV instrument. Reverse transcription and amplification primers, as well as the TaqMan probe, are targeted to the highly conserved 5′UTR. The assay procedure has been described previously (16).
The second-version prototypes, MMx1 and MMx2, differ from the present assay by requiring a smaller sample input volume (0.65 ml instead of 1 ml). In addition, a redesigned primer set was used. Finally, in light of known underestimation of HCV RNA levels, particularly in GT4 samples, a second TaqMan hydrolysis probe was introduced to the MMx2 prototype assay for better overall genotype coverage and the dedicated AmpliLink software was upgraded accordingly.
The Cobas Amplicor HCV Monitor Test v2.0 (CAM; Roche Molecular Systems) is a semiautomated assay, with manual sample preparation and automated amplification and detection on the Cobas Amplicor analyzer. The procedure has been described previously (7).
In the present study, all testing procedures were performed at one testing site, according to the manufacturer's instructions as described in the respective package inserts.
Analytical assay performance.
The limit of detection (LOD) of the prototype CAP/CTM v2/MMx2 assay was determined in serum and plasma using panels of the second WHO International Standard for HCV RNA (NIBSC code 96/798, 50,000 IU/vial). The standard was serially diluted to the following concentration levels: 30, 15, 7.5, 3, and 0 IU/ml.
High titer clinical specimens, including one serum and one plasma GT1 specimen (both with >106 IU/ml HCV RNA) and a high-titer GT1 armored HCV RNA control (3.08 × 1011 IU/ml HCV RNA) were used to assess the lower limit of quantification and linearity over the dynamic range. Serial dilutions to the following concentrations were produced for the clinical specimens to cover the expected range of the assay: 3.0 × 105, 3.0 × 104, 3.0 × 103, 300, 30, 15, and 7.5 IU/ml HCV RNA. The armored HCV RNA sample was diluted to the following concentrations: 2.0 × 108, 1.0 × 108, 3.0 × 107, 3.0 × 106, and 3.0 × 104 IU/ml HCV RNA. Linearity was evaluated by polynomial regression analysis in accordance with the Clinical and Laboratory Standards Institute guideline EP6-A (21).
HCV genotype inclusivity.
The agreement of CAP/CTM and CAP/CTM v2 with the reference assay CAM for the quantification of HCV RNA was assessed using clinical specimens harboring HCV genotypes 1 through 6. All titer values were log10 transformed, and the mean log titer values of each genotype were compared to the corresponding values obtained from the other tests.
Results were classified as moderately or severely under- or overquantified if they differed by 0.5 to 1.0 log10 IU/ml HCV RNA or >1.0 log10 IU under- or over, respectively, compared to the reference assay. For sequence analyses, amplification of the HCV 5′ noncoding region was performed by a nested reverse transcription-PCR to assess the frequency of mutations at positions A165, G145, and A142.
GT4 transcripts.
Genotype inclusivity was also assessed using synthetic transcripts harboring rare GT4 variants. In brief, three transcripts representing the complete 5′UTR of GT4 patient samples with known polymorphisms in comparison with a GT1 sample were investigated. The transcripts, containing substitutions A165T (transcript 1), A165T + G145A (transcript 2), and A165T + G145A + A142G (transcript 3), were obtained from Trenzyme (Constance, Germany) and from Roche Molecular Diagnostics. The RNA concentration of all transcripts was determined using the RiboGreen RNA Assay (Invitrogen, Carlsbad, CA). Readings were converted into copies/ml (cp/ml) and subsequently diluted in PCR-grade water to the desired concentrations. Aliquots were stored at −15°C to −25°C. All samples were thawed and refrozen the same number of times. On the day of testing, the transcripts were spiked into the lysis buffer included in the respective kits at a concentration level of 10,000 cp/ml and tested with CAP/CTM and the two prototype formulations, MMx1 and MMx2. The samples were taken through the entire process on the CAP/CTM system, including sample preparation, amplification, and detection.
Data analysis.
Probit analysis was performed to determine the LOD of the prototype CAP/CTM v2/MMx2 assay. The LOD was determined as the 95% probability of obtaining a positive HCV RNA result. Bland-Altman plot analysis was performed to assess the agreement level between each assay for the quantification of HCV RNA. In addition, Deming regression analyses were performed.
RESULTS
Analytical performance.
The LOD of the prototype CAP/CTM v2/MMx2 was determined in five different concentration levels of the second WHO International Standard for HCV RNA. Analytical sensitivities of 13.6 and 12.8 IU/ml for serum and EDTA plasma, respectively, were calculated by probit analysis (95% hit rate). In addition, three replicates each of high-titer specimens and cloned HCV RNA controls were used to determine a dynamic range from 15 IU/ml extending up to 1.0 × 108 IU/ml for serum and EDTA plasma, respectively. Standard deviations for these measurements varied from 0.46 to 0.05 log10 IU/ml across the dynamic range.
Genotype inclusivity.
A total of 232 clinical samples were analyzed in parallel by CAP/CTM, the two prototype CAP/CTM v2/MMx1 and v2/MMx2 assays, and CAM. These specimens included 108 samples with HCV GT1 (mean viral load assessed with CAM, 4.59 log10 IU of HCV RNA/ml; range, 690 to 650,000 IU/ml), 8 samples with HCV GT2 (mean, 4.91 log10 IU/ml; range, 1,000 to 510,000 IU/ml), 24 samples with HCV GT3 (mean, 4.93 log10 IU/ml; range, 2,300 to 520,000 IU/ml), 87 samples with HCV GT4 (mean, 4.48 log10 IU/ml; range, 610 to 660,000 IU/ml), and 3 and 2 samples with HCV GT5 (mean, 5.29 log10 IU/ml; range, 51,000 to 680,000 IU/ml) and HCV GT6 (mean, 4.94 log10 IU/ml; range, 61,000 to 120,000 IU/ml), respectively.
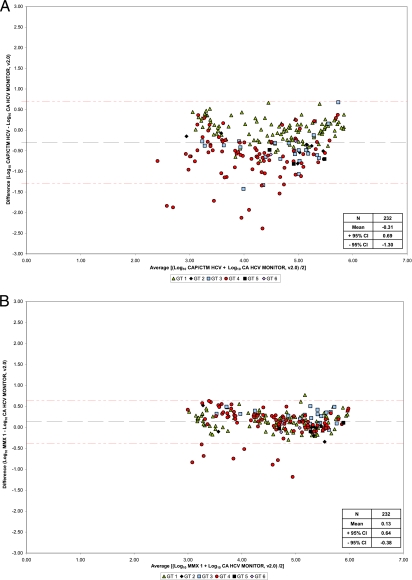
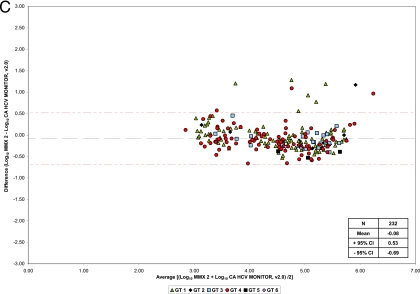
Bland-Altman analysis showed that mean differences between CAP/CTM, CAP/CTM v2/MMx1, CAP/CTM v2/MMx2, and the reference assay were −0.31, 0.13, and −0.08 log10 IU/ml HCV RNA, respectively (Fig. 1a A to C). No differences between serum and plasma samples were observed. Deming regression analysis yielded R2 regression coefficients of 0.68, 0.90, and 0.90 for CAP/CTM, CAP/CTM v2/MMx1, and CAP/CTM v2/MMx2 versus CAM, respectively (Fig. 2 A to C). Mean differences in log10 IU/ml HCV RNA for individual genotypes are shown in Table 1. When the prototype CAP/CTM v2/MMx2 and CAP/CTM were directly compared, mean log10 differences of 0.23 and 0.53 IU/ml were determined for all genotypes and GT4 specimens, respectively.
Fig. 1.
Bland-Altman analysis of CAP/CTM versus CAM (A), prototype CAP/CTM v2/MMx1 versus CAM (B), and prototype CAP/CTM v2/MMx2 versus CAM (C).
Fig. 2.
Regression analysis of CAP/CTM versus CAM (A), prototype CAP/CTM v2/MMx1 versus CAM (B), and prototype CAP/CTM v2/MMx2 versus CAM (C).
Table 1.
Performance comparison of CAM, CAP/CTM, and the prototype CAP/CTM v2/MMx1 and v2/MMx2 assays for the determination of HCV RNA concentrations in patients infected with genotypes 1 to 6
| GT | No. of samples | CAM (reference assay) |
CAP/CTM |
CAP/CTM v2/MMx1 |
CAP/CTM v2/MMx2 |
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean IU/ml | Mean log10IU/ml | Mean log10 | Difference to CAM | Mean log10 | Difference to CAM | Mean log10 | Difference to CAM | ||
| GT1 | 108 | 3.9 × 104 | 4.59 | 4.60 | 0.01 | 4.73 | 0.14 | 4.54 | -0.05 |
| GT2 | 8 | 8.1 × 104 | 4.91 | 4.49 | -0.42 | 4.89 | -0.02 | 4.96 | 0.05 |
| GT3 | 24 | 8.4 × 104 | 4.93 | 4.46 | -0.47 | 5.18 | 0.25 | 4.81 | -0.12 |
| GT4 | 87 | 3.0 × 104 | 4.48 | 3.85 | -0.63 | 4.59 | 0.11 | 4.38 | -0.10 |
| GT5 | 3 | 1.9 × 104 | 5.29 | 4.62 | -0.67 | 5.28 | -0.01 | 4.85 | -0.44 |
| GT6 | 2 | 8.6 × 104 | 4.94 | 4.51 | -0.43 | 4.87 | -0.07 | 4.65 | -0.29 |
In the present study, a reduction of moderately and severely under-quantified samples was observed with prototype CAP/CTM v2/MMx1 (n = 8) and prototype CAP/CTM v2/MMx2 (n = 8) compared to the CAP/CTM test (n = 73) (Table 2).
Table 2.
Overestimation and underestimation of HCV RNA by CAP/CTM and the prototype CAP/CTM v2/MMx1 and v2/MMx2 assays in comparison to CAMa
| GT (no. of samples) | Discrepancy | No. of discrepant samples |
||
|---|---|---|---|---|
| CAP/CTM | CAP/CTM v2/MMx1 | CAP/CTM v2/MMx2 | ||
| GT1 (108) | Moderate | 6 (3+/3−) | 3+ | 4 (3+/1−) |
| Severe | 0 | 0 | 3+ | |
| GT2 (8) | Moderate | 4− | 1+ | 0 |
| Severe | 0 | 0 | 1+ | |
| GT3 (24) | Moderate | 9 (1+/8−) | 1+ | 0 |
| Severe | 3− | 0 | 0 | |
| GT4 (87) | Moderate | 35− | 13 (6+/7−) | 8 (2+/6−) |
| Severe | 17− | 1− | 1+ | |
| GT5 (3) | Moderate | 2− | 0 | 1− |
| Severe | 0 | 0 | 0 | |
| GT6 (2) | Moderate | 1− | 0 | 0 |
| Severe | 0 | 0 | 0 | |
Discrepancies are classified as follows: moderate, titer value differences between ±0.5 log10 and ±1.0 log10 IU/ml; severe, titer value differences >1.0 log10IU/ml; +, overquantification compared to the reference assay.
Assessment of HCV isolates with sequence mismatches.
In 87 GT4 samples, sequencing of the HCV 5′ noncoding region revealed one sequence with a G-to-A substitution at position 145 and 7 sequences that contained an A-to-T substitution at nucleotide 165. Overall, a mean underquantification of 1.8 log10 IU/ml HCV RNA by CAP/CTM compared to CAM was observed in these eight isolates. In contrast, the mean difference between CAP/CTM v2/MMx2 and CAM for the same eight isolates was only −0.3 log10 IU/ml. One sample with an A165T substitution was not quantifiable by CAP/CTM, whereas CAP/CTM v2/MMx2 yielded a quantifiable viral load (161 IU/ml).
Interestingly, the G145A substitution was also observed in two non-GT4 samples (one GT1 and one GT2 sample). Comparable quantification of HCV RNA was observed in the GT1 sample. However, HCV RNA could not be detected by CAP/CTM in the GT2 sample that also contained an A142G substitution, whereas high levels of HCV RNA were detected with the prototype CAP/CTM v2/MMx2 and CAM (5.1 and 4.6 log10 IU/ml HCV RNA, respectively).
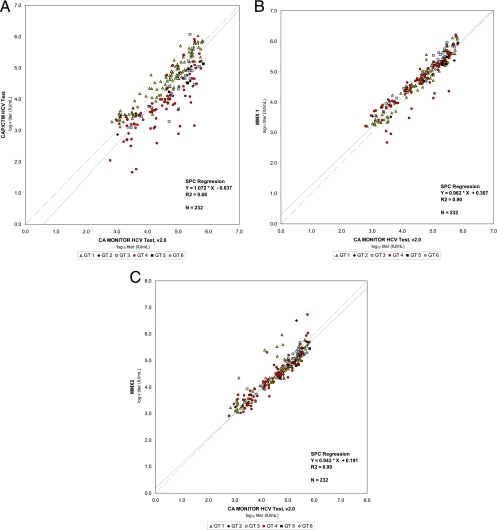
To further investigate the ability of the prototype CAP/CTM v2 assay to accurately quantify GT4 specimens harboring the described amino acid substitutions, three GT4 transcripts, as well as one GT1 transcript without mismatches, were tested in parallel by CAP/CTM and the prototype CAP/CTM v2/MMx1 and v2/MMx2 assays, respectively. The sample input was 10,000 cp/ml (4.0 log10 cp/ml) for all tested transcripts. Although quantification of the GT1 transcript was similar across the three assays, CAP/CTM could not detect two of three GT4 transcripts. In addition, the only GT4 transcript detected by CAP/CTM (transcript 1, containing the A165T substitution) was severely underquantified (>2 log10 cp/ml). CAP/CTM v2/MMx1 was readily able to detect all three GT4 transcripts. However, all three transcripts were severely underquantified, especially transcript 3 that contained three different amino acid substitutions (A165T + G145A + A142G). Finally, all three GT4 transcripts were readily and properly quantified by the prototype CAP/CTM v2/MMx2 assay, returning results close to the expected concentration of 4.0 log10 cp/ml (Fig. 3).
Fig. 3.
Detection and quantification of one GT1 transcript and three GT4 transcripts by CAP/CTM and the prototype CAP/CTM v2/MMx1 and v2/MMx2 assays. GT4 transcripts 1, 2, and 3 contain the following amino acid substitution(s): transcript 1, A165T; transcript 2, A165T + G145A; and transcript 3, A165T + G145A + A142G.
DISCUSSION
HCV RNA viral load monitoring plays a key role in the management of response-guided therapy in patients with chronic hepatitis C (18). This calls for a sensitive and accurate HCV RNA assay that offers both a low limit of detection and linear quantification over a broad dynamic range.
The CAP/CTM assay is widely used in clinical research and practice, and it has been evaluated in a number of studies (2, 12, 14, 16, 17, 20, 22). Among these, several report underestimation of HCV RNA levels in up to 30% of GT4 specimens compared to competitor assays (2, 16, 17, 20, 22). Recently, the hypothesis was established that these findings were linked to the occurrence of GT4 specimens harboring amino acid substitutions at positions 145 (G to A) and 165 (A to T) in the 5′UTR of the HCV genome (1, 3, 8, 10).
In the present study, we evaluated two formulations of a second-generation prototype real-time PCR-based assay, CAP/CTM v2/MMx1 and CAP/CTM v2/MMx2. Both formulations showed improved genotype inclusivity and tolerance for polymorphisms in GT4 isolates. While these polymorphisms are generally considered rare, a GenBank search revealed that the A substitution at position 145 was seen in 0.4%, and a T at position 165 was seen in 8.1% of patients with HCV GT4 (3). Indeed, within 87 HCV GT4 patient samples included in the present study, sequence analyses of the 5′UTR revealed a G-to-A-substitution at position 145 in one patient and an A-to-T substitution at position 165 in 7 patients. In line with previous findings on quantification of isolates with sequence mismatches, a mean underquantification of almost 2 log10 IU/ml was observed in these patient isolates when measured with CAP/CTM. However, these discrepancies were overcome when the prototype CAP/CTM v2/MMx2 assay was used (mean difference from the reference assay, −0.3 log10 IU/ml; mean difference in all genotype four samples, −0.10 log10 IU/ml). In addition, the improved prototype CAP/CTM v2/MMx2 assay properly quantified synthetic HCV RNA transcripts corresponding to GT1 and GT4, the latter engineered to express amino acid substitutions at positions 145, 165, and 142 of the 5′UTR.
Furthermore, the study demonstrated the ability of the prototype CAP/CTM v2/MMx1 and v2/MMx2 formulations to detect and quantify HCV RNA in a large collection of clinical samples representing GT1 to GT6 with a robust and linear correlation to CAM that we used as the reference assay. Bland-Altman analysis showed that the improved assay confirmed the overall good performance of CAP/CTM for the quantification of samples with HCV GT1 to GT3 and GT5 and GT6 based on three and two samples, respectively, while significantly improving GT4-specific performance. Since the best results for quantification of clinical specimens and GT4 transcripts were obtained with the MMx2-prototype, this formulation was selected for further development. A subsequent larger performance characteristics study is already under way.
The prototype CAP/CTM v2/MMx2 assay includes re-engineered TaqMan hydrolysis probes that bind within the same 5′UTR target region. This should ensure coverage of current and future HCV sequence variants, an aspect that remains crucial for expanding clinical applications of viral load monitoring in light of the changing face of anti-HCV therapeutic regimens in the years to come. Despite a general effort toward test harmonization and standardization, differences among assays are not likely to be completely overcome, and it will still be recommended to use the same test when assessing viral kinetics for response guided therapy in a single patient (5, 9, 13, 15).
In conclusion, our data indicate that a second-generation prototype of the CAP/CTM assay with MMx2 has improved genotype inclusivity, including GT4 samples that contain variants previously shown to be not detected or underestimated by the current CAP/CTM version. The assay's new formulation makes it better suited to detect evolving populations of HCV variants and may support the expanding applications of viral load monitoring in the clinical management of chronic hepatitis C.
ACKNOWLEDGMENTS
G.C., D.T., and H.Z. are employees of Roche Molecular Diagnostics. P.G. received a research grant from Roche Molecular Diagnostics. C.S. has been a consultant for and has received research grants from Roche Molecular Diagnostics. The remaining authors have no potential conflicts of interest to declare.
This study was funded by Roche Molecular Diagnostics, Rotkreuz, Switzerland.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Akhavan S., Ronsin C., Laperche S., Thibault V. 2011. Genotype 4 hepatitis C virus: beware of false-negative RNA detection. Hepatology 53: 1066–1067 [DOI] [PubMed] [Google Scholar]
- 2. Chevaliez S., Bouvier-Alias M., Brillet R., Pawlotsky J. M. 2007. Overestimation and underestimation of hepatitis C virus RNA levels in a widely used real-time PCR-based method. Hepatology 46: 22–31 [DOI] [PubMed] [Google Scholar]
- 3. Chevaliez S., Bouvier-Alias M., Castera L., Pawlotsky J. M. 2009. The Cobas AmpliPrep-Cobas TaqMan real-time PCR assay fails to detect hepatitis C virus RNA in highly viremic genotype 4 clinical samples. Hepatology 49: 1397–1398 [DOI] [PubMed] [Google Scholar]
- 4. Chevaliez S., Pawlotsky J. M. 2009. How to use virological tools for optimal management of chronic hepatitis C. Liver Int. 29 (Suppl. 1): 9–14 [DOI] [PubMed] [Google Scholar]
- 5. Craxi A. 2011. EASL clinical practice guidelines: management of hepatitis C virus infection. J. Hepatol. 55:245–264 [DOI] [PubMed] [Google Scholar]
- 6. Fried M. W., et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347: 975–982 [DOI] [PubMed] [Google Scholar]
- 7. Gerken G., et al. 2000. Performance of the COBAS Amplicor HCV Monitor test, version 2.0, an automated reverse transcription-PCR quantitative system for hepatitis C virus load determination. J. Clin. Microbiol. 38: 2210–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Germer J. J., Bommersbach C. E., Schmidt D. M., Bendel J. L., Yao J. D. 2009. Quantification of genotype 4 hepatitis C virus RNA by the COBAS AmpliPrep/COBAS TaqMan hepatitis C virus test. Hepatology 50: 1679–1680 [DOI] [PubMed] [Google Scholar]
- 9. Ghany M. G., Strader D. B., Thomas D. L., Seeff L. B. 2009. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49: 1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Halfon P., Martinot-Peignoux M., Khiri H., Marcellin P. 2010. Quantification of genotype 4 serum samples: impact of hepatitis C virus genetic variability. Hepatology 52: 401. [DOI] [PubMed] [Google Scholar]
- 11. Manns M. P., et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358: 958–965 [DOI] [PubMed] [Google Scholar]
- 12. Martinot-Peignoux M., et al. 2009. Clinical performances of two real-time PCR assays and bDNA/TMA to early monitor treatment outcome in patients with chronic hepatitis C. J. Clin. Virol. 46: 216–221 [DOI] [PubMed] [Google Scholar]
- 13. McCaughan G. W., et al. 2007. Asian Pacific Association for the Study of the Liver consensus statements on the diagnosis, management and treatment of hepatitis C virus infection. J. Gastroenterol. Hepatol. 22: 615–633 [DOI] [PubMed] [Google Scholar]
- 14. Pittaluga F., et al. 2008. Clinical evaluation of the COBAS AmpliPrep/COBAS TaqMan for HCV RNA quantitation in comparison with the branched-DNA assay. J. Med. Virol. 80: 254–260 [DOI] [PubMed] [Google Scholar]
- 15. Sarrazin C., et al. 2010. Prophylaxis, diagnosis and therapy of hepatitis C virus (HCV) infection: the German guidelines on the management of HCV infection. Z. Gastroenterol. 48: 289–351(In German.) [DOI] [PubMed] [Google Scholar]
- 16. Sarrazin C., et al. 2008. Evaluation of an automated, highly sensitive, real-time PCR-based assay (COBAS AmpliPrep/COBAS TaqMan) for quantification of HCV RNA. J. Clin. Virol. 43: 162–168 [DOI] [PubMed] [Google Scholar]
- 17. Sarrazin C., et al. 2006. Comparison of conventional PCR with real-time PCR and branched DNA-based assays for hepatitis C virus RNA quantification and clinical significance for genotypes 1 to 5. J. Clin. Microbiol. 44: 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sarrazin C., et al. 2010. Definition of rapid virologic response with a highly sensitive real-time PCR-based HCV RNA assay in peginterferon alfa-2a plus ribavirin response-guided therapy. J. Hepatol. 52: 832–838 [DOI] [PubMed] [Google Scholar]
- 19. Schutten M. 2008. Comparison of the Abbott RealTime HIV-1 and HCV viral load assays with commercial competitor assays. Expert Rev. Mol. Diagn. 8: 369–377 [DOI] [PubMed] [Google Scholar]
- 20. Sizmann D., et al. 2007. Fully automated quantification of hepatitis C virus (HCV) RNA in human plasma and human serum by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 38: 326–333 [DOI] [PubMed] [Google Scholar]
- 21. Tholen D. W., et al. 2003. Evaluation of the linearity of quantitative measurement procedures: a statistical approach: approved guideline. CLSI, Wayne, PA: http://www.clsi.org/source/orders/free/ep6-a.pdf [Google Scholar]
- 22. Vermehren J., et al. 2008. Differences between two real-time PCR-based hepatitis C virus (HCV) assays (RealTime HCV and Cobas AmpliPrep/Cobas TaqMan) and one signal amplification assay (Versant HCV RNA 3.0) for RNA detection and quantification. J. Clin. Microbiol. 46: 3880–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. World Health Organization 2009. Viral hepatitis: Report by the Secretariat. World Health Organization, Geneva, Switzerland: http://apps.who.int/gb/ebwha/pdf_files/EB126/B126_15-en.pdf [Google Scholar]