Abstract
We report here a PCR-based assay using a single primer pair targeting the RPL31 gene that allows discrimination between Candida glabrata, Candida bracarensis, and Candida nivariensis according to the size of the generated amplicon.
TEXT
Candida albicans remains the Candida species most commonly recovered from clinical specimens. However, the incidence of non-C. albicans Candida species has increased, notably Candida glabrata, which now ranks second among the etiologic agents of invasive candidiasis (7, 11, 13, 14, 16, 17).
Along with C. glabrata and belonging to the Nakaseomyces clade, two new species, Candida nivariensis and Candida bracarensis, have recently been reported to be emerging pathogens (1, 4, 6, 8, 12, 18). Differentiation of these species from C. glabrata is of major importance to understand their clinical and epidemiologic role in candidiasis.
Conventional routine identification using a combination of chromogenic media, rapid trehalose assimilation test, biochemical panels such as the ID32C (bioMérieux, Marcy l'Etoile, France) may fail to differentiate C. nivariensis and C. bracarensis from C. glabrata. Thus, to unambiguously identify these species, molecular methods, mainly sequencing of the internal transcribed spacer (ITS) or D1/D2 domain of the large subunit of the ribosomal DNA (rDNA) are required (1, 4, 5, 12). A multiplex PCR targeting the internal transcribed spacer 1 (ITS1) region and using specific forward primers for each of the three species has recently been proposed (15). A peptide nucleic acid fluorescence in situ hybridization method has been developed with specific probes for C. nivariensis and C. bracarensis (4). Overall, these methods, while reliable, are either time-consuming or have been evaluated on a rather low number of strains. In this work, we describe a PCR-based assay using a single primer pair to differentiate C. glabrata, C. nivariensis, and C. bracarensis with no need of sequencing.
We first performed an in silico search for highly conserved genes between C. glabrata and Saccharomyces cerevisiae that contain intron(s) on the Genolevures (http://www.genolevures.org/) and Saccharomyces Genome Database (http://downloads.yeastgenome.org/) (accessed 8 August 2010) websites. We postulated that conservation between Nakaseomyces species should be even higher than with S. cerevisiae and that because intron size is not being selected, it may differ between the considered species. We selected RPL31, a gene coding for a protein component of the large (60S) ribosomal subunit that fulfils our requirements. This gene exists in two copies in S. cerevisiae (RPL31A and RPL31B), but in a single copy in C. glabrata. A primer pair (RPL31cgF [cg stands for C. glabrata, and F stands for forward] [5′-GCCGGTTTGAAGGACGTTGTTACT-3′] and RPL31cgR [R stands for reverse] [5′-GAACAATGGGTTCTTGGCGT-3′]) was designed to amplify a fragment of 1,061 bp in C. glabrata (CBS 138). This pair, under the conditions described here, specifically amplifies RPL31B in S. cerevisiae and not RPL31A.
Seventy-five strains, including C. glabrata CBS 138, C. bracarensis CBS 10154, C. nivariensis CBS 9983 type strains, and closely related Nakaseomyces (syn. Kluyveromyces) delphensis CBS 2170 type strain (10), S. cerevisiae FY73, and C. albicans ATCC 90028 reference strains, and 68 clinical isolates (34 C. glabrata isolates, 26 C. nivariensis isolates, 5 C. bracarensis strains and one isolate of each C. parapsilosis, C. tropicalis, and C. krusei) were used in this study. The Candida isolates were obtained from different patients from three different hospitals in France. All C. glabrata, C. albicans, C. parapsilosis, C. tropicalis, C. krusei, and S. cerevisiae strains were phenotypically identified with the commercial ID32C strip. The C. bracarensis and C. nivariensis strains were identified by sequencing the ITS and the D1/D2 domain of the large subunit regions of rDNA by the method of White et al. (19) (data not shown). A monoclonal subculture of each was stored at −80°C before testing.
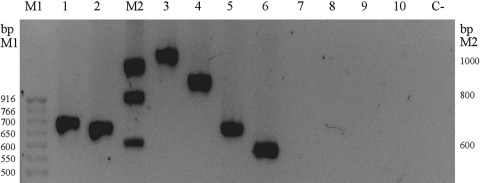
DNA was extracted using a rapid method with Chelex resin and heat shock (9). The PCR was done with 0.25 μg of DNA in a 50-μl reaction mixture volume containing 1× PCR buffer, 0.5 μM each primer (EuroGentec, Belgium), 0.25 mM each deoxynucleoside triphosphate (equimolar concentrations of dATP, dCTP, dGTP, and dTTP) (New England BioLabs, Evry, France) and 0.5 U of DreamTaq polymerase (Fermentas, Saint Rémy lès Chevreuse, France). A touch-down amplification program was performed as follows: 3 min at 95°C; 3 cycles, with 1 cycle consisting of 30 s at 95°C, 30 s at 62°C, and 30 s at 72°C; 3 cycles, with 1 cycle consisting of 30 s at 95°C, 30 s at 58°C, and 30 s at 72°C; 3 cycles, with 1 cycle consisting of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C; 28 cycles, with 1 cycle consisting of 30 s at 50°C and 30 s at 72°C; and a final extension step of 10 min at 72°C. The amplified products were separated by electrophoresis in an agarose gel (2% in Tris-borate-EDTA [TBE] buffer) stained with ethidium bromide (0.5 μg/ml) at 90 V for 2 h. Amplification products of C. nivariensis, C. bracarensis,. and N. delphensis type strains were directly sequenced and deposited under the accession numbers GenBank ID: JF690246, JF690247, and JN088223, respectively. Applied to our whole collection of strains, the method allowed DNA amplification for all C. glabrata, C. nivariensis, and C. bracarensis strains and the S. cerevisiae strain but not DNA amplification for C. albicans, C. parapsilosis, C. tropicalis, and C. krusei (Fig. 1).
Fig. 1.
Agarose gel electrophoresis of RPL31 PCR products. Lanes: M1, 50-bp ladder (New England BioLabs, Evry, France); M2, Smart ladder (Eurogentec, Liege, Belgium); 1, S. cerevisiae FY73; 2, C. nivariensis CBS 9983; 3, C. glabrata CBS 138, 4, C. bracarensis CBS 10154; 5, C. nivariensis CBS 9983; 6, N. delphensis CBS 2170; 7, C. albicans ATCC 90028; 8, C. parapsilosis KB10016244; 9, C. tropicalis KB10016966; 10, C. krusei KB10016563; and C−, control without a DNA template.
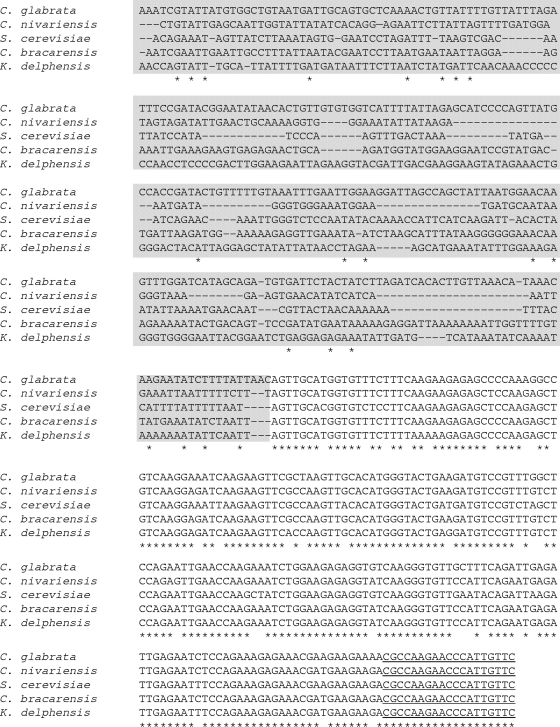
DNAs from C. albicans, C. parapsilosis, C. tropicalis, and C. krusei strains were checked by successful amplification using the ITS1 and ITS4 primer pairs (data not shown). Amplification products had expected sizes of 1,061 bp for C. glabrata and 691 bp for S. cerevisiae. All C. bracarensis and C. nivariensis strains and the N. delphensis CBS 2170 strain had an amplification product estimated at 900 bp, 670 bp, and 550 bp, respectively. These sizes were confirmed by directly sequencing amplicons from type strains (902 bp, 665 bp, and 591 bp). Indeed, alignment of homolog sequences shows that the differences in size are due to the intron (Fig. 2).
Fig. 2.
Multiple-sequence alignment of RPL31 orthologs in C. glabrata (CBS 138), C. nivarensis (CBS 9983), C. bracarensis (CBS 10154), N. delphensis (CBS 2170) and S. cerevisiae (FY73) (CLUSTALX 2.0.12). Primers RPL31cgF and RPL31cgR are underlined. The intronic portions of the genes are indicated by a gray background. Gaps introduced to maximize alignment are indicated by the dashes. Nucleotides that are identical in all five species are indicated by an asterisk below the sequences.
The recently described Candida species C. bracarensis and C. nivariensis are closely related to C. glabrata. Based on phenotypic characteristics, these species were initially misidentified as C. glabrata (2, 3). Molecular methods to differentiate the three species have been reported (1, 4, 5, 12, 15), but none used a simple PCR with a single pair of primers.
We report here a PCR-based assay using a single primer pair that allows us to distinguish species: 1,061 bp for C. glabrata, 902 bp for C. bracarensis, 665 bp for C. nivariensis, and 591 bp for N. delphensis. Experimental simplicity (a single primer pair, analysis on agarose gel electrophoresis) and the rapidity and accuracy of this method make it a powerful tool to both investigate collections of strains that possibly contain C. bracarensis and/or C. nivariensis strains and to confirm the identification of strains isolated from invasive infection. This should improve our knowledge on the respective epidemiology and pathogenic importance of these three species.
Nucleotide sequence accession numbers.
Amplification products of C. nivariensis, C. bracarensis, and N. delphensis type strains were sequenced and deposited in GenBank under accession numbers JF690246, JF690247, and JN088223, respectively.
Acknowledgments
This work was supported in part by grant “Attractivité” to C. Fairhead from the Université Paris Sud 11.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Alcoba-Florez J., et al. 2005. PCR protocol for specific identification of Candida nivariensis, a recently described pathogenic yeast. J. Clin. Microbiol. 43:6194–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alcoba-Florez J., et al. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 43:4107–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bishop J. A., Chase N., Lee R., Kurtzman C. P., Merz W. G. 2008. Production of white colonies on CHROMagar Candida medium by members of the Candida glabrata clade and other species with overlapping phenotypic traits. J. Clin. Microbiol. 46:3498–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bishop J. A., et al. 2008. Candida bracarensis detected among isolates of Candida glabrata by peptide nucleic acid fluorescence in situ hybridization: susceptibility data and documentation of presumed infection. J. Clin. Microbiol. 46:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borman A. M., et al. 2008. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J. Clin. Microbiol. 46:933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Correia A., Sampaio P., James S., Pais C. 2006. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int. J. Syst. Evol. Microbiol. 56:313–317 [DOI] [PubMed] [Google Scholar]
- 7. Fidel P. L., Jr., Vazquez J. A., Sobel J. D. 1999. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12:80–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujita S., et al. 2007. Catheter-related fungemia due to fluconazole-resistant Candida nivariensis. J. Clin. Microbiol. 45:3459–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hennequin C., et al. 1999. Identification of Fusarium species involved in human infections by 28S rRNA gene sequencing. J. Clin. Microbiol. 37:3586–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kurtzman C. P. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 4:233–245 [DOI] [PubMed] [Google Scholar]
- 11. Li L., Redding S., Dongari-Bagtzoglou A. 2007. Candida glabrata: an emerging oral opportunistic pathogen. J. Dent. Res. 86:204–215 [DOI] [PubMed] [Google Scholar]
- 12. Lockhart S. R., et al. 2009. Identification of Candida nivariensis and Candida bracarensis in a large global collection of Candida glabrata isolates: comparison to the literature. J. Clin. Microbiol. 47:1216–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malani A., et al. 2005. Candida glabrata fungemia: experience in a tertiary care center. Clin. Infect. Dis. 41:975–981 [DOI] [PubMed] [Google Scholar]
- 14. Pfaller M. A., Diekema D. J. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Romeo O., Scordino F., Pernice I., Lo Passo C., Criseo G. 2009. A multiplex PCR protocol for rapid identification of Candida glabrata and its phylogenetically related species Candida nivariensis and Candida bracarensis. J. Microbiol. Methods 79:117–120 [DOI] [PubMed] [Google Scholar]
- 16. Tortorano A. M., et al. 2004. Epidemiology of candidaemia in Europe: results of 28-month European Confederation of Medical Mycology (ECMM) hospital-based surveillance study. Eur. J. Clin. Microbiol. Infect. Dis. 23:317–322 [DOI] [PubMed] [Google Scholar]
- 17. Trick W. E., Fridkin S. K., Edwards J. R., Hajjeh R. A., Gaynes R. P. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35:627–630 [DOI] [PubMed] [Google Scholar]
- 18. Wahyuningsih R., et al. 2008. Candida nivariensis isolated from an Indonesian human immunodeficiency virus-infected patient suffering from oropharyngeal candidiasis. J. Clin. Microbiol. 46:388–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White T. J., Bruns T., Lee S., Taylor J. 1990. Amplification and direct sequencing of fungal rRNA genes for phylogenetics, p. 315–322In Innis M. A., Gelfand D. H., Sninsky J. J., Taylor T. J.(ed.), PCR protocols: a guide for methods and applications. Academic Press, New York, NY [Google Scholar]