Abstract
Early diagnosis of drug-resistant Mycobacterium tuberculosis is urgently needed to optimize treatment regimens and to prevent the transmission of resistant strains. Real-time PCR assays have been developed to detect drug resistance rapidly, but none of them have been widely applied due to their complexity, high cost, or requirement for advanced instruments. In this study, we developed a real-time PCR method based on melting curve analysis of dually labeled probes. Six probes targeting the rpoB 81-bp core region, katG315, the inhA promoter, the ahpC promoter, and embB306 were designed and validated with clinical isolates. First, 10 multidrug-resistant (MDR) strains with a wide mutation spectrum were used to analyze the melting temperature (Tm) deviations of different mutations by single real-time PCR. All mutations can be detected by significant Tm reductions compared to the wild type. Then, three duplex real-time PCRs, with two probes in each, were developed to detect mutations in 158 MDR isolates. Comparison of the results with the sequencing data showed that all mutations covered by the six probes were detected with 100% sensitivity and 100% specificity. Our method provided a new way to rapidly detect drug-resistant mutations in M. tuberculosis. Compared to other real-time PCR methods, we use fewer probes, which are labeled with the same fluorophore, guaranteeing that this assay can be used for detection in a single fluorescent channel or can be run on single-channel instruments. In conclusion, we have developed a widely applicable real-time PCR assay to detect drug-resistant mutations in M. tuberculosis.
INTRODUCTION
Tuberculosis (TB) is still one of the most serious threats to human health around the world. Recently, the dramatic increase of drug-resistant TB has caused an important problem for disease control due to the ineffectiveness of treatment and the transmission of drug-resistant strains (1, 17). The traditional phenotypic drug susceptibility tests pose serious delays in the detection of resistance due to the extremely slow growth of M. tuberculosis. Rapid diagnosis of drug resistance is essential in order to initiate effective antibiotic therapies and prevent the transmission of drug-resistant strains.
M. tuberculosis acquires drug resistance mainly through mutations in specific genes (2, 28, 29, 33). In our previous study, a set of drug-resistant mutation sites was indentified for the detection of multidrug-resistant (MDR) TB in Shanghai, China (19). Knowledge of drug-resistant mutations has led to the development of molecular methods to achieve the rapid diagnosis of drug-resistant TB (23). Among these methods, real-time PCR has been one of the most widely applied due to its rapidity, high sensitivity, reproducibility, and low risk of contamination (7, 10, 12, 14, 18, 31). The most common real-time PCR assays for detecting drug-resistant mutations have been developed through two main methods. In the first method, a fluorescent signal is generated by hybridization of a probe to the target sequence at the end of each PCR cycle. The TaqMan probe- and molecular beacon-based allele discrimination assays belong to this category. Because most of these assays require the use of two different fluorophore-labeled probes for differentiation of one allele (9, 32), the cost is relatively high and a real-time PCR instrument with multiple channels is required. In the second case, mutation detection is achieved by melting curve analysis. Two major assays, fluorescence resonance energy transfer (FRET) probe melting curve analysis and high-resolution melting curve (HRM) analysis, have been successfully applied to detect drug-resistant mutations in M. tuberculosis (5, 12, 16, 22, 24, 25). However, both of these assays have been applied to limited real-time PCR platforms: the FRET probe assay is restricted to the LightCycler instrument (Roche), and the HRM assay requires highly advanced real-time PCR instruments. More recently, several new probe-based melting curve analysis technologies, including unlabeled probes, dually labeled probes, and sloppy molecular beacons, have been developed for genotyping (3, 8, 34). However, limited studies have reported on their application to detection of drug-resistant mutations in M. tuberculosis (4, 15).
In the present study, we aim to develop a low-cost, widely applicable real-time PCR assay, based on melting curve analysis of dually labeled probes, to rapidly detect the drug-resistant mutations of M. tuberculosis.
MATERIALS AND METHODS
Bacterial isolates and DNA extraction.
The MDR M. tuberculosis isolates used for real-time PCR analysis were selected from the culture collection at the Shanghai Municipal Center for Disease Control and Prevention (Shanghai CDC) from March 2004 to November 2008. In total, 158 MDR isolates were randomly selected from 322 MDR TB cases (19). The sequencing analysis of drug-resistant genes (rpoB, katG, inhA, inhA promoter, ahpC promoter, and embB) indicated that the mutation patterns of these 158 strains represent the drug-resistant mutation types in Shanghai. The laboratory reference strain M. tuberculosis H37Rv, which is susceptible to all drugs and which has no mutation in any of the drug-resistant genes, was used as the wild-type control. DNA of the M. tuberculosis isolates was extracted by a rapid boiling method as previously described (26).
Probe and primer design.
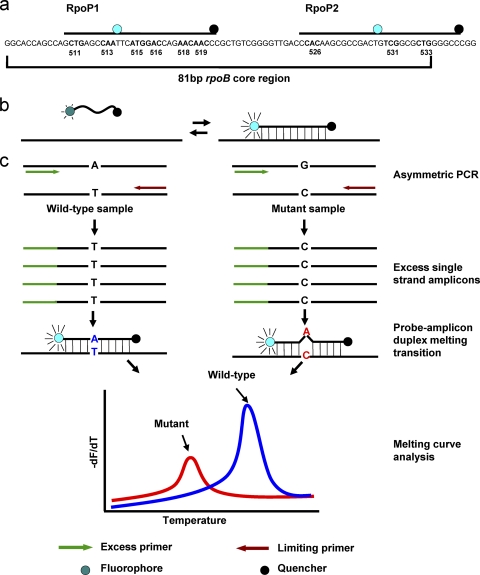
Six dually labeled probes and 10 primers were designed to detect mutations in five drug resistance-associated genes (Table 1). For the 81-bp rpoB core region, multiple probes (normally more than four) were usually required to cover the whole region in a PCR detection assay (7, 14, 31). In this study, we intended to simplify the procedure and reduce the cost by designing two long probes (more than 25 nucleotides) to detect mutations in this region. An internal fluorophore (6-carboxyfluorescein [FAM]) was attached to probes RpoP1 and RpoP2, and the quencher black hole quencher 1 (BHQ1) was attached at the 3′ ends of both probes (Table 1; Fig. 1A). Two mismatched nucleotides were introduced into probe rpoP2 to disrupt the palindrome sequence, which could also increase the sensitivity of discrimination (Table 1). Other normal dually labeled probes (in which the fluorophore and quencher were added to the 5′ and 3′ ends, respectively) were designed to detect mutations in katG315, the inhA promoter, the ahpC promoter, and embB306 (Table 1). All six probes are labeled with the same fluorophore, FAM.
Table 1.
Primers and probes used in this study
| Primer or probe | Targetd | Concn (μM) | Reactiona | Sequence | Oligonucleotide size (bp) |
|---|---|---|---|---|---|
| PCR primers | |||||
| rpobF | rpoB | 0.4 | I | GCCGCGATCAAGGAGTTCTTC | 21 |
| rpobR | 0.05 | CGGCACGCTCACGTGACAGAC | 21 | ||
| katF | katG | 0.05 | II | CGTATGGCACCGGAACCGGTAA | 22 |
| katR | 0.4 | GCTCCCACTCGTAGCCGTACA | 21 | ||
| inhF | inhA promoter | 0.05 | CACGTTACGCTCGTGGACATAC | 22 | |
| inhR | 0.4 | CTGTGGCAGTCACCCCGACAA | 21 | ||
| ahpF | ahpC promoter | 0.4 | III | GCGGCGATGCCGATAAATATGG | 22 |
| ahpR | 0.05 | CTCCTCATCATCAAAGCGGACAATG | 25 | ||
| embF | embB | 0.4 | GGTGATATTCGGCTTCCTGCTC | 22 | |
| embR | 0.05 | GCCGAACCAGCGGAAATAGTTGGA | 24 | ||
| Dually labeled probes | |||||
| rpobP1 | rpoB511 to rpoB519 | 0.2 | I | 5′-GGTTGTTCTGGTCCA8GAATTGGCTCAGC-BHQ1-3′b | 29 |
| rpobP2 | rpoB524 to rpoB533 | 0.2 | 5′-GCCCCAGCGtCGACAG8CGGtGCTTGTGGG-BHQ1-3′c | 30 | |
| katP | katG315 | 0.2 | II | 5′-FAM-TCACCAGCGGCATCG-BHQ1-3′ | 15 |
| inhP1 | inhA−15 | 0.2 | 5′-FAM-GCGAGACGATAGGTTG-BHQ1-3′ | 16 | |
| ahpP1 | ahpC−6, ahpC−9, ahpC−10, ahpC−12 | 0.2 | III | 5′-FAM-GCGACATTCCATCGTGCCG-BHQ1-3′ | 19 |
| embP | embB306 | 0.2 | 5′-FAM-TCGGCGACTCGGGCCATGCCC-BHQ1-3′ | 21 |
I, II, and III, the three duplex real-time PCRs.
8, the fluorophore (FAM)-labeled dT in probes rpoP1 and ropP2.
The two lowercase letters in this sequence indicate two mismatched nucleotides in probe rpoP2.
Negative numbers in designations indicate positions of mutations located before the start codon of the gene.
Fig. 1.
(a) Schematic representation of the rpoB 81-bp core region and two internal labeled long probes; (b) mechanism of the generation of a fluorescent signal by the dually labeled probe and targeted DNA sequence; (c) principle and process of dually labeled probe-based melting curve analysis. During the asymmetric amplification, the excess primer (green) generates excess copies of single-strand amplicons. These single-strand amplicons are complementary to the probe and increase the visibility of the probe-amplicon duplex melting transition. This melting transition appears as a peak on the derivative plot of melting curves. Mutations were detected as Tm deviations compared to the wild type.
Real-time PCR.
Both single and duplex real-time PCR assays were performed in this study. The real-time PCR mixture was prepared in a final volume of 20 μl. To avoid hydrolysis of probes, as occurs classically during the elongation steps in TaqMan real-time PCR, aTaq DNA polymerase (Promega), which lacks 5′ to 3′ exonuclease activity, was used for the PCRs. The final concentration of MgCl2 was 1.5 mM. The optimal concentrations of primers and probes for both single and duplex PCRs are listed in Table 1. To achieve asymmetric amplification, the final concentrations of the two primers from one primer pair were different, with the concentration of one primer being eightfold higher than that of the other one. One microliter of extracted DNA was used for each reaction. For condition optimization, betaine was introduced into one group of the single real-time PCR mixtures at a final concentration of 1 M. The real-time PCR was performed in capillary tubes in a Rotor-Gene Q apparatus (Qiagen). The cycling conditions were denaturation at 95°C for 1 min, followed by 40 cycles of amplification at 95°C for 6 s, 58°C for 30 s (with a single acquisition of fluorescence), 72°C for 10 s, 58°C for 15 s, and 72°C for 5 s. The melting program was 30 s at 95°C, 40°C for 0 s, and 85°C for 0 s. The rate of temperature increase was 1°C/s (or 0.5°C/s), and fluorescence was continuously acquired. A negative control without a DNA sample and a standard wild-type control with DNA of H37Rv were included for every real-time PCR experiment. The melting curve was analyzed using the Rotor-Gen (version 1.7.94) software.
Analytical sensitivity.
The three duplex reactions were performed with serially diluted genomic DNA (gDNA; range, 5.0 × 105 to 5.0 × 100 copies per reaction mixture) of the wild-type strain H37Rv and MDR isolate M1, which has three mutations, in rpoB531 (TCG → TTG), embB306 (ATG → ATA), and katG315 (AGC → ACC). The gDNA of these two isolates was extracted by the cetyltrimethylammonium bromide (CTAB) method as previously described (30). The concentration of each gDNA was determined by an ND-2000 UV-visible spectrophotometer (NanoDrop Technologies, Wilmington, DE).
RESULTS
Design of real-time PCR assay.
Dually labeled probes were usually used for TaqMan real-time PCR assays. In this study, six dually labeled probes were designed to detect drug-resistant mutations. In order to avoid the hydrolysis of the probe by Taq polymerase in TaqMan assays, the aTaq polymerase without 5′ nuclease activity was used in our assay to keep the probe intact during the amplification. The fluorescence detection relies on the difference in fluorescence emissions between the melted and hybridized configurations of the probe (Fig. 1B). Because the mean distance between the fluorophore and the quencher molecules of the melted single-strand probe is shorter than that of the hybridized probe-amplicon duplex, a difference in fluorescence emission will be readily detectable when the probe is released from its target sequence (6, 8). In order to increase the visibility of the probe-amplicon duplex melting transition, asymmetric PCR was employed to generate excess single-strand amplicons which are complementary to probes. At the end of the amplification, melting curve analysis was applied to detect mutations. In each run, melting curve analysis was performed with both clinical isolates and the wild-type control (H37Rv). Mutations were detected as the melting temperature (Tm) deviations compared to the wild type (Fig. 1C).
Tm deviations for mutations.
To test the ability of our real-time PCR design to rapidly detect drug-resistant mutations, six independent reactions with a single probe in each were performed using 10 MDR clinical strains with mutations which represented a broad spectrum of mutations on the basis of our previous sequencing data (Table 2). Of the two groups of reactions, the one with 1 M betaine showed a lower Tm (3 to 4°C) for the same probe and a narrower melting peak. Interestingly, the nonspecific peak at about 74°C was cleared by the addition of betaine (data not shown). So, 1 M betaine was always included in the following experiments.
Table 2.
Genotypic characteristics of 10 M. tuberculosis clinical MDR isolates and Tm deviations for different mutations obtained with the single real-time PCR assay
| MDR isolate no. | Mutation site | Nucleotide substitution | Amino acid substitution | Tm (°C) deviation from wild typea |
|||||
|---|---|---|---|---|---|---|---|---|---|
| rpoP1 (0.1)b | rpoP2 (0.2) | katP (0.1) | inhP (0.2) | ahpP (0.2) | embP (0.1) | ||||
| 125 | rpoB513 | CAA → AAA | Gln → Lys | 4.4 ± 0.2c | |||||
| ahpC−12 | C → T | 4.4 ± 0.1 | |||||||
| 264 | rpoB531 | TCG → TTG | Ser → Leu | 3.7 ± 0.2 | |||||
| embB306 | ATG → ATA | Met → Ile | 10.1 ± 0.1 | ||||||
| ahpC−6 | G → A | 8.8 ± 0.1 | |||||||
| 278 | rpoB511 | CTG → CCG | Leu → Pro | 9.1 ± 0.2 | |||||
| rpoB513 | CAA → CCA | Gln → Pro | |||||||
| katG315 | AGC → ACC | Ser → Thr | 6.6 ± 0.3 | ||||||
| embB306 | ATG → CTG | Met → Leu | 7.4 ± 0.1 | ||||||
| 280 | rpoB526 | CAC → TAC | His → Tyr | 2.6 ± 0.2 | |||||
| ahpC−10 | C → T | 5.5 ± 0.0 | |||||||
| 286 | rpoB531 | TCG → TTG | Ser → Leu | 3.7 ± 0.3 | |||||
| ahpC−9 | G → A | 8.8 ± 0.2 | |||||||
| embB306 | ATG → ATC | Met → Ile | 11.1 ± 0.1 | ||||||
| M31 | rpoB526 | CAC → AAC | His → Asn | 2.6 ± 0.1 | |||||
| katG315 | AGC → ACC | Ser → Thr | 6.4 ± 0.4 | ||||||
| M32 | embB306 | ATG → GTG | Met → Val | 4.0 ± 0.1 | |||||
| rpoB516 | GAC → GTC | Asp → Val | 4.4 ± 0.1 | ||||||
| katG315 | AGC → AAC | Ser → Asn | 8.2 ± 0.4 | ||||||
| M37 | embB306 | ATG → GTG | Met → Val | 3.6 ± 0.1 | |||||
| rpoB531 | TCG → TTG | Ser → Leu | 3.7 ± 0.1 | ||||||
| inhA−15 | C → T | 10.5 ± 0.1 | |||||||
| M54 | rpoB516 | GAC → GTC | Asp → Val | 4.8 ± 0.1 | |||||
| rpoB533 | CTG → CCG | Leu → Pro | 5.2 ± 0.1 | ||||||
| ahpC−9 | G → A | 8.6 ± 0.1 | |||||||
| M7 | rpoB526 | CAC → CTC | His → Leu | 3.1 ± 0.1 | |||||
| ahpC−6 | G → A | 8.8 ± 0.0 | |||||||
Tms of the probes were obtained with a temperature increase rate of 1°C/s during the melting analysis.
Standard deviations of the Tms for the six probes are given in parentheses.
Values are mean Tm deviations from three independent experiment repeats.
Negative numbers in designations indicate positions of mutations located before the start codon of the gene.
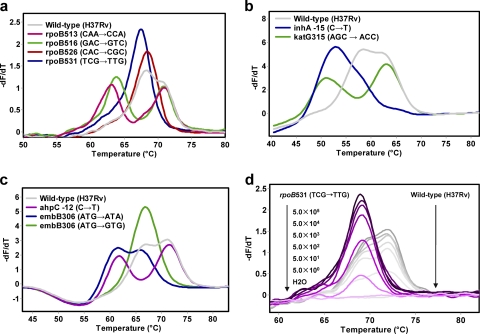
The average reference Tms for wild-type sequences were as follows: 69.8°C for rpoP1, 72.9°C for rpoP2, 59.2°C for katP, 64.6°C for inhP, 67.3°C for ahpP, and 72.5°C for embP (with very small standard deviations; Table 2). For all 10 MDR strains, Tms of all wild-type sequences were within the range obtained for reference strain H37Rv. All strains with mutations in the rpoB 81-bp core, katG315, inhA promoter, ahpC promoter, and embB306 were efficiently detected by measurement of the deviations in the Tms of the probes compared with the value for the wild-type reference strain H37Rv.
For rifampin (RIF) resistance, mutations in codons 513 and 516 in rpoB were detected as a result of the Tm reduction of probe rpoP1 (both of them had a 4.4°C Tm reduction). One strain with two mutations in codons 511 and 516 had a Tm reduction of 9.1°C (Table 2). The most common mutations in codons 526 and 531 were also detected as a result of the reductions in the Tms of probe rpoP2 (Table 2). Two different types of mutations in rpoB526 caused different Tm reductions (2.6 and 3.1°C for mutations CAC → TAC and CAC → CTC, respectively), and the most common mutation in rpoB codon 531 (TCG → TTG) caused a Tm reduction of 3.7°C. With regard to isoniazid (INH) resistance, all mutations in katG315, the inhA promoter, and the ahpC promoter can be efficiently detected by the corresponding probes. Mutations in katG315 were detected by a reduction in the Tm of probe katP (from 6.4 to 8.2°C; Table 2). The mutation inhA−15 (C → T) caused a 10.5°C reduction in the Tm of probe inhP. Mutations in the ahpC promoter region were detected by reductions in the Tm of probe ahpP (from 4.4 to 8.8°C), with different Tm reductions indicating mutations in different locations (Table 2). Mutations in embB306 were treated as a marker for MDR TB according to previous studies (13, 27). Among the 10 MDR strains, 5 of them with mutations in embB306 were detected by reductions in the Tm of probe embP (from 3.6 to 11.1°C) compared with the Tm of H37Rv, and the five different Tm reductions indicated four different mutation types (Table 2).
Thus, the detection of deviations in the Tms of the probes higher than 2.6°C (more than 10 times the standard deviation) indicated the presence of a mutation. These results were reproducible by three independent experiment repeats with very small standard deviations (Table 2).
Duplex real-time PCR and blind analysis of clinical isolates.
In order to achieve multiplex detection of resistance mutations, three duplex real-time PCRs with two probes in each reaction mixture (Table 1) were developed. The validity of this duplex assay was tested with all 158 MDR strains in a blinded manner. Reaction I, which contained probes rpoP1 and rpoP2, was designed to detect mutations in the rpoB 81-bp core region to predict RIF resistance. The melting curve analysis could clearly display the Tm reductions in probe rpoP1, and although the Tm reductions in probe rpoP2 always came with an overlap of the melting peaks of these two probes, the Tm reductions in rpoP2 could still be detected unambiguously (Fig. 2a). In comparison with the sequencing data, all mutations in the regions covered by the probe were detected with 100% sensitivity and specificity. In reaction II, probes katP and inhP were used to detect mutations in katG genes and the inhA promoter to predict INH resistance (Fig. 2b). The Tm reductions in both probes katP and inhP could be clearly detected by melting curve analysis, and the results indicated that 111 strains had mutations in katG315 and 11 strains had mutations in the inhA promoter region, findings that were in 100% agreement with the sequencing data. Reaction III, which contained probes ahpP and embP, was used to detect mutations in the ahpC promoter region and embB306 (Fig. 2c). In this reaction, the Tm reductions of both probes could be clearly detected, although a melting peak overlap of the two probes happened when the reduced Tm of probe embP was close to the Tm of probe ahpP. The melting curve analysis successfully detected all 22 strains with mutations in the ahpC promoter and 56 strains with mutations in embB306. Overall, mutations within the regions covered by the probes were detected with 100% sensitivity and 100% specificity by the three duplex real-time PCRs.
Fig. 2.
Detection of mutations in M. tuberculosis with the duplex reactions (the temperature increase rates were 0.5°C/s for reaction I and 1°C/s for reactions II and III). (a) Melting curve analysis from reaction I with probes rpoP1 and rpoP2; (b) melting curve analysis from reaction II with probes katP and inhP; (c) melting curve analysis from reaction III with probes embP and ahpP; (d) melting curves of reaction I with wild-type and rpoB531 (TCG → TTG) mutant M. tuberculosis strains with genomic DNA at from 5.0 × 105 to 5.0 × 100 copies per reaction mixture.
Analytical sensitivity of duplex assay.
To assess the sensitivity of our duplex assay, we performed three reactions on serially diluted DNA samples of wild-type H37Rv and MDR isolate M1. The analytical sensitivity study showed that reaction I could differentiate the wild type from the mutant when the concentration was as low as 5 DNA copies per reaction mixture (Fig. 2d). However, the sensitivities of reactions II and III, which contain four primers and were not extensively optimized, were about 100 times lower than the sensitivity of reaction I.
DISCUSSION
Rapid detection of the drug resistance of M. tuberculosis is essential for the effective treatment of patients and to prevent the dissemination of resistant strains. Among the molecular methods applied to detect-resistant mutations, real-time PCR has the advantages of rapidity, high sensitivity, reproducibility, and a low risk of contamination. Several real-time PCR-based assays, including both commercial and in-house assays, have been developed during the last few years (7, 9, 11, 14, 16, 18, 20, 21, 31). Most of them have been proved to have high sensitivities and specificities, but due to either cumbersomeness or a requirement for advanced instruments, none of them have been widely applied, especially in resource-limited settings.
In the present study, we developed a low-cost and widely applicable real-time PCR assay for rapid detection of drug-resistant M. tuberculosis. In this assay, dually labeled probes were designed to cover drug resistance-related regions and detect mutations on the basis of the melting curve analysis of the probes at the end of PCR amplification. In comparison to TaqMan- and molecular beacon-based allele discrimination real-time PCR methods, our assay has several advantages. In the TaqMan- or molecular beacon-based assay, two probes (one pair) are usually needed to achieve the allele differentiation (9, 32). The requirement to design individual probes for each resistance mutation not only increases the costs but also limits these assays to being suitable for detecting only the most common mutations, such as those in codon 315 of katG or codon 531 of rpoB (9). Both TaqMan- and molecular beacon-based methods were later modified to detect mutations in the rpoB 81-bp core region, and it has been reported that RIF-resistant M. tuberculosis can be detected in a one-tube reaction by using the beacons (7, 31). However, since both TaqMan probes and beacons can cover only short DNA regions, multiple probes (more than four) labeled with different fluorophores would be required to fully explore a region such as the rpoB 81-bp core, which would require a real-time PCR analyzer that can detect multiple fluorescent signals simultaneously in order to achieve multiple detection (7, 14, 31). In our methods, only one probe is needed to detect mutations in a specific region. For the rpoB core, two long internal fluorophore-labeled probes were enough to achieve the detection of mutations in this region, which reduced the cost that would be required for multiple probes. What is more, all probes in our assay were labeled with the same fluorophore (FAM), and the duplex detections were achieved from the Tm differences of two probes in each reaction, which guaranteed that this assay can be used for detection in a single fluorescent channel or run on single-channel instruments.
Similar methods based on melting curve analysis have been applied to FRET probes to detect drug-resistant mutations in M. tuberculosis (12, 16, 20). In those studies, two pairs of FRET probes (a sensor and an anchor) were designed to detect mutations in the rpoB core region, but two probes were still needed to detect mutations in codon 315 of katG, which caused the assay to have a higher cost than our assay. To our knowledge, FRET probe-based melting curve assays have been developed only for the LightCycler instrument (Roche), which is a large restriction to its use on other instruments. More recently, HRM analysis has been increasingly applied for indentifying resistant mutations (5, 22, 24, 25). HRM analysis can detect small Tm shifts between wild-type and mutant samples in the melting profiles of the amplicons. However, advanced real-time PCR instruments with a high temperature resolution and perfect temperature uniformity between samples are required. In our methods, the Tm deviations for different mutations are greater than 2.6°C, which can be detected by almost all real-time PCR platforms by melting analysis.
The major limitation of our assay is the overlapping of the melting peaks of two probes in the three duplex reactions, which increased the complexity of the melting curve analysis. However, as judgment of the results was made by comparing the melting curve of an unknown sample to that for the wild-type control, strain H37Rv, the mutations can still be detected unambiguously. Another limitation is the relatively low sensitivity of duplex reactions II and III, which may be caused by the low efficiency of the duplex asymmetric amplifications in these reactions. However, we believe that the sensitivities of these two reactions could be improved by modifying primer sequences and concentrations.
In conclusion, we have developed a widely applicable real-time PCR assay to detect drug-resistant mutations in M. tuberculosis. This new method has been proved to be efficient and reliable in detecting a wide variety of mutations in clinical isolates. Further studies are required to optimize the duplex assay and evaluate its performance with clinical sputum specimens.
ACKNOWLEDGMENTS
We are grateful to Gregory Dolganov (Stanford University School of Medicine) for providing assistance in this work.
This work was supported by the Key Project of Chinese National Programs (grant 2009ZX10004-313) and is a key project of the Science and Technology Commission of Shanghai Municipality (10JC1413700 and 10411955000). The first author (Tao Luo) is a trainee of the National Institutes of Health (NIH) under grant D43 TW007887.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Andrews J. R., et al. 2008. Exogenous reinfection as a cause of multidrug-resistant and extensively drug-resistant tuberculosis in rural South Africa. J. Infect. Dis. 198:1582–1589 [DOI] [PubMed] [Google Scholar]
- 2. Banerjee A., et al. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230 [DOI] [PubMed] [Google Scholar]
- 3. Chakravorty S., et al. 2010. Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique. J. Clin. Microbiol. 48:258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chakravorty S., et al. 2011. Rapid detection of fluoroquinolone-resistant and heteroresistant Mycobacterium tuberculosis by use of sloppy molecular beacons and dual melting-temperature codes in a real-time PCR assay. J. Clin. Microbiol. 49:932–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi G. E., et al. 2011. High-resolution melting curve analysis for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 49:1107–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Didenko V. V. 2001. DNA probes using fluorescence resonance energy transfer (FRET): designs and applications. Biotechniques 31:1106–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. El-Hajj H. H., Marras S. A., Tyagi S., Kramer F. R., Alland D. 2001. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 39:4131–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. El Housni H., Heimann P., Parma J., Vassart G. 2003. Single-nucleotide polymorphism genotyping by melting analysis of dual-labeled probes: examples using factor V Leiden and prothrombin 20210A mutations. Clin. Chem. 49:1669–1672 [DOI] [PubMed] [Google Scholar]
- 9. Espasa M., et al. 2005. Direct detection in clinical samples of multiple gene mutations causing resistance of Mycobacterium tuberculosis to isoniazid and rifampicin using fluorogenic probes. J. Antimicrob. Chemother. 55:860–865 [DOI] [PubMed] [Google Scholar]
- 10. Espy M. J., et al. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19:165–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garcia de Viedma D. 2003. Rapid detection of resistance in Mycobacterium tuberculosis: a review discussing molecular approaches. Clin. Microbiol. Infect. 9:349–359 [DOI] [PubMed] [Google Scholar]
- 12. Garcia de Viedma D., et al. 2002. New real-time PCR able to detect in a single tube multiple rifampin resistance mutations and high-level isoniazid resistance mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 40:988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hazbon M. H., et al. 2005. Role of embB codon 306 mutations in Mycobacterium tuberculosis revisited: a novel association with broad drug resistance and IS6110 clustering rather than ethambutol resistance. Antimicrob. Agents Chemother. 49:3794–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Helb D., et al. 2009. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang Q., et al. 2011. Multiplex fluorescence melting curve analysis for mutation detection with dual-labeled, self-quenched probes. PLoS One 6:e19206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kocagoz T., Saribas Z., Alp A. 2005. Rapid determination of rifampin resistance in clinical isolates of Mycobacterium tuberculosis by real-time PCR. J. Clin. Microbiol. 43:6015–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X., et al. 2007. Transmission of drug-resistant tuberculosis among treated patients in Shanghai, China. J. Infect. Dis. 195:864–869 [DOI] [PubMed] [Google Scholar]
- 18. Lin S. Y., Probert W., Lo M., Desmond E. 2004. Rapid detection of isoniazid and rifampin resistance mutations in Mycobacterium tuberculosis complex from cultures or smear-positive sputa by use of molecular beacons. J. Clin. Microbiol. 42:4204–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo T., et al. 2010. Selection of mutations to detect multidrug-resistant Mycobacterium tuberculosis strains in Shanghai, China. Antimicrob. Agents Chemother. 54:1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marin M., Garcia de Viedma D., Ruiz-Serrano M. J., Bouza E. 2004. Rapid direct detection of multiple rifampin and isoniazid resistance mutations in Mycobacterium tuberculosis in respiratory samples by real-time PCR. Antimicrob. Agents Chemother. 48:4293–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morgan M., Kalantri S., Flores L., Pai M. 2005. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect. Dis. 5:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ong D. C., Yam W. C., Siu G. K., Lee A. S. 2010. Rapid detection of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis by high-resolution melting analysis. J. Clin. Microbiol. 48:1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Palomino J. C. 2009. Molecular detection, identification and drug resistance detection in Mycobacterium tuberculosis. FEMS Immunol. Med. Microbiol. 56:103–111 [DOI] [PubMed] [Google Scholar]
- 24. Pietzka A. T., et al. 2009. Rapid identification of multidrug-resistant Mycobacterium tuberculosis isolates by rpoB gene scanning using high-resolution melting curve PCR analysis. J. Antimicrob. Chemother. 63:1121–1127 [DOI] [PubMed] [Google Scholar]
- 25. Ramirez M. V., et al. 2010. Rapid detection of multidrug-resistant tuberculosis using real-time PCR and high resolution melt analysis. J. Clin. Microbiol. 48:4003–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen G. M., et al. 2005. Evaluation of the mycobacterial interspersed repetitive units typing as a practical approach in molecular epidemiology of Mycobacterium tuberculosis. Zhonghua Jie He He Hu Xi Za Zhi 28:292–296 [PubMed] [Google Scholar]
- 27. Shen X., et al. 2007. Association between embB codon 306 mutations and drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 51:2618–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Slayden R. A., Barry C. E., III 2000. The genetics and biochemistry of isoniazid resistance in Mycobacterium tuberculosis. Microbes Infect. 2:659–669 [DOI] [PubMed] [Google Scholar]
- 29. Telenti A., et al. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647–650 [DOI] [PubMed] [Google Scholar]
- 30. van Embden J. D., et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wada T., et al. 2004. Dual-probe assay for rapid detection of drug-resistant Mycobacterium tuberculosis by real-time PCR. J. Clin. Microbiol. 42:5277–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yesilkaya H., et al. 2006. Evaluation of molecular-beacon, TaqMan, and fluorescence resonance energy transfer probes for detection of antibiotic resistance-conferring single nucleotide polymorphisms in mixed Mycobacterium tuberculosis DNA extracts. J. Clin. Microbiol. 44:3826–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y., Heym B., Allen B., Young D., Cole S. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591–593 [DOI] [PubMed] [Google Scholar]
- 34. Zhou L., Myers A. N., Vandersteen J. G., Wang L., Wittwer C. T. 2004. Closed-tube genotyping with unlabeled oligonucleotide probes and a saturating DNA dye. Clin. Chem. 50:1328–1335 [DOI] [PubMed] [Google Scholar]