Abstract
This study describes the attempt to trace the first Mycobacterium bovis outbreak in alpacas (Lama pacos) in Spain by spoligotyping and variable-number tandem-repeat (VNTR) analysis. Due to high genotype diversity, no matching source was identified, but local expansion of a clonal group was found and its significance for molecular tracing is discussed.
TEXT
Mycobacterium bovis is the etiological agent of bovine tuberculosis, but it can affect other domestic animals and wildlife. Molecular typing methods such as spoligotyping (17) and multilocus variable number tandem repeat (VNTR) analysis (10, 29) are wildly used to clarify epidemiological relations between outbreaks. Recently, three cases of M. bovis in alpacas (Lama pacos) have been reported as the first description of tuberculosis in this animal species in Spain (11). Since trade with camelids is increasing and scarce legislation addressing tuberculosis in these animals is given, a follow-up by molecular typing was deemed advisable to obtain insight into its epidemiology.
The alpacas had been held in two different, but epidemiologically linked herds: herd 1 (alpacas 1 and 2, with one animal imported from Peru and the other one from the United Kingdom) was located in the Ronda region (southern Spain), and herd 2 (alpaca 3, imported from Peru and moved from herd 1 to herd 2) was located 90 km away in the Antequera region (see Fig. S1 in the supplemental material). Spoligotyping (17) revealed an identical spoligotype profile, SB0295, for the three alpaca isolates (11). We excluded the possibility of infection in the country of origin in the case of the United Kingdom, because this spoligotype has not been reported (5, 14), and we ignore the situation in Peru due to the lack of molecular typing data from this country.
This study describes the findings of the attempt to trace back the outbreak by molecular characterization of M. bovis isolates from the greater surroundings of the alpaca farms.
In order to investigate the potential source of infection, a search of M. bovis spoligotypes, obtained according to the standard method (17), was performed in the Spanish Database of Animal Mycobacteriosis (http://www.vigilanciasanitaria.es/mycodb/index-en.php). For this purpose, we defined the geographical area within a radius of 150 km around the alpaca farms, considering Doñana National Park in the west, the Baetic Cordillera in the East, the Sierra Morena in the North, and the Mediterranean Sea in the South as natural geographical borders (see Fig. S1 in the supplemental material). In this study area, we gathered information from 171 cattle farms infected with M. bovis, two isolates from red deer (Cervus elaphus), and one from wild boar (Sus scrofa). These M. bovis isolates presented 36 different spoligotypes.
First, we considered that the most probable source could be an M. bovis isolate with a matching spoligotype (SB0295) obtained during the previous 5 years. This search retrieved 31 farms, and one isolate was randomly selected from each. Second, on the assumption that the evolution of the direct repeat (DR) locus is unidirectional by deletion of single or contiguous spacers (32), we also searched for related spoligotypes that could have led to or could have been derived from SB0295 and were present in the same farm or same municipality as M. bovis isolates with spoligotype SB0295. This search yielded 14 additional isolates with spoligotype SB0121 (which could generate SB0295 by loss of spacer 37) and one isolate with spoligotype SB1190 (which could derive from SB0295 by loss of spacer 2) (Table 1). In total, 44 cattle farms matched these criteria out of the 171 M. bovis-affected farms; also one isolate from red deer (SB0121) was found.
Table 1.
VNTR typing results from the Mycobacterium bovis isolates in this study
| Identification no.a | Farm | Spoligotypeb | MIRU-VNTR locus (VNTR no.)c |
VNTR type | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETR-A (2165) | ETR-B (2461) | ETR-D (580) | ETR-E (3192) | MIRU26 (2996) | QUB11a (2163a) | QUB11b (2163b) | QUB26 (4052) | QUB3232 (3232) | ||||
| MI05/00415 | 1 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI05/04818 | 2 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 8 | B |
| MI05/04819 | 3 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI05/04843 | 4 | SB0295 | 6 | 2 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | C |
| MI06/05219 | 5 | SB0295 | 6 | 4 | 3 | 4 | 5 | 11 | 2 | 5 | 7 | D |
| MI06/05242 | 6 | SB0295 | 6 | 4 | 3 | 3 | 5 | 12 | 2 | 5 | 7 | E |
| MI06/05243 | 7 | SB0295 | 6 | 4 | 3 | 3 | 5 | 12 | 2 | 5 | 7 | E |
| MI07/10875 | 8 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI07/11191 | 9 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI07/13777 | 10 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 8 | B |
| MI08/01247 | 11 | SB0295 | 4 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 13 | F |
| MI08/06612 | 12 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI08/08737 | 13 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 8 | B |
| MI08/09187 | 14 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI08/10025 | 15 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI08/13490 | 16 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 12 | G |
| MI08/13492 | 17 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 12 | G |
| MI08/13504 | 18 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/00035 | 19 | SB0295 | 3 | 3 | 3 | 3 | 5 | 11 | 2 | 4 | 7 | H |
| MI09/02177 | 20 | SB0295 | 6 | 4 | —d | 3 | 5 | 11 | 2 | 5 | 7 | NDe |
| MI09/02887 | 21 | SB0295 | 6 | 5 | 3 | 3 | 6 | 11 | 2 | 5 | 7 | I |
| MI09/02890 | 22 | SB0295 | 6 | 4 | 3 | 3 | 4 | 11 | 2 | 5 | 7 | J |
| MI09/02897 | 23 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 4 | 7 | K |
| MI09/05468 | 24 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/08242 | 25 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/09006 | 26 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/09018 | 27 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/09022 | 28 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/09027 | 29 | SB0295 | 6 | 4 | 3 | 3 | — | 11 | 2 | — | 8 | ND |
| MI09/09859 | 30 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/10345 | 31 | SB0295 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/08261 | A1f | SB0295 | 6 | 4 | 3 | 4 | 5 | 11 | 2 | 5 | 6 | L |
| MI09/09364 | A1 | SB0295 | 6 | 4 | 3 | 4 | 5 | — | 2 | 5 | 6 | ND |
| MI09/10805 | A2f | SB0295 | 6 | 4 | 3 | 4 | 5 | 11 | 2 | 5 | 6 | L |
| MI05/04829 | 5 | SB0121 | 6 | 2 | 3 | 3 | 5 | 11 | 1 | 5 | 6 | M |
| MI05/04844 | 32 | SB0121 | 5 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 9 | N |
| MI06/05234 | 33 | SB0121 | 5 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 9 | N |
| MI07/01182 | 34 | SB0121 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI08/01249 | 35 | SB0121 | 6 | 4 | 3 | 3 | 2 | 11 | 2 | 5 | 9 | O |
| MI08/06944 | 36 | SB0121 | 5 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 8 | P |
| MI08/09481 | 37 | SB0121 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI08/09485 | 38 | SB0121 | 4 | 4 | 3 | 3 | 5 | 9 | 2 | 5 | 9 | Q |
| MI08/13503 | 39 | SB0121 | 5 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 13 | R |
| MI09/00032 | 40 | SB0121 | 5 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 11 | S |
| MI09/02889 | 41 | SB0121 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/06628 | 42 | SB0121 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
| MI09/09232 | Wildlifeg | SB0121 | 5 | 4 | 3 | 3 | 5 | 10 | 2 | 5 | 8 | T |
| MI09/09274 | 43 | SB0121 | 5 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 8 | P |
| MI09/10347 | 31 | SB0121 | 5 | 4 | 3 | 3 | 5 | 10 | 2 | 5 | 8 | T |
| MI07/11145 | 44 | SB1190 | 6 | 4 | 3 | 3 | 5 | 11 | 2 | 5 | 7 | A |
The isolates are arranged regarding the spoligotype and in ascending order according to the year of isolation. The first two digits after MI in the strain reference correspond to the year of isolation.
International nomenclature according to Mbovis.org. ■ indicates the presence of a spacer, and □ indicates the absence of the spacer, as shown below.
SB0295: ■■□■■■■■□■■■■■■□■■■■□■■■■■■■■■■■■■■■□■□□□□□
SB0121: ■■□■■■■■□■■■■■■□■■■■□■■■■■■■■■■■■■■■■■□□□□□
SB1190: ■□□■■■■■□■■■■■■□■■■■□■■■■■■■■■■■■■■■□■□□□□□
Shown is the MIRU-VNTR locus with the corresponding VNTR number in parentheses.
—, not amplifiable.
Not determined because of the lack of at least one locus result.
Alpaca farms (11).
Isolate from red deer.
Mycobacterial interspersed repetitive-unit (MIRU)-VNTR typing was conducted on the selected 47 isolates with spoligotypes SB0295, SB0121, and SB1190 following the protocol described by Frothingham and Meeker-O'Connell (10), using nine VNTR markers: ETR-A (VNTR2165) (1, 26), ETR-B (VNTR2461) (10), ETR-D (MIRU4 [VNTR580]) (1, 26), ETR-E (MIRU31 [VNTR3192]), MIRU26 (VNTR2996) (28), QUB11a (VNTR2163a), QUB11b (VNTR2163b) (1, 26), QUB26 (VNTR4052) (23), and QUB3232 (VNTR3232) (26).
MIRU-VNTR typing of the cattle isolates SB0295 resulted in 11 different profiles (Table 1; see Fig. S1 in the supplemental material), but none of the profiles matched the profile of the alpaca isolates (VNTR type L). There was no variation at loci ETR-D and QUB11b. Locus QUB3232 was the most discriminatory locus, followed by ETR-B, which is consistent with other studies (1, 7, 9). Loci QUB3232 and QUB11a have been previously found to have a higher mutation rate regarding allelic diversity in M. tuberculosis and were therefore considered hypervariable (27); nevertheless, their use remains controversial as they may contribute to improved discrimination in determined settings (15). Conflicting findings may be due to variations between different lineages (31) or between species; e.g., allelic diversities may differ in M. bovis (18, 19). For the additional 16 isolates of different spoligotypes, QUB3232 was also the most discriminatory locus, followed by ETR-A and QUB11a. Loci ETR-B, MIRU26, and QUB11b achieved only poor discrimination, and loci ETR-D, MIRU31, and QUB26 did not vary at all. The index of discrimination (D) (16) was calculated using the In Silico website of the University of the Basque Country (http://www.insilico.ehu.es). The combination of spoligotyping and VNTR analysis yielded 22 different genotypes, and the discriminatory power was unexpectedly high (D = 0.889) given that isolates had been selected according to previously mentioned criteria.
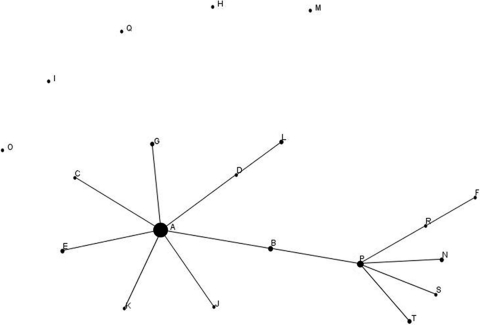
A population snapshot for the VNTR typing results (A to T) was performed by eBURST V3 (25), which has been suggested for analyzing populations with low or moderate recombination ratios (30), such as M. bovis (24). This analysis revealed a clonal group and a linked cluster consisting of related isolates sharing identical alleles at eight out of the nine loci with at least one other member of the group (Fig. 1). VNTR type A (6-4-3-3-5-11-2-5-7) was the most frequent type and was designated the putative founder. It was present in 20 farms and has been found throughout the years and all over the region. VNTR type P was the putative subgroup founder of the linked cluster. The software did not associate five of the VNTR types (H, I, M, O, and Q) with any group due to variations at more than one locus without link to any of the patterns. These findings could be compatible with expansion of a frequent pattern (VNTR type A) from which single-locus variants (SLVs) are derived. Such SLVs could be signs of genetic drift in clonal subpopulations of M. bovis (2, 22).
Fig. 1.
Population snapshot of allelic profiles of the 20 VNTR types (A to T) using eBURST V3 (25). Groups were defined as sets of related isolates sharing identical alleles at eight of nine loci with at least one other member of the group. Type A is designated the putative founder of a clonal group and type P as the subgroup founder of a linked cluster. Five VNTR types (H, I, M, O, and Q) were not associated with any of the clonal groups.
Relationships between the isolates using combined results of spoligotyping and MIRU-VNTR typing were calculated using the MIRU-VNTRplus tool to create an unweighted-pair group method with arithmetic mean (UPGMA) tree with default parameters (see Fig. S2 in the supplemental material). Consistent with eBURST analysis, the largest group clustered isolates with VNTR type A consisting of three subgroups of different spoligotypes (SB0295, SB0121, and SB1190). Due to the fact that SB0121 is ancestral to SB0295, which is derived by a single deletion step in the DR region, we hypothesize that an M. bovis strain with spoligotype SB0121 and VNTR type A is the putative founder of the main cluster.
According to a recent study, SB0295 is the fifth most frequent spoligotype in Spain, representing 4.1% of isolates (21). However, its prevalence is highest in the Autonomous Community Andalusia (18.7%). It has also been found in France (13) and Portugal (8). In Spain, M. bovis spoligotype SB0295 has been mainly isolated from cattle (96% of isolates), and it has also caused infection in deer, wild boars, and Iberian lynxes (3, 12, 22). Although no local wildlife isolates of spoligotype SB0295 or related spoligotypes were registered in the national database, we do not disregard the possibility of wildlife acting as a source of infection. In the Mediterranean habitats of central and southern Spain, wild boars and other ungulates are overabundant, with only little fencing being practiced (4, 20). Nowadays, cattle farms undergo strict control measures; however, extensive herd management is still widespread in Spain, and wildlife is known to intrude into livestock habitats despite fencing.
This study shows the importance of exhaustive collection of molecular typing data of both domestic and wildlife isolates for successful tracing. Ongoing outbreaks with continued transmission are more likely to show genetic diversity than are point source outbreaks (6); therefore, typing results also depend on the nature of the outbreak under investigation. Facing a high-diversity scenario, a less stringent interpretation of typing results may be necessary. Although we could not identify an identical genotype in this attempt to trace back the M. bovis outbreak in the alpacas, we suggest taking clonal groups of closely related strains into account when possible sources of infection need to be elucidated, especially in high-diversity settings.
Supplementary Material
Acknowledgments
This research was funded by the Spanish Ministry of the Environment and Rural and Marine Affairs and EU project TB-STEP (KBBE-2007-1-3-04, no. 212414). S. Rodriguez-Campos is a recipient of a Ph.D. studentship (AP2006-01630) of the Spanish Ministry of Education.
We appreciate the computational assistance of S. González and acknowledge the continuous efforts of the Official Veterinary Services of the Junta de Andalucía. We also thank the anonymous referees for helpful comments.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Allix C., et al. 2006. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J. Clin. Microbiol. 44:1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allix-Beguec C., Fauville-Dufaux M., Supply P. 2008. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive–unit–variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 46:1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aranaz A., et al. 2004. Bovine tuberculosis (Mycobacterium bovis) in wildlife in Spain. J. Clin. Microbiol. 42:2602–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ballesteros C., et al. 2009. First data on Eurasian wild boar response to oral immunization with BCG and challenge with a Mycobacterium bovis field strain. Vaccine 27:6662–6668 [DOI] [PubMed] [Google Scholar]
- 5. Barlow A. M., Mitchell K. A., Visram K. H. 1999. Bovine tuberculosis in llama (Lama glama) in the UK. Vet. Rec. 145:639–640 [DOI] [PubMed] [Google Scholar]
- 6. Barrett T. J., Ribot E., Swaminathan B. 2004. Molecular subtyping for epidemiology: issues in comparability of patterns and interpretation of data, p. 259–266.In Persing D. H., et al. (ed.), Molecular microbiology, diagnostic, principles and practice. ASM Press, Washington, DC [Google Scholar]
- 7. Boniotti M. B., et al. 2009. Molecular typing of Mycobacterium bovis strains isolated in Italy from 2000 to 2006 and evaluation of variable-number-tandem-repeats for a geographic optimized genotyping. J. Clin. Microbiol. 47:636–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duarte E. L., Domingos M., Amado A., Botelho A. 2008. Spoligotype diversity of Mycobacterium bovis and Mycobacterium caprae animal isolates. Vet. Microbiol. 130:415–421 [DOI] [PubMed] [Google Scholar]
- 9. Duarte E. L., Domingos M., Amado A., Cunha M. V., Botelho A. 2010. MIRU-VNTR typing adds discriminatory value to groups of Mycobacterium bovis and Mycobacterium caprae strains defined by spoligotyping. Vet. Microbiol. 143:299–306 [DOI] [PubMed] [Google Scholar]
- 10. Frothingham R., Meeker-O'Connell W. A. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189–1196 [DOI] [PubMed] [Google Scholar]
- 11. García-Bocanegra I., et al. 2010. Tuberculosis in alpacas (Lama pacos) caused by Mycobacterium bovis. J. Clin. Microbiol. 48:1960–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gortázar C., et al. 2005. Molecular characterization of Mycobacterium tuberculosis complex isolates from wild ungulates in south-central Spain. Vet. Res. 36:43–52 [DOI] [PubMed] [Google Scholar]
- 13. Haddad N., et al. 2001. Spoligotype diversity of Mycobacterium bovis strains isolated in France from 1979 to 2000. J. Clin. Microbiol. 39:3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hewinson R. G., Vordermeier H. M., Smith N. H., Gordon S. V. 2006. Recent advances in our knowledge of Mycobacterium bovis: a feeling for the organism. Vet. Microbiol. 112:127–139 [DOI] [PubMed] [Google Scholar]
- 15. Hilty M., et al. 2005. Evaluation of the discriminatory power of variable number tandem repeat (VNTR) typing of Mycobacterium bovis strains. Vet. Microbiol. 109:217–222 [DOI] [PubMed] [Google Scholar]
- 16. Hunter P. R., Gaston M. A. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamerbeek J., et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lari N., Bimbi N., Rindi L., Tortoli E., Garzelli C. 2011. Genetic diversity of human isolates of Mycobacterium bovis assessed by spoligotyping and variable number tandem repeat genotyping. Infect. Genet. Evol. 11:175–180 [DOI] [PubMed] [Google Scholar]
- 19. McLernon J., Costello E., Flynn O., Madigan G., Ryan F. 2010. Evaluation of mycobacterial interspersed repetitive-unit-variable-number tandem-repeat analysis and spoligotyping for genotyping of Mycobacterium bovis isolates and a comparison with restriction fragment length polymorphism typing. J. Clin. Microbiol. 48:4541–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muñoz P. M., et al. 2010. Spatial distribution and risk factors of Brucellosis in Iberian wild ungulates. BMC Infect. Dis. 10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodríguez S., et al. 2010. High spoligotype diversity within a Mycobacterium bovis population: clues to understanding the demography of the pathogen in Europe. Vet. Microbiol. 141:89–95 [DOI] [PubMed] [Google Scholar]
- 22. Romero B., et al. 2008. Persistence and molecular evolution of Mycobacterium bovis population from cattle and wildlife in Doñana National Park revealed by genotype variation. Vet. Microbiol. 132:87–95 [DOI] [PubMed] [Google Scholar]
- 23. Skuce R. A., et al. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519–528 [DOI] [PubMed] [Google Scholar]
- 24. Smith N. H., Gordon S. V., Rua-Domenech R., Clifton-Hadley R. S., Hewinson R. G. 2006. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat. Rev. Microbiol. 4:670–681 [DOI] [PubMed] [Google Scholar]
- 25. Spratt B. G., Hanage W. P., Li B., Aanensen D. M., Feil E. J. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol. Lett. 241:129–134 [DOI] [PubMed] [Google Scholar]
- 26. Supply P. Protocol and guidelines for multilocus variable number tandem repeat genotyping of M. bovis VENoMYC (Veterinary Network of Laboratories Researching into Improved Diagnosis and Epidemiology of Mycobacterial Diseases), p. 15–16. 2006 WP7 Workshop, 19 to 22 October 2006, Toledo, Spain. WP7 Workshop VENoMYC Coordination Action EU SSPE-CT-2004-501903. [Google Scholar]
- 27. Supply P., et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Supply P., et al. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Supply P., Magdalena J., Himpens S., Locht C. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991–1003 [DOI] [PubMed] [Google Scholar]
- 30. Turner K. M., Hanage W. P., Fraser C., Connor T. R., Spratt B. G. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Velji P., Nikolayevskyy V., Brown T., Drobniewski F. 2009. Discriminatory ability of hypervariable variable number tandem repeat loci in population-based analysis of Mycobacterium tuberculosis strains, London, UK. Emerg. Infect. Dis. 15:1609–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Warren R. M., et al. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.