Abstract
Entry into the host bacterial cell is one of the least understood steps in the life cycle of bacteriophages. The different envelopes of Gram-negative and Gram-positive bacteria, with a fluid outer membrane and exposing a thick peptidoglycan wall to the environment respectively, impose distinct challenges for bacteriophage binding and (re)distribution on the bacterial surface. Here, infection of the Gram-positive rod-shaped bacterium Bacillus subtilis by bacteriophage SPP1 was monitored in space and time. We found that SPP1 reversible adsorption occurs preferentially at the cell poles. This initial binding facilitates irreversible adsorption to the SPP1 phage receptor protein YueB, which is encoded by a putative type VII secretion system gene cluster. YueB was found to concentrate at the cell poles and to display a punctate peripheral distribution along the sidewalls of B. subtilis cells. The kinetics of SPP1 DNA entry and replication were visualized during infection. Most of the infecting phages DNA entered and initiated replication near the cell poles. Altogether, our results reveal that the preferentially polar topology of SPP1 receptors on the surface of the host cell determines the site of phage DNA entry and subsequent replication, which occurs in discrete foci.
INTRODUCTION
Bacterial viruses (phages or bacteriophages) are the most abundant biological entities in the biosphere (25). The vast majority (96% of the phages currently described [2]) use a tail device for specific recognition of the host, binding to its surface, and delivery of their genome from their icosahedral capsid to the bacterial cytoplasm. The first contact with the bacterial surface usually leads to reversible binding that is not saturable. It allows for dissociation of viable phages from the bacterium. In a second step, phages attach irreversibly to a specific cell envelope receptor committing to infection of the host. This process is normally saturable due to a limited number of active receptors accessible for the irreversible interaction at the cell surface. Reversible and irreversible adsorption can target the same receptor or involve different surface components (see reference 48 and references therein). The two strategies correlate with distinct phage adsorption machineries whose complexity can vary from a single receptor-binding protein (RBP) to complex baseplates with several RBPs (42, 47). Different phages of Gram-negative bacteria were shown to bind preferentially to cell poles at low multiplicities of infection (18). In case of phage lambda, this localization correlates with the polar topology of ManY, an inner membrane protein required for lambda DNA entry in the bacterium. The subsequent position of phage DNA replication appears to occur also at cytoplasmic positions close to the pole (18). However, the receptor for irreversible binding at the bacterial surface, LamB, is an abundant protein distributed throughout the Escherichia coli outer membrane following a helical pattern (20). It is thus likely that the preferential positioning of phage lambda at the cell poles results of fast two-dimensional diffusion of a subpopulation of LamB in the membrane (20). The complex lambda-LamB would then be immobilized at the pole for an as-yet-unknown mechanism to initiate infection. The situation is different in Gram-positive bacteria, where the distribution of surface receptors is defined by the cell wall dynamics, which is determined by the comparatively slower addition of new polymers to the growing cell wall sacculus (16, 46). The bacterial surface provided by the Gram-positive envelope imposes a different barrier to virus attack and thus distinct challenges for viral attachment and infection relative to Gram-negative bacteria. We sought to study the cell biology of viral infection initiation in the former group of bacteria.
Bacteriophage SPP1 infects the Gram-positive rod-shaped soil bacterium Bacillus subtilis. Infection is initiated by the reversible binding of SPP1 to glucosylated poly(glycerol phosphate) cell wall teichoic acids (WTA) (7). This reversible interaction allows for fast attachment of phages to the bacterium facilitating two-dimensional scanning of its surface for the SPP1 receptor YueB (7). The strong interaction between SPP1 and YueB commits the phage to infection (39, 41). YueB is an integral membrane protein with five transmembrane segments at its carboxyl terminus and a large ectodomain forming a 36.5-nm-long fiber that spans the thick B. subtilis cell wall to expose the SPP1 receptor domain at the bacterial surface (39, 41). Binding of the SPP1 adsorption apparatus to YueB triggers a sequence of conformational changes in the phage tail structure (34) that culminates in genome release from viral particles through the tail tube (40, 41). SPP1 DNA is found inside B. subtilis at 3 min postinfection (p.i.) at 37°C in rich medium (see reference 12 and references therein). Viral DNA transcription and replication initiates rapidly afterward. Concatemers of the phage genome are detected 8 min after infection (see references 12 and 30 and references therein). Expression of late genes coding for morphogenetic proteins initiates at 10 to 12 min p.i., followed by SPP1 DNA packaging and the formation of infective viral particles that are released by cell lysis occurring between 30 min and 60 min p.i. under optimal infection conditions (4). We studied here the spatial and temporal program of SPP1 infection of B. subtilis. We show that preferential polar interactions of the phage with its receptors determine the topology of DNA entry in the cell and the localization of phage multiplication foci in the bacterial cytoplasm.
MATERIALS AND METHODS
General methods.
B. subtilis strains were grown in Luria-Bertani (LB) medium with appropriate antibiotic concentrations (Table 1).
Table 1.
B. subtilis strains used in this studya
| Strain | Genotype and/or relevant featuresb | Source or reference |
|---|---|---|
| YB886 | B. subtilis 168 derivative freed of prophages PBSX and SPβ; wild-type strain; SPP1 indicator strain | 51 |
| YB886.Δ6 | YB886 derivative with deleted yueB (Δ6 deletion); contains an eryR marker inserted in the amyE locus; Eryr (0.5 μg ml−1) | This study |
| YB886.PspacYB | YB886 derivative expressing yueB under the control of the promoter Pspac (inducible with IPTG); Eryr (0.5 μg ml−1) | This study |
| YB886.Δ6.PxylYB | YB886.Δ6 derivative with PxylA-driven expression of yueB (inducible with xylose); Neor (7.5 μg ml−1) | This study |
| YB886.YBGFP | YB886 derivative driving native expression of yueB-gfp, yueBΩpCB-GFP; Spcr (100 μg ml−1) | This study |
| YB886.Δ6.PxylYBGFP | YB886.Δ6 derivative with PxylA-driven expression of yueB-gfp (inducible with xylose); Neor (7.5 μg ml−1) Spcr (100 μg ml−1) | This study |
| MMB357 | Expression of lacI-cfp; thrC::(Ppen-lacIΔ11-cfp (W7) mls Eryr (0.5 μg ml−1) Linr (12.5 μg ml−1) | A. D. Grossman |
| GSY10000 | YB886 derivative; expression of lacI-cfp; thrC::(Ppen-lacIΔ11-cfp(W7) mls Eryr (0.5 μg ml−1) Linr (12.5 μg ml−1) | This study |
See also Fig. 1.
Eryr, erythromycin resistance; Spcr, spectinomycin resistance; Neor, neomycin resistance; Linr, lincomycin resistance.
SPP1 phages were produced in B. subtilis YB886 (51) grown in LB medium and supplemented with 10 mM CaCl2 just before infection (38). Isolated phage plaques (105 to 106 PFU) were used to generate confluent plate lysates (108 to 5 × 108 PFU), and then the phages were amplified sequentially in small- and large-scale culture infections. Cell debris were pelleted at 8,000 × g for 15 min at 4°C. In large-scale preparations, the lysates were centrifuged overnight at 8,000 × g to sediment the phage particles. The phages were then purified by isopycnic centrifugation in a discontinuous gradient with layers of 1.7, 1.5, and 1.45 g of CsCl cm−3 prepared in TBT buffer (100 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2 [pH 7.5]) (9). Gradients were run in an SW41 Ti rotor (Beckman Coulter) at 32,000 rpm for 6 h at 20°C, and the phage blue band was collected with a syringe and a needle by lateral perforation of the tube (37). Phages were dialyzed against TBT and stocked in this buffer. Titrations were carried out using B. subtilis YB886 as the indicator strain unless stated otherwise.
Construction of bacteriophage SPP1delX110lacO64.
An array of lacO operators situated between NheI and XbaI restriction sites of plasmid pFX276, kindly provided by F. X. Barre (CGM, Gif-sur-Yvette, France) (27), were amplified by PCR using the primers 124-2 (5′-AAGAGCATGCATTACGTAATGGATCCACTTTATGCTTCCGGCTCG-3′; the restriction sites for SphI, SnaBI, and BamHI are underlined) and 125-2 (5′-AAGAGCATGCATCTCGAGGATGTGCTGCAAGGCG-3′; the sites for SphI and XhoI are underlined) and digested with SphI. The PCR fragment was cloned into the genome of the bacteriophage SPP1delX110 (45) cleaved at its unique SphI site at bp 39721 to 39726 (EMBL accession code X97918 [3]). The ligation reaction was transformed into B. subtilis competent cells (10). Single phage plaques were titrated to re-obtain single plaques corresponding to pure clones. Insertion of the lacO operators cassette was confirmed by PCR and DNA sequencing. PCR was routinely used to check for stability of the insert during multiplication of the SPP1delX110lacO64 phage.
Chemical modification of bacteriophages.
A 10 mM stock of EZ-Link Sulfo-NHS-LC-Biotin (sulfosuccinimidyl-6-[biotin-amido]hexanoate; Pierce) was made in Milli-Q water (Millipore) (18). The solution was mixed with CsCl-purified SPP1delX110lacO64 phage particles previously dialyzed against 20 mM HEPES-Na (pH 7.0)-100 mM NaCl-10 mM MgCl2 to yield a final concentration of 24 μM biotinylation reagent. After 30 min at room temperature the reaction was stopped by the addition of glycine to a final concentration of 0.025%. The concentration of Sulfo-NHS-LC-Biotin was optimized to have minimal effect on phage viability as tested by direct plaque assay (>80% phage viability). Determination of the irreversible adsorption constant (kads) of control and modified phages to B. subtilis YB886 was carried out as described previously (7, 39).
Bacterial strains construction.
YB886 strains expressing the different yueB forms were obtained by transformation of wild-type YB886 with genomic DNA constructs of B. subtilis L16601 (6, 39). The yueB Δ6 deletion was introduced in YB886 using CSJ4 DNA as a donor (39) and selecting for erythromycin and SPP1 resistance (Eryr at the amyE locus). The resulting strain was named YB886.Δ6 (Fig. 1 A). This strain served as the recipient for transformation with CSJ6 DNA (39), yielding strain YB886.Δ6.PxylYB that allows ectopic expression of yueB under the control of the PxylA promoter (Fig. 1A). Strain YB886.PspacYB was obtained by transformation of YB886 with DNA of CSJ3 (39), which expresses yueB from the Pspac promoter.
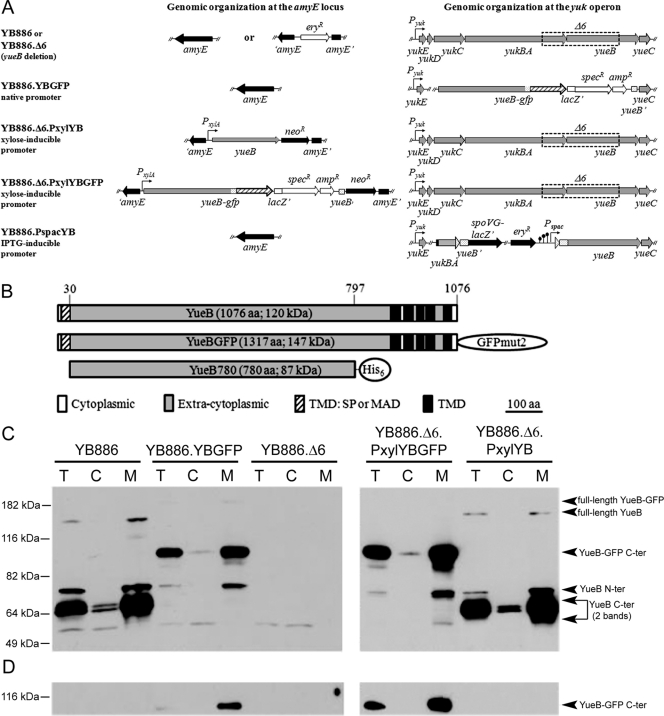
Fig. 1.
Expression of yueB and yueB-gfp fusions in B. subtilis. (A) Relevant genomic organization of strains used in the present study, with different levels of yueB or yueB-gfp expression (see Materials and Methods for details). The yueB-gfp fusion was cloned under the control of the native yukE promoter (strain YB886.YBGFP) or of the inducible PxylA promoter at the ectopic amyE locus (strain YB886.Δ6.PxylYBGFP). (B) Scheme of YueB, YueB-GFP, and YueB780 showing their putative signal peptide (SP) and transmembrane (TMD) segments, extracellular (ectodomain), and cytoplasmic regions. YueB780 is a dimeric elongated fiber active to trigger SPP1 DNA ejection in vitro whose sequence covers most of the ectodomain region of YueB (41). (C and D) Production of YueB and YueB-GFP in total cell extracts (T) and after fractionation into cytoplasmic (C) and membrane (M) fractions. Their composition was characterized by Western blotting with anti-YueB780 (C) and anti-GFP (D) antibodies. The amount of total protein loaded per lane was 30 μg (NanoDrop quantification) for YB886.Δ6.PxylYBGFP and YB886.Δ6.PxylYB (right panels), whereas 150 μg (5-fold) were loaded for YB886, YB886.YBGFP, and YB886.Δ6 (left panels) to increase sensitivity. Molecular mass markers are shown on the left. The assignment of YueB-derived bands is shown on the right.
For expression of yueB-gfp fusions the gfp mut2 gene was amplified by PCR from vector pEA18 (35) (using the primer pair gfp 30D and gfp 749R [AGTAAAGGAGAAGAACTTTTCACTGGAG and GATCCTCGAGGAATTCTTATTTGTATAGTTCATCCATGC; the XhoI and EcoRI sites are underlined]) and fused, by overlap extension PCR, to another PCR product that carried the 3′ end of yueB except for the stop codon (using the primer pair YB GFP-1 and YB GFP-2 [CGTTATTCGGAATTCCTCGAGTCCTTGTCGGACTTA] and [CTCCAGTGAAAAGTTCTTCTCCTTTACTGTTGTTGTTGTTCGCTTCATACGTTTCATCGCTTTCTGCTGT]; the sites XhoI and EcoRI are underlined [note that in the overlapping PCR only the flanking primers—YB GFP-1 and gfp 749R—carry restriction sites]). The fusion product was cleaved with EcoRI and cloned in vector pUS19 digested with the same enzyme (8). The resulting plasmid, pCB-GFP, was checked for sequence correctness and used to transform B. subtilis L16601 and CSJ6, yielding strains CBM-GFP and CBM191, with native or PxylA-dependent expression of yueB-gfp, respectively (6). Strains YB886.YBGFP, and YB886.Δ6.PxylYBGFP were obtained by transforming YB886 with CBM-GFP DNA and YB886.Δ6 with CBM191 DNA, respectively (Fig. 1A; Table 1) (6). The kads values of SPP1 to these strains were determined as described previously (7, 39).
B. subtilis MMB357 (28) (Table 1) producing the fusion protein LacI-CFP was kindly provided by A. D. Grossman and C. Lee (Massachusetts Institute of Technology [MIT], Cambridge, MA). The cassette coding for the fusion protein was transferred to B. subtilis YB886 by SPP1 transduction (50) to obtain the isogenic strain GSY10000 (Table 1). In brief, a lysate of SPP1 multiplied in MMB357 was used to infect 0.3 ml of a late-logarithmic-phase (A600 = 1.5; ∼4 × 108 CFU ml−1) culture of the receptor strain YB886 supplemented with 10 mM CaCl2. The culture infected with an input multiplicity of 1 was incubated for 10 min at 37°C, mixed with 2 ml of prewarmed LB medium, and further incubated with shaking for an additional 10 min. Bacteria were sedimented and resuspended in prewarmed medium containing 20 μl of anti-SPP1 serum to inactivate free phages. Bacteria were further incubated for 30 min, sedimented, and resuspended in 0.3 ml of LB medium. Serial dilutions of the culture were plated in solid medium supplemented with 12.5 μg of lincomycin and 0.5 μg of erythromycin ml−1 to select for transductants resistant to antibiotics of the macrolide, lincosamide, and streptogramin families (mls). The new strain was tested for tryptophan, methionine, and threonine auxotrophy, for phenylalanine prototrophy, for the absence of PBSX and SPβ bacteriophages (17), and for LacI-CFP fluorescence signal in SPP1delX110lacO64 infections (see below).
Fractionation of B. subtilis extracts and Western blotting of YueB engineered versions.
The method was adapted from São-José et al. (39). Strains YB886, YB886.Δ6, YB886.YBGFP, YB886.Δ6.PxylYBGFP, and YB886.Δ6.PxylYB (Fig. 1A) were grown overnight at 30°C in LB medium. Cultures were then diluted 1:100 in 500 ml of LB medium (containing 0.5% xylose in the case of Pxyl-driven expression) and grown at 37°C to an A600 of 0.8. Bacteria were sedimented and resuspended in 10 ml of lysis buffer (50 mM HEPES and 300 mM NaCl [pH 8] supplemented with 1 mg of lysozyme ml−1 and a protease inhibitor cocktail at the concentration described by the supplier [Complete EDTA-free; Roche Applied Science]). The sample was homogenized by sonication (45 s, 60%, 0.5-pulse with a 1-min interval, 10 cycles). An aliquot of 500 μl was taken (“total extract” sample), and the remaining volume was centrifuged at 20,000 × g for 20 min at 4°C to clear the lysate, removing large aggregates and nonlysed bacteria. After saving 500 μl of the supernatant (“soluble extract” fraction), 5 ml of supernatant was subjected to ultracentrifugation (120,000 × g, 90 min, 4°C). A 700-μl aliquot of the supernatant was carefully removed (“cytoplasmic extract” fraction), and the rest of the supernatant was discarded. The membrane-enriched pellet was resuspended in 250 μl of lysis buffer supplemented with 0.1% Triton and centrifuged at 10,000 × g for 20 min at 4°C to eliminate insoluble material. The supernatant was used as the “membrane extract” fraction.
The total protein in different fractions was quantified by using a NanoDrop ND-1000 spectrophotometer (Nuciber), and the amounts of protein specified in the legend of Fig. 1 were loaded into 8% SDS-PAGE gels, stained with Coomassie blue to monitor the reproducibility of the fractionation procedure (data not shown), or used for Western blot analyses (Fig. 1C and D) as described previously (39).
Fluorescence microscopy and image acquisition.
For phage binding localization, overnight cultures of B. subtilis YB886 and YB886.Δ6 strains were diluted 1:50 in fresh LB medium and grown at 37°C to exponential phase (A600 = 0.8). To stain the bacterial membranes the nontoxic vital membrane dye FM1-43FX (Invitrogen, Eugene, OR) was added to obtain the final concentration of 5 μg ml−1. To inhibit phage DNA synthesis, one of the culture samples was supplemented with 6 (p-hydroxyphenylazo)-uracil (HPUra; kindly provided by N. C. Brown, GLSynthesis, Worcester, MA) to obtain the final concentration of 200 μM, followed by incubation for 2 min at 37°C. Qdot655 streptavidin conjugate (Invitrogen) was added to the chemically modified bacteriophage SPP1delX110lacO64, immediately prior to use, to obtain the Qdot streptavidin conjugate concentration of 10 nM, followed by a 5-min incubation at room temperature and then centrifuged for 5 min at 5,000 × g. The supernatant of Qdot655-labeled bacteriophage SPP1delX110lacO64 was used to infect B. subtilis cultures supplemented with 10 mM CaCl2. The cultures infected with an input multiplicity of 6 infective phages per bacterium (PFU/CFU) were incubated for 2 min at 37°C, fixed with glutaraldehyde to a final concentration of 0.5%, and further incubated at room temperature for 2 min. To obtain only stably adsorbed phages, fixed bacterium-phage mixtures were diluted 100-fold with phosphate-buffered saline (PBS) (37), sedimented for 5 min at 5,000 × g, and resuspended in 10 μl of 1× PBS buffer. An aliquot of bacteria (2 μl) was immediately mounted on a thin film of 1.2% agarose (SeaKem GTG Agarose; Cambrex Bioscience, Rockland) in water on microscope slides (Marienfeld, Lauda-Königshofen, Germany) and then overlaid with a coverslip (Menzel-Gläser; Thermo Fisher Scientific). The cells were immediately imaged by contrast and fluorescence microscopy. The preparation for the microscopy was identical for all of the culture samples described below.
For YueB internal localization, strains producing YueB-GFP fusions were diluted 1:50 from an overnight culture in fresh LB medium. Strain YB886.Δ6.PxylYBGFP was grown in the presence of 0.5% xylose for induction. Cells were grown at 37°C to mid-exponential phase (A600 = 0.8). To stain the bacterial membranes, the vital membrane dye FM4-64FX was added to obtain the final concentration of 5 μg ml−1 and imaged as described above. For YueB external localization, overnight cultures of B. subtilis YB886.YBGFP and YB886.Δ6.PxylYBGFP strains were diluted 1:50 in fresh LB medium and grown at 37°C. Bacteria (500 μl) were collected during exponential growth (A600 = 0.8) by centrifugation, resuspended in 500 μl of prewarmed LB medium containing 2% bovine serum albumin (BSA), and then incubated for 10 min at 30°C with shaking. Rabbit polyclonal antibodies raised against YueB780 (Fig. 1B) (41) at 12 μg ml−1 were added, followed by incubation at 30°C for 10 min. The bacteria were centrifuged, washed once with 800 μl of LB, and resuspended in 400 μl of a 1:2,000 dilution of anti-rabbit IgG Alexa Fluor 546 fluorescent dye conjugate antibody (Molecular Probes) as a secondary antibody in BSA-LB. After incubation at 30°C for 5 min, the cells were washed with 800 μl of LB, resuspended in 500 μl of LB, and immediately imaged as described above. Alternatively, samples of exponentially growing bacteria (500 μl) were fixed for 15 min at room temperature with 4% (wt/vol) paraformaldehyde and 0.02% glutaraldehyde in PBS. Fixed cells were washed twice with 800 μl of PBS and resuspended in 400 μl of PBS containing 2% (wt/vol) BSA. After incubation at 4°C for 20 min, the cells were sedimented, suspended in 400 μl of BSA-PBS containing anti-YueB polyclonal antibodies (12 μg ml−1), and incubated at 4°C for 1 h. Cells were washed twice with 800 μl of PBS, followed by incubation with secondary antibody and imaging as described above for nonfixed cells.
For SPP1 phage DNA localization, an overnight culture of B. subtilis strain GSY10000 producing LacI-CFP was diluted 1:50 in fresh LB medium and grown at 37°C to an A600 of 0.8. The culture was supplemented with 10 mM CaCl2 and infected at an input multiplicity of 3 with SPP1delX110lacO64, followed by incubation for 2 min at 37°C and microscopy at room temperature.
Image acquisition of contrast and fluorescence observations was performed using a Leica DMRA2 microscope coupled to a Sony CoolSnap HQ cooled charge-coupled device camera (Roper Scientific). The digital images were acquired and analyzed with Metamorph version 6 software. Where indicated, images of fluorescent samples were deconvolved within Metamorph. Overlays of micrographs were assembled using Metamorph before exporting the images to Adobe Photoshop version CS3. Linear adjustment of the contrast was sometimes applied to entire images equally for better visualization of weak signals (see Fig. 4A and B) or to obtain a better signal-to-noise ratio in images taken at early times of postinfection (see Fig. 6B). No other electronic enhancement or manipulation was applied to our images.
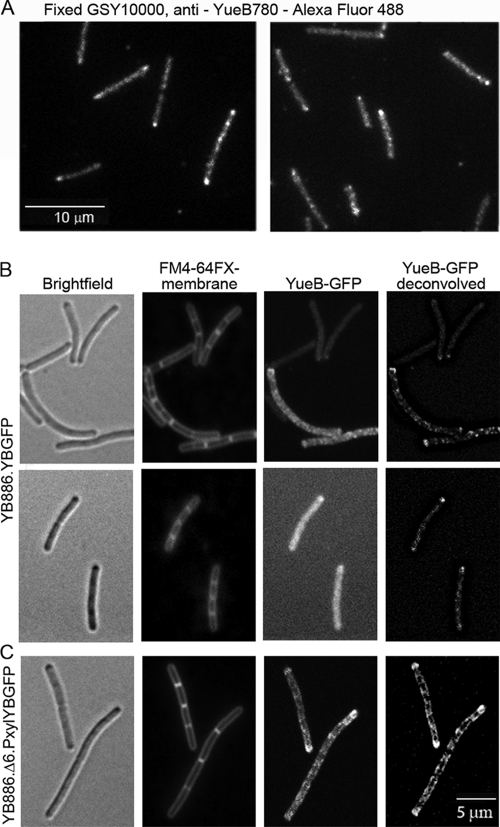
Fig. 4.
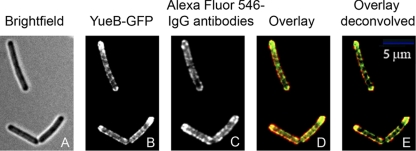
Localization of YueB in B. subtilis. (A) Labeling of YueB at the cell surface of fixed GSY10000 cells labeled with polyclonal anti-YueB780 antibodies. (B and C) Cytoplasmic localization of YueB in B. subtilis YB886.YBGFP (expression of the yueB-gfp fusion under the native Pyuk promoter; see Fig. 1A) (B) and YB886.Δ6.PxylYBGFP (yueB-gfp fusion under the control of a strong xylose-inducible promoter; see Fig. 1A) (C). The YueB cytoplasmic carboxyl terminus (39, 41) was fused to GFP (Fig. 1). The columns in panels B and C show, from left to right, bright-field images, FM4-64FX-stained membranes, YueB-GFP fluorescence, and two-dimensional deconvolution of the YueB-GFP signal.
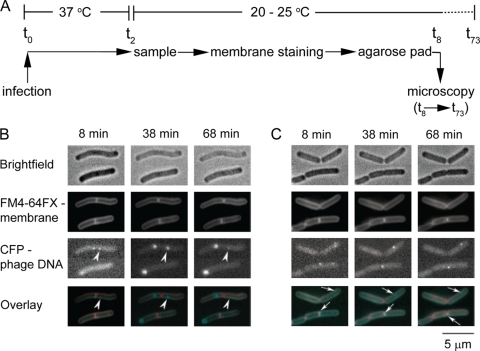
Fig. 6.
Time-lapse of bacteriophage SPP1delX110lacO64 infection. (A) Experimental setup: t0, infection point; t2, 2 min p.i., t8-73, 8 to 73 min p.i. (B and C) Images of B. subtilis producing LacI-CFP infected by phage SPP1delX110lacO64 (input multiplicity of 3) in the absence (B) or presence (C) of HPUra were taken at the postinfection times indicated. Rows: bright-field images, top row; FM4-64FX membrane staining, second row; LacI-CFP, third row. The bottom row shows overlays of membrane staining and CFP fluorescence. The arrowheads show a focus that disappears during infection. The white arrows show small LacI-CFP spots in infected cells treated with HPUra.
Electron microscopy.
The Qdot streptavidin conjugate stock solution (1 μM) was centrifuged at 5,000 × g for 3 min, followed by dilution of the supernatant in water to 5, 10, 20, and 50 nM. A 1-μl aliquot of each dilution was incubated with 9 μl of biotinylated SPP1delX110lacO64 for 5 min at room temperature. The reaction was centrifuged at 5,000 × g for 3 min, followed by negative staining of the supernatant with uranyl acetate (44) and electron microscopy.
RESULTS
SPP1 particles bind preferentially to B. subtilis cell poles.
Fluorescently labeled bacteriophage SPP1delX110lacO64 particles were produced by chemical attachment of biotin, followed by conjugation with streptavidin-coated quantum dots (Qdots; Qdot655 streptavidin), an adaptation of the method of Edgar et al. (18) used for phages of Gram-negative bacteria. More than 80% of the labeled SPP1 virions contained one or several Qdots, as assessed by electron microscopy (data not shown). No detectable difference in titer was found between phages chemically modified and control phages from the same virus preparation diluted in parallel during the labeling reaction (not shown). Biotinylation and Qdot attachment thus had no effect on viral infectivity. Irreversible adsorption of SPP1delX110lacO64 to B. subtilis YB886 (kads = 0.52 ± 0.04 min−1 A600−1) was slower than SPP1 wild-type (kads = 0.85 ± 0.03 min−1 A600−1) (Table 2). Biotinylation of SPP1delX110lacO64 particles led to a reduction of the kads to 0.43 ± 0.03 min−1 A600−1 and subsequent fixation of the Qdot655 streptavidin conjugate to modified phages caused and an additional decrease of the kads to 0.32 ± 0.06 min−1 A600−1 (Table 2). The latter effect on adsorption is probably due to steric hindrance of Qdots bound to the phage tail tip region, which affects its interaction with bacterial receptors (41). A subpopulation of phage particles with Qdots bound to the tip region was indeed observed by electron microscopy (not shown). The experimental conditions were then optimized for visualization of Qdot-phages bound to bacteria and to compensate for the ∼2.7-fold reduction of the kads of Qdot-SPP1delX110lacO64 particles relative to SPP1 wild type (see Materials and Methods and below).
Table 2.
SPP1 irreversible adsorption constants (kads) to host bacteria
| Phage | Strain | Mean kads (min−1A600−1) ± SD |
|---|---|---|
| SPP1 | YB886 | 0.85 ± 0.03 |
| SPP1 | YB886.Δ6.PxylYB | 2.71 ± 0.54 |
| SPP1 | YB886.YBGFP | 0.74 ± 0.01 |
| SPP1 | YB886.Δ6.PxylYBGFP | 2.80 ± 0.48 |
| SPP1delX110lacO64 | YB886 | 0.52 ± 0.04 |
| SPP1delX110lacO64 + Qdot | YB886 | 0.49 ± 0.10 |
| SPP1delX110lacO64-biotin | YB886 | 0.43 ± 0.03 |
| SPP1delX110lacO64-biotin-Qdot | YB886 | 0.32 ± 0.06 |
In order to determine the topology of SPP1 binding to B. subtilis cells, the bacterial membrane was stained with the vital green dye FM1-43FX to define individual cells contour (Fig. 2 A, top panels), and fluorescent virions were added at an input multiplicity of 6 PFU/CFU. This ratio was optimized to yield a majority of cells with one red fluorescent virion bound per cell (Fig. 2A, bottom panels). Infected bacteria were fixed with glutaraldehyde at 2 min p.i., diluted 100-fold 2 min later, sedimented, and resuspended for visualization of virions stably bound to the cell envelope. Control experiments showed that streptavidin-Qdots alone did not attach to B. subtilis cells (data not shown). The distribution of Qdot-labeled phages was determined according to the relative distance to the cell poles (d1 [the distance to the closest cell end] divided by d2 [the cell length] in Fig. 2B). No correction was made for the total surface available for phage binding in different regions of the cell (e.g., between poles and the bacilli rod) due to cell size variability.
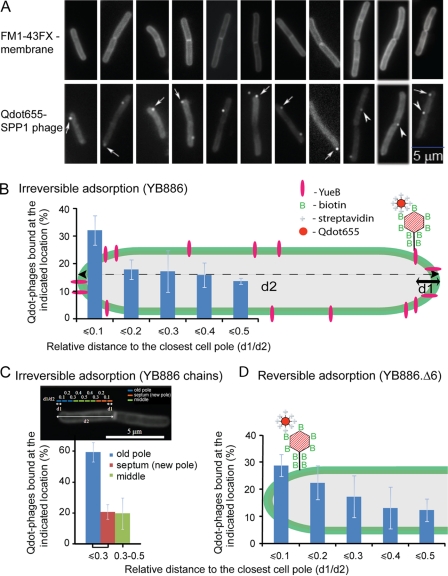
Fig. 2.
Localization of phage SPP1 particles on the surface of B. subtilis cells. (A) Snapshot gallery of wild-type B. subtilis YB886 membrane stained to define the cell contour (top row) and containing Qdot655-labeled SPP1delX110lacO64 phages (bottom row). Arrows and arrowheads show Qdot-phages at the cell poles and at septal regions, respectively. (B to D) Percentage of Qdot-labeled SPP1 particles at the indicated relative distance (d1/d2) to the closest cell pole in the wild-type strain YB886 (B), to the old cell pole or to the closed septum in chains of two or more YB886 bacteria (a subset of data from B) (C), and to the closest cell pole in the B. subtilis yueB deletion strain YB886.Δ6 (see Fig. 1A) (D). Infections were carried out at an input multiplicity of six phages per bacterium as described in Materials and Methods. The results are from three independent infection experiments in which a total number of 400 (B), 96 (C), and 125 (D) cells with a single Qdot phage were measured. Note that the lower number of counts for YB886.Δ6 is due to the lack of irreversible adsorption to these cells explaining why only 8% of the cells have a Qdot phage.
Infection of the B. subtilis wild-type strain YB886 yielded 35% of bacterial cells with bound Qdot-virions (n = 740, from three independent experiments), which localized preferentially at or close to the cell poles (d1/d2 ≤ 0.1, Fig. 2A and B). Analysis of a subset of cells growing in chains showed that binding to the old bacterial poles was strongly favored (Fig. 2C). Approximately 20% of SPP1 virions of this subset were found at closed septal regions corresponding to newly forming poles, while the remaining virions adsorbed to more central regions of the cell cylinder (Fig. 2C). We next visualized reversible binding alone by infecting the B. subtilis yueB-null mutant (strain YB886.Δ6). Only 8% of the cells lacking the SPP1 receptor YueB had Qdot-labeled phages bound (n = 767, from three independent experiments). The dilution step used in our experimental setup leads to dissociation of most of the reversibly bound phages (7). As in wild-type cells, Qdot-labeled phages preferentially bound at or near the cell poles (Fig. 2D). Phage binding gradually decreased from the poles toward mid-cell. The distributions shown in Fig. 2B and D were found to be similar in a Student t test (P of 0.997). However, this statistical analysis shall be taken cautiously due to the lower data sampling for phages bound to cells lacking YueB (Fig. 2D) compared to wild-type bacteria (Fig. 2B). Taken together, these findings suggested that both reversible and irreversible binding of SPP1 occur preferentially at the cell poles and thus that reversible adsorption may already target SPP1 for a polar distribution at the B. subtilis surface.
When a large input multiplicity (100 virions/cell) was used to infect wild-type B. subtilis cells, a few Qdot-labeled phages were found attached to the bacterial cells at 2 min p.i., again preferentially at their poles (Fig. 3, top row). In contrast, when cells overproducing YueB were infected, a large number of phages were found stably bound along the length of the bacterial cells, displaying a rather homogeneous punctate distribution on the cell surface (Fig. 3, bottom row). Since phage binding under our experimental conditions is essentially due to irreversible adsorption (see above), this further suggested that functional YueB available for interaction with SPP1 is present in limiting amounts at the surface of wild-type B. subtilis cells (7, 39).
Fig. 3.
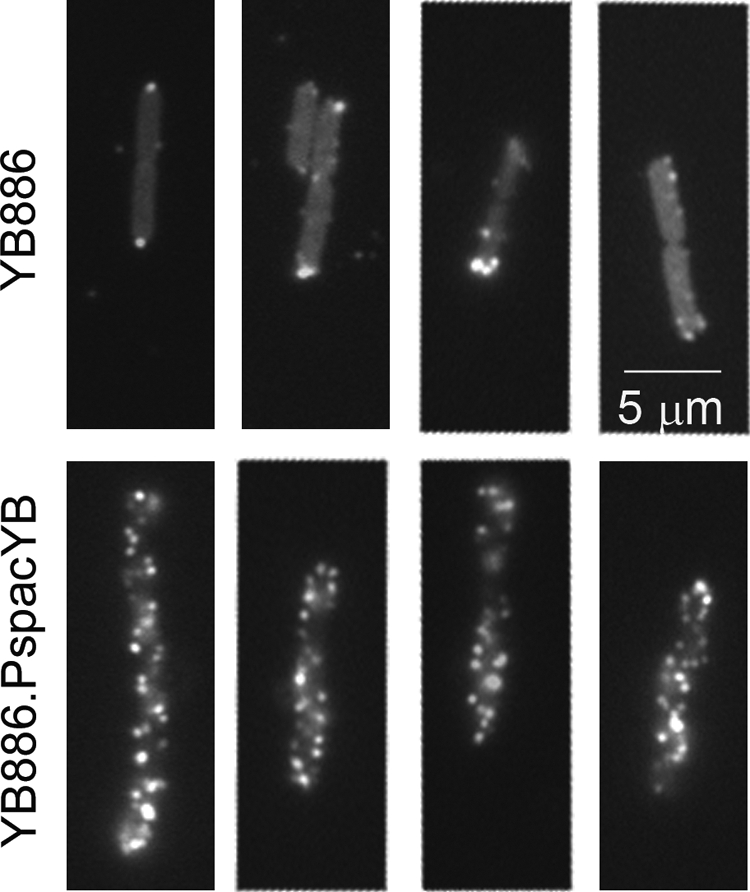
SPP1delX110lacO64 binding to B. subtilis cells at a high multiplicity of infection. Qdot-labeled phages (100 virions per bacterium) were incubated with B. subtilis YB886 (upper row) or with the YueB-overproducing strain YB886.PspacYB (bottom row) for 2 min, followed by fixation and dilution as described in Materials and Methods.
SPP1 receptor YueB exhibits a punctate pattern along the B. subtilis sidewalls and concentrates at the cell poles.
The polar localization of SPP1 particles at the B. subtilis surface might be imposed by the preferential reversible adsorption to bacterial poles (Fig. 2D) and/or result from the distribution of YueB in the bacterial envelope. YueB is an integral 120-kDa membrane protein found in low amounts in B. subtilis (Fig. 1B) (39). Its most abundant forms, as detected in extracts of wild-type B. subtilis cells by Western blotting with a polyclonal antibody raised against YueB780 (Fig. 1B) (41), are truncated polypeptides found associated with membranes after cell fractionation (39) (Fig. 1C). These forms did not result from sample degradation (proteolysis) since the same pattern of bands was found when cell extracts were taken directly into trichloroacetic acid to inactivate proteases (23; data not shown). These observations suggested that YueB is prone to proteolysis in vivo or at least immediately after bacterial death, originating the truncated forms observed in Fig. 1C. The dominant species in Western blots corresponded to a doublet of bands (only resolved in long runs of 8% polyacrylamide gels) with apparent masses of ∼65 kDa. A less intense band was observed at ∼70 kDa, and a faint band that likely corresponds to full-length YueB had the electrophoretic mobility expected for a 145-kDa protein (Fig. 1C). This abnormal migration behavior of YueB, whose subunit mass is 120 kDa (Fig. 1B), was attributed to its fiber shape (41). All of these species were absent in the ΔyueB strain YB886.Δ6 (Fig. 1C). In cells bearing a green fluorescent protein (GFP) fusion to the carboxyl terminus of YueB (Fig. 1), the immunodominant ∼65-kDa forms disappeared, and an ∼90-kDa band, which might correspond to a nonresolved doublet, appeared instead. This indicated that the ∼65-kDa species were truncated products of the YueB carboxyl terminus. Western blots developed with anti-GFP antibodies confirmed this assignment (Fig. 1D). Stronger signals for the same immunoreactive bands were obtained with YueB- and YueB-GFP-overproducing strains (Fig. 1C and D, right panels). Note that 5-fold less total protein was loaded onto these Western blots relative to those shown on the left panels of the figures. The region of YueB that binds to SPP1 is not known, and no function was assigned to the individual truncated forms of YueB that are likely present in the cell envelope (Fig. 1C and D).
We thus investigated both the localization of the cytoplasmic YueB-GFP fusion in live cells and the surface (extracellular) distribution of YueB by immunofluorescence microscopy (IFM) in nonpermeabilized cells to assess the SPP1 receptor topology. The efficiency of SPP1 plating in cells producing YueB-GFP was similar to that found for bacteria producing equivalent amounts of YueB in a titration plate assay (data not shown). Adsorption was only slightly reduced in the strain expressing the yueB-gfp fusion under the control of the native Pyuk promoter (Fig. 1A) relative to the control wild-type strain YB886, while no difference in kads was found between strains overproducing YueB and YueB-GFP (Table 2). We concluded that fusion of GFP to the carboxyl terminus of YueB did not affect receptor activity. Surface localization of YueB by IFM was performed using polyclonal antibodies raised against the extracellular YueB780 fiber region (Fig. 1B) (41). Preincubation of B. subtilis cells with anti-YueB780 serum reduced significantly the adsorption constant of SPP1 to bacteria (41), showing that the antibodies recognized regions of YueB at the cell surface that are necessary for its SPP1 receptor activity.
Expression of yueB driven from its native promoter as the single gene copy coding for YueB in the cell led to a relatively weak surface labeling in IFM of fixed cells (Fig. 4 A). A significant proportion of cells showed a peripheral spotted pattern along the cell cylinder and a strong fluorescence signal at the cell poles (Fig. 4A). A weak fluorescence signal displaying a similar distribution was also obtained for the YueB-GFP fusion whose gene was expressed under the control of the Pyuk native promoter (Fig. 4B). Membrane staining defining the contour of individual cells in bacterial chains revealed accumulation of YueB-GFP at old cell poles. The pattern of fluorescent protein distribution was particularly prominent when images were deconvolved (Fig. 4B, right panels). Although the fluorescence signals in both cases were weak, which is consistent with the low endogenous levels of YueB (39), the similarity of the localization pattern of YueB-GFP and extracellular YueB (IFM experiments) lent support to a discrete localization of YueB along the bacterium sidewalls and its accumulation at the old cell poles. It also indicated that the surface distribution of the YueB ectodomain is similar to that displayed by the transmembrane segments and the cytoplasmic tail at its carboxyl terminus.
Ectopic overexpression of yueB-gfp under the control of a xylose-inducible promoter (Fig. 1A and C) gave a more distinct and intense fluorescence signal in the ensemble of the bacterial population (Fig. 4C). The increased signal-to-noise ratio made the distribution of YueB-GFP along the sidewalls clearly visible, revealing a punctate/banded pattern and further confirming the enrichment of YueB at cell poles (Fig. 4C). Again, visualization of the signal in chains of cells showed that accumulation occurred at the old cell poles in particular. Colocalization of the surface labeling of YueB by IFM and of the YueB-GFP fusion (Fig. 5) suggested that the ectodomain and the cytoplasmic carboxyl terminus of the protein (despite the presence of several truncated forms, Fig. 1C) are physically associated. Integrity of the YueB dimer (41) might be maintained by inter- and intrasubunit interactions between the truncated polypeptides within the YueB quaternary structure explaining the observed similar distribution in the cell envelope.
Fig. 5.
Colocalization of YueB on the cytoplasmic side of the membrane and at the cell surface in B. subtilis YB886.Δ6.PxylYBGFP (see Fig. 1A) using a GFP fusion to the YueB cytoplasmic region (B, D, and E) and immunostaining with anti-YueB antibodies (surface labeling) (C to E), respectively. Overlays of the images are shown in panels D and E (deconvolved), where YueB-GFP is in green and anti-YueB antibodies are in red.
SPP1 DNA enters preferentially close to the bacterial poles and replicates in defined foci.
The preferential binding of SPP1 phages to the B. subtilis cell poles and the enrichment of the YueB receptor at these sites suggested that phage DNA entry might also be polar. In order to visualize SPP1 DNA entry, we cloned an array of lacO repeats at the unique SphI site of the bacteriophage SPP1delX110 genome. The repeat of lacO (21-bp) operators used was spaced by 10-bp random sequences to prevent genetic instability (27). SPP1delX110, which encapsidates viral chromosomes of wild-type length (45), carries a 3,417-bp delX deletion (14) within a nonessential region of its genome. Insertion of the lacO repeat array in a dispensable region of the phage genome combined with the delX deletion lead to no net increase in size of the genome and resulted in viable phages (SPP1delX110lacO64). The repetitive segment of DNA was maintained stably in the SPP1 genome and had no deleterious effect in phage multiplication (data not shown). The phage irreversible adsorption constant to host cells was reduced to 60% compared to SPP1 wild type (Table 2). Titration of SPP1delX110lacO64 with B. subtilis cells producing a LacI-CFP soluble fusion (strain GSY10000) was carried out in the presence or in the absence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The aim was to assess whether binding of multiple copies of LacI-CFP to the lacO tandem repeats in the SPP1 genome affects viral DNA replication or encapsidation into viral capsids. The titer obtained was identical (data not shown), revealing that the local clustering of LacI-CFP in the SPP1 DNA does not impair these processes.
Time-lapse observations of SPP1delX110lacO64 infecting strain GSY10000 were carried out according to the setup schematized in Fig. 6 A. This experiment showed the appearance of intracellular fluorescent foci upon infection (Fig. 6B). Their size increased with time, which correlated with an increase of their fluorescence intensity and a reduction in the cell background fluorescence (i.e., increased signal-to-noise ratio) (Fig. 6B and 7B). This presumably resulted from the clustering of the LacI-CFP fusion on phage DNA undergoing replication, which initiates at ∼3 min p.i. at 37°C (see references 12 and 30 and references therein). Consistently, no increase in phage DNA focus fluorescence was found in infected cells treated with HPUra (white arrows in Fig. 6C), which inhibits bacterial DNA polymerase III, the host enzyme that replicates the SPP1 DNA (36). In some cells, the SPP1 replication centers disappeared with time, possibly due to abortive phage multiplication processes (arrowhead in Fig. 6B). Taken together, these observations indicated that SPP1 genome replication initiates and proceeds at defined foci (“replication factories”).
Fig. 7.
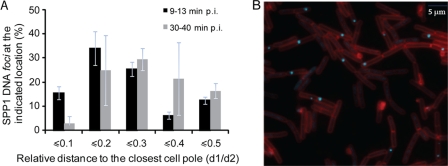
Localization of phage SPP1delX110lacO64 DNA in B. subtilis-infected cells. (A) Percentage of SPP1 DNA spots at the indicated distance relative (d1/d2) to the closest cell pole (old cell pole or closed new septum) on the bacterial cell, as observed by fluorescence microscopy (B). Measurements from three independent infections were carried out as schematized in Fig. 2B at early (▪; 200 cells with a single SPP1 DNA spot were measured from images taken between 9 and 13 min p.i.) and late ( ; 200 cells with a single SPP1 DNA spot were measured from images taken between 30 and 40 min p.i.) time points after infection. (B) Cells producing LacI-CFP (cyan) were infected by phage SPP1delX110lacO64 (input multiplicity of 3) and imaged at a late time point after infection (between 30 and 40 min p.i.). The cell outline was visualized with the vital membrane stain FM4-64FX (red).
; 200 cells with a single SPP1 DNA spot were measured from images taken between 30 and 40 min p.i.) time points after infection. (B) Cells producing LacI-CFP (cyan) were infected by phage SPP1delX110lacO64 (input multiplicity of 3) and imaged at a late time point after infection (between 30 and 40 min p.i.). The cell outline was visualized with the vital membrane stain FM4-64FX (red).
Quantitative analysis of the spatial distribution of SPP1 replication foci over time showed that they preferentially localized close to the cell poles, with a wide distribution centered at the pole-cylinder junction (d1/d2 ∼ 0.1 to 0.3) at early infection times (Fig. 7 A, black bars). This distribution was shifted toward a more central position in the cell as infection progressed (Fig. 7A, gray bars, and Fig. 7B). In chains of cells, a higher number of foci appeared to be present close to old cell poles relative to septal regions and to the middle cell at early times postinfection (data not shown). An accurate quantification was hampered by limited sampling of the small size and weakly fluorescent foci. Measurement of the lengths of individual cells (infected and noninfected) at different time points showed that cell growth was not affected by SPP1 infection (data not shown) and that cells kept growing on the microscope slide during the time-lapse experiments (see the elongation of the cells shown in Fig. 6B over time). However, we could not distinguish whether the increase in distance between foci and cell poles over time is a passive process due to cell elongation or whether it results from the active movement of the foci.
DISCUSSION
Virus binding and entry in the host cell are the critical steps for the initiation of infection. The results presented here show that during infection of the Gram-positive B. subtilis bacterium by bacteriophage SPP1 these events occur preferentially at or close to the old cell pole (Fig. 2) which correlates with the position of viral DNA entry and initiation of replication (Fig. 7).
Reversible adsorption, which targets glucosylated poly(glycerol phosphate) WTA, is independent of the SPP1 protein receptor YueB (7) but occurred also preferentially near the cell poles (Fig. 2D). This suggested a particular distribution of WTA, or at least of a particular (glucosylated) form of these polymers recognized by SPP1, at the cell surface. Proteins involved in several steps of WTA synthesis in B. subtilis have been reported to localize to division sites and, to a greater or lesser extent, along the lateral sides of the cells in a helix-like pattern but were particularly absent from polar regions (19). Since the putative WTA transporter TagGH was virtually absent from old cell poles, it was suggested that the export and hence the incorporation of WTA into the cross-walls occurs during division (septum formation) and is absent after cell separation and formation of the cell poles. This was consistent with the slower turnover of WTA at the cell poles relative to the cell cylinder (5, 15, 21, 26, 31). The resulting different properties of cell poles and their highly electronegative character suitable for cation ligation (43) might favor SPP1 binding. Alternatively (or simultaneously), if WTA remain at the old poles for several generation times, this might favor the modification (glucosylation) of the WTA, allowing SPP1 recognition at these sites.
Reversible binding at the cell pole may then facilitate virus particle search for its receptor YueB, which was found to localize predominantly at the cell poles too (Fig. 4 and 5). This can be of significant importance because in the absence of glucosylated WTA the time required for SPP1 to recognize and bind YueB increases drastically (7), probably due to the low abundance of this protein in the cell surface (Fig. 4). Consistently, the kinetic defect can be compensated by overproduction of YueB (7). The ensemble of the data suggests that SPP1 reversible binding to glucosylated WTA at the old poles provides a primary attachment and favorable positioning of SPP1 at the B. subtilis surface for rapid interaction with its low-abundance receptor YueB.
The localization of endogenous YueB by IFM (Fig. 4A) and of a functional YueB-GFP fusion expressed under the control of the native yueB promoter as the only copy of YueB in the cell (Fig. 4B) confirmed that the SPP1 receptor is produced in low amounts in wild-type bacteria and showed that it concentrates primarily at the old cell pole, although some fluorescence was also detected in the cell cylinder (Fig. 4B). Overproduction of YueB confirmed its polar localization and revealed an uneven distribution following a spotted, possibly helix-like, pattern along the cell length (Fig. 4 and 5). This organization was found for both the YueB cytoplasmic region fused to GFP and for the receptor region exposed to the cell surface that is accessible to antibody labeling (Fig. 5). Higher receptor density correlated with a marked increase in phage binding along the sidewalls (Fig. 3, bottom lane). We hypothesize that the localization along the cell cylinder of the integral membrane protein YueB, whose ectodomain spans across the entire B. subtilis cell wall, results from the mechanism of cell wall growth and structure. Indeed, insertion of new peptidoglycan along the sidewalls of in B. subtilis has been shown to follow a banded, helical pattern (16, 46), and a recent study using atomic force microscopy showed that the sacculus maintains a coiled-coil cabling architecture (24). We suggest that the large YueB ectodomain is found within the cell wall polymers and that YueB polar accumulation is facilitated by the low turnover of the polar caps of the bacterium.
YueB might be part of or associate with a putative T7SS machinery whose components are co-encoded by the yuk operon (39). The presence of YueB-related proteins is one of the features that distinguishes the T7SS of Firmicutes (type VIIb subfamily [1]). The subcellular localization of YueB in B. subtilis cells (Fig. 4 and 5) and that of one T7SS component in mycobacteria (13) suggest that this secretion apparatus might be preferentially located at cell poles, like other complex machineries such as the DNA uptake system during transformation (22), chemotaxis proteins complexes and components of the Tat secretion system (11, 29). The effective presence of a T7SS in B. subtilis, as well as the physiological role of YueB, remains to be elucidated. Since other components of the yuk operon are not essential for bacteriophage SPP1 infection (39), we conclude that SPP1 uses the YueB region exposed to the bacterial cell surface for specific docking but that the putative T7SS secretion machinery is not essential for the delivery of phage DNA to the bacterial cytoplasm.
We engineered the SPP1 genome with a set of lacO repeats to follow it in infected B. subtilis cells producing LacI-CFP. Interestingly, the phage DNA was found to localize in foci close to the cell poles early in infection (Fig. 7), which correlated with the preferential polar adsorption of SPP1 (Fig. 2). The majority of phage DNA foci were not at the tip of the poles but in the pole-cylinder junction region, suggesting that SPP1 might take advantage of particular features of this region of the cell wall for easier penetration toward the bacterial membrane to deliver its genome to the bacterial cytoplasm. A single, well-individualized fluorescent focus was found in individual bacteria infected at low input multiplicity (Fig. 7B). The increase in the fluorescence of each SPP1 DNA focus over time suggested that genome replication occurs at an intracellular location defined by the position of its entry (Fig. 6). This might involve an initial association of phage DNA to the membrane for its replication as previously suggested (12). It is interesting that plasmid pHP13 replisomes can also cluster in foci located predominantly close to the B. subtilis poles (49). In B. subtilis, the efficiency of phages SPP1 and φ29 genome replication was recently shown to be affected in mreB cytoskeleton mutants (33). The role of MreB was not established for SPP1, and we have shown that its genome replication occurs at a single focus. In contrast, φ29 DNA (and components of the φ29 replication machinery) localized in peripheral helix-like structures that were cytoskeleton dependent, and it was concluded that the MreB cytoskeleton organizes phage φ29 DNA replication at the membrane (32, 33). φ29 uses a terminal protein covalently attached to its linear double-stranded DNA molecule that is the primer for the initiation of phage DNA replication, whereas phage SPP1 replicates its DNA initially via the theta mode and later via a rolling-circle mode (4). Thus, organization of viral DNA replication by the bacterial MreB cytoskeleton, in multiple sites along the periphery of infected cells, might be specific to phages by using the protein-primed mechanism of DNA replication, while the replisomes of phages such as SPP1 and lambda remain confined to a single replication factory (18; the present study).
Edgar et al. (18) showed that different phages of Gram-negative bacteria adsorb selectively to cell poles and that coliphage lambda DNA entry targets a number of cell components at this subcellular localization. We showed here that the pole and possibly the pole-cylinder junction are also a preferential target for viral DNA delivery across the Gram-positive envelope. Phage SPP1 likely exploits particular features of WTA in this region for preferential adsorption to the bacterium and uses subsequently the locally enriched surface protein YueB for irreversible binding to the host. Assembly of a replication focus nearby the polar DNA entry region may spatially constrain the virus genome for optimal functioning of its replication factory. These findings provide further support to the growing evidence that phages take advantage of the host cell architecture for efficient spatial coordination of the sequential events occurring during infection.
ACKNOWLEDGMENTS
We thank François-Xavier Barre (CGM, Gif-sur-Yvette, France) for plasmid pFX276, the primers 124-2 and 125-2, and useful discussions. Alan D. Grossman and Cathy Lee (MIT, Cambridge, MA) are acknowledged for strain MMB357. HPUra was kindly provided by Neal C. Brown (GLSynthesis, Worcester, MA). We are indebted to Rudi Lurz (MPI-MG, Berlin, Germany) for electron microscopy observation of phages labeling with Qdots. We thank Adriano O. Henriques and Gonçalo Real (ITQB, Oeiras, Portugal) for sharing initial microscopy observations of YueB-GFP, the gift of pEA18, and useful discussions. We thank F.-X. Barre and A. O. Henriques for critical reading of the manuscript.
This study benefited from the facilities and expertise of the platform of the Microscopie Photonique of IMAGIF (Centre de Recherche de Gif). L.J. was supported by the University of Vilnius, the Université Paris-Sud, the French embassy in Lithuania, the program Egide Gilibert between France and Lithuania, the European Union Erasmus Program, and the Région Ile-de-France (SETCI). C.B. was supported by doctoral (SFRH/BD/19675/2004) and postdoctoral (SFRH/BPD/63392/2009) fellowships from the Fundação para a Ciência e a Tecnologia (FCT) of Portugal. This research was funded by the CNRS, INRA, IFR115, the ANR (grant ANR-08-JCJC-0024-01 to R.C.-L.) and the FCT (grant PTDC/BIA-MIC/66412/2006 to C.S.-J.).
Footnotes
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Abdallah A. M., et al. 2007. Type VII secretion: mycobacteria show the way. Nat. Rev. Microbiol. 5:883–891 [DOI] [PubMed] [Google Scholar]
- 2. Ackermann H.-W. 2009. Phage classification and characterization. Methods Mol. Biol. 501:127–140 [DOI] [PubMed] [Google Scholar]
- 3. Alonso J. C., et al. 1997. The complete nucleotide sequence and functional organization of Bacillus subtilis bacteriophage SPP1. Gene 204:201–212 [DOI] [PubMed] [Google Scholar]
- 4. Alonso J. C., Tavares P., Lurz R., Trautner T. A. 2006. Bacteriophage SPP1, p. 331–349 In Calendar R. (ed.), The bacteriophages. Oxford University Press, New York, NY [Google Scholar]
- 5. Archibald A. R., Coapes H. E. 1976. Bacteriophage SP50 as a marker for cell wall growth in Bacillus subtilis. J. Bacteriol. 125:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baptista C. 2009. Recognition of Bacillus subtilis receptors by bacteriophage SPP1: a genetic and biochemical approach. Ph.D. thesis Faculdade de Ciências da Universidade de Lisboa, Lisbon, Portugal [Google Scholar]
- 7. Baptista C., Santos M. A., São-José C. 2008. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J. Bacteriol. 190:4989–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benson A. K., Haldenwang W. G. 1993. Regulation of sigma B levels and activity in Bacillus subtilis. J. Bacteriol. 175:2347–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Biswal N., Kleinschmidt A. K., Spatz H. C., Trautner T. A. 1967. Physical properties of the DNA of bacteriophage SP50. Mol. Gen. Genet. 100:39–55 [DOI] [PubMed] [Google Scholar]
- 10. Boylan R. J., Mendelson N. H., Brooks D., Young F. E. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J. Bacteriol. 110:281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buist G., Ridder A. N., Kok J., Kulpers O. P. 2006. Different subcellular locations of secretome components of Gram-positive bacteria. Microbiology 152:2867–2874 [DOI] [PubMed] [Google Scholar]
- 12. Burger K. J. 1980. Membrane binding of bacteriophage SPP1 DNA. Mol. Gen. Genet. 179:373–376 [DOI] [PubMed] [Google Scholar]
- 13. Carlsson F., Joshi S. A., Rangell L., Brown E. J. 2009. Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog. 5:e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chai S., Szepan U., Lüder G., Trautner T. A., Alonso J. C. 1993. Sequence analysis of the left end of the Bacillus subtilis bacteriophage SPP1 genome. Gene 129:41–49 [DOI] [PubMed] [Google Scholar]
- 15. Clarke-Sturman A. J., et al. 1989. Cell wall assembly in Bacillus subtilis: partial conservation of polar wall material and the effect of growth conditions on the pattern of incorporation of new material at the polar caps. J. Gen. Microbiol. 135:657–665 [DOI] [PubMed] [Google Scholar]
- 16. Daniel R. A., Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776 [DOI] [PubMed] [Google Scholar]
- 17. Dröge A., Tavares P. 2000. In vitro packaging of DNA of the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol. 296:103–115 [DOI] [PubMed] [Google Scholar]
- 18. Edgar R., et al. 2008. Bacteriophage infection is targeted to cellular poles. Mol. Microbiol. 68:1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Formstone A., Carballido-López R., Noirot P., Errington J., Scheffers D.-J. 2008. Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis. J. Bacteriol. 190:1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gibbs K. A., et al. 2004. Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol. Microbiol. 53:1771–1783 [DOI] [PubMed] [Google Scholar]
- 21. Graham L. L., Beveridge T. J. 1994. Structural differentiation of the Bacillus subtilis 168 cell wall. J. Bacteriol. 176:1413–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hahn J., Maier B., Haijema B. J., Sheetz M., Dubnau D. 2005. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell 122:59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayashi S., Wu H. C. 1983. Biosynthesis of Bacillus licheniformis penicillinase in Escherichia coli and in Bacillus subtilis. J. Bacteriol. 156:773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayhurst E. J., Kailas L., Hobbs J. K., Foster S. J. 2008. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 105:14603–14608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hendrix R. W., Smith M. C. M., Burns R. N., Ford M. E., Hatfull G. F. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96:2192–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirchner G., Kemper M. A., Koch A. L., Doyle R. J. 1988. Zonal turnover of cell poles of Bacillus subtilis.. Ann. Inst. Pasteur Microbiol. 139:645–654 [DOI] [PubMed] [Google Scholar]
- 27. Lau I. F., et al. 2003. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 49:731–743 [DOI] [PubMed] [Google Scholar]
- 28. Lemon K. P., Grossman A. D. 2000. Movement of replicating DNA through a stationary replisome. Mol. Cell 6:1321–1330 [DOI] [PubMed] [Google Scholar]
- 29. Meile J.-C., Wu L. J., Ehrlich S. D., Errington J., Noirot P. 2006. Systematic localization of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory. Proteomics 6:2135–2146 [DOI] [PubMed] [Google Scholar]
- 30. Missich R., et al. 1997. The replisome organizer (G38P) of Bacillus subtilis bacteriophage SPP1 forms specialized nucleoprotein complexes with two discrete distant regions of the SPP1 genome. J. Mol. Biol. 270:50–64 [DOI] [PubMed] [Google Scholar]
- 31. Mobley H. L. T., Koch A. L., Doyle R. J., Streips U. N. 1984. Insertion and fate of the cell wall in Bacillus subtilis. J. Bacteriol. 158:169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muñoz-Espín D., Holguera I., Ballesteros-Plaza D., Carballido-López R., Salas M. 2010. Viral terminal protein directs early organization of phage DNA replication at the bacterial nucleoid. Proc. Natl. Acad. Sci. U. S. A. 107:16548–16553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muñoz-Espín D., et al. 2009. The actin-like MreB cytoskeleton organizes viral DNA replication in bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:13347–13352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Plisson C., et al. 2007. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 26:3720–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quisel J. D., Lin D. C., Grossman A. D. 1999. Control of development by altered localization of a transcription factor in B. subtilis. Mol. Cell 4:665–672 [DOI] [PubMed] [Google Scholar]
- 36. Rowley S. D., Brown N. C. 1977. Bacillus subtilis DNA polymerase III is required for the replication of DNA of bacteriophages SPP-1 and Ø105. J. Virol. 21:493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Santos M. A., De Lencastre H., Archer L. J. 1984. Homology between phages SPP1, 41c, 22a, ρ15, and SF6 of Bacillus subtilis. J. Gen. Virol. 65:2067–2072 [DOI] [PubMed] [Google Scholar]
- 39. São-José C., Baptista C., Santos M. A. 2004. Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J. Bacteriol. 186:8337–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. São-José C., De Frutos M., Raspaud E., Santos M. A., Tavares P. 2007. Pressure built by DNA packing inside virions: enough to drive DNA ejection in vitro, largely insufficient for delivery into the bacterial cytoplasm. J. Mol. Biol. 374:346–355 [DOI] [PubMed] [Google Scholar]
- 41. São-José C., et al. 2006. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J. Biol. Chem. 281:11464–11470 [DOI] [PubMed] [Google Scholar]
- 42. Sciara G., et al. 2010. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. U. S. A. 107:6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sonnenfeld E. M., Beveridge T. J., Doyle R. J. 1985. Discontinuity of charge on cell wall poles of Bacillus subtilis. Can. J. Microbiol. 31:875–877 [DOI] [PubMed] [Google Scholar]
- 44. Steven A. C., et al. 1988. Molecular substructure of a viral receptor-recognition protein: the gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 20:351–365 [DOI] [PubMed] [Google Scholar]
- 45. Tavares P., et al. 1992. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J. Mol. Biol. 225:81–92 [DOI] [PubMed] [Google Scholar]
- 46. Tiyanont K., et al. 2006. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. U. S. A. 103:11033–11038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Veesler D., et al. 2010. Crystal structure of bacteriophage SPP1 distal tail protein (gp19.1): a baseplate hub paradigm in gram-positive infecting phages. J. Biol. Chem. 285:36666–36673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vinga I., São-José C., Tavares P., Santos M. A. 2006. Bacteriophage entry in the host cell, p. 165–203 In Wegrzyn G. (ed.), Modern bacteriophage biology and biotechnology. Research Signpost, Kerala, India [Google Scholar]
- 49. Wang J. D., Rokop M. E., Barker M. M., Hanson N. R., Grossman A. D. 2004. Multicopy plasmids affect replisome positioning in Bacillus subtilis. J. Bacteriol. 186:7084–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yasbin R. E., Young F. E. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J. Virol. 14:1343–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yasbin R. E., Fields P. I., Andersen B. J. 1980. Properties of Bacillus subtilis 168 derivatives freed on their natural prophages. Gene 12:155–159 [DOI] [PubMed] [Google Scholar]