Abstract
NADPH:2-ketopropyl-coenzyme M oxidoreductase/carboxylase (2-KPCC), an atypical member of the disulfide oxidoreductase (DSOR) family of enzymes, catalyzes the reductive cleavage and carboxylation of 2-ketopropyl-coenzyme M [2-(2-ketopropylthio)ethanesulfonate; 2-KPC] to form acetoacetate and coenzyme M (CoM) in the bacterial pathway of propylene metabolism. Structural studies of 2-KPCC from Xanthobacter autotrophicus strain Py2 have revealed a distinctive active-site architecture that includes a putative catalytic triad consisting of two histidine residues that are hydrogen bonded to an ordered water molecule proposed to stabilize enolacetone formed from dithiol-mediated 2-KPC thioether bond cleavage. Site-directed mutants of 2-KPCC were constructed to test the tenets of the mechanism proposed from studies of the native enzyme. Mutagenesis of the interchange thiol of 2-KPCC (C82A) abolished all redox-dependent reactions of 2-KPCC (2-KPC carboxylation or protonation). The air-oxidized C82A mutant, as well as wild-type 2-KPCC, exhibited the characteristic charge transfer absorbance seen in site-directed variants of other DSOR enzymes but with a pKa value for C87 (8.8) four units higher (i.e., four orders of magnitude less acidic) than that for the flavin thiol of canonical DSOR enzymes. The same higher pKa value was observed in native 2-KPCC when the interchange thiol was alkylated by the CoM analog 2-bromoethanesulfonate. Mutagenesis of the flavin thiol (C87A) also resulted in an inactive enzyme for steady-state redox-dependent reactions, but this variant catalyzed a single-turnover reaction producing a 0.8:1 ratio of product to enzyme. Mutagenesis of the histidine proximal to the ordered water (H137A) led to nearly complete loss of redox-dependent 2-KPCC reactions, while mutagenesis of the distal histidine (H84A) reduced these activities by 58 to 76%. A redox-independent reaction of 2-KPCC (acetoacetate decarboxylation) was not decreased for any of the aforementioned site-directed mutants. We interpreted and rationalized these results in terms of a mechanism of catalysis for 2-KPCC employing a unique hydrophobic active-site architecture promoting thioether bond cleavage and enolacetone formation not seen for other DSOR enzymes.
INTRODUCTION
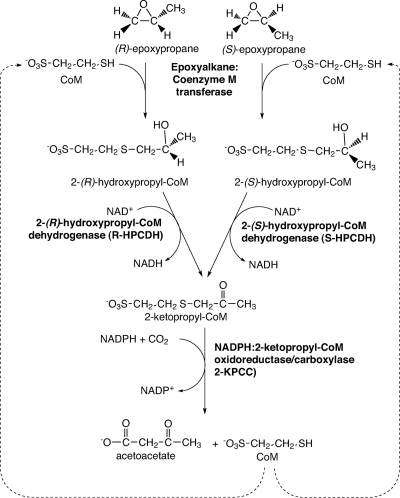
The bacterial metabolism of gaseous propylene by the proteobacterium Xanthobacter autotrophicus Py2 and the actinomycete Rhodococcus rhodochrous B276 is initiated by the insertion of a single oxygen atom into the olefin bond of propylene, forming (R)- and (S)-epoxypropane (19, 27, 35, 39). These epoxypropane enantiomers are further metabolized by a three-step linear pathway that uses four enzymes and the atypical cofactor coenzyme M (CoM) (2-mercaptoethanesulfonic acid) to catalyze the net carboxylation of epoxypropane to form acetoacetate as shown in Fig. 1 (1, 2, 4, 5, 18, 24). NADPH:2-ketopropyl-coenzyme M oxidoreductase/carboxylase (2-KPCC) is the CO2-fixing enzyme of this pathway, catalyzing the reductive cleavage and carboxylation of 2-ketopropyl-CoM (2-KPC) with the stoichiometry shown in equation 1 (1, 17, 18):
| (1) |
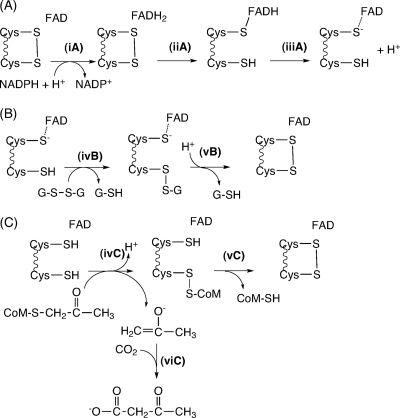
2-KPCC is the only known carboxylase that is a member of the disulfide oxidoreductase (DSOR) family of enzymes and, accordingly, employs a mechanistic strategy for organic substrate carboxylation not seen in any other known enzyme (10, 18). All members of the DSOR family follow the general mechanistic strategy shown in Fig. 2 A for the steps leading to the reduction of a cysteine disulfide bond, where the cysteine residues proximal and distal to flavin adenine dinucleotide (FAD) are termed the flavin and interchange thiols, respectively (29). For the DSOR enzymes catalyzing the reduction of an oxidized substrate containing a disulfide bond such as glutathione reductase, the interchange thiol attacks one of the sulfur atoms of the disulfide, leading to disulfide bond cleavage, reduction of one thiol of the substrate, and formation of a mixed disulfide between the interchange thiol and the second substrate thiol (29) (Fig. 2B). Reformation of the oxidized cysteine pair leads to the reduction and release of the second substrate thiol.
Fig. 1.
Pathway of aliphatic epoxide carboxylation in Xanthobacter autotrophicus Py2 and Rhodococcus rhodochrous B276.
Fig. 2.
Reactions of members of the DSOR family. (A) Steps resulting in the reduction of the redox-active cysteine disulfide for all DSOR enzymes; (B) steps resulting in reduction of a substrate with an oxidized disulfide bond, as illustrated for glutathione reductase; (C) steps resulting in reduction of the thioether bond of 2-KPC in 2-KPCC, resulting in the production of enolacetone, which undergoes carboxylation to form acetoacetate. Note that the reduced flavin cysteine is shown in the thiol rather than as the thiolate form for 2-KPCC for reasons described in the paper.
As shown in Fig. 2C, 2-KPCC catalyzes the reduction of a thioether rather than a disulfide bond, a feature not seen in any other known DSOR enzyme. Mechanistic (10) and structural (28, 30, 31) studies have provided evidence for a reaction mechanism where thioether bond cleavage results in the formation of a mixed disulfide between CoM and the interchange thiol with the formation of the enolacetone anion (Fig. 2C). The enolacetone anion then undergoes carboxylation to form the product acetoacetate, while the formation of the oxidized cysteine pair results in the release of free CoM (Fig. 2C).
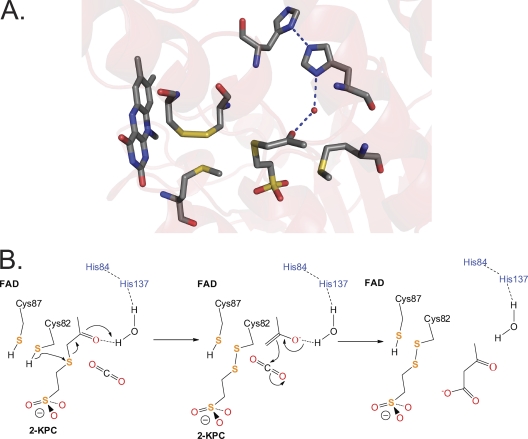
It is of interest to determine the unique features of 2-KPCC relative to other DSOR enzymes that allow the enzyme to catalyze the unique reactions of thioether bond cleavage, enolacetone anion formation, stabilization, and carboxylation. Significant insights into these features have come from X-ray crystallographic structures of 2-KPCC determined for various states, including the substrate-free enzyme (28), a 2-KPC-bound form of the enzyme (28), a form of the enzyme where the mixed disulfide of CoM was trapped (31), and a form in which the substrate CO2 is bound (30). Collectively, the structural characterization of 2-KPCC revealed an active-site architecture unique among DSOR enzymes, where substrate binding induces a conformational change that creates a hydrophobic pocket that encapsulates the substrate 2-KPC (22, 28). Of relevance to the question of how 2-KPCC stabilizes the enolacetone anion for subsequent carboxylation is that a hydrogen-bonding network was identified in the 2-KPC-bound enzyme, consisting of an ordered water molecule hydrogen bonded to both the carbonyl oxygen of 2-KPC and two histidine residues (H137 and H84) (Fig. 3 A). It has been proposed that this hydrogen-bonding network is responsible for stabilizing enolacetone formed from thioether bond cleavage, as the interchange thiol attacks the thioether bond, as shown in Fig. 3B (22, 28). An additional novel feature of the active site of 2-KPCC is a pair of methionine residues (M140 and M361), which flank the substrate 2-KPC. These methionine residues are not present in other DSOR enzymes and may contribute to the unique environment that promotes enolacetone stabilization and attack on CO2.
Fig. 3.
Active-site architecture and proposed mechanism for 2-KPCC. (A) Structure of 2-KPC bound to 2-KPCC, highlighting active-site residues believed to be key to catalysis (Protein Data Bank ID, 1MO9). (B) Proposed mechanism of thioether bond cleavage, enolacetone formation and stabilization, and carboxylation based on the structures solved for 2-KPCC and the results of the present work. The initial abstraction of a proton from C82 may be facilitated by a general base that has not yet been identified. The reduction of the mixed disulfide of CoM and Cys82 is not shown but will occur as for glutathione reductase (Fig. 2B).
The construction of site-directed substitutions of 2-KPCC would allow the tenets of this proposed mechanism to be tested directly. To date, however, all attempts to express 2-KPCC in an active state in a heterologous system have failed, in spite of the relative ease with which the other enzymes of the epoxypropane carboxylation pathway have been expressed and purified (11, 24, 34). In the present paper, we describe an expression system that solves the problems encountered previously and allows 2-KPCC to be expressed in a fully active and soluble state. Site-directed mutants were constructed in the redox-active disulfide, the histidine dyad, and one of the flanking methionine residues, and the effects of these mutations on various reactions catalyzed by 2-KPCC were determined in order to gain further insights into their catalytic roles.
MATERIALS AND METHODS
Materials.
Commercially available compounds used were of analytical grade and were purchased from either Sigma-Aldrich Chemicals or Fisher Scientific. 2-KPC was synthesized as described previously (1). All oligonucleotides were purchased from Integrated DNA Technologies.
Purification of native 2-KPCC from Xanthobacter autotrophicus strain Py2.
Native 2-KPCC was purified from propylene-grown X. autotrophicus Py2 as described previously (3).
Plasmid construction of recombinant 2-KPCC.
Total genomic DNA was isolated from propylene-grown cells of X. autotrophicus strain Py2 as described previously (24). The gene encoding 2-KPCC (xecC) was amplified by PCR using the primers 5′-CACCGTGAAAGTCTGGAACGCCC-3′ and 5′-TCACAGGCTCACCAGATTCT-3′ according to the Failsafe PCR protocol (Epicentre Biotechnologies, Madison, WI). PCR products were then ligated into a pBAD Directional TOPO vector (Invitrogen) according to the manufacturer's protocol to generate plasmid pDW1, which was then transformed into Escherichia coli One Shot Top10 cells (Invitrogen). pDW1 was isolated and the insert sequence was confirmed by DNA sequencing at the Center for Integrated BioSystems, Utah State University.
Site-directed mutagenesis.
Site-directed mutagenesis of pDW1 was carried out by utilizing a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocols. The sequences of the primer pairs used to create the desired mutations are as follows: C82A, 5′-TCC TGG GCG GCT CGG CCC CGC ACA ATG CGT-3′ and 5′-AAG GAC CCG CCG AGC CGG GGC GTG TTA CGC-3′; C87A, 5′-GTG CCC GCA CAA TGC GGC CGT GCC GCA CCA TAT GTT-3′ and 5′-GAA CAG ATG GTG CGG CAC GGC CGC ATT GTG CGG GCA-3′; H137A, 5′-GAA GTT CAT GAT GCC GGC CGG GCC GTT GCG CC-3′ and 5′-CGC AAC GGC CCG GCC GGC ATCATG AAC TTC CA-3′; H84A, 5′-CAC GCA CGC ATT GGC CGG GCA CGA GCC GCC-3′ and 5′-CCG CCG AGC ACG GGC CGG TTA CGC ACG CAC-3′; and M140A, 5′-CGG CCC GCA CGG CAT CGC GAA CTT CCA GTC CAA GG-3′ and 5′-CCT TGG ACT GGA AGT TCG CGA TGC CGT GCG GGC CG-3′. Mutations were confirmed by primer extension sequencing at SeqWright DNA Technology Services (Houston, TX).
Growth media.
E. coli Top10 cells were grown in Luria-Bertani rich (LB-Rich) broth containing ampicillin (100 μg/ml). The LB-Rich media contained the following components per liter: 20 g of tryptone, 15 g of yeast extract, 2 g of K2HPO4, 1 g of KH2PO4, and 8 g of NaCl.
Growth of bacteria.
All bacteria were grown at 37°C unless otherwise stated. E. coli Top10 cells that had been transformed with the pDW1 or corresponding mutant plasmid were plated and grown overnight. A single colony from this plate was used to grow a 25-ml liquid culture to an A600 of 0.6 for the preparation of 25% glycerol (vol/vol) stocks that were stored at −80°C until use. For use, cells from a frozen stock were inoculated into 125 ml of LB-Rich medium and grown to an A600 between 0.6 and 1.0, as measured on a Shimadzu UV160U spectrophotometer. This culture was used as the inoculum for a 15-liter capacity Microferm fermentor (New Brunswick Scientific) containing 12 liters of LB-Rich media supplemented with riboflavin (15 mg/liter) and antifoam A (0.005% [vol/vol]). Cells were allowed to grow at 37°C with agitation at 400 rpm and forced aeration to an A600 between 0.6 and 1.0. At this time, the temperature was reduced to 30°C, arabinose was added to 0.02%, and the cells were allowed to grow at this temperature for 6 h. Cells were concentrated using a tangential flow filtration system (Millipore) and were pelleted by centrifugation. Cell paste was drop frozen in liquid nitrogen and stored at −80°C.
Purification of recombinant 2-KPCC.
Cell paste was resuspended in 3 volumes of buffer A (50 mM Tris, 1 mM dithiothreitol [DTT], 0.1 mM EDTA, and 5% glycerol [vol/vol], prepared and buffered to pH 7.4 at 4°C) with DNase I (0.03 mg/ml) and lysozyme (0.03 mg/ml) and thawed at 30°C with shaking. All subsequent treatments were performed either on ice or at 4°C. The cell suspension was passed three times through a French pressure cell at a pressure of 1.1 × 105 kPa and clarified by centrifugation (184,000 relative centrifugal force for 30 min at 4°C). Clarified cell extract was applied to a 0.5- by 5.0-cm column of a Ni-NTA Superflow instrument (Pharmacia Biotech) at 7.0 ml/min. The column was then washed with 4-column volumes of buffer A, and the bound sample was eluted with a 15-column volume gradient from 0 to 400 mM imidazole. Purification was followed by SDS-PAGE. Appropriate fractions were pooled, and (NH4)2SO4 was added to 800 mM. The solution was incubated at 4°C with gentle stirring and then applied to a 2.6- by 5.5-cm column of phenyl Sepharose that had been preequilibrated with buffer A plus 800 mM (NH4)2SO4 (buffer C). The column was then washed with 4-column volumes of buffer C, and bound protein was eluted with a 15-column volume gradient from 0 to 100% of buffer A followed by an additional 5-column volume of buffer A. Appropriate fractions were pooled and concentrated by ultrafiltration using a YM30 membrane (Amicon).
For removal of the histidine patch (HP) thioredoxin leader acquired from the pBAD Directional TOPO vector, protein samples prepared as above were incubated with enterokinase (0.1 U EKMax; Invitrogen) at 4°C for 24 h. The protein was then reloaded onto the Ni-NTA Superflow column. Appropriate fractions were pooled, concentrated by ultrafiltration, and frozen drop-wise in liquid nitrogen for storage at −80°C.
Protein concentrations were determined using a modified biuret assay (9). 2-KPCC concentrations were also determined by using the previously determined extinction coefficient (ε450 of 11,828 M−1·cm− 1) (3).
SDS-PAGE and immunoblotting procedures.
SDS-PAGE (12% total acrylamide) was performed according to the Laemmli procedure (25). Electrophoresed proteins were visualized by staining with Coomassie blue R-250. The apparent molecular masses of polypeptides were determined by comparison with Rf values of standard proteins. Immunoblot analysis was conducted by electrophoretically transferring proteins from an SDS-polyacrylamide gel onto a polyvinylidene difluoride membrane. The membrane was incubated with polyclonal antiserum raised against purified 2-KPCC from X. autotrophicus strain Py2. Cross-reacting proteins were visualized using horseradish peroxidase conjugated to goat anti-rabbit immunoglobulin G (Promega).
Coupled spectrophotometric assay for 2-KPCC carboxylation activity.
A continuous spectrophotometric assay described previously (8) that couples acetoacetate production by 2-KPCC to acetoacetate reduction and NADH oxidation by β–hydroxybutyrate dehydrogenase (β-HBDH) was utilized. This assay relies on the fact that DTT can be used as an alternate reductant for 2-KPCC in place of NADPH while NADH cannot, so that loss of absorption at A340 due to NADH oxidation by β-HBDH as acetoacetate is reduced can be measured according to equations 2 and 3 (8):
| (2) |
| (3) |
where DTTred is reduced DTT and DTTox is oxidized DTT. By including a large excess of highly active β-HBDH (260 U/mg, where one unit is defined as 1 μmol/min) in assays with a fixed concentration of NADH, steady-state rates and kinetic parameters can be measured that cannot be measured in assays where NADPH is consumed directly by 2-KPCC in stoichiometric proportions to 2-KPC.
Assays were conducted in 2-ml anaerobic quartz cuvettes that contained a total reaction volume of 1 ml. Assays contained either 0.125 or 0.25 mg of 2-KPCC, 0.345 mg (90 units) of β-HBDH, 10 mM DTT, 0.2 mM NADH, and 60 mM carbonate species (added as 33.5 mM CO2 gas plus 26.5 mM KHCO3) in 100 mM Tris buffer, with the pH adjusted to 7.4 at 30°C. Reactions were allowed to equilibrate to 30°C, and assays were initiated by the addition of 2.5 μmol of 2-KPC.
Continuous spectrophotometric assay for 2-KPC formation (reverse reaction).
As described previously (10), 2-KPCC catalyzes the reverse of the physiologically important forward reaction shown in equation 1, a reaction that is dependent on NADP+ as the oxidant, as shown in equation 4:
| (4) |
Assays were conducted in 2-ml anaerobic quartz cuvettes with a total reaction volume of 1 ml. Each assay contained 0.125 mg of 2-KPCC, 5 mM NADP+, 5 mM CoM, and 100 mM acetoacetate in 100 mM Tris buffer [pH 7.4]. Reactions were allowed to equilibrate to 30°C, and assays were initiated by the addition of acetoacetate. Rates of reaction were measured by monitoring the increase in absorbance (A340) associated with the production of NADPH in a Shimadzu UV160U spectrophotometer containing a water-jacketed cell holder for temperature control.
2-Ketopropyl-CoM protonation assays.
In the absence of CO2, 2-KPCC catalyzes the cleavage and protonation of 2-KPC to form acetone, using either NADPH or DTT as the reductant, at rates about 25% of the rate of 2-KPC carboxylation, as shown using DTT as the reductant in equation 4 (10):
| (5) |
Assays for acetone formation were performed essentially as described previously (10). Assays were conducted in 9-ml sealed serum vials with a total reaction volume of 1 ml. Each assay contained either 0.125 or 0.25 mg of 2-KPCC and 10 mM DTT in 100 mM Tris buffer [pH 7.4]. Assay vials were incubated in a 30°C shaking water bath, and assays were initiated by the addition of 2.5 μmol of 2-KPC. Acetone formation was quantified as a function of time by removing samples and analyzing acetone by gas chromatography as described previously (13).
Single-turnover protonation assay.
Assays were performed in sealed 3-ml serum vials with a total reaction volume of 1 ml. Each assay contained 2-KPCC (0, 57, or 144 μM) in 100 mM Tris buffer [pH 7.4]. Assay vials were incubated in a 30°C shaking water bath (200 cycles min−1), and assays were initiated by the addition of 2.5 mM 2-KPC. After 10 min., the vials were transferred to a 65°C static water bath and incubated for 10 min to allow denaturation of the protein and volatilization of the acetone released. Acetone was quantified by gas chromatography as described above.
Acetoacetate decarboxylation assay.
Assays were performed as described previously (10). Briefly, assays were conducted in 9-ml sealed serum vials with a total reaction volume of 1 ml. Vials were depleted of carbonate species by including a KOH-containing trap as described previously (13). Each assay contained 250 mM acetoacetate and either 0 or 5 mM CoM in 100 mM Tris buffer [pH 7.4]. Assay vials were incubated in a 30°C shaking water bath, and assays were initiated by the addition of 0.125 mg 2-KPCC. Acetone was quantified by gas chromatography as described above.
Preincubation of 2-KPCC with 2-KPC.
For assays that used 2-KPCC preincubated with 2-KPC, 2-KPCC and 2-KPC were incubated together for 5 min at 30°C in GTP buffer (200 mM [each] glycine, Tris, and sodium phosphate) (pH 10). Following incubation, 2-KPCC was desalted by loading it onto a 2.3- by 5-cm column of Sephadex G-25 PD-10 that had been preequilibrated with 50 mM Tris buffer (pH 7.4).
Cys87-flavin charge transfer.
All spectra were obtained at room temperature in a quartz ultramicro-sized (120-μl) cuvette with a 1-cm light path. Aliquoted GTP buffer was adjusted to the desired pH and added to 2-KPCC for a final volume of 130 μl. Spectra were then obtained using a Shimadzu UV160U UV-Vis recording spectrophotometer interfaced with a personal computer running PC160 Personal Spectroscopy software, version 1.4. The reference cell contained protein-free GTP buffer. The actual pH of each solution used to obtain a spectrum was determined by mixing an appropriate amount of protein-free buffer A with each of the GTP buffers.
UV/visible spectral analysis of native 2-KPCC.
Samples of native 2-KPCC (3.6 mg/ml) were incubated anoxically in the presence of 10 mM DTT and 10 mM 2-bromoethanesulfonate (BES) for 4 h as described previously (8). The samples were then desalted using prepacked columns of Sephadex G-25 (PD-10; Pharmacia) equilibrated in 50 mM Tris buffer (pH 7.4). Desalted samples were air oxidized for 30 min. Desalted 2-KPCC was mixed with GTP buffer as described above.
Data analysis.
Kinetic constants (Km and Vmax) were calculated by fitting initial rate data to the Michaelis-Menten equation using the methods described by Cleland (14) using SigmaPlot software. pKa values were calculated by fitting A555 versus pH data to a four-parameter sigmoidal equation, also using SigmaPlot.
RESULTS
Cloning, expression, purification, and characterization of recombinant 2-KPCC (r-2-KPCC).
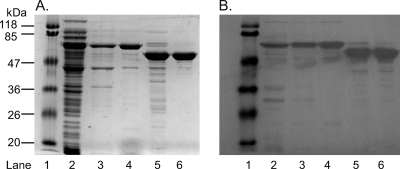
All previous attempts to express 2-KPCC in a recombinant form resulted in insoluble and consequently inactive protein. To increase the solubility and translation efficiency of 2-KPCC, a vector was chosen that included a thioredoxin (HP) fusion tag (26). Clarified cell extract from cultures of E. coli Top10 cells that had been transformed with pDW1 and induced with an optimized concentration of l-(+)-arabinose revealed highly soluble expression of 2-KPCC when subjected to SDS-PAGE and immunoblot analyses (Fig. 4). R-2-KPCC purified by two steps (Ni2+ affinity and phenyl Sepharose columns) was >90% homogeneous and migrated with the expected molecular mass of 68 kDa, (57 kDa for the enzyme plus 11 kDa for the fusion tag) (Fig. 4, lane 4). Cleavage of the HP tag using enterokinase resulted in the expected shift in migration to 57 kDa for 2-KPCC (Fig. 4, lanes 5 and 6). R-2-KPCC samples containing and lacking the HP tag had the characteristic UV/visible absorption spectrum expected for members of the DSOR family. A comparison of the absorption spectra of native 2-KPCC and r-2-KPCC (both with and without the HP tag) revealed that all three enzymes contained 1 mol of FAD/mol of protein monomer, demonstrating that the recombinant expression system results in incorporation of FAD into each active site.
Fig. 4.
SDS-PAGE and Western blotting of recombinant 2-KPCC. (A) SDS-polyacrylamide gel. Lanes: 1, molecular mass standards; 2, cell extract (13.3 μg); 3, first Ni2+ fraction (6.7 μg); 4, phenyl-Sepharose fraction (6.2 μg); 5, enterokinase digestion (15 μg); 6, the eluate from the second Ni2+ affinity column (15 μg). (B) Immunoblot-prepared blot from an identical SDS-polyacrylamide gel using antibodies raised to native 2-KPCC.
Purified 2-KPCC containing the HP tag from different enzyme preparations exhibited specific activities consistently in the range of 50 to 55 nmol of acetone produced/min/mg when assayed according to equation 4. This activity is in the same range as that measured for acetone formation with preparations of native 2-KPCC under identical assay conditions (note that rates of 2-KPC carboxylation and protonation when using DTT are 60 to 65% of the corresponding rates when NADPH is used as the reductant and that the rate of 2-KPC protonation is about 25% of the rate of 2-KPC carboxylation) (8, 10). Removal of the HP tag resulted in no change in 2-KPCC activity. Since 2-KPCC preparations containing and lacking the HP tag were both fully active, contained a full complement of FAD, and migrated on gel filtration columns at the expected positions (i.e., as dimers), subsequent experiments used the form of 2-KPCC retaining the HP tag to avoid the laborious, time-consuming, and expensive steps required to remove the tag for large preparations of enzyme.
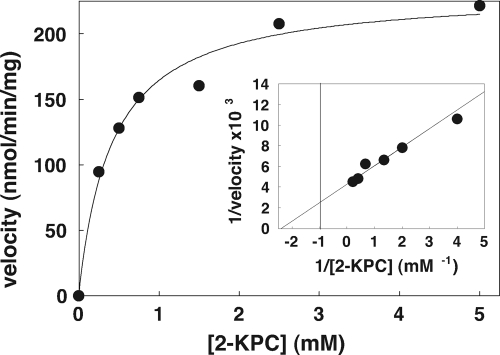
Kinetic parameters for r-2-KPCC-catalyzed substrate carboxylation were determined using the recently developed continuous spectrophotometric assay described in Materials an Methods (equations 2 and 3) by using saturating concentrations of CO2 and DTT and varying the concentration of 2-KPC, as was previously done for native 2-KPCC using a discontinuous assay that relies on measuring the incorporation of 14CO2 into acetoacetate (10). As shown in Fig. 5, the enzyme conformed to Michaelis-Menten kinetics, providing the following apparent kinetic parameters: Km = 0.42 ± 0.08 mM 2-KPC and Vmax = 233 ± 12 nmol acetoacetate formed·min−1·mg−1, which corresponds to a kcat of 13 min−1. In comparison, the apparent Km and Vmax values for native 2-KPCC determined previously by the discontinuous assay were 0.63 mM and 412 nmol/min/mg, respectively (10). The difference in Vmax to that reported here can be attributed to the use of DTT as the reductant in the discontinuous assay, which results in rates about 60% of the rates with the physiological reductant NADPH (8, 10).
Fig. 5.
Effect of 2-KPC concentration on the reductive cleavage and carboxylation of 2-KPC to acetoacetate and CoM. All assays were performed with 0.089 mg of 2-KPCC. The line through the data points was generated by nonlinear least-squares fitting to a rectangular hyperbola. Inset, double-reciprocal plot of the data. The line through the data points was also generated by nonlinear least-squares fitting of the data.
Redox-dependent activities for mutants in the redox-active disulfide (C87 and C82).
Cys87 and Cys82 have been identified by sequence analysis (37) and structural characterization (28) as the flavin and interchange thiols, respectively, of 2-KPCC (Fig. 3). Mutation of either cysteine residue to alanine resulted in complete loss of steady-state 2-KPC carboxylation (equations 1 and 2) and protonation (equation 5) activities, regardless of whether DTT or NADPH was used as the reductant in the assays (Table 1). Likewise, no activity was seen when the enzymes substituted for alanine were assayed in the reverse direction for 2-KPC production from acetoacetate (equation 4).
Table 1.
Effect of active-site mutations on the three redox-dependent activities of 2-KPCCa
| Enzyme | Specific activityb (nmol/min/mg) for: |
||
|---|---|---|---|
| 2-KPC protonation | 2-KPC carboxylation | Formation of 2-KPC from acetoacetate | |
| wt r-2-KPCC | 53.8 ± 6.4 | 221 ± 16 | 47 ± 4 |
| C87A r-2-KPCC | ND | ND | ND |
| C82A r-2-KPCC | ND | ND | ND |
| H84A r-2-KPCC | 22.7 ± 0.1 | 37.0 ± 4.2 | 12.0 ± 0.3 |
| H137A r-2-KPCC | ND | 18 ± 1 | 1.0 ± 0.1 |
All assays were repeated in duplicate. 2-KPC protonation and carboxylation assays were performed using DTT as the reductant, while formation of 2-KPC from acetoacetate was performed using NADP+ as the oxidant.
ND, no detectable activity.
It is noteworthy that the C87A variant was inactive for the forward steady-state reactions where DTT was used as the reductant in place of the physiologically important NADPH since, conceivably, this enzyme might be capable of reacting to form the mixed disulfide of C82 and CoM (Fig. 3B), which could then be reduced by DTT to allow further turnovers. To test the possibility that the C87A variant can undergo a single turnover, which would not be detected using the steady-state activity assays, we incubated a high concentration of the enzyme with the substrate 2-KPC and in the absence of reductant for a fixed time, and we then determined the amount of acetone that was produced. When 57 nmol or 144 nmol of 2-KPC was incubated in this fashion, 87 and 220 nmol of acetone, respectively, were produced once equilibrium was established. Since 2-KPCC is a dimer, this corresponds to an average production of 0.76 mol of acetone per enzyme-active site for both concentrations of enzyme assayed in this fashion, suggesting that C82 is active for a single turnover to cleave the thioether bond of 2-KPC to release enolacetone, which undergoes protonation to acetone. This reaction presumably occurs by formation of the C82-CoM mixed disulfide. As a control, this experiment was repeated with the C82A variant, and no acetone was observed within the detection limit of the assay. The addition of DTT to assays of the C87A variant did not increase the amount of acetone production.
UV/visible spectral properties of r-2-KPCC variants.
A distinguishing feature of DSOR enzymes is an interaction between the oxidized flavin and the deprotonated flavin thiol when the enzyme is in the two-electron reduced state (Fig. 2A, product of reaction iiiA) (38). This interaction results in a charge-transfer complex that alters the spectral features of FAD through a characteristic increase in absorbance above 525 nm and a decrease in absorbance at the maximal absorbance of 450 nm (38). The charge transfer complex can be observed in one of several ways for DSOR enzymes: (i) by titration of the enzyme under anoxic conditions to the two-electron reduced state; (ii) by rapid-scanning spectrophotometric techniques where the development of the charge transfer can be observed transiently for single-turnover catalysis; (iii) by site-directed mutagenesis of the interchange cysteine, in which case no disulfide can form and the oxidized enzyme exhibits the characteristic charge transfer; and (iv) by specific alkylation of the interchange thiol, which has the same effect as method iii. Methods iii and iv have routinely been used to determine the pKa values for the flavin thiol of DSOR enzymes (7, 33).
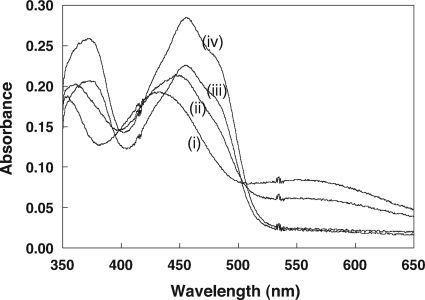
In order to characterize the interaction between C87 and FAD in 2-KPCC, we recorded UV/visible spectra for air-oxidized samples of native r-2-KPCC and the alanine substitutions in the interchange (C82) and flavin (C87) cysteine residues over a range of pH values from 5 to 11. The spectra of the native recombinant and C87A proteins were indistinguishable from that of the spectrum of oxidized native 2-KPCC, and no change in the spectra were observed over the range of pH values from 5 to 11 (data not shown). This is as expected, since for air-oxidized native r-2-KPCC, C82 and C87 will be in the disulfide form and C87A has no flavin thiol and hence no charge transfer can develop. As shown in representative spectra for C82A at different pH values in Fig. 6, the characteristic charge transfer due to interaction of the thiolate of C87 with FAD developed only at pH values substantially higher than physiological pH, reaching a maximum value at pH 11. This is in stark contrast to other DSOR enzymes where the flavin cysteine is largely in the deprotonated form in the microenvironment of the active sites of these enzymes at pH 7, since the microscopic pKa values are perturbed from the value of 8.2 for free cysteine to values around 5 (7, 15, 32, 33). The absorbance at 555 nm was plotted versus pH for a range of pH values to determine the pKa value for the flavin cysteine (C87) in C82A (Fig. 7, solid circles). A fit of this data provided a pKa of 8.76 ± 0.09, which is fully four units higher (four orders of magnitude less acidic) than that determined for the classical DSOR enzymes glutathione reductase (7) and mercuric reductase (33).
Fig. 6.
UV/visible absorption spectra of the r-2-KPCC C82A mutant at various pH values. Spectra were obtained using 0.15 mg of 2-KPCC diluted to a volume of 130 μl as described in Materials and Methods. The spectral noise at ∼A420 and ∼A535 is due to the ultramicro-sized cuvette that was used. The spectra shown are for pH values of (i) 11.0, (ii) 9.0, (iii) 7.5, and (iv) 5.0.
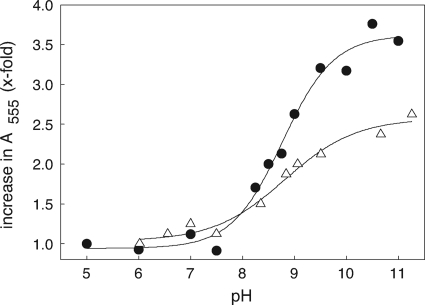
Fig. 7.
Determination of pKa values for the interchange thiol of 2-KPCC. The fold increase in A555 due to formation of charge transfer absorbance between FAD and the thiolate of C87 is plotted versus pH. The lines were derived from the four-parameter sigmoidal curve fit used to calculate pKa values. Symbols: •, C82A mutant of r-2-KPCC; ▵, native 2-KPCC alkylated on C82 with BES.
Recently, the CoM analog 2-bromoethanesulfonate was shown to be a highly specific affinity label for 2-KPCC that specifically alkylates the interchange thiol (C82) of 2-KPCC (8). As shown in Fig. 7 (open triangles), the charge transfer absorbance of BES-modified 2-KPCC developed in a similar fashion to that of C82A with increasing pH but to a lower maximal value than for the mutant. In spite of the lesser change in total absorbance, a fit of the titration data for BES-modified 2-KPCC yielded a pKa value that is identical within experimental error to that seen for the C82A mutant (8.74 ± 0.13).
Redox-dependent reactions of H84A and H137A 2-KPCC.
The structures of 2-KPCC show the presence of a “catalytic dyad” of histidine molecules (H137 and H84) that are hydrogen bonded to an ordered water molecule that is in turn within hydrogen-bonding distance of the carbonyl oxygen of 2-KPC in the 2-KPC-bound structure (Fig. 3) (28). This unique pair of histidine residues is not seen in any other member of the DSOR family and has been proposed to stabilize enolacetone formed from 2-KPC thioether bond cleavage (28) (Fig. 3B).
In order to test the proposed roles of H137 and H84 in the redox-dependent reactions of 2-KPCC, we individually mutated the residues to alanine residues and determined the effects on specific activities for 2-KPC carboxylation (equation 2), 2-KPC protonation (equation 5), and 2-KPC formation from acetoacetate (equation 4). As shown in Table 1, mutagenesis of the histidine proximal to the ordered water (H137) resulted in drastically reduced activities (92 to 100% loss of activity) for the three redox-dependent activities of 2-KPCC. Mutagenesis of the distal histidine resulted in more moderate losses of activity (58 to 76%).
Effect of mutations in the redox-active disulfide and catalytic dyad on acetoacetate decarboxylase activity of 2-KPCC.
2-KPCC was previously shown to catalyze a reaction that does not involve redox chemistry: the decarboxylation of acetoacetate to produce acetone and CO2 according to equation 6 (10):
| (6) |
Acetoacetate decarboxylase activity was very low when catalyzed by 2-KPCC alone but was stimulated 77-fold upon the addition of a saturating concentration (5 mM) of CoM (10).
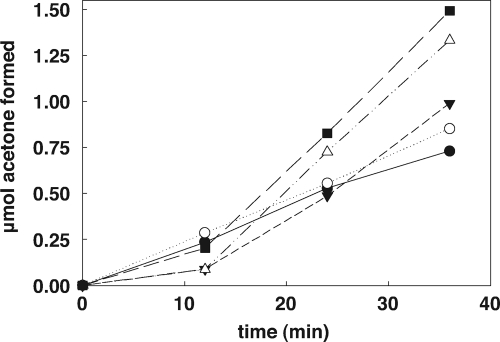
The rate of CoM-dependent acetoacetate decarboxylation by native r-2-KPCC was found to be comparable to that previously reported for native 2-KPCC (10). As shown in Fig. 8, the rates of CoM-dependent acetoacetate decarboxylation by H137A and H84A were comparable to that of the wild-type enzyme, while C82A and C87A exhibited rates that were somewhat higher than that of the native enzyme. As for native 2-KPCC, acetoacetate decarboxylase activity in each of the site-directed variants was approximately 70- to 80-fold lower when CoM was not included in the assays.
Fig. 8.
Acetoacetate decarboxylase activity of wild-type and mutant r-2-KPCC proteins. Assays contained 0.125 mg of 2-KPC, 250 mM acetoacetate, and 5 mM CoM. Data points represent the averages of duplicate experiments. Symbols: •, wild-type r-2-KPCC; ○, H84A r-2-KPCC; ▾, H137A r-2-KPCC; ▵, C87A r-2-KPCC; ▪, C82A r-2-KPCC.
Kinetic characterization of a mutation in one of the methionines flanking 2-KPC.
As shown in Fig. 3A, a distinguishing feature of 2-KPCC is the presence of two methionine residues (M140 and M361) that flank the substrate 2-KPCC. An alanine substitution was successfully created in M140, but unfortunately, attempts to make and express a corresponding substitution in M361 were unsuccessful. As shown in Table 2, the M140A substitution resulted in an enzyme with a 15-fold-higher Km for the substrate 2-KPC and a 3-fold-lower kcat, resulting in an overall catalytic efficiency for 2-KPC carboxylation that is 47-fold lower than that for the native enzyme.
Table 2.
Kinetic parameters for the wild-type and M140A r-2-KPCC mutanta
| Enzyme | Km for 2-KPC (mM) | Change in Km (x-fold) | kcat (min−1) | Change in kcat (x-fold) | kcat/Km (mM−1·min−1) |
|---|---|---|---|---|---|
| wt r-2-KPCC | 0.42 ± 0.08 | 1 | 13.3 | 1 | 32 |
| M140A r-2-KPCC | 6.37 ± 0.82 | 15.3 | 4.3 | 0.32 | 0.68 |
Apparent Km and kcat values for 2-KPC carboxylation were determined using the coupled spectrophotometric assay described in Materials and Methods.
DISCUSSION
In the present work, 2-KPCC has for the first time been expressed in a heterologous system. The biochemical and kinetic properties of r-2-KPCC containing the HP tag show that it is essentially identical to that of native 2-KPCC and thus suitable for construction of, and comparative studies with, site-directed variants. The characterization of five of these site-directed variants has allowed key tenets of the mechanism of thioether bond cleavage and substrate carboxylation proposed from mechanistic (8, 10) and structural (28, 30, 31) studies to be tested.
Catalytic roles and unique properties of the redox-active disulfide of 2-KPCC.
As expected, the redox-active cysteines of 2-KPCC are crucial for all steady-state reactions requiring thioether bond cleavage or formation (equations 1, 2, 4, and 5), consistent with the key intermediacy of the Cys82-CoM mixed disulfide in these reactions (Fig. 3B). The CoM-Cys82 mixed disulfide has previously been captured for native 2-KPCC by incubating the enzyme in the presence of 20 mM CoM prior to crystallization (31). The 2-KPCC with a C87A substitution catalyzed the single-turnover production of acetone from 2-KPC, demonstrating that the interchange thiol (C82) is still capable of thioether bond cleavage and presumably CoM-Cys82 mixed disulfide formation in the absence of the flavin thiol. The fact that the addition of DTT did not lead to an increase in acetone production in these assays indicates that DTT is not capable of directly reducing the CoM-C82 mixed disulfide, as by doing so C82 would be rereduced for additional rounds of catalysis.
These observations provide insights into how DTT mediates the redox-dependent reactions of 2-KPCC in place of NADPH. As noted above, the rates of DTT-dependent 2-KPC protonation or carboxylation are about 60% of the rates observed when NADPH is used as the reductant. It is presumed that DTT reduces the oxidized disulfide of C82 and C87 directly, i.e., without the intermediacy of FAD. Incubation of air-oxidized 2-KPCC in the absence of substrates results in reduction of the redox-active disulfide, as evidenced by the ability of the DTT-reduced enzyme to undergo stoichiometric alkylation on C82 by the CoM analog BES (8). Prolonged incubation of 2-KPCC with DTT results in bleaching of the flavin of 2-KPCC, but this occurs too slowly (on the order of 30 min to an h) to be a relevant pathway for electron transfer under steady-state conditions. In any event, the results presented herein show that DTT is a suitable reductant for 2-KPCC when both redox-active thiols are present but not when C87 has been replaced with alanine. This suggests that DTT does not directly reduce the mixed disulfide of C82 and CoM under steady-state turnover conditions but instead reduces the redox-active cysteine pair.
As shown in Fig. 6 and 7, the flavin thiol (C87) is in the fully protonated state at neutral pH for both the alanine substitution in the interchange thiol and for the native enzyme where the interchange thiol is alkylated by ethylsulfonate. C87 has a pKa value fully 4 units higher (four orders of magnitude more basic) than the perturbed value for the flavin thiols of other members of the DSOR family (7, 33). A key piece of structural information that explains this difference is the replacement of a histidine side chain that interacts with and lowers the pKa of the flavin thiol for all other DSOR enzymes (32) with a phenylalanine residue in 2-KPCC (28). It should be noted that DSOR enzymes are dimers, with each active site being at the interface between the two subunits and hence interacting with residues from the second subunit. The histidine that lowers the pKa of the interchange thiol in conventional DSOR enzymes and that is replaced with phenylalanine in 2-KPCC resides on the other subunit of the dimer (Phe501 of subunit 2 in this instance). All of the other active-site residues noted up until now reside within the primary subunit forming the active site of 2-KPCC (with two such active sites present on each dimeric enzyme). The replacement of histidine with phenylalanine in the environment of the flavin thiol is thought to help maintain the hydrophobicity of the enzyme-active site and to direct the immediate product of thioether bond cleavage (enolacetone) toward carboxylation to form acetoacetate (the physiologically important reaction) rather than protonation to form acetone (a fortuitous side reaction that occurs when CO2 is not present) (10, 28). The prevention of protonation of enolacetone to form acetone, an essentially irreversible reaction, is of paramount importance in 2-KPCC. In stark contrast, for glutathione reductase, a secondary (and major) role of the histidine that lowers the pKa of the flavin thiol is to protonate the first molecule of glutathione formed upon disulfide bond cleavage to drive the reaction forward and prevent the reverse reaction from occurring (32).
The lack of the histidine general base that deprotonates the flavin thiol would be expected to slow the rate of disulfide bond formation by 2-KPCC relative to that of glutathione reductase (compare steps in Fig. 2B and C). However, it is likely that a step other than disulfide bond formation or another step involving oxidation/reduction is rate limiting for 2-KPCC, which turns over much slower than glutathione reductase. In this context, it should be noted that 2-KPC carboxylation is approximately 40% faster when the physiological reductant NADPH is used instead of DTT, indicating that at least one redox step is partially rate limiting.
It should be noted that the steady state kcat we report here for 2-KPCC carboxylation (13.3 min−1) is fully 3,300 times lower than the turnover number reported for glutathione reduction by glutathione reductase from E. coli (44,000 min−1) (20). The large difference in steady-state turnover highlights the more difficult chemistry of thioether bond cleavage and substrate carboxylation relative to that of disulfide bond reduction catalyzed by classical DSOR enzymes. The turnover numbers for the other enzymes of the epoxypropane carboxylation pathway (Fig. 1) are significantly higher (6.5 s−1 for epoxyalkane:CoM transferase and 25 to 50 s−1 for the R- and S-hydroxypropyl-CoM dehydrogenases [12, 23, 34]). As a reflection of the low turnover of 2-KPCC, it is expressed at very high levels (∼25% of soluble cell protein) in propylene-grown cells of X. autotrophicus Py2 (3, 16). In spite of this high level of expression, 2-KPCC has the lowest specific activity of the enzymes of the epoxide carboxylation pathway in cell extracts, with an activity that is identical to the specific activity of epoxypropane consumption by whole-cell suspensions of X. autotrophicus Py2 growing with propylene as the carbon source (90 nmol of epoxypropane consumed/min/mg [36]). Clearly, the reaction catalyzed by 2-KPCC is the rate-limiting step of the epoxide carboxylation pathway shown in Fig. 1. Still, the very high level of expression of 2-KPCC allows X. autotrophicus and R. rhodochrous to grow fairly well (doubling times in exponential phase of about 8 h) with propylene as the carbon source (4, 36).
Role of the catalytic histidine dyad in stabilizing enolacetone in reactions involving thioether bond cleavage or breakage.
Site-directed mutagenesis of the histidines proximal (H137) and distal (H84) to the ordered water that interacts with the carbonyl oxygen of bound 2-KPC in the crystal structure (Fig. 3A) supports the hypothesis that these residues are critical in stabilizing enolacetone as an intermediate in reactions that make or break a thioether bond (equations 1, 2, 4, and 5). As expected, mutation of the proximal histidine resulted in much lower levels of activity than for the distal histidine, indicating that the proximal histidine can still provide some stabilization in the absence of the additional charge relay provided by H84.
Acetoacetate decarboxylase activity of 2-KPCC does not involve the redox-active disulfide or histidine dyad.
The decarboxylation of acetoacetate to acetone and CO2 by 2-KPCC must of necessity occur by C-C bond cleavage to form enolacetone, the same high-energy intermediate(s) formed in the redox-dependent reactions of 2-KPCC. The fact that the C82A, C87A, H84A, and H137A mutants all retain full (or slightly increased) CoM-dependent acetoacetate decarboxylase activity demonstrates that acetoacetate decarboxylation must occur by a fundamentally different mechanism than that for the reactions that involve redox chemistry and thioether bond cleavage. At the same time, acetoacetate decarboxylase activity still requires CoM for optimal activity. These results can be rationalized and explained by examining the structural features of the form of 2-KPCC captured in the presence of the mixed disulfide of CoM (31). This structure showed the presence of acetone bound to a distinct binding pocket 4.5 Å from the thiol sulfur of CoM (31). Within this pocket, which consists of residues from the second enzyme subunit, are Gln509, which formed a hydrogen bond with the carbonyl O of acetone, and His506. Based on this structure, it was proposed that this second pocket constitutes a binding pocket for the physiological product acetoacetate (31). By extrapolation, we propose now that this second pocket is the active site for acetoacetate decarboxylation. The stimulatory effect of CoM on acetoacetate decarboxylase activity can be rationalized in one or both of the following ways: (i) the thiol of CoM, when bound to the CoM binding pocket of 2-KPCC, is the proton donor for formation of acetone from enolacetone, i.e., CoM serves as a general acid to promote decarboxylation; and/or (ii), binding of CoM induces a conformational change similar to that induced by binding of the substrate 2-KPC that facilitates the binding of acetoacetate within the second binding pocket such that it can undergo decarboxylation.
Methionine140 modulates Km and kcat for 2-KPC carboxylation.
A distinctive feature of 2-KPCC relative to other DSOR enzymes is the pair of methionine residues that flank the substrate 2-KPC (28) (Fig. 3A). The step prior to that catalyzed by 2-KPCC in bacterial epoxide metabolism is catalyzed by a pair of stereoselective dehydrogenases that oxidize the R- and S-enantiomers of hydroxypropyl-CoM (HPC) to 2-KPC (1, 6). Interestingly, the crystal structure solved for R-hydroxypropyl-CoM dehydrogenase (R-HPCDH) in the presence of the product 2-KPC also shows a pair of methionine residues flanking 2-KPC in a fashion similar to that seen in 2-KPCC (21). Additionally, a homology model for the other dehydrogenase, S-HPCDH, also shows flanking methionines (22). Thus, methionine residues seem to be a crucial feature in the binding of CoM thioethers during the steps of bacterial epoxide carboxylation. The 2-KPCC mutant with an M140A substitution was kinetically characterized in the present work and found to exhibit both a marked increase in Km and a decrease in kcat (Table 2). Unfortunately, all attempts to make an alanine substitution in the other methionine (M361) were unsuccessful. Although the crystal structures of 2-KPCC and R-HPCDH do not indicate a catalytic role for the flanking methionines, this initial characterization shows that they appear to be crucial for substrate binding and catalysis.
Summary.
2-KPCC is distinct from all other known members of the DSOR family in catalyzing thioether bond cleavage and substrate carboxylation. The results of the present work establish the essential role of a novel catalytic dyad that facilitates enolacetone formation and stabilization and a unique active-site environment for the redox-active cysteine pair that is unlike that seen in any other DSOR enzyme. The combination of structural biology and site-directed mutagenesis, together with the characterization of redox-dependent and redox-independent reactions, provides a powerful complement for elucidating mechanistic details for this novel carboxylase.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant GM51805 to S.A.E. and by Department of Energy grant DE-FG02-04ER15563 to J.W.P.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Allen J. R., Clark D. D., Krum J. G., Ensign S. A. 1999. A role for coenzyme M (2-mercaptoethansulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc. Natl. Acad. Sci. U. S. A. 96:8432–8437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen J. R., Ensign S. A. 1996. Carboxylation of epoxides to β-keto acids in cell extracts of Xanthobacter strain Py2. J. Bacteriol. 178:1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allen J. R., Ensign S. A. 1997. Characterization of three protein components required for functional reconstitution of the epoxide carboxylase multienzyme complex from Xanthobacter strain Py2. J. Bacteriol. 179:3110–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allen J. R., Ensign S. A. 1998. Identification and characterization of epoxide carboxylase activity in cell extracts of Nocardia corallina strain B276. J. Bacteriol. 180:2072–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allen J. R., Ensign S. A. 1997. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J. Biol. Chem. 272:32121–32128 [DOI] [PubMed] [Google Scholar]
- 6. Allen J. R., Ensign S. A. 1999. Two short-chain dehydrogenases confer stereoselectivity for enantiomers of epoxypropane in the multiprotein epoxide carboxylating systems of Xanthobacter strain Py2 and Nocardia corallina B276. Biochemistry 38:247–256 [DOI] [PubMed] [Google Scholar]
- 7. Arscott L. D., Thorpe C., Williams C. H. J. 1981. Glutathione reductase from yeast. Differential reactivity of the nascent thiols in two-electron reduced enzyme and properties of a monoalkylated derivative. Biochemistry 20:1513–1520 [DOI] [PubMed] [Google Scholar]
- 8. Boyd J. M., Clark D. D., Kofoed M. A., Ensign S. A. 2010. Mechanism of inhibition of aliphatic epoxide carboxylation by the coenzyme M analog 2-bromoethanesulfonate. J. Biol. Chem. 285:25232–25242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chromý V., Fischer J., Kulhánek V. 1974. Re-evaluation of EDTA-chelated biuret reagent. Clin. Chem. 20:1362–1363 [PubMed] [Google Scholar]
- 10. Clark D. D., Allen J. R., Ensign S. A. 2000. Characterization of five catalytic activities associated with the NADPH:2-ketopropyl-coenzyme M [2-(2-ketopropylthio)ethanesulfonate] oxidoreductase/carboxylase of the Xanthobacter strain Py2 epoxide carboxylase system. Biochemistry 39:1294–1304 [DOI] [PubMed] [Google Scholar]
- 11. Clark D. D., Boyd J. M., Ensign S. A. 2004. The stereoselectivity and catalytic properties of Xanthobacter autotrophicus 2-[(R)-2-hydroxypropylthio]ethanesulfonate dehydrogenase are controlled by interactions between C-terminal arginine residues and the sulfonate of coenzyme M. Biochemistry 43:6763–6771 [DOI] [PubMed] [Google Scholar]
- 12. Clark D. D., Ensign S. A. 2002. Characterization of the 2-[(R)-2-hydroxypropylthio]ethanesulfonate dehydrogenase from Xanthobacter strain Py2: product inhibition, pH dependence of kinetic parameters, site-directed mutagenesis, rapid equilibrium inhibition, and chemical modification. Biochemistry 41:2727–2740 [DOI] [PubMed] [Google Scholar]
- 13. Clark D. D., Ensign S. A. 1999. Evidence for an inducible nucleotide-dependent acetone carboxylase in Rhodococcus rhodochrous B276. J. Bacteriol. 181:2752–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cleland W. W. 1979. Statistical analysis of enzyme kinetic data. Methods Enzymol. 63:103–138 [DOI] [PubMed] [Google Scholar]
- 15. Distefano M. D., Au K. G., Walsh C. T. 1989. Mutagenesis of the redox-active disulfide in mercuric ion reductase: catalysis by mutant enzymes restricted to flavin redox chemistry. Biochemistry 28:1168–1183 [DOI] [PubMed] [Google Scholar]
- 16. Ensign S. A. 1996. Aliphatic and chlorinated alkenes and epoxides as inducers of alkene monooxygenase and epoxidase activities in Xanthobacter strain Py2. Appl. Environ. Microbiol. 62:61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ensign S. A. 2001. Microbial metabolism of aliphatic alkenes. Biochemistry 40:5845–5853 [DOI] [PubMed] [Google Scholar]
- 18. Ensign S. A., Allen J. R. 2003. Aliphatic epoxide carboxylation. Annu. Rev. Biochem. 72:55–76 [DOI] [PubMed] [Google Scholar]
- 19. Gallagher S. C., Cammack R., Dalton H. 1997. Alkene monooxygenase from Nocardia corallina B-276 is a member of the class of dinuclear iron proteins capable of stereospecific epoxygenation reactions. Eur. J. Biochem. 247:635–641 [DOI] [PubMed] [Google Scholar]
- 20. Henderson G. B., et al. 1991. Engineering the substrate specificity of glutathione reductase toward that of trypanothione reduction. Proc. Natl. Acad. Sci. U. S. A. 88:8769–8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krishnakumar A. M., Nocek B. P., Clark D. D., Ensign S. A., Peters J. W. 2006. Structural basis for stereoselectivity in the (R)- and (S)-hydroxypropylthioethanesulfonate dehydrogenases. Biochemistry 45:8831–8840 [DOI] [PubMed] [Google Scholar]
- 22. Krishnakumar A. M., et al. 2008. Getting a handle on the role of coenzyme M in alkene metabolism. Microbiol. Mol. Biol. Rev. 72:445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krum J. G., Ellsworth H., Sargeant R. R., Rich G., Ensign S. A. 2002. Kinetic and microcalorimetric analysis of substrate and cofactor interactions in epoxyalkane:CoM transferase, a zinc-dependent epoxidase. Biochemistry 41:5005–5014 [DOI] [PubMed] [Google Scholar]
- 24. Krum J. G., Ensign S. A. 2000. Heterologous expression of bacterial epoxyalkane:coenzyme M transferase and inducible coenzyme M biosynthesis in Xanthobacter strain Py2 and Rhodococcus rhodochrous B276. J. Bacteriol. 182:2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 26. Lu Z., et al. 1996. Histidine patch thioredoxins. Mutant forms of thioredoxin with metal chelating affinity that provide for convenient purifications of thioredoxin fusion proteins. J. Biol. Chem. 271:5059–5065 [PubMed] [Google Scholar]
- 27. Miura A., Dalton H. 1995. Purification and characterization of the alkene monooxygenase from Nocardia corallina B-276. Biosci. Biotechnol. Biochem. 59:853–859 [Google Scholar]
- 28. Nocek B., et al. 2002. Structural basis for CO2 fixation by a novel member of the disulfide oxidoreductase family of enzymes, 2-ketopropyl-coenzyme M oxidoreductase/carboxylase. Biochemistry 41:12907–12913 [DOI] [PubMed] [Google Scholar]
- 29. Pai E. F. 1991. Variations on a theme: the family of FAD-dependent NAD(P)H-(disulphide)-oxidoreductases. Curr. Opin. Struct. Biol. 1:796–803 [Google Scholar]
- 30. Pandey A. S., Mulder D. W., Ensign S. A., Peters J. W. 2011. Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M oxidoreductase/carboxylase. FEBS Lett. 585:459–464 [DOI] [PubMed] [Google Scholar]
- 31. Pandey A. S., Nocek B., Clark D. D., Ensign S. A., Peters J. W. 2006. Mechanistic implications of the structure of the mixed-disulfide intermediate of the disulfide oxidoreductase, 2-ketopropyl-coenzyme M oxidoreductase/carboxylase. Biochemistry 45:113–120 [DOI] [PubMed] [Google Scholar]
- 32. Rietveld P., et al. 1994. Reductive and oxidative half-reactions of glutathione reductase from Escherichia coli. Biochemistry 33:13888–13895 [DOI] [PubMed] [Google Scholar]
- 33. Schultz P. G., Au K. G., Walsh C. T. 1985. Directed mutagenesis of the redox-active disulfide in the flavoenzyme mercuric ion reductase. Biochemistry 24:6840–6848 [DOI] [PubMed] [Google Scholar]
- 34. Sliwa D. A., Krishnakumar A. M., Peters J. W., Ensign S. A. 2010. Molecular basis for enantioselectivity in the (R)- and (S)-hydroxypropylthioethanesulfonate dehydrogenases, a unique pair of stereoselective short-chain dehydrogenases/reductases involved in aliphatic epoxide carboxylation. Biochemistry 49:3487–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Small F. J., Ensign S. A. 1997. Alkene monooxygenase from Xanthobacter strain Py2. Purification and characterization of a four-component system central to the bacterial metabolism of aliphatic alkenes. J. Biol. Chem. 272:24913–24920 [DOI] [PubMed] [Google Scholar]
- 36. Small F. J., Ensign S. A. 1995. Carbon dioxide fixation in the metabolism of propylene and propylene oxide by Xanthobacter strain Py2. J. Bacteriol. 177:6170–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Swaving J., Weijers C. A., van Ooyen A. J., de Bont J. A. M. 1995. Complementation of Xanthobacter Py2 mutants defective in epoxyalkane degradation, and expression and nucleotide sequence of the complementing DNA fragment. Microbiology 141:477–484 [DOI] [PubMed] [Google Scholar]
- 38. Walsh C. 1979. Enzymatic reaction mechanisms. W. H. Freeman and Co., New York, NY [Google Scholar]
- 39. Zhou N.-Y., Chan Kwo Chion C. K., Leak D. J. 1996. Cloning and expression of the genes encoding the propene monooxygenase from Xanthobacter, Py2. Appl. Microbiol. Biotechnol. 44:582–588 [Google Scholar]