Abstract
φA1122 is a T7-related bacteriophage infecting most isolates of Yersinia pestis, the etiologic agent of plague, and used by the CDC in the identification of Y. pestis. φA1122 infects Y. pestis grown both at 20°C and at 37°C. Wild-type Yersinia pseudotuberculosis strains are also infected but only when grown at 37°C. Since Y. pestis expresses rough lipopolysaccharide (LPS) missing the O-polysaccharide (O-PS) and expression of Y. pseudotuberculosis O-PS is largely suppressed at temperatures above 30°C, it has been assumed that the phage receptor is rough LPS. We present here several lines of evidence to support this. First, a rough derivative of Y. pseudotuberculosis was also φA1122 sensitive when grown at 22°C. Second, periodate treatment of bacteria, but not proteinase K treatment, inhibited the phage binding. Third, spontaneous φA1122 receptor mutants of Y. pestis and rough Y. pseudotuberculosis could not be isolated, indicating that the receptor was essential for bacterial growth under the applied experimental conditions. Fourth, heterologous expression of the Yersinia enterocolitica O:3 LPS outer core hexasaccharide in both Y. pestis and rough Y. pseudotuberculosis effectively blocked the phage adsorption. Fifth, a gradual truncation of the core oligosaccharide into the Hep/Glc (l-glycero-d-manno-heptose/d-glucopyranose)-Kdo/Ko (3-deoxy-d-manno-oct-2-ulopyranosonic acid/d-glycero-d-talo-oct-2-ulopyranosonic acid) region in a series of LPS mutants was accompanied by a decrease in phage adsorption, and finally, a waaA mutant expressing only lipid A, i.e., also missing the Kdo/Ko region, was fully φA1122 resistant. Our data thus conclusively demonstrated that the φA1122 receptor is the Hep/Glc-Kdo/Ko region of the LPS core, a common structure in Y. pestis and Y. pseudotuberculosis.
INTRODUCTION
Yersinia pestis, the etiologic agent of bubonic plague and pneumonic plague, is one of the most potent bacterial pathogens known and is transmitted by a bite from infected fleas to their mammalian hosts (56). The present number of human plague cases in the world is relatively stable, with approximately two thousand reported incidents each year. This number may, however, be largely underestimated due to inadequate diagnostics and reporting in some countries of disease endemicity.
φA1122 is a T7-related bacteriophage that is used as a diagnostic phage by the Centers for Disease Control and Prevention for the identification of Y. pestis. It infects most isolates of Y. pestis grown both at 20°C and at 37°C. Yersinia pseudotuberculosis is also infected but only when grown at 37°C (30). The receptor used by phage φA1122 for adsorption has not been identified yet.
Gram-negative bacteria are surrounded by two membranes, the inner membrane (IM) and the outer membrane (OM). The IM is a bilayer composed of phospholipids, and the OM is asymmetric, having phospholipids in its inner leaflet and, in most Gram-negative bacteria, lipopolysaccharides (LPS) in its outer leaflet (13, 14). Embedded in the OM are also a number of outer membrane proteins (OMPs) that function in solute and protein translocation, pathogenesis, structural linkers, and signal transduction (13, 43). Both OMPs and LPS may function as specific phage receptors (43, 77).
LPS is an amphipathic molecule that is anchored to the OM by lipid A (71). Core oligosaccharide is attached to lipid A, and the outermost structure of LPS is O-polysaccharide (O-PS). In Y. pestis, the LPS is rough, containing no O-PS (19, 21, 34, 37, 66); however, Y. pseudotuberculosis strains express O-PS (65), and the expression is optimal at temperatures below 30°C. For a long time it was believed that mutants of LPS-containing Gram-negative bacteria without a minimal core structure (i.e., one residue of 3-deoxy-d-manno-oct-2-ulosonic acid [Kdo]) are not viable (29, 63); thus, the core-lipid A unit was thought to be important for bacterial viability and membrane function. However, it has been proven in recent years that mutants synthesizing only lipid A or a precursor thereof are viable (49, 50, 74). Together with other LPS-free Gram-negative bacteria (33, 35, 36, 38, 47, 73, 78), Neisseria meningitidis can also survive without any LPS (72).
The T7 group phages T7 and T3, infecting Escherichia coli (45, 76), and Yersinia enterocolitica phage φYeO3-12 (2, 3), all use LPS as a receptor. The receptor recognition of these phages is dependent on the tail fiber protein gp17. Since the gp17 of φA1122 is highly similar to its counterparts in T7 and T3, it was assumed that φA1122 also may bind to LPS (30).
The lipid A-core oligosaccharide structures of Y. pestis and Y. pseudotuberculosis LPS are more or less identical, with both expressing several glycoforms that show temperature-dependent variations (41). The main core structure is heptasaccharide, which contains hexoses (Glc [d-glucopyranose] and Gal [d-galactopyranose]), heptoses (dd-Hep [d-glycero-d-manno-heptopyranose] and Hep [l-glycero-d-manno-heptopyranose]), and octulosonic acids (Kdo [3-deoxy-d-manno-oct-2-ulopyranosonic acid] and Ko [d-glycero-d-talo-oct-2-ulopyranosonic acid]). In bacteria grown at 25°C, the core main chain (from nonreducing end to lipid A) is a pentasaccharide, dd-Hep(IV)-Hep(III)-Hep(II)-Hep(I)-Kdo, with branching Ko from Kdo and Glc from Hep(I), while in bacteria grown at 37°C, dd-Hep is replaced by Gal, and Ko is replaced by Kdo (24). In this work, we present conclusive evidence demonstrating that the φA1122 receptor is the Hep/Glc-Kdo/Ko region of Y. pestis and Y. pseudotuberculosis LPS.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and media.
The bacteriophage, bacterial strains, and plasmids used in this work are described in Table 1. Unless otherwise stated, Yersinia and phage cultures were incubated at room temperature (RT; 22°C) and E. coli cultures at 37°C. Tryptone soya broth (TSB) medium (Oxoid) was used for bacterial liquid cultures, and soft agar medium included an additional 0.4% (wt/vol) agar (Biokar Diagnostics). For transconjugant selections, Yersinia selective agar (CIN agar; Oxoid) plates supplemented with the appropriate antibiotics were used. Luria agar (64) was used as solid medium for bacteria, and lambda agar (tryptone at 10 g/liter, NaCl at 2.5 g/liter, agar at 15 g/liter) for phage plates. Plates were supplemented with ampicillin (Amp; 100 μg/ml), kanamycin (Kan; 100 μg/ml), nalidixic acid (Nal; 100 μg/ml), or chloramphenicol (Clm; 30 μg/ml) when required.
Table 1.
Bacteriophages, bacterial strains, and plasmids used in this work
| Bacteriophage/strain/plasmid | Genotype/relevant featurea | Source/reference(s) |
|---|---|---|
| Bacteriophages | ||
| φA1122 | Reference phage used by the CDC to identify Y. pestis | 30 |
| φYeO3-12 | Phage using Y. enterocolitica serotype O:3 O-PS as receptor | 2, 54, 55 |
| φR1-37 | Phage using Y. enterocolitica serotype O:3 OC as receptor | 39, 70 |
| Bacterial strains | ||
| Yersinia pestis | ||
| KIM D27 | Nonpigmented isolate of KIM10.Lcr+ Pgm− Pst+ | 31 |
| KIM D27-ΔwabD | ΔwabD::nptII Kanr | This work |
| KIM D27-ΔwaaL | ΔwaaL::nptII Kanr | This work |
| KIM D27-ΔwaaQ | ΔwaaQ::nptII Kanr | This work |
| KIM D27-ΔwaaE | ΔwaaE::nptII Kanr | This work |
| KIM D27-ΔwaaA | ΔwaaA::nptII Kanr | This work |
| KIM D27Nar | Nalr | 39 |
| EV76-c | 70-kb virulence plasmid-cured derivative of EV76 | 10, 60 |
| 231 | pFra+ pCD+ pPst+; bv. antiqua, subsp. pestis; wild type | 6 |
| KM260(11) | pFra+ pCD− (Lcr−) pPst−; derived from wild-type bv. antiqua subsp. pestis strain 231; avirulent | 41, 42 |
| KM260(11)pKD46 | KM260(11) harboring plasmid pKD46; Ampr | 5 |
| KM260(11)ΔwabC | ΔwabC::nptII; derived from strain KM260(11)pKD46; Kanr | 5 |
| KM260(11)ΔwabD | ΔwabD::nptII; derived from strain KM260(11)pKD46; Kanr | 5 |
| KM260(11)ΔwabC/waaD | ΔwabC ΔwabD::nptII; derived from strain KM260(11)pKD46; Kanr | This work |
| KM260(11)ΔwabC/waaL | ΔwabC::nptII ΔwaaL::cat; derived from strain KM260(11)ΔwabCpKD46; Kanr Clmr | This work |
| KM260(11)ΔwabD/waaL | ΔwabD::nptII ΔwaaL::cat; derived from strain KM260(11)ΔwabDpKD46; Kanr Clmr | This work |
| KM260(11)ΔwaaL | ΔwaaL::nptII; derived from the strain KM260(11)pKD46; Kanr | 5 |
| KM260(11)ΔwaaQ | ΔwaaQ::nptII; derived from the strain KM260(11)pKD46; Kanr | 5 |
| KM260(11)ΔwaaQ/waaL | ΔwaaQ ΔwaaL::nptII; derived from the strain KM260(11)pKD46; Kanr | This work |
| KM260(11)ΔwaaE | ΔwaaE::nptII; derived from the strain KM260(11)pKD46; Kanr | 5 |
| KM260(11)ΔwaaF | ΔwaaF::nptII; derived from the strain KM260(11)pKD46; Kanr | 5 |
| KM260(11)ΔwaaC | ΔwaaC::nptII; derived from the strain KM260(11)pKD46; Kanr | 5 |
| KM260(11)ΔwaaA | ΔwaaA::nptII; derived from the strain KM260(11)pKD46; Kanr | 5 |
| Y. pseudotuberculosis | ||
| PB1 | Serotype O:1a | 59 |
| 1 | Serotype O:1a; Lcr− | 65 |
| 43 | Serotype O:3; Lcr− | 65 |
| 32 | Serotype O:4a; Lcr− | 65 |
| YPIII | Serotype O:3 | 12 |
| PB1Δwb | Serotype O:1a; O-PS-negative derivative of PB1; Kanr | This work |
| PB1Δwb-R4 | Spontaneous φA1122-resistant derivative of PB1Δwb | This work |
| PB1Δwb-R7 | Spontaneous φA1122-resistant derivative of PB1Δwb | This work |
| PB1Δwb-R12 | Spontaneous φA1122-resistant derivative of PB1Δwb | This work |
| Y. enterocolitica | ||
| 8081 | Serotype O:8 | 61 |
| 8081-R2 | Serotype O:8; O-PS-negative derivative of 8081 | 80 |
| 6471/76-c (YeO3-c) | Serotype O:3; virulence plasmid-cured derivative of 6471/76 | 68 |
| YeO3-R1 | Spontaneous O-PS-negative derivative of 6471/76-c | 70 |
| YeO3-c-OC | Δ(wzx-wbcQ) outer core-negative derivative of 6471/76-c | 11 |
| YeO3-OC-R | Δ(wzx-wbcQ) outer core- and O-PS-negative derivative of 6471/76 | 11 |
| E. coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG λ− tonA | Life Technologies |
| C600 | thi thr leuB tonA lacY supE | 7 |
| HB101 | F− Δ(gpt-proA)62 leuB6 glnV ara-14 galK2 lacY1 Δ(mcr-mrr) rpsL20 (Strr) xyl-5 mtl-1 recA13 | 15 |
| Sm10λpir | thi thr leuB tonA lacY supE recA::RP4-2-Yc::Mu-Kan (λpir) | 67 |
| Plasmids | ||
| pUC18 | Cloning vector; Ampr | 79 |
| pAY100.1 | O-PS gene cluster of YeO:3 cloned in pBR322; Ampr | 53 |
| pRV16NP | Outer core gene cluster of YeO:3 cloned in pTM100; Clmr | 39 |
| pRK2013 | Helper plasmid for conjugation; Kanr | 25 |
| p34S-Km | Cloning vector; Ampr Kanr | 23 |
| pCVD442 | Suicide vector; Ampr | 26 |
| pCVD442-wabD | Suicide vector with wabD::nptII; Ampr Kanr | 5 |
| pCVD442-waaL | Suicide vector with waaL::nptII; Ampr Kanr | 5 |
| pCVD442-waaQ | Suicide vector with waaQ::nptII; Ampr Kanr | 5 |
| pCVD442-waaE | Suicide vector with waaE::nptII; Ampr Kanr | 5 |
| pCVD442-waaA | Suicide vector with waaA::nptII; Ampr Kanr | 5 |
| pUCwbup | Upstream region of O-PS gene cluster of YPIII cloned in pUC18; Ampr | This work |
| pUCwbdel | Up- and downstream regions of the YPIII O-PS gene cluster cloned in pUC18; Ampr | This work |
| pUCwbGB | Kanr gene of p34S-Kan cloned between the up- and downstream regions of the YPIII O-PS gene cluster in pUCwbdel; Ampr Kanr | This work |
| pCVDwbGB | Suicide vector carrying the PvuII fragment of pUCwbGB, including the Kanr gene and the up- and downstream regions of O-PS gene cluster of YPIII; Ampr Kanr | This work |
Lcr, low calcium response; Pgm, pigmentation; Pst, pesticin resistance.
Construction of mutants.
Standard recombinant DNA techniques were applied as described previously (64). All the enzymes were used as recommended by the suppliers. To construct the strain PB1Δwb, a region upstream of the O-PS gene cluster was amplified by PCR using primers P1 (5′-CCGGAATTCGAGCTCATGCGTATCATTCTGCTGGGC-3′) and P2 (5′-CGGGGTACCTTATATATTATGTCGAAT-3′), and chromosomal DNA of the Y. pseudotuberculosis strain YPIII (Table 1) was used as template. This fragment was cloned into pUC18 digested with EcoRI-KpnI to obtain plasmid pUCwbup. A region downstream of the O-PS gene cluster was amplified by PCR using primers P3 (5′-CGGGGTACCTTTAGTTGAGCACTTTGT-3′) and P4 (5′-AAACTGCAGAGCTCTATGCTGTTCGACCCAATGAT-3′) and was cloned into pUCwbup digested with KpnI and PstI to give pUCwbdel. A kanamycin gene cassette was obtained by digestion of p34S-Km (Table 1) with KpnI and gel purified. This cassette was cloned into the KpnI site of pUCwbdel to give pUCwbGB. A PvuII fragment form pUCwbGB was cloned into the SmaI site of plasmid pCVD442 (Table 1) to give pCVDwbGB. Mutants were selected after mating E. coli Sm10λpir/pCVDwbGB with Y. pseudotuberculosis strain PB1 (Table 1) by following the protocol previously described (9). Mutant genotypes were confirmed by PCR and Southern blot hybridization with appropriate DNA probes (data not shown).
A series of Y. pestis D27 mutant strains with truncated LPS were generated by allelic exchange using pCVD442-based suicide vectors (Table 1). The suicide vectors were introduced into the parental Y. pestis D27 strain from E. coli S17-1λpir (Table 1) by conjugation, and Kanr Ampr merodiploid transconjugants were selected using CIN(Suc) agar for counterselection of the donor. The selected merodiploids were plated onto brain heart infusion (BHI) agar with 10% sucrose and grown at RT for 2 days. The correct allelic exchange in the resultant Sucr Kanr Amps colonies was confirmed using PCR with corresponding primers (Table 2). The Y. pestis KM260(11) LPS mutants, on the other hand, were generated by allelic exchange based on homologous recombination between genomic DNA and PCR products (5, 20).
Table 2.
Primers used for verification of the Y. pestis D27 LPS mutants
| Primer | Gene | Primer sequence |
|---|---|---|
| YPO0187-F | wabD | AATGCGGTACGTTGTGGTGA |
| YPO0187-R | wabD | TTTGCCGATGGTGATGATTG |
| YPO0417-F | waaL | AGTTGATTCCTGGCGAGTTG |
| YPO0417-R | waaL | CTCCCCTGCCTATCCTCACC |
| YPO0416-F | waaQ | GCTGCGTGTATGCTCCGTTGACTG |
| YPO0416-R | waaQ | ATCCGGGCCATAGCTGTTGTTTTG |
| YPO0054-F | waaE | TAGTAATGGGATCAAATGTC |
| YPO0054-R | waaE | ACTCTATCGCTGGTAAAAG |
| YPO0055-F | waaA | TTTGGCTGCGTTTACTATTA |
| YPO0055-R | waaA | ACACCGTGATTTCTTTTACC |
φA1122-resistant derivatives of Y. pestis D27 and Y. pseudotuberculosis PB1Δwb were isolated by spreading bacterial suspension on agar plates and pipetting drops of phage lysate on a dry bacterial lawn. Phage-resistant colonies were picked from within the lysis zone after 2 to 4 days.
Plasmid pRV16NP was mobilized to Y. pseudotuberculosis by triparental conjugation using helper strain HB101/pRK2013 (Table 1) (32).
Phage adsorption assays and calculation of efficiency of plating (EOP).
Approximately 8 × 105 PFU of φA1122 in 100 μl was mixed with a 500-μl sample of bacteria (A600 = 1.2). For Y. enterocolitica and Y. pseudotuberculosis overnight cultures and for Y. pestis, a 48-h culture was used. The suspension was incubated at RT for 5 min and centrifuged at 16,000 × g for 3 min, and the phage titer remaining in the supernatant, i.e., the residual PFU percentage, was determined. TSB was used as a nonadsorbing control in each assay, and the phage titer in the control supernatant was set to 100%. Each assay was performed in duplicate and repeated at least twice.
To assay the adsorption kinetics and eliminate the effect of reversible adsorption, the following protocol was used (1). The phage and bacteria were mixed in a total volume of 2 ml (ca. 2 × 106 PFU/108 CFU). A TSB tube without bacteria was used as a negative control. At different time points, duplicate 10-μl samples were withdrawn from the tube and mixed by vortexing with 990 μl of TSB in an Eppendorf tube to release reversibly adsorbed phage. After 2 min of centrifugation at 16,000 × g, 0.5 ml of the supernatant was withdrawn and stored on ice for titration of the PFU. The residual PFU percentage was calculated as described above. The adsorption rate constants were calculated as described previously (1).
To determine the efficiency of plating (EOP), 100 μl of wild-type (WT) and mutant Y. pestis cultured bacteria (A600 = 1.2) was mixed with 50 μl of φA1122 (2 × 103 PFU/ml) in 3 ml of 0.4% soft agar and poured onto LB plates. The number of PFU was counted after 24 to 48 h. Each strain was tested in triplicate. The EOP was calculated using the following formula: EOP = (number of PFU on mutant strain)/(number of PFU on the wild type).
Periodate and proteinase K treatments.
To test how proteinase K treatment affects φA1122 adsorption, 2 ml of D27 and PB1Δwb cultures was treated with proteinase K (0.2 mg/ml; Promega) at 37°C for 3 h and washed with 2 ml of TSB, and the phage adsorption assay was performed as described above. To confirm that the possible effect was not due to incubation at 37°C, a control without proteinase K addition was included.
In order to study whether periodate can destroy the phage receptor, 1.5 ml of D27 culture was centrifuged at 16,000 × g for 1 min, and the bacterial pellet was suspended into 1.5 ml sodium acetate (50 mM; pH 5.2) or sodium acetate containing either 10 or 100 mM IO4−. The cells were incubated for 2 h (protected from light), centrifuged as described above, washed with 1.5 ml TSB, centrifuged, and suspended in TSB. Finally, the A600 of the bacterial suspension was adjusted to 1, and the phage adsorption assay was carried out.
Isolation and analysis of lipopolysaccharide.
Small-scale isolation and deoxycholate-polyacrylamide gel electrophoresis (DOC-PAGE) analysis of LPS were performed as described earlier (70, 81). Large-scale isolation and compositional analyses of LPS were performed as described previously (22).
RESULTS
Effect of temperature on the expression of the φA1122 receptor.
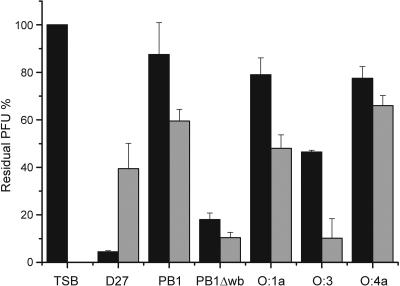
The earlier observation that φA1122 infects Y. pestis grown at 22°C and at 37°C but Y. pseudotuberculosis only when grown at 37°C (30) led us to study the effect of temperature on the expression of the phage receptors of these species more closely. To this end, the phage adsorption to Y. pestis D27 and different Y. pseudotuberculosis strains cultured at RT and 37°C was examined (Fig. 1). A slightly surprising finding was that D27 seemed to adsorb more phage at RT than at 37°C. All the smooth Y. pseudotuberculosis strains tested, representing serotypes O:1a, O:1b, O:3, and O:4a (Table 1), adsorbed more phage at 37°C than at RT, even though for serotype O:4a, the difference was not so apparent. On the other hand, strain PB1Δwb, a rough derivative of serotype O:1b strain PB1, adsorbed φA1122 equally well at both temperatures (Fig. 1). These results may suggest that the phage receptor is the LPS core that is sterically blocked by O-PS expression in Y. pseudotuberculosis at temperatures below 30°C. However, the possibility that the receptor is an OMP blocked by O-PS cannot be rigorously excluded.
Fig. 1.
Effect of temperature on φA1122 adsorption to Y. pestis and Y. pseudotuberculosis. Black bars show the residual PFU percentages after adsorption of phage on bacteria grown at RT, and gray bars show the residual PFU percentages after adsorption of phage on bacteria grown at +37°C. The control (TSB) and strains used for adsorptions are indicated below the columns. Error bars indicate ranges.
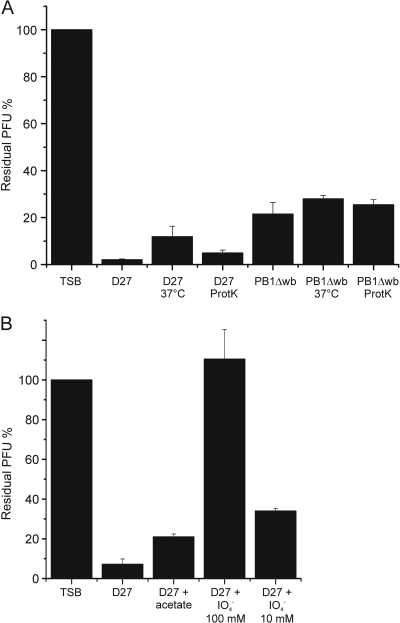
Periodate but not proteinase K destroys phage receptors.
Since bacteriophage can exploit both LPS and OMPs as receptors (43, 77), it was important to test whether the degradation of cell surface proteins or LPS could destroy the φA1122 receptor. For this reason, bacteria were treated with proteinase K or periodate prior to the phage adsorption assay. As seen in Fig. 2A, the proteinase K treatment of Y. pestis D27 or Y. pseudotuberculosis PB1Δwb did not reduce the adsorption capacity of the cells, suggesting that the functional receptor does not contain a protein structure. The possibility that the receptor is a protein resistant to proteinase K is unlikely, considering the broad substrate specificity of this enzyme (27).
Fig. 2.
Effects of different treatments of bacteria on φA1122 adsorption, shown as residual PFU percentages. (A) The effect of proteinase K treatment on adsorption of φA1122 to Y. pestis and Y. pseudotuberculosis. (B) The effect of periodate treatment on adsorption of φA1122 to Y. pestis. The control, strains, and treatments used for adsorptions are indicated below the columns. Error bars indicate ranges.
To study how the degradation of carbohydrates affects the phage adsorption, the effect of periodate (degrades carbohydrates containing a 1,2-diol motif in their structure) on the phage receptor was tested. Incubation of Y. pestis D27 in the presence of 100 mM periodate abolished φA1122 binding completely, whereas incubation in 10 mM periodate or acetate buffer alone did not (Fig. 2B). This result thus confirmed that a carbohydrate structure, most likely LPS, is the receptor for φA1122.
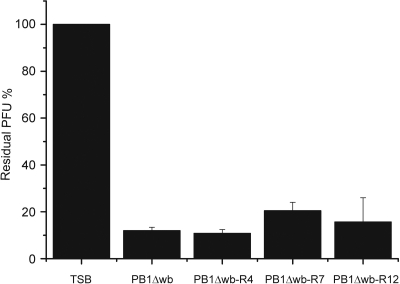
φA1122-resistant bacterial mutants.
In order to study φA1122 adsorption more thoroughly, we aimed to isolate phage-resistant mutants of Y. pestis D27 and Y. pseudotuberculosis PB1Δwb. LPS biosynthesis involves tens of genes; therefore, isolation of spontaneous LPS mutants is usually easy, but that was not the case here. After substantial efforts, we isolated a few D27 mutants but only with reduced infectivity; the EOP of φA1122 on the most resistant mutant obtained was still ca. 0.2 (data not shown). With PB1Δwb we were more successful; strain PB1Δwb-R7 was fully φA1122 resistant, and strains PB1Δwb-R4 and PB1Δwb-R12 were highly resistant, having EOPs of ca. 10−3 (Table 1). Surprisingly, when φA1122 adsorption to these mutants was measured, they all adsorbed the phage as efficiently as the parental strain PB1Δwb (Fig. 3). The same was true for the moderately resistant derivatives of D27 (data not shown). Thus, even though the phage propagation on these mutants was restricted, the phage receptor was not affected. In conclusion, these results indicated that under the experimental conditions applied, the intact phage receptor was essential for the growth of Y. pestis and Y. pseudotuberculosis. The LPS sugar compositions of the resistant mutants and those of wild-type bacteria were analyzed, and no significant differences were detected (data not shown).
Fig. 3.
φA1122 adsorption to phage-resistant derivatives of Y. pseudotuberculosis PB1Δwb, shown as residual PFU percentages. The control and strains used for adsorptions are indicated below the columns. Error bars indicate ranges.
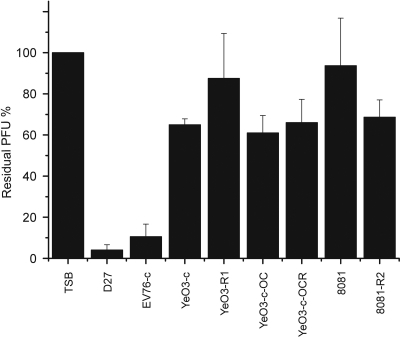
Absence of the φA1122 receptor in Y. enterocolitica.
The LPS core structures of Y. enterocolitica and Y. pestis are similar but not identical (16, 52, 75). To study whether Y. enterocolitica contains the φA1122 receptor, the phage adsorption assay was conducted with Y. enterocolitica strains belonging to serotypes O:3 and O:8 either expressing wild-type LPS or missing the O-PS, outer core (OC), or both (Table 1). Y. pestis strains D27 and EV76-c (Table 1) were used as positive controls. As seen in Fig. 4, none of the Y. enterocolitica strains, not even the rough or deep rough derivatives, specifically adsorbed φA1122. The phage receptor therefore is most likely a structure of the LPS cores of Y. pestis and Y. pseudotuberculosis that is not present in Y. enterocolitica.
Fig. 4.
φA1122 adsorption to Y. pestis and Y. enterocolitica serotype O:3 and O:8 wild-type and LPS mutant strains, shown as residual PFU percentages. The control and strains used for adsorptions are indicated below the columns. Error bars indicate ranges.
Blocking the φA1122 receptor.
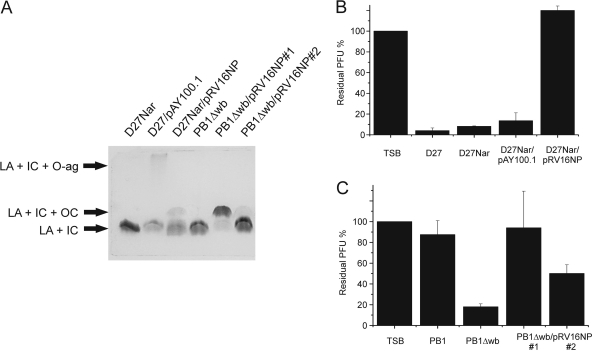
In Y. enterocolitica serotype O:3, O-PS and OC hexasaccharide are linked to the LPS inner core, with the latter specifically linked to Hep(II) (28, 57, 69). To obtain more information about the carbohydrate residues forming the φA1122 receptor, the effect of heterologous expression of Y. enterocolitica O:3 O-PS and OC on Y. pestis D27 and OC on Y. pseudotuberculosis PB1Δwb for phage adsorption was studied. To this end, plasmids pAY100.1 and pRV16NP (Table 1), containing Y. enterocolitica O:3 O-PS and OC gene clusters, respectively, were used. The expression of heterologous LPS was verified by DOC-PAGE analysis (Fig. 5A) and by using O-PS- and OC-specific bacteriophages φYeO3-12 and φR1-37 (Table 1), respectively (data not shown). The adsorption assay for these strains showed that the expression of Y. enterocolitica O:3 O-PS on Y. pestis had no effect on φA1122 binding, whereas Y. enterocolitica O:3 OC blocked the phage receptor completely (Fig. 5B). Consistently, strain D27/pAY100.1 was sensitive to φA1122, while strain D27Nar/pRV16NP was resistant (data not shown).
Fig. 5.
Expression of Y. enterocolitica O:3 O-PS and outer core hexasaccharide in Y. pestis and in Y. pseudotuberculosis. (A) Analysis of LPS by DOC-PAGE and silver staining. LA, lipid A; IC, inner core; OC, outer core; O-ag, O-PS. (B) Effect of Y. enterocolitica O:3 O-PS and outer core hexasaccharide expression on φA1122 adsorption to Y. pestis, shown as residual PFU percentages. (C) Effect of Y. enterocolitica O:3 outer core hexasaccharide expression on φA1122 adsorption to Y. pseudotuberculosis, shown as residual PFU percentages. The control and strains used for adsorptions are indicated below the columns. TSB was used as a no-bacteria control. Error bars indicate ranges.
In the case of Y. pseudotuberculosis PB1Δwb, two different clones were obtained after the conjugation, named PB1Δwb/pRV16NP#1 and PB1Δwb/pRV16NP#2. These clones differed from each other by the expression level of Y. enterocolitica O:3 OC, since OC was more strongly expressed in PB1Δwb/pRV16NP#1 than in PB1Δwb/pRV16NP#2 or in D27Nar/pRV16NP (Fig. 5A). Coherently with the Y. enterocolitica O:3 OC expression level, strain PB1Δwb/pRV16NP#1 was resistant to φA1122 and did not adsorb it at all, while PB1Δwb/pRV16NP#2 was sensitive and adsorbed the phage moderately (Fig. 5C). The OC overexpression of PB1Δwb/pRV16NP#1 is most probably due to a mutation in plasmid pRV16NP#1, since the overexpressing phenotype transfers with the plasmid (data not shown). The nature of this mutation is not yet known.
Mapping the φA1122 receptor by using LPS mutants.
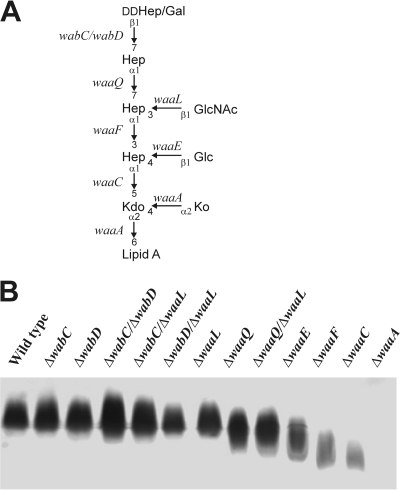
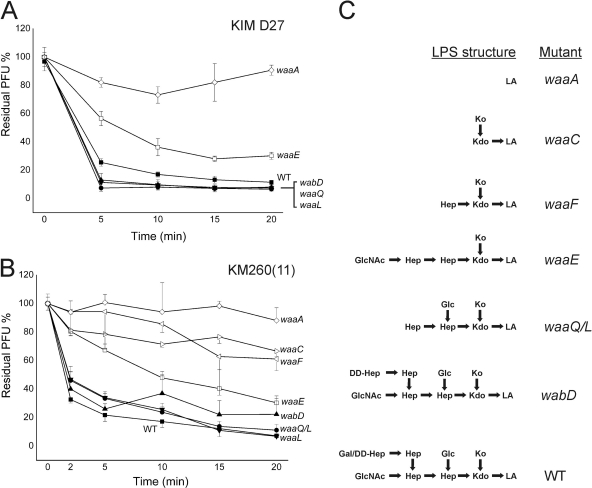
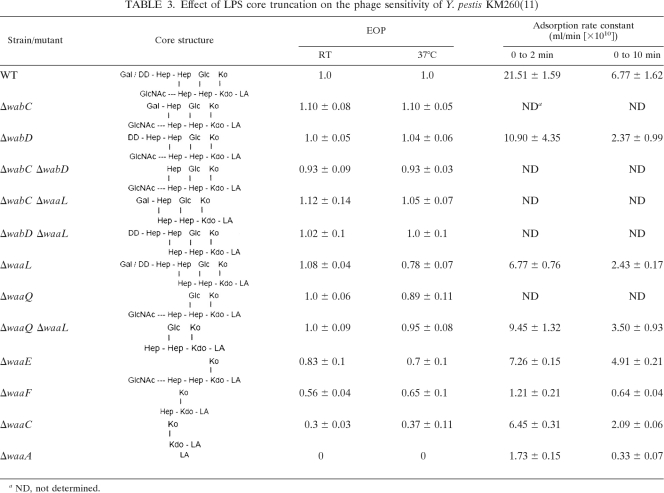
After establishing LPS as the probable receptor for phage φA1122, we deleted the wabD, waaL, waaQ, waaE, and waaA genes (Fig. 6A) of the LPS core biosynthetic pathway in Y. pestis D27 in order to pinpoint the role of LPS core in the phage receptor. The deletions caused the predicted LPS truncations (5), as verified by DOC-PAGE analysis (data not shown). Most of the deletion mutants grew normally under the applied conditions; however, the growth rate of the ΔwaaA mutant was severely decreased (data not shown). We measured the EOPs and adsorption kinetics of the D27 wild type and the mutant strains. Both approaches revealed that the phage infected the D27 ΔwabD, ΔwaaL, and ΔwaaQ strains at a level comparable to that of the wild-type strain; i.e., the EOPs were 0.97 to 0.98, and the residual PFU percentage in the adsorption kinetics assay at the 10-min time point was 8 to 10%, compared to 17% for the wild type (Fig. 7A). However, the D27 ΔwaaE strain was less sensitive (residual PFU percentage at 10 min was ∼40%, with an EOP of 0.28), and finally, the D27 ΔwaaA strain was completely resistant. These results provided direct evidence showing LPS as the receptor of the phage. To study the roles of the individual sugar residues in the Hep/Glc-Kdo/Ko region, we used a set of deep rough mutants of Y. pestis KM260(11) (Table 1; Fig. 6). In general, the Y. pestis KM260(11) mutants behaved similarly to the corresponding Y. pestis D27 mutants (Table 3; Fig. 7B). With the KM260(11) mutants, infection and adsorption defect became visible with the ΔwaaE strain missing the Glc residue, after which the phage sensitivity gradually decreased with further losses of the Hep(II) and Hep(I) residues (Table 3). As with D27, the KM260(11) ΔwaaA strain missing the Kdo/Ko region was completely resistant (Table 3).
Fig. 6.
Structure of Y. pestis LPS and characteristics of LPS mutants. (A) Schematic structure and the relevant genes involved in the biosynthesis of the core oligosaccharide. The glycosidic bonds between different residues are indicated. (Modified from reference 5.) (B) Silver-stained SDS-PAGE of LPS isolated from wild-type KM260(11) and LPS mutants. The deleted genes are indicated at the top of the gel.
Fig. 7.
Adsorption kinetics of φA1122 to Y. pestis LPS mutants, shown as residual PFU percentages. (A) Y. pestis KIM D27 derivatives. (B) Y. pestis KM260 (11) derivatives. Standard errors in panels A and B are indicated by vertical lines (in panel B, only in one direction). (C) LPS core structures of the mutants (Fig. 6). (Modified from reference 5.)
Table 3.
Effect of LPS core truncation on the phage sensitivity of Y. pestis KM260(11)
ND, not determined.
The adsorption kinetics assay (Fig. 7) demonstrated that irreversible adsorption to wild-type bacteria was extremely rapid; most of the adsorption (over 70%) took place within the first 2 min, and by 5 min, the adsorption rate leveled off. On the other hand, adsorption to the ΔwaaE, ΔwaaF, and ΔwaaC strains was much slower and continued over the duration of the experiment. The ΔwaaA bacteria adsorbed no or very few phage particles. The adsorption rate constants were calculated for the strains based on the data of the 2- and 10-min time points presented in Fig. 7B (Table 3). As expected, the wild-type strain had the highest adsorption rate constant, and the deep rough mutants had the lowest. Interestingly, for all strains, the 10-min constants were lower than the respective 2-min constants, indicating that a subfraction of phages adsorbed slower (44). Nevertheless, these numbers further demonstrated that the phage receptor was affected in the ΔwaaE, ΔwaaF, and ΔwaaC strains.
The above-described experiments were all carried out with bacteria grown at RT. Interestingly, when the experiments were carried out with bacteria grown at 37°C, we observed reduced adsorption efficiencies for ΔwabC, ΔwabD, ΔwabC ΔwabD, ΔwabC ΔwaaL, and ΔwabD ΔwaaL mutants (data not shown), while there were no significant differences in their plating efficiencies (Table 3). This could be attributed to some conformational changes occurring in the Y. pestis core structure, surface charge changes causing repulsion, or expression of loosely blocking surface structures. All these could be at least partially due to any of the temperature-dependent structural variations taking place in Y. pestis LPS structure at 37°C, i.e., replacement of Ko with Kdo(II) or of Gal with dd-Hep(IV), or to the change of lipid A acylation from penta/hexa acylated to tetra acylated and the decrease in 4-amino-4-deoxy-l-arabinose substitution of lipid A (40).
DISCUSSION
The susceptibility of a bacterium to bacteriophage infection is dependent primarily on whether or not the bacteriophage can find its specific attachment sites, i.e., the receptors on the cell. The recognition of the receptor is a highly specific process and is part of the natural mechanism of host recognition. During the last decade, the cell envelopes of both Gram-negative and Gram-positive bacteria have been studied intensively from structural, biosynthetic, genetic, and functional viewpoints. As a result of these studies, information of considerable value for investigations of phage receptor sites has emerged. Specifically, LPS, as a thoroughly studied component of the OM of Gram-negative bacteria, can give us detailed information on the structure of phage receptors. Different parts of LPS function as receptors for a number of phages in many different genera. For example, LPS is known to act as a receptor for bacteriophages T3, T4, and T7 in E. coli, for T2 and T4 in Shigella dysenteriae, for Sf6 in Shigella flexneri, for P22 in Salmonella enterica serovar Typhimurium, for K139 in Vibrio cholerae, and for φYeO3-12 and φR1-37 in Y. enterocolitica (2, 8, 17, 39, 45, 48, 51, 54, 58, 62, 70, 76).
φA1122 is a T7/T3/φYeO3-12-related diagnostic phage used by the CDC in identifying wild or clinical isolates of Y. pestis (30). In this work, we characterized the φA1122 receptor by studying the phage adsorption to different LPS mutants of Y. pestis and Y. pseudotuberculosis. We found that the phage adsorbed to Y. pestis D27 and rough Y. pseudotuberculosis strain PB1Δwb grown at RT and 37°C but to smooth PB1 only when grown at 37°C, indicating that the abundant Y. pseudotuberculosis O-PS expression at RT sterically blocked the phage receptor. The finding that periodate, but not proteinase K, destroyed the receptor (Fig. 2) pointed out the carbohydrate nature of the receptor. This carbohydrate is most likely LPS, since the enterobacterial common antigen cannot be destroyed by periodate treatment, owing to its structure (46). This was in agreement with the notion that the closely related phages T7 and T3 utilize the LPS core of E. coli as their receptors (45) and that the T3 and φA1122 tail fibers are 98.9% identical with 6 amino acid differences in 558 amino acids (30). The fact that selection of phage-resistant mutants was difficult and that all obtained phage-resistant derivatives of Y. pseudotuberculosis PB1Δwb retained the phage receptor, i.e., still adsorbed the phage as the wild type (Fig. 4), indicated that the receptor would be essential for bacterial growth under the experimental conditions used and, therefore, is deep in the core structure of LPS. Indeed, the growth rate of the fully resistant ΔwaaA mutant was significantly decreased, explaining why such mutants could not be isolated during the in vivo screening. These observations are corroborated by the findings that truncation of the LPS decreases the serum resistance, cationic antimicrobial peptide resistance, and virulence of Y. pestis (5). This also is an implication that phage receptor mutants would be efficiently eliminated by host defense mechanisms during infection. At present we can only speculate on the nature of the phage resistance mechanism in the isolated resistant derivatives; apparently a host factor(s) required for phage developmental cycle is mutated or missing. Since the virulence of such derivatives might not be compromised, this could be a concern if therapeutic use of phage φA1122 is considered (4).
As an attempt to further characterize which core sugar residues are needed for φA1122 adsorption, the potential of Y. enterocolitica serotype O:3 O-PS or OC to inhibit the phage binding when expressed on the Y. pestis or Y. pseudotuberculosis core oligosaccharide was studied. The O:3 O-PS expressed in Y. pestis had no effect on φA1122 adsorption, in contrast to OC, which blocked it efficiently (Fig. 5B). In fact, partial substitution of LPS molecules seemed to be sufficient, as the phage adsorption to both Y. pestis and Y. pseudotuberculosis was blocked even when unsubstituted LPS molecules were present (Fig. 5). An interesting finding was that while the Y. pseudotuberculosis O-PS blocked the phage receptor, the expression of Y. enterocolitica O:3 O-PS in Y. pestis had no effect on the phage binding. The location of the attachment site of Y. enterocolitica O:3 O-PS in the core is not known, but this study implies that it is different from that of Y. pseudotuberculosis, which is at O-3 of Hep(II).
Finally, to decipher the minimal portion of LPS required as a receptor for the phage, adsorption studies with truncated LPS mutants of Y. pestis D27 and Y. pestis strain KM260(11) were carried out. The mutants with outermost core truncations missing Hep(III), dd-Hep(IV), and GlcNAc had EOPs comparable to those of the wild type (WT) (Table 3) and supported the phage binding in the adsorption experiments (Fig. 7). The ΔwaaE mutants of both the D27 and KM260(11) strains showed a small defect, as both had decreased EOPs and reduced adsorption (Table 3; Fig. 7), and the defect increased with increasing truncation of the core (ΔwaaF and ΔwaaC mutants). The only fully φA1122-resistant derivatives were the ΔwaaA mutants of both strains. Effectively, it seems that since the removal of the Hep(I)-linked Glc residue in the ΔwaaE mutant reduced the phage adsorption by ∼30%, the Glc residue is part of the receptor. The phage infection assays (EOPs) with the ΔwaaE mutant strain point to the same conclusion. In summary, our data revealed that the complete phage receptor contains two Hep residues with a Glc branching from Hep(I) attached to lipid A via Kdo/Ko.
The φA1122 receptor was not present in any of the Y. enterocolitica strains studied (Fig. 3). The core structures of different Yersinia species are rather similar, and in fact, the Y. enterocolitica serotype O:3 and O:8 cores resemble those of Y. pestis and Y. pseudotuberculosis, with the major differences being an additional Glc residue β-(1→2) linked to Hep(II) and the presence of Kdo(II) instead of Ko α-(2→4) linked to Kdo(I). In addition, Y. enterocolitica O:3 deep rough mutants (hldE, waaF, and galU mutants) were fully φA1122 resistant (data not shown). In the hldE, waaF, and galU mutants, the core is truncated at Kdo(II), Hep(I), and Hep(II), respectively. As corresponding Y. pestis KM260(11) mutants were infected by φA1122, the presence of Ko appears to be of central importance to the phage receptor. Interestingly, the Kdo dioxygenase gene of Y. pestis involved in converting Kdo to Ko was recently identified (18). The homolog in Y. enterocolitica is a pseudogene, explaining the absence of Ko in this species. Finally, the distinct host range difference between T3 and φA1122 (30), despite the 98.9% identity between their tail fibers, could be due to Ko, which is also absent in E. coli; the presence of an additional hydroxyl group in Ko at C-3 instead of a hydrogen changes the chemical microenvironment from hydrophobic to more hydrophilic, and it might require a major change in the tail fiber binding pocket surface to accommodate this difference. Interestingly, out of the six replacements between the tail fibers of φA1122 and T3 (Y333H, L350V, K468M, G478S, L523S, S544A), the most dramatic change is provided by the K468M replacement. Future work will be needed to elucidate this question.
ACKNOWLEDGMENT
This work was funded by the Academy of Finland (grants 50441, 1104361, and 1114075 to M.S.).
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Adams M. H. 1959. Bacteriophages. Interscience Publishers, Inc., New York, NY [Google Scholar]
- 2. al-Hendy A., Toivanen P., Skurnik M. 1991. Expression cloning of Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb. Pathog. 10:47–59 [DOI] [PubMed] [Google Scholar]
- 3. al-Hendy A., Toivanen P., Skurnik M. 1992. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect. Immun. 60:870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anisimov A. P., Amoako K. K. 2006. Treatment of plague: promising alternatives to antibiotics. J. Med. Microbiol. 55:1461–1475 [DOI] [PubMed] [Google Scholar]
- 5. Anisimov A. P., et al. 2010. Yersinia pestis lipopolysaccharide in host-pathogen interactions, p. 77–87 In Shafferman A., Ordentlich A., Velan B. (ed.), The challenge of highly pathogenic microorganisms. Mechanisms of virulence and novel medical countermeasures. Springer, New York, NY [Google Scholar]
- 6. Anisimov A. P., Lindler L. E., Pier G. B. 2004. Intraspecific diversity of Yersinia pestis. Clin. Microbiol. Rev. 17:434–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Appleyard R. K. 1954. Segregation of new lysogenic types during growth of doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baxa U., et al. 1996. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys. J. 71:2040–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bengoechea J. A., Zhang L., Toivanen P., Skurnik M. 2002. Regulatory network of lipopolysaccharide O-antigen biosynthesis in Yersinia enterocolitica includes cell envelope-dependent signals. Mol. Microbiol. 44:1045–1062 [DOI] [PubMed] [Google Scholar]
- 10. Ben-Gurion R., Hertman I. 1958. Bacteriocin-like material produced by Pasteurella pestis. J. Gen. Microbiol. 19:289–297 [DOI] [PubMed] [Google Scholar]
- 11. Biedzka-Sarek M., Venho R., Skurnik M. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 73:2232–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bölin I., Norlander L., Wolf-Watz H. 1982. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect. Immun. 37:506–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bond P. J., Sansom M. S. 2004. The simulation approach to bacterial outer membrane proteins. Mol. Membr. Biol. 21:151–161 [DOI] [PubMed] [Google Scholar]
- 14. Bos M. P., Tommassen J. 2004. Biogenesis of the Gram-negative bacterial outer membrane. Curr. Opin. Microbiol. 7:610–616 [DOI] [PubMed] [Google Scholar]
- 15. Boyer H. W., Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459–472 [DOI] [PubMed] [Google Scholar]
- 16. Bruneteau M., Minka S. 2003. Lipopolysaccharides of bacterial pathogens from the genus Yersinia: a mini-review. Biochimie 85:145–152 [DOI] [PubMed] [Google Scholar]
- 17. Chua J. E., Manning P. A., Morona R. 1999. The Shigella flexneri bacteriophage Sf6 tailspike protein (TSP)/endorhamnosidase is related to the bacteriophage P22 TSP and has a motif common to exo- and endoglycanases, and C-5 epimerases. Microbiology 145:1649–1659 [DOI] [PubMed] [Google Scholar]
- 18. Chung H. S., Raetz C. R. 2011. Dioxygenases in Burkholderia ambifaria and Yersinia pestis that hydroxylate the outer Kdo unit of lipopolysaccharide. Proc. Natl. Acad. Sci. U. S. A. 108:510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Darveau R. P., Charnetzky W. T., Hurlbert R. E., Hancock R. E. W. 1983. Effects of growth temperature, 47-megadalton plasmid, and calcium deficiency on the outer membrane protein porin and lipopolysaccharide composition of Yersinia pestis EV76. Infect. Immun. 42:1092–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davies D. A. 1958. The smooth and rough somatic antigens of Pasteurella pseudotuberculosis. J. Gen. Microbiol. 18:118–128 [DOI] [PubMed] [Google Scholar]
- 22. De Castro C., Skurnik M., Molinaro A., Holst O. 2009. Characterization of the specific O-polysaccharide structure and biosynthetic gene cluster of Yersinia pseudotuberculosis serotype O:15. Innate Immun. 15:351–359 [DOI] [PubMed] [Google Scholar]
- 23. Dennis J. J., Zylstra G. J. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dentovskaya S. V., et al. 2008. Structural diversity and endotoxic activity of the lipopolysaccharide of Yersinia pestis. Biochemistry 73:192–199 [DOI] [PubMed] [Google Scholar]
- 25. Ditta G., Stanfield S., Corbin D., Helinski D. R. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. U. S. A. 77:7347–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Donnenberg M. S., Kaper J. B. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ebeling W., et al. 1974. Proteinase K from Tritirachium album Limber. Eur. J. Biochem. 47:91–97 [DOI] [PubMed] [Google Scholar]
- 28. Frirdich E., Whitfield C. 2005. Lipopolysaccharide inner core oligosaccharide structure and outer membrane stability in human pathogens belonging to the Enterobacteriaceae. J. Endotoxin Res. 11:133–144 [DOI] [PubMed] [Google Scholar]
- 29. Galloway S. M., Raetz C. R. 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 265:6394–6402 [PubMed] [Google Scholar]
- 30. Garcia E., et al. 2003. The genome sequence of Yersinia pestis bacteriophage φA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J. Bacteriol. 185:5248–5262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia E., Nedialkov Y. A., Elliott J., Motin V. L., Brubaker R. R. 1999. Molecular characterization of KatY (antigen 5), a thermoregulated chromosomally encoded catalase-peroxidase of Yersinia pestis. J. Bacteriol. 181:3114–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerhardt P., Murray R. G. E., Wood W. A., Krieg N. R. 1984. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 33. Hardy P. H., Jr., Levin J. 1983. Lack of endotoxin in Borrelia hispanica and Treponema pallidum. Proc. Soc. Exp. Biol. Med. 174:47–52 [DOI] [PubMed] [Google Scholar]
- 34. Hartley J. L., Adams G. A., Tornabene T. G. 1974. Chemical and physical properties of lipopolysaccharide of Yersinia pestis. J. Bacteriol. 118:848–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawahara K., et al. 2001. Structural analysis of a new glycosphingolipid from the lipopolysaccharide-lacking bacterium Sphingomonas adhaesiva. Carbohydr. Res. 333:87–93 [DOI] [PubMed] [Google Scholar]
- 36. Kawahara K., Moll H., Knirel Y. A., Seydel U., Zähringer U. 2000. Structural analysis of two glycosphingolipids from the lipopolysaccharide-lacking bacterium Sphingomonas capsulata. Eur. J. Biochem. 267:1837–1846 [DOI] [PubMed] [Google Scholar]
- 37. Kawahara K., Tsukano H., Watanabe H., Lindner B., Matsuura M. 2002. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 70:4092–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kawasaki S., et al. 1994. The cell envelope structure of the lipopolysaccharide-lacking gram-negative bacterium Sphingomonas paucimobilis. J. Bacteriol. 176:284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kiljunen S., et al. 2005. Yersiniophage φR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology 151:4093–4102 [DOI] [PubMed] [Google Scholar]
- 40. Knirel Y. A., et al. 2006. Structural features and structural variability of the lipopolysaccharide of Yersinia pestis, the cause of plague. J. Endotoxin Res. 12:3–9 [DOI] [PubMed] [Google Scholar]
- 41. Knirel Y. A., et al. 2008. New features of Yersinia lipopolysaccharide structures as revealed by high-resolution electrospray ionization mass spectrometry. Adv. Sci. Lett. 1:192–198 [Google Scholar]
- 42. Knirel Y. A., et al. 2005. Temperature-dependent variations and intraspecies diversity of the structure of the lipopolysaccharide of Yersinia pestis. Biochemistry 44:1731–1743 [DOI] [PubMed] [Google Scholar]
- 43. Koebnik R., Locher K. P., Van Gelder P. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239–253 [DOI] [PubMed] [Google Scholar]
- 44. Kropinski A. M. 2009. Measurement of the rate of attachment of bacteriophage to cells. Methods Mol. Biol. 501:151–155 [DOI] [PubMed] [Google Scholar]
- 45. Krüger D. H., Schroeder C. 1981. Bacteriophage T3 and bacteriophage T7 virus-host cell interactions. Microbiol. Rev. 45:9–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuhn H.-M., Meier-Dieter U., Mayer H. 1988. ECA, the enterobacterial common antigen. FEMS Microbiol. Rev. 54:195–222 [DOI] [PubMed] [Google Scholar]
- 47. Leone S., et al. 2006. The structures of the cell wall teichoic acids from the thermophilic microorganism Geobacillus thermoleovorans strain Fango. Carbohydr. Res. 341:2613–2618 [DOI] [PubMed] [Google Scholar]
- 48. Lindberg A. A. 1973. Bacteriophage receptors. Annu. Rev. Microbiol. 27:205–241 [DOI] [PubMed] [Google Scholar]
- 49. Mamat U., et al. 2008. Single amino acid substitutions in either YhjD or MsbA confer viability to 3-deoxy-d-manno-oct-2-ulosonic acid-depleted Escherichia coli. Mol. Microbiol. 67:633–648 [DOI] [PubMed] [Google Scholar]
- 50. Meredith T. C., Aggarwal P., Mamat U., Lindner B., Woodard R. W. 2006. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Biol. 1:33–42 [DOI] [PubMed] [Google Scholar]
- 51. Nesper J., Kapfhammer D., Klose K. E., Merkert H., Reidl J. 2000. Characterization of Vibrio cholerae O1 antigen as the bacteriophage K139 receptor and identification of IS1004 insertions aborting O1 antigen biosynthesis. J. Bacteriol. 182:5097–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oertelt C., Lindner B., Skurnik M., Holst O. 2001. Isolation and structural characterization of an R-form lipopolysaccharide from Yersinia enterocolitica serotype O:8. Eur. J. Biochem. 268:554–564 [DOI] [PubMed] [Google Scholar]
- 53. Oyston P. C., et al. 2003. Expression of heterologous O-antigen in Yersinia pestis KIM does not affect virulence by the intravenous route. J. Med. Microbiol. 52:289–294 [DOI] [PubMed] [Google Scholar]
- 54. Pajunen M., Kiljunen S., Skurnik M. 2000. Bacteriophage φYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pajunen M. I., Kiljunen S. J., Söderholm M.-E. L., Skurnik M. 2001. Complete genomic sequence of the lytic bacteriophage φYeO3-12 of Yersinia enterocolitica serotype O:3. J. Bacteriol. 183:1928–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Perry R. D., Fetherston J. D. 1997. Yersinia pestis--etiologic agent of plague. Clin. Microbiol. Rev. 10:35–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pinta E., et al. 2009. Identification and role of a 6-deoxy-4-keto-hexosamine in the lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3. Chemistry 15:9747–9754 [DOI] [PubMed] [Google Scholar]
- 58. Pinta E., et al. 2010. Characterization of the six glycosyltransferases involved in the biosynthesis of Yersinia enterocolitica serotype O:3 lipopolysaccharide outer core. J. Biol. Chem. 285:28333–28342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Porat R., McCabe W. R., Brubaker R. R. 1995. Lipopolysaccharide-associated resistance to killing of Yersiniae by complement. J. Endotoxin Res. 2:91–97 [Google Scholar]
- 60. Portnoy D. A., Falkow S. 1981. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J. Bacteriol. 148:877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Portnoy D. A., Moseley S. L., Falkow S. 1981. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect. Immun. 31:775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Prehm P., Jann B., Jann K., Schmidt G., Stirm S. 1976. On a bacteriophage T3 and T4 receptor region within the cell wall lipopolysaccharide of Escherichia coli B. J. Mol. Biol. 101:277–281 [DOI] [PubMed] [Google Scholar]
- 63. Rick P. D., Osborn M. J. 1977. Lipid A mutants of Salmonella typhimurium. Characterization of conditional lethal mutants in n 3-deoxy- d-mannooctulosonate-8-phosphate synthetase. J. Biol. Chem. 252:4895–4903 [PubMed] [Google Scholar]
- 64. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 65. Samuelsson K., Lindberg B., Brubaker R. R. 1974. Structure of O-specific side chains of lipopolysaccharides from Yersinia pseudotuberculosis. J. Bacteriol. 117:1010–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schütze H. 1932. Studies on B. pestis antigens. II. The antigenic relationship of B. pestis and B. pseudotuberculosis rodentium. Br. J. Exp. Path. 13:289 [Google Scholar]
- 67. Simon R., Priefer U., Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology (NY) 1:784–791 [Google Scholar]
- 68. Skurnik M. 1984. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Appl. Bacteriol. 56:355–363 [DOI] [PubMed] [Google Scholar]
- 69. Skurnik M. 2004. Lipopolysaccharides of Yersinia, p. 215–241 In Carniel E., Hinnebusch B. J. (ed.), Yersinia: molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom [Google Scholar]
- 70. Skurnik M., Venho R., Toivanen P., Al-Hendy A. 1995. A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol. Microbiol. 17:575–594 [DOI] [PubMed] [Google Scholar]
- 71. Skurnik M., Zhang L. 1996. Molecular genetics and biochemistry of Yersinia lipopolysaccharide. APMIS 104:849–872 [PubMed] [Google Scholar]
- 72. Steeghs L., et al. 1998. Meningitis bacterium is viable without endotoxin. Nature 392:449–450 [DOI] [PubMed] [Google Scholar]
- 73. Takayama K., Rothenberg R. J., Barbour A. G. 1987. Absence of lipopolysaccharide in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 55:2311–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tan L., Darby C. 2005. Yersinia pestis is viable with endotoxin composed of only lipid A. J. Bacteriol. 187:6599–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vinogradov E. V., et al. 2002. The core structure of the lipopolysaccharide from the causative agent of plague, Yersinia pestis. Carbohydr. Res. 337:775–777 [DOI] [PubMed] [Google Scholar]
- 76. Weidel W. 1958. Bacterial viruses; with particular reference to adsorption/penetration. Annu. Rev. Microbiol. 12:27–48 [DOI] [PubMed] [Google Scholar]
- 77. Wright A., McConnell M., Kanegasaki S. 1980. Lipopolysaccharide as a bacteriophage receptor. In Randall L. L., Philipson L. (ed.), Virus receptors. Chapmann and Hall, London, United Kingdom [Google Scholar]
- 78. Yang Y. L., et al. 2010. Structural variation of glycolipids from Meiothermus taiwanensis ATCC BAA-400 under different growth temperatures. Org. Biomol. Chem. 8:4252–4254 [DOI] [PubMed] [Google Scholar]
- 79. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 80. Zhang L., Radziejewska-Lebrecht J., Krajewska-Pietrasik D., Toivanen P., Skurnik M. 1997. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol. Microbiol. 23:63–76 [DOI] [PubMed] [Google Scholar]
- 81. Zhang L., Skurnik M. 1994. Isolation of an R− M+ mutant of Yersinia enterocolitica serotype O:8 and its application in construction of rough mutants utilizing mini-Tn5 derivatives and lipopolysaccharide-specific phage. J. Bacteriol. 176:1756–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]