Abstract
The DevR (DosR) response regulator initiates the bacterial adaptive response to a variety of signals, including hypoxia in in vitro models of dormancy. Its receiver domain works as a phosphorylation-mediated switch to activate the DNA binding property of its output domain. Receiver domains are characterized by the presence of several highly conserved residues, and these sequence features correlate with structure and hence function. In response regulators, interaction of phosphorylated aspartic acid at the active site with the conserved threonine is believed to be crucial for phosphorylation-mediated conformational change. DevR contains all the conserved residues, but the structure of its receiver domain in the unphosphorylated protein is strikingly different, and key threonine (T82), tyrosine (Y101), and lysine (K104) residues are placed uncharacteristically far from the D54 phosphorylation site. In view of the atypical location of T82 in DevR, the present study aimed to examine the importance of this residue in the activation mechanism. Mycobacterium tuberculosis expressing a DevR T82A mutant protein is defective in autoregulation and supports hypoxic induction of the DevR regulon only very weakly. These defects are ascribed to slow and partial phosphorylation and the failure of T82A mutant protein to bind cooperatively with DNA. Our results indicate that the T82 residue is crucial in implementing conformational changes in DevR that are essential for cooperative binding and for subsequent gene activation. We propose that the function of the T82 residue in the activation mechanism of DevR is conserved in spite of the unusual architecture of its receiver domain.
INTRODUCTION
Bacterial persistence is a hallmark of tuberculosis (TB). Most individuals exposed to Mycobacterium tuberculosis restrain the infection through an effective immune response that restricts the organisms within granulomas and leads to cessation of disease progression. However, bacilli located within granulomas are not killed and remain dormant in untreated individuals as a latent infection that can reactivate under conditions of immune compromise and cause active disease (14, 36). No drugs are available for the specific treatment of latent TB infection, and this presents a very serious challenge to the successful control of TB. It is believed that tubercle bacilli are exposed to oxygen limitation within granulomas, in response to which they switch to a state of metabolic dormancy and nonreplicative persistence. In vitro models of dormancy have provided us with valuable insights into the molecular mechanisms underlying the adaptation of mycobacteria to hypoxia (42, 43). The DevR-DevS two-component system, along with sensor kinase DosT, plays a key role in M. tuberculosis adaptation to hypoxia and to other signals likely to prevail in vivo, such as nitric oxide, carbon monoxide, and vitamin C (12, 16, 17, 21, 28, 35, 38). DevR (also called DosR) induces the expression of ∼48 genes that comprise the DevR/DosR regulon (28). The expression of the regulon is thought to be of importance for early adaptation to these stimuli as well as for long-term survival in the host (4, 15, 16, 17, 21, 34, 35, 41).
DevR/DosR is a member of the NarL subfamily of response regulators (12), and it is the best-characterized response regulator of M. tuberculosis. DevR is proposed by us and others as an attractive target for the development of inhibitors against dormant organisms (18, 26, 32, 40). Proof of concept for DevR as a dormancy target was established through inhibition of the DevR regulon, hypoxic survival, and reactivation of dormant M. tuberculosis bacilli using a phenylcoumarin (15). We are interested in understanding the activation mechanism of DevR, as these insights would facilitate the development of more potent inhibitors against this target. Of particular interest is the deciphering of the role of conserved amino acid residues implicated in the DevR activation mechanism. We and others have shown that phosphorylation of Asp54 (D54) serves as a switch to activate DevR (8, 29, 32, 45). DevR contains all the conserved residues that are implicated in the activation mechanisms of other response regulators, and these include Asp8 (D8), Asp9 (D9), Asp54 (D54), Thr82 (T82), Tyr101 (Y101), and Lys104 (K104) (12, 37, 45). We showed previously that the D8 and D9 residues together with D54, which likely form an acidic pocket (37) and coordinate Mg2+, were functionally important for DevR phosphorylation (33). The presence of this pocket at the expected location was confirmed with the DevR crystal structure (45).
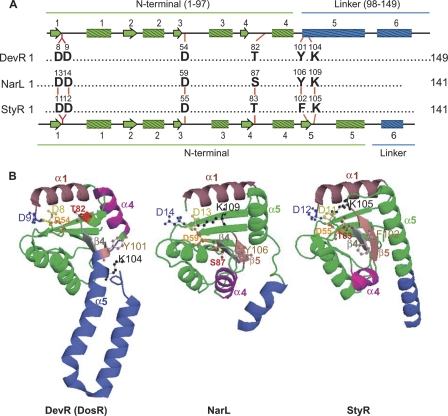
However, unphosphorylated DevR contains an unusual structural feature, which has not been seen before with other response regulators of the NarL subfamily and which is the presence of (βα)4 topology instead of the typical (βα)5 fold observed with the receiver domains of other response regulators (45). In this structure, the other conserved residues of the receiver domain, namely, T82, Y101, and K104, which are known to be important for the regulatory mechanism, are shifted away quite substantially compared to the equivalent residues in the structures of other NarL subfamily members, such as StyR and NarL. In particular, Y101 and K104, which are normally part of the β5 sheet, are moved to the α5 helix in the linker which extends away from the rest of the receiver domain. Thus, these residues are relatively far from the D54 phosphorylation site in DevR compared to their location in NarL and StyR (Fig. 1). Studies of activated receiver domains FixJ (5), CheY (1), and Spo0A (19) have shown that these residues, in particular T82, are crucial for generating and/or stabilizing the conformational change during activation. In the case of DevR (DosR), a helix rearrangement mechanism was proposed for generating the active conformation in the phosphorylated protein (45).
Fig. 1.
Activation pocket in DevR (DosR), NarL, and StyR. (A) Structure-based alignment of the conserved residues in the activation pocket of NarL subfamily members. A schematic representation of the secondary structure elements of N-terminal (green) and linker (blue) domains of DevR is shown on top of the alignment, and that for NarL and StyR is shown below (45). Residue numbers are indicated on the top of each residue. (B) Comparison of structures of unphosphorylated DevR (PDB3C3W), NarL (PDB1A04), and StyR (PDB1ZN2). The corresponding α-helices, β-sheets, and the conserved residues of the activation pocket in these proteins have been color matched and labeled. StyR and NarL display the classical (βα)5 fold of receiver domains. The structures were generated from the PDB files using program PyMol.
Thus, although sequence-based conservation was quite evident between DevR and other NarL family members, significant differences were noted in the location of the conserved residues in the structure, and an emergent question was whether the conserved amino acids in DevR were important for function. This study was designed to assess the functional role of the potentially key residue T82 in DevR activation. The T82 residue in the N-terminal domain of DevR was mutated to alanine (A), and an in vivo assessment of this mutation was first made. M. tuberculosis cultures expressing the DevR T82A mutant protein were found to be defective in DevR regulon gene expression. Analysis of the DevR T82A mutant protein established that the expression deficiency was caused by multiple defects, namely, a partial defect in phosphorylation, a failure to cooperatively recruit DevR to secondary binding sites at target promoters, and a lack of autoregulation. Our results establish that in spite of considerable differences in the arrangement of this residue in the unphosphorylated protein structure, with respect to other response regulators, it plays a key role in the DevR activation mechanism.
MATERIALS AND METHODS
Plasmids, bacterial strains, and culture conditions.
All plasmids and bacterial strains used in this study are described in Tables 1 and 2, respectively. M. tuberculosis strains were cultured at 37°C in DTA medium composed of Dubos medium containing 0.05% Tween 80 plus 0.5% albumin, 0.75% dextrose, and 0.085% NaCl. Escherichia coli strains and culture conditions were as described previously (2). Antibiotics were used at the following concentrations: hygromycin at 50 μg/ml for M. tuberculosis and 200 μg/ml for E. coli, and kanamycin at 20 μg/ml for M. tuberculosis and 50 μg/ml for E. coli.
Table 1.
Plasmids used in this study
| Plasmid | Relevant featuresa | Reference and/or source |
|---|---|---|
| pSD POperondevR | pJFR19 (E. coli-Mycobacterium integrating shuttle vector) containing devR (cloned at NdeI and XbaI sites); DevR is expressed from Rv3134c-devRS operon promoter (−608 to +998 [see reference 8]) cloned in NdeI and BstBI sites, Hygr | 7, 22 |
| pUS POperonT82A | pSD POperondevR harboring T82A mutation, Hygr | This study |
| pAV-DevR | pET28a overexpressing N-terminal His6-tagged WT DevR cloned in BamHI site, Kanr | 21 |
| pUS His6-T82A | pET28a overexpressing DevRT82A cloned in BamHI site, Kanr | This study |
| pSC-DevR | pGEX4T1 overexpressing N-terminal GST-tagged DevR WT cloned in BamHI site, Ampr | 8 |
| pUS GST-T82A | pGEX4T1 overexpressing DevRT82A cloned in BamHI site, Ampr | This study |
| p1738 | pFPV27 (E. coli-Mycobacterium shuttle plasmid with promoter less gfp [see reference 39]) containing Rv1738 promoter, Kanr | 9 |
The coordinates of the promoters (in parentheses) are with reference to the transcription start point (TSP) of Rv3134c. Hygr, hygromycin resistance; Kanr, kanamycin resistance.
Table 2.
Strains used in this study
| M. tuberculosis straina | Relevant features | Reference or source |
|---|---|---|
| ΔdevR mutant | 447-bp BalI deletion in M. tuberculosis H37Rv devR gene (deletes DevR amino acid residues from position 40 to 191) | 27 |
| Comp13* | ΔdevR mutant complemented with pSD POperondevR; expresses WT DevR protein | 22 |
| Comp17* | ΔdevR mutant complemented with pUS POperondevR T82A; expresses DevR T82A mutant protein | This study |
| Comp18 | Comp17 electroporated with p1738 (GFP reporter plasmid) | This study |
*, M. tuberculosis strains of similar genetic backgrounds that produce DevR T82A (Comp17) or WT protein (Comp13) from a single copy of devR integrated at identical chromosomal locations.
Site-directed mutagenesis of threonine to alanine in DevR (T82A).
Site-directed mutagenesis of the codon for T82 (ACG) to alanine (GCG) in devR was carried out in the integrative plasmid pSD POperon devR to generate pUS POperon T82A using Pfu Turbo DNA polymerase (Invitrogen) and primers T82AF and T82AR (Table 3) in a 50-μl PCR mixture that was supplemented with 8% dimethyl sulfoxide (DMSO). Reaction conditions were 95°C (2 min) followed by 35 cycles of 95°C (30 s), 65°C (30 s), and 72°C (12 min). The amplified products (50 μl) were subjected to restriction digestion using 10 U of DpnI for 1 h at 37°C. A portion of the digestion mix was used for transformation into E. coli XL-1 Blue. Transformants containing the integrative vector were selected on an LB agar plate containing hygromycin. pUS POperon T82A clone no. 5 (here called pUS POperon T82A) has a single desired mutation in the devR coding sequence, and this was used in further studies. Mutant and complemented strains were confirmed by DNA sequencing.
Table 3.
Primers used in this study
| Primer | Sequence (5′→3′)a | Application |
|---|---|---|
| T82A F | TGTCTGATCCTCGCGTCCTACACCTCT | Site-directed mutagenesis (this study) |
| T82A R | AGAGGTGTAGGACGCGAGGATCAGACA | |
| UGSTdevR F | GCCGGATCCATGGTAAAGGTCTTCTTGGTC | Cloning of devR with T82A mutation into pGEX4T1 (this study) |
| GSTdevR R | CCGGGATCCCTATCATGGTCCATCACCGG | |
| RTnarK2 F | CGGTTTGTACGGTGGTTCGGC' | Real-time RT-PCR (this study) |
| RTnarK2 R | TCACGAAGCACGACCATGGCC | |
| RT16S F | ATGACGGCCTTCGGGTTGTAA | Real-time RT-PCR (13) |
| RT16S R | CGGCTGCTGGCACGTAGTTG | |
| RT3134c F | CTGGCTGGGTCGGCCTTAGC | Real-time RT-PCR (13) |
| RT3134c R | TGACCTGGGAGGTTGTCG | |
| RTdevRC F5 | CGAGGATCCCTGTTGTCATGGTCCAT | Real-time RT-PCR (13) |
| RTdevR R | CGCGGCTTGCGTCCGACGTTC | |
| RT devS F | TACTGACCGACCGGGATCGT | Real-time RT-PCR (13) |
| RTdevS R | AGAGCCGCTGGATGACATGG | |
| RT1738 F | CGACGAACACGAAGGATTGA | Real-time RT-PCR (13) |
| RT1738 R | ACACCCACCAATTCCTTTTCC | |
| RT2031c F | CGCACCGAGCAGAAGGA | Real-time RT-PCR (13) |
| RT2031c R | ACCGTGCGAACGAAGGAA | |
| RTtgs1 F | CAGTGATTTGCGTCGCTACAG | Real-time RT-PCR (13) |
| RTtgs1 R | ACATCATTGATGGTGACGTCG | |
| RT3131 F | CGATCAGGCCGATGTCGCCTT | Real-time RT-PCR (13) |
| RT3131 R | TCACCTCCTGGCACCGGCC | |
| 3130F | TGGCTGCCGGGCCTTTCCCAT | DNase I footprinting (10) |
| 3131R | CATGGTCAGCGCCTTCCCCGG | |
| 0571c F | CGGCCGAAGTGAGCCACCACC | DNase I footprinting (11) |
| 0571c R | GCCAAGGACGACGACGGCCTT |
BamHI restriction enzyme sites are underlined.
Construction of M. tuberculosis strain expressing DevR T82A.
For DevR T82A expression from its native Rv3134c-devRS operon promoter, integrating plasmid pUS POperon T82A (Table 1) was electroporated into M. tuberculosis ΔdevR mutant bacteria to generate the Comp17 strain (Table 2). The reporter strain, Comp18, was generated by electroporation of the p1738 plasmid into Comp17. The presence of plasmid pUS POperon devR T82A in Comp17 and p1738 in Comp18 was confirmed by PCR.
Construction of DevR T82A-overexpressing plasmid and purification of DevR T82A and WT proteins from E. coli.
The devR T82A coding region was amplified from the integrative plasmid pUS POperon devR T82A by PCR (Table 1) and cloned into the expression plasmids pET28a and pGEX4T1 to generate pUS-His6T82A and pUS-GSTT82A, respectively, which express N-terminal His6- and N-terminal glutathione S-transferase (GST)-tagged DevR T82A (here referred to as His-T82A and GST-T82A, respectively). His-T82A, GST-T82A, His-wild type (WT), and GST-WT proteins were overexpressed in E. coli C43(DE3) using standard procedures. The recombinant proteins were purified by standard techniques and used in phosphorylation assays and for DNase I footprinting.
Western blotting of M. tuberculosis lysates.
Frozen M. tuberculosis stocks were revived in DTA medium, subcultured thrice, grown in a shaker incubator at 220 rpm (160 ml in airtight 500-ml Teflon screw-cap flasks) till an ∼A595 of 0.2 to 0.3 was reached, and subsequently processed for immunoblotting and RNA analysis (below). Briefly, a 20-ml aliquot was chilled on ice (aerobic) and centrifuged immediately at 5,000 rpm for 10 min at 4°C, and the pellet was stored at −20°C. Sixty-milliliter aliquots of these cultures were distributed (10-ml aliquots in 50-ml tubes that were tightly closed) and kept standing for 5 days to generate hypoxic cultures as described previously (32). The cells were harvested from dedicated culture tubes after appropriate incubation, and whole-cell lysates were prepared as described previously (31). HspX and SigA proteins were detected in the lysates (containing ∼15 μg protein) by Western blotting using polyclonal anti-HspX and anti-SigA antibodies as described previously (2). SigA protein was used as a loading control.
M. tuberculosis RNA isolation.
The remaining culture (80 ml from the cultures described above) was harvested, and RNA was isolated. Briefly, a 20-ml aliquot was snap-chilled on ice and centrifuged immediately as described above (aerobic), and the remaining culture was kept standing for 5 days (hypoxic) as described above. The harvested cell pellets were each resuspended in 1 ml of TRI reagent (Molecular Research Center) and lysed in a mini-bead beater using 0.1-mm zirconium/silica beads (Biospec). RNA was purified as described previously (8).
RT and real-time PCR.
DNA-free RNA (200 ng) was reverse transcribed into cDNA using Multi Scribe reverse transcriptase (50 U) and random hexamer primers per the manufacturer's instructions (Applied Biosystems). The cDNA (2 μl) was subjected to real-time PCR using gene-specific primers (Table 3) and Power SYBR green PCR master mix in a MyiQ thermal cycler (Bio-Rad). Reaction conditions were 94°C (10 min), followed by 40 cycles of 94°C (30 s), 56 to 65°C (45 s), and 72°C (30 s). A reverse transcription (RT)-negative (without reverse transcriptase) reaction was used to account for residual DNA, if any, and transcript numbers were normalized to that of 16S rRNA. The normalized copy number values were then used to determine the relative quantities (RQ) of individual gene transcripts. Three independent cultures were each analyzed in duplicate, and the results are expressed as the mean ± the standard deviation (SD).
In vitro phosphorylation assays.
Time course phosphorylation assays were performed per standardized procedures. Briefly, 2 units of acetate kinase (Sigma) was incubated with 5 μCi [γ-32P]ATP (3,500 Ci/mmol; Brit, Hyderabad, India) in a 10-μl reaction mix containing 25 mM Tris-Cl, pH 7.5, 60 mM potassium acetate, and 10 mM MgCl2 at 25°C for 20 min. The mutant or wild-type protein (6 μM each) was then added to this reaction mix, and the mixture was added to buffer containing 40 mM Tris-Cl, pH 8.0, 20 mM NaCl, 0.2 mM EDTA, and 0.2 mM dithiothreitol (DTT) and incubated at room temperature for 15 to 30 min. The reaction was terminated with 4 μl of stop solution containing 300 mM Tris-Cl, pH 6.8, 60% glycerol, 12% SDS, 7.5% β-mercaptoethanol, and 0.6% bromophenol blue and subsequently analyzed by electrophoresis on 15% SDS-polyacrylamide gel and phosphorimaging.
Phosphorylation with sensor kinase DevS201 (cytoplasmic C-terminal fragment of DevS containing 201 amino acids [DevS201], kindly provided by Kohinoor Kaur), was carried out as described previously (32). Briefly, DevS201 (15 μM) was incubated in buffer containing 50 mM Tris-Cl, pH 8.0, 50 mM KCl, 10 mM MgCl2, 500 μM ATP, and 0.1 μCi [γ-32P]ATP at 25°C for 60 min. DevR (WT or mutant) proteins (20 μM) were added to the reaction mix described above and incubated for 30 s to 32 min. The reaction was terminated with 4 μl of stop buffer and analyzed by SDS-PAGE and phosphorimaging.
DNase I footprinting.
DNase I footprinting was carried out as described previously (13) to compare the DNA binding patterns of WT and mutant DevR proteins. The sequences of the primers used in DNA fragment preparation by PCR are shown in Table 3. Briefly, the DNA fragments were generated by PCR using [γ-32P]ATP end-labeled primers (∼3,000 Ci/mmol; Brit, Hyderabad, India). DevR was phosphorylated by incubating it with 50 mM acetyl phosphate for 20 min at 25°C in 40 mM Tris-Cl (pH 8.0) and 5 mM MgCl2. Binding of 10 to 15 ng of 32P-labeled DNA (75,000 to 100,000 cpm) to phosphorylated DevR was performed on ice for 30 min in a 50-μl reaction mixture in binding buffer (25 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 20 mM KCl, 6 mM MgCl2, 5% glycerol). After DNase I treatment (0.2 U; Promega) for 4 min at 22°C in the presence of 2.5 MgCl2 and 5 mM CaCl2, the reaction was stopped by the addition of 90 μl of 2× stop solution (200 mM NaCl, 30 mM EDTA, 1% sodium dodecyl sulfate, and 66 μg of yeast tRNA/ml). The reaction products were extracted with phenol-chloroform, precipitated with 3 volumes of ethanol at −80°C for 1 h, washed with 70% ethanol, and air dried. DNA was dissolved in formamide-urea loading dye, loaded onto 6% denaturing polyacrylamide gel that was prerun in 0.5× Tris-borate-EDTA buffer till it attained 50 to 55°C at 70 W. The gel was dried, exposed, and visualized by phosphorimager (Bio-Rad). The phosphorimage was analyzed by Quantity One software (Bio-Rad).
GFP reporter assay.
Green fluorescent protein (GFP) reporter assays were conducted in DTA medium as described previously (8). The promoter activity is expressed in relative fluorescence units (RFU)/optical density at 595 nm (OD595) of GFP (mean values of RFU/OD ± standard deviation).
RESULTS
T82 in DevR is essential for hypoxic induction of HspX.
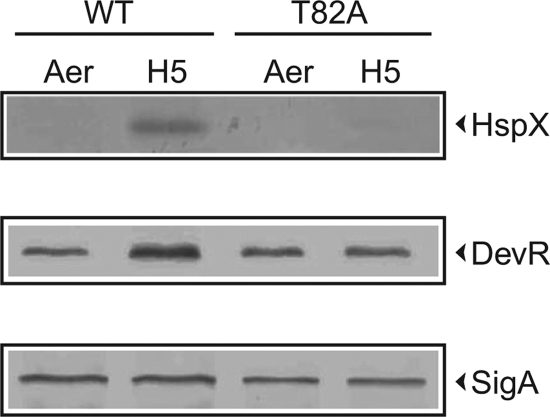
DevR is known to mediate the induction of the DevR regulon under hypoxia, and the expression of HspX is considered a reliable marker for activation of this regulon. To obtain a quick assessment of whether T82 was important in the activation mechanism of DevR, the induction of HspX was monitored in an M. tuberculosis strain expressing the DevR T82A mutant protein (Fig. 2). This strain was constructed by introducing an integrative plasmid carrying a copy of the gene encoding the DevR T82A mutant protein expressed from its own operon promoter into M. tuberculosis ΔdevR bacteria. Immunoblotting revealed that HspX expression was severely decreased in hypoxic M. tuberculosis cultures expressing DevR T82A compared to good induction in an isogenic strain expressing the WT DevR protein.
Fig. 2.
Immunoblotting analysis. M. tuberculosis DevR T82A and WT lysates. M. tuberculosis lysates (15 μg) were fractionated by SDS-PAGE, subjected to immunoblotting, and probed with anti-HspX, anti-DevR, or anti-SigA rabbit sera. The blots were analyzed densitometrically using Quantity One software (Bio-Rad). The intensities of HspX- and DevR-derived signals in DevR T82A- expressing bacteria were normalized with respect to those of SigA and were ∼15% and 40%, respectively, with respect to those in hypoxic M. tuberculosis cultures expressing WT DevR protein. Aer, aerobic; H5 refers to 5-day hypoxic cultures.
M. tuberculosis DevR T82A is defective in autoregulation and DevR regulon induction.
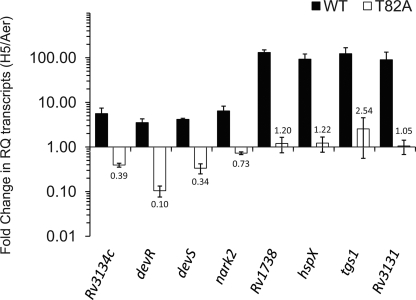
It is well established that the DevR protein is expressed at basal levels in aerobic M. tuberculosis cultures and that the protein level is elevated under hypoxia by a positive autoregulation mechanism (2, 8). Immunoblotting revealed that basal production of mutant DevR occurred at a level that is equivalent to that of WT protein in wild-type M. tuberculosis cultures. However, in contrast to an ∼2.5-fold increase in the DevR protein level in hypoxic cultures of WT bacteria, no similar increase was observable with mutant cultures under similar conditions, suggesting a defect in autoregulation (Fig. 2). The defect in autoregulation was confirmed by quantitative analysis of RNA; an ∼4-fold induction in devR transcripts was noted for WT bacteria, in contrast to an ∼10-fold reduction in mutant bacteria under hypoxia (Fig. 3).
Fig. 3.
Real-time RT-PCR analysis of DevR regulon transcripts. The fold change in the relative quantity (RQ) of transcripts under hypoxic versus aerobic conditions (fold decrease in DevR T82A-expressing Comp17 and fold increase in DevR WT- expressing Comp13 bacteria) is shown.
We next extended our analysis to determine the role of the T82 residue in the induction of the DevR regulon under hypoxia by comparing the relative quantities of selected DevR regulon transcripts in M. tuberculosis strains expressing mutant or WT DevR protein. In contrast to transcriptional induction (∼3.5- to 130-fold) noted for the WT DevR-expressing strain, very weak induction of tgs1, Rv1738, and hspX transcripts (∼1.2- to 2.5-fold) occurred in mutant bacteria under hypoxia (Fig. 3). The weak induction of Rv1738 in the T82 mutant strain was confirmed by means of the GFP reporter assay using p1738 (mean hypoxic GFP fluorescence, ∼432 RFU/OD versus ∼16,890 RFU/OD in the presence of WT DevR). The weak induction of hspX transcripts paralleled the minimal induction of HspX protein detected in mutant bacteria during hypoxia.
A possible reason for the DevR regulon expression defect under hypoxia is that decreased transcription fails to sustain mutant DevR protein expression at a level necessary for autoregulation and target genes induction. However, this explanation appears unlikely as the DevR T82A protein level was maintained at aerobic levels even during hypoxia despite a significant decrease in devR transcript levels (Fig. 2 and 3). Therefore, we infer from these findings that the defect in regulon induction in DevR T82A mutant bacteria may be a consequence of an activation defect. Toward understanding the underlying basis of this defect, the biochemical properties of the DevR T82A mutant protein were analyzed next.
T82A mutant protein autophosphorylates slowly compared to wild-type DevR protein.
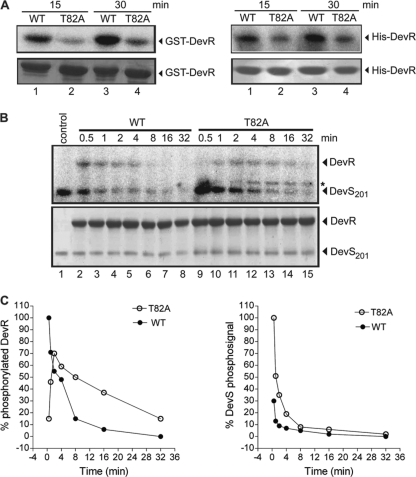
Phosphorylation of full-length WT DevR is essential for sequence-specific interaction with the DNA of target promoters and subsequent gene activation (8–11, 13). Toward exploring a possible phosphorylation defect in DevR T82A, the mutant protein was cloned and overexpressed in E. coli (see Materials and Methods for details), and its properties were compared to those of the WT DevR protein using an acetyl phosphate-based assay (Fig. 4 A). Because the fusion protein may dimerize via the GST tag and the tag may potentially interfere with the conformation of the protein, we used His6-tagged mutant protein as well as GST-tagged protein to examine the properties of the T82 mutant protein. A phosphorylation defect was observed with the mutant protein irrespective of the tag it carried at its amino terminus (a GST or His6 tag); phosphorylation occurred at ∼35 to 40% efficiency compared to that of the WT DevR protein during a 15- to 30-min assay. Thus, the alanine substitution of T82 resulted in a significant decrease in DevR phosphorylation.
Fig. 4.
Time course of phosphorylation and dephosphorylation of DevR T82A and WT proteins. (A) In vitro phosphorylation of GST- (left) and His6- (right) tagged DevR proteins was carried out using [32P]acetyl phosphate for 15 min (lanes 1 and 2) and 30 min (lanes 3 and 4). (B) Phosphorylation of GST-DevR WT (lanes 2 to 8) and GST-DevR T82A (lanes 9 to 15) in the presence of DevS. Lane 1 contains ∼15 μM DevS autophosphorylated in the presence of [γ-32P]ATP. In panels A and B, the top and bottom portions represent phosphorimage and Coomassie stained gel, respectively. * in panel B represents a minor contaminant in GST-DevR T82A that was not detectable by Coomassie staining. (C) Data (described above for panel B) of phosphorylation and dephosphorylation of mutant and WT proteins (left) and the residual phophosignal in DevS after phosphotransfer to DevR (right). Each experiment was repeated at least thrice. and representative figures are shown.
DevR T82A is defective in receiving phosphosignal from DevS sensor kinase.
Since DevR activation in vivo occurs by a phosphotransfer mechanism from its cognate sensor kinase (29, 32, 33), the relative ability of the DevR T82A mutant protein to receive a phosphosignal from DevS was examined next. In a time course phosphorylation assay (30 s to 32 min), the WT DevR protein was rapidly phosphorylated in the presence of phosphorylated DevS (DevS∼32P) (Fig. 4B, lanes 2 to 8). In contrast, the DevR T82A protein is phosphorylated at a lower rate (∼15 to 25%, lane 9) than that of the WT protein (100%, lane 2) at 0.5 min. The extent of phosphorylation was also lower; peak phosphorylation of ∼70% was noted for the mutant protein at 2 min compared to 100% peak phosphorylation for the WT protein at 0.5 min (Fig. 4C).
The phosphorylation defect was confirmed by examining the rate of loss of the phosphosignal from DevS. Note that DevS∼P is very stable in the absence of DevR (Fig. 4B, lane 1), and the loss of the phosphosignal from DevS is related to its transfer to DevR. Approximately 90% of the DevS-associated phosphosignal is lost within 1 min in the presence of WT DevR, whereas in the presence of DevR T82A, >50% of the phosphosignal is detected in association with DevS at 2 min (Fig. 4C).
Phosphorylated DevR T82A is relatively more stable and does not readily lose the phosphosignal.
After establishing that T82 is required for optimum phosphorylation (described above), the levels of stability of phosphorylated T82A and WT DevR were compared. The phosphosignal was dissipated more rapidly and completely from the WT protein than from the mutant protein (Fig. 4B and C). The decrease in the DevR phosphosignal is attributed to DevS-associated phosphatase activity (32), because of which WT DevR∼P is not visualized beyond ∼16 min. In comparison, the dephosphorylation of the DevR T82A protein is slower and incomplete. Collectively, the phosphorylation assays indicate that DevR T82A is phosphorylated slowly, but once it receives the phosphosignal, it appears to remain in the phosphorylated state for a relatively longer period of time than WT DevR (Fig. 4B, lanes 6, 7, 13, and 14). Since the mutant protein exhibits a phosphorylation defect and altered phosphorylation kinetics and phosporylation is essential for binding to DNA, we examined next the DNA binding property of DevR T82A by DNase I footprinting.
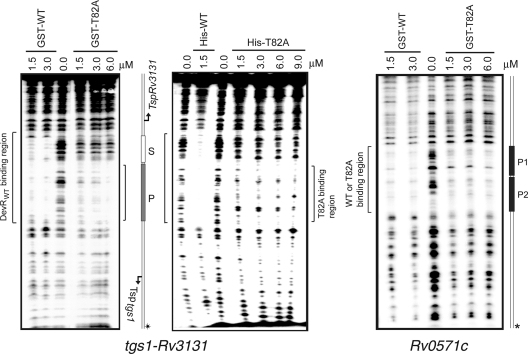
DevR T82A protein is defective in cooperative binding at a target promoter.
The tgs1-Rv3131 intergenic promoter region was chosen to assess DevR T82A interaction with DNA by DNase I footprinting (Fig. 5). This region is well characterized, and it possesses a primary binding site, P, and a secondary binding site, S, for phosphorylated DevR. Binding to the secondary site occurs by cooperative interaction between DevR molecules, and importantly, binding to both sites is essential for robust induction of both genes (10, 13). Densitometric analysis of DNase I footprints shows that the T82A mutant protein is defective in binding to the P site compared to the WT protein (99% and 90% protection by His- and GST-tagged WT proteins versus 60% and 74% protection by mutant proteins, respectively) (Table 4). The defect in DNA binding at the primary P site can be attributed to a partial defect in protein phosphorylation of the T82A protein. It is also evident that the relative protection of the S and P sites is significantly different between the WT and mutant proteins. Thus, while ∼98% and 82% protection at the S site was detected with His- and GST-tagged WT proteins, respectively, protection of only 22% and 26% was observed at this site with the mutant protein. This amounts to an S/P protection ratio of approximately 1 for the WT protein and 0.37 and 0.35 for the mutant proteins at a 1.5 μM protein concentration. The relatively poor occupancy of the S site by T82A supports a role for phosphorylation-induced conformational changes in enabling cooperative binding of DevR to DNA. We infer that these conformational changes are attenuated in the mutant protein (as phosphorylation is slow and partial in comparison to that of the WT protein).
Fig. 5.
DevR T82A protein is defective in cooperative binding to DNA. DNase I footprinting of DevR (T82A or WT) on tgs1-Rv3131 and Rv0571c promoter DNA. 32P-radiolabeled DNA strand is indicated by an asterisk.
Table 4.
Densitometric analysis of DNase I footprints for tgs1-Rv3131 target promotera
| Protein (tag, concn) | Protection at P box (ASI) | Protection at S box (ASI) | Ratio of S/P site protection |
|---|---|---|---|
| WT (His, 1.5 μM) | 99 | 98 | 0.99 |
| T82A mutant (His, 1.5 μM) | 60 | 22 | 0.37 |
| WT (GST, 1.5 μM) | 85 | 82 | 0.96 |
| WT (GST, 3.0 μM) | 90 | 88 | 0.98 |
| T82A mutant (GST, 1.5 μM) | 74 | 26 | 0.35 |
| T82A mutant (GST, 3.0 μM) | 80 | 33 | 0.41 |
ASI, arbitrary signal intensity units (average of 3 experiments).
The defect in cooperative binding of the mutant protein at the S site was confirmed by footprinting analysis of another DevR target, the Rv0571c promoter, which contains two adjacent primary DevR binding sites, P1 and P2, and no secondary sites. All the other known promoters harbor at least one primary P site and one or more secondary S sites (11). Based on the binding property of the mutant protein in tgs1-Rv3131c promoter DNA, it was predicted that it would bind independently to both the P1 and P2 primary sites in Rv0571c DNA. Densitometric analysis demonstrates that the ratios of P2/P1 site protection were indeed quite similar for both the WT and mutant proteins, although the latter exhibited a similar partial defect in binding to each of the sites (Fig. 5 and Table 5). Thus, footprint analysis of the tgs1-Rv3131 and Rv0571c target promoters support the statement that the T82 residue is crucial for cooperative binding of DevR.
Table 5.
Densitometric analysis of DNase I footprints for Rv0571c target promotera
| Protein | GST tag concn (μM) | Protection at P1 box (ASI) | Protection at P2 box (ASI) | Ratio of P2/P1 site protection |
|---|---|---|---|---|
| WT | 1.5 | 90 | 87 | 0.97 |
| 3.0 | 92 | 82 | 0.89 | |
| T82A mutant | 1.5 | 76 | 75 | 0.99 |
| 3.0 | 76 | 79 | 1.04 |
ASI, arbitrary signal intensity units.
The cooperativity defect was noted for T82A mutant proteins carrying either a GST or His6 fusion protein tag. We have previously characterized binding of DevR to several target promoters using the GST-tagged DevR protein and also shown that sequence-specific binding does not occur in the presence of unphosphorylated DevR (8, 10). Any potential interference by the fusion tag present in the recombinant protein in binding to DNA (Fig. 5) or during phosphorylation (Fig. 4) was ruled out, as the defects were evident in the mutant DevR protein harboring either a His6 or GST tag. Together, the results of phosphorylation assays and DNase I footprinting suggest that T82 plays a key role and is required for phosphorylation-mediated cooperative DNA binding.
DISCUSSION
In the present study, we have used in vivo and in vitro approaches to demonstrate the functional importance of T82 in the activation mechanism of DevR. We show that this residue is essential for optimum phosphorylation, cooperative binding of DevR to DNA and subsequent autoregulation, and robust DevR regulon activation in M. tuberculosis.
Phosphorylation of DevR at D54 (32, 33) is essential for activating its DNA binding function (8, 10). The DevR T82A mutant protein was partially defective in phosphorylation in the presence of both acetyl phosphate and its cognate sensor kinase DevS. Because the phosphorylation defect was associated with defects in DNA binding and gene activation, we confirm that phosphorylation is intimately connected to downstream events and propose that T82 promotes the cascade of downstream functions. In other response regulators, the threonine or serine residue at a position in the sequence corresponding to T82 in DevR is described to be essential for transducing the signal to the output domain on phosphorylation. In this activation mechanism it is proposed that the conserved threonine residue associates via hydrogen bonding with phosphoaspartate and triggers conformational changes and rearrangements involving the conserved Tyr/Phe and Lys residues in the receiver domains of response regulators (6, 30). As mentioned earlier, the present study was designed to examine whether T82 played this role in DevR, in view of its unusual structural topology compared to other response regulators (Fig. 1). Our analysis of the T82 mutant protein suggests that despite the differences in structures between DevR and other regulators of the same subfamily, such as NarL (3) and StyR (25), replacement of threonine with alanine adversely affected DevR function, and this substitution appears to hamper the conformational transition associated with phosphorylation, as observed with other response regulators.
Comparison of the DNA binding and gene activation properties of DevR T82A and those of the isolated WT DevR C-terminal domain (DevRC) (13) reveals interesting parallels and establishes the essential function of cooperativity in binding to DNA for DevR function. The full-length DevR T82 mutant and isolated DevRC exhibit similar defects in cooperative binding to DNA and autoregulation, and both proteins support only very weak gene activation. In DevR T82A-expressing M. tuberculosis cultures, poor target gene induction is attributed to the dual defects in phosphorylation and cooperative binding to DNA. Slow and partial phosphorylation of DevR T82A is associated with an inability to bind the low affinity site in promoter DNA. In this respect, the T82 mutant protein resembles the isolated DevRC protein which is recruited to the high-affinity site and not to the low-affinity site (13). The similar binding patterns of DevR T82A and DevRC, which are altogether different from that of WT protein, suggest that while phosphorylation-induced conformation definitely occurs in DevR T82A to unmask its DNA binding activity, propagation of conformational transitions between the domains is most likely different from that occurring in the WT protein. Thus, while the adopted conformation of the phosphorylated T82A mutant protein supports its interaction with the high-affinity site, it does not appear to permit recruitment of a second molecule of DevR to the low-affinity site. On this basis we conclude that the T82A mutant protein is defective in protein-protein interactions essential for cooperative recruitment. In the absence of a complete active conformation, DevR T82A mimics DevRC. In this respect, DevR differs from NarL, whose determinants for DNA recognition and binding reside in the C-terminal portion and whose N terminus does not contribute to the ability of the NarL C-terminal domain (NarLC) to bind DNA (23). It is relevant to recall here that the transcription start point (TSP)-proximal binding site overlaps with the −35 promoter element (8, 9, 10). This conserved feature of DevR target promoters suggests the possible mechanism underlying the activation defect; the failure to cooperatively bind DevR at the TSP-proximal site may preclude interactions between DevR T82A/DevRC and RNA polymerase that are necessary for robust gene induction. Further studies are required to confirm this hypothesis. A noteworthy difference between DevR T82A and DevRC, however, is that of DevR stability; the T82 mutant protein (Fig. 2) but not DevRC (13) is sustained during hypoxia. In contrast to both, WT bacteria accumulate the DevR protein under hypoxia (Fig. 2) due to positive autoregulation (8). In M. tuberculosis DevR T82A bacteria, the mutant protein overrides the autoregulation defect to maintain the DevR protein at basal levels of expression. The sustained level of the mutant protein supports the hypothesis proposed earlier that the N-terminal domain confers stability to intact DevR (13).
Mutational studies of several distantly and closely related response regulators, including FixJ, CheY, OmpR, and CovR, support the key role of the Thr residue in the phosphorylation activation mechanism of DevR. Threonine/serine mutant proteins of these regulators display one or more of the following defects: in phosphorylation kinetics, as in FixJ (44); in phosphorylation efficiency and stability, as in CheY (1); in cooperative DNA binding and gene activation, as in OmpR (24); or in decreased phosphorylation and binding to promoter DNA, as in CovR (20).
Analysis of the structures of unphosphorylated and phosphorylated forms of the FixJ receiver domain may provide some insights into the functional relevance of T82 in DevR activation. In FixJ, the conserved T82 residue interacts with the phosphoryl group by a hydrogen bond and induces rearrangement of the β4-α4 loop and flipping of the T82 and F101 residues (5). In the absence of the structure of phosphorylated DevR, the precise role of T82 in DevR activation is not known, but the findings of the present study strongly suggest that it participates in conformational rearrangement to generate the DNA binding species. Crystal structure of StyR suggests that phosphorylation-mediated activation is transmitted by the β4-α4 loop through a T83-phosphate hydrogen bond (25). However, the consequences of the mutation of T83 in StyR and S87 in NarL (corresponding to T82 of DevR) are yet unknown.
We have recently observed that a minimum of two DevR binding sites characterize all DevR regulon promoters (11). Interestingly, phosphorylation unmasks the DNA binding property of the mutant protein but only to the high-affinity site at a target promoter, and substitution of T82 significantly attenuates binding to the secondary site. Thus, the partial defect in phosphorylation of the T82A mutant protein is not drastic enough to abrogate its binding to the P site (Fig. 5). Our findings provide new and important insight into the role of the domain containing T82 in the DevR activation mechanism. The properties of the DevR T82A mutant protein highlight the essential role of cooperativity in DevR-mediated gene activation. In functional terms, therefore, active DevR may be defined as that conformational species which binds cooperatively at target promoters. We propose that disruption of cooperative DevR-DNA interactions can be an effective strategy for blocking DevR function and DevR-mediated gene activation during adaptation to dormancy in M. tuberculosis. In conclusion, our results support the hypothesis that the function of T82 in the activation mechanism of DevR is conserved in spite of the unusual topology of its receiver domain.
ACKNOWLEDGMENTS
We thank Neil G. Stoker, Royal Veterinary College, London, United Kingdom, for the generous gift of the M. tuberculosis ΔdevR (dosR) deletion mutant strain, and Malini Rajagopalan, University of Texas Health Center, for the generous gift of the integrating plasmid pJFR19.
This work was supported by a grant from the Department of Biotechnology, Government of India, to J.S.T. J.S.T. thanks the Department of Biotechnology, Government of India, for a Tata Innovation Fellowship. U.S.G. and K.S. thank the Council for Scientific and Industrial Research (CSIR) for a Senior Research Associateship (Scientist's Pool Scheme) and a Junior Research Fellowship, respectively.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Appleby J. L., Bourret R. B. 1998. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J. Bacteriol. 180:3563–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bagchi G., Chauhan S., Sharma D., Tyagi J. S. 2005. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology 151:4045–4053 [DOI] [PubMed] [Google Scholar]
- 3. Baikalov I., et al. 1996. Structure of the Escherichia coli response regulator NarL. Biochemistry 35:11053–11061 [DOI] [PubMed] [Google Scholar]
- 4. Balazsi G., Heath A. P., Shi L., Gennaro M. L. 2008. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 4:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birck C., et al. 1999. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure 7:1505–1515 [DOI] [PubMed] [Google Scholar]
- 6. Bourret R. B. 2010. Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 13:142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chauhan A., et al. 2006. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J. Bacteriol. 188:1856–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chauhan S., Tyagi J. S. 2008. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J. Bacteriol. 190:4301–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chauhan S., Tyagi J. S. 2008. Interaction of DevR with multiple binding sites synergistically activates divergent transcription of narK2-Rv1738 genes in Mycobacterium tuberculosis. J. Bacteriol. 190:5394–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chauhan S., Tyagi J. S. 2009. Powerful induction of divergent tgs1-Rv3131 genes in Mycobacterium tuberculosis is mediated by DevR interaction with a high-affinity site and an adjacent cryptic low-affinity site. J. Bacteriol. 191:6075–6081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chauhan S., Sharma D., Singh A., Surolia A., Tyagi J. S. 7 June 2011. Comprehensive insights into Mycobacterium tuberculosis DevR (DosR) regulon activation switch. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkr375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dasgupta N., et al. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 80:141–159 [DOI] [PubMed] [Google Scholar]
- 13. Gautam U. S., Chauhan S., Tyagi J. S. 2011. Determinants outside the DevR C-terminal domain are essential for cooperativity and robust activation of dormancy genes in Mycobacterium tuberculosis. PLoS One 6:e16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grange J. M. 1992. The mystery of the mycobacterial ‘persistor.’ Tuber. Lung Dis. 73:249–251 [DOI] [PubMed] [Google Scholar]
- 15. Gupta R. K., Thakur T. S., Desiraju G. R., Tyagi J. S. 2009. Structure-based design of DevR inhibitor active against nonreplicating Mycobacterium tuberculosis. J. Med. Chem. 52:6324–6334 [DOI] [PubMed] [Google Scholar]
- 16. Kumar A., Toledo J. C., Patel R. P., Lancaster J. R., Jr., Steyn A. J. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U. S. A. 104:11568–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar A., et al. 2008. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283:18032–18039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lamichhane G. 2010. Novel targets in M. tuberculosis: search for new drugs. Trends Mol. Med. 17:25–33 [DOI] [PubMed] [Google Scholar]
- 19. Lewis R. J., Brannigan J. A., Muchova K., Barak I., Wilkinson A. J. 1999. Phosphorylated aspartate in the structure of a response regulator protein. J. Mol. Biol. 294:9–15 [DOI] [PubMed] [Google Scholar]
- 20. Lin W. J., et al. 2009. Threonine phosphorylation prevents promoter DNA binding of the Group B Streptococcus response regulator CovR. Mol. Microbiol. 71:1477–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Majumdar S. D., et al. 2010. Expression of DevR and DevR(N)-Aph proteins is associated with hypoxic adaptation defect and virulence attenuation of Mycobacterium tuberculosis. PLoS One 5:e9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Majumdar S. D., et al. 2010. Ph.D. thesis All India Institute of Medical Sciences, New Delhi, India [Google Scholar]
- 23. Maris A. E., et al. 2002. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 9:771–778 [DOI] [PubMed] [Google Scholar]
- 24. Mattison K., Oropeza R., Byers N., Kenney L. J. 2002. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J. Mol. Biol. 315:497–511 [DOI] [PubMed] [Google Scholar]
- 25. Milani M., et al. 2005. An active-like structure in the unphosphorylated StyR response regulator suggests a phosphorylation-dependent allosteric activation mechanism. Structure 13:1289–1297 [DOI] [PubMed] [Google Scholar]
- 26. Murphy D. J., Brown J. R. 2007. Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect. Dis. 7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parish T., et al. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park H. D., et al. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roberts D. M., Liao R. P., Wisedchaisri G., Hol W. G., Sherman D. R. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279:23082–23087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robinson V. L., Buckler D. R., Stock A. M. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7:626–633 [DOI] [PubMed] [Google Scholar]
- 31. Rodrigue S., et al. 2007. Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 189:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saini D. K., et al. 2004. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150:865–875 [DOI] [PubMed] [Google Scholar]
- 33. Saini D. K., Malhotra V., Tyagi J. S. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 565:75–80 [DOI] [PubMed] [Google Scholar]
- 34. Sherman D. R., et al. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. U. S. A. 98:7534–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shiloh M. U., Manzanillo P., Cox J. S. 2008. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3:323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stead W. W. 1967. Pathogenesis of a first episode of chronic pulmonary tuberculosis in man: recrudescence of residuals of the primary infection or exogenous reinfection? Am. Rev. Respir. Dis. 95:729–745 [DOI] [PubMed] [Google Scholar]
- 37. Stock A. M., Robinson V. L., Goudreau P. N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 38. Taneja N. K., Dhingra S., Mittal A., Naresh M., Tyagi J. S. 2010. Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One 5:e10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valdivia R. H., Hromockyj A. E., Monack D., Ramakrishnan L., Falkow S. 1996. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene 173:47–52 [DOI] [PubMed] [Google Scholar]
- 40. Vohra R., Gupta M., Chaturvedi R., Singh Y. 2006. Attack on the scourge of tuberculosis: patented drug targets. Recent Pat. Antiinfect. Drug Discov. 1:95–106 [DOI] [PubMed] [Google Scholar]
- 41. Voskuil M. I., et al. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wayne L. G., Hayes L. G. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wayne L. G., Sohaskey C. D. 2001. Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139–163 [DOI] [PubMed] [Google Scholar]
- 44. Weinstein M., Lois A. F., Monson E. K., Ditta G. S., Helinski D. R. 1992. Isolation of phosphorylation-deficient mutants of the Rhizobium meliloti two-component regulatory protein, FixJ. Mol. Microbiol. 6:2041–2049 [DOI] [PubMed] [Google Scholar]
- 45. Wisedchaisri G., Wu M., Sherman D. R., Hol W. G. 2008. Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation. J. Mol. Biol. 378:227–242 [DOI] [PMC free article] [PubMed] [Google Scholar]