Abstract
Staphylococcus lugdunensis is often found as part of the normal flora of human skin but has the potential to cause serious infections even in healthy individuals. It remains unclear what factors enable S. lugdunensis to transition from a skin commensal to an invasive pathogen. Analysis of the complete genome reveals a putative iron-regulated surface determinant (Isd) system encoded within S. lugdunensis. In other bacteria, the Isd system permits the utilization of host heme as a source of nutrient iron to facilitate bacterial growth during infection. In this study, we establish that S. lugdunensis expresses an iron-regulated IsdG-family heme oxygenase that binds and degrades heme. Heme degradation by IsdG results in the release of free iron and the production of the chromophore staphylobilin. IsdG-mediated heme catabolism enables the use of heme as a sole source of iron, establishing IsdG as a pathophysiologically relevant heme oxygenase in S. lugdunensis. Together these findings offer insight into how S. lugdunensis fulfills its nutritional requirements while invading host tissues and establish the S. lugdunensis Isd system as being involved in heme-iron utilization.
INTRODUCTION
Iron acquisition is a critical process for pathogenic bacteria during infection. This is due to the numerous fundamental cellular processes that require iron, such as DNA replication, electron transport, and protection from reactive oxygen damage (5). Vertebrates exploit the iron requirement of bacterial pathogens by creating an extracellular environment virtually devoid of free iron (5). The most abundant source of iron within the human body is found in the form of heme, a tetrapyrrole ring with a coordinated iron center. Heme is further bound by hemoproteins, the most abundant of which is hemoglobin (7). To circumvent this sequestration, pathogens have evolved complex iron acquisition systems that are expressed upon entry into host tissues. In Gram-positive bacteria, one such system is the iron-regulated surface determinant (Isd) system, which binds hemoglobin, removes the heme, and transports it into the bacterial cytoplasm. Upon entry into the cell, heme can then be degraded by heme oxygenases, resulting in the release of nutrient iron (28, 32). Interestingly, heme is the preferred source of iron for Staphylococcus aureus during infection, and heme acquisition by the Isd system is critical for full virulence (27, 31).
Staphylococcus lugdunensis is a coagulase-negative staphylococcus (CNS) that is often found as part of the normal skin flora and has the potential to cause aggressive infections similar to those caused by S. aureus. Emerging as an important human pathogen, S. lugdunensis has an elevated level of virulence that makes it stand out among the CNS (15, 33). Unlike other CNS, S. lugdunensis can cause a wide array of serious infections, such as acute endocarditis, brain abscesses, meningitis, prosthetic joint infections, pneumonia, and toxic shock (15). The mortality rate associated with S. lugdunensis endocarditis can reach 50%, which presents a serious public health problem considering the propensity of S. lugdunensis to infect the heart (2). This is in striking contrast to the 14.5% mortality rate associated with S. aureus endocarditis and the 20% mortality rate associated with Staphylococcus epidermidis endocarditis (3). Additionally, the majority of infections caused by S. lugdunensis occur in healthy adults, primarily in the outpatient setting (20). In fact, nearly half of patients infected with S. lugdunensis do not exhibit identifiable comorbidities (20). This indicates that S. lugdunensis, unlike other CNS, is not confined to being an opportunistic pathogen (20). Although S. lugdunensis remains highly susceptible to the majority of antimicrobial therapies, antibiotic resistance is evolving (20). The proportion of S. lugdunensis isolates that are susceptible to all antibiotics has decreased from 68% in 1993 to 45% in 2010 (20, 38). Additionally, the prevalence of ampicillin-resistant strains is increasing, and mecA, the genetic locus conferring resistance to β-lactam antibiotics, has been identified in S. lugdunensis isolates (4, 35). This increase in antibacterial resistance, coupled with the substantial virulence of this organism, presents a clinical challenge that necessitates a thorough understanding of the molecular determinants of virulence within S. lugdunensis.
The availability of the S. lugdunensis genome sequence permits in-depth analysis of the genes underlying the virulence of this dynamic pathogen (36). In this regard, we have determined that S. lugdunensis encodes a complete Isd system, including an IsdG heme oxygenase. This observation suggests that S. lugdunensis is capable of using heme as a source of iron during infection. Here we use biochemical and genomic techniques to characterize this new member of the IsdG family of heme-degrading oxygenases and provide insight into the nutrient acquisition pathways that contribute to the pathogenesis of S. lugdunensis.
MATERIALS AND METHODS
Bacterial strains and construction of expression vectors.
All S. lugdunensis experiments were carried out using S. lugdunensis strain N920143. All S. aureus experiments were carried out using the clinical isolate Newman. S. aureus isogenic variants lacking isdG and isdI have been described previously (27). Bacteria were grown overnight in tryptic soy broth (TSB) at 37°C with shaking at 180 rpm unless otherwise stated.
Protein purification.
Recombinant S. lugdunensis IsdG was purified from Escherichia coli BL21(DE3) as described previously (31). Expression vectors were created through PCR amplification of genomic S. lugdunensis isdG and subsequent cloning into pCR2.1 (Invitrogen). Successful transformants were sequenced for accuracy. Correct inserts were then subcloned into pET15b (Novagen), creating an inducible expression vector expressing N-terminally histidine tagged IsdG. Protein concentrations were determined using a bicinchoninic acid (BCA) assay, and purity was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining.
Heme degradation and product purification.
Heme degradation reactions were performed in Tris-buffered saline (50 mM Tris [pH 7.5], 150 mM NaCl) at room temperature. Wild-type IsdG or mutant IsdG at a concentration of 40 μM was incubated with 40 μM heme (Sigma) for 30 min at 4°C to allow heme-protein complexes to form. Catalase (from bovine liver; Sigma) was added to the sample at a 0.5:1 molar ratio of catalase to hemoprotein; ascorbic acid was added to a final concentration of 1 mM; and spectral changes between 300 and 800 nm were measured at 15, 30, 60, 90, and 120 min after the addition of ascorbate. Heme degradation products were purified as described previously (28).
High-performance liquid chromatography (HPLC) analysis was performed on a Varian ProStar system using a Microsorb-MV C18 column. Analysis was performed with a flow rate of 1 ml min−1 using 95% water–5% acetonitrile with 0.1% trifluoroacetic acid (TFA) as the mobile phase. The mobile phase increased linearly from 5% to 80% over a 45-min period. Staphylobilin derived from both S. aureus IsdG and S. lugdunensis IsdG eluted at approximately 32 min, or a mobile phase of 60%.
Difference absorption spectroscopy.
All absorption spectra were obtained using a Varian Cary 50 Bio spectrophotometer. Heme binding analyses were performed using difference absorption spectroscopy at 413 nm. Aliquots of heme (2 μM to 28 μM) were added both to a sample aliquot (10 μM IsdG) and to a reference aliquot of Tris-buffered saline (50 mM Tris [pH 7.5], 150 mM NaCl). Samples were incubated at 4°C on a rotisserie for 30 min to allow ample time for protein-heme complexes to form.
Heme binding assay.
The Soret band of a 10 μM sample of purified IsdG was measured as heme was added incrementally. The heme concentrations measured ranged from 2 to 40 μM. After each addition of heme, the sample was incubated at 4°C on a rotisserie to allow time for heme-protein complexes to form.
Heme utilization assay.
To evaluate the ability of S. lugdunensis to utilize heme as a nutrient iron source, a liquid growth assay was used. S. lugdunensis was grown overnight in RPMI medium with 1% Casamino Acids. Cultures were normalized to an optical density at 600 nm (OD600) of 1.8 and were subcultured 1:50 into the various media. RPMI medium with 1% Casamino Acids was used for the high-iron condition, RPMI medium with 5 μM ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDHA) for the iron-depleted condition, and RPMI medium with 5 μM EDDHA and 5 μM heme for the heme utilization condition. The OD600 was measured over 24 h, and the assay was repeated with three biological replicates in triplicate.
To assess the ability of S. lugdunensis IsdG to degrade heme and allow for its use as an iron source, a liquid growth assay was utilized. The S. aureus clinical isolate strain Newman was used in all experiments (11). All strains were transformed with the pOS1plgt vector (6, 29). For phenotypic complementation experiments, strains were created by transforming S. aureus ΔisdGI with the pOS1-derived vector containing a full-length copy of S. lugdunensis isdG under the control of the constitutive promoter of the S. aureus lipoprotein diacylglycerol transferase gene (lgt). Inserts from successful PCR2.1 transformants (see “Protein purification” above) were subcloned into pOS1plgt, making pisdG. Strains were grown overnight in RPMI medium with 1% Casamino Acids, the appropriate antibiotic, and 0.5 mM EDDHA. Cultures were normalized to an OD600 of 0.6 and were subcultured 1:100 into Chelex-treated RPMI medium with 0.5 mM EDDHA, 100 μM CaCl2, 25 μM ZnCl2, 1 mM MgCl2, 25 μM MnCl2, 1 μM heme, and the appropriate antibiotic. The OD600 was measured over 55 h. The assay was repeated to test three biological replicates in triplicate.
Quantitative immunoblotting.
Cultures were grown overnight in TSB alone or TSB with 100 to 350 μM 2,2′-dipyridyl (Dip). Bacteria were sedimented through centrifugation at 6,000 × g for 10 min. Cells were then resuspended in TSM (100 mM Tris [pH 7.0], 500 mM sucrose, 10 mM MgCl2) and were incubated in the presence of lysostaphin (10 μg) at 37°C for 30 min. Protoplasts were then collected by centrifugation at 16,000 × g for 10 min and were lysed by sonication. Total-protein concentrations were normalized using a BCA assay. Samples were run on a 15% SDS-PAGE gel and were then transferred to a nitrocellulose membrane. Membranes were probed with an anti-IsdG (α-IsdG) polyclonal antibody raised against S. aureus IsdG and were then blotted with a conjugated goat anti-rabbit antibody (Invitrogen). Membranes were imaged using an Odyssey infrared imager. To analyze the effect of heme on cytoplasmic IsdG levels, cultures were grown overnight in TSB with 350 μM Dip and increasing concentrations of heme. Samples were prepared and probed as described. IsdG levels were quantified using an Odyssey infrared imager. The fold change in IsdG levels as a result of increasing Dip concentrations is representative of six biological replicates, while the fold change in response to exogenous heme is representative of eight biological replicates.
IsdG-family phylogenetic tree.
We reconstructed the evolutionary relationships of IsdG and IsdI (IsdG/I) using amino acid sequences obtained from bacterial species with annotated IsdG-family heme oxygenases. We looked for IsdG proteins in NCBI's GenBank by using a tBLASTx search of the nonredundant (nr) nucleotide database restricted to bacteria. Staphylococcus aureus, Bacillus anthracis, Mycobacterium tuberculosis, and Bradyrhizobium japonicum isdG and isdI nucleotide sequences were used for this tBLASTx search, and amino acid sequences associated with tBLASTx hits that had an E value of ≤1e−05 were retrieved. Retrieved amino acid sequences were further screened by removing all sequences lacking the IsdG catalytic triad or a functional annotation. We also searched UniProt for all reviewed Isd proteins with known heme oxygenase function. A single representative operational taxonomic unit from each retrieved species was used to reconstruct the tree.
Prior to tree building, amino acid sequences were aligned with MUSCLE (13). The final aligned data set had 22 IsdG-family amino acid sequences with a length of 133 amino acids. The Isd phylogeny was reconstructed using maximum likelihood (ML) (21). The best-fit model of amino acid substitution used in the ML reconstruction was determined with ProtTest, version 2.4 (1). According to the Akaike information criterion (AIC), the best-fit substitution model was WAG+I+G+F (38). The ML tree was reconstructed with PhyML, version 3.0 (16, 17), by maximizing the topology likelihood of 10 random starting trees from the best of nearest-neighbor interchange (NNI) and subtree pruning and regrafting (SPR) branch rearrangements. The gamma shape parameter (four rate categories) and the proportion of invariable sites were optimized via ML. Amino acid equilibrium frequencies were set at empirical levels. Node statistical support was determined using a nonparametric bootstrap with 500 replicates.
RESULTS
S. lugdunensis encodes an Isd system.
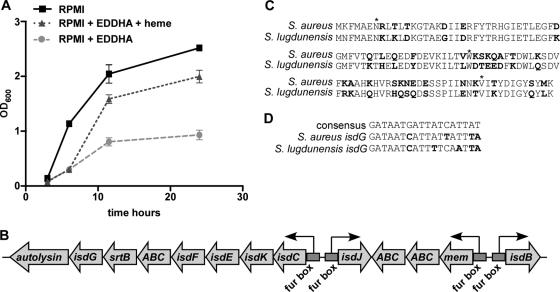
S. lugdunensis requires iron for growth and can use heme to satisfy its iron requirement (Fig. 1A). This suggests that systems dedicated to the acquisition of heme are encoded within the S. lugdunensis genome. Using the nucleotide sequences of the S. aureus isd operon as queries, a putative Isd system was identified in S. lugdunensis (Fig. 1B). The S. aureus Isd system comprises 10 genes encoding proteins that function in concert to bind hemoglobin, remove the heme cofactor, and transport heme into the cell, where it is degraded to release nutrient iron. Several proteins of the S. aureus Isd system are conserved within S. lugdunensis, but important differences exist. For example, the Isd system of S. lugdunensis does not encode an isdA gene but rather an isdA-like gene, isdJ, which has two heme-binding near transporter (NEAT) domains as opposed to the single NEAT domain of S. aureus isdA. Additionally, the S. lugdunensis Isd system includes a gene encoding a putative ABC transporter, SLUG_00980, as well as an insertion between isdJ and isdB of three genes that are not present in the S. aureus Isd system. Moreover, the S. aureus Isd system utilizes two heme oxygenases, IsdG and IsdI, while the S. lugdunensis Isd system appears to employ a single heme oxygenase. We refer to the S. lugdunensis heme oxygenase as IsdG, because it is 68% identical to the amino acid sequence of S. aureus IsdG and only 62% identical to the amino acid sequence of S. aureus IsdI. Importantly, the active site of S. aureus IsdG, comprising residues N7, W67, and H77 (NWH), is conserved within S. lugdunensis IsdG (Fig. 1C) (40). Analysis of the region upstream of the S. lugdunensis isdCKEF-SLUG_00980-srtB-isdG-SLUG_01010 transcriptional start site revealed sequences with a high degree of similarity to the consensus Fur box sequence from S. aureus (Fig. 1D). Fur boxes are nucleotide sequences to which the iron-dependent repressor Fur binds. Therefore, the presence of putative Fur boxes indicates that these genes are likely to be iron regulated, becoming maximally expressed under iron-depleted conditions (14). These observations suggest that S. lugdunensis encodes an iron-regulated Isd system containing an IsdG-family heme oxygenase.
Fig. 1.
S. lugdunensis can utilize heme for growth and encodes a putative Isd system. (A) Growth comparison of S. lugdunensis grown in a medium alone, an iron-depleted medium, or an iron-depleted medium supplemented with 5 μM heme. (B) Genomic organization of the isd locus within S. lugdunensis, with four predicted transcriptional start sites designated by bent arrows. Putative Fur binding sites are marked by stippled boxes. All assignments are based on the annotated S. lugdunensis N920143 genome (18a) (accession number FR870271). (C) Amino acid alignment of S. aureus IsdG and S. lugdunensis IsdG, with nonconserved amino acids shown in boldface. Amino acids within the conserved catalytic triad are indicated by asterisks. (D) Nucleotide sequence alignment of the S. aureus Fur box consensus sequence, the S. aureus isdG Fur box sequence, and the S. lugdunensis isdG Fur box sequence. Nucleotides that differ from the consensus sequence are shown in boldface.
S. lugdunensis IsdG binds and degrades heme.
Hemin binding proteins have a distinct absorption spectrum defined by peak absorbance in the range of 390 to 430 nm, referred to as a Soret band. Hemin, the oxidized form of heme, was used in all experiments; however, for simplicity, we refer to both of these molecules as heme. S. lugdunensis IsdG purified from Escherichia coli grown in the absence of heme does not exhibit absorption patterns indicative of a heme binding protein. However, reconstitution of IsdG with heme produces an optical absorption spectrum with a peak absorbance at 413 nm, which increases incrementally upon addition of heme (Fig. 2A). The Soret peak of heme-protein complexes is distinct from that of free heme at a neutral pH. This difference allows for the spectrophotometric titration of IsdG with heme, from which the stoichiometry of IsdG and heme can be determined. As heme is added incrementally to purified IsdG (10 μM), the change in absorbance measured at 413 nm has a discrete inflection point at approximately 10 μM heme, indicating a 1:1 stoichiometry of protein and heme (Fig. 2A, inset). Together, these data establish S. lugdunensis IsdG as a heme binding protein.
Fig. 2.
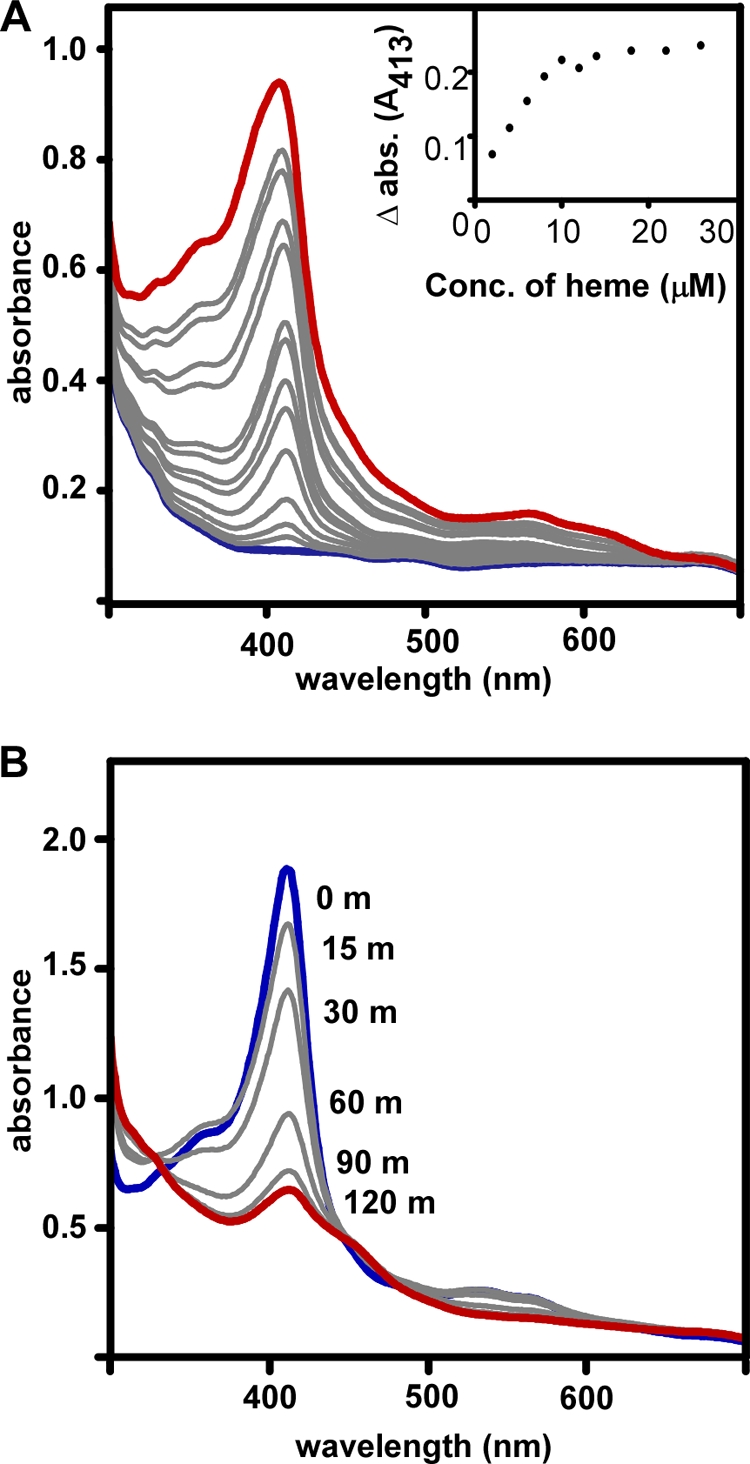
S. lugdunensis IsdG binds and degrades heme. (A) Increasing amounts of heme (2 to 40 μM) were added to a sample of purified IsdG (10 μM) and to a reference sample. (Inset) The difference between the absorbances (Δabs.) of the protein-heme complex and free heme at 413 nm is plotted against the total heme concentration. A 10 μM protein sample was used. (B) Spectra of 40 μM IsdG-heme complex were taken at the time of addition of 1 mM ascorbate (0 m; curve shown in blue) and 15, 30, 60, 90, and 120 min (curve shown in red) thereafter. Reactions were performed in the presence of catalase at a 0.5:1 (catalase to hemoprotein) molar ratio.
The ability of S. lugdunensis IsdG to degrade heme was evaluated using optical absorption spectroscopy. Purified recombinant IsdG was incubated with heme, and protein-heme complexes were allowed to form. Next, ascorbate was added as a reductant, and heme degradation was monitored spectrophotometrically. Spectral analysis of the reaction was performed prior to the addition of ascorbate as well as at 15, 30, 60, 90, and 120 min following the addition of ascorbate. This reaction resulted in almost complete elimination of the peak at 413 nm, indicating opening of the macrocyclic conjunction of heme (Fig. 2B). All reactions were performed in the presence of catalase to distinguish between coupled oxidation of heme and enzymatic heme degradation. These results demonstrate that S. lugdunensis IsdG catalyzes the oxidative degradation of heme in the presence of an electron donor, consistent with the assignment of S. lugdunensis IsdG as a heme oxygenase. Notably, the rate of degradation is similar to that observed with the S. aureus IsdG and IsdI enzymes (31).
A defining characteristic of IsdG-family heme oxygenases is the presence of an NWH catalytic triad required for heme catabolism (40). We evaluated the functional requirement for these three residues in S. lugdunensis IsdG by using mutational analysis. Each residue within the predicted catalytic triad was mutated to alanine, and the heme binding properties of purified mutant proteins were assessed. All three mutant proteins (N7A, W67A, and H77A) retained the ability to bind heme, indicating that these mutations do not disrupt the overall fold or heme binding properties of IsdG (Fig. 3). However, inactivation of any residue within the catalytic triad ablated enzymatic function, as demonstrated by the retention of the peak at 413 nm 90 min after the addition of an electron donor (Fig. 3). These results indicate that the catalytic NWH triad is required for S. lugdunensis IsdG enzymatic activity.
Fig. 3.
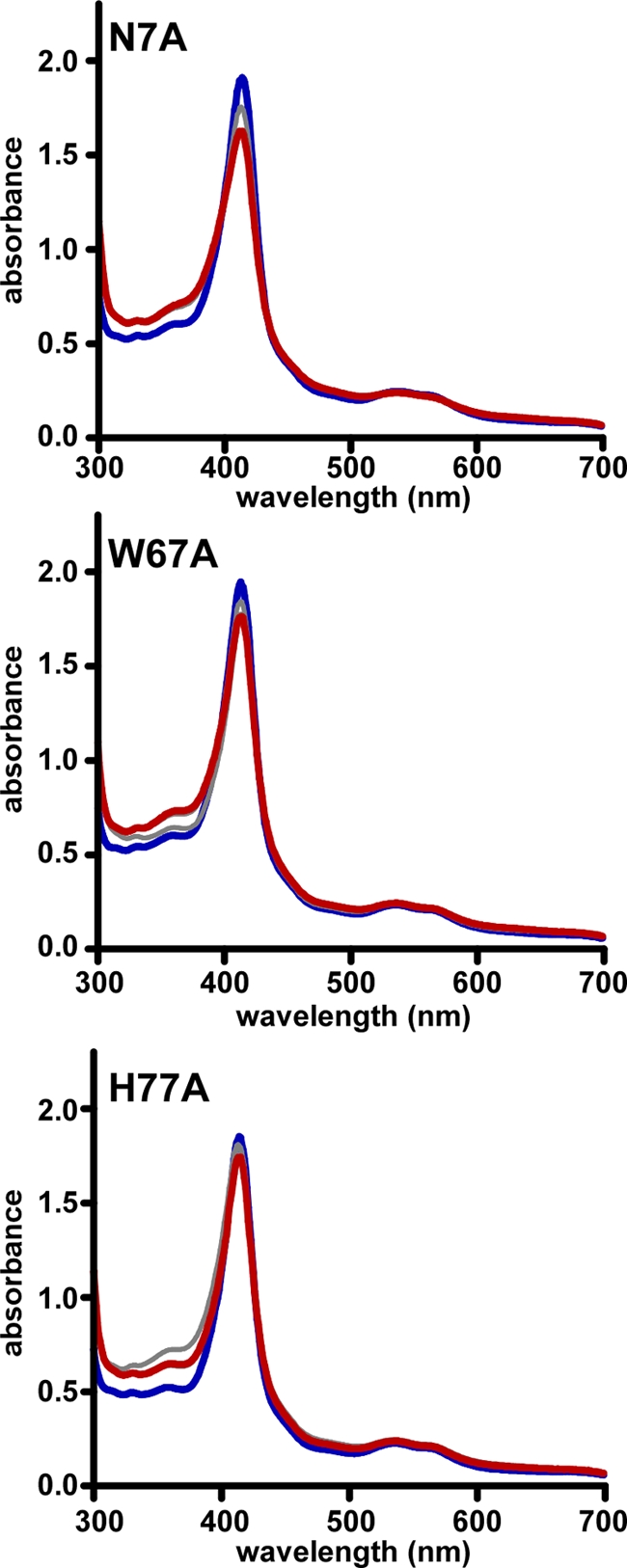
The catalytic triad is functionally conserved in S. lugdunensis IsdG. Point mutations were made within the NWH catalytic triad of S. lugdunensis IsdG by converting each of the three amino acids to alanine. Proteins were expressed and purified similarly to those of the wild type and were assessed for enzymatic activity. Spectra of 40 μM IsdG-heme complex were taken at the time of addition of 1 mM ascorbate (0 m; curve shown in blue) and 15, 30, 60, and 90 min (curve shown in red) thereafter.
S. lugdunensis IsdG degrades heme to staphylobilin.
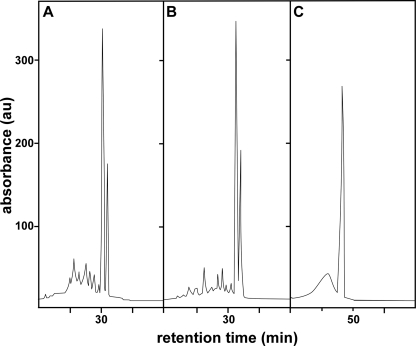
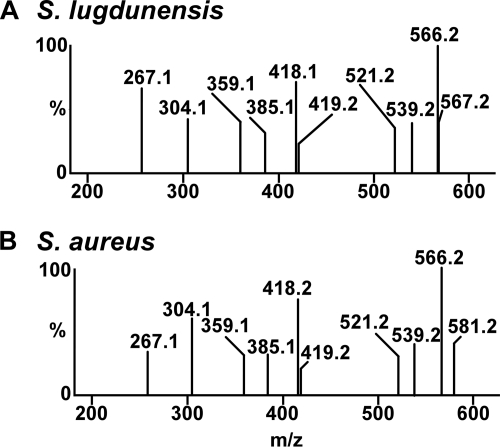
Two structurally distinct families of heme oxygenases have been identified; they are referred to as the HO-1 family and the IsdG family (30, 31). While the HO-1 family of heme oxygenases is ubiquitous across kingdoms, the IsdG family has been identified only in bacteria (8, 25, 31, 32). The mechanism of heme catabolism in the HO-1 family is highly conserved and typically results in the conversion of heme to the blue-green molecule α-biliverdin (37). Within eukaryotes, biliverdin is further reduced to the yellow molecule α-bilirubin by biliverdin reductase (23). In contrast, S. aureus IsdG family members degrade heme directly to a yellow oxo-bilirubin molecule that has been named staphylobilin (28). Based on this, we sought to determine if the yellow product of S. lugdunensis IsdG-mediated heme degradation is also staphylobilin. Initially, we analyzed the product of IsdG-mediated heme degradation using high-performance liquid chromatography (HPLC) and compared its retention time to that of staphylobilin and biliverdin (Fig. 4). When analyzed by HPLC, the heme degradation product generated by S. lugdunensis IsdG eluted in a series of two peaks with retention times identical to those of staphylobilin (Fig. 4A and B). The two HPLC peaks represent distinct isomers of staphylobilin generated by IsdG-family heme oxygenases (28). Importantly, the heme degradation product of S. lugdunensis IsdG eluted earlier than either biliverdin or bilirubin, indicating a higher degree of polarity (Fig. 4B and C). After HPLC purification, the products of S. lugdunensis and S. aureus IsdG-mediated heme degradation were subjected to high-resolution electrospray ionization-mass spectrometry (HR-ESI-MS). This analysis allowed for the molecular composition of each HPLC peak to be determined and subsequently compared. Both HPLC peaks from S. aureus staphylobilin and the S. lugdunensis IsdG heme degradation product were determined to have a mass of 598.25 Da (m/z 599.2605 [M+H], Δ = 0.5 to 4.2 ppm), corresponding to a molecular formula of C33H34N4O7 (Fig. 5). Importantly, the molecular masses of biliverdin and bilirubin are 582.6 and 584.7 Da, respectively, and thus are clearly distinct from that of staphylobilin. Together these data demonstrate that S. lugdunensis IsdG degrades heme to the yellow molecule staphylobilin.
Fig. 4.
HPLC analysis of the S. lugdunensis IsdG heme degradation product. HPLC comparison of established heme degradation products indicates that S. lugdunensis IsdG degrades heme to staphylobilin. (A) S. aureus staphylobilin purification monitored at 465 nm. (B) S. lugdunensis IsdG-mediated heme degradation product monitored at 465 nm. (C) Biliverdin purification monitored at 405 nm.
Fig. 5.
Tandem LC-HR-ESI-MS analysis of the S. lugdunensis IsdG heme degradation product and S. aureus staphylobilin. (A) ESI-MS-MS of fragment ion selection for m/z 599.3 for the S. lugdunensis IsdG-mediated heme degradation product. (B) ESI-MS-MS of fragment ion selection for m/z 599.3 for S. aureus staphylobilin.
S. lugdunensis IsdG is iron regulated.
To ensure proper metal homeostasis, bacteria strictly regulate the uptake and metabolism of metals from the environment (10, 18, 23, 24, 26). A putative Fur-binding sequence upstream of the predicted isdG operon suggests that isdG expression increases upon iron starvation (Fig. 1D). To test this hypothesis, we used immunoblotting to assess the abundance of IsdG within cells grown in increasing concentrations of the iron chelator 2,2′-dipyridyl (Dip). These experiments revealed that IsdG expression increases under iron-depleted conditions, establishing S. lugdunensis IsdG as an iron-regulated enzyme (Fig. 6A and B).
Fig. 6.
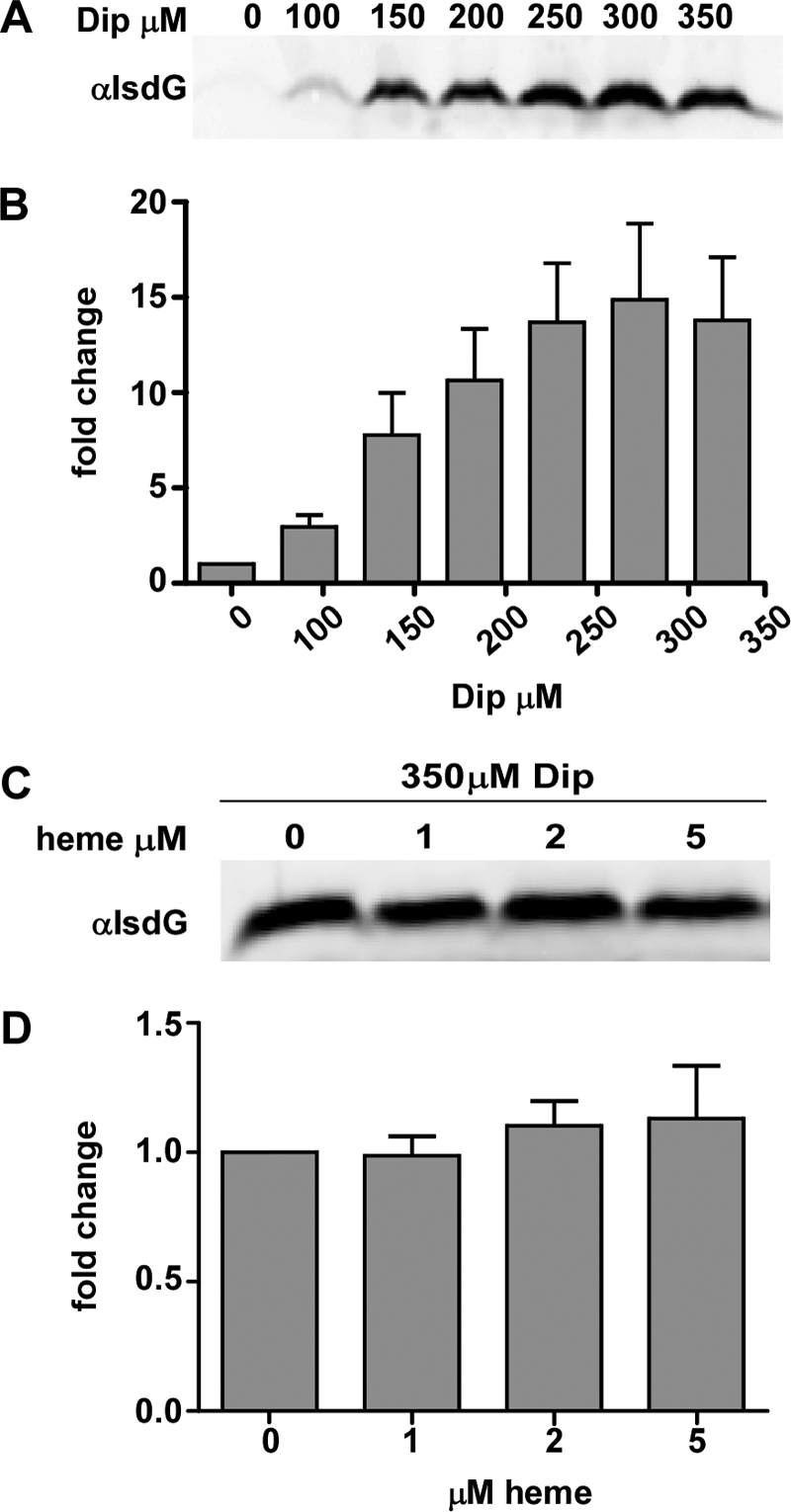
S. lugdunensis isdG is iron regulated. S. lugdunensis was grown overnight with the indicated supplements, and protoplasts were lysed and normalized by total-protein concentration. (A) Immunoblot analysis of the effect of iron chelation by 2,2′-dipyridyl (Dip) on the IsdG expression level. (B) Difference (expressed as the fold change) between cytoplasmic IsdG levels in increasing concentrations of Dip and those in medium alone. (C) Immunoblot analysis of the effect of exogenous heme on the expression of IsdG when S. lugdunensis is grown in an iron-depleted medium. (D) Difference (expressed as the fold change) between the IsdG levels of cells grown in an iron-depleted medium with increasing concentrations of exogenous heme and those in cells grown in an iron-depleted medium alone.
S. aureus encodes two IsdG-family enzymes, IsdG and IsdI, which are differentially regulated in response to iron and heme (27). While both IsdG and IsdI are highly expressed under iron-depleted conditions, IsdG is targeted for proteolytic degradation in the absence of heme. Consequently, maximal IsdI levels occur under low-iron conditions, while IsdG is most abundant under iron-depleted conditions in the presence of heme. We therefore sought to determine the effect of heme exposure on IsdG levels in S. lugdunensis by using immunoblotting. S. lugdunensis was grown under iron-depleted conditions, and the medium was supplemented with increasing amounts of heme; then the level of intracellular IsdG was analyzed using S. aureus IsdG antisera as a probe. This experiment revealed that the presence of heme has no effect on the intracellular abundance of IsdG in S. lugdunensis (Fig. 6C and D). Together these experiments show that S. lugdunensis IsdG is upregulated under iron-depleted conditions and is not affected by the addition of heme. This is consistent with a model whereby S. lugdunensis upregulates its heme utilization machinery during times of nutrient iron starvation.
S. lugdunensis IsdG facilitates the use of heme as an iron source.
Heme degradation results in the release of free iron for use as a nutrient source. To determine the biological function of S. lugdunensis IsdG, we measured the ability of S. lugdunensis IsdG to complement the heme utilization defect of the S. aureus heme oxygenase mutant (S. aureus ΔisdGI) (31). Heme utilization was measured by comparing the growth of plasmid-containing strains of S. aureus in an iron-depleted medium supplemented with heme. To eliminate complications associated with iron-dependent effects on transcription, isdG was cloned into the p-OS1plgt vector, resulting in the constitutive expression of isdG under the control of the lgt promoter (6, 29). The expression of S. lugdunensis IsdG completely restored the ability of S. aureus ΔisdGI to utilize heme as an iron source for growth (Fig. 7). In fact, the complemented strain grew better than the wild type at early time points, demonstrating that overexpression of IsdG provides an enhanced ability to grow on heme as a sole source of iron. These results demonstrate that S. lugdunensis IsdG degrades heme to release free iron and enhance bacterial growth.
Fig. 7.
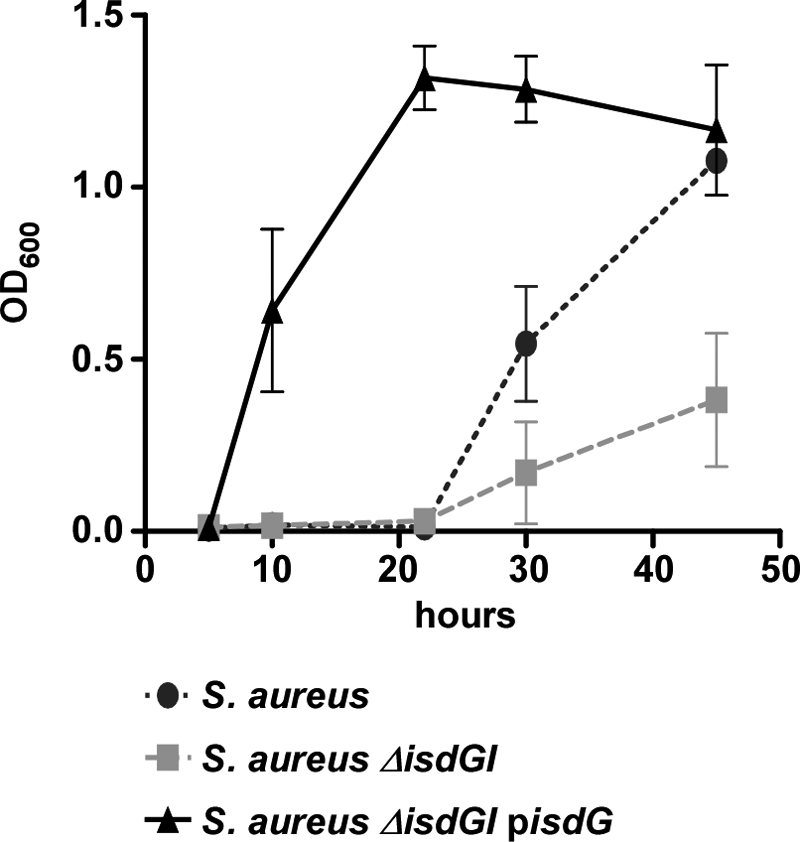
S. lugdunensis IsdG complements the heme utilization defect of an S. aureus heme oxygenase mutant. Shown is a comparison of the growth of S. aureus, S. aureus ΔisdGI, and S. aureus ΔisdGI expressing S. lugdunensis isdG in trans from the pOS1plgt plasmid. All strains contain a pOS1plgt plasmid and were grown in minimal medium with supplemental heme as the only iron source.
DISCUSSION
While the types of infections caused by S. lugdunensis are well described, the molecular mechanisms employed during pathogenesis have not been identified. As with most bacterial pathogens, S. lugdunensis cannot survive and replicate in the absence of iron. Moreover, free iron within host tissues is found at concentrations significantly lower than that required to sustain bacterial growth (5). To combat this barrier to growth, S. lugdunensis presumably encodes systems dedicated to the acquisition of nutrient iron during infection. Consistent with this, we report here that S. lugdunensis encodes an Isd system that includes an IsdG-family heme oxygenase that degrades heme to staphylobilin and free iron. IsdG is upregulated under conditions of iron starvation, ensuring that the heme-catabolizing machinery is abundant during times of nutrient stress. Moreover, S. lugdunensis IsdG promotes the growth of an S. aureus heme oxygenase mutant on heme, supporting the placement of this enzyme within the heme-iron acquisition pathway. Based on these findings, it is likely that the S. lugdunensis Isd system is utilized under low-iron conditions to bind host hemoproteins, remove heme, and transport it into the cytoplasm, where it can be degraded to release free iron.
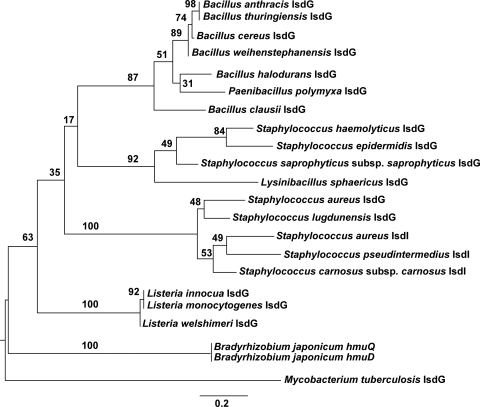
Functional IsdG-family heme oxygenases have been identified in S. aureus, Bacillus anthracis, Bradyrhizobium japonicum, and Mycobacterium tuberculosis (8, 25, 31, 32). To gain insight into the evolutionary relationships between these IsdG family members, we interrogated bacterial genomes to identify putative IsdG orthologs and to create a phylogenetic tree using annotated IsdG family members (Fig. 8) (22, 39). Interestingly, staphylococcal IsdG members segregated into two distinct clades, with S. aureus and S. lugdunensis in one clade and S. epidermidis and Staphylococcus haemolyticus in a separate clade (Fig. 8). Moreover, S. aureus IsdG appears to be more closely related to S. lugdunensis IsdG than to any other IsdG family member, including its intrachromosomal paralog, IsdI. The analysis of all sequenced bacterial genomes revealed predicted IsdG orthologs across diverse classes of bacteria, such as Alphaproteobacteria, Actinobacteria, Betaproteobacteria, Chloroflexi, Corynebacterineae, and Deinococcus-Thermus. The broad conservation of IsdG across diverse classes of bacteria implicates heme degradation as a conserved mechanism by which bacteria meet their iron requirements. The number of species with a putative IsdG heme oxygenase suggests that the use of IsdG to catabolize heme may be pervasive among bacteria. This observation, combined with the bacterium-specific production of staphylobilin, raises many questions regarding the full function of this enzyme. For example, many species with a predicted IsdG heme oxygenase have no known association with any plant or animal from which they could acquire heme. Organisms such as Salinispora tropica, Oligotropha carboxidovorans, and Thermus thermophilus occupy unique niches, thriving in inhospitable environments such as thermal hot springs, ocean sediments, and pyrite deposits. Further investigation of the genomes of these bacteria living in extreme environments revealed that they all possess the genes required to synthesize heme endogenously. It is possible that these bacteria encode an IsdG-family heme oxygenase to convert endogenously synthesized heme to staphylobilin, adding support to the hypothesis that staphylobilin fulfills an important biological function. Alternatively, IsdG enzymes may exist to degrade excess heme that has been synthesized endogenously as a means to prevent heme-associated toxicity.
Fig. 8.
Two distinct staphylococcal clades within a phylogenetic tree of annotated IsdG-family heme oxygenases. Shown is a midpoint-rooted phylogenetic tree of IsdG/I amino acid sequences from bacterial taxa with characterized heme oxygenases. The phylogeny was reconstructed using ML, and the tree with the highest log likelihood (lnL = −3,131.92126) is shown. A nonparametric bootstrap (500 replicates) was used to determine node support. Bootstrap values are shown at each node. Branch lengths are drawn to scale and are measured in amino acid substitutions per site.
The S. aureus Isd system includes three cell wall-anchored proteins, IsdA, IsdB, and IsdC, that bind heme. Genes encoding putative orthologs of IsdB and IsdC have been identified within the S. lugdunensis Isd system (Fig. 1). Further, the S. aureus Isd system encodes IsdF (a polytopic transmembrane protein), IsdE (a lipoprotein ATPase), and a sortase, SrtB, responsible for anchoring IsdC to the cell wall (21). Each of these proteins is conserved within the S. lugdunensis Isd system. However, important differences between the two systems exist. First, while S. lugdunensis does not encode an IsdA protein, it does encode an IsdA-like protein, IsdJ, which has an additional heme-binding NEAT domain not present in S. aureus IsdA. Second, there is a putative ABC transporter within the S. lugdunensis Isd system, situated between isdF and srtB. Third, the Isd operon within S. lugdunensis includes the isdK gene, located between isdC and isdE, which is predicted to encode a hypothetical protein that has no homology to any Isd protein but contains a NEAT domain. The ABC transporter and IsdK may be involved in iron homeostasis, since their location within the predicted Isd operon suggests that they are also iron regulated. Alternatively, they may facilitate heme uptake by acting as accessory proteins to the IsdEF heme transporter. Fourth, S. lugdunensis has an insertion of three genes that appear to encode a membrane transporter between isdJ and isdB. While it is not known if these genes are involved in heme-iron acquisition, the identification of a Fur box upstream of the transcriptional start site of these genes suggests that they are upregulated in low-iron environments. Finally, there is no identifiable IsdI ortholog within S. lugdunensis.
IsdG and IsdI are differentially regulated within S. aureus (27). Both heme oxygenases are transcriptionally regulated by Fur in response to iron levels; however, in the absence of heme, IsdG is targeted for proteolytic degradation (27). The biological significance of this differential regulation is not fully understood. One possibility is that by differentially regulating IsdG and IsdI, S. aureus can more precisely control heme degradation. S. aureus gains access to host heme through the expression of hemolysins, which lyse erythrocytes. Hemolysin expression is regulated in response to bacterial density by the global virulence regulator agr (12). The two heme oxygenases in S. aureus may function such that IsdI is required early in infection to degrade the small amount of available heme present prior to the secretion of hemolysins. After the colonization of host organs, hemolysin-mediated erythrocyte lysis leads to an influx of heme, necessitating the activity of a second heme oxygenase, IsdG. An iron-regulated heme oxygenase may be sufficient within S. lugdunensis, because the amount of heme available for degradation may be more consistent throughout infection. This model is supported by the finding that hemolytic activity due to alpha-, beta-, or gamma-hemolysins, present in S. aureus, has not been detected in S. lugdunensis (15). Additionally, most isolates of S. lugdunensis have been found to produce a delta-like hemolysin, encoded not in the agr locus but in the slush locus (38). The location of these genes outside the agr locus suggests that they are regulated by an as yet undetermined mechanism. Together these observations indicate that the hemolytic capacity of S. lugdunensis is lower than that of S. aureus and that the expression of the only hemolysin of S. lugdunensis may be constant throughout the course of infection. Consequently, cytoplasmic heme levels in S. lugdunensis may not differ considerably throughout the course of infection. Regulation of IsdG through Fur alone may allow for sufficient control of the heme-catabolizing machinery within the cell. Alternatively, the S. lugdunensis Isd system may be less efficient than that of S. aureus, resulting in comparatively lower cytoplasmic heme levels, which eliminate the need for a second heme oxygenase.
S. lugdunensis IsdG degrades heme to staphylobilin (Fig. 4 and 5). This is the first evidence that the production of staphylobilin is conserved among bacterial species encoding an IsdG-family heme oxygenase. Bilirubin, biliverdin, carbon monoxide, and iron are the products of vertebrate heme catabolism, and all of these molecules have important functions within mammalian cells (19). For example, biliverdin and carbon monoxide have beneficial anti-inflammatory functions, and all three molecules act as antioxidants in serum (19, 34). The biological roles of heme degradation products in bacteria remain unknown. Bacterial HO-1-family heme oxygenases degrade heme to biliverdin; however, no biliverdin reductase ortholog has been identified in bacteria. Furthermore, it remains unclear if bacterially derived biliverdin has a physiological function. Carbon monoxide is produced during heme catabolism by heme oxygenases (19, 28). It is plausible that CO production affects bacteria; however, the impact of endogenously produced CO has yet to be determined. The establishment of staphylobilin as a conserved heme degradation product in bacteria supports the idea that this molecule may serve a valuable biological function.
This work establishes the role of S. lugdunensis IsdG in heme-iron utilization, a process that is required for the full virulence of S. aureus (27). S. aureus strains lacking isdG or isdI show severely attenuated growth within murine hearts following systemic challenge (27). This supports the idea that bacterial heme degradation is vital to cardiac colonization. Future experiments will aim to develop a murine model of S. lugdunensis infection in order to elucidate the contribution of IsdG to the pathogenesis of S. lugdunensis endocarditis. The significant structural differences between IsdG family members and human HO-1 heme oxygenases highlight the potential of IsdG as a therapeutic target. In keeping with this, further investigations into the contribution of the Isd system to S. lugdunensis pathogenesis may facilitate the development of antimicrobials that inhibit the Isd system and treat S. lugdunensis infections.
ACKNOWLEDGMENTS
We thank the members of the Skaar laboratory for critical reading of the manuscript and Marion Calcutt of the Vanderbilt Mass Spectrometry Research Center for technical assistance.
This research was supported by U.S. Public Health Service grants AI69233 and AI073843 from the National Institute of Allergy and Infectious Diseases. E.P.S. is a Burroughs Wellcome Fellow in the Pathogenesis of Infectious Diseases. K.P.H. was funded by the Cellular and Molecular Microbiology Training Grant Program 5 T32 A107611-10. Work in Ireland was supported by IRCSET and by Science Foundation Ireland (08/IN.1/B1845).
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Abascal F., Zardoya R., Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 [DOI] [PubMed] [Google Scholar]
- 2. Anguera I., et al. 2005. Staphylococcus lugdunensis infective endocarditis: description of 10 cases and analysis of native valve, prosthetic valve, and pacemaker lead endocarditis clinical profiles. Heart 91:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arias M., et al. 2010. Skin and soft tissue infections caused by Staphylococcus lugdunensis: report of 20 cases. Scand. J. Infect. Dis. 42:879–884 [DOI] [PubMed] [Google Scholar]
- 4. Becker K., et al. 2006. Does nasal cocolonization by methicillin-resistant coagulase-negative staphylococci and methicillin-susceptible Staphylococcus aureus strains occur frequently enough to represent a risk of false-positive methicillin-resistant S. aureus determinations by molecular methods? J. Clin. Microbiol. 44:229–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun V., Günter K., Hantke K. 1991. Transport of iron across the outer membrane. Biol. Met. 4:14–22 [DOI] [PubMed] [Google Scholar]
- 6. Bubeck Wardenburg J., Williams W. A., Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 103:13831–13836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bullen J. J., Griffiths E. 1999. Iron and infection: molecular, physiological, and clinical aspects, 2nd ed. John Wiley and Sons, New York, NY. [Google Scholar]
- 8. Chim N., Iniguez A., Nguyen T. Q., Goulding C. W. 2010. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J. Mol. Biol. 395:595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chipperfield J. R., Ratledge C. 2000. Salicylic acid is not a bacterial siderophore: a theoretical study. Biometals 13:165–168 [DOI] [PubMed] [Google Scholar]
- 10. Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86:1637–1645 [DOI] [PubMed] [Google Scholar]
- 11. Duthie E. S., Lorenz L. L. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95–107 [DOI] [PubMed] [Google Scholar]
- 12. Dunman P. M., et al. 2001. Transcription profiling based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Escolar L., Pérez-Martín J., de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frank K. L., Del Pozo J. L., Patel R. 2008. From clinical microbiology to infection pathogenesis: how daring to be different works for Staphylococcus lugdunensis. Clin. Microbiol. Rev. 21:111–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 17. Guindon S., et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 18. Hantke K. 2001. Bacterial zinc transporters and regulators. Biometals 14:239–249 [DOI] [PubMed] [Google Scholar]
- 18a.Heilbronner S.Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. [17 June 2011.]. doi: 10.1111/j.1574-6968.2011.02339.x. [DOI] [PMC free article] [PubMed]
- 19. Kirkby K. A., Adin C. A. 2006. Products of heme oxygenase and their potential therapeutic applications. Am. J. Physiol. Renal Physiol. 290:F563–F571 [DOI] [PubMed] [Google Scholar]
- 20. Kleiner E., Monk A. B., Archer G. L., Forbes B. A. 2010. Clinical significance of Staphylococcus lugdunensis isolated from routine cultures. Clin. Infect. Dis. 51:801–803 [DOI] [PubMed] [Google Scholar]
- 21. Mazmanian S. K., et al. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science (New York, NY) 299:906–909 [DOI] [PubMed] [Google Scholar]
- 22. Nei M., Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY. [Google Scholar]
- 23. Noguchi M., Yoshida T., Kikuchi G. 1979. Purification and properties of biliverdin reductases from pig spleen and rat liver. J. Biochem. 86:833–848 [DOI] [PubMed] [Google Scholar]
- 24. Osman D., Cavet J. S. 2008. Copper homeostasis in bacteria. Adv. Appl. Microbiol. 65:217–247 [DOI] [PubMed] [Google Scholar]
- 25. Puri S., O'Brian M. R. 2006. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J. Bacteriol. 188:6476–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reniere M. L., Torres V. J., Skaar E. P. 2007. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals 20:333–345 [DOI] [PubMed] [Google Scholar]
- 27. Reniere M. L., Skaar E. P. 2008. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol. Microbiol. 69:1304–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reniere M. L., et al. 2010. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol. Microbiol. 75:1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schneewind O., Model P., Fischetti V. A. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267–281 [DOI] [PubMed] [Google Scholar]
- 30. Schuller D. J., Wilks A., Ortiz de Montellano P. R., Poulos T. L. 1999. Crystal structure of human heme oxygenase-1. Nat. Struct. Biol. 6:860–867 [DOI] [PubMed] [Google Scholar]
- 31. Skaar E. P., Gaspar A. H., Schneewind O. 2004. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436–443 [DOI] [PubMed] [Google Scholar]
- 32. Skaar E. P., Gaspar A. H., Schneewind O. 2006. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J. Bacteriol. 188:1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sotutu V., Carapetis J., Wilkinson J., Davis A., Curtis N. 2002. The “surreptitious Staphylococcus”: Staphylococcus lugdunensis endocarditis in a child. Pediatr. Infect. Dis. J. 21:984–986 [DOI] [PubMed] [Google Scholar]
- 34. Stocker R., Yamamoto Y., McDonagh A. F., Glazer A. N., Ames B. N. 1987. Bilirubin is an antioxidant of possible physiological importance. Science (New York, NY) 235:1043–1046 [DOI] [PubMed] [Google Scholar]
- 35. Tee W. S. N., Soh S. Y., Lin R., Loo L. H. 2003. Staphylococcus lugdunensis carrying the mecA gene causes catheter-associated bloodstream infection in premature neonate. J. Clin. Microbiol. 41:519–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tse H., et al. 2010. Complete genome sequence of Staphylococcus lugdunensis strain HKU09-01. J. Bacteriol. 192:1471–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Unno M., et al. 2004. Crystal structure of the dioxygen-bound heme oxygenase from Corynebacterium diphtheriae: implications for heme oxygenase function. J. Biol. Chem. 279:21055–21061 [DOI] [PubMed] [Google Scholar]
- 38. Vandenesch F., Etienne J., Reverdy M. E., Eykyn S. J. 1993. Endocarditis due to Staphylococcus lugdunensis: report of 11 cases and review. Clin. Infect. Dis. 17:871–876 [DOI] [PubMed] [Google Scholar]
- 39. Whelan S., Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18:691–699 [DOI] [PubMed] [Google Scholar]
- 40. Wu R., et al. 2005. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem. 280:2840–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]