Abstract
In Salmonella enterica, ThiI is a bifunctional enzyme required for the synthesis of both the 4-thiouridine modification in tRNA and the thiazole moiety of thiamine. In 4-thiouridine biosynthesis, ThiI adenylates the tRNA uridine and transfers sulfur from a persulfide formed on the protein. The role of ThiI in thiazole synthesis is not yet well understood. Mutational analysis described here found that ThiI residues required for 4-thiouridine synthesis were not involved in thiazole biosynthesis. The data further showed that the C-terminal rhodanese domain of ThiI was sufficient for thiazole synthesis in vivo. Together, these data support the conclusion that sulfur mobilization in thiazole synthesis is mechanistically distinct from that in 4-thiouridine synthesis and suggest that functional annotation of ThiI in genome sequences should be readdressed. Nutritional studies described here identified an additional cysteine-dependent mechanism for sulfur mobilization to thiazole that did not require ThiI, IscS, SufS, or glutathione. The latter mechanism may provide insights into the chemistry used for sulfur mobilization to thiazole in organisms that do not utilize ThiI.
INTRODUCTION
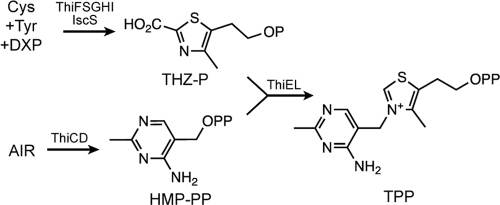
Thiamine pyrophosphate (TPP) is an essential coenzyme used by pyruvate dehydrogenase, α-ketoacid decarboxylase, α-ketoacid oxidase transketolase, and other enzymes, where it stabilizes acyl carbanion intermediates through dissociation of the proton at C-2 of the thiazolium ring (9). Though thiamine was isolated in the 1930s and its chemical synthesis was achieved 4 years later, significant questions remain about the biochemical details of TPP synthesis. Genetic studies, primarily in Salmonella enterica and Escherichia coli, have defined 10 gene products involved in TPP synthesis in bacteria (Fig. 1) (10). With the exception of ThiI, a role for each of these gene products in TPP synthesis has been reconstituted in vitro (3, 7, 12, 13, 20, 24, 29). Microbes and plants synthesize thiamine de novo with separate pathways for the formation of the thiazole (2-carboxy-4-methyl-5-β-hydroxyethylthiazole phosphate [THZ-P]) and pyrimidine (4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate [HMP-PP]) moieties (1, 5, 18, 22). In bacteria, the two moieties are joined by thiamine phosphate synthase, the product of the thiE gene, to form thiamine monophosphate. The biologically active form of the coenzyme, thiamine pyrophosphate (TPP), is then generated by a single phosphorylation catalyzed by thiamine monophosphate kinase (ThiL) (14, 29).
Fig. 1.
Thiamine biosynthesis in S. enterica. A schematic of the thiamine biosynthetic pathway in S. enterica is shown. The enzymes involved in each step are indicated above the relevant arrow. Abbreviations: Cys, cysteine; DXP, deoxyxylulose phosphate; Tyr, tyrosine; AIR, 5-aminoimidazole ribotide; THZ-P, 2-carboxy-4-methyl-5-β-hydroxyethylthiazole phosphate; HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate.
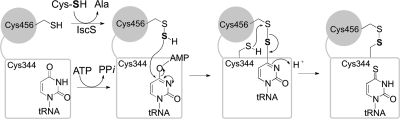
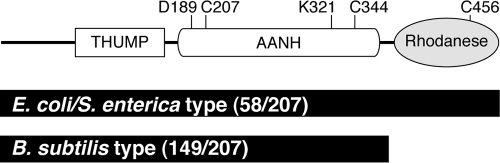
The thiI locus was defined by lesions that caused a thiazole auxotrophy (28) and was subsequently found to be allelic with nuvA, a locus required for 4-thiouridine modification of tRNA (15). Synthesis of 4-thiouridine has been reconstituted in vitro, and our current understanding is summarized in Fig. 2. Experiments in vitro demonstrated the transfer of sulfur from the source cysteine via an IscS persulfide to form a ThiI persulfide (11) located at residue Cys456 (19). In 4-thiouridine biosynthesis, ThiI activates the tRNA uridine as its adenylate (30, 32), which reacts with the Cys456 persulfide (11, 19). Residue Cys344 then acts as a nucleophile, forming a Cys456-Cys344 disulfide and releasing 4-thiouridine. The ThiI disulfide is then reduced by cellular reductants to complete the reaction cycle (17, 27). The ThiI protein in E. coli/S. enterica contains three distinct domains, and each has been implicated in the mechanism of 4-thiouridine synthesis (Fig. 3). The THUMP domain is thought to bind the tRNA, the adenine nucleotide alpha-hydrolase domain (AANH) is responsible for uridine adenylation, and the C-terminal rhodanese domain participates in sulfur transfer (2, 6, 27, 32). Five ThiI residues have been shown to be important for 4-thiouridine biosynthesis: Asp189, Lys321, Cys207, Cys344, and Cys456. Asp189 and Lys321 are conserved residues in the AANH domain and are required for adenylation of the uridine (16). Cys207 is also in the AANH domain, and the ThiIC207A variant showed intermediate activity in vivo and in vitro (17). Cys456 is conserved in the rhodanese domain and carries the nucleophilic cysteine persulfide that is essential for the transfer of sulfur to uridine (19). Cys344 is a highly conserved residue located in the AANH domain that forms a disulfide bond with Cys456, releasing 4-thiouridine (17).
Fig. 2.
Sulfur mobilization by ThiI in 4-thiouridine biosynthesis. Events depicted occur in the active site of ThiI. The boxes represent the THUMP and AANH domains of ThiI, while the gray circles represent the rhodanese domain. Critical cysteine residues 344 and 456 are shown. tRNA represents the rest of the tRNA molecule containing the uridine being acted on by ThiI.
Fig. 3.
Structural domains of S. enterica ThiI. Motifs in the S. enterica ThiI are based on those suggested previously (27), and the residue numbers are those of E. coli. Residues 1 to 163 contain an N-terminal ferredoxin-like (NFLD) domain and a THUMP domain (2), both of which are thought to interact with the tRNA (27). Residues 173 to 400 contain the adenylation domain (AANH). The C-terminal portion contains the rhodanese-like domain.
Sulfur mobilization in thiazole synthesis shares many features with those described above for 4-thiouridine synthesis. In thiazole synthesis, ThiFS forms a stable complex, and ThiF catalyzes the adenylation of the terminal glycine residue of ThiS (31). It is the acyladenylate on ThiS that accepts sulfur from a persulfide donor (24). It has been suggested that sulfur mobilization in thiazole synthesis utilizes ThiI as a persulfide carrier between IscS and the ThiS-acyladenylate (17, 24). Indeed, in vivo results suggest that thiI is required for the formation of the ThiS thiocarboxylate (24), although ThiI was not required for the in vitro formation of the ThiS thiocarboxylate (31). The role of ThiI in sulfur mobilization in thiazole synthesis has not been directly tested by mutational analyses or in vitro studies.
This study was initiated to probe the role of ThiI in thiazole synthesis in the context of what is known about sulfur mobilization in 4-thiouridine biosynthesis. The results here showed that ThiI residues critical for 4-thiouridine biosynthesis were not required for thiazole synthesis and that the rhodanese domain of ThiI was sufficient for synthesis of thiazole in vivo. Nutritional studies showed that there was an alternative mechanism of sulfur mobilization to thiazole that depends on exogenous cysteine. These results provide insights into sulfur mobilization in thiazole synthesis and described important criteria for the appropriate annotation of ThiI in genomic data.
MATERIALS AND METHODS
Strains, media, and chemicals.
Minimal medium was NCE (26) supplemented with MgSO4 (1 mM), trace minerals (0.1×) (4), and glucose (11 mM). The final concentration of thiamine or thiazole was 100 nM. Rich medium was Difco nutrient broth (NB; 8 g/liter) with NaCl (5 g/liter). Solid medium contained 1.5% agar. Antibiotics were added at the following concentrations in rich and minimal media, respectively: chloramphenicol (Cm), 20 and 5 g/liter; ampicillin (Ap), 150 and 30 g/liter; tetracycline (Tc), 20 and 10 g/liter; kanamycin (Km), 50 and 12.5 g/liter. All chemicals were purchased from Sigma-Aldrich, St. Louis, MO. The strains used in this study were derivatives of S. enterica strain LT2, and their genotypes are listed in Table 1. All strains were generated for this study or were part of the laboratory collection.
Table 1.
Strain and plasmid list
| Strain or plasmid | Genotype (strain) or vector and protein encoded (plasmid) |
|---|---|
| Strains | |
| DM10000 | Wild type |
| DM269 | thiI887::Tn10d(Tc) |
| DM11537 | ΔthiF |
| DM12225 | phnU::Tn10d(Tc) |
| DM12260 | thiI887::Tn10d(Tc) pET-14b |
| DM11774 | thiI887::Tn10d(Tc) pDM1322 (pET-ThiI) |
| DM11544 | thiI887::Tn10d(Tc) pDM1324 (pET-ThiID189A) |
| DM11545 | thiI887::Tn10d(Tc) pDM1326 (pET-ThiIC207S) |
| DM11546 | thiI887::Tn10d(Tc) pDM1327 (pET-ThiIK321M) |
| DM11528 | thiI887::Tn10d(Tc) pDM1328 (pET-ThiIC344S) |
| DM11547 | thiI887::Tn10d(Tc) pDM1329 (pET-ThiIC456S) |
| DM13436 | thiI887::Tn10d(Tc) pSU19 |
| DM13437 | thiI887::Tn10d(Tc) pDM1319 (pSUthiI) |
| DM13438 | thiI887::Tn10d(Tc) pDM1320 (pSUthiI-Cter) |
| DM13439 | thiI887::Tn10d(Tc) pDM1321 (pSUthiI-CterC456S) |
| DM13477 | thiI887::Tn10d(Tc) pDM1331 (pSUthiI-CterR414H) |
| DM10959 | thiI1205::Tn10d(Tc) |
| DM13162 | ΔiscS |
| DM13304 | thiI1205::Tn10d(Tc) ΔiscS |
| DM13483 | thiI1205::Tn10d(Tc) gshA102::MudJ(Kn) |
| DM13484 | thiI1205::Tn10d(Tc) sufS::Cat |
| Plasmids | |
| pDM1322 (pET-ThiI) | pET-14b (Apr); ThiI |
| pDM1324 (pET-ThiID189A) | pET-14b (Apr); ThiID189A |
| pDM1326 (pET-ThiIC207S) | pET-14b (Apr); ThiIC207S |
| pDM1327 (pET-ThiIK321M) | pET-14b (Apr); ThiIK321M |
| pDM1328 (pET-ThiIC344S) | pET-14b (Apr); ThiIC344S |
| pDM1329 (pET-ThiIC456S) | pET-14b (Apr); ThiIC456S |
| pDM1318 (pSCthiI) | pSC-B (Apr Knr); ThiI |
| pDM1316 (pSCthiI-Cter) | pSC-B (Apr Knr); ThiI-Cter (residues 385-482) |
| pDM1317 (pSCthiI-CterC456S) | pSC-B (Apr Knr); ThiI-CterC456S (residues 385-482) |
| pDM1330 (pSCthiI-CterR414H) | pSC-B (Apr Knr); ThiI-CterR414H (residues 385-482) |
| pDM1319 (pSUthiI) | pSU19 (Cmr); ThiI |
| pDM1320 (pSUthiI-Cter) | pSU19 (Cmr); ThiI-Cter (residues 385-482) |
| pDM1321 (pSUthiI-CterC456S) | pSU19 (Cmr); ThiI-CterC456S (residues 385-482) |
| pDM1331 (pSCthiI-CterR414H) | pSU19 (Cmr); ThiI-CterR414H (residues 385-482) |
Mutant construction. (i) Local mutagenesis.
A P22 lysate (HT int201 [21, 23]) was generated on strain DM12255, which contains a Tn10d (Tc) insertion in phnU, linked to the thiI locus. This lysate was mutagenized by hydroxylamine (8) and used to transduce a wild-type strain (DM10000) to Tcr. Tcr transductants were screened for those that required exogenous thiamine when grown on glucose minimal medium (Thi−). The causative mutations were transduced into a new genetic background to confirm causation of the thiamine requirement, and the lesions in thiI were identified by sequence analysis. All DNA sequencing was performed at the University of Wisconsin—Madison Biotechnology Center.
(ii) Site-directed mutagenesis.
A plasmid (pDM1322) containing thiI ligated into the NdeI and BamHI sites of pET-14b (Novagen) was constructed. Relevant thiI mutants were generated by site-directed mutagenesis with the QuikChange XL site-directed mutagenesis kit according to the manufacturer's protocol (Agilent Technologies, Santa Clara, CA). Sequences of primers used are available from the corresponding author upon request. The presence of the relevant alleles in the resulting constructs was confirmed by sequence analysis.
Expression of the C-terminal domain of ThiI.
The C-terminal domain of ThiI (residues 385 to 482) was amplified by PCR using chromosomal DNA of wild-type S. enterica as a template and using primers SalthiI-R385for and SalthiI-endrev. The primers introduced an initiating methionine and restriction sites (BamHI and HindIII) to the amplified sequence. The amplified DNA was cloned as a blunt-end fragment into plasmid pSC-B-amp/kan (Stratagene) to give plasmid pDM1316. Plasmid pDM1316 was digested with BamHI and HindIII at the sites engineered in the primers, and the resulting 312-bp C-terminal thiI fragment was cloned into pSU19 to give plasmid pDM1320. Plasmids containing the C-terminal variants, C456S and R414H, were constructed by cloning a PCR amplification product from pDM1329 DNA or DM12302 chromosomal DNA, respectively. In each case, the constructs were confirmed by sequence analyses.
Nutritional analysis.
Nutritional requirements were determined by liquid growth analysis and in solid medium by using soft agar overlays. For liquid growth analysis, strains were grown overnight in NB (antibiotic was present for plasmid-containing strains). Cells were pelleted and resuspended in NaCl (0.85%). A 0.2-ml aliquot of this suspension was used to inoculate 5 ml minimal medium containing glucose. Cell growth was measured by monitoring absorbance at 650 nm (A650) while strains were incubated with shaking (200 rpm) at 37°C. Alternatively, 5 μl of cell suspension was used to inoculate 195 μl of the appropriate minimal medium in each well of a 96-well microtiter plate. Growth at 37°C with shaking at intensity level 2 was monitored using a microplate spectrophotometer (Bio-Tek Instruments). The specific growth rate was determined as μ = ln(X/X0)/T, where X is the A650 value during the exponentially linear portion of growth (routinely between A650 0.2 and 0.7) and T is time in hours. For soft agar overlays, cells were grown overnight in NB. An 0.2-ml aliquot of the overnight culture was used to seed 3.5 ml of 0.75% agar, which was overlaid on a minimal medium plate. Nutritional supplements were then spotted, and growth was assessed after 18 h of incubation at 37°C.
Bioinformatic analysis.
The TIGRfam TIGR00342 was downloaded from the Comprehensive Microbial Resource (CMR; http://cmr.jcvi.org), using the CMR Gene Attribute Download tool and the JCVI/CMR's OMNIOME.pep database. Using CMR, blastp (WU-BLAST version 2.0MP) was run on TIGR00342 using E. coli K-12 ThiI residues 404 to 482 as the query. Default parameters were used, except that the E-value cutoff was lowered to 0.001. Members of TIGR00342 containing the rhodanese domain were separated from those without a rhodanese domain, and duplicate species entries were removed. Bioinformatic analysis was performed by the Advanced Genome Analysis Resource facility at the University of Wisconsin Biotechnology Center.
RESULTS AND DISCUSSION
The functions of ThiI in 4-thiouridine biosynthesis and thiamine synthesis are genetically separable.
Five residues of ThiI have been shown to be important for synthesis of the 4-thiouridine modification of tRNA in vitro (Fig. 3) (16, 17, 19). These residues were independently changed in S. enterica ThiI, and plasmids carrying each of the variants (C344S, C456S, D189A, K321M, and C207S) or the wild-type gene were constructed. The resulting derivatives of plasmid pET-14b expressing ThiI proteins were transformed into a mutant strain lacking thiI, DM269 (thiI887::Tn10d). In the resulting strains, the plasmid-encoded protein was the only source of ThiI. Each strain was monitored for growth in the absence of thiamine to assess complementation of the thiI lesion. All variants except ThiIC456S restored growth of a thiI mutant strain in minimal medium. Some of these data are shown in Fig. 4. These results indicated that of the five ThiI residues required for 4-thiouridine synthesis, only residue Cys456 was required for thiazole synthesis.
Fig. 4.
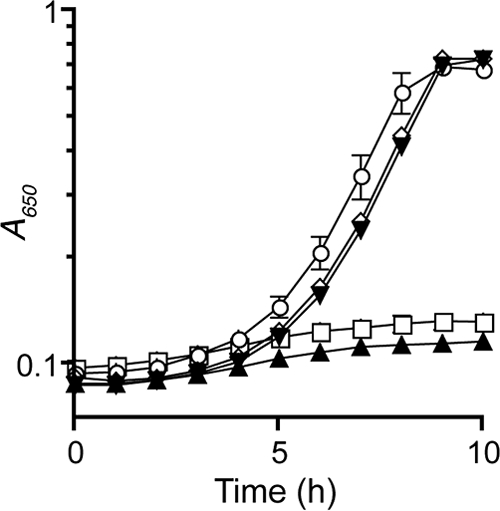
ThiI variants defective in 4-thiouridine biosynthesis complement a thiI mutant strain. Growth of strains at 37°C with shaking was monitored over time as absorbance at 650 nm. Strains were grown in minimal NCE medium with glucose and ampicillin; all strains grew with wild-type rate and yield if thiamine was added to the medium. Strains were DM269 (thiI887::Tn10d) with plasmids pET-14b (□), pET-ThiI (○), pET-ThiIC344S (⋇), pET-ThiID189A (▼), and pET-ThiIC456S (▲). Data shown are the averages and standard deviations of three independent cultures.
The ThiI variants that complemented a thiI mutant strain provided insights into the mechanism of ThiI in thiazole biosynthesis. ThiI residues Asp189, Lys321, and Cys207 lie in the adenylation domain. Substitution at any of these residues eliminated or decreased tRNA modification, consistent with the role of ThiI in adenylating the tRNA uridine in synthesis of the 4-thiouridine (17). In contrast, ThiI variants with substitutions at any of these residues were proficient for thiazole synthesis in vivo. These data allow the conclusion that the adenylation activity of ThiI is not required for thiazole synthesis. Similarly, residues Cys456 and Cys344 were directly involved in sulfur mobilization for the 4-thiouridine modification of tRNA in vitro (17, 19). In contrast, Cys456, but not Cys344, was required for thiazole synthesis in vivo. These data support the conclusion that sulfur mobilization in thiazole synthesis is mechanistically distinct from that in 4-thiouridine synthesis.
The C-terminal rhodanese domain of ThiI is sufficient for thiazole synthesis.
Local mutagenesis of the thiI gene generated 16 independent mutants that required thiamine for growth. The distribution of causative mutations was striking, with 14 of the 16 mutations generating a nonsense codon, two of which were isolated twice (Table 2 ). All of the lesions affected the rhodanese domain (residues 385 to 482). Of the two missense mutations isolated, one caused a substitution at residue Cys456 (C456Y) and the other resulted in an R414H substitution in the rhodanese domain.
Table 2.
ThiI variants that result in a thiamine requirement in vivo
| Strain | Allele | Base change(s) | Protein variant(s)a |
|---|---|---|---|
| DM12300 | thiI1209 | C1172T, C1414T | T391I, Q472* |
| DM12302 | thiI1210 | G1241A | R414H |
| DM12304 | thiI1211 | C1342T | Q448* |
| DM12355 | thiI1212 | C1255T | Q419* |
| DM12357 | thiI1213 | C1255T | Q419* |
| DM12359 | thiI1214 | G1356A | W452* |
| DM12361 | thiI1215 | C886T | Q296* |
| DM12422 | thiI1216 | C175T | Q59* |
| DM12424 | thiI1217 | G1397A | C456Y |
| DM12426 | thiI1218 | C1342T | Q448* |
| DM12428 | thiI1219 | C1393T | Q465* |
| DM12430 | thiI1220 | C1342T | Q448* |
| DM12432 | thiI1221 | C1342T | Q448* |
| DM12883 | thiI1222 | C886T | Q296* |
| DM12885 | thiI1223 | G1355A | W452* |
| DM12889 | thiI1224 | C886T | Q296* |
Asterisks indicate the generation of nonsense codons.
The above results raised the question of whether the rhodanese domain of ThiI was sufficient for synthesis of thiazole. An algorithm to detect linker regions in proteins defined such a region in ThiI from residues 381 to 390 (25). A plasmid expressing only the rhodanese domain of ThiI (residues 385 to 482) was constructed and used to test complementation of a thiI mutant. This construct was sufficient to restore thiamine-independent growth of a thiI mutant strain (Fig. 5). As shown in Fig. 5, complementation by the rhodanese domain was eliminated when residue Cys456 was changed to Ser, or when Arg414 was changed to His. Together, these results support the hypothesis that the rhodanese domain of ThiI was sufficient for thiazole synthesis. These data further demonstrate that ThiI mediated sulfur transfer in 4-thouridine synthesis via a different mechanism than that in the synthesis of thiazole.
Fig. 5.
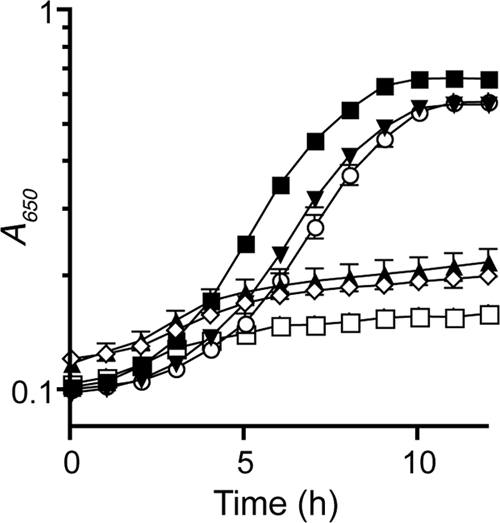
The rhodanese domain of ThiI is sufficient for thiamine-independent growth. Strains were grown in NCE medium supplemented with glucose. The following plasmids were introduced into DM269 (thiI): pSU19 (□), pSUThiI (○), pSUThiI-Cter (▾), pSUThiI-CterC456S (▲), and pSUThiI-CterR414H (⋇). Strain DM269 with pSU19 was also grown in medium supplemented with thiamine (■).
A possible role for the rhodanese domain in sulfur mobilization to thiazole.
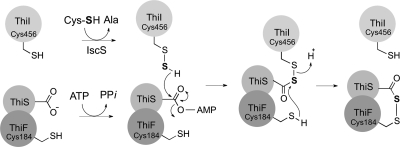
The accepted mechanistic model for the role of ThiI in 4-thiouridine synthesis is shown in Fig. 2. In this model, the THUMP domain binds the tRNA and the AANH domain activates the tRNA uridine by adenylation (11, 16, 32). The ThiI persulfide on Cys456 then displaces the adenosyl group on the uridine (19, 30). Finally, Cys344 acts as a nucleophile, forming a Cys456-Cys344 disulfide and releasing the product 4-thiouridine. The resulting disulfide in ThiI can then be reduced to begin the next reaction cycle (17, 27, 30). Previously, it was proposed that ThiI served a similar role in sulfur transfer in thiazole synthesis (17, 24). Figure 6 represents a mechanism for ThiI in sulfur mobilization to thiazole that is both consistent with previous studies and supported by the data described above. The mechanism depicted implies analogous chemistry in 4-thiouridine biosynthesis and thiazole biosynthesis. In both cases, the substrate (i.e., tRNA uridine or ThiS terminal glycine) is first activated by adenylation. In the case of 4-thiouridine biosynthesis, ThiI catalyzes the substrate adenylation reaction. In thiazole synthesis, ThiF has been shown to adenylate the terminal glycine residue of ThiS, generating an acyladenylate (24). The data showing that the adenylation activity of ThiI is not required for thiazole synthesis support this assignment. In both pathways, IscS transfers a sulfur atom to ThiI Cys456, forming a persulfide, a proposal consistent with the requirement for Cys456 in both pathways. This persulfide then attacks the substrate (uridine or ThiS) adenylate, displacing the adenosyl group and forming a disulfide between ThiI and the 4 position of uridine or an acyl persulfide with the C terminus of ThiS. In the final step, a second cysteine (ThiI Cys344 or ThiF Cys184, respectively) attacks the disulfide bond to release either the final product (4-thiouridine biosynthesis) or ThiI (in the case of thiazole biosynthesis). The last step is consistent with the in vivo data showing that an E. coli strain with a ThiFC184S variant is a thiamine auxotroph (31), which implicated the ThiFS acylpersulfide as either an intermediate or a final sulfur donor in thiazole synthesis (24, 31).
Fig. 6.
Proposed model for ThiI mechanism in thiazole biosynthesis. Sulfur mobilization is proposed to proceed as described in the text. The gray circle represents the ThiI rhodanese domain. The terminal glycine carboxyl group of ThiS is depicted.
Evaluation of ThiI annotations in genome sequences.
Search of the JCVI/CMR's OMNIOME.pep database for proteins with the TIGR00342 multidomain (THUMP and AANH) identified 207 proteins annotated as thiamine biosynthesis/4-thiouridine biosynthesis proteins. Of these, 58 (28%) contained a rhodanese domain, exemplified by the E. coli/S. enterica-type ThiI discussed herein. However, 159 (72%) had only the THUMP and AANH domains, exemplified by Bacillus subtilis. The THUMP domain, the AANH domain, and the rhodanese domain are implicated in the synthesis of 4-thiouridine (2, 16, 17). However, data in the above sections showed that the rhodanese domain alone is sufficient for ThiI to function in thiazole synthesis. The B. subtilis ThiI is among the 72% of the TIGR00342-containing proteins without a rhodanase domain annotated as thiamine biosynthetic proteins, and yet this protein is not required for thiazole synthesis (J. Perkins, personal communication; P. Dos Santos, personal communication). A role for the ThiI protein of B. subtilis in the synthesis of 4-thiouridine is presumed by the annotation but has not been reported.
S. enterica ThiI is a bifunctional enzyme that (i) binds and adenylates tRNA and (ii) mobilizes sulfur. Both functions are required for 4-thiouridine biosynthesis, while only sulfur mobilization is required for thiazole synthesis, as shown here. These findings suggest that the current annotation of the ThiI family of proteins should be revisited to better reflect their separable roles in the synthesis of 4-thiouridine and thiazole synthesis. Specifically, proteins annotated as ThiI based on containing only the AANH and THUMP domains are unlikely to be involved in thiazole synthesis and would be better annotated as 4-thiouridine sulfurtransferases. Based on the results here, only those proteins with the THUMP, AANH, and rhodanese domain are likely to be involved in both pathways. At this point, it is unclear how organisms with ThiI proteins with only two domains mobilize sulfur for 4-thiouridine synthesis in vivo. It has been suggested that an orphan rhodanese domain acts with the ThiI protein in B. subtilis and other organisms that contain a short ThiI protein (27).
Exogenous cysteine can restore thiamine-independent growth of a thiI mutant strain.
The result that the rhodanese domain of ThiI was sufficient for thiazole synthesis, coupled with the fact that not all thiamine-producing organisms have a rhodanese domain associated with a ThiI enzyme, suggested that another paradigm(s) for sulfur mobilization to thiazole synthesis might exist. For instance, the IscS persulfide could serve as the direct sulfur donor in some metabolic networks, or organisms may employ a different mechanism altogether. It seemed plausible that both the redox environment of the cell and the intracellular concentration of cysteine or other free thiols could influence these possibilities, perhaps in organismal or environmentally specific ways. This idea is supported by the observation that in vitro cysteine and IscS could serve as the sole sulfur source to ThiS (12). Further, sulfide can provide the source of sulfur in vitro without ThiI, ThiF, or ThiS (12, 20) (N. C. Martinez-Gomez, unpublished data), suggesting that thiazole synthase, ThiG, could utilize a variety of sulfides.
In vivo, a thiI mutant requires thiazole, and efforts to isolate suppressor mutations by spontaneous or chemical mutagenesis were unsuccessful. As might be predicted from the in vitro results, sulfide provided in the form of Li2S or Na2S restored growth of a thiI mutant strain in the absence of thiamine, although this result was not considered physiologically relevant (data not shown). Unexpectedly, nutrient studies found that addition of cysteine (0.2 mM) to the medium allowed growth of a thiI mutant strain in the absence of thiazole (Table 3). Restoration of thiamine-independent growth was specific to the addition of cysteine; no other physiologically relevant sulfur source or reducing compound tested restored growth to a thiI mutant strain (data not shown). Significantly, the ability of cysteine to allow growth did not require IscS. Neither did this effect of cysteine require SufS (a cysteine desulfurase) or GshA (glutathione biosynthetic enzyme). These conclusions were based on the data in Table 3, which showed that the thiazole requirement of each double mutant strain (thiI iscS, thiI gshA, and thiI sufS) was satisfied by exogenous cysteine. The slightly reduced growth rate of the double mutants compared to the thiI mutant is consistent with the deleterious effects of these mutations on other aspects of metabolism. Significantly, the addition of cysteine fully satisfied the thiazole requirement as reflected by the similar growth rates allowed by cysteine and thiazole. Finally, cysteine did not allow growth of a thiF mutant in the absence of thiazole (data not shown), indicating that cysteine did not serve as a direct sulfur source to ThiG.
Table 3.
Cysteine allows growth of a thiI mutant in minimal mediuma
| Strain | Relevant genotype | Specific growth rate |
Final cell yield |
||||
|---|---|---|---|---|---|---|---|
| Min | Cys | THZ | Min | Cys | THZ | ||
| DM10959 | thiI | 0.35 ± 0.01 | 0.74 ± 0.03 | 0.78 ± 0.04 | 0.45 ± 0.09 | 1.45 ± 0.03 | 1.56 ± 0.00 |
| DM13304 | thiI iscS | 0.12 ± 0.01 | 0.31 ± 0.01 | 0.36 ± 0.01 | 0.36 ± 0.03 | 0.98 ± 0.03 | 1.45 ± 0.02 |
| DM13483 | thiI gshA | 0.27 ± 0.01 | 0.50 ± 0.01 | 0.53 ± 0.01 | 0.39 ± 0.02 | 1.44 ± 0.00 | 1.53 ± 0.04 |
| DM13484 | thiI sufS | 0.30 ± 0.01 | 0.69 ± 0.07 | 0.69 ± 0.01 | 0.43 ± 0.01 | 1.45 ± 0.04 | 1.56 ± 0.02 |
Strains were grown in NCE medium supplemented with glucose (11 mM) and the indicated additions. Min, minimal medium. Growth rate is reported as μ [μ = ln(X/X0)/T], and the final cell yield is A650 after 25 h of growth. Data shown are the averages and standard deviations of three independent cultures.
Together, these data suggest that an alternative mechanism for sulfur mobilization to ThiS existed in the cell that depends on exogenous cysteine. The finding that cysteine satisfied the thiazole requirement of iscS, gshA, and sufS derivatives of a thiI mutant indicates that the alternative mechanism of sulfur mobilization does not depend on either of the dominant cysteine desulfurases in the cell or the primary thiol reductant, glutathione. It is possible that an additional cysteine desulfurase, possibly via an orphan rhodanese in the cell, can mediate sulfur transfer to ThiS. The requirement for exogenous cysteine might indicate a need to accumulate an appropriate persulfide. Such a scenario may reflect and/or mimic a paradigm for thiazole synthesis in organisms that do not require ThiI for thiazole biosynthesis.
Conclusions.
In S. enterica and E. coli, ThiI is involved in the synthesis of both thiazole and 4-thiouridine (15, 28). The data here showed that the THUMP and AANH domains of this protein, which are the basis for annotation of proteins as ThiI in genomes, are not required for thiazole synthesis. This work emphasizes that caveats can exist when annotation of gene function is propagated from a single experimental organism. Finding an additional cysteine-dependent mechanism of sulfur mobilization to thiazole in vivo suggests that there may be diverse processes for sulfur mobilization to thiazole. Further characterization of this mechanism could improve understanding of the relationship between the thiol environment in the cell and mobilization of sulfur to thiazole synthesis.
ACKNOWLEDGMENTS
We acknowledge undergraduates James Vasta and Patrick Borchert for isolating several of the thiI mutants.
The National Institutes of Health grant GM47296 and the 21st Century Scientists Scholars program from the J. M. McDonnell Foundation to D.M.D. supported this work. L.D.P. was supported by NSF through Graduate Research Fellowship grant DGE-0718123.
Footnotes
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Ajjawi I., Rodriguez Milla M. A., Cushman J., Shintani D. K. 2007. Thiamin pyrophosphokinase is required for thiamin cofactor activation in Arabidopsis. Plant Mol. Biol. 65:151–162 [DOI] [PubMed] [Google Scholar]
- 2. Aravind L., Koonin E. V. 2001. THUMP—a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 26:215–217 [DOI] [PubMed] [Google Scholar]
- 3. Backstrom A. D., Austin R., McMordie S., Begley T. P. 1995. Biosynthesis of thiamin. I. The function of the thiE gene product. J. Am. Chem. Soc. 117:2351–2352 [Google Scholar]
- 4. Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Begley T. P., et al. 1999. Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171:293–300 [DOI] [PubMed] [Google Scholar]
- 6. Bordo D., Bork P. 2002. The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep. 3:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chatterjee A., et al. 2008. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat. Chem. Biol. 4:758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hong J. S., Ames B. N. 1971. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc. Natl. Acad. Sci. U. S. A. 68:3158–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jordan F. 2003. Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat. Prod. Rep. 20:184–201 [DOI] [PubMed] [Google Scholar]
- 10. Jurgenson C. T., Begley T. P., Ealick S. E. 2009. The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78:569–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kambampati R., Lauhon C. T. 2000. Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J. Biol. Chem. 275:10727–10730 [DOI] [PubMed] [Google Scholar]
- 12. Kriek M., et al. 2007. Thiazole synthase from Escherichia coli: an investigation of the substrates and purified proteins required for activity in vitro. J. Biol. Chem. 282:17413–17423 [DOI] [PubMed] [Google Scholar]
- 13. Martinez-Gomez N. C., Downs D. M. 2008. ThiC is an [Fe-S] cluster protein that requires AdoMet to generate the 4-amino-5-hydroxymethyl-2-methylpyrimidine moiety in thiamin synthesis. Biochemistry 47:9054–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCulloch K. M., Kinsland C., Begley T. P., Ealick S. E. 2008. Structural studies of thiamin monophosphate kinase in complex with substrates and products. Biochemistry 47:3810–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mueller E. G., Buck C. J., Palenchar P. M., Barnhart L. E., Paulson J. L. 1998. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 26:2606–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mueller E. G., Palenchar P. M. 1999. Using genomic information to investigate the function of ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis. Protein Sci. 8:2424–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mueller E. G., Palenchar P. M., Buck C. J. 2001. The role of the cysteine residues of ThiI in the generation of 4-thiouridine in tRNA. J. Biol. Chem. 276:33588–33595 [DOI] [PubMed] [Google Scholar]
- 18. Nosaka K. 2006. Recent progress in understanding thiamin biosynthesis and its genetic regulation in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 72:30–40 [DOI] [PubMed] [Google Scholar]
- 19. Palenchar P. M., Buck C. J., Cheng H., Larson T. J., Mueller E. G. 2000. Evidence that ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis, may be a sulfurtransferase that proceeds through a persulfide intermediate. J. Biol. Chem. 275:8283–8286 [DOI] [PubMed] [Google Scholar]
- 20. Park J. H., et al. 2003. Biosynthesis of the thiazole moiety of thiamin pyrophosphate (vitamin B1). Biochemistry 42:12430–12438 [DOI] [PubMed] [Google Scholar]
- 21. Roberts G. P. 1978. Isolation and characterization of informational suppressors in Salmonella typhimurium. Ph.D. thesis. University of California, Berkeley, CA [Google Scholar]
- 22. Roje S. 2007. Vitamin B biosynthesis in plants. Phytochemistry 68:1904–1921 [DOI] [PubMed] [Google Scholar]
- 23. Schmieger H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75–88 [DOI] [PubMed] [Google Scholar]
- 24. Taylor S. V., et al. 1998. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem. 273:16555–16560 [DOI] [PubMed] [Google Scholar]
- 25. Udwary D. W., Merski M., Townsend C. A. 2002. A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a type I polyketide synthase. J. Mol. Biol. 323:585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vogel H. J., Bonner D. M. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97–106 [PubMed] [Google Scholar]
- 27. Waterman D. G., Ortiz-Lombardia M., Fogg M. J., Koonin E. V., Antson A. A. 2006. Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 356:97–110 [DOI] [PubMed] [Google Scholar]
- 28. Webb E., Claas K., Downs D. M. 1997. Characterization of thiI, a new gene involved in thiazole biosynthesis in Salmonella typhimurium. J. Bacteriol. 179:4399–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Webb E., Downs D. 1997. Characterization of thiL, encoding thiamin-monophosphate kinase, in Salmonella typhimurium. J. Biol. Chem. 272:15702–15707 [DOI] [PubMed] [Google Scholar]
- 30. Wright C. M., Palenchar P. M., Mueller E. G. 2002. A paradigm for biological sulfur transfers via persulfide groups: a persulfide-disulfide-thiol cycle in 4-thiouridine biosynthesis. Chem. Commun. (Camb.) 2002:2708–2709 [DOI] [PubMed] [Google Scholar]
- 31. Xi J., Ge Y., Kinsland C., McLafferty F. W., Begley T. P. 2001. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: identification of an acyldisulfide-linked protein-protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proc. Natl. Acad. Sci. U. S. A. 98:8513–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. You D., Xu T., Yao F., Zhou X., Deng Z. 2008. Direct evidence that ThiI is an ATP pyrophosphatase for the adenylation of uridine in 4-thiouridine biosynthesis. Chembiochem 9:1879–1882 [DOI] [PubMed] [Google Scholar]