Abstract
Myxobacteria are predatory and are prolific producers of secondary metabolites. Here, we tested a hypothesized role that secondary metabolite antibiotics function as weapons in predation. To test this, a Myxococcus xanthus Δta1 mutant, blocked in antibiotic TA (myxovirescin) production, was constructed. This TA− mutant was defective in producing a zone of inhibition (ZOI) against Escherichia coli. This shows that TA is the major M. xanthus-diffusible antibacterial agent against E. coli. Correspondingly, the TA− mutant was defective in E. coli killing. Separately, an engineered E. coli strain resistant to TA was shown to be resistant toward predation. Exogenous addition of spectinomycin, a bacteriostatic antibiotic, rescued the predation defect of the TA− mutant. In contrast, against Micrococcus luteus the TA− mutant exhibited no defect in ZOI or killing. Thus, TA plays a selective role on prey species. To extend these studies to other myxobacteria, the role of antibiotic corallopyronin production in predation was tested and also found to be required for Corallococcus coralloides killing on E. coli. Next, a role of TA production in myxobacterial fitness was assessed by measuring swarm expansion. Here, the TA− mutant had a specific swarm rate reduction on prey lawns, and thus reduced fitness, compared to an isogenic TA+ strain. Based on these observations, we conclude that myxobacterial antibiotic production can function as a predatory weapon. To our knowledge, this is the first report to directly show a link between secondary metabolite production and predation.
INTRODUCTION
Microorganisms produce a wide range of structurally diverse small molecules called secondary metabolites. These molecules, which by definition are not part of the primary metabolism, have great therapeutic value. For example, most clinically used antibiotic classes are derived from microbial secondary metabolites (27, 44). Although these natural products are important for human health, their physiological functions to the producer organisms are mostly unknown. In the case of antibiotics, it has long been assumed they might play a fitness advantage to producer organisms by killing or inhibiting the growth of competitors. However, this notion has received renewed criticisms (9, 48). A primary issue is that the concentration of antibiotics that exhibits antimicrobial activity is typically high, e.g., in the micromolar range. Thus, how microbes could produce an effective antibiotic concentration for killing in their natural habitats, such as soil, is unclear and unsubstantiated. Recently, it has been shown that antibiotics at much lower and sublethal concentrations can significantly alter microbial gene expression (10). These and other observations have highlighted the role of secondary metabolite antibiotics as signaling molecules (39). Elucidating the biological role of secondary metabolites to the producers is a fundamental concern to understand their evolution and regulation, to develop informed strategies to optimize/engineer strains, and to find new isolates that make therapeutically useful products (8, 12, 14).
Myxobacteria have interesting properties, including their rich production of secondary metabolites and predatory behaviors (4, 45). Predation involves their ability to move or glide to establish prey contact. Killing by myxobacteria appears to involve both secreted diffusible factors and direct cell-cell contact (28, 30, 37). Killed and lysed preys are digested into small molecules for consumption. Although myxobacterial predation was first described over 6 decades ago, little is known about its molecular mechanism (1). Interestingly, of the known myxobacterial secondary metabolites, about 20% have antibiotic activity (40). Thus, a possible logical link between antibiotic production and a role in microbial predation exists (37). In contrast, actinomycetes, prolific producers of secondary metabolites, including antibiotics, are not known to consume other microbes and are thus are not considered predatory. The biological roles for these thousands of actinomycetes metabolites are mostly unknown and bewildering.
Myxobacteria are ubiquitous, soil-dwelling, Gram-negative bacteria. Upon starvation, thousands of vegetative cells coalesce to build fruiting bodies in which cells differentiate into spores. When vegetative myxobacteria encounter prey they are postulated to neutralize them by the secretion of antibiotics and hydrolytic enzymes (34, 37). Extracellular digestion of prey into small molecules allows nutrient uptake by myxobacteria. In support of this, the myxobacterial growth rate increases as cell density increases when myxobacteria are grown on macromolecular substrates, e.g., proteins (35). This observation led to the notion that myxobacteria feed as microbial “wolfpacks”; i.e., higher cell density increases extracellular hydrolytic enzyme and antibiotic concentrations, thus allowing more efficient prey killing and digestion. Although this hypothesis seems reasonable, it is also striking that a single myxobacterium can penetrate a prey microcolony and rapidly kill, digest, and consume it (30). This form of predation appears to depend on cell-cell contact. Thus, myxobacteria are effective predators in packs or as lone cells.
The best-studied and genetically trackable myxobacterium species is Myxococcus xanthus. The genome sequence of the common laboratory strain (DK1622) reveals 18 gene clusters for secondary metabolite synthesis (17, 46). Currently, 5 of the potential 18 compounds have been identified. One of these compounds, TA, has antibacterial activity. Rosenberg and colleagues first discovered TA activity in an environmental isolate from Tel Aviv, Israel (36). Independently, Reichenbach's group discovered and solved the structure of myxovirescin, which turned out to be identical to that of TA (16, 40). In DK1622, antibiotic TA and numerous hydrolytic enzymes are thus candidate weapons for predation (17). However, to date little is known about host factors involved in predation (4, 33). Here, we investigated the role that antibiotic production might play in predation. Our underlying hypothesis was that myxobacteria produce antibiotics as small-molecule weapons to penetrate prey cells to halt metabolism or kill them for subsequent enzymatic digestion. Lytic antibiotics may also facilitate digestion. We further postulate that in the absence of antibiotics, live and metabolically active prey might fend off predation by repairing cellular damage caused by impermeable macromolecular enzymes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
M. xanthus strain DK1622 (wild type [WT]) and Corallococcus coralloides c127 (WT) were used in this study as predators (22, 43). Micrococcus luteus (University of Wyoming microbiology collection), Escherichia coli MG1655 (WT; obtained from ATCC), and a permeable strain of E. coli LBB928 (D21f2; rfa-1 rfa-31 tolC::Tn10) (15) with or without plasmids were used as prey species. E. coli Top10 and DH5α were used for routine cloning. Plasmid p-lspA (pCA24N::lspA) was obtained from the ASKA library (25). Plasmid pRL706 (p-rpoB+) and pRL706-1275F (p-rpoB-V1275F) were described previously (31).
M. xanthus was routinely grown at 33°C in CTT (1% Casitone, 10 mM Tris-HCl [pH 7.6], 8 mM MgSO4, 1 mM KH2PO4). C. coralloides was grown at 33°C in PT medium (0.4% Casitone, 0.4% yeast extract, 0.2% MgSO4·7H2O, 0.1% CaCl2·2H2O). E. coli and Micrococcus luteus were routinely grown in Luria-Bertani medium (LB). Antibiotic concentrations used were kanamycin (Kan) at 40 μg/ml, ampicillin (Amp) at 100 μg/ml, and chloramphenicol (Cm) at 25 μg/ml. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added at 1 mM for cells carrying plasmid p-rpoB+ and p-rpoB-V1275F and 20 μM for cells carrying plasmid p-lspA.
Strain construction.
A markerless deletion of the ta1 gene (mxan_3935), which encodes a megasynthetase for TA biosynthesis, was constructed in DK1622. The mutation was made by a gene replacement method with the pBJ113 vector that contains a positive-negative (Kanr-galK) selection cassette (23). To construct this deletion, an upstream 1,234-bp fragment and a downstream 1,210-bp fragment were PCR amplified and directly cloned into the pCR2.1 vector following the TOPO TA cloning instructions (Invitrogen). These fragments were then ligated together and then cloned into pBJ113 at the KpnI and XbaI restriction sites, creating pXW12. The TA deletion primers cln2-F (5′-GAGGTCCACGAGGTTCCAT-3′), cln2-R (5′-AGTGCCCCTCGTAGCTCA-3′), Mx3934-cln-FW (5′-TATGTCTCGGGTGCCATGTA-3′), and Mx3934-cln-RS (5′-GGTTGCTGAAGGACTCCAAT-3′) were used to amplify the upstream and downstream regions, respectively. pXW12 was electroporated into DK1622 and homologously recombined into the genome by selecting Kanr (24). Subsequent plasmid resolution and loss were counterselected for on 2% galactose CTT agar (1.5%) plates. This markerless deletion removed 25.5 kbp of the ta1 gene sequence and was verified by PCR with primers flanking the deleted region, generating strain DW1034 (Δta1).
Zone of inhibition (ZOI) assay.
Overnight myxobacterium cultures were collected by centrifugation and resuspended in TPM buffer (10 mM Tris-HCl [pH 7.6], 1 mM KH2PO4 [pH 7.6], 8 mM MgSO4) to a calculated Klett unit measurement of 100 (∼3 × 108 CFU/ml). Then 5 μl of cells were spotted on 1/2 CTT (0.5% Casitone) or PT 1.5% agar plates. Plates were incubated at 33°C for 48 h prior to indicator strain overlay (100 μl of cells at an optical density at 600 nm [OD600] of 1 mixed into 3 ml of molten 1/2 CTT, 0.7% agar). Plate overlays were then incubated overnight at 33°C before observation.
Killing assay.
Assay conditions were optimized so that M. xanthus was prespotted on agar plates 2 h prior to prey spotting. These sequential steps helped to minimize rapid prey lysis that occurs when both strains were premixed prior to spotting (data not shown). Thus, 10 μl of M. xanthus cells at 100 Klett units were spotted on the described plates (see figure legends) and preincubated at 33°C. Subsequently, 5 μl of prey cells (OD600, 10) were carefully (i.e., avoiding splashing) pipetted directly on top of the nascent M. xanthus swarm. Plates were incubated at 33°C, and at various times cell spots were scraped, washed, and centrifuged for 2 min; the pellets were then resuspended in 0.5 ml TPM. Cells were then serially diluted and grown on LB agar plates to count surviving prey cells. We note that myxobacterial growth was blocked by the high salt concentration in LB. C. coralloides predation was done in a similar manner except that PT agar plates were supplemented with 1 mM IPTG. To define experimental conditions and reproducibility and to ensure that phenotypes were attributed to engineered mutations, all assays were repeated at least twice and up to six times.
MIC.
To determine MIC values, 2-fold serial broth dilutions of spectinomycin were done in 96-well microtiter dishes. MG1655 (LB) and DK1622 (CTT) were tested over dilution ranges from 0.25 to 32 μg/ml and 4 to 512 μg/ml, respectively. The inoculums contained 5 ×105 CFU/ml (160 μl of culture in a final volume of 180 μl). Both spectinomycin-containing and strain-only controls were included. Microtiter dishes were scored after incubations at 33°C for 18 h for E. coli and 2.5 days for M. xanthus. The MIC was scored as the lowest concentration of spectinomycin that resulted in no visual detection of growth.
Swarm assay.
M. xanthus swarms were monitored on TPM or 1/2 CTT (1.5% agar) plates with or without prey lawns. E. coli lawns were made by top spreading 100 μl of cells (OD600 = 10 or 60) on TPM plates or 100 μl (OD600 = 1) on 1/2 CTT plates. Since E. coli cannot grow on TPM agar, heavier or more opaque lawn inoculums were used to allow easy visual detection of swarm expansion. Then 5 μl of 100-Klett unit M. xanthus cells were spotted, swarm diameters were measured daily, and the swarm areas were calculated [π × 1/2(l × w)]. In described experiments (see the figure legends), 20 μM IPTG was added to E. coli cultures 1 h prior to harvest and IPTG was also included in the agar plates. To ensure experimental accuracy and reproducibility, measurements were done in triplicate and were repeated at least twice.
RESULTS
Antibiotic TA plays a selective and conditional role in prey killing.
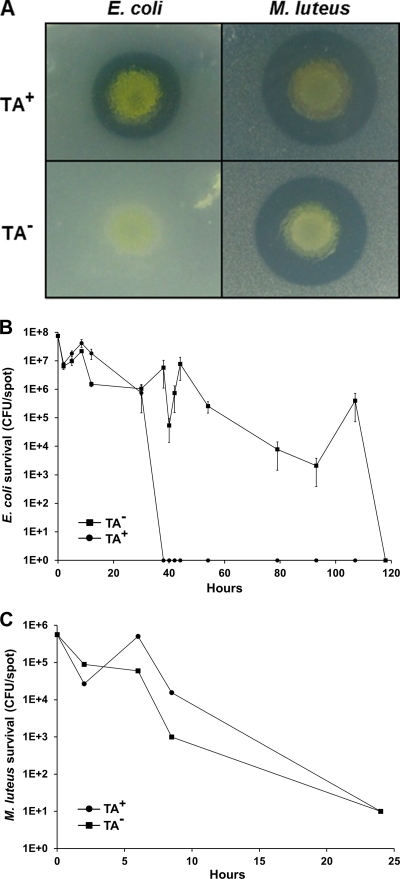
Myxobacteria are known to produce a variety of antibiotics. The best-characterized antibiotic produced by the lab strain DK1622 is TA, and we thus sought to test its role in predation (41). Consequently, a TA− mutant was made and the production of diffusible antibacterial activity was compared to that of the isogenic parent (DK1622). For these studies, E. coli and Micrococcus luteus were chosen because they are susceptible to predation (19), and they are found in the natural habitats of M. xanthus (soil and/or dung) (40). A ZOI assay found that a TA− mutant (DW1034) produced no clearing halo against E. coli compared to that of the parental strain (Fig. 1A). Therefore, under these conditions TA represents the major diffusible factor that blocks E. coli growth. Moreover, since TA is bactericidal, cells are likely lysed. In contrast, the clearing halo of the TA− mutant against Micrococcus luteus was nearly identical to that of the parental strain (Fig. 1A), showing that TA is not the primary antibacterial agent against this Gram-positive bacterium. Therefore, TA exhibits select activity toward bacterial species (16).
Fig. 1.
M. xanthus produces diffusible factors that inhibit bacterial growth and are involved in prey killing. (A) ZOI comparison of isogenic DK1622 (TA+) to DW1034 (TA−; Δta1) on 1/2 CTT agar. Unlike DK1622, DW1034 produced no clearing halo against E. coli (MG1655). Against Micrococcus luteus the ZOIs were similar between M. xanthus strains. (B) E. coli (MG1655) kill kinetics by DK1622 (TA+) and DW1034 (TA−) predators on 1/2 CTT agar. At various times, prey survival was measured in triplicate and standard errors were plotted. (C) Similar to panel B, except that Micrococcus luteus was substituted as prey.
Next we sought to test the role of TA production in prey killing. Assays were designed to quantify killing in a temporally relevant manner. As prey killing is central to predation, we view this assay as a reasonable approximation of predation. In our first experiments, killing was monitored under conditions where prey cells were growing and thus had the metabolic capacity to respond and repair cellular damage caused by predation. 1/2 CTT medium was chosen, and since M. xanthus grows relatively slowly (∼5-h doubling time), the fast-growing prey has the capacity to outgrow the predator (e.g., E. coli by ∼10-fold). In these assays we found that a TA− strain had a major defect in E. coli killing compared to that of the TA+ parent (Fig. 1B). At times a 7-log difference in prey killing was observed. Over prolonged incubations (5 days) the E. coli prey eventually succumbed to complete killing by the TA− strain. Delayed killing may result from the eventual accumulation of alternative lytic agents at concentrations sufficiently high to substitute for the missing TA activity (34, 37). These assays were repeated (six times) and were reproducible. In particular, we note that the sudden and dramatic decline in the E. coli population occurred in a predictable manner at ∼36 h (Fig. 1B, TA+ strain). We interpret this result to mean that by ∼36 h the TA+ M. xanthus population has produced a critical concentration of TA (in the context of other, unknown factors) to kill the entire E. coli population. We also note that the recovery in the E. coli population around 8 h (Fig. 1B) was reproducible. Thus, between 2 and 8 to 10 h E. coli growth must outcompete predation by an ∼10-fold margin. We further note that the differences in kill kinetics were not the result of idiosyncratic strain differences, as we constructed another strain with a different ta1 (insertion) mutation and found a similar defect in killing (data not shown). Lastly, the killing dynamics of the TA− mutant were more variable than those of the TA+ strain. This variability is reflective in an irregularly shaped TA− kill curve and large standard errors (Fig. 1B, plotted on log scale).
In contrast to the case with E. coli, the kill kinetics of the TA− mutant against Micrococcus luteus was nearly identical to that of the parental DK1622 strain (Fig. 1C). These results showed a clear correlation between the production of a diffusible antibacterial factor and prey killing (Fig. 1). That is, against E. coli the TA− strain was defective in producing a ZOI and killing, while against Micrococcus luteus these phenotypes were nearly identical to those of DK1622. These results suggest that TA plays an important role in E. coli killing, including lysis, and consequently predation. In contrast, TA plays no discernible role in Micrococcus luteus killing. Therefore, other unknown cellular factors must be involved in Micrococcus luteus killing.
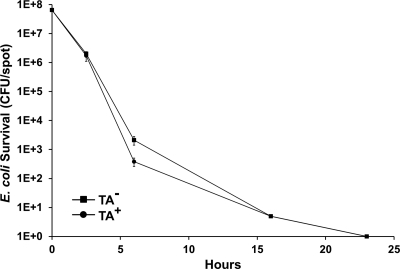
TA is bactericidal, and, as such, its killing capacity is likely dependent on the metabolic activity of the prey cells (49). Consequently, we tested whether TA production was involved in killing when prey cells were metabolically quiescent. In these experiments, killing was measured on TPM agar that lacks added nutrients. When E. coli was spotted alone on TPM agar, the cells did not grow but remained fully viable for at least 5 days (data not shown). As Fig. 2 shows, no significant difference in E. coli killing was found on TPM agar between the TA+ and TA− strains. We also note that the killing dynamics of E. coli were significantly shorter and less variable on TPM than 1/2 CTT agar (compare Fig. 1B to Fig. 2). These differences are likely explained by the lack of E. coli growth on TPM agar, thus causing total CFU to decrease faster and perhaps at a more constant rate. We conclude that TA production was important for killing metabolically active and growing E. coli but not against static cells. Thus, TA plays a conditional role in killing. We also note that these results again show that M. xanthus produces other, unknown agents that kill E. coli (Fig. 1B and Fig. 2).
Fig. 2.
TA production was not required for E. coli killing under nutrient free conditions. Predation assays were done in triplicate, and standard errors were plotted. When incubated alone, MG1655 shows no viability loss (data not shown). When incubated with isogenic TA+ (DK1622) or TA− (DW1034) strains, the E. coli (MG1655) kill kinetics were nearly identical.
Addition of exogenous antibiotic can substitute for missing TA activity.
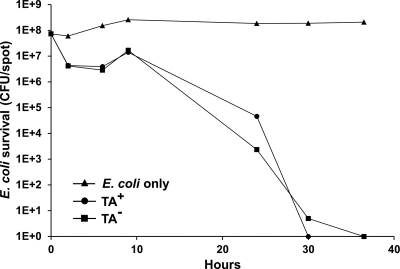
We postulated that antibiotic production serves as a small-molecule weapon to penetrate and kill or neutralize prey metabolism to allow subsequent macromolecular enzymatic digestion. We thus tested whether the killing deficiency of a TA− mutant could be corrected by the addition of an exogenous antibiotic. We selected spectinomycin as such an antibiotic because it is bacteriostatic and has poor activity against M. xanthus. Broth dilution tests found that the MIC of spectinomycin against MG1655 was 32 μg/ml, while against DK1622 it was >512 μg/ml. In a preliminary experiment we added spectinomycin to 1/2 CTT agar plates at concentrations of 4, 8, 16 or 32 μg/ml and conducted E. coli killing assays against TA+ and TA− M. xanthus strains. We found that all four concentrations of spectinomycin significantly blocked E. coli growth and rescued the killing defect of the TA− strain (data not shown). We then repeated this experiment using spectinomycin at 8 μg/ml. As shown, spectinomycin blocks prey growth but not viability (Fig. 3). As found in the prior experiment, when spectinomycin was present in 1/2 CTT agar, no significant difference was seen in E. coli killing kinetics between the TA+ and TA− strains (Fig. 3). In addition, the E. coli kill kinetics was similar, though faster, than that found for the TA+ strain in the absence of spectinomycin (compare Fig. 1B to Fig. 3). This modest increase in kill kinetics may reflect that functional levels of spectinomycin are present at the beginning of the experiment, while in the absence of spectinomycin M. xanthus needs time to produce sufficient concentrations of TA for killing. In sum, this result shows that exogenous addition of a bacteriostatic antibiotic can rescue or substitute for the killing defect of a TA− strain.
Fig. 3.
Exogenous antibiotic addition corrects predation defect of TA− mutant. Kill assays on E. coli MG1655 were done as described in Fig. 1, except that spectinomycin (8 μg/ml) was added to 1/2 CTT agar. Spectinomycin-only treatment was not bactericidal toward E. coli. In the presence of spectinomycin, the TA− (DW1034) kill kinetics were similar to those of TA+ (DK1622) cells.
Antibiotic resistance toward TA confers resistance toward predation.
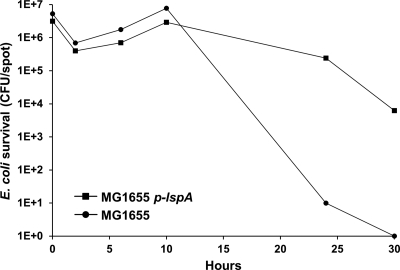
The above finding reveals a clear correlation between antibiotic production and predation. To further investigate this relationship we tested whether an E. coli TAr strain was resistant toward prey killing. For this experiment we used an E. coli strain that contains an expression plasmid for LspA (type II signal peptidase), the molecular target for TA, and thus confers resistance against TA when overexpressed (Y. Xiao and D. Wall, unpublished data). Killing assays were conducted with this strain under LspA inducing conditions on 1/2 CTT agar, and the results were compared to those for the parental E. coli strain. Fig. 4 indeed shows that LspA overexpression confers resistance toward DK1622 (TA+) predation. At times, a >4-log increase in resistance was observed. Therefore, an engineered prey strain that was TA resistant resulted in resistance to killing and thus predation by a M. xanthus TA producer.
Fig. 4.
Prey resistance toward antibiotic TA confers predation resistance. Kill kinetics against E. coli (MG1655) was compared to an isogenic TAr strain (MG1655 + p-lspA). Kill assays with DK1622 (TA+) were done as described in Fig. 1, except 20 μM IPTG was included in the 1/2 CTT agar.
Corallopyronin production by Corallococcus plays a role in predation.
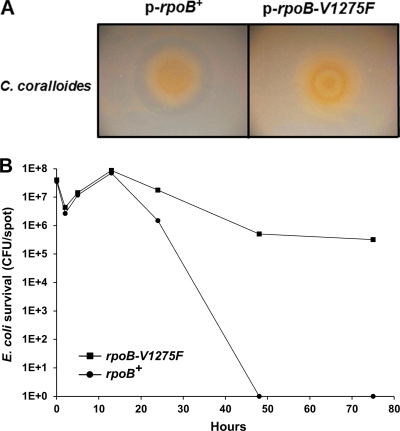
We next sought to test whether a different antibiotic produced by another species of myxobacteria might also be involved in predation. For these studies we selected Corallococcus coralloides as a myxobacterium because it produces the antibiotic corallopyronin (Cor), which targets bacterial RNA polymerase (22). Recent studies have shown Cor binds to the “switch region” of RNA polymerase, which is a binding site distinct from those used by the rifamycin antibiotics (2, 31). To test the role that Cor might play in predation, we obtained an E. coli rpoB mutation that confers Cor resistance (31). This RpoB mutant has a Val-to-Phe substitution at amino acid position 1275 (rpoB-V1275F) and showed a >8-fold increase in the MIC toward Cor (31). First, in a ZOI assay against C. coralloides, we found that an E. coli strain overexpressing the rpoB+ gene exhibited a clearing halo, while in contrast the isogenic rpoB-V1275F-overexpressing strain did not produce a halo (Fig. 5 A). We conclude that Cor is the major diffusible antibacterial factor produced by C. coralloides against this E. coli strain and that the rpoB-V1275F allele indeed confers Corr. Next, a killing assay was conducted to test whether an E. coli strain harboring rpoB-V1275F was resistant toward predation. As indicated in Fig. 5B, E. coli overexpressing rpoB-V1275F was resistant to C. coralloides killing compared to the isogenic rpoB+ overexpressing strain. At times, an ∼6-log difference in killing was observed. Thus, similar to our TA findings, these results indicate that antibiotic Cor is a weapon that can be used by C. coralloides on E. coli prey.
Fig. 5.
Corallococcus coralloides predation involves antibiotic corallopyronin production. (A) C. coralloides produced a ZOI against a control E. coli strain (LBB928 harboring p-rpoB+). In contrast, against an isogenic E. coli strain harboring a corallopyronin resistance allele, p-rpoB-V1275F, no clearing halo was observed. (B) The same strains were used in a kill assay. As shown, LBB928 carrying p-rpoB-V1275F was resistant to C. coralloides killing compared to a LBB928 p-rpoB+ strain. All assays were done on PT agar plates with 1 mM IPTG.
TA activity contributes toward predator fitness.
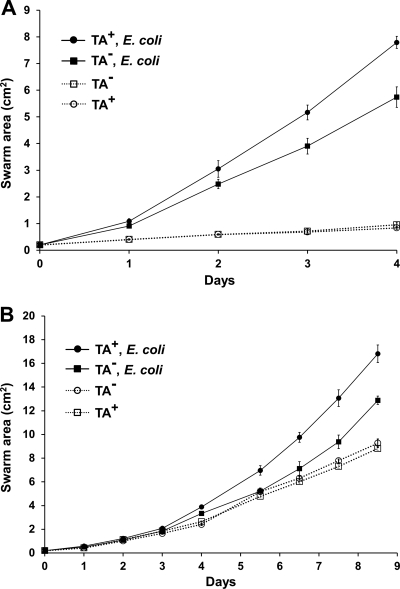
Another approach to study predation is swarm assays, which indirectly measure prey killing and consumption (21, 33). In contrast to the killing assay, this method allows predator fitness to be assessed. In these studies, we first measured swarm expansion rates of TA+ and TA− strains on TPM agar with or without an E. coli lawn. Consistent with above results, the TA+ strain swarmed at a modestly higher rate than the TA− strain over an E. coli lawn (Fig. 6 A). These findings were found in three independent experiments, one of which is shown in Fig. 6. On control plates without prey, the two M. xanthus strains swarmed at identical rates (Fig. 6A). These results thus show the TA− mutant was not defective in gliding motility per se but instead was less effective in prey handling (killing and consumption) (21, 33). The lower degree of swarming observed on TPM starvation agar, in the absence of prey, likely reflects a lack of nutrients for M. xanthus growth. Thus, for M. xanthus to exhibit robust swarm expansion on TPM agar it must kill and feed on the E. coli lawn. Swarm assays were also conducted on 1/2 CTT agar (moderate nutrient levels), which supports prey growth. It was similarly found that the TA+ strain showed a reproducibly higher swarm expansion rate than the TA− mutant on prey lawns but not in the absence of prey (Fig. 6B). These results again indicate that the TA+ strain was more proficient at handling prey than the TA− mutant. As found on TPM agar, M. xanthus swarm expansion on 1/2 CTT agar was enhanced by prey lawns (Fig. 6B), perhaps owing to richer nutrients (prey), which likely increase growth and/or swarm rates. We also note that in the presence of prey lawns the overall swarm rate was nearly 2-fold higher on TPM agar than on 1/2 CTT agar. The reason for this difference is not obvious, but it may reflect available nutrient levels (20). In sum, these results suggest that the TA− mutant has a reduced fitness to handle E. coli prey.
Fig. 6.
TA production increases the swarm expansion rate specifically over prey lawns. (A) M. xanthus swarm expansion was measured on TPM starvation agar without (dashed lines/open markers) or top spread with a thick E. coli lawn (MG1655; solid lines/filled markers). To these plates 5 μl of TA+ (DK1622) or TA− (DW1034) M. xanthus cells were spotted. Swarm diameters were measured daily, and the swarm areas were calculated. Experiments were done in triplicate, and swarm areas were averaged and standard errors plotted. (B) Same as panel A, except that 1/2 CTT agar was used and the duration of the experiment was over 2-fold longer.
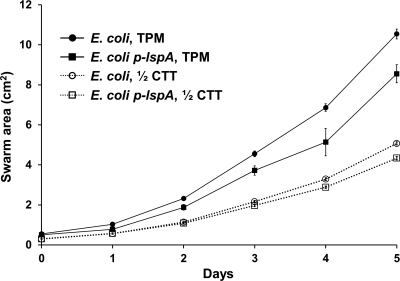
In reciprocal experiments, the fitness of DK1622 on E. coli prey either overexpressing LspA (TAr) or not (TAs) was again assessed by monitoring swarm expansion. It was found that the DK1622 swarm expansion rate was moderately reduced on TAr E. coli compared to that of the parental control on TPM agar (Fig. 7). Similar, but more modest differences were also found on 1/2 CTT agar (data not shown). Thus, under the described conditions, TA production or potency toward prey contributes toward M. xanthus fitness as judged by swarm expansion.
Fig. 7.
Prey resistance toward antibiotic TA reduces swarm expansion. E. coli MG1655 lawns were made on indicated plates. As indicated, MG1655 either harbored plasmid p-lspA or it did not. All plates contained 20 μM IPTG, and strain DK1622 was used for swarm expansion. Experiments were done in triplicate, and swarm areas were averaged and standard errors plotted.
DISCUSSION
In this work we provide evidence that secondary metabolites produced by myxobacteria can play an important role in microbial predation. Specifically, we compared predation of TA+ and TA− strains in a series of conditions. First, a ZOI test found that a TA− strain lacks diffusible antibiotic activity toward E. coli. This phenotype correlates with defects in E. coli killing (Fig. 1). In contrast, we found no discernible defect of the TA− mutant toward Micrococcus luteus killing. These results are consistent with the findings that antibiotic TA (myxovirescin) exhibits potent antibacterial activity against E. coli and is not active against Micrococcus luteus (16). We further tested the role of TA production on other bacterial species. For species that were susceptible to predation, we found phenotypes that were either similar or intermediate to those reported here (unpublished results). In general terms, as one might expect, we found a correlation between the degree of strain sensitivity to antibiotic TA and the extent TA plays a role in predation to the producer organism. E. coli and Micrococcus luteus represent two opposite extremes for the role that TA can play in predation.
The physiological state of prey cells was also found to be important for the utility of TA in predation. Against metabolically quiescent E. coli (starvation), TA's role was diminished (Fig. 2). We interpret this result to mean that static prey cells are more vulnerable to M. xanthus extracellular hydrolytic enzyme attack, because they have a reduced metabolic capacity to repair cellular damage caused by these agents. In addition, the predation defect of a TA− mutant was corrected by exogenous addition of a bacteriostatic antibiotic (Fig. 3). This finding corroborates our hypothesis that antibiotics serve as small-molecule weapons to penetrate and kill or neutralize prey metabolism. Prey neutralization thus facilitates their digestion by impermeable macromolecular enzymes. The digestion of prey into small molecules thus allows nutrient uptake and utilization by myxobacteria. In this scenario both static and bactericidal antibiotics would facilitate prey digestion. In addition, TAr E. coli strains were found to confer resistance toward predation by a TA+ strain (Fig. 4). Similar results were obtained with C. coralloides and corallopyronin production (Fig. 5). Thus, in general terms, our results provide direct evidence to support the hypothesis that myxobacterial antibiotics play a biological and presumed ecological role in predation.
Swarm expansion assays independently found that TA activity contributes toward M. xanthus predatory fitness (Fig. 6 and 7). An advantage this assay offers is that it indirectly measures prey killing and consumption; however, a significant disadvantage is that it provides poor temporal resolution of predation, as swarm rates are slow and measured daily. In contrast, the killing assay offers high temporal resolution, as hourly measurements can easily be made. Thus, we believe, the killing assay provides a more sensitive means to differentiate predation phenotypes. In any event, our data suggest that the TA− mutant grows at a lower rate on E. coli prey, as swarm rate expansion is a function of growth and motility (Fig. 6) (6). Consistent with this, Rosenberg and colleagues reported that compared to a TA+ strain, a M. xanthus TA− mutant was defective in growing and competing against E. coli, although experimental details were not provided (34, 37).
Our findings begin to elucidate the molecular mechanism of myxobacterial predation, whereby antibiotics serve as a front line weapon. However, we recognize myxobacteria encode a multitude of agents to attack prey (33, 37). Consequently, some of these agents might have redundant functions. As a case in point, the M. xanthus genome is predicted to encode ∼70 secreted proteases, many of which are likely involved in predation (17). If these agents are indeed redundant, then elucidating their functions in predation could be problematic. Myxobacteria belong to the Deltaproteobacteria and thus are phylogenetically related to the relatively well characterized microbial predator Bdellovibrio bacteriovorus (29). Genomic analysis reveals these predators do share some gene products likely involved in predation, such as type IV pili and a large array of hydrolytic enzymes, but notably B. bacteriovorus does not produce antibiotics (3, 11). The latter difference may reflect that B. bacteriovorus is a more specialized predator whereby it solely binds the Gram-negative cell envelope followed by penetration and host-dependent growth. Myxobacteria, on the other hand, might be viewed as a generalized predator, perhaps due in part to their ability to produce numerous antibiotics and to digest a broad range of Gram-positive and -negative bacteria. Other predatory bacteria, such as Lysobacter spp. and Aristabacter necator, produce antibiotics, but their role in predation is unknown (7, 18, 29).
The biological significance of secondary metabolite production has largely remained elusive. In the case of antibiotics, however, an obvious presumed role exists, namely, that antibiotics provide a competitive advantage to the producer organism by inhibiting competitor growth in natural habitats. However, shortly after the inception of this hypothesis and more recently, this idea has been brought into question (9, 42, 47). To our knowledge this report first shows that secondary metabolites from producer organisms can serve as “antibiotics” in microbial predation. To conserve material and increase effective concentrations, we speculate that myxobacteria can sense their environment to regulate the production and secretion of antibiotics for judicious use. In contrast, nonpredatory bacteria, such as the prolific secondary metabolite-producing genus Streptomyces, the biological role of their antibiotics remains a puzzle. However, one plausible explanation that takes into count environmentally relevant concentrations suggests that these molecules could serve as intercellular signals (10). In other cases, the biological function of antibiotics has been delineated. For instance, bacteria residing in the intestinal tract of insects, such as beetles or ants, form symbiotic relationships whereby these actinobacteria produce antibiotics that kill pathogenic microbes and consequently protect the insects' fungal gardens or symbionts (32, 38, 39). However, to our knowledge, antibiotics produced by actinobacteria do not play a predatory role.
Understanding the biology of antibiotic production may lead to improved strategies to discover new antibiotics and perhaps optimize production yields and variant discovery. Thus, for example, isolating environmental microbial predators or microbes from insect intestines may facilitate the discovery of new antibiotics, which are sorely needed (13). Moreover, understanding natural product functions can be exploited in the laboratory to evolve strains with improved yields and potencies (12). The use of predatory assays may also provide a means to select mutants that induce cryptic antibiotic pathways. This is relevant as, for instance, the M. xanthus laboratory strain has the genomic potential to produce 15 unique secondary metabolites that have not been described (26). Lastly, understanding antibiotic regulation, secretion, and producer resistance may lead to engineered strains with optimized yields. These research areas should be pursued, as myxobacteria are well endowed to produce secondary metabolites, some of which have therapeutic potential (5).
ACKNOWLEDGMENTS
We thank Joe Fralick for providing strains.
This work was supported by the University of Wyoming, NCRR, and the Wyoming INBRE (2P20RR016474) grant.
Footnotes
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Anscombe F. J., Singh B. N. 1948. Limitation of bacteria by micro-predators in soil. Nature 161:140. [DOI] [PubMed] [Google Scholar]
- 2. Belogurov G. A., et al. 2009. Transcription inactivation through local refolding of the RNA polymerase structure. Nature 457:332–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berleman J. E., Chumley T., Cheung P., Kirby J. R. 2006. Rippling is a predatory behavior in Myxococcus xanthus. J. Bacteriol. 188:5888–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berleman J. E., Kirby J. R. 2009. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol. Rev. 33:942–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bode H. B., Muller R. 2006. Analysis of myxobacterial secondary metabolism goes molecular. J. Ind. Microbiol. Biotechnol. 33:577–588 [DOI] [PubMed] [Google Scholar]
- 6. Burchard R. P. 1974. Growth of surface colonies of the gliding bacterium Myxococcus xanthus. Arch. Microbiol. 96:247–254 [DOI] [PubMed] [Google Scholar]
- 7. Cain C. C., et al. 2003. Synergistic antimicrobial activity of metabolites produced by a nonobligate bacterial predator. Antimicrob. Agents Chemother. 47:2113–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clardy J., Fischbach M. A., Currie C. R. 2009. The natural history of antibiotics. Curr. Biol. 19:R437–R441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davies J. 2006. Are antibiotics naturally antibiotics? J. Ind. Microbiol. Biotechnol. 33:496–499 [DOI] [PubMed] [Google Scholar]
- 10. Davies J., Spiegelman G. B., Yim G. 2006. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 9:445–453 [DOI] [PubMed] [Google Scholar]
- 11. Evans K. J., Hobley L., Lambert C., Sockett R. E. 2008. Bdellovibrio: lone hunter “cousin” of the “pack hunting” myxobacteria, p. 351–362 In Whitworth D. E. (ed.), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC [Google Scholar]
- 12. Fischbach M. A., Lai J. R., Roche E. D., Walsh C. T., Liu D. R. 2007. Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes. Proc. Natl. Acad. Sci. U. S. A. 104:11951–11956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fischbach M. A., Walsh C. T. 2009. Antibiotics for emerging pathogens. Science 325:1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fischbach M. A., Walsh C. T., Clardy J. 2008. The evolution of gene collectives: how natural selection drives chemical innovation. Proc. Natl. Acad. Sci. U. S. A. 105:4601–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fralick J. A., Burns-Keliher L. L. 1994. Additive effect of tolC and rfa mutations on the hydrophobic barrier of the outer membrane of Escherichia coli K-12. J. Bacteriol. 176:6404–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerth K., Irschik H., Reichenbach H., Trowitzsch W. 1982. The myxovirescins, a family of antibiotics from Myxococcus virescens (Myxobacterales). J. Antibiot. (Tokyo) 35:1454–1459 [DOI] [PubMed] [Google Scholar]
- 17. Goldman B. S., et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashizume H., et al. 2001. Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. I. Taxonomy, isolation and biological activities. J. Antibiot. (Tokyo) 54:1054–1059 [DOI] [PubMed] [Google Scholar]
- 19. Hillesland K. L., Lenski R. E., Velicer G. J. 2007. Ecological variables affecting predatory success in Myxococcus xanthus. Microb. Ecol. 53:571–578 [DOI] [PubMed] [Google Scholar]
- 20. Hillesland K. L., Velicer G. J. 2005. Resource level affects relative performance of the two motility systems of Myxococcus xanthus. Microb. Ecol. 49:558–566 [DOI] [PubMed] [Google Scholar]
- 21. Hillesland K. L., Velicer G. J., Lenski R. E. 2009. Experimental evolution of a microbial predator's ability to find prey. Proc. Biol. Sci. 276:459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Irschik H., Jansen R., Hofle G., Gerth K., Reichenbach H. 1985. The corallopyronins, new inhibitors of bacterial RNA synthesis from myxobacteria. J. Antibiot. (Tokyo) 38:145–152 [DOI] [PubMed] [Google Scholar]
- 23. Julien B., Kaiser A. D., Garza A. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 97:9098–9103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kashefi K., Hartzell P. L. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483–494 [DOI] [PubMed] [Google Scholar]
- 25. Kitagawa M., et al. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 26. Krug D., et al. 2008. Discovering the hidden secondary metabolome of Myxococcus xanthus: a study of intraspecific diversity. Appl. Environ. Microbiol. 74:3058–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J. W., Vederas J. C. 2009. Drug discovery and natural products: end of an era or an endless frontier? Science 325:161–165 [DOI] [PubMed] [Google Scholar]
- 28. Margalith P. 1962. Bacteriolytic principles of Myxococcus fulvus. Nature 196:1335–1336 [Google Scholar]
- 29. Martin M. O. 2002. Predatory prokaryotes: an emerging research opportunity. J. Mol. Microbiol. Biotechnol. 4:467–477 [PubMed] [Google Scholar]
- 30. McBride M. J., Zusman D. R. 1996. Behavioral analysis of single cells of Myxococcus xanthus in response to prey cells of Escherichia coli. FEMS Microbiol. Lett. 137:227–231 [DOI] [PubMed] [Google Scholar]
- 31. Mukhopadhyay J., et al. 2008. The RNA polymerase “switch region” is a target for inhibitors. Cell 135:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oh D. C., Poulsen M., Currie C. R., Clardy J. 2009. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 5:391–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pham V. D., Shebelut C. W., Diodati M. E., Bull C. T., Singer M. 2005. Mutations affecting predation ability of the soil bacterium Myxococcus xanthus. Microbiology 151:1865–1874 [DOI] [PubMed] [Google Scholar]
- 34. Rosenberg E., Dworkin M. 1996. Autocides and a paracide, antibiotic TA, produced by Myxococcus xanthus. J. Ind. Microbiol. 17:424–431 [Google Scholar]
- 35. Rosenberg E., Keller K. H., Dworkin M. 1977. Cell density-dependent growth of Myxococcus xanthus on casein. J. Bacteriol. 129:770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rosenberg E., Vaks B., Zuckerberg A. 1973. Bactericidal action of an antibiotic produced by Myxococcus xanthus. Antimicrob. Agents Chemother. 4:507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosenberg E., Varon M. 1984. Antibotics and lytic enzymes, p. 109–125 In Rosenberg E. (ed.), Myxobacteria. Development and cell interactions. Springer-Verlag, New York, NY [Google Scholar]
- 38. Scott J. J., et al. 2008. Bacterial protection of beetle-fungus mutualism. Science 322:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shank E. A., Kolter R. 2009. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 12:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shimkets L. J., Dworkin M., Reichenbach H. 2006. The myxobacteria, p. 31–115 In Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E. (ed.), The prokaryotes, 3rd ed., vol. 7 Springer Verlag, Heidelberg, Germany [Google Scholar]
- 41. Simunovic V., et al. 2006. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. Chembiochem 7:1206–1220 [DOI] [PubMed] [Google Scholar]
- 42. Waksman S. 1961. The role of antibiotics in nature. Perspect. Biol. Med. 4:271–286 [Google Scholar]
- 43. Wall D., Kolenbrander P. E., Kaiser D. 1999. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J. Bacteriol. 181:24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walsh C. 2003. Where will new antibiotics come from? Nat. Rev. Microbiol. 1:65–70 [DOI] [PubMed] [Google Scholar]
- 45. Weissman K. J., Muller R. 2009. A brief tour of myxobacterial secondary metabolism. Bioorg. Med. Chem. 17:2121–2136 [DOI] [PubMed] [Google Scholar]
- 46. Wenzel S. C., Muller R. 2009. Myxobacteria—′microbial factories' for the production of bioactive secondary metabolites. Mol. Biosyst. 5:567–574 [DOI] [PubMed] [Google Scholar]
- 47. Williams S., Vicker J. 1986. The ecology of antibiotic production. Microb. Ecol. 12:43–52 [DOI] [PubMed] [Google Scholar]
- 48. Yim G., Wang H. H., Davies J. 2006. The truth about antibiotics. Int. J. Med. Microbiol. 296:163–170 [DOI] [PubMed] [Google Scholar]
- 49. Zafriri D., Rosenberg E., Mirelman D. 1981. Mode of action of Myxococcus xanthus antibiotic TA. Antimicrob. Agents Chemother. 19:349–351 [DOI] [PMC free article] [PubMed] [Google Scholar]