Abstract
Vfr, a transcription factor homologous to the Escherichia coli cyclic AMP (cAMP) receptor protein (CRP), regulates many aspects of virulence in Pseudomonas aeruginosa. Vfr, like CRP, binds to cAMP and then recognizes its target DNA and activates transcription. Here we report that Vfr has important functional differences from CRP in terms of ligand sensing and response. First, Vfr has a significantly higher cAMP affinity than does CRP, which might explain the mysteriously unidirectional functional complementation between the two proteins (S. E. H. West et al., J. Bacteriol. 176:7532–7542, 1994). Second, Vfr is activated by both cAMP and cGMP, while CRP is specific to cAMP. Mutagenic analyses show that Thr133 (analogous to Ser128 of CRP) is the key residue for both of these distinct Vfr properties. On the other hand, substitutions that cause cAMP-independent activity in Vfr are similar to those seen in CRP, suggesting that a common cAMP activation mechanism is present. In the course of these analyses, we found a remarkable class of Vfr variants that have completely reversed the regulatory logic of the protein: they are active in DNA binding without cAMP and are strongly inhibited by cAMP. The physiological impact of Vfr's ligand sensing and response is discussed, as is a plausible basis for the fundamental change in protein allostery in the novel group of Vfr variants.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic pathogen of immunocompromised individuals, typically infecting the pulmonary tract, urinary tract, burns, and wounds (14, 28). For successful infection, P. aeruginosa relies on many extracellular and cell-associated virulence factors whose production, in turn, is controlled by multiple regulatory proteins (17). Vfr (virulence factor regulator) is one of these regulators and is involved in the expression of a set of genes for extracellular virulence factors (11, 15, 26, 43), the type III secretion system (7, 37, 44, 45), quorum sensing (2, 22), and flagellar biosynthesis (8).
Vfr belongs to the CRP/FNR superfamily of transcription factors, one of the largest groups of bacterial environmental sensors (named for the Escherichia coli cyclic AMP [cAMP] receptor protein/fumarate nitrate reductase regulator). Vfr, like CRP, requires cAMP binding to be activated in vitro (12, 38) and in vivo (37, 45). The notion of cAMP as the physiological ligand for the protein is generally accepted, and this view was further supported by the presence of that ligand in the crystal structure of an active form of Vfr (6a). The overall structure of the cAMP-bound Vfr is superimposable on the structure of active cAMP-bound CRP. While CRP is specific to cAMP, Vfr has been proposed to additionally respond to cGMP, based on either structural modeling (3) or host-pathogen interaction models (30, 41). However, no definitive physiological role of cGMP has been identified, mainly because its concentration in cells is very low (13, 41). Here we report that Vfr's ligand sensing and response differ biochemically from those of CRP in two ways: (i) Vfr has a significantly higher cAMP affinity than does CRP, and (ii) Vfr can be activated by cGMP in addition to cAMP. We note that our data for cGMP activation of Vfr are in disagreement with a recent report that cGMP cannot activate Vfr and actually blocks cAMP activation of Vfr (12). Further analysis of Vfr variants altered at the cAMP pocket indicated that Thr133, a C-helix residue, plays an important role in both of these ligand properties.
Vfr is highly similar to CRP in both sequence and structure. The Vfr sequence is 67% identical and 91% similar to that of CRP (43). Structurally, as with CRP, the functional form is a homodimer, and each monomer has two distinct domains (an N-terminal ligand-binding domain and a C-terminal DNA-binding domain) connected by a long C-helix dimerization component (6a). It is therefore a reasonable hypothesis that, like CRP, Vfr exists in equilibrium between active and inactive forms, and cAMP shifts the equilibrium toward the active form. Figure 1 A illustrates the key structural components in an active form of Vfr (Protein Data Bank [PDB] identification [ID] code 2OZ6). Importantly, the cAMP-binding site is far away from the DNA-binding site (>20 Å), and therefore, cAMP binding cannot directly affect the DNA-binding site. Instead, Vfr most likely undergoes a global conformational change when binding to cAMP, as does CRP. In the case of CRP, the notion of C-helix repositioning as the activation mechanism has been well established by several mutagenic studies, along with the recently solved inactive-form structures (29, 35). This C-helix repositioning mechanism is also well established for other family members, such as CooA and FNR (16, 24). Given the high similarity of Vfr to CRP, it is plausible that Vfr also utilizes the C-helix repositioning mechanism, but this has never been tested. We provide mutagenic evidence that this is the case. Unexpectedly, our mutagenic approach led to the identification of a fundamentally new class of Vfr variants in which the response to cAMP is qualitatively reversed. Taken together, the evidence demonstrates that the C-helix plays a central role in the ligand-sensing function of Vfr.
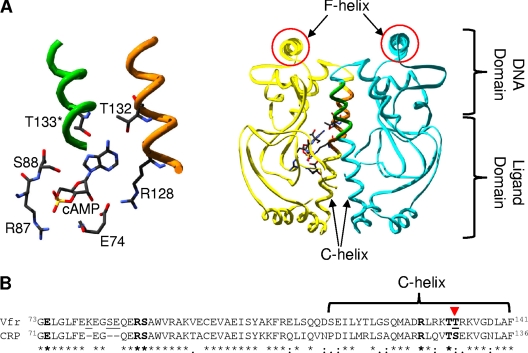
Fig. 1.
(A) cAMP-bound structure of Vfr (PDB ID 2OZ6) with cAMP-contacting residues highlighted. The right side shows the overall structure, while the cAMP pocket is enlarged and rotated slightly in the left panel. The cAMP-contacting residues shown are all from one subunit except for Thr133 (T133). The protein functions as a dimer (one part in yellow and the other in blue). The two F-helices (the DNA-contacting regions) are circled in red. The figure was visualized using Swiss-PdbViewer, version 4.01, and POV-Ray, version 3.6.2. (B) Alignment of ligand-binding domains of Vfr (NP_249343.1) and CRP (NP_417816.1), which cover the known primary cAMP-binding pocket residues. The known cAMP-contacting residues in both proteins are shown in boldface, and the two proteins differ only in one residue that is indicated by the inverted triangle. The T-Coffee program was used to generate the alignment. In the alignment, an asterisk (*) indicates the same amino acid in the two proteins, a colon (:) indicates a conservative substitution, and a period (.) indicates a semiconservative substitution.
MATERIALS AND METHODS
Materials.
The compounds cAMP and cGMP were purchased from Sigma (St. Louis, MO).
Strains, plasmids, and recombinant DNA methodology.
Standard methods were used for the isolation and manipulation of DNA (32). Plasmid DNA isolation was carried out using QIAquick plasmid purification kits from Qiagen Inc. (Chatsworth, CA). Restriction enzymes were purchased from New England BioLabs (Beverly, MA) and were used as recommended. Synthetic oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Bacterial strains carrying different plasmids were propagated in 1% tryptone, 0.5% yeast extract, and 0.5% NaCl (LC medium) with 15 μg/ml tetracycline, 25 μg/ml chloramphenicol, or 50 μg/ml ampicillin as appropriate.
Cloning of vfr, generation of site-directed and randomized Vfr variants, and in vivo screening for Vfr* (constitutively active Vfr) variants.
P. aeruginosa PAO1 vfr was PCR amplified and cloned into EcoRI-HindIII-digested pEXT20 (9). For a histidine-tagged version of Vfr, the vfr gene was cloned similarly, but a reverse primer containing seven additional histidine codons between the last amino acid codon and the stop codon was used. The wild-type Vfr and the Vfr variants used in the present study were all His tagged unless stated otherwise. Site-directed mutagenesis was carried out by PCR amplification with mutagenic primers (5). The method used for codon randomization was the same as that used for site-directed mutagenesis except that the primers contained randomized codons at the desired positions. In this study, the codons for positions 132 and 133 of Vfr were randomized, and the resultant plasmid pool was transformed into UQ3811 for cAMP-independent activity: UQ3811 is an E. coli cya crp reporter strain harboring a chromosomally encoded lacZ gene under the control of a CRP consensus class I promoter (48). Then the resultant transformants were screened for increased β-galactosidase activity relative to that of wild-type Vfr in the absence of cAMP, as indicated by bluer coloration. The vfr genes from the isolated blue colonies were then sequenced to reveal the causative mutation.
Overexpression and purification of Vfr proteins.
Overexpression of the His-tagged wild-type Vfr and Vfr variants was carried out in the strain background of UQ3809, an E. coli cya crp strain, and purification was carried out by using a nickel-nitrilotriacetate column (Novagen, Madison, WI). The final purity of the proteins was >95%. Because the protein was never exposed to cAMP either during growth or during purification, we believe Vfr was isolated as “an apo-protein.”
Hydroxyapatite batch purification of untagged wild-type Vfr.
Non-His-tagged Vfr was purified from UQ6049 (Table 1) using the hydroxyapatite batch purification method as follows. A 50-ml culture of the cells was harvested after protein overexpression with 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG), resuspended in 5 ml of 25 mM morpholinepropanesulfonic acid (MOPS) buffer-0.2 M NaCl–10% glycerol (pH 7.4), broken with a French pressure cell (∼120 MPa), and centrifuged for 30 min at 11,700 × g. The supernatant was then mixed with 0.3 g of solid hydroxyapatite resin. After unbound materials were removed from the resin, a high-salt buffer containing 25 mM MOPS (pH 7.4), 10 mM potassium phosphate, 1.2 M KCl, and 5% glycerol was added, and the resin was washed twice. Vfr was then eluted with a high-phosphate buffer containing 25 mM MOPS (pH 7.4), 160 mM potassium phosphate, 50 mM KCl, and 5% glycerol. The eluent was precipitated with ammonium sulfate with a final saturation of 50%, dissolved in 50 mM Tris-HCl (pH 8)–0.5 M KCl, and stored at −80°C until use. Vfr prepared by this procedure resulted in an enrichment of the protein to ∼20% of total protein.
Table 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Brief description | Source or reference |
|---|---|---|
| Bacterial strains | ||
| UQ3740 | M182 carrying λ prophage with CC(−41.5)::lacZ fusion (RLG4649) | 33 |
| UQ3741 | M182 carrying λ prophage with CC(−61.5)::lacZ fusion (RLG4650) | 34 |
| UQ3809 | UQ3740 with ilv::Tn10 cya and crp::cam | 48 |
| UQ3811 | UQ3741 with ilv::Tn10 cya and crp::cam | 48 |
| UQ4249 | UQ3740 with crp::cam | 47 |
| UQ4246 | UQ3741 with crp::cam | This work |
| UQ5284 | pUX2679 in UQ3809 | This work |
| UQ6049 | pUX3251 in UQ3809 | This work |
| Plasmids | ||
| pEXT20 | Escherichia coli expression vector | 9 |
| pUX2679 | pEXT20 plasmid bearing the P. aeruginosa vfr allele (+7 His tag codons) | This work |
| pUX3251 | pEXT20 plasmid bearing the P. aeruginosa vfr allele (without a His tag) | This work |
Measurement of in vitro DNA-binding activity of wild-type Vfr and Vfr variants.
In vitro DNA-binding assays were carried out by using a fluorescence polarization method with a Beacon 2000 fluorescence polarization detector (Invitrogen Corp., Carlsbad, CA). The target DNA for this was a 26-mer CRP consensus sequence (5′-GTAAATGTGATGTACATCACATGGAT-3′) labeled with a fluorescent dye, Texas Red, as described previously (48). We chose this probe for the DNA-binding assay of Vfr for the following three reasons. (i) Vfr and CRP have almost identical F-helix residues directly involved in DNA binding, suggesting the presence of a common target DNA sequence. (ii) The CRP probe is very close to the proposed consensus DNA sequence of Vfr (15). (iii) The use of the probe allowed us to directly compare Vfr with CRP in terms of DNA binding, since the probe has been routinely used in our analysis of CRP (46, 47, 48). Binding assays were performed in 50 mM Tris-HCl (pH 8.0), 50 mM KCl, and 1 mM EDTA with a probe concentration of 5 nM in the presence of 6.4 μM salmon sperm DNA (nonspecific competitor).
Quantitative analysis of the ligand-protein equilibrium.
The Vfr-DNA binding isotherms were analyzed in terms of an equilibrium shift model previously proposed for CRP (46). The model was implemented with commercial software (Maplesoft), which solved the coupled equilibrium-constant equations numerically for trial values of the unknowns. Values were optimized to fit observed isotherms by the downhill simplex method (30a).
Measurement of in vivo β-galactosidase activity.
The in vivo activities of Vfr and CRP were measured in appropriate E. coli reporter strains. UQ3811 was used for cAMP-independent activities, and UQ4249 was used for cAMP-dependent activities (Table 1). Typically the cells were fully grown overnight at 37°C in LC medium containing 50 μg/ml ampicillin. The next day, cells were diluted to an A600 of 0.1 in fresh LC medium containing 50 μg/ml ampicillin and were grown at 37°C and 220 rpm. Cells at an A600 of 1 to 1.5 were then used for the measurement of β-galactosidase activity according to a standard method (23).
RESULTS
DNA affinities of apo-Vfr and cAMP-bound Vfr in comparison with their CRP counterparts.
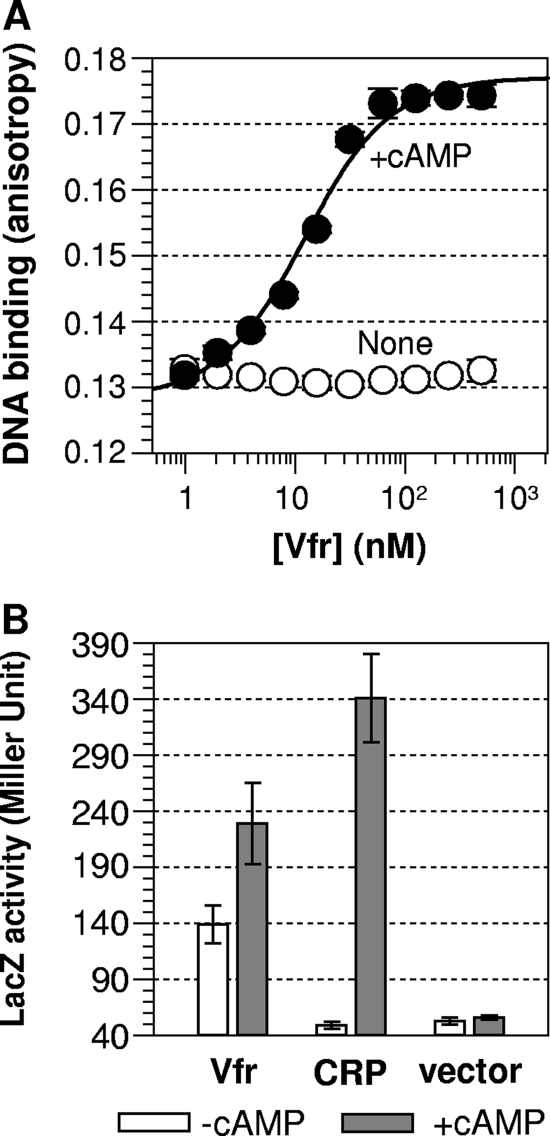
For the measurement of DNA binding, we used a fluorescence polarization method. The DNA-Vfr interaction causes an increase in fluorescence polarization (anisotropy) due to slowed Brownian motion of the labeled DNA. For this assay, we used purified His-tagged apo-Vfr, as described in Materials and Methods. When saturated with cAMP (10 μM), the purified Vfr showed a high affinity for the target DNA, corresponding to a dissociation constant (Kd) of 9.7 nM (Fig. 2 A). In the absence of cAMP, the protein failed to show any detectable DNA binding up to a protein concentration of 500 nM (Fig. 2A). Under the same buffer condition and with the same DNA probe used for Vfr, cAMP-bound CRP showed a similar DNA affinity (Kd, ≈10 nM) (48), suggesting that the DNA-binding characteristic of Vfr is similar to that of CRP. Although Vfr's DNA binding behavior in response to cAMP might be substantially different in the context of a longer and suboptimal native Vfr DNA binding sequence, our result concurs with published reports that Vfr requires cAMP for DNA binding (12, 38) and additionally proves that our His-tagged Vfr was functionally intact in terms of cAMP sensing and response.
Fig. 2.
Vfr is cAMP responsive. (A) In vitro DNA-binding activity. Activities were measured in the absence (open circles) and presence (filled circles) of cAMP by using the fluorescence anisotropy method. In each case, fluorescence anisotropy values were measured up to a protein concentration of 500 nM. Each data point is the average of three independent measurements, and solid lines show the best fit of the data to an equation described by Lundblad et al. (20). (B) In vivo activity. The activities of Vfr and CRP were measured in appropriate E. coli cells (for strains, see Table 1). Open and shaded bars indicate in vivo β-galactosidase activities in the absence of cAMP (in UQ3811) and in the presence of cAMP (in UQ4249), respectively.
We then compared Vfr's in vivo transcriptional activities in the presence and absence of cAMP. These in vivo data are consistent with the in vitro DNA-binding data but also suggested another property of Vfr. The plasmid containing vfr (pUX2679 [Table 1]) was introduced via transformation into an E. coli cya crp (UQ3811) or crp (UQ4249) lacZ reporter strain (Table 1), and then the ability of the Vfr protein to promote in vivo transcriptional activation (and therefore β-galactosidase production) was measured. While Vfr displayed high β-galactosidase activity in the cAMP-producing cells (Fig. 2B), Vfr displayed in vivo transcriptional activity significantly above the background level even in the absence of cAMP (Fig. 2B). A recent study found that lasR promoter activity in vivo is Vfr dependent but cAMP independent in P. aeruginosa (12), and our measurement of Vfr activity in an E. coli cya strain is apparently consistent with that report. Nonetheless, the detection of significant Vfr activity in the E. coli cya reporter is surprising, given that we were unable to detect DNA affinity for Vfr in the absence of cAMP (Fig. 2A). It is possible that the Vfr activity measured in an E. coli cya strain is not all cAMP independent: Even a very low level of cAMP in E. coli cya (1) might be enough to activate Vfr, especially given the substantially higher affinity of Vfr for cAMP (see below). Also, adenosine is a possible adventitious ligand of Vfr, a matter discussed elsewhere in the context of the interpretation of CRP* (constitutively active CRP) activity (46). On the other hand, even in our in vivo transcriptional assay, Vfr activity was still cAMP dependent (Fig. 2B). Therefore, we believe that Vfr requires cAMP for binding to its DNA targets and for transcriptional activation, especially for the suboptimal natural Vfr targets, as demonstrated previously by Fuchs et al. (12).
For comparison, CRP's in vivo transcriptional activity was measured in the same reporter strain, which showed no observable activity in E. coli cya (Fig. 2B). Several mutually nonexclusive explanations for the constitutive in vivo activity of Vfr are possible. (i) In the absence of cAMP, Vfr is more shifted toward the active form than is CRP, although the cAMP-independent DNA affinity of Vfr was not measurable in vitro. (ii) Vfr has a better interaction with the E. coli RNA polymerase than does CRP. (iii) The concentration of Vfr in the E. coli cya reporter is higher than that of CRP. Nonetheless, the last possibility is unlikely for the following reason. The in vivo activities of both Vfr and CRP were measured in the absence of IPTG, and CRP's activity in the E. coli cya reporter was not increased even when the protein was overexpressed by 100 μM IPTG (data not shown).
Vfr has a higher affinity for cAMP than does CRP.
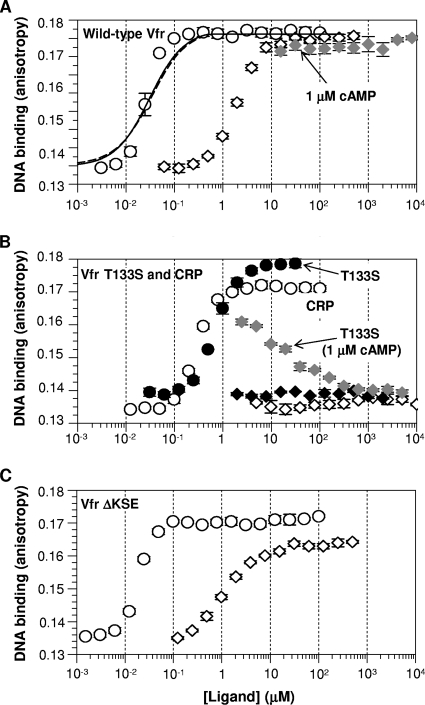
If Vfr has an equilibrium shift toward the active form relative to CRP (that is, there is a higher fraction of active Vfr than of active CRP in the absence of cAMP), we reasoned that this would result in a higher apparent cAMP affinity for the Vfr protein than for CRP, based on our earlier work on CRP and CRP* variants (46). Therefore, we first tested if this was the case. For the assessment of cAMP affinity, we used a functional approach of measuring the effective range of cAMP concentrations required for the DNA binding of Vfr. As shown in Fig. 3 A, the binding isotherm revealed that at 100 nM cAMP, Vfr was able to saturate the probe DNA. This level of cAMP is about 10-fold lower than the concentration (1 μM) required for full activation of CRP (Fig. 3B). This in turn indicates that Vfr indeed has a higher cAMP affinity than does CRP. Several research groups have measured the cAMP affinity of CRP using direct cAMP-binding assays, and the Kd value of about 20 μM is generally accepted (19, 21, 31, 39). Nonetheless, significant deviations from this value have been noted (10, 38). Thus, some degree of discrepancy exists in the literature, possibly due to methodological differences: some investigators measured physical cAMP affinity, while others assessed cAMP binding by monitoring the protein's conformational change (albeit without DNA). On the other hand, the cAMP affinity of Vfr has been measured by two groups, which found Kd values in the range of 1.0 to 5.5 μM (6a, 38). Notably, the two Vfr research groups used different methods for directly measuring the cAMP affinity of Vfr. This suggests that methodological difference alone cannot fully explain the discrepancy in reported cAMP affinities. Given this situation, our new “coupled functional” assay provides a unique comparison of cAMP affinity between Vfr and CRP.
Fig. 3.
Ligand concentration-dependent activation of histidine-tagged Vfr and Vfr variants altered at the cAMP pocket in comparison with that of CRP (the cAMP receptor protein of Escherichia coli). The DNA affinity of each protein was measured in various concentrations of the ligands cAMP (open circles) and cGMP (open diamonds). (A) Wild-type Vfr. The solid line (if Kc is fixed to 3.2 × 1010) and dotted line (if ka is fixed to 4.3 × 108 M−1) indicate the best fits for the cAMP isotherm (see Table 2 for more information). (B) Vfr T133S (open symbols) and CRP (filled symbols). The data for CRP were excerpted from the work of Youn et al. (46). The shaded diamonds in panels A and B indicate cGMP titration of wild-type Vfr (A) or Vfr T133S (B) in the presence of 1 μM cAMP. (C) Vfr ΔKSE. The concentration of each Vfr protein used was 200 nM; CRP was used at 100 nM. The scales of the x axis (ligand concentration) and y axis (anisotropy value) are the same in the three panels.
The cAMP isotherm of Vfr was further analyzed using the published scheme of the equilibrium shift model (46), which dissects the apparent cAMP affinity into two distinct factors: the protein's conformational equilibrium (Kc) and the intrinsic cAMP affinity of the protein (ka). However, neither the Kc nor the ka of Vfr could be independently determined, due to the lack of measurable DNA affinity of cAMP-free Vfr. Therefore, we were able only to estimate Vfr's upper limit for each parameter in reference to the reported CRP parameters: First, when we assumed that Vfr and CRP have the same intrinsic cAMP affinity (ka), Vfr was found to have a 1.2 × 103-fold greater active population in the absence of cAMP than does CRP (Table 2). Second, when we assumed that Vfr and CRP have the same protein equilibrium (Kc), Vfr was found to have a 74-fold greater intrinsic cAMP affinity (Table 2). The relative contributions of these parameters to the overall higher cAMP affinity of Vfr have yet to be determined. Nevertheless, the analysis quantitatively demonstrates that Vfr possesses an intrinsic capacity for higher cAMP binding. Therefore, the analysis is consistent with the hypothesis that Vfr is shifted further toward the active form than is CRP.
Table 2.
Estimated boundary of Kc and ka values of Vfr relative to CRP values
| Protein | ka (M−1)a | Kcb |
|---|---|---|
| CRP | 4.3 × 108c | 1.1 × 10−6c |
| Vfr (fixed ka) | 4.3 × 108 | 1.3 × 10−3 |
| Vfr (fixed Kc) | 3.2 × 1010 | 1.1 × 10−6 |
The intrinsic affinity of each protein for cAMP. The ka of Vfr was assumed to be identical to that of CRP.
The protein's conformational equilibrium. The protein equilibrium shift is defined by [proteinactive]/[proteininactive] in the absence of cAMP. The Kc of Vfr was assumed to be identical to that of CRP.
From the report of Youn et al. (46).
Vfr responds to cGMP.
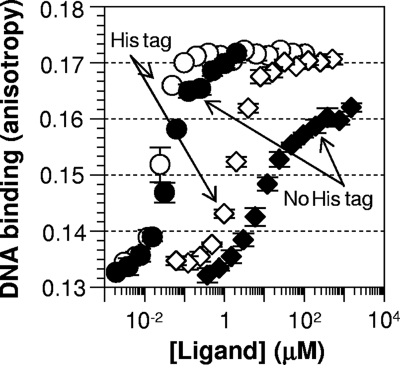
As mentioned in the introduction, there has been a controversy over the cGMP responsiveness of Vfr. As predicted by several researchers (3, 30, 41), Vfr in our hands can be activated by cGMP (Fig. 3A). However, cGMP was required at concentrations much higher than those of cAMP to promote Vfr's DNA binding, implying that the affinity of cGMP is much lower than that of cAMP. We also titrated cGMP up to 8 mM in the presence of 1 μM cAMP, but cGMP had no effect on the DNA-binding behavior of cAMP-bound wild-type Vfr (Fig. 3A, shaded diamonds). This result is in contrast to the recent report of Fuchs et al., who reported cGMP interference with the cAMP activation of Vfr (12). We further tested whether the histidines at the C terminus of our Vfr protein would explain the apparent discrepancy in Vfr's cGMP response between our work and that of Fuchs et al. For this purpose, we recloned vfr without the codons for a His tag into an expression plasmid, transformed it into an E. coli cya crp strain (deficient in cAMP production), and partially purified the untagged Vfr protein (∼20% purity) via hydroxyapatite batch purification. We favored hydroxyapatite over cAMP-agarose for cleaner apo-Vfr preparation. We reason that a cAMP-agarose column would produce very pure Vfr, which, however, would unavoidably be the cAMP-bound form. Then it would be a challenge to get high-quality apo-Vfr without perturbing the protein (given the very high cAMP affinity of Vfr). Notably, the partially purified untagged Vfr protein showed a cAMP isotherm for activation very similar to that of His-tagged Vfr (Fig. 4). The cGMP isotherm of untagged Vfr did not reach full activation, which may imply that the cGMP-bound active form is slightly different from the cAMP-bound active form. Nonetheless, it is clear that the untagged Vfr continued to be activated by cGMP (Fig. 4). This result suggests that the presence or absence of a His tag is not the basis for the discrepancy between the two groups. We note that the results in reference 12 depended on a very different assay, with different buffer conditions, at a single high concentration of cGMP. We also note that only a single consensus DNA target of Vfr was examined in this study, whereas Fuchs et al. (12) examined several natural DNA targets of Vfr.
Fig. 4.
Both histidine-tagged Vfr and untagged Vfr are activated by cGMP as well as cAMP. The purity of the His-tagged Vfr was >95%, and the untagged Vfr was partially purified (∼20%) for this assay. In both cases, 200 nM total protein was used, so the actual concentration of untagged Vfr in the reaction mixture was about 10-fold lower than that of His-tagged Vfr. Circles and diamonds indicate cAMP and cGMP titrations, respectively.
In summary, our purified Vfr protein (both His tagged and untagged) is activated by cGMP and in this regard is fundamentally different from CRP and from the Vfr protein investigated by Fuchs et al. We agree with their view that cGMP is physiologically irrelevant because of its poor affinity for Vfr (Fig. 3) and its low concentration in cells (13, 41). In contrast, we interpret the difference in cGMP response as a fundamental biochemical difference in the ligand response of Vfr.
Thr133 is important for both higher cAMP affinity and the cGMP response of Vfr.
In order to find a sequence determinant for Vfr's distinct properties, we targeted differences in the known cAMP-contacting residues between Vfr and CRP. Our rationale for this was that CRP's cAMP pocket is known to bind cGMP as well as cAMP (29, 46). Sequence alignment of CRP and Vfr (Fig. 1B) revealed two main differences between the two proteins in the pocket region: (i) Vfr has three additional residues (Lys80, Ser83, and Glu84) in the cAMP-contacting loop, and (ii) Vfr has Thr at position 133 instead of Ser at the analogous position 128 of CRP. Based on the assumption that one or both of these differences is responsible for the differential cAMP and/or GMP response in Vfr, we changed each region of Vfr to mimic CRP, resulting in two separate Vfr variants: Vfr T133S and Vfr ΔKSE (lacking the three residues Lys80, Ser83, and Glu84). Vfr T133S showed exactly the properties one would expect from a substitution of a critical residue. The Vfr T133S protein displayed much poorer cAMP affinity (a higher cAMP requirement) for activation than did wild-type Vfr (Fig. 3B). Importantly, Vfr T133S was also totally unresponsive to cGMP up to a 10 mM cGMP concentration (Fig. 3B). Then we titrated cGMP in the presence of 1 μM cAMP and found that cGMP was capable of interfering with the activation of Vfr T133S by cAMP (Fig. 3B). While cAMP-saturated Vfr T133S displayed a slightly reduced DNA-protein interaction (Kd, 44.3 nM), the ligand properties of Vfr T133S were nearly identical to those of E. coli CRP (Fig. 3B). Our results therefore strongly suggest that Thr133 is a key determinant of Vfr's ligand properties. In contrast, the Vfr ΔKSE variant showed little deviation from wild-type Vfr in either cAMP or cGMP responses (Fig. 3C), suggesting a minimal role for these residues in Vfr's ligand property.
The simultaneous impact of the T133S substitution on both cAMP affinity and the cGMP response by Vfr is interesting. It might be coincidental, such that Thr133 affords Vfr a better cAMP pocket and at the same time a more relaxed pocket to accommodate cGMP for activation. However, we prefer a simpler hypothesis for the simultaneous impact of the substitution. We propose that Thr133 is superior to the substituted Ser in terms of the protein's equilibrium shift toward the active form, and the resulting shifted protein equilibrium in wild-type Vfr is the common molecular mechanism for both its higher cAMP affinity and its additional cGMP responsiveness. A similar scenario has been demonstrated with CRP* variants (46). In short, the results suggest that both of the distinct Vfr properties originate from a common sequence basis (Thr133) and therefore potentially from a single underlying molecular mechanism.
cAMP activates Vfr through the C-helix repositioning mechanism.
We have demonstrated that CRP and CooA share a ligand activation mechanism (16, 48), and this view has been supported by other reports (24, 29, 35). We therefore asked if Vfr's activation by cAMP occurs via a similar C-helix repositioning. For this purpose, we randomized the codons for Thr132 and Thr133, two C-helix residues that, by analogy to CRP (48), are likely to be crucial; then we screened for constitutively active Vfr variants by monitoring β-galactosidase activity in E. coli lacking cAMP. Sequence analysis showed that most of the selected Vfr variants have Leu or a β-branched amino acid at both positions 132 and 133 (Table 3). In addition, aromatic residues such as Tyr, Phe, and Trp were also effective at position 133 (Table 3). The selected Vfr* variants are somewhat reminiscent of the CRP* variants reported previously (48), but they differ in two respects: (i) aromatic amino acids at the second position are new and were never found among the relevant CRP* variants; and (ii) Vfr T132L T133I, analogous to the best CRP* variant (CRP T127L S128I), was not found in our hunt. We thought the latter result was especially surprising and formed the following hypothesis: if Vfr T132L T133I has extremely high constitutive activity, it might cause severe growth inhibition of the host E. coli and therefore prevent the Vfr T132L T133I variant from surviving. The correlation between unusually high CRP activity and toxicity has been established previously (48). To test this hypothesis and to circumvent the hypothesized toxicity, we performed a similar randomization and screening using a vfr mutant with a reduced capacity to interact with RNA polymerase. To reduce the transcriptional activity of Vfr without altering DNA binding or the ligand pocket, we constructed Vfr H164L, a Vfr variant with a defect in the “activating region 1” surface that interacts with RNA polymerase (34, 42). Based on the precedent of an analogous CRP variant, Vfr H164L would have reduced transcriptional activity (42) and presumably would not kill the host. Under the new experimental condition, the expected Vfr T132L T133I H164L variant was identified multiple times as Vfr* (Table 3). The very high proportion of the Vfr T132L T133I H164L variant among our constitutive Vfr variants in this new background (50%) further suggests that Vfr T132L T133I H164L is the highest-activity Vfr*. Further, the other Vfr* variants identified under this scheme closely resemble Vfr T133L T133I H164L, as well as CRP* variants reported previously (48). Overall, the substitution pattern at the key C-helix residues for Vfr* is highly similar to that for CRP*, which is strongly suggestive of a common activation mechanism for Vfr and CRP.
Table 3.
Screening results for constitutively active Vfr variants randomly altered at Thr132 and Ser133a
| vfr background | Amino acid at position: |
No. of different nucleotides at other positionsb | |
|---|---|---|---|
| 132 | 133 | ||
| Wild type | Leu | Tyr | 7 |
| Leu | Leu | 2 | |
| Ala | Tyr | 2 | |
| Val | Leu | 2 | |
| Thr | Ile | 2 | |
| Ile | Leu | 2 | |
| Ala | Trp | 1 | |
| Leu | Phe | 1 | |
| Ile | Thr | 1 | |
| Phe | Phe | 1 | |
| Leu | Thr | 1 | |
| Leu | His | 1 | |
| Val | Ile | 1 | |
| Thr | Leu | 1 | |
| Val | Leu | 1 | |
| Ile | Val | 1 | |
| Ala | Ile | 1 | |
| H164L mutant | Leu | Ile | 5 |
| Leu | Val | 2 | |
| Leu | Leu | 1 | |
| Val | Ile | 1 | |
| Ile | Ile | 1 | |
The positions were randomized in the two vfr backgrounds: the wild-type vfr and the vfr H164L background. The reporter strain used was UQ3811.
Amino acid sequences are the same, but DNA sequences (codons) are different.
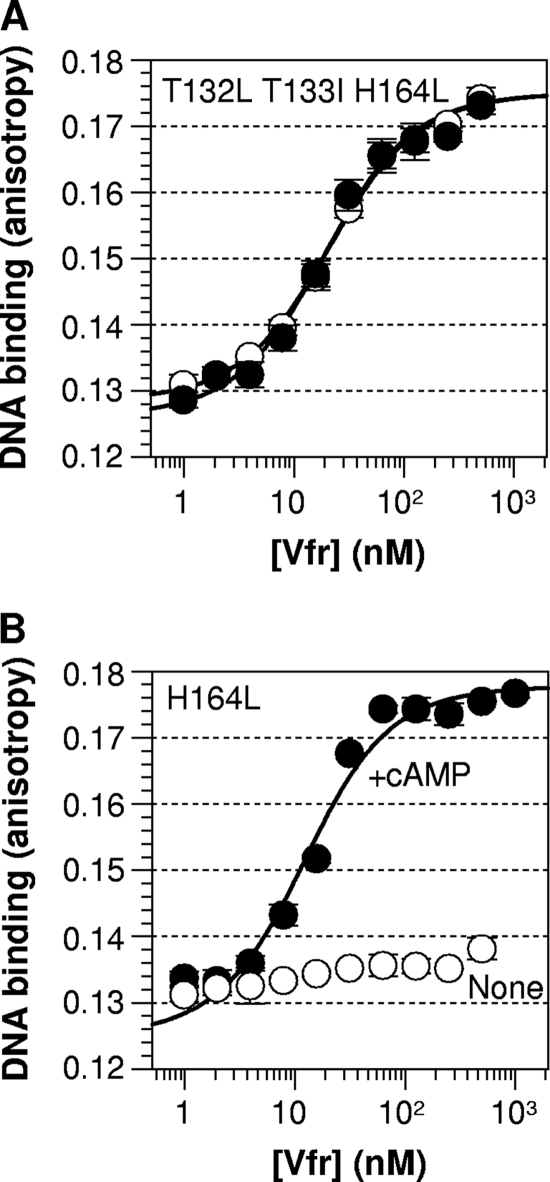
In vitro DNA-binding analysis further confirmed the high cAMP-independent activity of Vfr T132L T133I H164L. We tried to build Vfr T132L T133I but continuously failed to construct the variant in E. coli, indirectly supporting the hypothesized toxicity. This is also consistent with our inability to find the variant in the wild-type vfr background. Therefore, we compared the DNA-binding activities of Vfr T132L T133I H164L and Vfr H164L in order to evaluate the DNA affinity of Vfr T132L T133I. While Vfr H164L behaved like wild-type Vfr, Vfr T132L T133I H164L showed very high cAMP-free DNA binding (Fig. 5). The result demonstrates that Vfr T132L T133I H164L (and therefore Vfr T132L T133I) is a strongly constitutive variant and suggests a conserved leucine zipper mechanism for the activation of both Vfr and CRP. In summary, the in vivo Vfr* activity pattern, confirmed in vitro, indicates that the C-helix region is important for activation and suggests a C-helix repositioning mechanism similar to that of CRP.
Fig. 5.
T132L T133I substitution of Vfr results in constitutive DNA binding. The in vitro DNA affinity of Vfr T132L T133I H164L was measured and compared with that of Vfr H164L. Open circles, no ligand; filled circles, the presence of 100 μM cAMP. Solid lines show the best fit of the data to an equation described by Lundblad et al. (20).
Some Vfr variants, altered at positions 132 and 133, completely reverse the normal response of Vfr to cAMP.
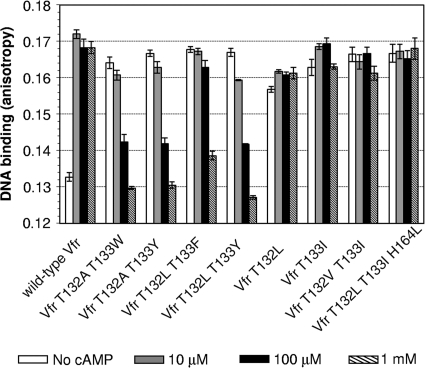
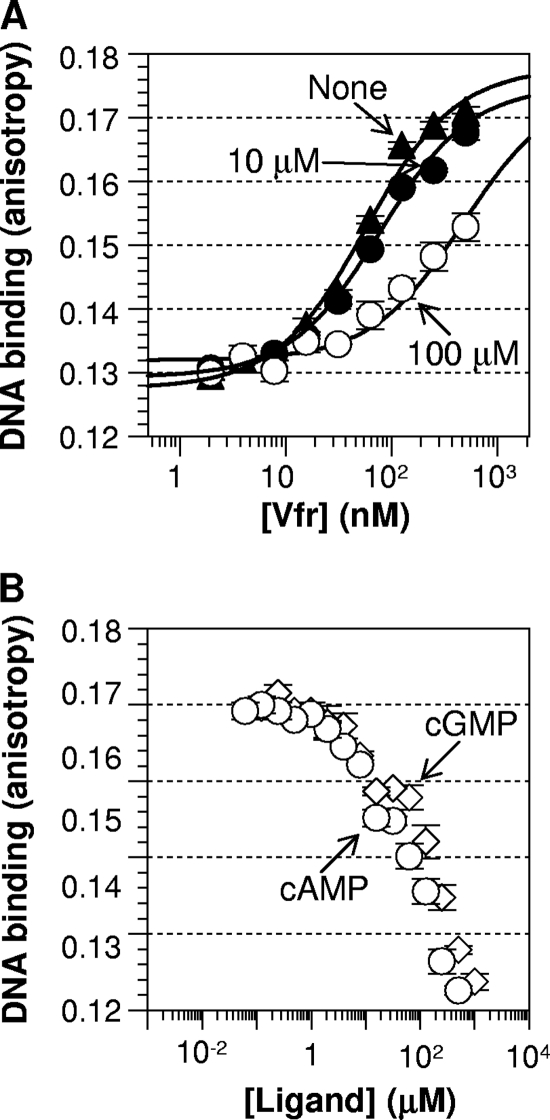
While screening for cAMP-independent Vfr variants, we observed several Vfr variants that showed higher activity in the cAMP-deficient strain than in the cAMP-producing strain (data not shown). We interpreted this to mean that this group of Vfr variants had significant cAMP-independent activity and that cAMP actually inhibited their activity. To test this hypothesis, we purified several of these cAMP-inhibited Vfr variants along with other Vfr* variants and then monitored the effect of cAMP on their DNA binding in vitro. As expected, cAMP indeed reduced the DNA-binding activities of the selected Vfr variants (Fig. 6, left side). While all the Vfr* variants with an improved leucine zipper interaction (with Leu, Ile, or others) showed cAMP-independent activity, those with aromatic substitutions showed a negative response to cAMP. Note that this group of variants is a subset of cAMP-independent variants and that the Vfr variants of the reversed polarity phenotype usually had Ala or Leu at position 132 and Phe, Tyr, or Trp at position 133. Thus, the sequence basis for this novel property of reversed cAMP action can be attributed to the presence of an aromatic acid at position 133. Although the data set was limited, Tyr and Trp had similar efficacy, and Phe was inferior. We then examined the effective range of cAMP for inhibition using a representative Vfr variant, Vfr T132L T133Y. As expected, Vfr T132L T133Y showed less DNA binding with the addition of an increased level of cAMP (Fig. 7 A). The cAMP titration shows that the inhibition started at 10 μM and was almost complete at 1 mM (Fig. 7B). Notably, Vfr T132L T133Y was inhibited by cGMP as well, with an affinity surprisingly similar to that of cAMP (Fig. 7B). Thus, the Vfr variant (and perhaps this class of Vfr variants) is different from wild-type Vfr in three ways. (i) It has a cAMP-free constitutive activity. (ii) Ligands such as cAMP and cGMP shift the protein's equilibrium toward the inactive form. (iii) There is no affinity difference between cAMP and cGMP for the inhibitory role. While the underlying molecular mechanism has yet to be resolved, this result demonstrates the versatility of the structurally conserved CRP/FNR family of proteins in sensing diverse ligands.
Fig. 6.
Reversed cAMP responsiveness among some Vfr variants altered at positions 132 and 133. The cAMP response of these Vfr variants is fundamentally different both from that of wild-type Vfr and from that of constitutively active Vfr variants. Open bars, no ligand; shaded bars, 10 μM cAMP; filled bars, 100 μM cAMP; hatched bars, 1 mM cAMP. The protein concentration for each Vfr sample was 200 nM.
Fig. 7.
Both cAMP and cGMP inhibit the DNA-binding activity of Vfr T132L T133Y. (A) Protein titration without a ligand and in the presence of 10 μM or 100 μM cAMP. (B) The DNA affinities of 200 nM Vfr T132L T133Y were measured in various concentrations of the ligands cAMP (circles) and cGMP (diamonds).
DISCUSSION
In several members of the CRP/FNR superfamily of transcription factors, the C-helix repositioning mechanism is well established for transmitting ligand-binding signals to the DNA-binding domain (16, 24, 27, 35, 48). Our current data suggest that the same protein motif (C-helix) is critical for the activation of Vfr. Therefore, in analogy to CRP, a similar cAMP activation mechanism for Vfr can be posited. Thr132 at the critical “d” position in the heptad repeat of the coiled-coil C-helix provides a suboptimal leucine zipper interaction in the dimerization interface. The suboptimal leucine zipper interaction is strengthened by cAMP binding to assume an active protein conformation, which is competent to bind DNA. The stronger leucine zipper interaction by cAMP could be mimicked by amino acid substitutions in the region.
The important differences between Vfr and CRP lie in the higher cAMP affinity and cGMP responsiveness of Vfr. Our result with the Vfr T133S variant shows that the presence of Thr133 is responsible for both properties of Vfr. We believe that Thr133 affords Vfr a greater active population in the absence of any ligand than Ser128 does for CRP. Consistent with this proposal is the frequent observation of Thr133 among the Vfr* variants altered at positions 132 and 133, while Ser133 was never found (Table 3). Then, as demonstrated with CRP* variants (46), such a shifted protein equilibrium of Vfr relative to that of CRP can simultaneously explain both Vfr's higher cAMP affinity and its cGMP responsiveness. Another obvious consequence of this relatively shifted equilibrium would be the presence of Vfr activity even in the absence of cAMP. This view is consistent with our observation of some constitutive activity of wild-type Vfr in vivo (Fig. 2B). The Vfr target DNA sequence in our reporter strain is a CRP consensus, highly similar to the sequence upstream of lasR, one of Vfr's physiological target DNA sequences. Thus, our result is in agreement with the previous report that Vfr does not require cAMP for the expression of lasR in P. aeruginosa (12). The cAMP-independent expression of lasR might provide a physiological rationale for Vfr's constitutive activity.
It is clear that cAMP is the physiological ligand of Vfr, a notion extensively supported by in vivo and in vitro data (37, 38, 45). What, then, is implied by the higher biochemical cAMP affinity of Vfr? One possibility is that P. aeruginosa utilizes a lower in vivo cAMP concentration than does E. coli. This, in turn, would explain the paradoxical observation that Vfr could complement the E. coli crp deletion mutant, yet CRP was not able to fully complement the P. aeruginosa vfr deletion mutant (43). It is possible that CRP would not be cAMP saturated in P. aeruginosa if P. aeruginosa cells utilize a lower concentration of cAMP for signaling. Although the intracellular concentrations of cAMP in P. aeruginosa and E. coli have been found to be similar (13, 25, 36), it has been challenging to measure this compound accurately (4).
Next, why does Vfr respond to cGMP biochemically at all? We believe this phenomenon is an inevitable side effect of Vfr's relatively shifted protein equilibrium. As discussed above, such a shifted protein equilibrium could provide Vfr either with cAMP-independent activity or with higher cAMP affinity. While the cGMP response of Vfr is biochemically interesting, it is mostly likely physiologically irrelevant. This is because (i) our in vitro data indicate that Vfr requires a very high cGMP concentration for activation and (ii) such a high cGMP accumulation has never been detected under conditions under which Vfr is supposed to be functional (12, 13, 41). For the same reason, we speculate that Vfr's shifted equilibrium is an effective way to afford high cAMP affinity to Vfr.
The class of Vfr variants displaying a reversed response to cAMP is quite remarkable and holds both biochemical and evolutionary interest. Such a reversed polarity requires two conditions: (i) cAMP-free activity and (ii) cAMP inhibition. Simultaneous acquisition of these distinct traits by a simple substitution at the C-helix region of Vfr demonstrates that the region is critical for the ligand response and the function of the protein. It has been reported that the Clp proteins of Xanthomonas species have such a reversed polarity with cyclic di-GMP as the inhibitory ligand (6, 18, 40). We note, however, that Clp utilizes a different mechanism for reversed polarity, because it does not contain C-helix residues reminiscent of those of the Vfr variants reported here. These examples imply that the CRP/FNR family of proteins might be more versatile in ligand sensing and response than previously thought.
Mechanistically, the constitutive activity of these reversed-polarity Vfr variants can be explained by the creation of a stronger leucine zipper interaction around the C-helix. How cAMP binding might shift the protein toward the inactive form is less obvious. The ambiguity is partly due to uncertainty as to the site to which that inhibitory cAMP binds. Vfr has two cAMP-binding sites, one around the C-helix with high affinity, and the other around the E-/F-helices with low affinity (6a). For inhibitory cAMP binding, we prefer the high-affinity site, mainly because the substitutions in the novel Vfr variants are in the C-helix region. Assuming this to be the case, modeling of the active Vfr structure, using Swiss-PdbViewer, version 4.0.1, suggests that an aromatic residue at position 133 could cause steric hindrance of the bound cAMP, the β4/β5 loop, and/or the C-helix. Thus, cAMP binding would destabilize the interactions involving those regions which, in CRP, play a central role in stabilizing the active form (48). On the other hand, we disfavor the secondary cAMP pocket because of its low affinity for cAMP, at least in wild-type Vfr, which does not coincide well with the surprisingly high inhibitory cAMP affinity among the Vfr variants.
In summary, Vfr displays a conserved cAMP-sensing mechanism of C-helix repositioning for the transition from the inactive form to the active form. What is unique to Vfr is its extremely high cAMP affinity and its cGMP responsiveness. Further study to determine whether there is any physiological demand for Vfr's high cAMP affinity and/or for its responsiveness to cGMP is required.
ACKNOWLEDGMENTS
This work was supported by NSF MCB-1020498 (to H.Y.) and by start-up funds from California State University, Fresno (to H.Y.). This work was also supported by the College of Agricultural and Life Sciences at UW—Madison and by NIH GM53228 (to G.P.R.).
We thank Katrina Forest for providing the template DNA for the PCR amplification of the vfr gene and for critical reading of the manuscript. We thank Sang-Jin Suh and Matthew Wolfgang for critical reading of the manuscript. We thank Samyuktha Dasari for technical assistance.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Aiba H., Kawamukai M., Ishihama A. 1983. Cloning and promoter analysis of the Escherichia coli adenylate cyclase gene. Nucleic Acids Res. 11:3451–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albus A. M., Pesci E. C., Runyen-Janecky L. J., West S. E. H., Iglewski B. H. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beatson S. A., Whitchurch C. B., Sargent J. L., Levesque R. C., Mattick J. S. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bettenbrock K., et al. 2007. Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J. Bacteriol. 189:6891–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chiang L. W., Kovari I., Howe M. M. 1993. Mutagenic oligonucleotide-directed PCR amplification (Mod-PCR): an efficient method for generating random base substitution mutations in a DNA sequence element. PCR Methods Appl. 2:210–217 [DOI] [PubMed] [Google Scholar]
- 6. Chin K.-H., et al. 2010. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J. Mol. Biol. 396:646–662 [DOI] [PubMed] [Google Scholar]
- 6a. Cordes T. J., Worzalla G. A., Ginster A. M., Forest K. T. 2011. Crystal structure of the Pseudomonas aeruginosa virulence factor regulator. J. Bacteriol. 193:4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dasgupta N., Ashare A., Hunninghake G. W., Yahr T. L. 2006. Transcriptional induction of the Pseudomonas aeruginosa type III secretion system by low Ca2+ and host cell contact proceeds through two distinct signaling pathways. Infect. Immun. 74:3334–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dasgupta N., Ferrell E. P., Kanack K. J., West S. E. H., Ramphal R. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 184:5240–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dykxhoorn D. M., St. Pierre R., Linn T. 1996. A set of compatible tac promoter expression vectors. Gene 177:133–136 [DOI] [PubMed] [Google Scholar]
- 10. Emmer M., deCrombrugghe B., Pastan I., Perlman R. 1970. Cyclic AMP receptor protein of E. coli: its role in the synthesis of inducible enzymes. Proc. Natl. Acad. Sci. U. S. A. 66:480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferrell E., Carty N. L., Colmer-Hamood J. A., Hamood A. N., West S. E. 2008. Regulation of Pseudomonas aeruginosa ptxR by Vfr. Microbiology 154:431–439 [DOI] [PubMed] [Google Scholar]
- 12. Fuchs E. L., et al. 2010. The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and -independent mechanisms. J. Bacteriol. 192:3553–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fuchs E. L., et al. 2010. In vitro and in vivo characterization of the Pseudomonas aeruginosa cAMP phosphodiesterase CpdA required for cAMP homeostasis and virulence factor regulation. J. Bacteriol. 192:2779–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holder I. A. 1993. P. aeruginosa burn infections: pathogenesis and treatment, p. 275–295 In Campa M., Bendinelli M., Friedman H. (ed.), Pseudomonas aeruginosa as an opportunistic pathogen. Plenum Press, New York, NY [Google Scholar]
- 15. Kanack K. J., Runyen-Janecky L. J., Ferrell E. P., Suh S. J., West S. E. 2006. Characterization of DNA-binding specificity and analysis of binding sites of the Pseudomonas aeruginosa global regulator, Vfr, a homologue of the Escherichia coli cAMP receptor protein. Microbiology 152:3485–3496 [DOI] [PubMed] [Google Scholar]
- 16. Kerby R. L., Youn H., Thorsteinsson M. V., Roberts G. P. 2003. Repositioning about the dimer interface of the transcription regulator CooA: a major signal transduction pathway between the effector- and DNA-binding domains. J. Mol. Biol. 325:809–823 [DOI] [PubMed] [Google Scholar]
- 17. Lau G. W., Hassett D. J., Britigan B. E. 2005. Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 13:389–397 [DOI] [PubMed] [Google Scholar]
- 18. Leduc J. L., Roberts G. P. 2009. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J. Bacteriol. 191:7121–7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin S.-H., Lee J. C. 2002. Communications between the high-affinity cyclic nucleotide binding sites in E. coli cyclic AMP receptor protein: effect of single site mutations. Biochemistry 41:11857–11867 [DOI] [PubMed] [Google Scholar]
- 20. Lundblad J. R., Laurance M., Goodman R. H. 1996. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 10:607–612 [DOI] [PubMed] [Google Scholar]
- 21. Małecki J., Polit A., Wasylewski Z. 2000. Kinetic studies of cAMP-induced allosteric changes in cyclic AMP receptor protein from Escherichia coli. J. Biol. Chem. 275:8480–8486 [DOI] [PubMed] [Google Scholar]
- 22. Medina G., Juárez K., Diaz R., Soberón-Chávez G. 2003. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 149:3073–3081 [DOI] [PubMed] [Google Scholar]
- 23. Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Moore L. J., Mettert E. L., Kiley P. J. 2006. Regulation of FNR dimerization by subunit charge repulsion. J. Biol. Chem. 281:33268–33275 [DOI] [PubMed] [Google Scholar]
- 25. Notley-McRobb L., Death A., Ferenci T. 1997. The relationship between external glucose concentration and cAMP levels inside Escherichia coli: implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology 143:1909–1918 [DOI] [PubMed] [Google Scholar]
- 26. Nouwens A. S., et al. 2003. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology 149:1311–1322 [DOI] [PubMed] [Google Scholar]
- 27. Passner J. M., Schultz S. C., Steitz T. A. 2000. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 Å resolution. J. Mol. Biol. 304:847–859 [DOI] [PubMed] [Google Scholar]
- 28. Pollack M. 2000. Pseudomonas aeruginosa, p. 2310–2327 In Mandell G. L., Bennett J. E., Dolin R. (ed.), Mandell, Douglas and Bennett's principles and practice of infectious diseases. Churchill Livingstone, Philadelphia, PA [Google Scholar]
- 29. Popovych N., Tzeng S.-R., Tonelli M., Ebright R. H., Kalodimos C. G. 2009. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc. Natl. Acad. Sci. U. S. A. 106:6927–6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poschet J. F., et al. 2007. Pharmacological modulation of cGMP levels by phosphodiesterase 5 inhibitors as a therapeutic strategy for treatment of respiratory pathology in cystic fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 293:L712–L719 [DOI] [PubMed] [Google Scholar]
- 30a. Press W. H., Teukolsky S. A., Vetterling W. T., Flannery B. P. 1989. Numerical recipes, 1st ed. Cambridge University Press, New York, NY [Google Scholar]
- 31. Ren Y. L., Garges S., Adhya S., Krakow J. S. 1990. Characterization of the binding of cAMP and cGMP to the CRP* 598 mutant of the Escherichia coli cAMP receptor protein. Nucleic Acids Res. 18:5127–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Savery N. J., et al. 1998. Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 17:3439–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Savery N. J., et al. 2002. Determinants of the C-terminal domain of the Escherichia coli RNA polymerase α subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J. Bacteriol. 184:2273–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma H., Yu S., Kong J., Wang J., Steitz T. A. 2009. Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding. Proc. Natl. Acad. Sci. U. S. A. 106:16604–16609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Siegel L. S., Hylemon P. B., Phibbs P. V., Jr 1977. Cyclic adenosine 3′,5′-monophosphate levels and activities of adenylate cyclase and cyclic adenosine 3′,5′-monophosphate phosphodiesterase in Pseudomonas and Bacteroides. J. Bacteriol. 129:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith R. S., Wolfgang M. C., Lory S. 2004. An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun. 72:1677–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suh S., et al. 2002. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology 148:1561–1569 [DOI] [PubMed] [Google Scholar]
- 39. Takahashi M., Blazy B., Baudras A. 1989. Ligand-modulated binding of a gene regulatory protein to DNA: quantitative analysis of cyclic-AMP induced binding of CRP from Escherichia coli to nonspecific and specific DNA targets. J. Mol. Biol. 207:783–796 [DOI] [PubMed] [Google Scholar]
- 40. Tao F., He Y. W., Wu D. H., Swarup S., Zhang L. H. 2010. The cNMP domain of Xanthomonas campestris global regulator Clp defines a new class of c-di-GMP effectors. J. Bacteriol. 192:1020–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Veron W., et al. 2007. Natriuretic peptides affect Pseudomonas aeruginosa and specifically modify lipopolysaccharide biosynthesis. FEBS J. 274:5852–5864 [DOI] [PubMed] [Google Scholar]
- 42. West D., et al. 1993. Interactions between the Escherichia coli cyclic AMP receptor protein and RNA polymerase at class II promoters. Mol. Microbiol. 10:789–797 [DOI] [PubMed] [Google Scholar]
- 43. West S. E. H., Sample A. K., Runyen-Janecky L. J. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532–7542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Whitchurch C. B., et al. 2005. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol. Microbiol. 55:1357–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wolfgang M. C., Lee V. T., Gilmore M. E., Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253–263 [DOI] [PubMed] [Google Scholar]
- 46. Youn H., Koh J., Roberts G. P. 2008. Two-state allosteric modeling suggests protein equilibrium as an integral component for cyclic AMP (cAMP) specificity in the cAMP receptor protein of Escherichia coli. J. Bacteriol. 190:4532–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Youn H., Kerby R. L., Koh J., Roberts G. P. 2007. A C-helix residue, Arg123, has important roles in both the active and inactive forms of the cAMP receptor protein. J. Biol. Chem. 282:3632–3639 [DOI] [PubMed] [Google Scholar]
- 48. Youn H., Kerby R. L., Conrad M., Roberts G. P. 2006. Study of highly constitutively active variants suggests how cAMP activates cAMP receptor protein. J. Biol. Chem. 281:1119–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]